Abstract
Tyrosine phosphatase non-receptor type substrate-1 (SHPS-1), a transmembrane protein, plays a vital role in cell migration and proliferation. Our previous studies have shown that insulin-like growth factor-I (IGF-I) stimulates SHPS-1 phosphorylation, leading to recruitment of SHP-2, c-Src, Shc, and Grb2·p85 to phosphorylated SHPS-1. Assembly of this signaling complex is required for optimal stimulation of both mitogen-activated protein and phosphatidylinositol 3-kinase pathways. The main aim of the present study was to identify novel proteins that interacted with the cytoplasmic domain of SHPS-1 (SHPS-1/CD) in response to IGF-I stimulation and define the role of these interactions in mediating specific biological functions. We performed a functional proteomic screening to identify SHPS-1 binding partners using combination of mRNA display and the tandem affinity purification-tag methods. Screening identified a number of proteins not previously known to interact with phosphorylated SHPS-1/CD. These novel SHPS-1 binding partners represent several functional categories including heat shock proteins, protein kinases and phosphatases, and proteins that regulate transcription or translation. In Vivo and in vitro studies suggested that most of the proteins bound to SHPS-1 via binding to one of the four SH2 domain containing proteins, SHP-2, CTK, SUPT6H, and STAT1, that directly bound to SHPS-1. Although the binding of most of these proteins to SHPS-1 was positively regulated by IGF-I, a few were negatively regulated, suggesting differential regulation of protein complexes assembled on SHPS-1/CD in response to IGF-I. Further studies showed that truncation of SHPS-1/CD significantly impaired IGF-I-dependent AKT signal transduction and subsequent biological functions including cell survival, protein synthesis, protein aggregation, and prevention of apoptosis. The results emphasize the importance of formation of SHPS-1 signaling complex induced by IGF-I and provide novel insights into our knowledge of the role of this molecular scaffold in regulation of IGF-I-stimulated signal transduction and biological actions.
Src homology 2 domain containing protein tyrosine phosphatase (SHP) substrate-1 (SHPS-1) is a transmembrane protein that plays an important role in mediating various cellular responses, including cell migration and proliferation. Insulin-like growth factor-I (IGF-I)1 stimulates proliferation and migration in vascular smooth muscle cells (VSMCs) (1). In VSMCs IGF-I stimulation leads to recruitment of the signaling protein Shc to SHPS-1 allowing sustained MAPK activation (2). The cytoplasmic domain of SHPS-1 contains four tyrosine residues located within YXXL/I/V motifs that can undergo phosphorylation after growth factor stimulation or cell adhesion and then bind to SH2 domain containing proteins such as SHP-2 (3–5). SHPS-1 has been shown to function as a scaffold protein that is capable of binding to various proteins and recruit them to the plasma membrane (6). These proteins include protein tyrosine phosphatases (4, 7), Src family kinases (8), focal adhesion kinase-related cytosolic kinase, PYK2 (9), and Janus kinase 2 as well as adapter proteins, e.g. Grb2 (7). Transfer of SHP-2 to SHPS-1 is required for growth factor-mediated cell proliferation, DNA synthesis, and MAPK activation (4, 10, 11). SHPS-1 recruitment of signaling proteins has been implicated in the cellular response to hyperglycemic stress. Our lab has shown that recruitment of several proteins to SHPS-1 including SHP-2, c-Src, Shc, and Grb2 following hyperglycemia is essential for smooth muscle cells to respond to IGF-I (2, 12–14).
Many cellular processes are carried out by multiprotein complexes (15, 16). Increasing evidence has shown that SHPS-1 is involved in various biological phenomena, including suppression of anchorage-independent cell growth, negative regulation of immune cell responses, self-recognition of red blood cells, mediation of macrophage multi-nucleation, skeletal muscle differentiation, neuronal survival, and synaptogenesis (6). However, there is still insufficient information about SHPS-1 and its interacting partners to understand the role of SHPS-1 in modulating many of these cellular functions. Therefore, identification of new SHPS-1-binding proteins has a potential to significantly improve our understanding of the IGF-I-mediated signal transduction pathways.
Two approaches recently used to identify protein binding partners are mRNA display techniques, such as mRNA-protein fusions (17–21) and in vitro virus, based on cell-free co-translation and affinity selection (22–24) and tandem affinity purification (TAP)-mass spectrometry methods. These methods are well suited for identifying the components of complexes that form in near physiological conditions. It has been successfully applied in the analysis of protein-protein interaction networks in both prokaryotic and eukaryotic cells (25–33). Therefore in the present study, we employed a novel combination of mRNA display and TAP tag to identify proteins that interact with SHPS-1/cytoplasmic domain (CD). Candidates identified by this method were confirmed by co-immunoprecipitation and pulldown assays. The identification of these proteins provides an opportunity to determine their roles in mediating physiological events that occur in VSMCs in response to hyperglycemic stress and IGF-I stimulation.
MATERIALS AND METHODS
Human IGF-I was a gift from Genentech (South San Francisco, CA). Immobilon P membranes were purchased from Millipore Corp. (Bedford, MA). Dulbecco's modified Eagle's medium (DMEM) containing 4500 mg of glucose per liter (DMEM-H, 25 mm) or DMEM containing 900 mg glucose per liter (DMEM-N, 5 mm) was purchased from Invitrogen, and penicillin and streptomycin were purchased from Invitrogen. Blasticidin was obtained from Invivogen (San Diego, CA). Antibodies against pAKT (Ser-473), pAKT (Thr-308), pSTAT1 (Tyr-701), pSTAT1 (Ser-727), AKT, STAT1, GRP78, cleaved caspase-3, β-actin, and HA were from Cell Signaling Technology (Danvers, MA). Antibodies against HSC70, APG2, HSP70, HSP90, ILK, NOX4, RPTPα, VIM, CALR, GRP75, GRP94, TSC2, CTK, and the monoclonal anti-phosphotyrosine antibody (pY99) were purchased from Santa Cruz Biotechnology (Santa Cruz, CA). Antibodies against CAPZβ, DDX6, FTH1, and SUPT6H were from Abcam (Cambridge, MA). The Grb2 monoclonal antibody and the caveolin antibody were purchased from BD Biosciences (San Diego, CA). The anti-pGSK-3β (Ser-9), anti-SHP-2 antibody, and active Src protein were purchased from Upstate Cell Signaling Solutions/Millipore (Lake Placid, NY). The anti-Src antibody was purchased from Calbiochem (San Diego, CA). A polyclonal antibody for the cytoplasmic domain or extracellular domain of SHPS-1 or SHP-2 was generated in our laboratory. Two synthetic peptides (peptide no. 217: YARAAARQARATLTYADLDM and control no. 136: YARAAARQARAVQLYAVVSEE) were prepared as described previously (2, 12). Benzonase nuclease, Insect GeneJuice Transfection Reagent, Insect PopCulture Reagent, and nickel-nitrilotriacetic acid (Ni-NTA) His·Bind resin were purchased from EMD Biosciences (Novagen brand). All other reagents were purchased from Sigma chemical company unless stated otherwise.
Cell Culture
VSMCs were prepared from porcine aortas and maintained in culture as described previously (34).
Construction of TAP Tag and the SHPS-1/CD with TAP Tag
The SHPS-1/CD (amino acids 394–503) was amplified from pcDNA-SHPS-1 by PCR using forward primer (5′-CCGCGGGATCCCGAATCAGACAGAAGAAAG-3′) containing a BamHI site and a reverse primer (5′-CACCCCTCGAGCTTCCTCGGGACCTGGAC-3′) containing an XhoI site and digested with BamHI and XhoI. The fragment was subcloned into the BamHI and XhoI site of pCMV-CBPzz vector (23) (a gift from Drs. H. Yanagawa and E. Miyamoto-Sato, Keio University, Japan), which contained a C-terminal TAP tag coding sequence (25, 26) as pCMV-SHPS-1/CDCBPzz.
The TAP tag (without bait protein) and SHPS-1/CD with TAP tag as shown Fig. 1A were amplified individually from pCMV-SHPS-1/CDCBPzz by PCR using the same reverse primer (5′-GAGGCCAAGCTTTTAGTGATGGTGATGGTGATGGG-3′) containing a HindIII site and two independent forward primers, forward primer 1 (TGACCATGGGCGAGCTCAAGAGAAGATGG) and forward primer 2 (5′-TGACCATGGGCCGAATCAGACAGAAGAAAG-3′) containing a NcoI site. The PCR products were digested with NcoI and HindIII. The fragments were subcloned into the NcoI/HindIII site of pIEx-5 vector, which encoded His tag and adiopkinetic hormone signal sequence (EMD Biosciences, Novagen brand) termed pIEx-5- CBPzz or pIEx-5-SHPS-1/CDCBPzz. pIEx-5-SHPS-1/CD-4FCBPzz (Tyr-428/Tyr-452/Tyr-469/Tyr-495/4Phe) was also prepared as described above.
Fig. 1.
Selection of SHPS-1/CD-interacting partners from SMCs proteome library. A, construction of the bait SHPS-1/CD (cytoplasmic domain of SHPS-1) for selection. The fragment containing the cytoplasmic domain of SHPS-1 is fused with the TAP affinity selection tag, which contains the IgG binding domain (z domain) of protein A, a binding domain protease cleavage site, and the calmodulin binding peptide (CBP) sequence (25). B, schematic diagram of SHPS-1/CD phosphorylation by Src in vitro. C, selection procedure for SHPS-1/CD-interacting partners. The selection is composed of transcription, cell-free translation, purification, and reversed transcription of mRNA-protein fusion library, followed by two-step affinity selection, IgG beads for protein A tag selection and Calmodulin beads for CBP tag, and PCR to amplify the enriched library.
Expression and Purification of TAP Tag and SHPS-1/CD with TAP Tag
Sf9 cells in 10-ml suspension cultures (1 × 106 cells/ml culture) were transfected with 20 μg of the indicated plasmids (pIEx-5-CBPzz, pIEx-5-SHPS-1/CDCBPzz, or pIEx-5-SHPS-1/CD-4FCBPzz) using Insect GeneJuice Transfection Reagent following the manufacturer's instructions. Total culture extracts were prepared 48 h later by the addition of Insect PopCulture Reagent (500 μl) to the medium followed by the addition of benzonase nuclease (5 μl). The TAP tag or SHPS-1/CD with TAP tag or SHPS-1/CD-4F with TAP tag protein was purified using Ni-NTA His·Bind resin following the manufacturer's instructions. The protein was dialyzed against 10 mm Tris-HCl (pH 8.0), 150 mm NaCl, 0.1% Nonidet P-40, 10% glycerol, and 100 μm soduim orthovanadate (Na3O4). The purity of both proteins was verified by SDS-PAGE with Coomassie Brilliant Blue staining. The protein concentration was determined by BCA assay (Pierce).
Phosphorylation of SHPS-1/CD Expressed from Insect Cell by Src
The TAP tag fusion protein containing the cytoplasmic domain of SHPS-1, purified as described above, was immobilized on IgG-agarose (Sigma) in IPP150 buffer (10 mm Tris-HCl, pH 8.0, 150 mm NaCl, 0.1% Nonidet P-40, 10% glycerol, 0.1 mm Na3O4). The SHPS-1/CD (or SHPS-1/CD-4F) was phosphorylated by active Src in a buffer containing 50 mm HEPES-NaOH (pH 7.6), 3 mm MnCl2, 10 mm MgCl2, 0.1 mm EGTA, 1 mm dithiothreitol, 0.1 mm Na3O4, and 0.2 mm ATP and incubating at 30 °C for 30 min. To determine the phosphorylation of SHPS-1/CD (or SHPS-1/CD-4F), the phosphorylated SHPS-1/CD (SHPS-1/CD-4F) was cleaved from the agarose beads using TEV protease in TEV buffer (Invitrogen) at 16 °C for 4 h. The cleavage products were analyzed by immunoblotting using an anti-pY99 antibody (Fig. 1B and supplemental Fig. S1).
Construction of cDNA Library and Generation of mRNA-displayed Proteome Library from VSMCs
Total RNA was isolated by TRIzol (Invitrogen) from VSMCs. Thereafter, poly(A)+ mRNA purified from total RNA with Oligotex mRNA kit (Qiagen) was used to generate the cDNA library. First strand cDNA synthesis was performed using a degenerate primer, TTN6 (Novagen, Madison, WI), which allowed non-biased coverage of all genes and all possible domains of the open reading frames. After second strand synthesis, directional EcoRI/HindIII linker (Novagen) was ligated to both ends. The resulting double-stranded DNAs were restricted and directionally ligated to the left and right consensus fragments. The left consensus fragment contained a T7 promoter and a tobacco mosaic virus 5′ un-translated region to provide efficient in vitro transcription and translation with rabbit reticulocyte lysate followed by a sequence coding an E tag for affinity purification. The right consensus fragment contained the coding sequence for FLAG/His6 tags followed by a short sequence for hybridizing, and cross-linking with the puromycin-containing DNA linker. The resulting double-stranded DNAs were PCR amplified and fractionated. The mRNA-displayed VSMC proteome domain library was generated according to the previously reported procedures (18, 19, 35). The resulting mRNA-displayed proteome library was successively purified on the basis of the E and FLAG affinity tags at the N- and C termini, respectively, using 10-fold excess of free E and FLAG peptides for competitive elution from the corresponding affinity chromatography resins (termed mRNA display-based preselection). The purified library was used in the SHPS-1/CD-interacting selection.
Selection of Phosphorylated SHPS-1/CD-binding Proteins from mRNA-displayed Proteome Library
The general selection scheme is given in Fig. 1C. An excess (e.g. 60 μg) of purified TAP tag without bait protein (TAP tag alone) was immobilized on IgG-agarose beads and phosphorylated by Src as described above. The purified mRNA-displayed proteome library (1.5 pmol one round) was first diluted in the selection buffer containing 50 mm Tris-HCl, pH 7.6, 150 mm NaCl, 0.1% Nonidet P-40, 1 mm EDTA, 1 mm EGTA, 1 mm dithiothreitol, 10% glycerol, 0.1 mm Na3O4, 0.1 mg/ml bovine serum albumin, 0.1 mg/ml salmon sperm DNA and protease inhibitor mixture (Roche) and then added to IgG-agarose beads with TAP tag incubated for 30 min at 4 °C. After centrifugation, the supernatant that contained the pre-cleared mRNA-displayed proteome library was added to IgG-agarose beads with phosphorylated SHPS-1/CD containing TAP tag, which was prepared using 60 μg of purified SHPS-1/CD with TAP tag immobilized on IgG-agarose beads and phosphorylated by Src as described above. After incubation for 1 h at 4 °C, the mixture was washed three times with the selection buffer and washed once with TEV buffer (Invitrogen). Tagged proteins were cleaved from the beads using TEV protease in TEV buffer at 16 °C for 4 h. Supernatants from the TEV reactions containing cleaved SHPS-1/CD and its interacting partners, were incubated with calmodulin-Sepharose (Stratagene) in the calmodulin binding buffer (10 mm Tris-HCl, pH 8.0, 150 mm NaCl, 5 mm β-mercaptoethanol, 1 mm magnesium acetate, and 2 mm CaCl2) followed by loading into an empty column (25). The SHPS-1/CD and its interacting partners were eluted by using the same buffer containing 2 mm EGTA. The selected molecules were PCR-amplified for cloning into a TOPO vector (Invitrogen) for sequencing and analysis.
SHPS-1/CD IgG-agarose Pulldown Assay and in Vitro Binding Assay
VSMCs with or without IGF-I treatment were lysed in the RIPA buffer. The purified SHPS-1/CD was immobilized on IgG-agarose beads and phosphorylated by Src as described above followed by incubation with cell lysate for 4 h at 4 °C in a buffer containing 10 mm Tris-HCl, pH 7.6, 150 mm NaCl, 0.1% Nonidet P-40, 10% glycerol, 1 mm EDTA, 1 mm EGTA, 1 mm dithiothreitol, 0.5 mm Na3VO4, and protease inhibitors (Roche). The SHPS-1/CD-interacting proteins were cleaved from the beads using TEV protease in TEV buffer at 16 °C for 4 h and analyzed by immunoblotting using an antibody against the protein of interest.
To investigate the interaction between SHPS-1/CD and SH2 domain containing proteins, SHPS-1/CD binding assays were performed using both radiolabeled SH2 domain containing proteins generated by TNT (coupled in vitro transcription and translation) with rabbit reticulocyte lysate and the purified TAP-tagged SHPS-1/CD overexpressed in insect cells. For the TNT approach, radiolabeled SH2 domain containing proteins were synthesized in the presence of 10 μCi of [35S]methionine (PerkinElmer Life Sciences) in a total volume of 25 μl for 90 min at 30 °C as described previously (19). The other steps were the same as the pulldown assay described above.
pLenti-HA-SHPS-1/WT and pLenti-HA-SHPS-1/−CD
Full-length human SHPS-1 cDNA was generated by reverse transcription-PCR from mRNA that had been derived from human fibroblasts (GM10; Human Genetic Cell Repository, Camden, NJ) (36). The full-length SHPS-1 sequence was PCR-amplified with HA containing at the 3′-end of primers using previously generated pcDNA-SHPS-1 as a template and cloned into the expression vector pLenti6/V5-D-TOPO; the truncated cytoplasmic domain of SHPS-1 mutant (deleting amino acid from 399 to 503) was generated by PCR amplification containing the HA sequence. The construction was confirmed by DNA sequencing and cloned into pLenti viral transduction system as explained previously (2).
Generation of Virus Stocks and Establishment of SMCs Expressing pLenti Construct
293FT cells (Invitrogen) were prepared for generation of virus stocks and VSMC expressing SHPS-1 WT-HA and SHPS-1/−CD−HA were established using procedures that were described previously (2). The expression of the HA-tagged SHPS-1 WT or SHPS-1/−CD (MT-CD) protein was detected by immunoblotting with an anti-HA (1:1000) or anti-SHPS-1 antibodies followed by a horseradish peroxidase-conjugated anti-rabbit secondary antibody.
Immunoprecipitation
All immunoprecipitations were carried out as described previously unless otherwise noted (2).
Isolation Cytoplasmic and Membrane Fraction Proteins
Isolation of cytoplasmic and membrane fraction proteins was performed as described previously (37).
Measurement of Cell Death: Trypan Blue Staining
VSMCs were seeded at 5,000 cells per well and grown to 70–80% confluence in 24-well culture plates (Falcon 3036). The cultures were incubated in serum-free DMEM-H or DMEM-N for overnight before the addition of IGF-I for indicated times. Cells were collected from each well and then spun for 10 min at 2,000 rpm at 4 °C. A sample was taken from cell suspension following the cell pellet was resuspended in medium and mixed with an equal volume of 0.4% trypan blue in phosphate-buffered saline. After 5 min of incubation at room temperature the extent of cell death was quantitated by measuring the percentage of viable cells able to exclude trypan blue.
In Vitro Caspase-3 Activity Assay
VSMCs were seeded at 5,000 cells per well and grown to 70–80% confluence in 24-well culture plates (Falcon 3036). The cultures were incubated in serum-free DMEM-H or DMEM-N for overnight prior to the addition of IGF-I. After incubation for 36 h, the caspase-3 activity was measured by caspase-3 colorimetric activity assay kit (Chemicon International, Inc.) following the manufacturer's instructions with minor modification. Aliquots of 75 μg of crude cell lysate were incubated with caspase-3 substrate Ac-DEVD-pNA (p-nitroaniline) at 37 °C for 8 h. The chromophore p-nitroaniline (pNA), the product of caspase-mediated cleavage, as a relative caspase-3 activity was quantified at 405 nm in a microtiter plate reader.
Measurement of Protein Aggregation
Protein aggregation was measured as described previously (38, 39). In brief, cells were incubated in serum-free medium for overnight and then exposed to 0 or 50 ng/ml IGF-I for the appropriate length of time prior to heat shock, which was induced by incubating the cultures at 42 °C for 45 min before lysis. The ratio of HSP70 in the Triton X-100 insoluble to the soluble fraction is used as a marker of denaturation and aggregation (39). The images obtained were also scanned using an Duoscan T1200 scanner (Agfa, Brussels, Belgium). Densitometric analyses of the images were performed using NIH Image, version 1.38v.
Measurement of Protein Synthesis
The measurement of protein synthesis was performed employing SHPS-1 WT and SHPS-1/−CD (MT-CD) as described previously (13, 40).
Statistical Analysis
The Student's t test was used to compare differences between treatments. The results that are shown in all experiments are the representative of at least three separate experiments and expressed as the mean ± S. E. of three independent experiments.
RESULTS
Selection of SHPS-1/CD-interacting Partners from SMCs Proteome Library
To identify SHPS-1/CD-interacting partners, we combined the TAP tag purification technique with mRNA-display as shown in Fig. 1. A summary of the candidate SHPS-1/CD-interacting proteins is listed in supplemental Table 1. We identified a number of previously uncharacterized binding partners that interact with phosphorylated SHPS-1/CD. The proteins that were identified could be classified into functional groups including heat shock proteins, protein kinases and phosphatases, GTP-binding proteins, cytoskeletal proteins, and proteins that regulate transcription or translation. This suggests that localization of these interacting partners on SHPS-1/CD could modulate several different types of signaling interactions and downstream events.
Interaction of SHPS-1/CD with Its Binding Partners Is Regulated by IGF-I
To determine whether SHPS-1 that was present in intact cells could interact with binding partners of interest, and whether these interactions were regulated by IGF-I, we performed co-immunoprecipitation and pulldown assays using lysates prepared from intact VSMC. As shown in Fig. 2, A–C and summarized in E, 24 proteins associated with the SHPS-1 regardless of whether the SMC had been stimulated by IGF-I. Of these, 12 proteins showed increased association with phosphorylated SHPS-1 following IGF-I stimulation (Fig. 2A). Three proteins showed minimal increase and four showed no increase (Fig. 2B). The fact that these proteins could be co-immunoprecipitated with SHPS-1 suggests that they can assemble into the SHPS-1 complex in vivo. In contrast, five proteins showed decreased binding to SHPS-1 following IGF-I stimulation (Fig. 2C). To confirm that these proteins were binding to the tyrosine phosphorylated CD of SHPS-1, the results were confirmed by using a pulldown assay in which a peptide containing SHPS-1/CD that had been phosphorylated by Src in vitro was used as bait. SMC lysates that had been stimulated by IGF-I were incubated with the SHPS-1/CD and then analyzed for the presence of selected proteins. The results in Fig. 2D show the proteins that were associated with endogenous SHPS-1 either increased or decreased and in the same direction as had been detected using intact cells. Interestingly, GRP78 and calreticulin (CALR) required both IGF-I exposure and SHPS-1 phosphorylation suggesting that either IGF-I was stimulating a cofactor that mediated their binding to phosphorylated SHPS-1 or that it stimulated their release from an associated intracellular protein that was acting to prevent SHPS-1 binding. In contrast, APG2 and HSC70 associated with the non-phosphorylated SHPS-1/CD and both showed decreased binding to SHPS-1 in response to SHPS-1 phosphorylation, confirming the results of co-immunoprecipitation.
Fig. 2.
SHPS-1 interaction with its binding proteins is modulated in response to IGF-I. A–C, co-immunoprecipitation of SHPS-1 and its binding partners. Nontransfected confluent cultures were serum-starved overnight and then stimulated with IGF-I for 5 or 10 min. The cells were lysed in the RIPA buffer. Sequential immunoprecipitation was performed using an anti-SHPS-1 antibody (or an antibody indicated) followed by immunoblotting with an antibody against the protein of interest. SHPS-1 phosphorylation was detected by immunoprecipitating with an anti-SHPS-1 antibody and immunoblotting with anti-pY99 antibody. To control the loading, the blot was stripped and reprobed with anti-SHPS-1 antibody (or an antibody indicated). The interaction between SHP-2 and SHPS-1 is used as a positive control. The negative controls are shown in supplemental Fig. S3. D, confirmation of phosphorylated SHPS-1/CD associated to the binding partners using pulldown assays. Cells with or without IGF-I treatment were lysed in the RIPA buffer. The purified SHPS-1/CD was immobilized on IgG-agarose beads and phosphorylated by Src, followed by incubation with lysate for 4 h at 4 °C. The SHPS-1/CD-interacting proteins were cleaved from the beads using TEV protease and then analyzed by immunoblotting as described under “Materials and Methods.” E, summary of interactions of SHPS-1 with its associated proteins in response to IGF-I stimulation. The SHPS-1/CD-interacting proteins belong to several protein groups such as heat shock proteins, protein kinases, and transcription regulatory proteins.
To determine whether the SHPS-1-interacting proteins that contained SH2 domains could bind directly to SHPS-1, an in vitro binding assay was employed. The results show that all four SH2 domain-containing proteins bound directly to the SHPS-1/CD (Fig. 3 A). Of the four, SHP-2 and CTK bound to phosphorylated SHPS-1/CD both in vivo and in vitro (Fig. 2A and Fig. 3A), suggesting that they directly bound to SHPS-1 via SH2 domain-phosphorylated tyrosine interactions. In contrast, the other two interacting partners, SUPT6H and STAT1 exhibited differences between in vivo and in vitro binding analyses. In the in vitro binding assay both SUPT6H and STAT1 showed no difference between phosphorylated and non-phosphorylated SHPS-1/CD (Fig. 3A). In the in vivo assay SUPT6H binding to SHPS-1/CD was increased whereas STAT1 was decreased in response to IGF-I (Fig. 2, A and C). DDX6, a protein that lacks SH2 domain, was used as negative control for in vitro binding assay. In addition to DDX6, other proteins such as GRP78, GRP75, HSP70, and KTN1 that lack SH2 domains did not directly bind to SHPS-1/CD in vitro (supplemental Fig. S6).
Fig. 3.
Interactions of SHPS-1 with the associated proteins is primarily mediated by SH2 domain containing partners in response to IGF-I stimulation. A, proteins labeled with [35S]methionine were generated by a TNT reaction with rabbit reticulocyte lysate at 30 °C for 90 min. An aliquot of the TNT mixture was incubated with SHPS-1/CD, which was immobilized on IgG-agarose beads and phosphorylated by Src. The SHPS-1/CD-binding proteins were cleaved from the beads using TEV protease and then analyzed by SDS-PAGE, and images were obtained using a Storm 870 PhosphorImager as described under “Materials and Methods.” T, the expressed protein; W, the last wash; E, the eluent (supernatant after cleaved from beads). DDX6 that does not contain an SH2 domain was used as a negative control; SHP-2 is positive control. Interactions between SH2 domain containing SHP-2 (B) or STAT1 (C) or CTK (D) or SUPT6H (E) and SHPS-1 associated proteins after stimulation with IGF-I. Nontransfected confluent cultures were serum-starved overnight and then stimulated with IGF-I for 5 or 10 min. The cells were lysed in the RIPA buffer. Sequential immunoprecipitation was performed using each indicated antibody followed by immunoblotting with an antibody to the protein of interest. To control the loading, the blot was stripped and reprobed with a corresponding antibody. F, confluent cultures were serum-starved overnight and incubated with or without the SHPS-1 cell-permeable peptide (peptide no. 217; 10 μg/ml) or control peptide (control no.136) for 1 h followed by with or without IGF-I treatment for 5 min. The cell lysates were immunoprecipitated with anti-SHPS-1 antibody (or an antibody indicated) followed by immunoblotting for SHP-2 and phosphorylated SHPS-1 (pY99) or other antibodies indicated. For loading control, the membrane was reprobed with a corresponding antibody. G, pSMC lysate was directly immunoblotted using anti-pSTAT1 Tyr-701 or Ser-727 phosphospecific antibodies. For loading control, the membrane was reprobed with anti-STAT1 antibody. H, diagrammatic representation of the dynamic interactions of SHPS-1 with binding partners that are mediated by SH2 domain containing proteins in response to IGF-I stimulation.
Because many of the proteins that were shown to increase their binding to SHPS-1 following SHPS-1 phosphorylation or IGF-I stimulation do not have SH2 domains, it is possible that their association with the SHPS-1 complex is mediated by binding to one of these four intermediary proteins. Therefore, we further investigated the role of these four proteins within the group that contained SH2 domains to determine whether they could mediate the binding of non-SH2 domain containing proteins to SHPS-1. Fig. 3B shows protein complexes that were analyzed following immunoprecipitation of SHP-2. The binding of ILK, TSC2, GRP75, GRP78, GRP94, AKT, HSP60, and NOX4 to SHP-2 were significantly increased following IGF-I stimulation whereas calreticulin showed no change. To determine whether SHP-2 could function as an intermediary for the association of these proteins with SHPS-1, a cell permeable peptide that had been shown to inhibit SHP-2 binding to SHPS-1 was used (2). Exposure to this peptide inhibited the ability of SHPS-1 to bind to GRP78, TSC2, ILK, GRP75, NOX4, and HSP60 (Fig. 3F). Similarly, the possible roles of CTK, SUPT6H, and STAT1 are investigated in detail individually in Fig. 3, C–E, and the results are summarized in H. Several SHPS-1 associated, non-SH2 domain-containing proteins, which showed increased SHPS-1 association following IGF-I treatment, were found to associate with CTK or SUPT6H (Fig. 3, D and E). GRP78 and GRP75 association with CTK was increased after IGF-I stimulation, but others such as AKT association with CTK or SUPT6H (Fig. 3, D and E) were not regulated by IGF-I. The results suggest that CTK mediates interaction of GRP78 and GRP75 with SHPS-1. In contrast a few SHPS-1 associated, non-SH2 domain-containing proteins, which were downregulated by IGF-I treatment, were found to associate with STAT1 (Fig. 3C). HSC70 showed reduced binding to SHPS-1 following IGF-I stimulation but its binding to STAT1 increased as shown in Fig. 3C suggesting that STAT1 could modulate HSC70/SHPS-1 association. Because STAT1 was dissociated from SHPS-1 upon IGF-I stimulation, we investigated whether IGF-I induces STAT1 phosphorylation at Tyr-701 and Ser-727, which is a prerequisite for transport of STAT1 to the nucleus from the cytoplasm (41, 42). As shown in Fig. 3G, IGF-I stimulated STAT1 phosphorylation on Tyr-701 and Ser-727 in vivo; however, STAT1 was not phosphorylated in vitro in the rabbit reticulocyte lysate cell-free system (supplemental Fig. S5A). Because STAT1 association with SHPS-1 is decreased and STAT1 phosphorylation is increased in response to IGF-I it is possible that STAT1 phosphorylation inhibits binding to SHPS-1 and thus enhances its dissociation. Because phosphorylation regulates subsequent nuclear localization of STAT1 it may be that SHPS-1 disassociation and nuclear localization are coordinated. To demonstrate the validity and specificity of the interacting proteins with SHPS-1, we performed experiments using reverse co-immunoprecipitation wherever possible, and we also used negative controls containing IgG (supplemental Fig. S2). To further evaluate the specificity of the co-immunoprecipitation results, we utilized both positive controls (SHP-2) and negative controls, e.g. SUG1 and BAG1 in addition to the IgG controls. Immunoblotting of lysates for total levels of specific proteins with and without IGF-I stimulation was performed to demonstrate that the change in SHPS-1/CD binding was not because of a change in protein expression. The total protein level remained unchanged as shown in supplemental Fig. S3.
Overexpression of SHPS-1 Enhances the Localization of SHPS-1/CD-binding Proteins to the Cell Membrane and SHPS-1/CD Complexes Mediates IGF-I Signaling
To determine whether loss of binding of known signaling intermediates to the cytoplasmic domain of SHPS-1 would alter IGF-I-stimulated signaling events, we expressed HA-tagged wild type SHPS-1 (SHPS-1 WT) and a mutant form of SHPS-1 that lacks most of the cytoplasmic domain (MT-CD) in VSMCs (Fig. 4 A). SHPS-1 WT expression was increased 20 ± 4.5-fold compared with endogenous levels of SHPS-1 (data not shown). Because SHPS-1 recruitment of several signaling molecules and adaptor proteins, such as SHP-2, Shc, Grb2, and p85 in response to IGF-I stimulation has been reported previously, we investigated the role of recruitment of AKT using these two cell types. As shown in Fig. 4B, the localization of these proteins to the transmembrane protein SHPS-1 in SHPS-1 WT cells was increased significantly compared with MT-CD cells in response to IGF-I stimulation. Both the HA-tagged SHPS-1 WT and MT-CD are contained in the membrane fraction (supplemental Fig. S4).
Fig. 4.
IGF-I-stimulated SHPS-1-associated protein recruitment to the plasma membrane is SHPS-1/CD-dependent. A, SMCs expressing HA-SHPS-1 WT, SHPS-1/−CD (MT-CD), were lysed. The blots were probed with an anti-SHPS-1 antibody (antigen from the extracellular domain of SHPS-1 in house-made) to detect expression of SHPS-1 WT or MT-CD. B, cells that were expressing SHPS-1 WT or MT-CD were serum-starved overnight and stimulated with IGF-I for the indicated times. The cells were lysed in the RIPA buffer. Sequential immunoprecipitation was performed using anti-HA antibody followed by immunoblotting with an antibody against the protein of interest. The blots were stripped and reprobed with anti-ΗΑ antibody as a loading control. C, membrane proteins were isolated as described under “Materials and Methods” and immunobloted with anti-pan-AKT antibody, anti-phospho-AKT at Thr-308 or Ser-473 antibodies. The blots were stripped and reprobed with anti-caveolin antibody as a membrane fraction marker and anti-14–3-3β antibody as a cytoplasm fraction marker.
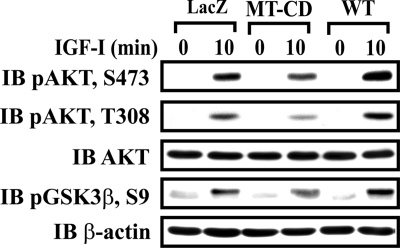
AKT localizes in the cell membrane in response to PI3K activation. To determine whether SHPS-1 modulates AKT membrane recruitment in response to IGF-I stimulation, the cellular membrane fraction was analyzed. The truncation of the CD of SHPS-1 impaired the localization of total AKT in the membrane fraction compared with cells expressing SHPS-1 WT (Fig. 4C). Additionally phosphorylation of both Thr-308 and Ser-473 within AKT was significantly increased in SHPS-1 WT cells compared with MT-CD cells following IGF-I stimulation. Because AKT plays a regulatory role in controlling cellular survival and apoptosis and is regulated by IGF-I, we wished to investigate whether the SHPS-1-mediated AKT localization was involved in the ability of IGF-I to inhibit apoptosis. IGF-I induced significant increase in AKT Ser-473 phosphorylation at 10 min in the cells expressing SHPS-1 WT (20.5 ± 2.2-fold increase; n = 3, p < 0.05) compared with LacZ cells (16.5 ± 1.8-fold increase; n = 3) (Fig. 5 ). In contrast the MT-CD showed only 9.4 ± 1.4-fold (n = 3, p < 0.01) increase in response to IGF-I stimulation (Fig. 5). Similarly, phosphorylation of AKT at Thr-308 was significantly impaired in MT-CD cells in response to IGF-I stimulation as compared with cells expressing either LacZ or SHPS-1 WT. Because GSK-3β is a known downstream physiological target of AKT and has been implicated in cellular apoptosis, GSK-3β phosphorylation was also investigated. As shown in Fig. 5, IGF-I-induced GSK-3β phosphorylation on Ser-9 was significantly enhanced in SHPS-1 WT cells (7.1 ± 0.4-fold increase; n = 3, p < 0.01) compared with LacZ cells (5.5 ± 0.3-fold increase; n = 3); whereas phosphorylation of GSK-3β on Ser-9 was impaired in cells expressing MT-CD (3.6 ± 0.3-fold increase; n = 3, p < 0.05). The increased phosphorylation of GSK-3β at Ser-9 inhibits GSK-3β activation.
Fig. 5.
IGF-I-stimulated AKT phosphorylation and inactivation of GSK-3β are SHPS-1/CD-dependent. The SHPS-1 WT or MT-CD-expressing cells were serum-starved overnight and stimulated with IGF-I for indicated times. Twenty microliters of cell lysate from the same experiment was used for detection of phospho-AKT and phospho-GSK-3β (Ser-9). The blots were stripped and reprobed with anti-AKT and β-actin antibodies as loading controls.
SHPS-1/CD Modulates the Ability of IGF-I to Inhibit Apoptosis
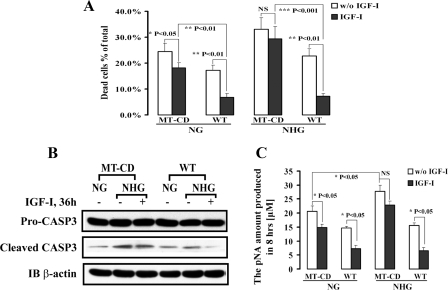
IGF-I promotes cell survival and prevents apoptosis in a number of cell types (43–45). Because AKT and GSK-3β activation were impaired in cells expressing MT-CD, we determined whether through its ability to recruit these interacting proteins, SHPS-1 was playing a role in the ability of IGF-I to prevent cell death. Because we have shown that the AKT response to IGF-I is augmented in hyperglycemia and that this reaction is dependent upon SHPS-1 recruitment, the apoptotic responses of cells expressing either SHPS-1 WT or MT-CD were compared in cells maintained in medium containing normal (5.0 mm) or high (25 mm) glucose. Cells expressing SHPS-1 WT that were maintained in serum-free conditions with high glucose exhibited increased cell death (e.g. 21.1 ± 2.8% compared with cells maintained in normal glucose 17.2 ± 2.1% (p < 0.05)) (Fig. 6 A). Cell death was increased to a greater extent in cells expressing MT-CD 33.5 ± 4.5% (high glucose) versus 21.1 ± 2.8% (low glucose); n = 3, p < 0.01 (Fig. 6A). In normal glucose IGF-I was more effective in preventing cell death in SHPS-1 WT cells compared with MT-CD cells (n = 3, p < 0.001). In high glucose IGF-I had no effect on cell death in the MT-CD cells but was very effective in SHPS-1 WT cells. To further characterize cellular apoptosis, we measured cleaved caspase-3 and its enzymatic activity in vitro in both cell types. In SHPS-1 WT cells IGF-I was more effective in preventing caspase-3 cleavage than in MT-CD cells (Fig. 6B). Similarly the enzymatic activity of caspase-3 was reduced to a greater extent in the SHPS-1 WT cells (2.2 ± 0.3-fold decrease; n = 3, p < 0.05), whereas in MT-CD cells the effect of IGF-I on caspase-3 was significantly less (Fig. 6C). In addition, MT-CD cells had increased basal enzymatic activity of caspase-3, which was consistent with increased cleaved caspase-3 compared with SHPS-1 WT (Fig. 6C).
Fig. 6.
Hyperglycemia-induced cell death and caspase-3 activity are attenuated by IGF-I treatment mediated by SHPS-1/CD. The cell death assay was conducted as described under “Materials and Methods.” VSMCs expressing SHPS-1 WT or MT-CD were grown to 70–80% confluence in normal glucose with 10% fetal bovine serum in 24-well culture plates and then starved overnight in serum-free DMEM-N (NG; 5 mm) or serum-free DMEM-H (NHG; 25 mm). NHG, normal switched to high glucose; NG, normal glucose. Forty-eight hours after the addition of IGF-I, cell number was determined by trypan blue staining and counting, and error bars indicate the standard error. NS, not significant. (A); or thirty-six hours after the addition of IGF-I, cells were lysed and analyzed by immunoblotting using anti-cleaved caspase-3 (CASP3) antibody (B) or for measurement of CASP3 activation, and error bars represent S. E. (C) as described under “Materials and Methods.” The blots were stripped and reprobed with anti-β-actin antibody as a loading control. The results shown are the mean ± S. E. of three separate experiments.
SHPS-1/CD Modulates the Ability of IGF-I to Regulate Protein Aggregation
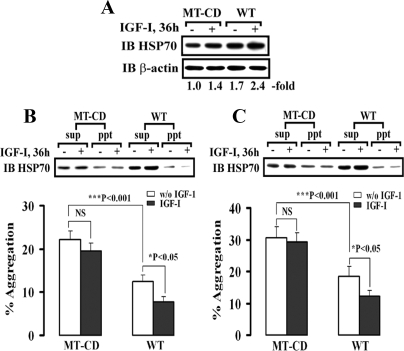
To determine whether other SHPS-1-associated proteins that were activated during the cellular response to stress might have altered activity in cells expressing SHPS-1/CD we examined protein aggregation. HSP70 is known to protect cells from both necrotic and apoptotic cell death due to its ability to assist in protein folding and to reduce protein aggregation (accumulation of unfolded or misfolded proteins). SHPS-1 WT cells expressed significantly higher levels of HSP70 basally compared with MT-CD cells, and following IGF-I stimulation the increase in HSP70 was significantly greater in SHPS-1 WT cells compared with cells expressing MT-CD (Fig. 7 A). Because serine 9 phosphorylation and subsequent inactivation of GSK-3β has been shown to be the mechanism by which AKT alters HSP70 expression (46), these results suggest that the SHPS-1 cytoplasmic domain is required for IGF-I-stimulated HSP70 induction. Similarly GRP75, 78, and 94 have been implicated in regulating the unfolded protein response. To determine whether their increased association with SHPS-1 following IGF-I stimulation is associated with changes in protein aggregation we measured protein aggregation in cells expressing both forms of SHPS-1 following IGF-I treatment. The ratio of HSP70 in the Triton X-100 insoluble to the soluble fraction was used as an indicator of aggregation. In cells expressing MT-CD, basal protein aggregation was significantly increased compared with cells expressing SHPS-1 WT (Fig. 7, B and C). In SHPS-1 WT cells, IGF-I prevented protein aggregation for up to 36 h (n = 3, p < 0.05), whereas in cells expressing MT-CD, IGF-I had no effect. Protein aggregation significantly increased in cells expressing both forms of SHPS-1 after heat shock treatment (Fig. 7C) but the degree of increase was attenuated if intact SHPS-1 was present. These results suggest that increased association of GRP75, 78, and 94 with SHPS-1 as well as increased HSP70 expression reduces protein aggregation and that the cytoplasmic domain of SHPS-1 plays a role in the ability of these proteins to modulate this response.
Fig. 7.
Expression of HSP70 and reduced protein aggregation in response to IGF-I are SHPS-1/CD-dependent. VSMCs expressing SHPS-1 WT (WT) or MT-CD were grown to 70–80% confluence and serum-starved overnight. The total HSP70 protein from cell lysate for thirty-six hours after the addition of IGF-I was analyzed by immunoblotting using antibody against HSP70 (A). The blots were stripped and reprobed with anti-β-actin antibody as a loading control. The protein aggregation assay was conducted as described under “Materials and Methods.” VSMCs expressing SHPS-1 WT or MT-CD were grown to 70–80% confluence and serum-starved overnight. Thirty-six hours after the addition of IGF-I prior to with (B) or without (C) heat shock at 42 °C for 45 min, cells were lysed for immunoblotting with anti-HSP70 antibody (upper panel). The graphs show the mean results from three independent experiments displayed as aggregation calculated from arbitrary scanning units, and the error bars represent S. E. NS, indicates not significant. (lower panel). The ratio of HSP70 in the Triton X-100 insoluble to the soluble fraction is used as a marker of protein denaturation and aggregation.
SHPS-1/CD Is Required for IGF-I-dependent Protein Synthesis
CALR, SUPT6H, and STAT1 were involved in translational regulation and were identified as SHPS-1 interacting partners. Hence we wished to investigate the role of SHPS-1/CD in protein synthesis. Because p70S6K is a direct downstream target of AKT and its phosphorylation is linked to stimulation of protein synthesis, we measured the phosphorylation of p70S6K and protein synthesis. As shown in Fig. 8 A, overexpression of SHPS-1 WT significantly enhanced IGF-I-dependent phosphorylation of p70S6K at Thr-389 compared with cells expressing MT-CD. When the cells expressing SHPS-1 WT were analyzed, IGF-I stimulated a 2.4 ± 0.02 (mean ± S. E.; n = 3)-fold increase in protein synthesis after 6 h. In contrast, IGF-I stimulated a 1.72 ± 0.03 (mean ± S. E.; n = 3)-fold increase in protein synthesis after 6 h in cells expressing MT-CD (Fig. 8B). Further, compared with LacZ cells, overexpression of SHPS-1 WT enhanced IGF-I-dependent protein synthesis (n = 3; p < 0.01), whereas cells expressing MT-CD had an impaired response to IGF-I (n = 3, p < 0.05).
Fig. 8.
SHPS-1/CD enhances IGF-I-stimulated p70S6K phosphorylation and protein synthesis. A, the SHPS-1 WT (WT) or MT-CD-expressing cells were serum-starved overnight and stimulated with IGF-I for indicated times. Twenty microliters of cell lysate was used for detection of phospho-p70S6K (Thr-389). The blots were stripped and reprobed with anti-|gb-actin antibody as a loading control. B, the protein synthesis was measured as described under “Materials and Methods.” The graph shows the mean-fold increase in protein synthesis of SHPS-1 WT or MT-CD or LacZ in response to IGF-I, and error bars represent S. E. The results shown are the mean ± S. E. of three separate experiments.
DISCUSSION
IGF-I has diverse biological actions, including regulation of cellular proliferation, differentiation, migration, and survival (1). The biological effects of IGF-I are mediated through its receptor tyrosine kinase, which phosphorylates specific substrates to activate downstream signaling pathway (47, 48). Prior studies from our group have shown that IGF-I stimulates SHPS-1 phosphorylation resulting in localization of SHP-2, which then recruits c-Src·Shc and Grb2·p85 complexes to phosphorylated SHPS-1. This recruitment is necessary for IGF-I-stimulated Shc phosphorylation and subsequent MAPK and AKT activation as well as cell migration and proliferation (2, 12–14). However, very little is known about additional proteins that are recruited to SHPS-1/CD and how their interaction with SHPS-1 alters IGF-I-mediated biological responses. The goal of the present study is to 1) identify novel SHPS-1-interacting partners to define the interaction networks that modulate IGF-I signaling pathways; and 2) determine whether SHPS-1/CD has a role in mediating cellular responses to IGF-I stimulation other than cell migration and proliferation.
In the present study we employed a novel approach using mRNA display combined with the TAP tag method, which is well suited for studying protein complexes assembled in physiological conditions. This method has added advantage of post-translation modification of the bait protein, such as tyrosine phosphorylation of SHPS-1, prior to selection as compared with other methods such as in vitro virus (24). We determined that SHPS-1 interacts with several hitherto uncharacterized binding partners, which represent several functional categories including stress response proteins, protein kinases and phosphatases, GTP-binding proteins, cytoskeletal proteins, and transcription regulatory proteins. This suggests multiple roles for SHPS-1 in VSMC function. Although SHPS-1 has been proposed to have multiple functions (6) our study is the first to comprehensively analyze the SHPS-1 interactome. Most of the proteins identified during screening were found to be differentially regulated in response to IGF-I demonstrating the specificity of these protein-protein interactions. Furthermore, we identified a few chaperone proteins that bound to SHPS-1/CD. The fact that the chaperone proteins failed to directly bind to SHPS-1/CD in vitro suggests that these chaperone proteins associated with SHPS-1/CD through intermediary proteins rather than via SHPS-1 misfolding and that their binding may be functionally significant (supplemental Fig. S6). Although a number of novel SHPS-1/CD binding partners were identified in the present study using the mRNA display and TAP tag method and the interactions were confirmed with co-immunoprecipitation, previous studies have shown that the evaluation of in vitro protein-protein interactions by multiple orthogonal techniques is essential (49). Thus, these interactions need to be validated using other orthogonal technique. Additionally, due to the limitation of this method, such as the abundance of specific sequences in the initial mRNA library and the efficiency of expression of specific proteins, some SHPS-1/CD-binding proteins might not have been detected. Therefore, additional orthogonal techniques might identify new binding partners.
Our screening revealed four proteins containing SH2 domains that had the ability to bind directly to SHPS-1 including SHP-2, STAT1 and SUPT6H, and CTK. While these proteins bind directly to SHPS-1 via SH2 domain-phosphorylated tyrosine interaction, several other proteins that did not bind directly to SHPS-1 did associate with SHP-2 or CTK. Previously we and others have established the importance of SHP-2 recruitment to SHPS-1 for assembly of signaling complexes (2–4, 12). Recently CTK has been implicated in phosphorylation of SHPS-1 (50). These results suggest that the direct interaction between these two proteins and SHPS-1 may have a significant role in mediating IGF-I-stimulated signaling events and that SHP-2 or CTK could mediate the transfer of some of the proteins that were detected with mRNA display and TAP tag method to SHPS-1. However, our results do not exclude the possibility that other unidentified proteins could mediate these interactions, and silencing of SHP-2 or CTK would be required to prove that they are the only proteins that mediate their transfer. The role of STAT-1 and SUPT6H is less clear. The differences in the interactions of STAT1 and SUPT6H with SHPS-1 in vitro and in vivo could be explained by the intracellular concentrations of these proteins or by the presence of other co-factors that are present in vivo that regulate their recruitment (19, 21, 24). However, further study will be required to determine the significance of their association with or dissociation from SHPS-1 for downstream signaling.
SHPS-1 is a transmembrane protein, and overexpression of wild type SHPS-1 increases the assembly of SHPS-1-interacting proteins at membrane in response to IGF-I stimulation. This is in agreement with our and other studies, which showed that overexpression of wild type SHPS-1 increases the amount of SHP-2 that associates with the plasma membrane in response to insulin, thereby positively regulating the Ras-mitogen-activated protein kinase signaling cascade and that truncation of SHPS-1 leads to failure of SHP-2 transfer to SHPS-1 and IGF-IR (4, 36). Other studies analyzing the interactions between SHPS-1 and a small number of binding partners have suggested a role for the cytoplasmic domain of SHPS-1 as an important regulatory region (3, 7, 9, 10).
The ability of IGF-I to promote cell survival has been attributed to PI3K/AKT pathway activation (51–58). Our results indicate that following IGF-I treatment, apoptosis was more pronounced in MT-CD cells compared with cells expressing wild type SHPS-1. This was accompanied by disruption of AKT recruitment to SHPS-1, impaired AKT, and GSK-3β phosphorylation. This suggests that IGF-I-induced cell survival signaling via the AKT-GSK-3β-caspase-3 pathway requires the presence of SHPS-1/CD. This conclusion is consistent to our finding that optimal PI3K activation is dependent on assembly of a Shc·Grb2·p85 complex on SHPS-1 in response to IGF-I (14). PDK1 phosphorylates AKT at Thr-308 (59, 60), and NOX4 modulates PDK1 tyrosine phosphorylation, which increases its activity (61). Because NOX4 and PDK1 interact with SHPS-1, it is possible that this interaction is required for optimal PDK1 activity.
AKT acts on several downstream targets such as GSK-3β and p70S6K evoking multiple cellular responses (62, 63). In our study, MT-CD-expressing cells had impaired AKT activation and showed attenuation in GSK-3β activity in response to IGF-I. Inactivation of GSK-3β has been shown to be the mechanism by which AKT alters HSP70 expression (46). The identification of chaperone proteins, HSP70, HSP90, GRP75, GRP78, and GRP94, and CALR, as SHPS-1-interacting partners suggests a potential role of SHPS-1 as a component of the cellular response to hyperglycemia stress that may be important for regulating protein folding, aggregation, and degradation. Because hyperglycemia induces endoplasmic reticulum stress (64, 65), which in turn increases protein aggregation (66–68), localization of these chaperone proteins to SHPS-1 could play a role in the protection of cellular proteins from aggregation and thus enhance cellular survival. Upon IGF-I stimulation, the reduction of protein aggregation was more pronounced in SHPS-1 WT-expressing cells compared with MT-CD-expressing cells, and SHPS-1 WT was associated with a major increase in HSP70 protein levels. Because HSP70 prevents protein misfolding (69, 70), SHPS-1 may be functioning to inhibit aggregation (68) and caspase-dependent apoptosis (71). Our data suggest that prevention of apoptosis by SHPS-1/CD may be mediated by multiple events including AKT·GSK-3β·caspase-3 and HSP70 inhibition of caspase-3 activation (72).
In this study GRPs were shown to associate with SHPS-1/CD in response to IGF-I suggesting they may also have a role in IGF-I-mediated inhibition of protein aggregation and subsequent apoptosis. GRP78, 75, and 94 have also been shown to function to prevent aggregation by binding to folding intermediates that are incompletely folded, or assembled, thus helping to solubilize aggregates and prevent transport of incompletely assembled, misfolded, or aggregated proteins from the endoplasmic reticulum (73).
In conclusion, in this study we used a novel combination of mRNA display and TAP tag method to identify SHPS-1 interacting partners, which encompass several protein families including heat shock proteins, protein kinases, and transcription regulatory proteins. The identification and characterization of novel binding partners of SHPS-1 underline the significance of the cytoplasmic domain of the SHPS-1 in its ability to recruit and localize the interacting proteins initiating a plethora of signaling cascades that play multifunctional roles in response to IGF-I-mediating cell survival, protein aggregation, and protein synthesis.
Acknowledgments
We thank Dr. Walker H. Busby, Jr., for his help in the peptides and SHPS-1 antibody; Drs. H. Yanagawa and E. Miyamoto-Sato (Keio University) for providing pCMV-CBPzz vector; and Dr. Laura A. Maile for comments. We also thank Ms. Laura Lindsey for her help in preparing the manuscript.
Footnotes
* This work was supported, in whole or in part, by National Institutes of Health Grant HL56850.
 The on-line version of this article (available at http://www.mcponline.org) contains supplemental material.
The on-line version of this article (available at http://www.mcponline.org) contains supplemental material.
1 The abbreviations used are:
- IGF-I
- insulin-like growth factor-I
- VSMC
- vascular smooth muscle cell
- MAPK
- mitogen-activated protein kinase
- TAP
- tandem affinity purification
- CD
- cytoplasmic domain
- DMEM
- Dulbecco's modified Eagle's medium
- RIPA
- radioimmune precipitation assay buffer
- TNT
- coupled in vitro transcription and translation
- WT
- wild type
- PI3K
- phosphatidylinositol 3-kinase
- HSP
- heat shock protein
- CALR
- calreticulin
- HA
- heamagglutinin
- TEV
- tobacco etch virus
- MT
- mutant type
- CTK
- Csk-type protein tyrosine kinase
- GTP
- guanosine triphosphate
- GRP
- glucose-regulated protein
- ATP
- adenosine triphosphate.
REFERENCES
- 1.Clemmons D. R. ( 2007) Modifying IGF1 activity: an approach to treat endocrine disorders, atherosclerosis and cancer. Nat. Rev. Drug Discov. 6, 821– 833 [DOI] [PubMed] [Google Scholar]
- 2.Ling Y., Maile L. A., Lieskovska J., Badley-Clarke J., Clemmons D. R. ( 2005) Role of SHPS-1 in the regulation of insulin-like growth factor I-stimulated Shc and mitogen-activated protein kinase activation in vascular smooth muscle cells. Mol. Biol. Cell 16, 3353– 3364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fujioka Y., Matozaki T., Noguchi T., Iwamatsu A., Yamao T., Takahashi N., Tsuda M., Takada T., Kasuga M. ( 1996) A novel membrane glycoprotein, SHPS-1, that binds the SH2-domain-containing protein tyrosine phosphatase SHP-2 in response to mitogens and cell adhesion. Mol. Cell. Biol. 16, 6887– 6899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Takada T., Matozaki T., Takeda H., Fukunaga K., Noguchi T., Fujioka Y., Okazaki I., Tsuda M., Yamao T., Ochi F., Kasuga M. ( 1998) Roles of the complex formation of SHPS-1 with SHP-2 in insulin-stimulated mitogen-activated protein kinase activation. J. Biol. Chem. 273, 9234– 9242 [DOI] [PubMed] [Google Scholar]
- 5.Oh E. S., Gu H., Saxton T. M., Timms J. F., Hausdorff S., Frevert E. U., Kahn B. B., Pawson T., Neel B. G., Thomas S. M. ( 1999) Regulation of early events in integrin signaling by protein tyrosine phosphatase SHP-2. Mol. Cell. Biol. 19, 3205– 3215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Oshima K., Ruhul Amin A. R., Suzuki A., Hamaguchi M., Matsuda S. ( 2002) SHPS-1, a multifunctional transmembrane glycoprotein. FEBS Lett. 519, 1– 7 [DOI] [PubMed] [Google Scholar]
- 7.Kharitonenkov A., Chen Z., Sures I., Wang H., Schilling J., Ullrich A. ( 1997) A family of proteins that inhibit signalling through tyrosine kinase receptors. Nature 386, 181– 186 [DOI] [PubMed] [Google Scholar]
- 8.Gresham H. D., Dale B. M., Potter J. W., Chang P. W., Vines C. M., Lowell C. A., Lagenaur C. F., Willman C. L. ( 2000) Negative regulation of phagocytosis in murine macrophages by the Src kinase family member, Fgr. J. Exp. Med. 191, 515– 528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Timms J. F., Swanson K. D., Marie-Cardine A., Raab M., Rudd C. E., Schraven B., Neel B. G. ( 1999) SHPS-1 is a scaffold for assembling distinct adhesion-regulated multi-protein complexes in macrophages. Curr. Biol. 9, 927– 930 [DOI] [PubMed] [Google Scholar]
- 10.Ling Y., Maile L. A., Clemmons D. R. ( 2003) Tyrosine phosphorylation of the beta3-subunit of the alphaVbeta3 integrin is required for embrane association of the tyrosine phosphatase SHP-2 and its further recruitment to the insulin-like growth factor I receptor. Mol. Endocrinol. 17, 1824– 1833 [DOI] [PubMed] [Google Scholar]
- 11.Maile L. A., Badley-Clarke J., Clemmons D. R. ( 2003) The association between integrin-associated protein and SHPS-1 regulates insulin-like growth factor-I receptor signaling in vascular smooth muscle cells. Mol. Biol. Cell 14, 3519– 3528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lieskovska J., Ling Y., Badley-Clarke J., Clemmons D. R. ( 2006) The role of Src kinase in insulin-like growth factor-dependent mitogenic signaling in vascular smooth muscle cells. J. Biol. Chem. 281, 25041– 25053 [DOI] [PubMed] [Google Scholar]
- 13.Maile L. A., Capps B. E., Ling Y., Xi G., Clemmons D. R. ( 2007) Hyperglycemia alters the responsiveness of smooth muscle cells to insulin-like growth factor-I. Endocrinology 148, 2435– 2443 [DOI] [PubMed] [Google Scholar]
- 14.Radhakrishnan Y., Maile L. A., Ling Y., Graves L. M., Clemmons D. R. ( 2008) Insulin-like growth factor-I stimulates Shc-dependent phosphatidylinositol 3-kinase activation via Grb2-associated p85 in vascular smooth muscle cells. J. Biol. Chem. 283, 16320– 16331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Alberts B. ( 1998) The cell as a collection of protein machines: preparing the next generation of molecular biologists. Cell 92, 291– 294 [DOI] [PubMed] [Google Scholar]
- 16.Papin J. A., Hunter T., Palsson B. O., Subramaniam S. ( 2005) Reconstruction of cellular signalling networks and analysis of their properties. Nat. Rev. Mol. Cell Biol. 6, 99– 111 [DOI] [PubMed] [Google Scholar]
- 17.Roberts R. W., Szostak J. W. ( 1997) RNA-peptide fusions for the in vitro selection of peptides and proteins. Proc. Natl. Acad. Sci. U. S. A. 94, 12297– 12302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu R., Barrick J. E., Szostak J. W., Roberts R. W. ( 2000) Optimized synthesis of RNA-protein fusions for in vitro protein selection. Methods Enzymol. 318, 268– 293 [DOI] [PubMed] [Google Scholar]
- 19.Shen X., Valencia C. A., Szostak J. W., Szostak J., Dong B., Liu R. ( 2005) Scanning the human proteome for calmodulin-binding proteins. Proc. Natl. Acad. Sci. U. S. A. 102, 5969– 5974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Takahashi T. T., Austin R. J., Roberts R. W. ( 2003) mRNA display: ligand discovery, interaction analysis and beyond. Trends Biochem. Sci. 28, 159– 165 [DOI] [PubMed] [Google Scholar]
- 21.Shen X., Valencia C. A., Gao W., Cotten S. W., Dong B., Huang B. C., Liu R. ( 2008) Ca(2+)/Calmodulin-binding proteins from the C. elegans proteome. Cell Calcium 43, 444– 456 [DOI] [PubMed] [Google Scholar]
- 22.Nemoto N., Miyamoto-Sato E., Husimi Y., Yanagawa H. ( 1997) In vitro virus: bonding of mRNA bearing puromycin at the 3′-terminal end to the C-terminal end of its encoded protein on the ribosome in vitro. FEBS Lett. 414, 405– 408 [DOI] [PubMed] [Google Scholar]
- 23.Miyamoto-Sato E., Takashima H., Fuse S., Sue K., Ishizaka M., Tateyama S., Horisawa K., Sawasaki T., Endo Y., Yanagawa H. ( 2003) Highly stable and efficient mRNA templates for mRNA-protein fusions and C-terminally labeled proteins. Nucleic Acids Res. 31, e78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Miyamoto-Sato E., Ishizaka M., Horisawa K., Tateyama S., Takashima H., Fuse S., Sue K., Hirai N., Masuoka K., Yanagawa H. ( 2005) Cell-free cotranslation and selection using in vitro virus for high-throughput analysis of protein-protein interactions and complexes. Genome Res. 15, 710– 717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rigaut G., Shevchenko A., Rutz B., Wilm M., Mann M., Séraphin B. ( 1999) A generic protein purification method for protein complex characterization and proteome exploration. Nat. Biotechnol. 17, 1030– 1032 [DOI] [PubMed] [Google Scholar]
- 26.Puig O., Caspary F., Rigaut G., Rutz B., Bouveret E., Bragado-Nilsson E., Wilm M., Séraphin B. ( 2001) The tandem affinity purification (TAP) method: a general procedure of protein complex purification. Methods 24, 218– 229 [DOI] [PubMed] [Google Scholar]
- 27.Gavin A. C., Bösche M., Krause R., Grandi P., Marzioch M., Bauer A., Schultz J., Rick J. M., Michon A. M., Cruciat C. M., Remor M., Höfert C., Schelder M., Brajenovic M., Ruffner H., Merino A., Klein K., Hudak M., Dickson D., Rudi T., Gnau V., Bauch A., Bastuck S., Huhse B., Leutwein C., Heurtier M. A., Copley R. R., Edelmann A., Querfurth E., Rybin V., Drewes G., Raida M., Bouwmeester T., Bork P., Seraphin B., Kuster B., Neubauer G., Superti-Furga G. ( 2002) Functional organization of the yeast proteome by systematic analysis of protein complexes. Nature 415, 141– 147 [DOI] [PubMed] [Google Scholar]
- 28.Ho Y., Gruhler A., Heilbut A., Bader G. D., Moore L., Adams S. L., Millar A., Taylor P., Bennett K., Boutilier K., Yang L., Wolting C., Donaldson I., Schandorff S., Shewnarane J., Vo M., Taggart J., Goudreault M., Muskat B., Alfarano C., Dewar D., Lin Z., Michalickova K., Willems A. R., Sassi H., Nielsen P. A., Rasmussen K. J., Andersen J. R., Johansen L. E., Hansen L. H., Jespersen H., Podtelejnikov A., Nielsen E., Crawford J., Poulsen V., Sørensen B. D., Matthiesen J., Hendrickson R. C., Gleeson F., Pawson T., Moran M. F., Durocher D., Mann M., Hogue C. W., Figeys D., Tyers M. ( 2002) Systematic identification of protein complexes in Saccharomyces cerevisiae by mass spectrometry. Nature 415, 180– 183 [DOI] [PubMed] [Google Scholar]
- 29.Kemmeren P., van Berkum N. L., Vilo J., Bijma T., Donders R., Brazma A., Holstege F. C. ( 2002) Protein interaction verification and functional annotation by integrated analysis of genome-scale data. Mol. Cell 9, 1133– 1143 [DOI] [PubMed] [Google Scholar]
- 30.Butland G., Peregrín-Alvarez J. M., Li J., Yang W., Yang X., Canadien V., Starostine A., Richards D., Beattie B., Krogan N., Davey M., Parkinson J., Greenblatt J., Emili A. ( 2005) Interaction network containing conserved and essential protein complexes in Escherichia coli. Nature 433, 531– 537 [DOI] [PubMed] [Google Scholar]
- 31.Bürckstümmer T., Bennett K. L., Preradovic A., Schütze G., Hantschel O., Superti-Furga G., Bauch A. ( 2006) An efficient tandem affinity purification procedure for interaction proteomics in mammalian cells. Nat. Methods 3, 1013– 1019 [DOI] [PubMed] [Google Scholar]
- 32.Gavin A. C., Aloy P., Grandi P., Krause R., Boesche M., Marzioch M., Rau C., Jensen L. J., Bastuck S., Dümpelfeld B., Edelmann A., Heurtier M. A., Hoffman V., Hoefert C., Klein K., Hudak M., Michon A. M., Schelder M., Schirle M., Remor M., Rudi T., Hooper S., Bauer A., Bouwmeester T., Casari G., Drewes G., Neubauer G., Rick J. M., Kuster B., Bork P., Russell R. B., Superti-Furga G. ( 2006) Proteome survey reveals modularity of the yeast cell machinery. Nature 440, 631– 636 [DOI] [PubMed] [Google Scholar]
- 33.Daulat A. M., Maurice P., Froment C., Guillaume J. L., Broussard C., Monsarrat B., Delagrange P., Jockers R. ( 2007) Purification and identification of G protein-coupled receptor protein complexes under native conditions. Mol. Cell Proteomics 6, 835– 844 [DOI] [PubMed] [Google Scholar]
- 34.Parker A., Gockerman A., Busby W. H., Clemmons D. R. ( 1995) Properties of an insulin-like growth factor-binding protein-4 protease that is secreted by smooth muscle cells. Endocrinology 136, 2470– 2476 [DOI] [PubMed] [Google Scholar]
- 35.Kurz M., Gu K., Lohse P. A. ( 2000) Psoralen photo-crosslinked mRNA-puromycin conjugates: a novel template for the rapid and facile preparation of mRNA-protein fusions. Nucleic Acids Res. 28, E83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Maile L. A., Clemmons D. R. ( 2002) Regulation of insulin-like growth factor I receptor dephosphorylation by SHPS-1 and the tyrosine phosphatase SHP-2. J. Biol. Chem. 277, 8955– 8960 [DOI] [PubMed] [Google Scholar]
- 37.Xi G., Shen X., Clemmons D. R. ( 2008) p66shc Negatively regulates insulin-like growth factor I signal transduction via inhibition of p52shc binding to Src homology 2 domain-containing protein tyrosine phosphatase substrate-1 leading to impaired growth factor receptor-bound protein-2 membrane recruitment. Mol. Endocrinol. 22, 2162– 2175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mao H., Wang Y., Li Z., Ruchalski K. L., Yu X., Schwartz J. H., Borkan S. C. ( 2004) Hsp72 interacts with paxillin and facilitates the reassembly of focal adhesions during recovery from ATP depletion. J. Biol. Chem. 279, 15472– 15480 [DOI] [PubMed] [Google Scholar]
- 39.Zhou W., Freed C. R. ( 2004) Tyrosine-to-cysteine modification of human alpha-synuclein enhances protein aggregation and cellular toxicity. J. Biol. Chem. 279, 10128– 10135 [DOI] [PubMed] [Google Scholar]
- 40.Zheng B., Clemmons D. R. ( 1998) Blocking ligand occupancy of the alphaVbeta3 integrin inhibits insulin-like growth factor I signaling in vascular smooth muscle cells. Proc. Natl. Acad. Sci. U. S. A. 95, 11217– 11222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nguyen H., Ramana C. V., Bayes J., Stark G. R. ( 2001) Roles of phosphatidylinositol 3-kinase in interferon-gamma-dependent phosphorylation of STAT1 on serine 727 and activation of gene expression. J. Biol. Chem. 276, 33361– 33368 [DOI] [PubMed] [Google Scholar]
- 42.Sadzak I., Schiff M., Gattermeier I., Glinitzer R., Sauer I., Saalmüller A., Yang E., Schaljo B., Kovarik P. ( 2008) Recruitment of Stat1 to chromatin is required for interferon-induced serine phosphorylation of Stat1 transactivation domain. Proc. Natl. Acad. Sci. U. S. A. 105, 8944– 8949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Harrington E. A., Bennett M. R., Fanidi A., Evan G. I. ( 1994) c-Myc-induced apoptosis in fibroblasts is inhibited by specific cytokines. EMBO J. 13, 3286– 3295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang L., Ma W., Markovich R., Chen J. W., Wang P. H. ( 1998) Regulation of cardiomyocyte apoptotic signaling by insulin-like growth factor I. Circ. Res. 83, 516– 522 [DOI] [PubMed] [Google Scholar]
- 45.Wang L., Ma W., Markovich R., Lee W. L., Wang P. H. ( 1998) Insulin-like growth factor I modulates induction of apoptotic signaling in H9C2 cardiac muscle cells. Endocrinology 139, 1354– 1360 [DOI] [PubMed] [Google Scholar]
- 46.Kim H. S., Skurk C., Maatz H., Shiojima I., Ivashchenko Y., Yoon S. W., Park Y. B., Walsh K. ( 2005) Akt/FOXO3a signaling modulates the endothelial stress response through regulation of heat shock protein 70 expression. FASEB J. 19, 1042– 1044 [DOI] [PubMed] [Google Scholar]
- 47.LeRoith D., Neuenschwander S., Wood T. L., Hennighausen L. ( 1995) Insulin-like growth factor-I and insulin-like growth factor binding protein-3 inhibit involution of the mammary gland following lactation: studies in transgenic mice. Prog. Growth Factor Res. 6, 433– 436 [DOI] [PubMed] [Google Scholar]
- 48.Jones J. I., Prevette T., Gockerman A., Clemmons D. R. ( 1996) Ligand occupancy of the alpha-V-beta3 integrin is necessary for smooth muscle cells to migrate in response to insulin-like growth factor. Proc. Natl. Acad. Sci. U. S. A. 93, 2482– 2487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Howell J. M., Winstone T. L., Coorssen J. R., Turner R. J. ( 2006) An evaluation of in vitro protein-protein interaction techniques: assessing contaminating background proteins. Proteomics 6, 2050– 2069 [DOI] [PubMed] [Google Scholar]
- 50.Mitsuhashi H., Futai E., Sasagawa N., Hayashi Y., Nishino I., Ishiura S. ( 2008) Csk-homologous kinase interacts with SHPS-1 and enhances neurite outgrowth of PC12 cells. J. Neurochem. 105, 101– 112 [DOI] [PubMed] [Google Scholar]
- 51.Hayashi K., Takahashi M., Kimura K., Nishida W., Saga H., Sobue K. ( 1999) Changes in the balance of phosphoinositide 3-kinase/protein kinase B (Akt) and the mitogen-activated protein kinases (ERK/p38MAPK) determine a phenotype of visceral and vascular smooth muscle cells. J. Cell Biol. 145, 727– 740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Imai Y., Clemmons D. R. ( 1999) Roles of phosphatidylinositol 3-kinase and mitogen-activated protein kinase pathways in stimulation of vascular smooth muscle cell migration and deoxyriboncleic acid synthesis by insulin-like growth factor-I. Endocrinology 140, 4228– 4235 [DOI] [PubMed] [Google Scholar]
- 53.Liu Z. P., Wang Z., Yanagisawa H., Olson E. N. ( 2005) Phenotypic modulation of smooth muscle cells through interaction of Foxo4 and myocardin. Dev. Cell 9, 261– 270 [DOI] [PubMed] [Google Scholar]
- 54.Romano F., Chiarenza C., Palombi F., Filippini A., Padula F., Ziparo E., De Cesaris P. ( 2006) Platelet-derived growth factor-BB-induced hypertrophy of peritubular smooth muscle cells is mediated by activation of p38 MAP-kinase and of Rho-kinase. J. Cell. Physiol. 207, 123– 131 [DOI] [PubMed] [Google Scholar]
- 55.Laurino L., Wang X. X., de la Houssaye B. A., Sosa L., Dupraz S., Cáceres A., Pfenninger K. H., Quiroga S. ( 2005) PI3K activation by IGF-1 is essential for the regulation of membrane expansion at the nerve growth cone. J. Cell Sci. 118, 3653– 3662 [DOI] [PubMed] [Google Scholar]
- 56.Radcliff K., Tang T. B., Lim J., Zhang Z., Abedin M., Demer L. L., Tintut Y. ( 2005) Insulin-like growth factor-I regulates proliferation and osteoblastic differentiation of calcifying vascular cells via extracellular signal-regulated protein kinase and phosphatidylinositol 3-kinase pathways. Circ. Res. 96, 398– 400 [DOI] [PubMed] [Google Scholar]
- 57.Zaka M., Rafi M. A., Rao H. Z., Luzi P., Wenger D. A. ( 2005) Insulin-like growth factor-1 provides protection against psychosine-induced apoptosis in cultured mouse oligodendrocyte progenitor cells using primarily the PI3K/Akt pathway. Mol. Cell Neurosci. 30, 398– 407 [DOI] [PubMed] [Google Scholar]
- 58.Matthews L. C., Taggart M. J., Westwood M. ( 2008) Modulation of caveolin-1 expression can affect signalling through the phosphatidylinositol 3-kinase/Akt pathway and cellular proliferation in response to insulin-like growth factor I. Endocrinology 149, 5199– 5208 [DOI] [PubMed] [Google Scholar]
- 59.Cohen P., Alessi D. R., Cross D. A. ( 1997) PDK1, one of the missing links in insulin signal transduction? FEBS Lett. 410, 3– 10 [DOI] [PubMed] [Google Scholar]
- 60.Toker A., Newton A. C. ( 2000) Cellular signaling: pivoting around PDK-1. Cell 103, 185– 188 [DOI] [PubMed] [Google Scholar]
- 61.Block K., Eid A., Griendling K. K., Lee D. Y., Wittrant Y., Gorin Y. ( 2008) Nox4 NAD(P)H oxidase mediates Src-dependent tyrosine phosphorylation of PDK-1 in response to angiotensin II: role in mesangial cell hypertrophy and fibronectin expression. J. Biol. Chem. 283, 24061– 24076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Shen W. H., Boyle D. W., Wisniowski P., Bade A., Liechty E. A. ( 2005) Insulin and IGF-I stimulate the formation of the eukaryotic initiation factor 4F complex and protein synthesis in C2C12 myotubes independent of availability of external amino acids. J. Endocrinol. 185, 275– 289 [DOI] [PubMed] [Google Scholar]
- 63.Jefferies H. B., Fumagalli S., Dennis P. B., Reinhard C., Pearson R. B., Thomas G. ( 1997) Rapamycin suppresses 5′TOP mRNA translation through inhibition of p70s6k. EMBO J. 16, 3693– 3704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wang H., Kouri G., Wollheim C. B. ( 2005) ER stress and SREBP-1 activation are implicated in beta-cell glucolipotoxicity. J. Cell Sci. 118, 3905– 3915 [DOI] [PubMed] [Google Scholar]
- 65.Werstuck G. H., Khan M. I., Femia G., Kim A. J., Tedesco V., Trigatti B., Shi Y. ( 2006) Glucosamine-induced endoplasmic reticulum dysfunction is associated with accelerated atherosclerosis in a hyperglycemic mouse model. Diabetes 55, 93– 101 [PubMed] [Google Scholar]
- 66.Kaufman R. J. ( 1999) Stress signaling from the lumen of the endoplasmic reticulum: coordination of gene transcriptional and translational controls. Genes Dev. 13, 1211– 1233 [DOI] [PubMed] [Google Scholar]
- 67.Mori K. ( 2000) Tripartite management of unfolded proteins in the endoplasmic reticulum. Cell 101, 451– 454 [DOI] [PubMed] [Google Scholar]
- 68.Bence N. F., Sampat R. M., Kopito R. R. ( 2001) Impairment of the ubiquitin-proteasome system by protein aggregation. Science 292, 1552– 1555 [DOI] [PubMed] [Google Scholar]
- 69.Pilon M., Schekman R. ( 1999) Protein translocation: how Hsp70 pulls it off. Cell 97, 679– 682 [DOI] [PubMed] [Google Scholar]
- 70.Hartl F. U., Hayer-Hartl M. ( 2002) Molecular chaperones in the cytosol: from nascent chain to folded protein. Science 295, 1852– 1858 [DOI] [PubMed] [Google Scholar]
- 71.Kristiansen M., Messenger M. J., Klöhn P. C., Brandner S., Wadsworth J. D., Collinge J., Tabrizi S. J. ( 2005) Disease-related prion protein forms aggresomes in neuronal cells leading to caspase activation and apoptosis. J. Biol. Chem. 280, 38851– 38861 [DOI] [PubMed] [Google Scholar]
- 72.Beere H. M. ( 2001) Stressed to death: regulation of apoptotic signaling pathways by the heat shock proteins. Sci. STKE 2001, re1 [DOI] [PubMed] [Google Scholar]
- 73.Cho D. Y., Yang G. H., Ryu C. J., Hong H.J. ( 2003) Molecular chaperone GRP78/BiP interacts with the large surface protein of hepatitis B virus in vitro and in vivo. J. Virol. 77, 2784– [DOI] [PMC free article] [PubMed] [Google Scholar]