Abstract
Water deficit or dehydration is the most crucial environmental constraint on plant growth and development and crop productivity. It has been postulated that plants respond and adapt to dehydration by altering their cellular metabolism and by activating various defense machineries. The nucleus, the regulatory hub of the eukaryotic cell, is a dynamic system and a repository of various macromolecules that serve as modulators of cell signaling dictating the cell fate decision. To better understand the molecular mechanisms of dehydration-responsive adaptation in plants, we developed a comprehensive nuclear proteome of rice. The proteome was determined using a sequential method of organellar enrichment followed by two-dimensional electrophoresis-based protein identification by LC-ESI-MS/MS. We initially screened several commercial rice varieties and parental lines and established their relative dehydration tolerance. The differential display of nuclear proteins in the tolerant variety under study revealed 150 spots that showed changes in their intensities by more than 2.5-fold. The proteomics analysis led to the identification of 109 differentially regulated proteins presumably involved in a variety of functions, including transcriptional regulation and chromatin remodeling, signaling and gene regulation, cell defense and rescue, and protein degradation. The dehydration-responsive nuclear proteome revealed a coordinated response involving both regulatory and functional proteins, impinging upon the molecular mechanism of dehydration adaptation. Furthermore a comparison between the dehydration-responsive nuclear proteome of rice and that of a legume, the chickpea, showed an evolutionary divergence in dehydration response comprising a few conserved proteins, whereas most of the proteins may be involved in crop-specific adaptation. These results might help in understanding the spectrum of nuclear proteins and the biological processes they control under dehydration as well as having implications for strategies to improve dehydration tolerance in plants.
Environmental stress is a primary cause of crop loss worldwide, resulting in average yield losses of more than 70% for major crops every year (1–3), and plays a major role in determining the geographic distribution of plant species. Several environmental stresses are united by the fact that at least part of their detrimental effect on plant performance is caused by the disruption of the water status of the plants. Among unfavorable environmental conditions, water deficit or dehydration is the most crucial factor that adversely affects plant growth, development, and productivity. Of the 1.5 billion hectares of global cropland, only 250 million hectares (17%) are irrigated. Nevertheless this irrigated land provides about 40% of the world's food production, whereas the remaining 60% comes from rain-fed agriculture (4). There is hardly a physiological process in plants that is not impaired by water deficit or dehydration. However, very few plants have been subjected to biochemical and molecular studies to analyze the mechanisms of dehydration tolerance. In recent years, the physiological and molecular basis for plant responses to dehydration tolerance has been a subject of intense research (5, 6). Dehydration response in plants is a complex phenomenon, and the exact structural and functional modifications caused by dehydration are poorly understood. Thus, the identification of novel genes, determination of their differential expressions, and understanding of their functions are of crucial importance in improving plants' levels of tolerance against such stress. The dehydration-responsive genes are presumed to function not only in protecting cells from water deficit but also in regulating genes for signal transduction (5). The transcriptome analyses of gene expression have greatly contributed to our understanding of the dehydration response in plants (7, 8); however, there has been a lack of correlation between mRNA levels and protein abundance (9, 10). It is thus insufficient to predict protein expression levels from quantitative mRNA data. Furthermore the primary sequences of proteins undergo significant levels of post-translational modifications and are readily subjected to targeted proteolysis (11). In contrast, proteome studies aim at identifying the complete set of proteins encoded by the genome, thereby complementing transcriptome studies. The new generation of proteomics techniques facilitates the investigation of the global protein expression profile using efficient protein extraction methods coupled with protein identification by mass spectrometry. Nevertheless a large number of proteins with varying levels of abundance and diverse isoelectric points, hydrophobicity, and relative molecular mass limit the characterization of the complete proteome of a cell. In this context, organellar proteomics is a promising strategy that reduces the complexity of the total cellular proteome enabling the visualization of low abundance proteins and allowing the study of a specific group of proteins that are central to the biological problem under investigation. In addition, the subcellular proteome is important because a fractionated subset of proteins can provide suitable information regarding where and how these proteins exert their particular functions (12–14).
The eukaryotic nucleus is a highly organized organelle that contains specific functional domains essential for the regulated expression of proteins; thus, it is an attractive target for the study of cellular homeostasis and the determination of the genomic response to stress tolerance. The identification and characterization of the nuclear proteins are thus important for a better understanding of genome regulation and function and multiple signaling events (14–21) dictating cellular adaptation under stress. Compared with the intensive research on the nuclear proteome in the model plant Arabidopsis, information on its monocot counterpart is lacking.
The use of crop plants of agricultural importance is a potential approach in investigating dehydration tolerance because different cultivars are available with differing degrees of tolerance. This approach provides correlative evidence for genes putatively involved in the dehydration response. Furthermore the transient and moderate dehydration represented in studies of crop species probably describes the most common form of dehydration encountered by plants. Rice is the most important food source with nearly half of the world's population relying on its successful harvest, and it is also a model plant for biological research. Currently rice is grown in nearly 114 countries, and more than 50 countries have an annual production of 100,000 tons or more. The current annual rice production is 651 million tons (22), and more than 90% of the rice is grown and consumed in Asia, which houses 60% of the world's population. For most rice-producing countries, rice is the staple food, and it provides more than 50% of the total calories consumed (Internation Rice Research Institute). It is now known that the global growth yield of rice is not only stagnating but is negatively affected by dehydration (23), highlighting the need for a greater understanding of how plants respond to such stress. Nonetheless the plant nuclear proteome is still in its infancy particularly with respect to stress tolerance (15, 20). In rice, most proteomics studies to date have primarily focused on developing proteome maps and constructing databases of expressed proteins for various organs, tissues, and subcellular components (16, 18, 24, 25), whereas there is far less information available on stress-induced temporal changes.
Earlier we reported the development of a nucleus-specific proteome reference map and identified many novel dehydration-responsive nuclear proteins of a food legume, the chickpea (20). Here we developed the nucleus-specific comparative proteome of a tolerant rice variety subjected to progressive dehydration to identify crop-specific mechanisms of dehydration response and organellar candidates involved in dehydration tolerance. As a first step, various physiological and biochemical parameters (i.e. cellular water status, proline accumulation, stability of the cell membrane, and photosynthetic machinery) were investigated to screen eight rice varieties for their relative tolerance in response to dehydration. The differential display of nuclear proteome revealed 150 protein spots whose intensities changed significantly by more than 2.5-fold at one or more time points during dehydration. A total of 109 differentially expressed nuclear proteins were identified during the course of dehydration using LC-ESI-MS/MS. The differential expression profiles of the candidate proteins may provide new insights into the underlying mechanisms involved in dehydration tolerance. These may also provide the basis for targeted alteration of metabolic routes for effective engineering strategies in rice for agricultural benefits.
EXPERIMENTAL PROCEDURES
Plant Growth, Maintenance, and Dehydration Treatment
Seeds of eight rice (Oryza sativa L.) varieties (Azucena, Anjali, Buddha, IR-64, IR-20, Moroberekan, Rasi, and Vandana) were obtained from the University of Agricultural Sciences, Bangalore, India and the Directorate of Rice Research, Hyderabad, India and grown in a mixture of soil and Soilrite (2:1, w/w; 10 plants/5.6-liter-capacity pot) in an environmentally controlled growth room. The seedlings were maintained at 28 ± 2 °C, 70 ± 5% relative humidity under a 16-h photoperiod (270 μmol m−2 s−1 light intensity). The pots were provided with 300 ml of water everyday that maintained the soil moisture content at ∼30%. A gradual dehydration condition was applied on the 4-week-old seedlings by withdrawing water, and tissues were harvested up to 120 h. The unstressed and stressed plants were kept in parallel in the same growth room. The samples from the unstressed plants were collected at each time point over the course of the dehydration experiment and were finally pooled to normalize the growth and developmental effects. The harvested unstressed and the stressed tissues were instantly frozen in liquid nitrogen and stored at −80 °C unless otherwise described.
Determination of Dehydration-induced Physiological and Biochemical Changes
The relative water content (RWC),1 lipid peroxidation, and electrolyte leakage were measured as described previously (26). In addition, the contents of free proline, total protein, photosynthetic pigments, and carotenoids were estimated as described by Bhushan et al. (26). The experiments were carried out in triplicate.
Isolation of Pure Nuclei
The nuclei were isolated as described earlier (14) with some modifications. About 40 g of seedling tissue from the Rasi variety was ground into powder in liquid nitrogen with 0.3% (w/w) polyvinylpolypyrrolidone and immediately transferred into an ice-cold 1000-ml beaker containing 400 ml of ice-cold 1× homogenization buffer (HB) (10 mm Trizma (Tris base), 80 mm KCl, 10 mm EDTA, 1 mm spermidine, 1 mm spermine, 0.5 m sucrose, pH 9.5) with 0.15% β-mercaptoethanol and 0.5% Triton X-100. The contents were gently stirred for 30 min for the complete lysing of the organellar membranes. This suspension was filtered through four layers of cheesecloth and two layers of Miracloth into an ice-cold 500-ml centrifuge bottle. The homogenate was pelleted by centrifugation with a fixed-angle rotor (1200 × g) at 4 °C for 20 min. The supernatant was separated, and the pellet was gently resuspended in 30 ml of ice-cold wash buffer (1× HB without Triton X-100). To remove the particulate matter remaining in the suspension, the resuspended nuclei were filtered into a 50-ml centrifuge tube through two layers of Miracloth by gravity. The content was centrifuged (57 × g) at 4 °C for 2 min to remove intact cells and tissue residues. The supernatant was transferred into a fresh centrifuge tube, and the nuclei were pelleted by centrifugation (1200 × g) at 4 °C for 15 min in a swinging bucket centrifuge. The pellet was washed two additional times by resuspension in wash buffer followed by centrifugation (1200 × g) at 4 °C for 15 min.
Assessment of the Purity of the Nuclear Fractions
The integrity of the isolated nuclei was analyzed by staining with 4′,6′-diamidino-2-phenylindole hydrochloride (DAPI). The nuclear fraction was stained for 15 min with 0.1 μg/ml DAPI in 0.1 m PBS (pH 7.4) and then washed twice with PBS. For microscopy, a small volume of the suspension was placed on a slide and covered with a cover glass. Images were taken with and without a UV filter. The chlorophyll content in the cell extracts, the supernatant, and the nuclei-enriched fraction was determined as described earlier (14).
Protein Blot Analysis
Immunoblotting was carried out by resolving nuclear proteins by uniform 12.5% SDS-PAGE followed by electrotransfer onto nitrocellulose membrane at 150 mA for 2 h. The membranes were subsequently blocked with 5% (w/v) nonfat milk for 1 h and incubated with the respective primary antibodies for 2 h. The blots were then incubated with alkaline phosphatase-conjugated secondary antibody for 1 h, and the signal was detected using the nitro blue tetrazolium/5-bromo-4-chloro-3-indolyl phosphate method.
Enzyme Assays
The activities of the marker enzymes catalase (EC 1.11.1.6) (27), alcohol dehydrogenase (EC 1.1.1.1) (28), fumarate hydratase (EC 4.2.1.2) (29), and glucose-6-phosphate dehydrogenase (EC 1.1.1.49) (30) were determined spectrophotometrically. The assays were done in triplicate.
Nuclear Protein Extraction and 2-DE
Nuclear proteins were prepared from the nuclei-enriched fraction using TriPure reagent (Roche Applied Science) according to the manufacturer's instructions. The protein pellet was resuspended in IEF sample buffer (8 m urea, 2 m thiourea, 4% (w/v) CHAPS). The protein concentration was determined using the 2-D Quant kit (GE Healthcare). IEF was carried out with 300 μg of protein. Aliquots of proteins were diluted with 2-D rehydration buffer (8 m urea, 2 m thiourea, 4% (w/v) CHAPS, 20 mm DTT, 0.5% (v/v) Pharmalyte (pH 4–7), and 0.05% (w/v) bromphenol blue), and 250 μl of solution was used to rehydrate the immobilized pH gradient strips (13 cm; pH 4–7). Protein was loaded by an in-gel rehydration method onto IEF strips, and electrofocusing was performed using an IPGphor system (GE Healthcare) at 20 °C for 45,000 V-h. The focused strips were subjected to reduction with 1% (w/v) DTT in 10 ml of equilibration buffer (6 m urea, 50 mm Tris-HCl (pH 8.8), 30% (v/v) glycerol, and 2% (w/v) SDS) followed by alkylation with 2.5% (w/v) iodoacetamide in the same buffer. The strips were then loaded on top of 12.5% polyacrylamide gels for SDS-PAGE. The electrophoresed proteins were stained with the Silver Stain Plus kit (Bio-Rad).
Image Acquisition and Data Analysis
Gel images were scanned with a Bio-Rad FluorS system equipped with a 12-bit camera. PDQuest version 7.2.0 (Bio-Rad) was used to assemble the first level match set (master image) from three replicate 2-DE gels. For each time point, at least three 2-DE gels, representing three biological replicates, were used for the data analysis. The detailed data analyses were carried out as described previously (26). The filtered spot quantities from the standard gels were assembled into a data matrix of high quality spots from the seven time points for further analysis.
Protein Identification and Expression Clustering
Protein samples were excised mechanically using pipette tips, destained, and in-gel digested with trypsin, and the peptides were extracted according to standard techniques (31). Peptides were analyzed by electrospray ionization LC/MS/TOF using an Ultimate 3000 HPLC system (Dionex) coupled to a Q-Trap 4000 mass spectrometer (Applied Biosystems). Tryptic peptides were loaded onto a C18 PepMap100 (3 μm; LC Packings) and separated with a linear gradient of water, acetonitrile, and 0.1% formic acid (v/v). The MS/MS data were extracted using Analyst Software version 1.4.1 (Applied Biosystems). Peptides were identified by searching the peak list against the Mass Spectrometry Protein Sequence Database (MSDB) version 20060831 (3,239,079 sequences; 1,079,594,700 residues) using Mascot version 2.1 (Matrix Science). The database search parameters were: taxonomy, O. sativa (rice) (66,430 sequences); peptide tolerance, ±1.2 Da; fragment mass tolerance, ±0.8 Da; maximum allowed missed cleavage, 1; instrument type, ESI-TRAP. Protein scores were derived from ion scores as a non-probabilistic basis for ranking protein hits, and the protein scores were derived as the sum of a series of peptide scores. The score threshold to achieve p < 0.05 is set by Mascot algorithm and is based on the size of the database used in the search. Wherein there were more than one accession number for the same peptide, the match was considered in terms of the putative function. In the case of the same protein being identified in multiple spots in which several peptides were found to be shared by the isoforms, differential expression pattern was observed for each of the candidate, and the proteins were thus listed as independent entities. The function of each of the identified protein was analyzed in view of the metabolic role of the candidate protein in the nucleus. The protein functions were assigned using a protein function database, Pfam or InterPro. As the functional annotation is based on Pfam and InterPro, the functional redundancy, if any, is thus greatly minimized. Self-organizing tree algorithm (SOTA) clustering was performed on the log-transformed -fold induction expression values across seven time points using Multi Experiment Viewer (MEV) software (The Institute for Genomic Research). The clustering was done with the Pearson correlation as distance with 10 cycles and a maximum cell diversity of 0.8 (32).
Subcellular Localization
The coding region of α7 (A7P) and β6 (B6S) subunits of 20 S proteasome and putative WD repeat-containing protein (WD) was amplified by PCR using forward and reverse (A7PF, 5′-CATGCCATGGATGAGCAGCATAGGCACAGG-3′; A7PR, 5′-GGACTAGTATCAGCATCCATCTCCTCAAGCGC-3′; B6SF, 5′-CCACATGTATGTCGAGGCGCGGCGACTG-3′; B6SR, 5′-GGACTAGTGTCCTTCCTCAGGTCGATGTAC-3′; WDF, 5′-CCACATGTATGATCTGCGCAATCTCCGGCG-3′; WDR, 5′-GGA-CTAGTCTCTTCCGATGGCTTTGCGTC-3′) primers, respectively. The amplified fragments were cloned into binary vector pCAM35S-GFP by fusing the coding region of the candidate genes in-frame. The plasmids were introduced into onion epidermal cells by particle bombardment (PDS-1000/He; Bio-Rad) using 1-μm gold particles. The localization of green fluorescent protein (GFP) was observed using a confocal laser scanning microscope (Leica Microsystems).
RESULTS AND DISCUSSION
Effect of Dehydration and Screening of Rice Varieties for Dehydration Tolerance
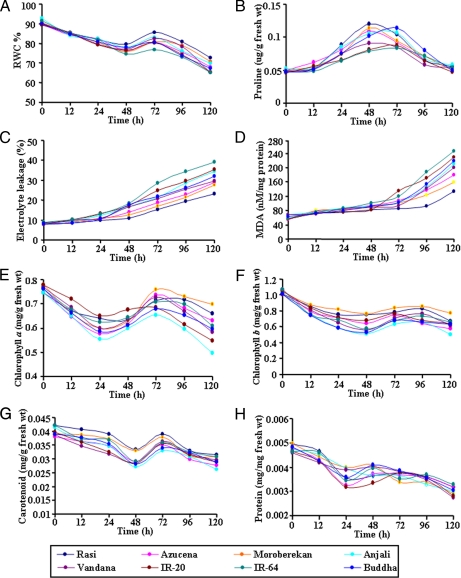
We subjected 4-week-old Rasi seedlings to gradual dehydration over 120 h. There were no visible changes in the seedlings until 48 h of dehydration, but the leaves rolled after 72 h, and the damage was further aggravated at 96–120 h (Fig. 1) during which the seedlings exhibiting pronounced wilting. Dehydration leads to a host of biochemical, physiological, and metabolic changes in plants. These changes are apparently adaptive and include various biochemical pathways associated with signal perception, transduction, and gene regulation in a temporal and spatial manner. Plant tissues can maintain turgor by avoiding dehydration and/or tolerating dehydration (33). Osmotic adjustment (OA) mostly involves the accumulation of ions and compatible solutes in the cell in response to a decrease in the water potential. Th e accumulated compatible solutes may also protect specific cellular functions irrespective of turgor (34). OA is recognized as an effective component of drought resistance in many crop plants, and RWC is considered to be the best integrated measure of plant water status (35). To find the most tolerant variety of rice, eight different varieties were subjected to progressive dehydration. Among the rice varieties studied, Rasi showed the smallest decline (∼20%) in RWC as compared with the other varieties (up to 30%) (Fig. 2A). Varietal differences in RWC during a stress period may be due to different rates of stress development and thus do not necessarily reflect different strategies of stress response. All of the varieties showed a steady increase in RWC around 48–72 h after a gradual decline during the initial stages. This could be substantiated by the observation that the proline content increased during 24–48 h causing probable OA. When compared with the other varieties, the maximum endogenous free proline content was observed in Rasi during dehydration that can be attributed to the maximum (∼90%) regain in RWC (Fig. 2, A and B). The rapid accumulation of free proline within the tissue is the most significant of the parameters responding to dehydration. Many studies have shown the important osmoprotective role of proline under stress conditions (36, 37). Changes in the cell membrane integrity are closely linked with membrane lipid peroxidation and electrolyte leakage. The conductance measurement of solute leakage from leaf cells was used as a parameter to assess the extent of membrane damage. Dehydration stress increased the amount of electrolyte leakage in all varieties of rice studied here (Fig. 2C). The maximum increase occurred in IR-64 and IR-20 by ∼4.5- and 4.2-fold, respectively, as compared with a 3-fold increase in Rasi. The electrolyte leakage was found to be maintained at an almost constant level in Rasi until 48 h and showed a marginal increase in later stages, whereas IR-64 showed a sharp rise. There was a significant increase in malondialdehyde (MDA) (∼4.0-fold) in both IR-64 and IR-20, whereas Rasi did not show much increase in MDA content (Fig. 2D). Membrane stability has been widely used to correlate abiotic stress tolerance in plants (38, 39).
Fig. 1.
The morphological responses induced by dehydration in rice. Four-week-old seedlings were subjected to progressive dehydration up to 120 h. The photographs of the same sample were taken from unstressed seedlings and at 48-, 72-, and 120-h time points. The framed region on the right in each panel depicts the leaf from plants under dehydration.
Fig. 2.
Effect of dehydration on water status, proline accumulation, cell membrane stability, photosynthetic pigments, and protein content. Four-week-old seedlings from eight different rice varieties were used for comparative estimation of RWC (A), proline accumulation (B), electrolyte leakage (C), MDA (D), chlorophyll a (E), chlorophyll b (F), carotenoid (G), and total protein (H) in a time-dependent manner under dehydration. All experiments were done in triplicates (n = 3), and average mean values were plotted against time.
With consideration to electrolyte leakage and lipid peroxidation, Rasi displayed the maximum maintenance of cell membrane integrity, which is considered to be a direct indicator of dehydration tolerance. The status of photosynthetic pigments (i.e. chlorophyll a, chlorophyll b, and carotenoid) was studied to elucidate the photosynthetic efficiencies during dehydration. The Moroberekan and Rasi varieties maintained significantly higher chlorophyll (Fig. 2, E and F) and carotenoid levels (Fig. 2G) in addition to protein content (Fig. 2H) than the Anjali and Buddha varieties. The higher carotenoid levels in Rasi and Moroberekan might result in improved protection from damage caused by dehydration. Carotenoids prevent the formation of ROS, namely singlet oxygen, by intercepting the triplet chlorophyll state (40). Initially there was a decline in the photosynthetic pigments, whereas an increase in pigment was observed during 48–72 h in all of the varieties. Interestingly during this period, there was an increase in RWC presumably due to proline accumulation. The maintenance of higher RWC in Rasi can be attributed to OA at lowered water potential, which might help in maintaining the metabolic activities and physiological processes. A decline in RWC and proline accumulation was exhibited after 72 h as the OA was unable to maintain the turgor in the affected tissues. Taken together, these results suggest that Rasi is potentially more dehydration-tolerant than the remaining varieties studied.
Evaluation of Nuclear Integrity and Purity of the Nuclear Fraction
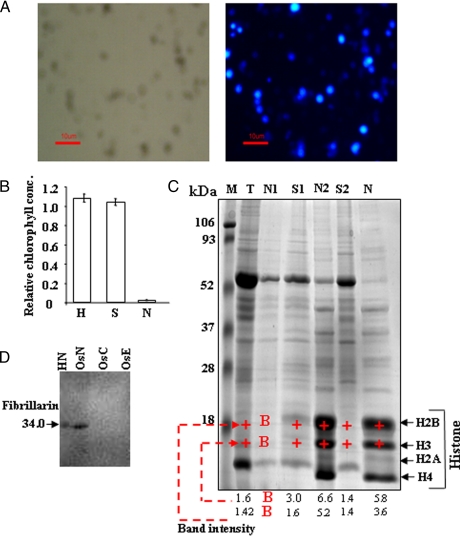
The ability to purify cellular organelles is central to a range of biological, biochemical, and proteomics studies. The separation of high purity nuclei from plant is a difficult task as it might compromise the yield, and thus nuclear isolation has been optimized to obtain highly pure nuclei. The nuclei were isolated using hyperosmotic sucrose buffer, and the nuclei-enriched pellet was obtained. The integrity of the isolated nuclei was analyzed by staining with DAPI and was examined by fluorescence microscopy. The nuclei were uniform spheres with an average diameter of ∼3–5 μm. The DAPI-stained fluorescent image of the nuclei is perfectly superimposable with their phase-contrast image, indicating integrity and absence of other contaminating organelles (Fig. 3A). Possible chloroplast contamination, if any, in the nuclear fraction was examined by spectrophotometric analysis. As shown in Fig. 3B, the supernatant retained most of the chlorophyll, and less than 2.5% of the chlorophyll was present in the nuclei fraction. Furthermore major plastid proteins were not identified in our proteome data sets suggesting that the fraction was not contaminated with detectable plastid contamination.
Fig. 3.
Analysis of rice nuclear fraction. A, the purified nuclear fraction was stained with DAPI and visualized by fluorescence microscopy. The phase-contrast micrograph of the nuclei is shown in the left panel; the DAPI-stained nuclei are shown in the right panel. B, determination of chlorophyll content at different stages of purification of nuclear fraction. The amounts of chlorophyll present in whole cell extracts (H), supernatant (S), and nuclear (N) fraction was estimated and compared. Data represent means ± S.D. of three replicates. C, representative analytical 1-D electrophoresis profile (12.5% SDS-PAGE, Coomassie Blue) obtained from rice protein extract prepared using HB. M, molecular mass protein standards; T, total cell extract; N1, nuclear protein without wash; S1, supernatant protein; N2 and S2, nuclear protein and supernatant protein, respectively, after first wash; N, final nuclear protein after another wash. The four abundant bands at around 10–18 kDa analyzed by MS were shown to contain histones H2B, H3, H2A, and H4. The intensity of the bands corresponding to histones H2B, H3 (+) was quantified by using the densitometry Quantity One 1-D software (Bio-Rad); B indicates a signal close to background levels. D, immunoblot analysis of extracted nuclear proteins with anti-fibrillarin antibodies. Aliquots of 100 μg of protein, each from nuclear (OsN), chloroplast (OsC), and extracellular matrix (OsE) fractions of rice as well as HeLa nuclear extract (HN), were separated by 12.5% SDS-PAGE. HeLa nuclear extract was used as positive control, whereas chloroplast and extracellular matrix fractions were used as negative controls. The 1-D gel was electroblotted onto Hybond-C membrane, and fibrillarin was detected using alkaline phosphatase-conjugated secondary antibody.
The nuclear proteins were prepared from the nuclei-enriched fraction using TriPure reagent to remove the contaminating nucleic acids, which might interfere in the IEF process. The enrichment of nuclear proteins was established by analyzing 1-D protein profiling wherein the nuclear fraction was clearly distinct from that of the crude extract and the supernatant recovered during the purification steps (Fig. 3C). The band of ∼56 kDa, representing Rubisco, the most abundant protein in the crude extract, was diminished with successive steps of purification, reflecting a high degree of purity. To confirm the enrichment of the nuclear proteins, we analyzed four abundant protein bands (10–18-kDa regions) of the nuclear fraction by nano-LC-MS/MS. These bands contained the histone proteins H2A, H2B, H3, and H4 of the nucleosome core particles. The histone profile of the nuclear fraction showed 5–7-fold purification for histones H2B and H3, suggesting the enrichment of the nuclear proteins in the fractions. The enrichment of the nuclear proteins was further evaluated by immunoblot analysis using specific antibody for the nuclear marker protein fibrillarin. As expected, this nucleus-resident protein was detected in the nuclear fraction but not in the extracellular matrix or chloroplast fraction (Fig. 3D).
In a separate set of experiments, the purity of the nuclear fraction was examined by assaying different organellar marker enzyme activities that could possibly contaminate the nuclear preparation. Catalase was used as a marker enzyme for peroxisome, alcohol dehydrogenase was used as a marker enzyme for cytosol, fumarate hydratase was used as a marker enzyme for mitochondria, and glucose-6-phosphate dehydrogenase was used as a marker enzyme for plastids. The whole cell extracts showed high catalase, alcohol dehydrogenase, fumarate hydratase, and glucose-6-phosphate dehydrogenase activity, whereas the nuclear proteins did not show any significant activities of these enzymes (Table I). The distinct 1-D gel profile, signal present in nuclear extracts, and enzyme activity together demonstrated that the protein had been enriched with the purification and enrichment of the nuclei and also that the nuclear preparation had no appreciable level of peroxisomes, chloroplasts, mitochondria, or other cytosolic contamination.
Table I. Biochemical analysis of nuclear purity.
Nuclear purity was determined on the basis of marker enzyme activities specific for cytosol (alcohol dehydrogenase), microbody (catalase), mitochondria (fumarase hydratase), and plastid (glucose-6-phosphate dehydrogenase). Experiments were done in triplicates. CE, crude extract; N1, nuclear fraction without wash; N2, nuclear fraction after first wash; N, final purified nuclear fraction.
| Fraction | Catalase |
Alcohol dehydrogenase |
Glucose-6-phosphate dehydrogenase |
Fumarate hydratase |
||||
|---|---|---|---|---|---|---|---|---|
| Activity | % of CE | Activity | % of CE | Activity | % of CE | Activity | % of CE | |
| μmol/mg/min | μmol/mg/min | μmol/mg/min | μmol/mg/min | |||||
| Crude extract | 28.02 ± 0.58 | 100 | 3140 × 10−5 ± 0.00004 | 100 | 158 × 10−4 ± 0.00018 | 100 | 6920 × 10−3 ± 0.17 | 100 |
| N1 | 5.01 ± 0.025 | 17.88 | 470 × 10−5 ± 0.00002 | 14.96 | 25.9 × 10−4 ± 0.00015 | 16.39 | 1090 × 10−3 ± 0.020 | 15.75 |
| N2 | 1.08 ± 0.011 | 3.85 | 70 × 10−5 ± 0.00001 | 2.22 | 4.5 × 10−3 ± 0.00012 | 2.848 | 172.50 × 10−3 ± 0.0018 | 2.49 |
| N | 0.035 ± 0.004 | 0.12 | 3.3 × 10−5 ± 0.00003 | 0.1 | 0.36 × 10−4 ± 0.00014 | 0.22 | 12.58 × 10−3 ± 0.0019 | 0.18 |
Dehydration-induced Changes and 2-DE Analysis
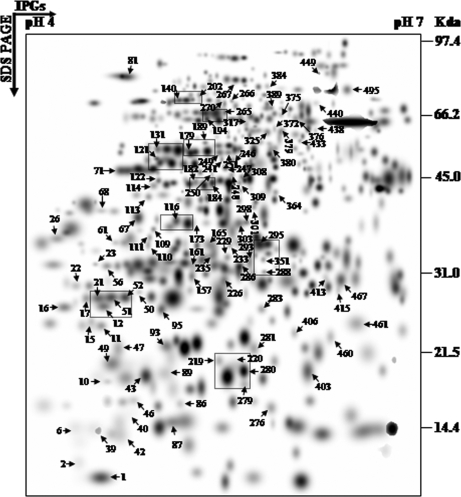
The central focus of this study was to characterize global protein expression patterns in the nucleus of a tolerant rice variety during dehydration. The dehydration-induced temporal changes in the nuclear proteome were monitored using high resolution 2-DE of nuclear extracted proteins from unstressed and dehydrated rice seedlings up to 120 h. For each time point, gels were performed at least in triplicate, which showed a high level of reproducibility with correlation coefficient values of at least 0.8, and the gels were then computationally combined into a representative standard gel (supplemental Fig. 1). The spots that survived several stringent criteria (classified as “high quality” spots) were used to estimate the spot quantities; otherwise a large number of protein spots were included in the match set. For example, 335 spots were detected in the unstressed gel, but 317 were classified as high quality (supplemental Table 1). The spot densities at the lower level were normalized against the total density present in the respective gels to overcome the experimental errors introduced due to differential staining. To compare the different time points, a second level match set was created (Fig. 4). A second normalization was carried out against the densities of unaltered protein spots. From the higher level match set, the filtered spot quantities were assembled into a data matrix that consisted of 523 unique spots, indicating changes in intensity for each spot during dehydration. Nearly 94% of the spots on the standard gels were of high quality, reflecting the reproducibility of the experimental replicates (supplemental Table 1).
Fig. 4.
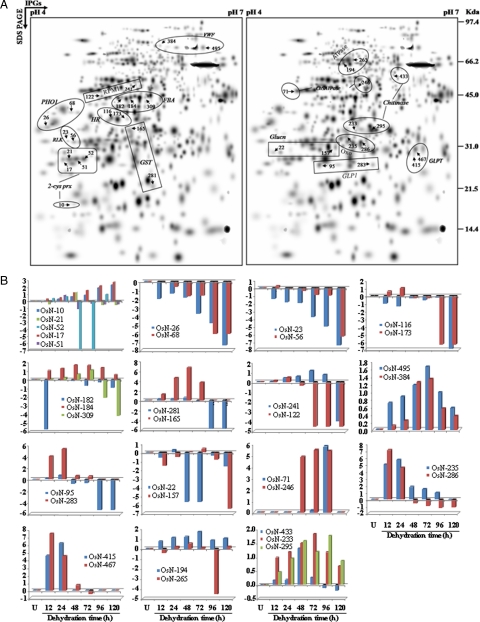
Higher level match set of protein spots detected by 2-D electrophoresis. The match set was created in silico from seven standard gels for each of the time points as depicted in supplemental Fig. 1. For the sake of clarity the protein spots within the dashed frames represent the enlarged sections in Fig. 5. The numbers indicate protein spots listed in supplemental Table 2 that were significantly identified by MS/MS analysis.
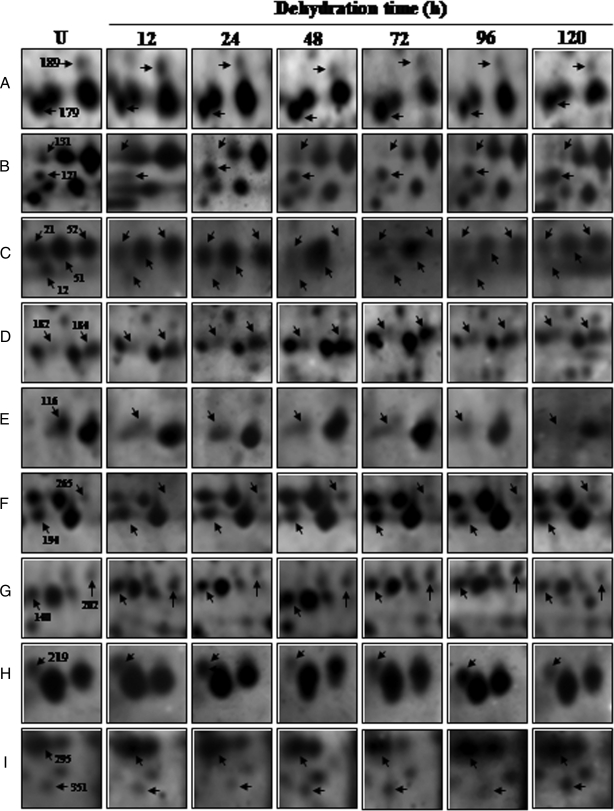
The 2-DE patterns of all protein spots under study were compared and resulted in 150 differentially expressed protein spots that showed changes of significance in their intensities, by more than 2.5-fold, at one or more time points. Although most of the spots showed quantitative changes, some spots also showed qualitative changes. Nine typical gel regions are enlarged and shown in Fig. 5. MS/MS analysis was carried out for 115 dehydration-responsive proteins (DRPs), resulting in 109 proteins with a significant match. These 109 protein spots actually account for 87 distinct proteins suggesting 80% unique protein identifications, whereas the remaining 20% of the identified DRPs either correspond to post-translationally modified forms or may be members of multigene families. Of the 109 DRPs, 45 proteins were clearly up-regulated, and 14 proteins were down-regulated, whereas 50 of the proteins showed a mixed pattern of time-dependent expression. A number of proteins involved in genome regulation and stress responses identified in our study, e.g. high mobility group I/Y (OsN-50), putative DNA helicase (OsN-113), and actin (OsN-179), have been shown previously to be nucleus-resident proteins. However, we also identified many non-canonical DRPs, such as WIP1 (OsN-87) and von Willebrand factor type A domain-containing protein as being associated with this organelle.
Fig. 5.
Time-dependent changes of the differentially expressed proteins. Four-week-old seedlings were exposed to dehydration up to 120 h. Nuclear proteins were extracted and separated by 2-DE. Enlarged sections A–I correspond to the dashed frames in Fig. 4 and are representative of selected differentially expressed nuclear protein spots across the time points under dehydration. U, unstressed.
Some of the identified proteins showed discrepancies with their theoretical Mr or pI. However, these kinds of phenomena are commonly found in 2-DE gels for several reasons. Increasing evidence suggests that protein turnover under stress is one such common adaptive phenomenon. Furthermore our data revealed 15 proteins that appeared as 35 identities (isoforms) as indicated in Fig. 6A. Interestingly four proteins representing eight isoforms showed that each set of isoforms shares similar up- or down-regulated expression patterns in response to dehydration (Fig. 6B). These include putative PHO1-like protein (OsN-26 and -68), von Willebrand factor type A domain-containing protein (OsN-384 and -495), chromosome segregation ATPases (OsN-71 and -246), and the receptor-like protein kinase (OsN-23 and -56). The second category consists of 16 identities including putative β-1,3-glucanase (OsN-22 and -157), disease resistance protein RPM1-like (OsN-122 and -241), hypersensitive induced response protein (OsN-116 and -173), glutathione S-transferase (OsN-165 and -281), germin-like protein 1 (OsN-95 and -283), germin-like protein subfamily T member 1 precursor (OsN-415 and -467), putative oxalate oxidase (OsN-235 and -286), and H+-transporting two-sector ATPase (OsN-194 and -265) that appeared as eight proteins with opposite patterns of expression. However, putative chitinase (OsN-233, -295, and -433), fructose-bisphosphate aldolase (OsN-182, -184, and -309), and 2-Cys peroxiredoxin (OsN-10, -17, -21, -51, and -52) showed 11 isoforms with mixed patterns of expression. Likewise many similar phenomena were also observed in other previously reported proteomics studies (41–44). Proteins can resolve into multiple spots due to proteolytic processing, multiple isoforms, or changes in charge state resulting from post-translational modifications (45–47). Isoforms are highly related gene products that can differ in their biological activity, regulatory properties, temporal and spatial expression, intracellular location, or any combination thereof. Isoforms are almost always either the products of one gene or of multiple genes that evolved from a single ancestor gene mostly as splice variants. The significance of their existence can be attributed to their additive function, which ultimately culminates to a cumulative and rapid response to stress (48).
Fig. 6.
Highlighted possible isoforms detected by 2-DE and their expression profile patterns. A, all 35 differentially expressed protein spots matched to 15 unique proteins are shown. The numbered spots correspond to proteins listed in Tables II B, log-transformed expression values were plotted against different hours of dehydration stress for their expression pattern profiles. U, unstressed. OxO, oxalate oxidase; GLP1, germin-like protein1; 2-cys prx, 2-Cys peroxiredoxin; RLK, receptor like protein kinase; PHO1, PHO1-like protein; ChSATPase, chromosome segregation ATPases; ATPase, H+-transporting two-sector ATPase (EC 3.6.3.14); VWF, von Willebrand factor type A domain-containing protein; Glunc, β-1,3-glucanase; RPM1, disease resistance protein RPM1-like; HR, hypersensitive induced response protein; GLPT, germin-like protein subfamily T member 1 precursor, putative, expressed; Chitinase, putative chitinase; FBP, fructose-bisphosphate aldolase, putative, expressed.
Functional Distribution of Dehydration-responsive Nuclear Proteins
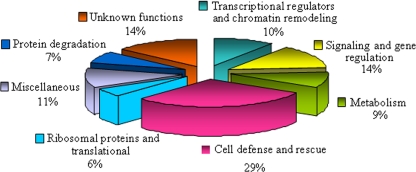
The nucleus-resident proteins are expected to act in a variety of functions during cellular adaptation to dehydration. Of all the differentially expressed nuclear proteins identified, 109 DRPs could be assigned to eight functional classes of which 15 were designated as unknown proteins based on their putative roles in the dehydration response (Fig. 7 and supplemental Table 2). Among the identified DRPs, the major functional category corresponded to proteins involved in defense and rescue (29%), and the second category corresponded to functions involved in signaling and gene regulation (14%). In addition, another category (10%) consisted of proteins involved in transcriptional regulation and chromatin remodeling, and a significant fraction of the proteins were involved in metabolic functions (9%). Proteins were also found to play a role in protein degradation (7%), others were ribosomal proteins and proteins involved in translation (6%), and a large number of proteins belong to the miscellaneous class (11%). Proteins whose function could not be ascertained and that were designated as proteins of “unknown functions” constituted 14% of the total and are therefore considered to be nucleus-specific proteins in rice. The proteomics analysis covered a wide range of nuclear functions in rice during dehydration (Fig. 7).
Fig. 7.
Functional cataloging of dehydration-responsive proteins in rice nucleus. The identified dehydration-responsive proteins were assigned a putative function using Pfam and InterPro databases and functionally categorized as represented in the pie chart.
Dynamics of the Dehydration-responsive Nuclear Protein Network
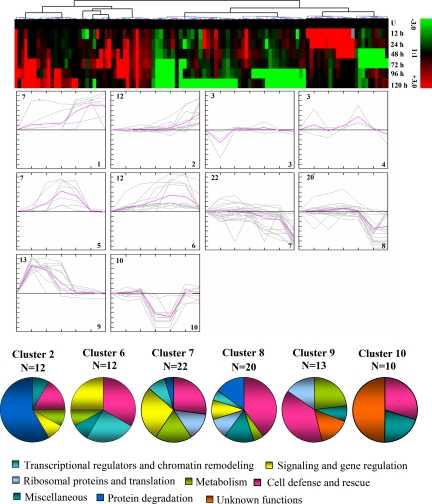
To achieve a comprehensive overview of the expression profile of the nuclear proteins that are co-expressed at different time points during dehydration, SOTA clustering was performed. The data were taken in terms of -fold expression with respect to the unstressed expression value. Furthermore the data sets were log-transformed to base 2 to level the scale of expression and to reduce the noise. The analysis yielded 10 expression clusters where only the clusters with n > 7 were taken into consideration for the study of co-expression patterns for functionally similar proteins (Fig. 8). Detailed information on the proteins within each cluster can be found in supplemental Fig. 2.
Fig. 8.
Clusterogram of expression profiles of nucleus-specific dehydration-responsive proteins in rice. A time course of dehydration-responsive protein expression in rice nucleus was constructed using 2-D electrophoresis. The differentially expressed 109 proteins were grouped into 10 clusters based on their expression profiles. The SOTA cluster tree is shown at the top, and the expression profiles in clusters are shown below. Each protein is represented by a single column of colored boxes, and each time point is represented by a single row. Induction (or repression) ranges from pale to saturated red (or green). The expression profile of each individual protein in the cluster is depicted by gray lines; the mean expression profile is marked in pink for each cluster. The number of proteins in each cluster is given in the left upper corner, and the cluster number is given in the right lower corner. The clusters with n > 7 were taken into consideration for the study of co-expression patterns for functionally similar proteins. Detailed information on proteins within each cluster can be found in supplemental Fig. 2. U, unstressed.
The most abundant groups, Clusters 7 and 8, comprised proteins showing down-regulation during the later hours of dehydration. These groups were found to be dominated by proteins involved in cell defense and rescue, signaling, and gene regulation. The other major groups are Clusters 6 and 9. The proteins in Cluster 6, maintaining up-regulated expression during 48–72 h of dehydration, were involved in signaling, gene regulation, and chromatin remodeling. However, Cluster 9 included proteins that were induced in the early dehydration response and involved in cell defense and rescue followed by metabolism. Cluster 2 consisted of proteins showing up-regulation during 96–120 h, and those in Cluster 10 showed down-regulation during 48–72 h of dehydration. The major functional classes represented by the proteins in Cluster 2 were involved in protein degradation, and Cluster 10 consisted mostly of proteins of “unknown function.” The cellular defense strategy seems to be dependent on the advancement of dehydration, and thus both early and late responses could be seen in these clusters. Furthermore the analysis of the hierarchical clustering of proteins according to their functional classes provides a detailed view of the functionality of dehydration-responsive components (supplemental Fig. 3).
Immunoblot Analysis and Subcellular Localization of a Few Nuclear Proteins
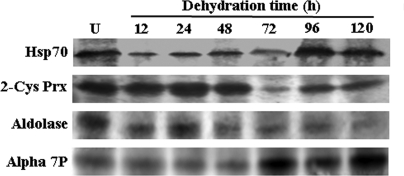
Several dehydration-responsive proteins with the same molecular weights but with different pI values were identified as the same proteins, for example 2-Cys peroxiredoxin and aldolase. Because 2-Cys peroxiredoxin and aldolase migrate in multiple spots, we used 1-D Western blot analysis to determine whether changes in individual isoelectric variants were representative of the entire protein population along with the α7 subunit of 20 S proteasome and Hsp70. 2-Cys peroxiredoxin and aldolase, represented by five and three spots, respectively, on the 2-DE gel, showed a different 1-D Western blot profile (supplemental Table 2 and Fig. 9). In contrast, a high level of similarity was observed in the expression profiles of the α7 subunit of 20 S proteasome and Hsp70 as determined by the two different techniques. However, there were differences in -fold induction and time specificity of expression for these proteins that may be due to different isoforms and different roles in dehydration tolerance.
Fig. 9.
Immunoblot analysis for a few of the representative dehydration-responsive proteins. The expression profile for Hsp70, 2-Cys peroxiredoxin (2-Cys Prx), aldolase, and α7 subunit of 20 S proteasome (Alpha 7P) was detected using 1-D Western blot analysis. An equal amount (100 μmg) of nuclear protein for all seven time points was separated by 12.5% SDS-PAGE and blotted onto Hybond-C membrane (GE Healthcare). The blots were probed with respective primary antibodies, and signal was detected. U, unstressed.
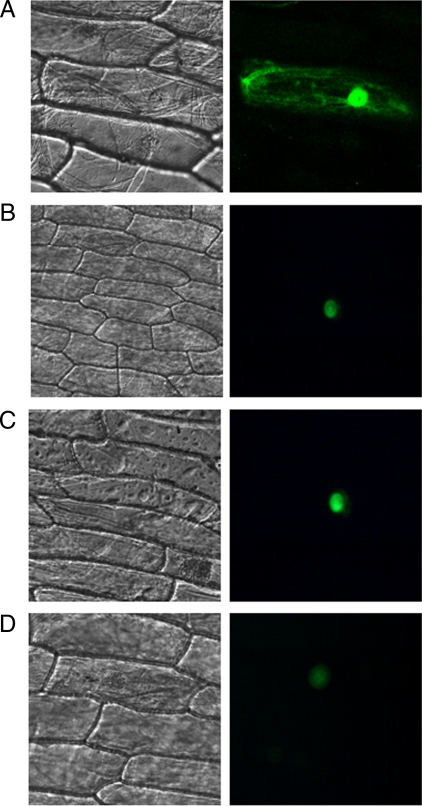
Proteasomes are soluble, but they can also be found in association with subcellular organelles. We found several proteins that were involved in proteasome degradation pathway. To confirm that these proteins were localized to the nucleus, we fused GFP to the carboxyl termini of three different proteins as described under “Experimental Procedures.” The fluorescence distribution in the cells transformed with GFP constructs was consistent with the expected nuclear localization of these proteins (Fig. 10) and was in agreement with the data from proteomics analysis.
Fig. 10.
Verification of subcellular localization of dehydration stress-regulated nuclear proteins. The cDNA of three dehydration-responsive nuclear proteins identified in rice were fused in-frame to the 5′-end of GFP and transiently expressed in Allium epidermal cells upon cell bombardment. After incubation for 24 h, Allium cells were visualized by using confocal microscopy. The phase-contrast micrograph of the Allium cells is shown in the left panel; the fluoresced image of the same is shown in the right panel. The GFP images represent localization of the following proteins fused to GFP: pCAM35S-GFP (A), α7 subunits of 20 S proteasome (A7P), A7P-GFP (B); β6 (B6S) subunits of 20 S proteasome, B6S-GFP (C); and putative WD repeat-containing protein, WD-GFP (D).
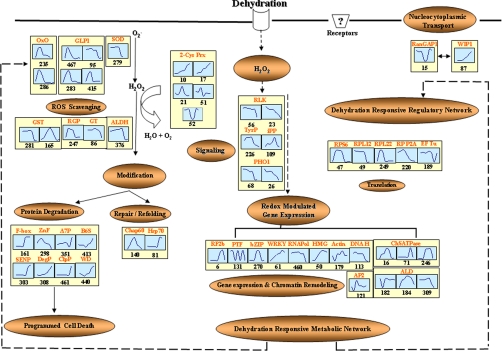
Regulatory and Metabolic Networks of Dehydration-responsive Nuclear Proteins
Our data reveal two types of dehydration responses, i.e. functional and regulatory, that help in protection of the nuclei themselves as well as the alteration in the nuclear proteome for cell survival. A model representing these responses in the nucleus under dehydration is depicted in Fig. 11. It is understood that the complex plant response to dehydration involves many proteins associated with different physiological, biochemical, and molecular processes. Acquired stress tolerance is often a result of various stress response mechanisms that may act in a coordinated fashion or synergistically to prevent cellular damage and to reestablish homeostasis (1). Accordingly the functionally classified proteins are listed in supplemental Table 2 and discussed in the following section.
Fig. 11.
Pathway involved in dehydration-dependent functional and regulatory networks in rice nucleus. Proteins identified in this study are indicated in the box. Graphs are representative of expression profiles of individual proteins, and the number given below each graph indicates the protein identification number. OxO, oxalate oxidase; GLP1, germin-like protein 1; SOD, superoxide dismutase; ALDH, aldehyde dehydrogenase; 2-Cys Prx, 2-Cys peroxiredoxin; RLK, receptor-like protein kinase; TyrP, tyrosine phosphatase; iPP, inorganic pyrophosphatase; PHO1, PHO1-like protein; RPS6, ribosomal protein S6; RPL12, ribosomal protein L12; RPL22, ribosomal protein L22; RPP2A, ribosomal protein P2A; Chap60, chaperonin 60; Hsp70, 70-kDa heat shock protein; RGP, reversibly glycosylated peptide; GT, glycosyltransferase; F-box, F-box-like protein; ZnF, zinc finger, C3HC4 type (RING finger); A7P, α7 subunit of 20 S proteasome; B6S, β6 subunit of 20 S proteasome; DegP, putative DegP protease; SENP, putative sentrin-specific protease; ClpP, ATP-dependent Clp protease proteolytic subunit; WD, WD repeat-containing protein, putative; RF2b, bZIP transcription factor RF2b; PTF, putative transcription factor; bZIP, putative bZIP protein; WRKY, WRKY transcription factor; RNApol, putative RNA polymerase II subunit 5; HMG, putative high mobility group I/Y; Actin, putative actin; ALD, fructose-bisphosphate aldolase; DNA H, putative DNA helicase; ChSATPase, chromosome segregation ATPases; AP2, AP2 domain-containing protein; WIP, WPP domain-interacting protein 1; RanGAP1, Ran GTPase-activating protein 1.
Transcriptional Regulators and Chromatin Remodeling
Regulatory proteins, such as transcription factors, are low abundance proteins and have a high turnover rate; as a result, many of these would have escaped detection by 2-DE. Nevertheless several transcription factors such as RF2b (OsN-6), WRKY (OsN-61), putative transcription factor (OsN-131), and putative bZIP (OsN-270) were identified. RF2b and putative bZIP showed up-regulation at 72–96 h of dehydration, whereas WRKY displayed up-regulation across all the time points, and putative transcription factor showed a mixed pattern of regulation. RF2b is known to be involved in the biotic stress response (49), but its role in abiotic stress has not yet been established. The bZIP transcription factor, which binds to the G-box regulatory element, is found in many plant promoters. It has been shown to be involved in developmental and physiological processes in response to major environmental cues. The WRKY factors act by directing the temporal and spatial expression of specific genes, thereby ensuring proper cellular responses to internal and external stimuli. The transcription of WRKY is strongly and rapidly up-regulated in response to wounding, pathogen infection, or abiotic stress (50). OsN-121, an AP2 domain-containing protein, showed a mixed pattern of expression under dehydration. CBF1 encodes an AP2 domain-containing transcriptional activator that stimulates transcription in response to low temperature and water deficit in Arabidopsis (51). The RNA polymerase II subunit 5 (OsN-460) is one of the communicating subunits of RNA polymerase II that interacts with transcriptional regulators (52), and it showed a mixed pattern of regulation in our study.
It is known that abiotic stress affects the cellular gene expression machinery and molecules involved in nucleic acid processing, including DNA helicase (OsN-113) (53, 54). Chromosome segregation ATPases (OsN-16, -246, and -271), high mobility group I/Y (OsN-50), and actin (OsN-179) were up-regulated in response to dehydration, suggesting their involvement in chromatin remodeling and gene regulation (55–57). The identification of the transcription factors and proteins involved in chromatin remodeling and signaling suggests their possible role in dehydration-dependent regulation of gene expression.
Signaling and Gene Regulation
Protein phosphorylation and dephosphorylation regulate numerous biological processes and are catalyzed by protein kinases and phosphatases. Several receptor-like protein kinases (OsN-23 and -53), 33-kDa secretory protein (serine/threonine kinase-containing domain; OsN-229), tyrosine phosphatase-like protein (OsN-226), and putative inorganic pyrophosphatase (OsN-109) were found to be early responsive proteins, suggesting their role in the recognition of signaling molecules and the subsequent activation of an intracellular defense response. During dehydration, putative PHO1-like proteins (OsN-26 and -68), regulatory factors of Pi transport, might be involved in Pi homeostasis and G-protein-associated signaling (58). A number of the glycolytic enzymes present in the nucleus are thought to be modified forms of those found in the cytoplasm, for example aldolase (OsN-182, -184, and -309). These proteins are known to bind to DNA and may play a role in transcription and replication (59).
Various sets of proteins are involved in transcriptional and translational processes in the cell, indicating the possible translocation of many such molecules in and out of the nucleus at any given time (60). The major regulator in this translocation is an intracellular signaling protein called RanGAP1 (OsN-15), which stands for Ran GTPase-activating protein 1, in the nucleus. RanGAP1 was up-regulated during the later phases of dehydration (supplemental Table 2). This protein is known to play a crucial role in nucleocytoplasmic transport (61). WPP domain-interacting protein 1 (WIP1; OsN-87) is a plant-specific zinc finger protein that interacts with RanGAP1 (OsN-15), co-localizes at the nuclear envelope and cell plate (62), and shows up-regulation in the later phases of dehydration; however, its definite role in abiotic stress is not yet known.
Metabolism
It has become increasingly evident that plants have an extraordinary capacity to perceive changes in their external environment and adapt rapidly to maximize opportunity and minimize risk. This plasticity depends on the evolution of diverse mechanisms to regulate cellular homeostasis where glycosylation is one of these mechanisms. Reversible glycosylated polypeptides (OsN-247) are highly conserved plant-specific proteins that are believed to be involved in the process of delivering sugar residues to glycosyltransferases (OsN-86) (63). Glycosyltransferases are able to recognize hormones, secondary metabolites, and biotic and abiotic chemicals and toxins in the environment; thus, their differential expression suggests their potential role in the defense response against dehydration.
During dehydration, proteins involved in amino acid metabolism, such as aminotransferase (OsN-250), methylenetetrahydrofolate reductase (OsN-266), methyltransferases (OsN-380), and 5-methyltetrahydropteroyltriglutamate-homocysteine methyltransferase (OsN-449), were found to be up-regulated. The aminotransferase (OsN-250) reaction has been reported to be involved in diverse metabolic pathways in addition to plant stress responses (64).
The regulation of methylenetetrahydrofolate reductase (OsN-266) activity is crucial for maintaining cellular concentrations of methionine and S-adenosylmethionine. Methyl transfer from the ubiquitous S-adenosyl-l-methionine to nitrogen, oxygen, or carbon atoms is catalyzed by methyltransferases (OsN-380) and modifies nucleic acids and proteins. DNA methylation has been implicated in the regulation of several cellular processes. One such interesting finding was the up-regulation of 5-methyltetrahydropteroyltriglutamate-homocysteine methyltransferase (OsN-449), which is implicated in the synthesis of methionine and glutamate dehydrogenase, suggesting their possible role in dehydration tolerance.
Cell Defense and Rescue
The nucleus is the reservoir of ROS accumulation, but recent reports have suggested that it may also be an active source of ROS production (20, 65). It is becoming evident that ROS, generated during pathogen attack and abiotic stress, are recognized by plants as a signal for triggering defense responses. A significant proportion of the identified proteins were found to be linked to antioxidative/detoxifying reactions. Germin and germin-like proteins (OsN-95, -283, -415, and -467) are early dehydration-responsive proteins. These proteins are involved in many developmental and stress-related processes and exhibit oxalate oxidase activity because their production results in stress-related release of hydrogen peroxide (H2O2) (66). Probable superoxide dismutase (Cu-Zn) precursor (OsN-279) showed an early dehydration response, indicating its role in scavenging the stress-induced generation of ROS. We identified five spots as 2-Cys peroxiredoxin (OsN-10, -17, -21, -51, and -52) of which three showed mixed patterns of expression (supplemental Table 2) and two showed up-regulation during dehydration. This protein is known to be regulated by drought in rice seedlings (67). The ascorbate peroxidase is highly sensitive to inactivation by ROS and is often insufficient to protect the proteins during severe drought stress (68). Therefore, the 2-Cys peroxiredoxin can be an important alternative for the detoxification of H2O2 under normal conditions as well as under dehydration-induced oxidative stress. ROS interact with the lipid membranes that can amplify cellular toxicity by the formation of lipid hydroperoxides and their aldehyde degradation products (69). Aldehyde dehydrogenase (OsN-376) is responsible for the degradation of these aldehydes arising from reactions of ROS with lipids and proteins (70), and levels of aldehyde dehydrogenase were significantly increased during the early time periods.
Glutathione S-transferases (OsN-165 and -281) with enzymatic GSH peroxidase activity catalyze the conjugation of glutathione to a variety of toxic substrates arising from oxidative stress (71). The ROS can act as messengers either directly or by using an oxidized product as a secondary messenger (72, 73). Typically cellular damage is observed under stress when the ROS levels are increased, although many oxidative modification products are involved in metabolic regulation and signal transduction. These indicate a functional justification for the possible involvement of ROS in the stressed tissues and clarify the potential role of detoxifying enzymes under dehydration. In adapting to such adverse conditions, the cells may undergo altered metabolic processes that result in the differential expression of diverse sets of proteins, including a variety of well characterized antioxidant enzymes (supplemental Table 2). It is most likely that oxidized proteins are degraded relatively rapidly and are readily recognized by proteases (74, 75). Several differentially expressed proteins appeared to be the products of degradation because their observed molecular mass was much smaller than the theoretical mass. We observed this phenomenon in a few proteins, such as ternary complex factor MIP1 (OsN-93), chromosome segregation ATPases (OsN-16), and putative chitinase (OsN-42) among others. The above mentioned disparities may be attributed to the stress-responsive ROS-mediated degradation of proteins (76–78).
Furthermore we identified Hsp70 (OsN-81) and putative chaperonin 60β (OsN-140), which prevent protein misfolding and random aggregation in the cell. Under normal conditions, Hsp70 is present mainly in the cytosol, but it translocates to the nucleus and nucleolus during physiological stress (79). This movement of Hsp70 in and out of the nucleolus has been associated with the repair of stress-induced nucleolar damage (80) by storing non-nucleolar unfolded proteins. The concentration of unfolded proteins at one locus in the nucleus reduces the damage by preventing their random aggregation with other nuclear proteins and macromolecular structures.
The expression of a number of genes from different pathways can affect the response of rice to dehydration. Pathogenesis-related protein PR10a (OsN-43), chitinase (OsN-42, -233, -295, and -433) and endo-1,3-glucanase (OsN-22, -157, and -202) are expressed by most plants in response to pathogen infection and, in many cases, in response to abiotic stress (81, 82).
Ribosomal Proteins and Translation
Several spots representing ribosomal proteins were identified; putative S6 (RPS6; OsN-47) and 50 S L22-like (OsN-249) ribosomal proteins were up-regulated, whereas L12 (OsN-49) showed down-regulation, and 60 S acidic ribosomal protein P2A (OsN-220) showed a mixed pattern of regulation. L12 may contribute to a mechanism for regulating ribosome synthesis and/or maturation (83). There is evidence that the phosphorylation of RPS6 may be regulated in plants during development and in response to changes in the environment (84). Although the significance of the modulation of the ribosomal protein expression in response to dehydration is not clear, it can be linked to the differences in protein composition between unstressed and stressed plants. In the unstressed cells, the protein synthesis elongation factor (EF)-Tu (OsN-189) catalyzes the GTP-dependent binding of aminoacyl-tRNA (OsN-372) to the A-site of the ribosome during the elongation phase of protein synthesis. However, EF-Tu acts as a molecular chaperone during stress, and it may be involved in protein folding and protection.
Protein Degradation
During stress adaptation, protein degradation is necessary for the removal of abnormal or damaged proteins and for altering the balance of proteins. A significant proportion of the nuclear proteins identified in this study are involved in the ubiquitin-proteasome pathway. Although the cytoplasmic proteasome has been reported to be involved in the ubiquitin-mediated turnover of misfolded proteins and in signal transduction (74), recent studies have indicated the important role of nuclear proteasomes in chromatin-associated protein degradation (75) and gene expression (85). We observed the mixed expression pattern of the β6 (OsN-413) and α7 subunits (OsN-351) of the 20 S proteasome during dehydration. It has been shown that several RING fingers act as E3 enzymes in the ubiquitination process. The C3HC4 type RING zinc finger protein (OsN-298) is related to post-translational modification, protein turnover, or protein-protein interaction (86) and may play an important role in defense against dehydration. The WD repeat-containing putative protein (OsN-440) is a U-box domain-containing protein, which is known to have in vitro E3 ubiquitin ligase activity (87). As components of the Skp1p-cullin-F-box complex, F-box proteins (OsN-161) are critical for the controlled degradation of cellular proteins.
Proteases play key roles in plants, maintaining strict protein quality control and degrading specific sets of proteins in response to diverse environmental and developmental stimuli. In this study, putative DegP (OsN-308), sentrin-specific (OsN-303), and ATP-dependent proteolytic subunit of Clp (OsN-461) proteases were found to be up-regulated at later time points of dehydration. The functions of these proteases in protein degradation in the nucleus can now be efficiently studied by reverse genetics approaches. Furthermore the identification of these proteases allows us to decipher the hierarchy of the putative proteolytic cascade in plant nucleus under dehydration.
Miscellaneous
A total of 11% of the proteins did not fall into the easily categorized groups discussed above. Proteins such as ATP synthase CF1 α chain (OsN-389) were down-regulated, whereas H+-transporting two-sector ATPase α chain (OsN-265) and ATP synthase δ subunit (OsN-12) showed mixed patterns, and putative vacuolar proton-ATPases (OsN-267) and H+-transporting two-sector ATPase β chain (OsN-194) were up-regulated. Emerging evidence suggests that the nuclear envelope itself is responsible for transport and signaling activities quite distinct from those associated with the nuclear pore (88). Nuclear monovalent cation transporters and channels are likely to play a role in the modulation of chromatin structure and gene expression (89), although it is premature to speculate on the possible physiological functions of putative nuclear ion transporters during dehydration. The protein spot OsN-2 was identified as a putative retroelement, which may play an important role in reorganization of genes and in the optimization of the gene expression in response to various environmental stimuli.
Proteins of Unknown Functions
Of the 109 proteins we identified, 14% were annotated as hypothetical proteins (supplemental Table 2). To determine the putative function of these proteins, we submitted this group of proteins to functional domain analysis (supplemental Table 3). These proteins were examined for conserved domains using the InterPro database and queried for domains in the SMART (Simple Modular Architecture Research Tool), Panther, and Pfam databases. These analyses identified the conserved domain of seven proteins with known activities as shown in supplemental Table 3. Hypothetical protein (OsN-15) having Rna1p (RanGAP1) amino-terminal domain is involved in nuclear transport. Furthermore hypothetical protein (OsN-280) having conserved domain Alba (acetylation lowers binding affinity) is uniformly and abundantly distributed on the chromosome. Alba is proposed to play a role in transcriptional regulation by maintaining chromatin architecture (90). Hypothetical protein (OsN-46) with SPT2 chromatin protein domain (91) and OsN-317 with a cyclin-like F-box domain are known to be involved in transcriptional regulation.
Comparative Biology of Dehydration-responsive Nuclear Proteome
Although a number of proteomics studies have been reported in plants, comparative crop-specific organellar proteomics studies describing evolutionary significance are limited. To investigate the comparative nuclear proteome at the organismal level, we compared the nuclear proteome of rice with that of Arabidopsis (15), Medicago (21), and chickpea (14, 20), revealing 32 proteins as common residents in these species (supplemental Table 4). Proteins involved in chromatin remodeling and gene regulation, DNA replication and transcription, RNA processing and export, signaling, nucleocytoplasmic transport, and protein synthesis and degradation were found to be the most abundant classes across all nuclear proteomes cited above. Interestingly although there exists a certain level of similarity among the functional classes of nuclear proteins, a large number of proteins were found to be species-specific (supplemental Table 4). The differential protein networks in different crop species might signify the evolutionary species-specific dynamism of the nuclear proteome. Currently there is only one report available on dehydration-responsive nuclear proteome in chickpea (20), and its comparison with that of rice has provided valuable insights into the dehydration response in two major lineages of economically important food crops. There is a wide divergence between the two crops; the chickpea grows on residual soil moisture, whereas rice requires plenty of water. Furthermore they belong to two distantly related taxonomic groups; the chickpea is a eudicot, and rice is a monocot. More than 90% of the DRPs identified were unique to each crop, whereas only 16 were found to be identical (Table II). In addition, there is a high degree of variance in their protein classes. For example, the highest number of glycine-rich RNA-binding proteins was identified from chickpea (20), whereas 2-Cys peroxiredoxin proteins were most predominant in rice. These differences may be attributed to different cellular environments and ecological habitats. Despite the evolutionary relevance of variation, our study on the differential display of nuclear proteome shows similarities in the case of a few of the essential functional protein classes. It has become increasingly evident that functionally important proteins are more evolutionarily conserved than less vital proteins (92, 93). As expected, proteins involved in signaling and gene regulation and in cell defense and rescue were found to be the most abundant in both the crops. The conservation of differentially expressed nuclear protein orthologues and paralogues between the crops suggests a possible ubiquitous mechanism of cell defense strategy as well as a common layout of the signaling network. These data together provide evidence for molecular diversity as opposed to commonality in the differential protein profile at the organismal level. Nevertheless the higher percentage of crop-specific DRPs signifies the necessity of studying the nuclear proteome of different crops and especially the major lineages of higher plants to better understand the critical role of this organelle in stress tolerance.
Table II . List of common dehydration-responsive nuclear proteins in two different plants.
| Functional category and identification | O. sativaa | Cicer arietinum |
|---|---|---|
| Signaling and gene regulation | ||
| Receptor-like protein kinase | OsN-56 | Similarity to receptor-like protein kinase |
| 33-kDa secretory protein, putative, expressed (serine/threonine kinase domain) | OsN-229 | Serine/threonine kinase |
| Protein-tyrosine phosphatase-like protein | OsN-226 | Hypothetical protein OSJNBa0011L09.21 (protein-tyrosine phosphatase, putative) |
| Fructose-bisphosphate aldolase, putative, expressed | OsN-184 | Fructose-bisphosphate aldolase (EC 4.1.2.13) |
| Transcriptional regulators and chromatin remodeling | ||
| WRKY DNA binding domain-containing protein | OsN-61 | AY045676 NID (WRKY DNA binding domain) |
| Putative transcription factor | OsN-131 | Putative transcription factor |
| Putative actin | OsN-179 | Actin |
| Cell defense and rescue | ||
| Putative thioredoxin peroxidase (2-Cys peroxiredoxin) | OsN-17 | 2-Cys peroxiredoxin-like protein (fragment) |
| Probable superoxide dismutase (Cu-Zn) precursor | OsN-279 | Superoxide dismutase (EC 1.15.1.1) (manganese) 1 (similarity) |
| 70-kDa heat shock-related protein, putative, expressed | OsN-81 | dnaK-type molecular chaperone CSS1 precursor |
| Putative chaperonin 60β | OsN-140 | Probable chaperonin 60β chain |
| AP2 domain-containing protein | OsN-121 | AP2/EREBP transcription factor baby boom |
| Ribosomal proteins and translation | ||
| Translational elongation factor Tu | OsN-189 | EF-Tu precursor, chloroplast |
| Protein degradation | ||
| α7 subunit of 20 S proteasome | OsN-351 | ATP dependent 26 S proteasome regulatory subunit |
| F-box protein family-like | OsN-161 | Cyclin-like F-box; agenet hypothetical protein (kelch repeat-containing F-box family protein) |
| Miscellaneous | ||
| H+-transporting two-sector ATPase (EC 3.6.3.14) β chain | OsN-194 | H+-transporting two sector ATPase (EC 3.6.3.14) β chain |
a The first letters (Os) signify the source plant, O. sativa, followed by the subcellular fraction, nucleus (N). The numerals indicate the spot number in this study.
In summary, the present study represents the first effort to delineate the molecular basis of the acquisition of tolerance under water deficit conditions in the rice nucleus. Although biological processes, including the dehydration response, are controlled by intricate regulatory networks of protein expression that are thought to be regulated by many regulatory genes, only a few such genes have been reported to date. The exploration of the dehydration-responsive protein expression patterns in rice uncovered novel regulatory proteins, including bZIP transcription factor RF2b, WIP1, von Willebrand factor type A, and Alba domain-containing protein, strongly suggesting putative functions for them in plant dehydration response. These results support the existence of different regulatory systems that operate in dehydration tolerance. The identification of the differentially regulated proteins in the rice nucleus under progressive water loss provides a foundation for further functional studies to determine their precise biochemical roles in dehydration tolerance. In addition, it may serve as a basis to compare nucleus-specific key regulatory processes at the genotype level.
Acknowledgments
We thank Dr. E. A. Shiddiqi, Directorate of Rice Research, Hyderabad, India and Dr. H. E. Shashidhar, University of Agricultural Sciences, Bangalore, India for providing the seeds of different rice varieties. We also thank Jasbeer Singh for illustrations and graphical representation in the manuscript.
Footnotes
* This work was supported by grants from the Council of Scientific and Industrial Research (CSIR), Government of India and the National Institute of Plant Genome Research, New Delhi, India.
 The on-line version of this article (available at http://www.mcponline.org) contains supplemental material.
The on-line version of this article (available at http://www.mcponline.org) contains supplemental material.
1 The abbreviations used are:
- RWC
- relative water content
- 2-DE
- two-dimensional electrophoresis
- DAPI
- 4′,6′-diamidino-2-phenylindole hydrochloride
- SOTA
- self-organizing tree algorithm
- DRPs
- dehydration-responsive proteins
- ROS
- reactive oxygen species
- MDA
- malondialdehyde
- OA
- osmotic adjustment
- HB
- homogenization buffer
- 2-D
- two-dimensional
- GFP
- green fluorescent protein
- 1-D
- one-dimensional
- Rubisco
- ribulose-bisphosphate carboxylase/oxygenase
- bZIP
- basic leucine zipper
- EF
- elongation factor
- E3
- ubiquitin-protein isopeptide ligase
- Alba
- acetylation lowers binding affinity
REFERENCES
- 1.Wang W., Vinocur B., Altman A. ( 2003) Plant responses to drought, salinity and extreme temperatures: towards genetic engineering for stress tolerance. Planta 218, 1– 14 [DOI] [PubMed] [Google Scholar]
- 2.Bray E. A. ( 2004) Genes commonly regulated by water-deficit stress in Arabidopsis thaliana. J. Exp. Bot. 55, 2331– 2341 [DOI] [PubMed] [Google Scholar]
- 3.Boyer J. S. ( 1982) Plant productivity and environment. Science 218, 443– 448 [DOI] [PubMed] [Google Scholar]
- 4.Food and Agricultural Organization of the United Nations ( 2002) Crops and Drops: Making the Best Use of Water for Agricultural, Food and Agricultural Organization of the United Nations, Rome [Google Scholar]
- 5.Shinozaki K., Yamaguchi-Shinozaki K., Seki M. ( 2003) Regulatory network of gene expression in the drought and cold stress responses. Curr. Opin. Plant Biol. 6 410– 417 [DOI] [PubMed] [Google Scholar]
- 6.Xiong L., Schumaker K. S., Zhu J. K. ( 2002) Cell signaling during cold, drought, and salt stress. Plant Cell 14, S165– 183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kreps J. A., Wu Y., Chang H. S., Zhu T., Wang X., Harper J. F. ( 2002) Transcriptome changes for Arabidopsis in response to salt, osmotic, and cold stress. Plant Physiol. 130, 2129– 2141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rabbani M. A., Maruyama K., Abe H., Khan M. A., Katsura K., Ito Y., Yoshiwara K., Seki M., Shinozaki K., Yamaguchi-Shinozaki K. ( 2003) Monitoring expression profiles of rice genes under cold, drought, and high-salinity stresses and abscisic acid application using cDNA microarray and RNA get-blot analyses. Plant Physiol. 133, 1755– 1767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tian Q., Stepaniants S. B., Mao M., Weng L., Feetham M. C., Doyle M. J., Yi E. C., Dai H., Thorsson V., Eng J., Goodlett D., Berger J. P., Gunter B., Linseley P. S., Stoughton R. B., Aebersold R., Collins S. J., Hanlon W. A., Hood L. E. ( 2004) Integrated genomic and proteomic analyses of gene expression in mammalian cells. Mol. Cell. Proteomics 3, 960– 969 [DOI] [PubMed] [Google Scholar]
- 10.Gygi S. P., Rochon Y., Franza B. R., Aebersold R. ( 1999) Correlation between protein and mRNA abundance in yeast. Mol. Cell. Biol. 19, 1720– 1730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pradet-Balade B., Boulmé F., Beug H., Müllner E. W., Garcia-Sanz J. A. ( 2001) Translation control: bridging the gap between genomics and proteomics? Trends Biochem. Sci. 26, 225– 229 [DOI] [PubMed] [Google Scholar]
- 12.Dreger M. ( 2003) Subcellular proteomics. Mass Spectrom. Rev. 22, 27– 56 [DOI] [PubMed] [Google Scholar]
- 13.Bhushan D., Pandey A., Chattopadhyay A., Choudhary M. K., Chakraborty S., Datta A., Chakraborty N. ( 2006) Extracellular matrix proteome of chickpea (Cicer arietinum L.) illustrates pathway abundance, novel protein functions and evolutionary perspect. J. Proteome Res. 5, 1711– 1720 [DOI] [PubMed] [Google Scholar]
- 14.Pandey A., Choudhary M. K., Bhushan D., Chattopadhyay A., Chakraborty S., Datta A., Chakraborty N. ( 2006) The nuclear proteome of chickpea (Cicer arietinum L.) reveals predicted and unexpected proteins. J. Proteome Res. 5, 3301– 3311 [DOI] [PubMed] [Google Scholar]
- 15.Bae M. S., Cho E. J., Choi E. Y., Park O. K. ( 2003) Analysis of the Arabidopsis nuclear proteome and its response to cold stress. Plant J. 36, 652– 663 [DOI] [PubMed] [Google Scholar]
- 16.Khan M. M., Komatsu S. ( 2004) Rice proteomics: recent developments and analysis of nuclear proteins. Phytochemistry 65, 1671– 1681 [DOI] [PubMed] [Google Scholar]
- 17.Calikowski T. T., Meulia T., Meier I. ( 2003) A proteomic study of the Arabidopsis nuclear matrix. J. Cell. Biochem. 90, 361– 378 [DOI] [PubMed] [Google Scholar]
- 18.Li G., Nallamilli B. R., Tan F., Peng Z. ( 2008) Removal of high abundance proteins for nuclear subproteome studies in rice (Oryza sativa) endosperm. Electrophoresis 29, 604– 617 [DOI] [PubMed] [Google Scholar]
- 19.Pendle A. F., Clark G. P., Boon R., Lewandowska D., Lam Y. W., Andersen J., Mann M., Lamond A. I., Brown J. W., Shaw P. J. ( 2005) Proteomics analysis of the Arabidopsis nucleolus suggests novel nucleolar functions. Mol. Biol. Cell 16, 260– 269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pandey A., Chakraborty S., Datta A., Chakraborty N. ( 2008) Proteomics approach to identify dehydration responsive nuclear proteins from chickpea (Cicer arietinum L). Mol. Cell. Proteomics 7, 88– 107 [DOI] [PubMed] [Google Scholar]
- 21.Repetto O., Rogniaux H., Firnhaber C., Zuber H., Küster H., Larré C., Thompson R., Gallardo K. ( 2008) Exploring the nuclear proteome of Medicago truncatula at the switch towards seed filling. Plant J. 56, 398– 410 [DOI] [PubMed] [Google Scholar]
- 22.Food and Agricultural Organization of the United Nations ( 2007) FAOSTAT: Agricultural Database, Food and Agricultural Organization of the United Nations, Rome [Google Scholar]
- 23.Babu R. C., Zhang J. X., Blum A., Ho T. H. D., Wu R., Nguyen H. T. ( 2004) HVA1, a LEA gene from barley confers dehydration tolerance in transgenic rice (Oryza sativa L.) via cell membrane protection. Plant Sci. 166, 855– 862 [Google Scholar]
- 24.Abbasi F. M., Komatsu S. ( 2004) A proteomic approach to analyze salt responsive proteins in rice leaf sheath. Proteomics 4, 2072– 2081 [DOI] [PubMed] [Google Scholar]
- 25.Heazlewood J. L., Howell K. A., Whelan J., Millar A. H. ( 2003) Towards an analysis of the rice mitochondrial proteome. Plant Physiol. 132, 230– 242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bhushan D., Pandey A., Choudhary M. K., Datta A., Chakraborty S., Chakraborty N. ( 2007) Comparative proteomic analysis of differentially expressed proteins in chickpea extracellular matrix during dehydration stress. Mol. Cell. Proteomics 6, 1868– 1884 [DOI] [PubMed] [Google Scholar]
- 27.Luck H. ( 1965) Catalase, in Methods of Enzymatic Analysis ( Bergmeyer H., ed) pp. 885– 894Academic Press, New York [Google Scholar]
- 28.Widholm J. M., Kishinami I. ( 1988) Allyl alcohol selection for lower alcohol dehydrogenase activity in Nicotiana plumbaginifolia cultured cells. Plant Physiol. 86, 266– 269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hatch M. D. ( 1978) A simple spectrophotometric assay for fumarate hydratase in crude tissue extracts. Anal. Biochem. 85, 271– 275 [DOI] [PubMed] [Google Scholar]
- 30.Simcox P. D., Reid E. E., Canvin D. T., Dennis D. T. ( 1977) Enzymes of the glycolytic and pentose phosphate pathways in proplastids from the developing endosperm of Ricinus communis. Plant Physiol. 59, 1128– 1132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Casey T. M., Arthur P. G., Bogoyevitch M. A. ( 2005) Proteomic analysis reveals different protein changes during endothelin-1- or leukemic inhibitory factor-induced hypertrophy of cardiomyocytes in vitro. Mol. Cell. Proteomics 4, 651– 661 [DOI] [PubMed] [Google Scholar]
- 32.Romijn E. P., Christis C., Wieffer M., Gouw J. W., Fullaondo A., van der Sluijs P., Braakman I., Heck A. J. ( 2005) Expression clustering reveals detailed co-expression patterns of functionally related proteins during B cell differentiation. Mol. Cell. Proteomics 4, 1297– 1310 [DOI] [PubMed] [Google Scholar]
- 33.Kramer P. J., Boyer J. S. ( 1995) Water Relations of Plants and Soils, pp. 193– 195Academic Press, San Diego, CA [Google Scholar]
- 34.Paleg L. G., Stewart G. R., Starr R. ( 1985) The effect of compatible solutes on proteins. Plant Soil 89, 83– 94 [Google Scholar]
- 35.Blum A. ( 1996) Developing drought and low N tolerant maize, in Proceedings of a Symposium at the International Maize and Wheat Improvement Center (CIMMYT), El-Batan, March 25–29, 1996 ( Edmeades G. O., Banziger M., Mickelson H. R., Pena-Valdivia C. B., eds) pp. 131– 135CIMMYT, El-Batan, Mexico [Google Scholar]
- 36.Jain R. K., Dhawan R. S., Sharma D. R., Chowdhry J. B. ( 1987) Salt tolerance and proline accumulation: a comparative study in salt tolerant and wild type cultured cells of eggplant. Plant Cell Rep. 6, 382– 384 [DOI] [PubMed] [Google Scholar]
- 37.Demiral T., Turkan I. ( 2006) Exogenous glycinebetaine affects growth and proline accumulation and retards senescence in two rice cultivars under NaCl stress. Environ. Exp. Bot. 56, 72– 79 [Google Scholar]
- 38.Blum A., Ebercon A. ( 1981) Cell membrane stability as a measure of drought and heat tolerance in wheat. Crop Sci. 21, 43– 47 [Google Scholar]
- 39.Premachandra G. S., Saneoka H., Kanaya M., Ogata S. ( 1991) Cell membrane stability and leaf surface wax content as affected by increasing water deficits in maize. J. Exp. Bot. 42, 167– 171 [Google Scholar]
- 40.Young A. J. ( 1991) The photoprotective role of carotenoids in higher plants. Physiol. Plantarum 83, 702– 708 [Google Scholar]
- 41.Kim D. W., Rakwal R., Agrawal G. K., Jung Y. H., Shibato J., Jwa N. S., Iwahashi Y., Iwahashi H., Kim D. H., Shim Ie. S., Usui K. ( 2005) A hydroponic rice seedling culture model system for investigating proteome of salt stress in rice leaf. Electrophoresis 26, 4521– 4539 [DOI] [PubMed] [Google Scholar]
- 42.Salekdeh G. H., Siopongco J., Wade L. J., Ghareyazie B., Bennett J. ( 2002) Proteomic analysis of rice leaves during drought stress and recovery. Proteomics 2, 1131– 1145 [DOI] [PubMed] [Google Scholar]
- 43.Gallie D. R., Le H., Caldwell C., Tanguay R. L., Hoang N. X., Browning K. S. ( 1997) The phosphorylation state of translation initiation factors is regulated developmentally and following heat shock in wheat. J. Biol. Chem. 272, 1046– 1053 [DOI] [PubMed] [Google Scholar]
- 44.Wan X. Y., Liu J. Y. ( 2008) Comparative proteomics analysis reveals an intimate protein network provoked by hydrogen peroxide stress in rice seedling leaves. Mol. Cell. Proteomics 7, 1469– 1488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Colvis C., Garland D. ( 2002) Posttranslational modification of human alphaA-crystallin: correlation with electrophoretic migration. Arch. Biochem. Biophys. 397, 319– 323 [DOI] [PubMed] [Google Scholar]
- 46.Hanson S. R., Hasan A, Smith D. L., Smith J. B. ( 2000) The major in vivo modifications of the human water-insoluble lens crystallins are disulfide bonds, deamidation, methionine oxidation and backbone cleavage. Exp. Eye Res. 71, 195– 207 [DOI] [PubMed] [Google Scholar]
- 47.Ueda Y., Duncan M. K., David L. L. ( 2002) Lens proteomics: the accumulation of crystallin modifications in the mouse lens with age. Invest. Ophthalmol. Vis. Sci. 43, 205– 215 [PubMed] [Google Scholar]
- 48.Anensen N., Oyan A. M., Bourdon J. C., Kalland K. H., Bruserud O., Gjertsen B.T. ( 2006) A distinct p53 protein isoform signature reflects the onset of induction chemotherapy for acute myeloid leukemia. Clin. Cancer Res. 12, 3985– 3992 [DOI] [PubMed] [Google Scholar]
- 49.Dai S., Zhang Z., Chen S., Beachy R. N. ( 2004) RF2b, a rice bZIP transcription activator, interacts with RF2a and is involved in symptom development of rice tungro disease. Proc. Natl. Acad. Sci. U.S.A. 101, 687– 692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Eulgem T., Rushton P. J., Robatzek S., Somssich I. E. ( 2000) The WRKY superfamily of plant transcription factors. Trends Plant Sci. 5, 199– 206 [DOI] [PubMed] [Google Scholar]
- 51.Stockinger E. J., Gilmour S. J., Thomashow M. F. ( 1997) Arabidopsis thaliana CBF1 encodes an AP2 domain-containing transcriptional activator that binds to the C-repeat/DRE, a cis-acting DNA regulatory element that stimulates transcription in response to low temperature and water deficit. Proc. Natl. Acad. Sci. U.S.A. 94, 1035– 1040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Dorjsuren D., Lin Y., Wei W., Yamashita T., Nomura T., Hayashi N., Murakami S. ( 1998) RMP, a novel RNA polymerase II subunit 5-interacting protein, counteracts transactivation by hepatitis B virus X protein. Mol. Cell. Biol. 18, 7546– 7555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lohman T. M., Bjornson K. P. ( 1996) Mechanism of helicase catalyzed DNA unwinding. Annu. Rev. Biochem. 65, 169– 214 [DOI] [PubMed] [Google Scholar]
- 54.Matson S. W., Bean D. W., George J. W. ( 1994) DNA helicases; enzymes with essential roles in all aspects of DNA metabolism. BioEssays 16, 13– 22 [DOI] [PubMed] [Google Scholar]
- 55.Reeves R. ( 2001) Molecular biology of HMGA proteins: hubs of nuclear function. Gene 227, 63– 81 [DOI] [PubMed] [Google Scholar]
- 56.Jessberger R. ( 2002) The many functions of SMC proteins in chromosome dynamics. Nat. Rev. Mol. Cell Biol. 3, 767– 778 [DOI] [PubMed] [Google Scholar]
- 57.Rando O. J., Zhao K., Crabtree G. R. ( 2000) Searching for a function for nuclear actin. Trends Cell Biol. 10, 92– 97 [DOI] [PubMed] [Google Scholar]
- 58.Wang Y., Ribot C., Rezzonico E., Poirier Y. ( 2004) Structure and expression profile of the Arabidopsis PHO1 gene family indicates a broad role in inorganic phosphate homeostasis. Plant Physiol. 135, 400– 411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ronai Z. ( 1993) Glycolytic enzymes as DNA binding proteins. Int. J. Biochem. 25, 1073– 1076 [DOI] [PubMed] [Google Scholar]
- 60.Raikhel N. ( 1992) Nuclear targeting in plants. Plant Physiol. 100, 1627– 1632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Traglia H. M., O'Connor J. P., Tung K. S., Dallabrida S., Shen W. C., Hopper A. K. ( 1996) Nucleus-associated pools of Rna1p, the Saccharomyces cerevisiae Ran/TC4 GTPase activating protein involved in nucleus/cytosol transit. Proc. Natl. Acad. Sci. U.S.A. 93, 7667– 7672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Morgan X. X., Tea M., Iris M. ( 2007) The kingdom- and tissue-specific anchorage of plant RanGAP to the nuclear envelope involves a novel family of plant nuclear pore associated transmembrane proteins, in Botany and Plant Biology Joint Congress, Chicago, July 8, 2007, No. P22007 Abstr. 223, The Botanical Society of America, Rockville, MD [Google Scholar]
- 63.Drakakaki G., Zabotina O., Delgado I., Robert S., Keegstra K., Raikhel N. ( 2006) Arabidopsis reversibly glycosylated polypeptides 1 and 2 are essential for pollen development. Plant Physiol. 142, 1480– 1492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Liepman A. H., Olsen L. J. ( 2004) Genomic analysis of aminotransferases in Arabidopsis thaliana. Crit. Rev. Plant Sci. 23, 73– 89 [Google Scholar]
- 65.Ashtamker C., Kiss V., Sagi M., Davydov O., Fluhr R. ( 2007) Diverse subcellular locations of cryptogein-induced reactive oxygen species production in tobacco bright yellow-2 cells. Plant Physiol. 143, 1817– 1826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Dunwell J. M., Khuri S., Gane P. J. ( 2000) Microbial relatives of the seed storage proteins of higher plants: Conservation of structure and diversification of function during evolution of the cupin superfamily. Microbiol. Mol. Biol. Rev. 64, 153– 179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Dietz K. J., Horling F., König J., Baier M. J. ( 2002) The function of the chloroplast 2-cysteine peroxiredoxin in peroxide detoxification and its regulation. J. Exp. Bot. 53, 1321– 1329 [PubMed] [Google Scholar]
- 68.Shikanai T., Takeda T., Yamauchi H., Sano S., Tomizawa K. I., Yokota A., Shigeoka S. ( 1998) Inhibition of ascorbate peroxidase under oxidative stress in tobacco having bacterial catalase in chloroplasts. FEBS Lett. 428, 47– 51 [DOI] [PubMed] [Google Scholar]
- 69.Oberschall A., Deák M., Török K., Sass L., Vass I., Kovács I., Fehér A., Dudits D., Horváth G. V. ( 2000) A novel aldose/aldehyde reductase protects transgenic plants against lipid peroxidation under chemical and drought stresses. Plant J. 24, 437– 446 [DOI] [PubMed] [Google Scholar]
- 70.Sunkar R., Bartels D., Kirch H. H. ( 2003) Overexpression of a stress-inducible aldehyde dehydrogenase gene from Arabidopsis thaliana in transgenic plants improves stress tolerance. Plant J. 35, 452– 464 [DOI] [PubMed] [Google Scholar]
- 71.Edwards R, Dixon D. P., Walbot V. ( 2000) Plant glutathione S-transferases: enzymes with multiple functions in sickness and in health. Trends Plant Sci. 5, 193– 198 [DOI] [PubMed] [Google Scholar]
- 72.Apel K., Hirt H. ( 2004) Reactive oxygen species: Metabolism, oxidative stress and signal transduction. Annu. Rev. Plant Biol. 55, 373– 399 [DOI] [PubMed] [Google Scholar]
- 73.Mittler R., Vanderauwera S., Gollery M., Van Breusegem F. ( 2004) Reactive oxygen gene network of plants. Trends Plant Sci. 9, 490– 498 [DOI] [PubMed] [Google Scholar]
- 74.DiDonato J., Mercurio F., Rosette C., Wu-Li J., Suyang H, Ghosh S., Karin M. ( 1996) Mapping of the inducible IkappaB phosphorylation sites that signal its ubiquitination and degradation. Mol. Cell. Biol. 16, 1295– 1304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Gardner R. G., Nelson Z. W., Gottschling D. E. ( 2005) Degradation-mediated protein quality control in the nucleus. Cell 120, 803– 815 [DOI] [PubMed] [Google Scholar]
- 76.Hajduch M., Rakwal R., Agrawal G. K., Yonekura M., Pretova A. ( 2001) High resolution two-dimensional electrophoresis separation of proteins from metal-stressed rice (Oryza sativa L.) leaves: drastic reductions/fragmentation of ribulose-1,5-bisphosphate carboxylase/oxygenase and induction of stress-related proteins. Electrophoresis 22, 2824– 2831 [DOI] [PubMed] [Google Scholar]
- 77.Agrawal G. K., Rakwal R., Yonekura M., Kubo A., Saji H. ( 2002) Proteome analysis of differentially displayed proteins as a tool for investigating ozone stress in rice (Oryza sativa L. seedlings. Proteomics 2, 947– 959 [DOI] [PubMed] [Google Scholar]
- 78.Desimone M., Henke A., Wagner E. ( 1996) Oxidative stress induces partial degradation of the large subunit of ribulose-1,5-bisphosphate carboxylase/oxygenase in isolated chloroplasts of barley. Plant Physiol. 111, 789– 796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Nollen E. A., Salomons F. A., Brunsting J. F., van der Want J. J., Sibon O. C., Kampinga H. H. ( 2001) Dynamic changes in the localization of thermally unfolded nuclear proteins associated with chaperone-dependent protection. Proc. Natl. Acad. Sci. U.S.A. 98, 12038– 12043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Pelham H. R. ( 1984) HSP70 accelerates the recovery of nucleolar morphology after heat shock. EMBO J. 3, 3095– 3100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Hashimoto M., Kisseleva L., Sawa S., Furukawa T., Komatsu S., Koshiba T. ( 2004) A novel rice PR10 protein, RSOsPR10, specifically induced in roots by biotic and abiotic stresses, possibly via the jasmonic acid signaling pathway. Plant Cell Physiol. 45, 550– 559 [DOI] [PubMed] [Google Scholar]
- 82.Akiyama T., Pillai M. A. ( 2001) Molecular cloning, characterization and in vitro expression of a novel endo-1,3-beta-glucanase up-regulated by ABA and drought stress in rice (Oryza sativa L.). Plant Sci. 161, 1089– 1098 [Google Scholar]
- 83.Plafker S. M., Macara I. G. ( 2002) Ribosomal protein L12 uses a distinct nuclear import pathway mediated by importin 11. Mol. Cell. Biol. 22, 1266– 1275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Scharf K. D., Nover L. ( 1982) Heat-shock-induced alterations of ribosomal protein phosphorylation in plant cell cultures. Cell 30, 427– 437 [DOI] [PubMed] [Google Scholar]
- 85.Gillette T. G., Gonzalez F., Delahodde A., Johnston S. A., Kodadek T. ( 2004) Physical and functional association of RNA polymerase II and the proteasome. Proc. Natl. Acad. Sci. U.S.A. 101, 5904– 5909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Borden K. L., Freemont P. S. ( 1996) The RING finger domain: a recent example of a sequence-structure family. Curr. Opin. Struct. Biol. 6, 395– 401 [DOI] [PubMed] [Google Scholar]
- 87.Hatakeyama S., Yada M., Matsumoto M., Ishida N., Nakayama K. I. ( 2001) U box proteins as a new family of ubiquitin-protein ligases. J. Biol. Chem. 276, 33111– 33120 [DOI] [PubMed] [Google Scholar]
- 88.Garner M. H. ( 2002) Na, K-ATPase in the nuclear envelope regulates Na+:K+ gradients in hepatocyte nuclei. J. Membr. Biol. 187, 97– 115 [DOI] [PubMed] [Google Scholar]
- 89.Mansharamani M., Hewetson A., Chilton B. S. ( 2001) An atypical nuclear P-type. ATPase is a RING-finger binding protein. J. Biol. Chem. 276, 3641– 3649 [DOI] [PubMed] [Google Scholar]
- 90.Aravind L., Iyer L. M., Anantharaman V.The two faces of Alba: the evolutionary connection between proteins participating in chromatin structure and RNA metabolism. Genome Biol. 20034R64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Nourani A., Robert F., Winston F. ( 2006) Evidence that spt2/sin1, an HMG-like factor, plays roles in transcription elongation, chromatin structure, and genome stability in Saccharomyces cerevisiae. Mol. Cell. Biol. 26, 1496– 1509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Hirsh A. E., Fraser H. B. ( 2001) Protein dispensability and rate of evolution. Nature 411, 1046– 1049 [DOI] [PubMed] [Google Scholar]
- 93.Jordan I. K., Rogozin I. B., Wolf Y. I., Koonin E. V. ( 2002) Essential genes are more evolutionarily conserved than are nonessential genes in bacteria. Genome Res. 12, 962– 968 [DOI] [PMC free article] [PubMed] [Google Scholar]