Abstract
Neurons and endocrine cells have the regulated secretory pathway (RSP) in which precursor proteins undergo proteolytic processing by prohormone convertase (PC) 1/3 or 2 to generate bioactive peptides. Although motifs for PC-mediated processing have been described ((R/K)Xn(R/K) where n = 0, 2, 4, or 6), actual processing sites cannot be predicted from amino acid sequences alone. We hypothesized that discovery of bioactive peptides would be facilitated by experimentally identifying signal peptide cleavage sites and processing sites. However, in vivo and in vitro peptide degradation, which is widely recognized in peptidomics, often hampers processing site determination. To obtain sequence information about peptides generated in the RSP on a large scale, we applied a brief exocytotic stimulus (2 min) to cultured endocrine cells and analyzed peptides released into supernatant using LC-MSMS. Of note, 387 of the 400 identified peptides arose from 19 precursor proteins known to be processed in the RSP, including nine peptide hormone and neuropeptide precursors, seven granin-like proteins, and three processing enzymes (PC1/3, PC2, and peptidyl-glycine α-amidating monooxygenase). In total, 373 peptides were informative enough to predict processing sites in that they have signal sequence cleavage sites, PC consensus sites, or monobasic cleavage sites. Several monobasic cleavage sites identified here were previously proved to be generated by PCs. Thus, our approach helps to predict processing sites of RSP precursor proteins and will expedite the identification of unknown bioactive peptides hidden in precursor sequences.
The generation of peptide hormones or neuropeptides involves the proteolytic processing of precursor proteins by specific proteases. In neurons and endocrine cells, most, if not all, of these bioactive peptides are generated within the RSP1 in which the processing enzymes PC1/3 or PC2 cleave precursors at basic residues (1, 2). The PC-mediated cleavage most often occurs at consecutive basic residues, but not all basic residues serve as PC recognition sites (2). This is partly because the secondary structure of a precursor also affects the substrate recognition (3). Identification of processing sites is hence a prerequisite for locating unknown peptides hidden in a precursor sequence.
Peptidomics has been advocated to comprehensively study peptides cleaved off from precursor proteins by endogenous proteases (4–6). These naturally occurring peptides are beyond the reach of current proteomics and should be analyzed in their native forms. Unlike proteomics, peptidomics has the potential to uncover processing sites of precursor proteins. Most peptidomics studies, which target tissue peptidomes from brain or endocrine organs (7–11), have provided limited information about secretory peptides that could help to identify processing sites; they are too often blurred by subsequent actions of exopeptidases (cutting off a single amino acid or dipeptide from either end of a peptide).
In MS-based identification of bioactive peptides present in biological samples, their relative low abundance in a total pool of naturally occurring peptides should be considered. Once extracted from cultured cells or tissues, bona fide secretory peptides and nonsecretory peptides or peptide fragments caused by degradation of abundant cytosolic proteins cannot be discriminated, and therefore we need to analyze samples rich in secretory peptides to facilitate the identification of bioactive peptides. Several attempts have been made to isolate secretory proteins or peptides, such as subcellular fractionation for harvesting secretory granules (12, 13). With all these efforts, a limited number of secretory peptides have been identified, and many known bioactive peptides still escape analysis.
We took advantage of the fact that peptides processed in the RSP are enriched in secretory granules of neurons and endocrine cells and released on exocytosis. Here we applied a brief exocytotic stimulus (2 min) to cultured human endocrine cells and identified peptides released into supernatant using LC-MSMS on an LTQ-Orbitrap mass spectrometer. Nearly 97% of the identified peptides arose from precursor proteins known to be recruited to the RSP, such as peptide hormone precursors and granin-like secretory proteins. Our approach was validated by the identification of previously known processing sites of peptide hormone precursors. In addition, a majority of the identified peptides retained cleavage sites that agree with consensus cleavage sites for PCs, which are informative enough to deduce the processing sites of RSP proteins. This peptidomics approach will expedite the identification of unknown bioactive peptides.
EXPERIMENTAL PROCEDURES
Peptide Preparation
Monolayer cultures of TT cells (14, 15) were rinsed three times with Hanks' medium (Invitrogen). Culture supernatants of the cells incubated for 2 min before and after stimulation with 10 μm forskolin plus 10 μm carbachol were harvested and rapidly extracted at 4 °C using an RP-1 solid phase extraction cartridge (GL Sciences) without centrifuging the supernatants. Bound substances were eluted in 60% ACN, 0.1% formic acid. After lyophilization of a small aliquot, samples were reconstituted in 10 μl of 2% ACN, 0.1% formic acid. In the one-dimensional analysis, solid phase-extracted analytes were subjected to LC-MSMS without gel filtration using an aliquot equivalent to 5 × 105 cells. Peptide fractions were obtained by HPLC on a gel filtration column equilibrated with 60% ACN, 0.1% TFA at a flow rate of 1.5 ml/min (G2000SWXL, 21.5 × 300 mm, Tosoh Corp.). For gel-filtrated fractions, an aliquot corresponding to 1.5 × 106 cells was used for LC-MSMS.
LC-MSMS
Nano-LC-MSMS experiments were performed with a Chorus nanoflow system (CS Analytics) connected to an LTQ-Orbitrap mass spectrometer (ThermoFisher Scientific) equipped with a nanoelectrospray emitter (MonoSpray C18 Nano, 100 μm × 50 mm, GL Sciences). Samples were dissolved in solvent A (2% ACN, 0.1% formic acid). The nanoflow system was run at a flow rate of 500 nl/min with a gradient from 5 to 45% solvent B (89% ACN, 0.1% formic acid) in 40 min and then to 95% B in 1 min. A protonated ion of polycyclodimethylsiloxane with m/z 445.120025 was used for internal calibration throughout. The mass spectrometer was operated in a data-dependent mode to automatically switch between MS and MSMS acquisitions. Survey full-scan spectra were acquired in the m/z range 400–1500 with five most intense ions (intensity threshold, 2e+05) sequentially isolated for MSMS in the linear ion trap using collision-induced dissociation with dynamic exclusion onward throughout the following scans. The resultant product ions were recorded in the Orbitrap.
Data Analysis and Peptide Identification
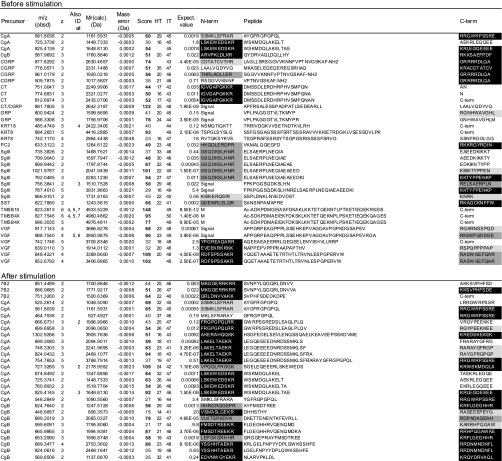
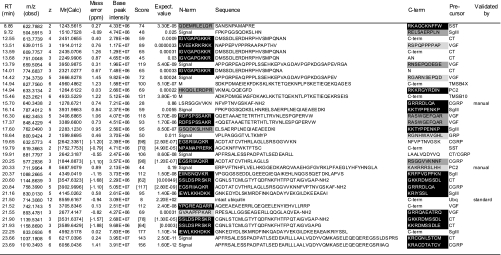
Peak picking, deisotoping, and deconvolution of MSMS spectra were preformed using Mascot Distiller (version 2.1.1.0) with the default parameters for Orbitrap. Peak lists were searched against IPI human (72,079 entries on July 2, 2008) using Mascot (version 2.2) with no enzyme specification. Pyroglutamination, C-terminal amidation, N-terminal acetylation, and methionine oxidation were simultaneously allowed as variable modifications. Peptide tolerance was set to 2 ppm, and MSMS tolerance was 25 millimass units. The significance threshold was the Mascot default setting of 5%. Each MSMS spectrum was checked manually to confirm or contradict the Mascot assignment. The false discovery rate for the identity threshold was in all cases 0% as estimated by using the Mascot decoy database function. The signals corresponding to intact calcitonin (CT), calcitonin gene-related peptide (CGRP), and somatostatin (SST) (with 1-ppm mass tolerance) underwent a mass shift of 116.01 Da after reductive alkylation with iodoacetamide and were sequenced as such by MSMS in a separate LC-MSMS analysis. Table I lists peptides that were identified with a score above the Mascot homology threshold. In the supplemental table, peptides with a score above the identity threshold (corresponding to an expectation value below 0.05) are listed and were considered identified.
Table I. Peptides identified before and after stimulation.
Data were obtained without gel filtration chromatography and summarized from three runs using an identical LC-MSMS setting and peptides whose scores (column 7) exceeded homology thresholds (HT, column 8) in at least two runs are listed. For peptides identified in multiple runs, a higher score is listed. Mr(Calc) represents the theoretical monoisotopic molecular mass (Da) based on the peptide sequence. The score value beyond an identity threshold (IT, column 9) is indicated in bold. In column 1, “CT/CGRP” indicates that the peptides are shared by CT and CGRP precursors. If the peptide was identified at different charges, the charge states are also shown in column 4. Expectation values are indicated in column 10. The N- (N-term) and C-terminal (C-term) flanking 10 amino acids (columns 11 and 13) are shown and marked as follows: closed boxes with white letters, typical PC cleavage sites containing consecutive basic residues and sites having C-terminal amidation motifs; dark gray boxes, cleavage sites containing basic residues at P4, P6 or P8 position; pale gray boxes, sites having basic residues at P1 but not at P2, P4, P6 or P8 position. “Signal” indicates that the peptide flanks its signal sequence. “C-term” indicates that the peptide C-terminus is the end of the precursor protein. Ac-, N-terminal acetylation; -NH2, C-terminal amidation; <Q, pyroglutamic acid. Oxidized methionine residues are underlined in column 12. Sequences are based on the following IPI accession numbers: CgA, 00746813; CgB, 00006601, CGRP, 00027855; CT, 00000914; GRP, 00011722; KRT18, 00554788; KRT8, 00554648; PC2, 00029131; SgIII, 00292071; SST, 00000130; TMSB10, 00220827; TMSB4X, 00220828; VGF, 00069058; 7B2, 00008944; NPW, 00853190; PACAP, 00000027; PENK, 00000828; SgII, 00009362. PACAP, pituitary adenylate cyclase-activating polypeptide; NPW, neuropeptide W; PENK, proenkephalin A; KRT, cytokeratin; TMSB4X, thymosin β-4 X-linked; TMSB10, thymosin β-10.
RESULTS
Comprehensive Analysis of Peptides Released on Exocytosis
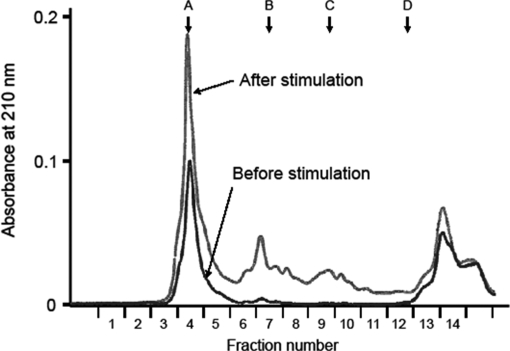
As a model system, we used the human medullary thyroid carcinoma cell line TT that stores peptide hormones including CT and CGRP in secretory granules (14, 15). A combination of forskolin and carbachol was used to induce exocytosis. Media from cells incubated for 2 min before and after stimulation were separately harvested and solid phase-extracted for peptide analysis. Total peptide amounts were assessed by gel filtration HPLC in which 1000–10,000-Da molecules are eluted in fractions labeled 7–10 (Fig. 1). This exocytotic stimulus elicited a 5.5-fold increase in secreted peptide amounts as assessed by the absorbance at 210 nm.
Fig. 1.
Gel filtration profiles of culture supernatant extracts from TT cells before (black trace) and after stimulation (gray trace). Arrows indicate molecular mass markers: A, 66,500 Da; B, 4,271 Da; C, 1,673 Da; D, 556 Da.
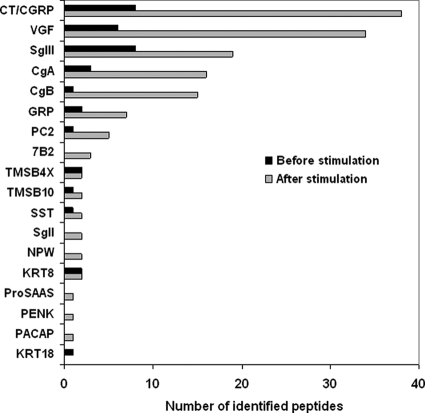
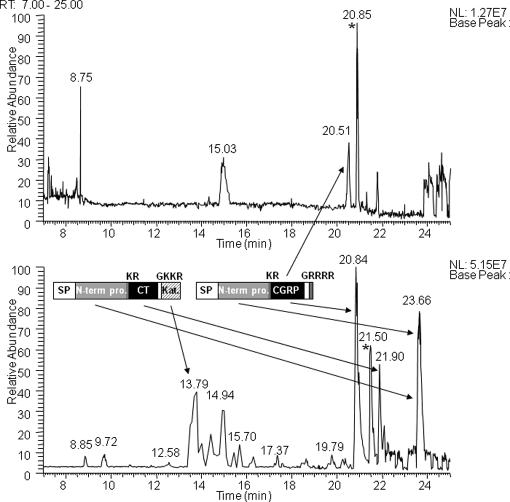
The solid phase-extracted samples were directly analyzed by LC-MSMS without gel filtration. We first examined a basal level secretion of peptides. In the medium conditioned by TT cells for 2 min, 36 peptides were identified from 13 precursors of which 30 peptides arose from nine secretory proteins including four peptide hormone precursors (CT/CGRP, gastrin-releasing peptide (GRP), and SST), four granin-like proteins (chromogranin A (CgA), chromogranin B (CgB), secretogranin III (SgIII), and VGF), and the processing enzyme PC2 (Fig. 2 and Table I). Because this cell line is known as a hyperproducer of CT and CGRP (14, 16), we tried to locate signals with mass values (within a mass tolerance of 2 ppm from a theoretical value) corresponding to bioactive CT (3415.58 Da) and CGRP (3786.96 Da). Signals from CGRP were observed, but no signals for CT were detected in the base peak chromatogram (Fig. 3).
Fig. 2.
The numbers of peptides identified before and after stimulation sorted by precursor names. CT and CGRP are grouped as they arise from alternatively spliced exons. Peptide sequences are indicated in Table I. PACAP, pituitary adenylate cyclase-activating polypeptide; NPW, neuropeptide W; PENK, proenkephalin A; KRT, cytokeratin; TMSB4X, thymosin β-4 X-linked; TMSB10, thymosin β-10.
Fig. 3.
Representative base peak chromatograms of the secretopeptidome from unstimulated (top) and stimulated (bottom) cells. Samples without gel filtration chromatography were analyzed. Major processing products of CT and CGRP precursors are illustrated along with arrows pointing at their peaks in the chromatogram. The base peaks marked with asterisks at 20.85 min (unstimulated) and 21.50 min (stimulated) are intact ubiquitin. SP, signal peptide; N-term pro., N-terminal propeptide; Kat., katacalcin. Base peaks at 8.75 and 15.03 min in the top panel were unrelated to peptide signals. RT, retention time; NL, normalized ion intensity.
In contrast, this stimulation facilitated identification of larger numbers of peptides, which resulted in 152 peptides being identified from 18 precursors (Fig. 2 and Table I). The six additional precursors all belonged to secretory proteins known to be processed in the RSP, including three peptide hormone precursors (pituitary adenylate cyclase-activating polypeptide, neuropeptide W, and proenkephalin A) and three granin-like proteins (7B2, SgII, and pro-SAAS). In total, 146 of 152 peptides arose from the 15 RSP precursors. The remaining six peptides were derived from thymosins and cytokeratin 8. According to the stimulus-induced increase in total peptide amounts released to culture supernatant, more peptides were identified from the former nine precursor proteins (Figs. 1 and 2). Across the CT and CGRP precursor sequences, known major processing products (17) were identified, namely CT N-terminal propeptide (6217.04 Da), bioactive CT (3415.58 Da), katacalcin (2435.07 Da), CGRP N-terminal propeptide (6056.04 Da), and bioactive CGRP (3786.96 Da) (Fig. 3).
Investigation of Cleavage Sites through Identified Peptides
Regarding the processing of peptide hormone precursors, it has long been known that PC1/3 or PC2 cleaves the precursors at sites containing consecutive basic amino acids following N-terminal signal peptide cleavage (1, 2). Cameron et al. (18) recently studied the specificity of PCs and drew a conclusion that the PC-mediated cleavage occurs at sites containing pairs of basic amino acids separated by 0, 2, 4, or 6 residues. However, some peptide hormone precursors are processed at monobasic residues although at much less frequency (2). In any case, the resultant C-terminal basic residues are subsequently removed by carboxypeptidase E. If the exposed C-terminal residue is glycine, peptidyl-glycine α-amidating monooxygenase catalyzes peptide α-amidation, a common post-translational modification often required for a peptide to be fully bioactive (19).
Having confirmed that almost all the sequenced peptides arose from RSP precursors (Table I), we extracted 10 amino acids N- or C-terminally flanking the sequenced peptides to analyze their cleavage sites. Any glycine immediately followed by a basic residue(s) that creates an amidation site was also counted as a PC consensus site. Monobasic sites were defined as those that do not harbor basic residues except for P1 position and considered potential processing sites as well. PC consensus cleavage sites, signal sequence cleavage sites, and monobasic sites, referred to as informative cleavage sites in the present study, were found in 120, 26, and 29 peptides, respectively (72 peptides were counted twice under this definition). Overall 142 of 152 had such informative cleavage sites at either or both ends.
Identity of Major Peptides in the TT Secretopeptidome
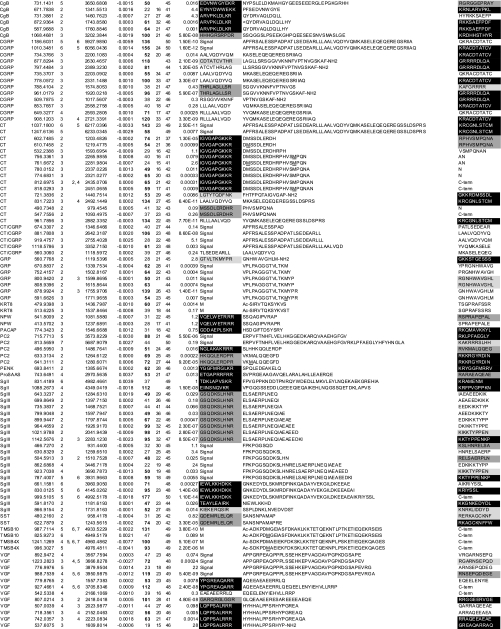
In the LC-MSMS setting used throughout this study, any signal that transcended a given intensity threshold was automatically subjected to MSMS and ignored thereafter if it persisted within a precursor mass tolerance of 5 ppm with the aim of sequencing as many peptides as possible. The signal is not always subjected to MSMS at its maximum intensity in LC-MS profiles, and therefore this setting could return relatively low scores even for abundant peptides, which may not be included in Table I. To identify intense signals in LC-MSMS base peak chromatograms, we examined peptide peak intensities in all MS spectra. Table II provides the list of 35 peptides that were detected at the indicated monoisotopic m/z and charge state with a base peak intensity beyond 2e+06 (see also Fig. 3 and supplemental Fig. 1). Three peptide sequences had expectation values (above 0.05) that did not exceed the Mascot significance threshold. First, the 1278.67-Da peptide was qualified as a CGRP-derived peptide based on the observation that CGRP-derived peptides are most abundantly expressed in the TT secretopeptidome (Fig. 3 and supplemental Fig. 3). Second, the 5687.91-Da peptide also yielded suboptimum MSMS spectra but was identified as a PC2-derived peptide because of matches for eight consecutive b-ions (supplemental Fig. 3). Third, the 8559.61-Da peptide was qualified using MSMS spectral comparison with commercially available human ubiquitin (supplemental Fig. 2).
Table II. Identity of the major peptides released on exocytosis (base peak intensities beyond 2e+06 in the Fig. 3 base peak chromatogram).
Values in brackets represent those obtained by peptides after reductive alkylation. For the ubiquitin MSMS spectrum, see supplemental Fig. 2. Note that the MSMS spectra of the 1278.67- and 5687.91-Da peptides (supplemental Fig. 3) did not meet the homology threshold criteria but were considered identified as described in the text. Grayscale boxes are defined in Table I legend. Oxidized methionine residues are underlined in column 10. TMSB4X, thymosin β-4 X-linked; TMSB10, thymosin β-10; RT, retention time; obsd, observed; Mr(Calc), theoretical monoisotopic molecular mass (Da) based on the peptide sequence; Expect., Expectation; N-term, the N-terminal flanking 10 amino acids; C-term, the C-terminal flanking 10 amino acids.
This list covered all the major processing products of CT and CGRP. Intact CGRP continued to be observed at multiple charged ions (+3 to +6) over a retention time of 10 min in the mass chromatogram, suggesting that it represents one of the most abundant peptides in this secretopeptidome (Fig. 3). The CGRP-derived peptide (ACDTATCVTHRLAGLLSRSGGVVKN) appeared to be generated from endoproteolytic cleavage of intact CGRP because the C-terminal cleaved half (1278.67 Da) was detected as well (Table II). Except for this peptide, all the N-terminal cleavage sites of CT/CGRP-derived peptides are known as major processing sites (17). Similar findings were obtained with C-terminal cleavage sites except for four non-basic sites. It remains to be clarified whether these non-basic cleavage sites point to the processing that actually occurred in the RSP.
With regard to the SST precursor, the 1243.56-Da peptide corresponds to the first 12 amino acids of SST-28. The dibasic sites RK (position 78–79) located downstream of this 12-residue peptide (Table II) is the known cleavage site for SST-14 (AGCKNFFWKTFTSC) (20). The single arginine (position 65) flanking the 1243.56-Da peptide and SST-28 is also an established processing site (20). At position −4 relative to this scissible bond (referred to as the P4 position), a single arginine (position 62) exists and forms a consensus PC cleavage site. As for the GRP precursor, the 1599.87-Da peptide is C-terminally flanked by an atypical single arginine, which is followed by intact neuromedin C (21). This arginine is known as an established processing site, although responsible enzymes remain to be identified. Thus, a majority of the cleavage sites of peptide hormone precursors were consistent with the previously identified processing sites. Except for precursor C termini, all the peptides derived from granin-like precursors (SgII, SgIII, and VGF) retained informative cleavage sites defined in this study. The 2677.41-Da VGF-derived amidated peptide was recently identified and designated NERP-1 (16).
Integrity of the Secretopeptidome Demonstrated by an In-depth Analysis
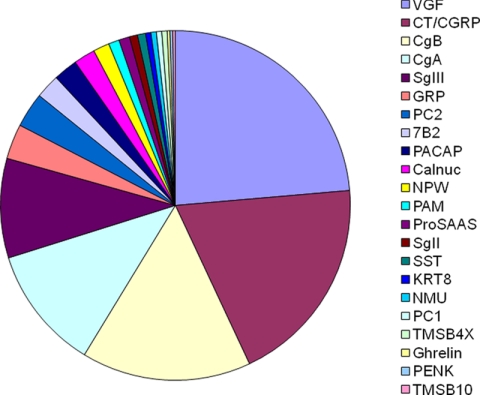
The secretopeptidome was separated into four fractions using gel filtration HPLC to perform an in-depth analysis. This analysis contributed to a substantial increase in sequenced peptides, and thus we were able to identify a total of 400 peptides from 23 precursors (Fig. 4 and the supplemental table). Some peptides arose from precursors not identified by the one-dimensional analysis; these included peptide hormone precursors neuromedin U and ghrelin, processing enzymes PC1 and peptidyl-glycine α-amidating monooxygenase, and the calcium-binding protein calnuc. The identification of PC1- and peptidyl-glycine α-amidating monooxygenase-derived peptides confirmed the integrity of this secretopeptidome as they are enzymes involved in the RSP proteolytic processing. Four calnuc-derived peptides had typical cleavage sites suggestive of the PC function (supplemental table). Recently calnuc was identified in a soluble fraction of bovine adrenal secretory granules (13). Altogether it is likely that nearly 99% of sequenced peptides were released upon exocytosis, which again demonstrates that this secretopeptidome is extremely rich in peptides stored in secretory granules.
Fig. 4.
Pie representation of the 400 peptides (sorted by names of 23 precursors) secreted after stimulation. PACAP, pituitary adenylate cyclase-activating polypeptide; NPW, neuropeptide W; PENK, proenkephalin A; KRT, cytokeratin; TMSB4X, thymosin β-4 X-linked; TMSB10, thymosin β-10; PAM, peptidyl-glycine α-amidating monooxygenase; NMU, neuromedin U.
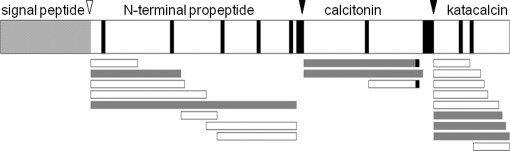
We examined a total of 400 sequenced peptides to see whether they meet the criteria for the PC consensus sites (supplemental table). PC consensus sites were found in 299 peptides, and signal sequence cleavage sites were found in 43 peptides. Monobasic sites were found in 98 peptides. Overall a total of 373 peptides were shown to have informative cleavage sites to predict the precursor processing in the RSP. As an example, the sequences of CT-derived peptides listed in Tables I and II were mapped to the precursor sequence (Fig. 5). The map consists of the known major processing products and intermediate products sharing N or C termini, which correspond to the established PC1/3 or PC2 cleavage sites. Consistent with the reported length of its signal peptide (17), no peptide was identified in the first 25 amino acids. Overall this map is reminiscent of CT precursor processing previously elucidated in parafollicular cells of the thyroid gland (17).
Fig. 5.
CT precursor processing deduced by a panel of identified peptides. The established signal peptide cleavage site (open arrowhead) and known processing sites (closed arrowheads) are shown across the top of the CT precursor with basic residues (thin black boxes) and the signal peptide (hatched box) indicated. Sequences of identified peptides (bars) are detailed in Tables I and II. Major peptides, defined in Table II, are indicated by gray bars. Black boxes denote C-terminally amidated residues.
DISCUSSION
The most outstanding finding in the present study is that an exocytotic stimulus applied to cultured endocrine cells is highly effective in identifying secretory peptides. Peptide profiles identified with this protocol strongly suggest that most peptides were released from secretory granules on exocytosis (Fig. 4). It should be noted that in a total of 400 identified peptides nearly 97% arose from previously known RSP precursor proteins. This non-invasive approach dispenses with time-consuming procedures such as subcellular fractionation and can be extended to different cell culture models. To the best of our knowledge, this is the first study ever to conduct a comprehensive analysis focused on peptides from the RSP proteins.
Most peptidomics studies have dealt with endocrine organs or brains, which are considered major sources of peptide hormones or neuropeptides (4, 7–11). It is now recognized that endogenous proteases must be inactivated before tissue extraction to prevent massive production of peptide fragments caused by degradation of abundant intracellular proteins, which hampers MS detection of endogenous peptides (7, 8). For tissue peptidome studies, microwave irradiation before or after decapitation has been proposed to prevent protease activation that is thought to occur immediately after sacrifice (7, 8). Despite these efforts, a limited number of secretory peptides were identified (4, 7–11).
Regarding MSMS of naturally occurring peptides, precursor mass acquisition with conventional mass spectrometers (mass accuracy, ∼50 ppm) and subsequent filtering in the setting of “no enzyme” very often lead to ambiguous peptide identification (22). To cope with this issue, many peptidomics studies have used in-house databases containing a limited number of entries to identify peptides that would otherwise remain elusive (11, 23). On the other hand, we used the public IPI human database and took into account four variable modifications simultaneously for MSMS interpretation. Because of the mass accuracy of Orbitrap (∼2 ppm), a false discovery rate using a decoy database was minimized to 0% for peptide matches above identity thresholds in the Mascot MSMS ion search. This MSMS identification scheme may inevitably miss many peptides, which could be considered identified if a specific database with limited entries was used as in previous peptidomics studies (11, 23). The peptides listed in the supplemental table are all beyond identity thresholds; accepting peptides beyond homology thresholds will allow more than 200 additional secretory peptides to enter the table, with only two different peptides from keratin 8 turning up (data not shown). These examples include neuromedin U 25 (3018.52 Da) and pancreastatin (5076.36 Da) with a Mascot expectation value of 0.063 and 0.43, respectively. In the supplemental table, they are not considered identified because an expectation value for accepting MSMS spectra is the Mascot default significant threshold value of 0.05. Nonetheless we used the stringent setting (described under “Experimental Procedures”) to preclude misleading assignments and to demonstrate that this secretopeptidome shows little contamination by non-secretory components.
In the present study, we made every effort to prevent peptide degradation or chemical modifications (deamidation, methionine oxidation, and pyroglutamination) that may occur during sample preparation. As described under “Experimental Procedures,” peptides released during 2 min were immediately subjected to solid phase extraction. Peptide extraction was performed at 4 °C and completed within 20 min after harvesting the supernatant. In addition, lyophilized samples were analyzed by LC-MSMS immediately after reconstitution. Even with these attentive procedures, several clusters of N- or C-terminally truncated peptides that share cleavage sites at the other end were sequenced as reported in previous peptidomics studies on tissue peptidomes (4, 7 –11). However, most peptides (30 of 35) dominantly detected in LC-MSMS (Table II, Fig. 3, and supplemental Fig. 1) did not have cleavage sites suggestive of exopeptidase digestion aside from the five CT- or CGRP-derived peptides mentioned under “Results.” These five peptides appeared to be N- or C-terminally truncated peptides of major processing products. They have not been reported as major processing products in previous biochemical studies to the best of our knowledge. However, the possibility that their C termini or N termini were generated by unknown endopeptidases (cutting within a peptide) cannot be excluded.
It was unexpected that thymosins and ubiquitin represented major peptides in the TT secretopeptidome. Given a previous report of its storage in adrenal secretory granules and exocytosis-induced secretion (24), ubiquitin may be localized in TT secretory granules and secreted upon exocytosis. The secretory nature of thymosin β-4 has also been reported (25); however, thymosins are not regarded as peptides localized in secretory granules. In any case, the successful identification of intact peptide forms indicates that a majority of peptides may not be affected by exopeptidase digestion.
Gel filtration-based separation caused an increase in the number of sequenced peptides among which several peptides suggestive of the PC-mediated cleavage were identified, such as those from the calcium-binding protein calnuc (supplemental table). Calnuc was identified in a soluble fraction of bovine adrenal secretory vesicles (13), and hence our finding suggests that it is a precursor to unknown bioactive peptides.
To identify unique cleavage sites of RSP precursor proteins, we examined N- and C-terminal flanking sequences of the 400 peptides identified. Overall 152 unique cleavage sites that match PC consensus sites were elucidated of which 105 cleavage sites were conserved consecutive dibasic residues. This finding appears to support the contention revealed by previous studies that the most often encountered PC cleavage sites are conserved paired dibasic sites (1, 2, 19). The observation that a majority of cleavage sites were consistent with PC consensus sites ((R/K)Xn(R/K) where n = 0, 2, 4, or 6) should not be overestimated. For instance, the C-terminal cleavage site of the CGRP precursor (RLAGLLS↓RSG) matches this rule but is not known as a processing site. At present its abundance relative to the longer major product of intact CGRP was unavailable, and therefore it remains to be clarified whether the peptide would represent a major processing product secreted by TT cells. We should also consider that shorter peptides tend to be better ionized and readily detected in mass spectrometry schemes.
Conversely cleavages at non-consensus monobasic sites could represent bona fide initial endoproteolytic cleavage sites. In the present study, CgA-derived peptides (1048.51 and 1161.59 Da) sharing the N-terminal cleavage site (FRAR↓AYGF) were identified (supplemental table), suggesting that the single arginine (position 356) is a processing site. The C-terminal cleavage site of the 1914.01-Da peptide (VGF residues 463–481) had an arginine at the P10 position. Indeed they have both been shown to be an authentic recognition site for PC2 using PC2 knock-out mice (26). Another example is the N-terminal cleavage site (LSFR↓ARAY) of the CgA 1275.65- and 1388.73-Da peptides known as the processing site for CgA LF-19 peptide (27). Thus atypical monobasic sites should also be considered potential processing sites for a precursor whose processing remains largely unknown. In this context, the single arginine of VGF (position 212) in FQAR↓MPDS may represent a cleavage site as shown by six peptides (supplemental table). It is envisaged that further detailed analysis of the secretopeptidome could identify processing sites with higher confidence. Although the peptide repertoire from a cancer cell line does not necessarily reflect the in vivo processing pattern of RSP precursor proteins, this will not detract from the significance of our study. Indeed the peptides identified with our approach retain cleavage sites created in the RSP to a degree that allows the accurate prediction of processing sites in known peptide hormone precursors (Fig. 5). In summary, we showed that peptidomics has the potential to identify processing sites of precursors processed in the RSP. By dissecting the secretopeptidome we should have a clearer picture of the precursor processing that actually occurs in the RSP that would also facilitate the discovery of bioactive peptides.
Acknowledgments
We thank Gary S. Goldberg (University of Medicine and Dentistry of New Jersey) for critical reading of the manuscript.
Footnotes
* This work was supported in part by the Program for Promotion of Fundamental Studies in Health Sciences of the National Institute of Biomedical Innovation and by grants-in-aid for scientific research from the Japanese Society for the Promotion of Science, Japan.
 The on-line version of this article (available at http://www.mcponline.org) contains supplemental material.
The on-line version of this article (available at http://www.mcponline.org) contains supplemental material.
- RSP
- secretory pathway
- CgA
- chromogranin A
- CgB
- chromogranin B
- CT
- calcitonin
- CGRP
- calcitonin gene-related peptide
- GRP
- gastrin-releasing peptide
- PC
- prohormone convertase
- SgII
- secretogranin II
- SgIII
- secretogranin III
- SST
- somatostatin
- LTQ
- linear trap quadrupole
- IPI
- International Protein Index
REFERENCES
- 1.Zhou A., Webb G., Zhu X., Steiner D. F. ( 1999) Proteolytic processing in the secretory pathway. J. Biol. Chem. 274, 20745– 20748 [DOI] [PubMed] [Google Scholar]
- 2.Fricker L. D. ( 2005) Neuropeptide-processing enzymes: applications for drug discovery. AAPS J. 7, E 449– 455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brakch N., Rholam M., Boussetta H., Cohen P. ( 1993) Role of beta-turn in proteolytic processing of peptide hormone precursors at dibasic sites. Biochemistry 32, 4925– 4930 [DOI] [PubMed] [Google Scholar]
- 4.Clynen E., Baggerman G., Veelaert D., Cerstiaens A., Van der Horst D., Harthoorn L., Derua R., Waelkens E., De Loof A., Schoofs L. ( 2001) Peptidomics of the pars intercerebralis-corpus cardiacum complex of the migratory locust, Locusta migratoria. Eur. J. Biochem. 268, 1929– 1939 [DOI] [PubMed] [Google Scholar]
- 5.Schrader M., Schulz-Knappe P. ( 2001) Peptidomics technologies for human body fluids. Trends Biotechnol. 19, S 55– 60 [DOI] [PubMed] [Google Scholar]
- 6.Sasaki K., Sato K., Akiyama Y., Yanagihara K., Oka M., Yamaguchi K. ( 2002) Peptidomics-based approach reveals the secretion of the 29-residue COOH-terminal fragment of the putative tumor suppressor protein DMBT1 from pancreatic adenocarcinoma cell lines. Cancer Res. 62, 4894– 4898 [PubMed] [Google Scholar]
- 7.Svensson M., Sköld K., Svenningsson P., Andren P. E. ( 2003) Peptidomics-based discovery of novel neuropeptides. J. Proteome Res. 2, 213– 219 [DOI] [PubMed] [Google Scholar]
- 8.Che F. Y., Lim J., Pan H., Biswas R., Fricker L. D. ( 2005) Quantitative neuropeptidomics of microwave-irradiated mouse brain and pituitary. Mol. Cell. Proteomics 4, 1391– 1405 [DOI] [PubMed] [Google Scholar]
- 9.Boonen K., Baggerman G., D'Hertog W., Husson S. J., Overbergh L., Mathieu C., Schoofs L. ( 2007) Neuropeptides of the islets of Langerhans: a peptidomics study. Gen. Comp. Endocrinol. 152, 231– 241 [DOI] [PubMed] [Google Scholar]
- 10.Cape S. S., Rehm K. J., Ma M., Marder E., Li L. ( 2008) Mass spectral comparison of the neuropeptide complement of the stomatogastric ganglion and brain in the adult and embryonic lobster, Homarus americanus. J. Neurochem. 105, 690– 702 [DOI] [PubMed] [Google Scholar]
- 11.Bora A., Annangudi S. P., Millet L. J., Rubakhin S. S., Forbes A. J., Kelleher N. L., Gillette M. U., Sweedler J. V. ( 2008) Neuropeptidomics of the supraoptic rat nucleus. J. Proteome Res. 7, 4992– 5003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wegrzyn J., Lee J., Neveu J. M., Lane W. S., Hook V. ( 2007) Proteomics of neuroendocrine secretory vesicles reveal distinct functional systems for biosynthesis and exocytosis of peptide hormones and neurotransmitters. J. Proteome Res. 6, 1652– 1665 [DOI] [PubMed] [Google Scholar]
- 13.Brunner Y., Couté Y., Iezzi M., Foti M., Fukuda M., Hochstrasser D. F., Wollheim C. B., Sanchez J. C. ( 2007) Proteomics analysis of insulin secretory granules. Mol. Cell. Proteomics 6, 1007– 1017 [DOI] [PubMed] [Google Scholar]
- 14.Gkonos P. J., Born W., Jones B. N., Petermann J. B., Keutmann H. T., Birnbaum R. S., Fischer J. A., Roos B. A. ( 1986) Biosynthesis of calcitonin gene-related peptide and calcitonin by a human medullary thyroid carcinoma cell line. J. Biol. Chem. 261, 14386– 14391 [PubMed] [Google Scholar]
- 15.Zabel M., Seidel J., Kaczmarek A., Surdyk-Zasada J., Grzeszkowiak J., Górny A. ( 1994) Hybridocytochemical and immuno-ultrastructural study of calcitonin gene expression in cultured medullary carcinoma cells. Histochemistry 102, 323– 327 [DOI] [PubMed] [Google Scholar]
- 16.Yamaguchi H., Sasaki K., Satomi Y., Shimbara T., Kageyama H., Mondal M. S., Toshinai K., Date Y., González L. J., Shioda S., Takao T., Nakazato M., Minamino N. ( 2007) Peptidomic identification and biological characterization of neuroendocrine regulatory peptide-1 and -2. J. Biol. Chem. 282, 26354– 26360 [DOI] [PubMed] [Google Scholar]
- 17.Jacobs J. W., Goodman R. H., Chin W. W., Dee P. C., Habener J. F., Bell N. H., Potts J. T., Jr ( 1981) Calcitonin messenger RNA encodes multiple polypeptides in a single precursor. Science 213, 457– 459 [DOI] [PubMed] [Google Scholar]
- 18.Cameron A., Apletalina E. V., Lindberg I. ( 2002) The enzymology of PC1 and PC2. In The Enzymes ( Dalby R. E., Sigman D. S., eds) 3rd Ed., pp. 291– 328, Academic Press, NY [Google Scholar]
- 19.Eipper B. A., Stoffers D. A., Mains R. E. ( 1992) The biosynthesis of neuropeptides: peptide alpha-amidation. Annu. Rev. Neurosci. 15, 57– 85 [DOI] [PubMed] [Google Scholar]
- 20.Shen L. P., Pictet R. L., Rutter W. J.Human somatostatin I: sequence of the cDNA. Proc. Natl. Acad. Sci. U. S. A. (1982)79, 4575–4579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Minamino N., Kangawa K., Matsuo H. ( 1984) Neuromedin C: a bombesin-like peptide identified in porcine spinal cord. Biochem. Biophys. Res. Commun. 119, 14– 20 [DOI] [PubMed] [Google Scholar]
- 22.Brosch M., Swamy S., Hubbard T., Choudhary J. ( 2008) Comparison of Mascot and X!Tandem performance for low and high accuracy mass spectrometry and the development of an adjusted mascot threshold. Mol. Cell. Proteomics 7, 962– 970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fäth M., Sköld K., Norrman M., Svensson M., Fenyö D., Andren P. E. ( 2006) SwePep, a database designed for endogenous peptides and mass spectrometry. Mol. Cell. Proteomics 5, 998– 1005 [DOI] [PubMed] [Google Scholar]
- 24.Kieffer A. E., Goumon Y., Ruh O., Chasserot-Golaz S., Nullans G., Gasnier C., Aunis D., Metz-Boutigue M. H. ( 2003) The N- and C-terminal fragments of ubiquitin are important for the antimicrobial activities. FASEB J. 17, 776– 778 [DOI] [PubMed] [Google Scholar]
- 25.Bock-Marquette I., Saxena A., White M. D., Dimaio J. M., Srivastava D. ( 2004) Thymosin beta4 activates integrin-linked kinase and promotes cardiac cell migration, survival and cardiac repair. Nature 432, 466– 472 [DOI] [PubMed] [Google Scholar]
- 26.Pan H., Che F. Y., Peng B., Steiner D. F., Pintar J. E., Fricker L. D. ( 2006) The role of prohormone convertase-2 in hypothalamic neuropeptide processing: a quantitative neuropeptidomic study. J. Neurochem. 98, 1763– 1777 [DOI] [PubMed] [Google Scholar]
- 27.Orr D. F., Chen T., Johnsen A. H., Chalk R., Buchanan K. D., Sloan J. M., Rao P., Shaw C. ( 2002) The spectrum of endogenous human chromogranin A-derived peptides identified using a modified proteomic strategy. Proteomics . 21586– 1600 [DOI] [PubMed] [Google Scholar]