Abstract
The advent of quantitative proteomics opens new opportunities in biomedical and clinical research. Although quantitative proteomics methods based on stable isotope labeling are in general preferred for biomolecular research, biomarker discovery is a case example of a biomedical problem that may be better addressed by using label-free MS techniques. As a proof of concept of this paradigm, we report the use of label-free quantitative LC-MS to profile the urinary peptidome of kidney chronic allograft dysfunction (CAD). The aim was to identify predictive biomarkers that could be used to personalize immunosuppressive therapies for kidney transplant patients. We detected (by LC-M/MS) and quantified (by LC-MS) 6000 polypeptide ions in undigested urine specimens across 39 CAD patients and 32 control individuals. Although unsupervised hierarchical clustering differentiated between the groups when including all the identified peptides, specific peptides derived from uromodulin and kininogen were found to be significantly more abundant in control than in CAD patients and correctly identified the two groups. These peptides are therefore potential biomarkers that might be used for the diagnosis of CAD. In addition, ions at m/z 645.59 and m/z 642.61 were able to differentiate between patients with different forms of CAD with specificities and sensitivities of 90% in a training set and, significantly, of ∼70% in an independent validation set of samples. Interestingly low expression of uromodulin at m/z 638.03 coupled with high expression of m/z 642.61 diagnosed CAD in virtually all cases. Multiple reaction monitoring experiments further validated the results, illustrating the power of our label-free quantitative LC-MS approach for obtaining quantitative profiles of urinary polypeptides in a rapid, comprehensive, and precise fashion and for biomarker discovery.
A major goal of clinical proteomics is to identify biomarkers that can aid in the diagnosis and prognosis of different conditions. In their ideal form, these biomarkers will not only assist the clinician in the diagnosis of a disease, but they will also give directions as to which therapy may be more appropriate for each patient, thus contributing to the development of personalized medicine. In this regard, urine represents an ideal, but yet largely unexplored, source of biomarkers because of the presence of large numbers of small peptides in this biological fluid and because it can be obtained non-invasively.
However, although proteomics techniques are instrumental for increasing our understanding of molecular cell biology (1) the impact of proteomics in clinical practice has not yet reached initial expectations perhaps because of technological limitations (2, 3). Using hyphenated methods such as novel LC-MS techniques for quantitative proteomics (4, 5) may prove advantageous for the identification and validation of biomarkers (3, 6). This is because LC-MS allows the detection of proteomes with greater depth, dynamic range, and enhanced accuracy of quantization than when using one-dimensional profiling techniques that record all ions in a single mass spectrum, such as MALDI-TOF MS or SELDI-TOF MS (7). On-line LC-ESI-MS is quantitative in nature because the initial LC separation step contributes to reducing the amount of analytes that are simultaneously ionized, thus reducing the possibility of ion suppression, and because ion formation by electrospray ionization is proportional to analyte concentration (8, 9). Initial reports that used LC-MS for the analysis of the urinary proteome provided proof of principle of the use of this technique for the analysis of urinary polypeptides (10–12), and recently, using new generation LC-MS/MS instrumentation, more than 1500 proteins have been detected in urine (13). Nevertheless despite these advances in our understanding of the qualitative composition of the urinary proteome, precise and comprehensive quantification of urinary polypeptides to discover potential biomarkers remains a challenge.
The ideal, and more widely used, strategies to derive quantitative information from LC-MS experiments are based on differential stable isotope labeling of proteins or peptides, which are then mixed and quantified relative to each other in single multidimensional LC-LC-MS experiments (14). This technique, however, is not ideal for biomarker discovery because of problems associated with protein derivatization in a clinical setting, because of its limited throughput, and because, although not impossible, isotope labeling techniques make it difficult to compare a large number of specimens; at present labeling reagents can be used for simultaneous comparison of up to eight protein samples (15).
Novel analytical strategies for quantitative proteomics that do not require isotope labeling have been reported (4, 5, 16). These techniques can quantify polypeptides with precisions and accuracies comparable to those based on isotope labeling (17). In addition, such label-free quantitative LC-MS approaches can compare an unlimited number of samples, and it is therefore ideal for biomarker discovery as experimental designs normally involve comparing a large number of specimens to statistically validate the results. Thus, label-free quantitative LC-MS would clearly assist in analyzing the full potential of urine clinical samples as a source of disease biomarkers. The aim of the study presented herein was to prove this concept taking chronic allograft dysfunction (CAD)1 as a paradigm.
During the last years, the incidence and prevalence of end stage renal disease has increased worldwide (18). Successful renal transplantation improves the patients' quality of life and increases survival as compared with long term dialysis treatment (19). However, despite these improvements, a substantial portion of grafts develop progressive dysfunction and fail within a decade even with the use of appropriate doses of immunosuppressive drugs to prevent acute rejection (20). CAD is responsible for more than 50% of graft losses and remains a central clinical challenge. Although patients can return to dialysis after transplant failure, loss of a functioning graft is associated with a 3-fold increase in the risk of death, a substantial decrease in quality of life for those who survive, and a 4-fold increase in healthcare costs (21).
CAD is mediated by a combination of immune, ischemic, and inflammatory stimuli, and multiple pathways and mediators lead to cumulative structural damage to all compartments of the transplanted kidney. Sclerosing changes associated with tubulointerstitial injury are mediated by the processes of active fibrogenesis, resulting in epithelial loss and the phenotype of tubular atrophy and chronic interstitial fibrosis (22). Available diagnostic methods include clinical presentation, biochemical parameters, and biopsies. Currently the only non-invasive biomarker of CAD is serum creatinine and glomerular filtration rate (GFR), but neither is particularly sensitive or specific and may not reflect early alterations (20, 22). At present, biopsy allograft is regarded as the gold standard for the diagnosis of CAD allowing its early detection; however, this is a costly procedure that is associated with clinical complications (23).
Clinicians are hence faced with a dilemma. On the one hand, protocol biopsies may detect rejection at an earlier subclinical stage and allow prompt initiation of treatment, which may translate into improved long term graft survival (24). On the other hand, this also implies that patients with preserved graft function, i.e. without CAD, undergo this invasive procedure unnecessarily. Therefore, identification of non-invasive biomarkers for the early diagnosis of CAD would be invaluable for alleviating the major health and economic burden that this condition causes to western countries (25).
The aim of the present study was to evaluate whether the urinary peptidome, as analyzed by a novel analytical strategy based on label-free quantification of urinary polypeptides by LC-MS, would differentiate between patients with CAD, those showing stable renal transplant (SRT), and a group of living donors. To our knowledge, this represents the first study reporting urine polypeptide signatures and individual biomarkers that group patients according to their underlying renal phenotype and hence represent potential candidates for non-invasive diagnosis of CAD.
EXPERIMENTAL PROCEDURES
Study Cohorts
Seventy-one individuals were included in the present study: an initial training experiment analyzed 18 patients with clinical and histopathological characterization of CAD and 14 controls. CAD patients were recipients of a renal graft that received immunosuppressive treatment with levels in the therapeutic range of a calcineurin inhibitor, mycophenolate mofetil, and prednisone. These patients fell into two groups: 1) eight patients (five men and three women) with interstitial fibrosis and tubular atrophy (IF/TA) and no evidence of any specific etiology (IF/TA group) and 2) 10 patients (seven men and three women) with chronic active antibody-mediated rejection (CAAR) defined by morphological features including transplant glomerulopathy (TG), IF/TA with or without peritubular capillary loss, fibrous intimal thickening in arteries without duplication of the internal elastica, diffuse C4d deposition in peritubular capillaries, and the presence of donor-specific antibody (CAAR group). There were no significant differences between the IF/TA and CAAR groups with respect to age, gender, diabetes duration, arterial blood pressure, body mass index, and GFR.
The controls fell into two groups: 1) stable renal transplant recipients: five (three men and two women) live donor recipients of a first renal graft who were at the end of the 1st month after surgery with normal transplant biopsy and followed immunosuppressive treatment with a calcineurin inhibitor, mycophenolate mofetil, and prednisone; and 2) healthy controls: nine volunteers (six men and three women) with normal blood pressure (systolic blood pressure <130 mm Hg and diastolic blood pressure <80 mm Hg) and no history of diabetes mellitus, ischemic heart disease, stroke, or peripheral vascular disease). Base-line clinical parameters are shown in Table I, training set. Nephroangiosclerosis was the cause of kidney failure in 60% of transplant patients, 20% had IgA nephropathy, and 20% had no evidence of any specific etiology. There were no significant differences among the etiology of renal disease between transplants patients with CAD and stable renal function.
Table I. Clinical characteristic of training set and validation set cohorts and controls groups.
IF/TA, IF/TA with no other etiology; GFR, GFR from serum creatinine estimate by Modification of Diet in Renal Disease (MDRD) Study (49) equation (eGFR = 186 × (SCr)−1.154 × (age)−0.203 × (0.742 if female) × (1.21 if African American)).
| CAD (mean ± S.D.) |
Control (mean ± S.D.) |
|||
|---|---|---|---|---|
| IF/TA group | CAAR group | Stable renal transplant recipients | Healthy controls | |
| Training set | ||||
| Sample number (n) | 8 | 10 | 5 | 9 |
| Age (yr) | 51 ± 10.69 | 47.22 ± 17.07a | 36.2 ± 8 | 43 ± 10 |
| Creatinine (mg/dl) | 3.2 ± 1.68 | 2.98 ± 1.64a | 1.08 ± 0.3 | 0.91 ± 0.3 |
| GFR (ml/min/1.73 m2) | 28.88 ± 17.65 | 33.44 ± 12.05a | 82.22 ± 4 | 110 ± 10 |
| Proteinuria (g/24 h) | 2.67 ± 2.90 | 3.11 ± 3.33a | 0.20 ± 0.05 | 0.11 ± 0.02 |
| Validation set | ||||
| Sample number (n) | 10 | 11 | 9 | 9 |
| Age (yr) | 53.7 ± 17.8 | 54.09 ± 12.08a | 50.1 ± 13 | 41 ± 9.5 |
| Creatinine (mg/dl) | 2.49 ± 0.74 | 2.05 ± 0.37a | 0.96 ± 0.1 | 0.78 ± 0.11 |
| GFR (ml/min/1.73 m2) | 28.34 ± 8.76 | 31.6 ± 9.5a | 83.9 ± 14.5 | 103 ± 12 |
| Proteinuria (g/24 h) | 1.37 ± 1.44 | 1.84 ± 2.47a | 0.17 ± 0.10 | 0.11 ± 0.51 |
a t test not significant between IF/TA versus CAAR groups.
A second validation set of samples included a population of 21 CAD patients (10 IF/TA and 11 CAAR) unrelated to the training set of samples and 18 controls (nine SRT and nine unaffected individuals). Criteria for diagnosis were as in the training set, and base-line clinical parameters are shown in Table I, validation set. These studies were approved by the institutional review board at the Hospital Clinic in Barcelona, and both patients and controls gave informed consent for the collection and analysis of their urine.
Histopathology
Transplant biopsies consisted of two cores obtained with 18-gauge needles using ultrasound guidance because of clinical indication. Paraffin sections were prepared and stained with hematoxylin-eosin, trichrome, periodic acid-Schiff, and periodic acid-Schiff-methenamine silver. The biopsies were analyzed and were scored according to the Banff classification by a pathologist who was blinded to the results of molecular studies (26). TG was diagnosed by light microscopy based on double contours of glomerular basement membranes (27) and was supported by immunofluorescence studies, which showed mesangial IgM and/or C3 or negative immunofluorescence findings. Peritubular capillaritis in TG biopsies was graded according to the quantitative criteria of the last Banff update (26). The sections from the frozen biopsies were incubated with the murine monoclonal anti-human C4d 100 IL (Quidel Corp., San Diego, CA) and stained with fluorescent antiserum (CyTM2-conjugated AffiniPure goat anti-mouse IgG, Jackson ImmunoResearch Laboratories, West Grove, PA).
Sample Preparation and Purification for MS Analysis
Fifty milliliters of early morning urine were collected immediately prior to renal biopsy. Protease inhibitor mixture (Complete Mini, Roche Applied Science) was added, and specimens were rapidly frozen in dry ice and stored at −80 °C until analyzed. Urine samples were concentrated, and polypeptides were separated from inorganic salts by solid phase extraction using a reversed phase hydrophile-liphophile balance (HLB) Oasis 94226 (Waters, Milford, MA) as the stationary phase essentially as described previously (11) with minor modifications. Briefly cartridges were conditioned with 10 ml of 100% ACN and equilibrated with 10 ml of 0.1% TFA, 5% ACN. After loading 1 ml of sample (acidified to 0.1% TFA at pH 3 and a final concentration of 5% ACN by adding 10 μl of 10% TFA and 50 μl of ACN), the cartridge bed was washed with 10 ml of 0.1% TFA, 5% ACN, and peptides were subsequently eluted with 2 ml of 0.1% TFA, 60% ACN. Separation from organic salts was by strong cation exchange using magnetic beads (Dynabeads, Invitrogen) as follows. Beads were conditioned with 1 m NaCl, 50 mm ammonium bicarbonate, pH 8.8 and equilibrated with loading solution (0.1% TFA, 20% ACN). After application of the sample from the reversed phase step, bound peptides were washed three times with loading solution. Elution was with 500 mm ammonium acetate in 20% ACN. Eluted peptides were dried in a SpeedVac andstored at −80 °C.
Mass Spectrometry
Dry peptides were dissolved in 10 μl of 0.1% TFA, 2% ACN, and 10% of this solution was analyzed in an LC-MS/MS system that consisted of a nanoflow ultrahigh pressure liquid chromatograph (Acquity, Waters/Micromass) connected on line with a Q-TOF Premier mass spectrometer (Waters/Micromass) equipped with a nano-ESI ion source. Separations were performed in a bridged ethyl hybrid (BEH) 100-μm × 100-mm column (Waters/Micromass) at a 400 nl/min flow rate with an operating back pressure of about 3000 p.s.i. Gradient runs were from 2% B to 30% B in 30 min followed by a 5-min wash at 80% B and a 7-min equilibration step at 2% B. Solvent A was 0.1% formic acid in LC-MS grade water (Optigrade, LGC), and solvent B was 0.1% formic acid in LC-MS grade ACN (Optigrade, LGC). For LC-MS/MS experiments, survey MS scans of 500 ms were followed by three MS/MS scans (500 ms each), which were triggered in data-dependent mode when multiply charged ions in the MS survey scans were above 15 counts/s. Peptide quantification was performed by LC-MS analysis, acquiring 1-s survey scans. No MS/MS functions were included in the quantification protocol, but otherwise the settings were the same as for the LC-MS/MS experiments.
Data Analysis
Quantification
Lists of ions selected for MS/MS were fed into PESCAL, a program written in-house for the automation of label-free LC-MS data analysis (4). PESCAL uses the m/z and retention time of the ions detected to construct extracted ion chromatograms (XICs) across the LC-MS runs of individual urine samples. Windows for XIC construction were 25 ppm and 2 min for m/z and retention time, respectively. The intensity values (peak areas and heights) of these XICs were parsed into Excel files for normalization, statistical manipulation, and analysis. Peak intensity values were normalized to the mean intensity of all peaks within a sample and then to the mean of the individual peptide ions across the samples. These values were further normalized to creatinine content in the respective urine sample.
Statistical Analyses
For unsupervised clustering analysis, normalized ion intensity values were log-converted and fed into Cluster Eisein software (28) for clustering and TreeView Eisein software for visualization of the clustering results.
The non-parametric Kuskal-Wallis test was used to infer the statistical significance of LC-MS results when more than two sample groups were compared, whereas Mann-Whitney U test was for two-group comparisons. These analyses were performed using a commercial statistical software package (GraphPad Prism 4.03).
Statistical significance of clinical chemistry tests was inferred by the Student's t test. Relative levels of polypeptide ions were also analyzed using discriminant analysis (Systat, version 10.2, Systat Software Inc., Richmond, CA) to identify combinations of these polypeptides that best discriminate between disease states. A logistic regression model was also built using the same polypeptide ions to calculate prediction scores for each sample, allowing us to construct a receiver operating characteristic curve based on these values (29).
MS/MS Data Analysis
The identity of a subset of peptides detected was determined by searching MS/MS spectra against the Swiss-Prot database version 5.1.6 restricted to human entries (15,720 sequences) using the Mascot search engine. Searches were restricted to 50 and 100 ppm for parent and fragment ions, respectively. No enzyme restriction was selected. Hits were considered significant when they were above the statistical significant threshold (as returned by Mascot) and at least two peptides matched a protein entry. Selected MS/MS spectra were also searched by MS-Tag and MS-Homology (Protein Prospector v5.1.8 Basic) against NCBI (November 25, 2008) and Swiss-Prot (June 10, 2008) databases.
LC-Multiple Reaction Monitoring (MRM)-MS/MS
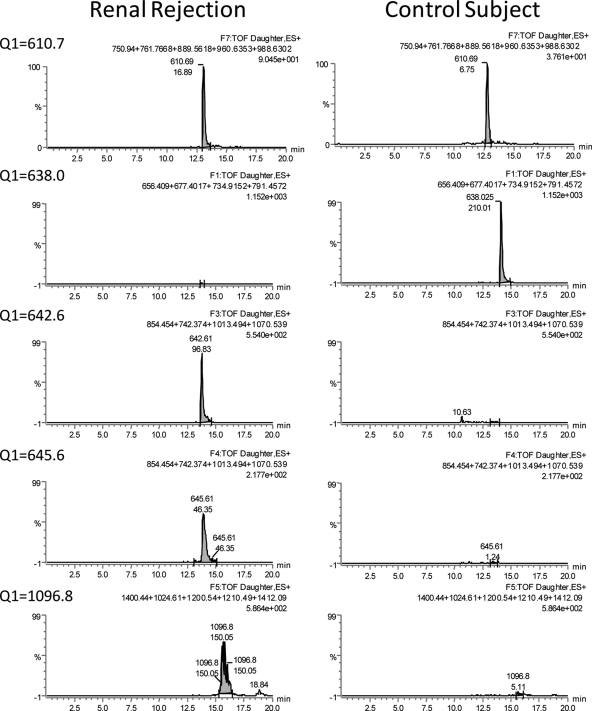
“pseudoMRM” was performed in the Q-TOF Premier/nano-ultraperformance LC system described above using a 10-min gradient (2–35% B in 10 min). Five ions were monitored simultaneously (at m/z 610.7, 638.0, 642,6, 645.6, and 1096.8) in alternative scans at a rate of 150 ms/scan. Typical chromatographic peak widths were 20–40 s at the base, which ensured collection of >10 points per chromatographic peak. The transitions monitored are shown in Fig. 8, and this was done by calculating peak areas of XICs of added ion intensities of the most intense fragment ions. CID was performed with a ramp collision energy of 22–30 eV in the 150-ms scan. The Quantitate function in MassLynx was used to automate these analyses. The integration of each chromatographic peak was corrected manually to ensure accurate quantification. Values were normalized to creatinine content.
Fig. 8.
Examples of chromatograms for each of the ions monitored by targeted LC-MS/MS. The m/z values of the parent ions monitored are shown on the left. The sum of fragments that were then added in XICs are shown in the right top corner of each chromatogram.
RESULTS
Patients
We aimed to identify urinary peptides that could serve as biomarkers of CAD. For this purpose, 14 controls and 18 specimens with well defined clinical features were included in an initial training set; the results were then validated in a second set of samples using an unrelated patient population consisting of 21 CAD and 18 control individuals.
The control groups showed normal blood pressure (systolic blood pressure <130 mm Hg and diastolic blood pressure <80 mm Hg) and no history of diabetes mellitus, ischemic heart disease, stroke, or peripheral vascular disease. Table I shows the base line of the control clinical parameters in training and validation set experiments.
The IF/TA and CAAR groups did not present significant differences with respect to age, gender, diabetes duration, arterial blood pressure, body mass index, and GFR. However, they both were classified into IF/TA and CAAR (CAD subgroups) according to histopathological criteria. Table II (left) summarizes Banff scores in the IF/TA and CAAR groups of the training set. IF/TA patient histopathology showed evidence of neither CAAR nor TG, and C4d was negative. Mean glomerular double contour score was 1.89, and C4d was positive in all patients in the CAAR group. Evidence of chronic active T cell-mediated rejection was excluded in all samples. Criteria for diagnosis of the second validation subset of patients were as in the training set of samples, and Table II (right) shows the Banff scores in the IF/TA and CAAR groups of this validation study.
Table II. Histopathology of allograft biopsies of the training set and validation set.
IF/TA, IF/TA with no other etiology. Transplant biopsies were normal in stable renal recipients.
| Training set (mean ± S.D.) |
Validation set (mean ± S.D.) |
|||
|---|---|---|---|---|
| IF/TA group (n = 8) | CAAR group (n = 10) | IF/TA group (n = 10) | CAAR group (n = 11) | |
| Glomerular double contours (cg) | 0 | 1.89 ± 0.7a | 0 | 1.82 ± 0.75a |
| Interstitial fibrosis (ci) | 1.88 ± 0.9 | 2.11 ± 0.7b | 1.9 ± 0.56 | 2.18 ± 0.6b |
| Tubular atrophy (ct) | 1.88 ± 0.9 | 2.11 ± 0.7b | 1.9 ± 0.56 | 2.18 ± 0.6b |
| Arterial fibrous intimal thickening (cv) | 0.88 ± 0.6 | 1.56 ± 1.0a | 1.6 ± 0.69 | 1.36 ± 0.67a |
| Hyaline arteriolar thickening (ah) | 0.75 ± 1.1 | 1.56 ± 1.1b | 1.5 ± 0.7 | 1.73 ± 0.46b |
| Peritubular capillaritis (ptc) | 0.75 ± 0.4 | 1.33 ± 0.7b | 0.4 ± 0.69 | 1.64 ± 0.5a |
a Significant Mann-Whitney test (p < 0.05).
b Non-significant Mann-Whitney test.
Analytical Strategy
To increase the probabilities of identifying useful biomarkers of CAD, we aimed at quantifying urinary peptides across samples as comprehensively and accurately as possible. For this, we used an analytical strategy that consists of using the LC-MS elution profiles of individual peptide ions that had been detected previously in LC-MS/MS experiments. PESCAL software was used for the automation of this analysis. This is therefore a targeted quantification strategy because we only quantified (by LC-MS) those ions that had been detected previously (although not necessary identified) in urine by data-dependent LC-MS/MS. To generate a list of quantifiable urinary peptides, we pooled undigested urinary peptides from the same patient group and analyzed them by LC-MS/MS. These analyses were performed in triplicate (three times per sample group), generating in each replicate LC-MS/MS experiment a list of identified peptides that we then used as an exclusion list in the following LC-MS/MS run as reported before (11). Approximately the same numbers of MS/MS spectra were obtained per sample group. These experiments resulted in the selection of 6250 multiply charged ions for MS/MS. It should be noted that this was an unfiltered list, and many of these ions were detected in more than one sample pool. Targeted quantification of the 6250 ions that had been identified by data-dependent MS/MS acquisition was then performed by integration of the extracted ion chromatogram peak areas from LC-MS data of individual samples (without pooling); this was achieved by using PESCAL software.
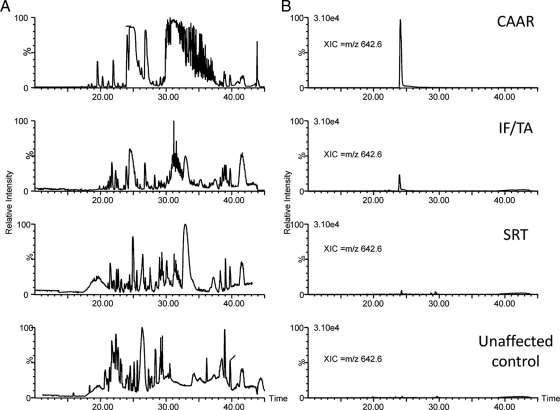
To exemplify how this analytical strategy works, Fig. 1 A shows representative LC-MS ion chromatograms from urine peptide mixtures of IF/TA, CAAR, SRT, and control donors. Each of the peaks in these chromatograms was formed by the elution of several polypeptides, and therefore, by themselves, they cannot be used to quantify individual peptides. For quantification, we calculated the area under the curve of a chromatogram obtained from the elution profile of each individual peptide, i.e. an XIC, an example of which is shown in Fig. 1B. To make this approach practical, we used PESCAL, an in-house computer program to automate the creation of these XICs and calculate their intensities; PESCAL also allows matching the XICs with those from the peptides identified previously by LC-MS/MS (4). In the illustrative example shown in Fig. 1B, a molecular ion at m/z 642.6 was present with about 5-fold enrichment in CAAR urine compared with IF/TA and at least 15-fold enhancement when compared with SRT and control samples.
Fig. 1.
Label-free quantitative strategy for the identification of urinary biomarkers of CAD. Representative base peak ion chromatograms (A) and representative XIC of an ion at m/z 642.6 across the indicated sample group (B) are shown.
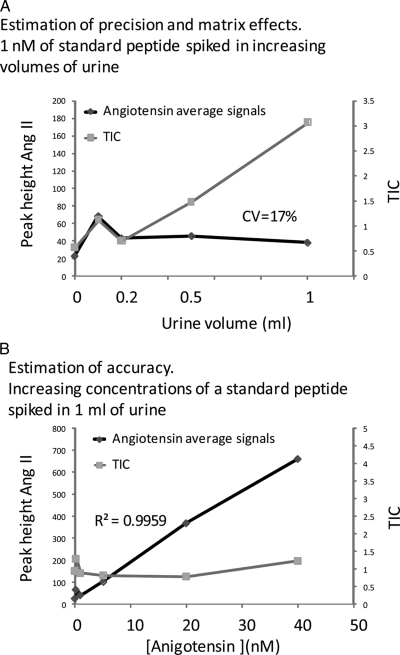
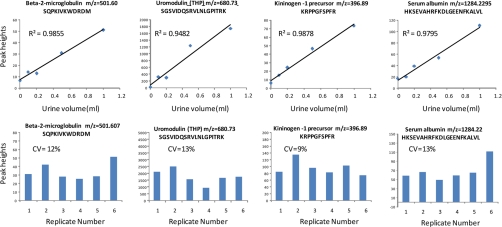
To investigate the linearity of the technique and the potential of matrix effects contributing to variability, we spiked control urine with standard peptides, which were then analyzed by our LC-MS methods. As Fig. 2 A shows the signals of the standard peptide angiotensin II were not affected by increasing amounts of matrix in the sample, arguing against the occurrence of ion suppression with these settings. Similarly these signals were linear over at least a 40-fold range (Fig. 2B). We also analyzed endogenous urinary peptides at different urine volumes and found that the signals of these were linear over a 10-fold volume range (Fig. 3 A). Replicate experiments demonstrated good reproducibility (coefficients of variation (CVs) between 9 and 13%; Fig. 3B). Thus, the data in Figs. 2 and 3 indicate that signals obtained with our LC-MS technique are linear over a useful working range and that matrix effects are not significant probably because of the extensive extraction and separation used (reversed phase and strong cation exchange extraction followed by a 30-min gradient in an ultrahigh pressure LC instrument) and the low probability of ion suppression to occur when using nanoelectrospray as the ionization source (30, 31). This is in line with previous reports demonstrating accurate quantification of polypeptides by label-free LC-MS (4, 5).
Fig. 2.
Linearity of label-fee LC-MS of a peptide spiked in urine. A, the signal of angiotensin II (Ang II), a standard peptide spiked in increasing volumes of urine, was not affected by increasing concentrations of matrix ions. B, the signal of increasing amounts of the same peptide as in A was linear in a constant background of matrix ions. TIC, total ion chromatogram.
Fig. 3.
Precision and accuracy of signals of endogenous urinary peptides as determined by label-free LC-MS. The top panel graphs show that the intensities of representative urinary peptides were linear relative to the amount of sample analyzed by LC-MS. The bottom graphs show the reproducibilities of signal intensities generated in replicate LC-MS runs. CVs were defined as mean/S.D. × 100. THP, Tamm-Horsfall glycoprotein
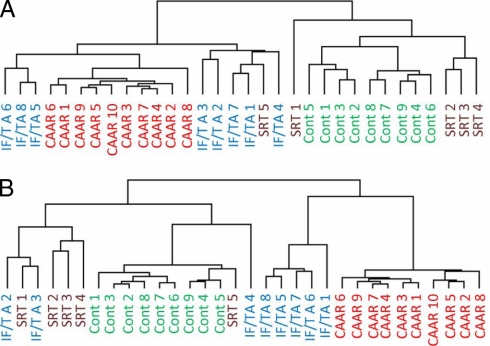
Hierarchical Clustering of Label-free Quantitative LC-MS Data Classifies Urinary Peptidomes According to Their Underlying Pathological Phenotype
As mentioned above, using this strategy we detected ∼6250 ions in our analyses before filtering and about 2300 after deleting duplicates; and for all of them we obtained XICs and calculated their intensities. Initial experiments demonstrated that it was possible to differentiate control and CAAR patients based on unsupervised hierarchical clustering using the intensities of all the identified peptides with SRT and IF/TA showing some overlap (Fig. 4A). Filtering of the data to include only the 500 peptide ions showing the least statistical variability in their peak intensities resulted in a similar clustering pattern (Fig. 4B). Unlike Fig. 4A where one SRT patient was placed in the CAAR/IF/TA cluster, when considering only 500 peptides the SRT and control were completely separated from CAAR. Further filtering to include lower numbers of peptides for hierarchical clustering gave results similar to those in Fig. 4B (data not shown). These results indicate that the polypeptide composition in the urine of CAD patients is significantly different from that of SRT and control subjects and that our label-free quantitative LC-MS strategy can detect these differences.
Fig. 4.
Unsupervised hierarchical clustering analysis classifies individual patients according to their underlying pathological phenotype. The figure shows hierarchical clustering trees based on quantitative urinary polypeptide data from CAD subtypes (CAAR and IF/TA) and control specimens (SRT and control (Cont)) considering the whole data set (A) and the least variable 500 molecular ions (B).
Identification of Peptides Derived as Specific Biomarkers for the Diagnosis of CAD
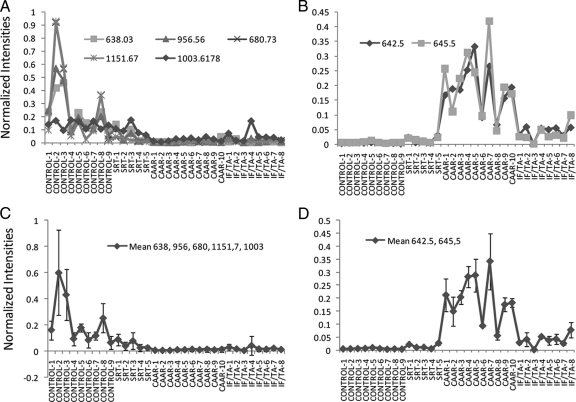
To characterize in more detail the molecular differences between these specimens, we compared the intensities of the subset of peptides that produced statistically significant differences. Peptides at m/z 638.03, m/z 956.56, and 1003.62 were significantly more abundant in control patients than in the other groups (Fig. 5A). Importantly these peptides showed greater intensities in SRT than in CAAR samples with a significant -fold change from 4.0 to 8.9, a fact of great clinical relevance because these data indicate that these biomarkers may allow monitoring the evolution of the transplanted kidney. MS/MS data demonstrated that these peptides are derived from uromodulin and kininogen (supplemental data). Their expression in individual patients revealed that, although the differences were more apparent in the kinins, uromodulin peptides were consistently more abundant in control and SRT than in CAD groups, showing a lower variability (Fig. 5, A and C). The individual peptide ions that best discriminated between controls and CAD groups were those derived from uromodulin at m/z 638 (SGSVIDQSRVLNLGPITR) and kininogen at m/z 1003 (DLIATMMPPISPAPIQSDDDWIPDIQI), resulting in correct discrimination between control and CAD patients of 84% (14 of 18). These results were corroborated by a logistic regression analysis, which resulted in selection of the same ions. A receiver operating characteristic curve constructed from the logistic regression scores gave an area under the curve value of 0.82.
Fig. 5.
Specific peptides are potential biomarkers for the diagnosis of chronic allograft dysfunction. A, normalized ionic intensities of individual peptides derived from uromodulin and kinin present higher levels in control than in CAD urines. B, normalized intensities of individual unknown ions show higher levels in control than in CAD urines. C, mean ± S.D. expression of peptides shown in A. Kruskal-Wallis test was significant at p < 0.0001. The difference between control subtypes versus CAD subtypes was significant at p < 0.0001. The means within control and CAD subgroups were not significant (p > 0.05). D, means of ions shown in B. The data were significantly different (p < 0001, Kruskal-Wallis test). The differences between CAD and control means were significant at p < 0.0001. The means of CAAR versus IF/FA were also significantly different (p < 0.0001).
A greater sensitivity was achieved by considering the mean intensity of all the peptides derived from uromodulin. Indeed this value could correctly identify 94% (17 of 18) of all CAD patients with 100% specificity (none of the control group had low levels of uromodulin peptides; Fig. 5C). In addition, the statistical analysis performed using all the uromodulin peptides reached significant differences not only between SRT and CAAR (6.3 ± 1.9-fold change) but also between SRT and IF/TA (3.8 ± 1.3-fold change). Therefore, a combination of biomarkers had more discriminatory power than any of the single biomarkers we identified and clearly showed a potential to follow the progression stage of renal transplant. This fact argues that using the mixture of peptide biomarkers identified herein would increase the specificity and sensitivity of CAD diagnosis.
Other peptides could be identified in these specimens, but they did not provide discriminatory information. As an example, peptides derived from β2-microglobulin did not show significant differences between the studied groups in most peptides but one, which could be assigned as potential biomarker (data not shown). Thus, our data demonstrate that a specific combination of peptides derived from uromodulin and bradykinin represents a potential excellent and specific biomarker cluster that reflects kidney health. Measurement of this cluster of biomarkers could potentially differentiate between patients with CAD and those with a stable transplant, a finding of great clinical interest.
Identification of Urinary Molecular Features for the Classification of CAD into Clinical Subtypes
The peptides discussed above, although useful for the diagnosis of CAD, could not further discriminate between the two CAD groups, namely CAAR and IF/TA groups. We therefore mined our data further by considering the ions that could be detected but for which it was not possible to derive sufficient structural information for identification. These ions are likely to be peptides, but they could also be carbohydrates or other organic bases. Fig. 5B shows that ions at m/z 645.59 and m/z 642.61 were significantly more abundant in CAAR patients than in IF/TA (4.4-2.7-fold change) and SRT (5.3–14.6-fold change) specimens, allowing us to differentiate between the two different forms of CAD. These biomarkers correctly identified 100% of the pure IF/TA group and 90% (nine of 10) of the CAAR patients.
Quantitative analysis of these ions in individual patients indicated that these were consistently more abundant in CAAR patients than in the IF/TA group both when they were considered in isolation (Fig. 5B) and when the mean of these potential biomarkers was calculated (Fig. 5D). Indeed the mean expression of ions at m/z 645.59 and m/z 642.61 was greater in 10 of 10 CAAR patients than in six of seven patients in the IF/TA group (Fig. 5D). These results indicate that the mean expression of these two ions can discriminate between patients with different forms of CAD.
Investigation Potential CAD Biomarkers in a Validation Patient Population
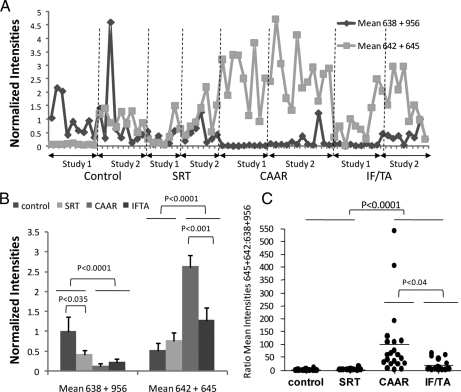
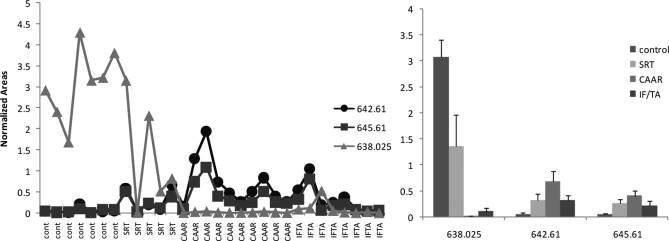
To verify our training set findings in a second validation set of samples, we analyzed urinary peptides consisting of 21 CAD patients and 18 controls (Fig. 6A). As with the initial experiment (Fig. 5), the data of this validation study indicated that the mean intensities of two uromodulin ions at m/z 638 and m/z 956 were statistically significantly higher in the control groups than in the CAD groups (Fig. 6B). The ions at m/z 642 and m/z 645 could also distinguish between CAD and control subjects (p < 0.0001, n = 39 CAD and 32 controls; Fig. 6B), and in addition, their intensities were significantly higher in CAAR than in the IF/TA subtype (p < 0.0004, n = 20 CAAR and 19 IF/TA).
Fig. 6.
A combination of biomarkers better discriminates CAD and controls subjects and classifies chronic allograft dysfunction into clinical subtypes. A, the mean intensities of the molecular ions with the indicated m/z values were plotted for each of the 39 patients and 32 controls in training (Study 1) and validation (Study 2) experiments. B, these means were statistically different in CAD and control subjects. Graphs represent mean ± S.E. of the means shown in A with statistical significances shown for pairwise comparisons. Kruskal-Wallis analysis returned a p < 0.0001. C, ratio of the means of 638–956 to 642–645 is also a useful indicator of CAD. Values were obtained by obtaining the ratio of the values shown in A for each patient and were statically significant by Kruskal-Wallis test at p < 0.0001. p values obtained by Dunn's multiple comparison tests are shown in the figure.
This combination of biomarkers can assist in the diagnosis of CAD as indicated in Fig. 6 (A and C), which shows that low intensities of ions at m/z 638 and 956 together with a high concentration of ions at m/z 642 and 645 are characteristic of CAD subtypes. The ratios of these two groups of biomarkers were also of diagnostic value (Fig. 6D).
Verification Using an Orthogonal Technique
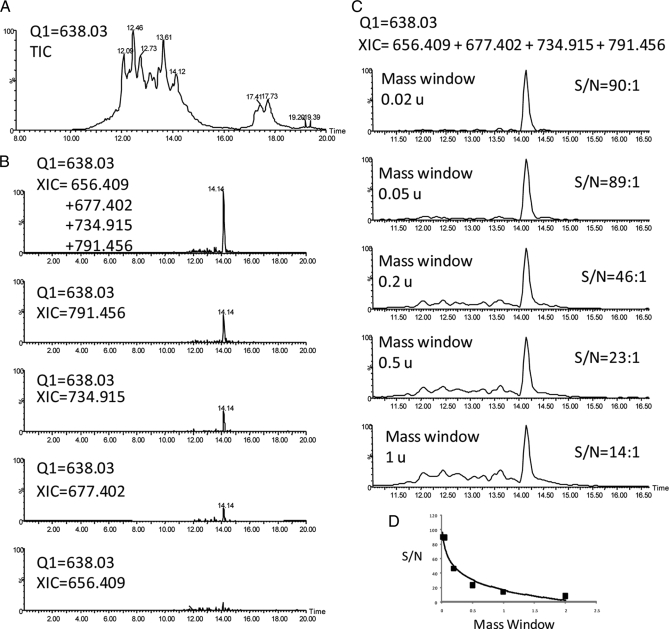
We investigated whether MRM could be used to monitor and validate the putative biomarkers found in the experiments shown above. Although MRM is often performed in triple quadrupole instruments because of their increased sensitivities relative to hybrid instruments such as Q-TOF mass spectrometers, performing peptide MRM in Q-TOF mass spectrometers has the advantage that it is possible to add the ionic intensities of peptide fragments produced during CID MS/MS, which splits the signal of the peptide into several fragments and thus compromises sensitivity. Thus we designed experiments in which the Q1 of the Q-TOF selects for the m/z of the peptide to be monitored with all the fragments being analyzed and recorded by TOF MS. This is exemplified with m/z 638.03 in Fig. 7A, which shows that several ions had m/z 638 in a urine analysis over a 10-min gradient. We then obtained XICs of fragment ions produced by CID MS/MS. As shown in Fig. 7B (and as expected), adding four of these fragment ions produced peaks with greater signal to noise ratios (S/N) than when only one fragment was monitored as when using triple quadrupole. Monitoring the addition of several fragments could in principle result in a decrease in specificity because nearly coeluting species could produce isobaric fragment ions upon CID. This potential problem was minimized by using narrow mass windows when constructing the XICs of fragment ions. Thus, as Fig. 7C shows, a mass window of 0.02 mass unit (u) produced a peak with S/N = 90, whereas allowing a 1-u mass window produced a peak of just S/N = 14. Fig. 7D indicates that at a window of 0.05 u the S/N enhancement by narrowing mass windows reached a plateau.
Fig. 7.
Targeted quantification and validation of potential biomarkers by pseudoMRM LC-MS/MS in a Q-TOF mass spectrometer. A, total ion chromatogram (TIC) produced when Q1 was set to transmit m/z 638 only in an LC-MS/MS analysis of urine. B, XICs of the indicated fragment ions produced upon CID of uromodulin peptide with m/z 638.03. C, XICs as in B with different mass window settings. D, plot of S/N as a function of XIC mass window.
Using this pseudoMRM approach we were able to obtain specific and “clean” transitions for five of the potential biomarkers identified above (Fig. 8 ). These techniques were then used to monitor the amounts of these biomarkers in a representative series of samples of the validation group. These results, shown in Fig. 9 , confirmed that ions at m/z 638.03, m/z 642.61, and m/z 645.61 are promising potential biomarkers of CAD and validated the LC-MS approach (compare graphs in Figs. 5 and 9, bearing in mind that the LC-MS approach in Fig. 5 provides relative quantification whereas the LC-MS/MS approach in Fig. 9 provides a readout in absolute units).
Fig. 9.
Results of targeted LC-MS/MS experiment. Results of urine analysis, as obtained by the pseudoMRM approach shown in Figs. 7 and 8, are shown in the left graph for individual samples. The right graph shows the mean ± S.D. of data on the left grouped by phenotypes.
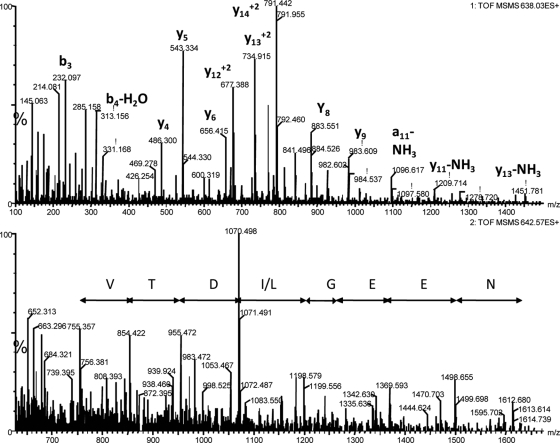
MS/MS Sequencing of Potential Biomarkers of CAD
The potential biomarker ions at m/z 638 and 956 were triply and doubly charged versions of the same peptide and were identified by Mascot searchers as being derived from uromodulin. This identity was confirmed by manual assignment of all the fragment ions in the MS/MS spectrum (Fig. 10A). Ions at m/z 645 and m/z 642 had four charges, and they are related in that many of their fragment ions were common between the two species (supplemental Fig. 1). Although the MS/MS spectra were of good quality, they did not return a positive match by Mascot or Protein Prospector searches against NCBI or Swiss-Prot databases. Manual interpretation produced a partial putative sequence (VTD(I/L)GEEN; Fig. 10B and supplemental Fig. 2) that did not match any entry in actualized NCBI and Swiss-Prot protein databases.
Fig. 10.
MS/MS spectra of potential biomarkers. The top panel shows an MS/MS spectrum of m/z 638.03 (3+) that matched to a uromodulin fragment with sequence SGSVIDQSRVLNLGPITR. The bottom panel shows a spectrum of m/z 642.3 (4+) that did not produce a positive match by database searches. A potential amino acid sequence is shown.
DISCUSSION
Despite its limited initial success, MS still offers great potential for biomarker discovery. The most popular techniques for biomarker discovery among clinicians are single spectrum profiling techniques such as SELDI-TOF MS and MALDI-TOF MS, both of which have been successfully used to identify potential biomarkers (32).
However, there is little overlap between the biomarkers identified by these techniques in different laboratories, and those abundant and reproducible features are unspecific. Because of their limited dynamic range, these methods may not be directly interfaced with MS/MS instruments for sequencing useful biomarkers for diagnosis. This contrasts with the reproducibility and dynamic range of hyphenated techniques such as LC-MS and capillary electrophoresis-MS with the former being routinely used in diagnostic and forensic applications (33). Because of its larger dynamic range and depth of analysis, LC-MS can detect and quantify hundreds/thousands of polypeptides in biological fluids (6), and the use of this technique for the analysis of the urinary proteome and peptidome is well documented (10–13), a fact of great interest because of the potential of urine as a non-invasive source of biomarkers. However, because of their recent development, the use of quantitative LC-MS for comprehensive quantification of polypeptides in urine for biomarker discovery is not significant.
Here we used a label-free LC-MS approach for the discovery of urine biomarkers able to diagnose CAD, the main cause of kidney transplant loss. Previous studies (11, 12) demonstrated that urine contains a large number of small peptides; these urinary components may represent a rich source of biomarkers that is not fully explored. For this reason and because we also wanted to avoid the variability that a proteolytic step may introduce to our analyses, we chose not to digest the sample prior to LC-MS analysis. We also chose to perform a targeted quantification approach because we reasoned that this would lead to less variability and decided to quantify all the entries in the redundant list of >6000 multiply charged ions that we identified across samples. We did not initially filter this list because the redundancy of quantitative data should not have adverse effects on the outcome of the analysis and may, in some cases, add confidence to the results and help to rule out potential artifacts.
By performing this targeted approach by LC-MS, we circumvented problems with lengthy duty cycles imposed when quantifying by LC-MS/MS (quantification of >2000 peptides in a single 30-min gradient run, as in the present study, by multiple reaction monitoring is not possible even with the new generation of fast scan instruments), which can lead to undersampling (as in data-dependent experiments needed to perform quantitative strategies based on isobaric tags for relative and absolute quantitation (iTRAQ) and similar labeling reagents); thus the need for extensive separation was obviated in the present study. As a result, we reduced the LC-MS run time (total cycle) to about 45 min per sample, which allowed us to have a throughput of about 30 samples per day per LC-MS system. The LC-MS run time could be shortened further for studies where fewer analytes need to be followed for validation purposes as in the pseudoMRM approach presented here (Figs. 7 and 8). The precision and accuracy of this LC-MS method has been demonstrated before (4). In addition spiking experiments (Fig. 2) and correlation to targeted MRM (Fig. 9) showed that this LC-MS technique provides adequate quantitation (CVs of about 10–15% on average).
There are several urine potential biomarkers for CAD reported in the literature that were identified by radioimmunoassay or gene expression analysis (34, 35), but to our knowledge, this is the first report on the use of this noninvasive peptidomics approach for the identification of polypeptide biomarkers in urine of CAD patients. Indeed using this approach we identified specific peptides derived from uromodulin and kinins (bradykinin) as specific biomarkers of a healthy kidney whose presence could be used with statistically significant confidence to discriminate between CAD patients and unaffected individuals. Of special interest are those that can distinguish between SRT and CAD specimens. Moreover MRM experiments monitoring the putative biomarkers found in the training sample set confirmed that the best discriminatory markers were the combination of ions at m/z 638.0 and m/z 642.5. Thus high 642.5 and low 638.0 is characteristic of CAD urine and can also discriminate CAD subtypes.
The ability of discriminating between CAD subtypes is another important outcome of this study because CAAR (a specific form of CAD) has a unique pathogenesis that distinguishes it from other chronic pathologic conditions of kidney allografts. Two features highlight the importance of an early diagnosis of this disease: CAAR is associated with very poor allograft survival, and CAAR is the first example of a pathologic entity distinct from the generic and nonspecific histological forms of CAD such as IF/TA (36). The ability of easily and non-invasively distinguishing these two forms of CAD opens new opportunities for further clinical studies and for assessing clinical outcomes based on specific therapeutic regimens.
Effective strategies to prevent renal function deterioration in kidney transplant patients focus on the early detection and treatment of patients developing CAD. Specific and sensitive biomarkers are needed to identify patients at risk of developing CAD or initial lesions that do not show changes in the current available biomarkers such as serum creatinine or GFP. This feature is of particular importance because these non-invasive biomarkers have been proved to have poor predictive value (most confidence intervals included 0.5, indicating no predictive ability) for 10 individual histological measurements (Banff 97 scores) and the Chronic Allograft Damage Index. Thus, serum creatinine and GFR have a limited clinical role in predicting the early histopathological changes that precede CAD and should not be used for this purpose (37).
Renal biopsy has proved to be an effective diagnostic method for CAD; nevertheless the implied risk for the patients limits it use and, by implication, its success. There is thus an urgent need for a simple and non-invasive method of risk assessment that can be applied to all renal transplant patients to identify those few at greatest risk of CAD development. As the transplant community continues to struggle with the challenge of reducing late allograft loss due to CAD, the new biomarkers identified in this study will undoubtedly emerge as surrogates to develop a renal risk score applicable to a more general population of patients with a renal allograft.
Uromodulin is exclusively expressed in the kidney and constitutes the most abundant protein in human urine. The exact physiological role of uromodulin, however, remains to be elucidated (38). Nevertheless it has been proposed that this protein is involved in the development of cast nephropathy, in the formation of renal stones, in immunologic defense in the kidney, and in the modulation of systemic immunologic events (39). More than 2 decades ago, in a study of renal allografts with cellular acute rejection, extratubular uromodulin was identified in 63.6% of the specimens, representing 76.1% of the patients whose tissues were examined (40). In the kidney transplantation field, uromodulin has been suggested as being suitable for monitoring the functional state of transplanted kidneys. In a very preliminary study from the late 1990s (41), the urinary uromodulin after uninephrectomy and transplantation among relatives was determined by ELIAS SYNELISA-uromodulin immunoassay to study the influence of the acute reduction in renal mass on the excretion of this protein; uninephrectomy in healthy man was associated with a marked increase of around 40% in the excretion of uromodulin from the kidney that remained in the donor. In the kidney that was transplanted, the uromodulin excretion rate was unchanged. The uromodulin excretion rate was correlated with GFR, but mechanisms underlying this association were unknown as uromodulin does not undergo glomerular filtration (41, 42). The present study indicates that the best biomarker may not be full-length uromodulin and has identified specific peptides derived from uromodulin in donors and recipients of kidney transplantation more suitable for monitoring the functional state of transplanted kidneys. Full-length uromodulin protein forms oligomers that tend to precipitate (43), and therefore monitoring the full-length protein in urine is prone to artifacts. In contrast, small peptides are soluble, easy to handle, and amenable to detection by different biophysical and immunological techniques.
Besides the immunologically mediated damage on the graft, the intrarenal renin-angiotensin system (RAS) is viewed as an additional mechanism in the development and progression of CAD (44). RAS blocking agents efficiently control post-transplant hypertension and are useful to reduce proteinuria and partially exert their beneficial cardiovascular effects by potentiating endogenous kinins (45–47). However, because of the nature of the studies and in particular the duration of treatment and the numbers of patients evaluated, we cannot establish whether these interventions have a long term beneficial effect on cardiovascular outcomes and the prevention of progressive allograft dysfunction. Moreover the optimal approach for the provision of more complete RAS blockade has yet to be identified (48). From this point of view, peptides derived from kinins (in particular bradykinin) could be evaluated in the future not only for the diagnosis of CAD but also as a tracing biomarker of the renoprotector effects of RAS blockage and in the progression of CAD.
In view of the complex relationship among clinical and biological CAD factors and between early histopathological changes and graft outcome, the identification of non-invasive biomarkers of CAD stages and subtypes is imperative to improve long term graft survival. The biomarkers reported here, by themselves or in combination, could form the basis of a urine test for the early diagnosis of CAD and its subtypes without the need for invasive biopsies. This will facilitate a more rapid introduction of targeted and personalized immunosuppressive regimens to improve long term graft outcomes. Confirmation of these results in larger scale cross-center studies should lead to a significant improvement in techniques for monitoring renal transplant progression.
Acknowledgments
We are grateful to John Timms for critical reading of the manuscript, to Robert Chalkley for help with MS/MS interpretation, to Bart Vanhaesebroeck for support, and to Federico Oppenheimer for clinical advice.
Footnotes
* This work was supported by Fondo de Investigaciones Sanitarias Grants FIS 03/0557 and FIS 06/1222, by an Emili Letang Fellowship award (to L. F. Q.) from Hospital Clinic in Barcelona, andn by the Bart's and the London Charity (to P. R. C. and M. P. A.).
 The on-line version of this article (available at http://www.mcponline.org) contains supplemental material.
The on-line version of this article (available at http://www.mcponline.org) contains supplemental material.
1 The abbreviations used are:
- CAD
- chronic allograft dysfunction
- CAAR
- chronic active antibody-mediated rejection
- GFR
- glomerular filtration rate
- IF/TA
- interstitial fibrosis and tubular atrophy
- MRM
- multiple reaction monitoring
- RAS
- renin-angiotensin system
- SRT
- stable renal transplant
- TG
- transplant glomerulopathy
- XIC
- extracted ion chromatogram
- PESCAL
- Peak Statistic Calculator
- NCBI
- National Center for Biotechnology Information
- CV
- coefficient of variation
- S/N
- signal to noise ratio
- u
- mass unit
References
- 1.Cox J., Mann M. ( 2007) Is proteomics the new genomics? Cell 130, 395– 398 [DOI] [PubMed] [Google Scholar]
- 2.Norden A. G., Rodriguez-Cutillas P., Unwin R. J. ( 2007) Clinical urinary peptidomics: learning to walk before we can run. Clin. Chem. 53, 375– 376 [DOI] [PubMed] [Google Scholar]
- 3.Rifai N., Gillette M. A., Carr S. A. ( 2006) Protein biomarker discovery and validation: the long and uncertain path to clinical utility. Nat. Biotechnol. 24, 971– 983 [DOI] [PubMed] [Google Scholar]
- 4.Cutillas P. R., Vanhaesebroeck B. ( 2007) Quantitative profile of five murine core proteomes using label-free functional proteomics. Mol. Cell. Proteomics 6, 1560– 1573 [DOI] [PubMed] [Google Scholar]
- 5.Silva J. C., Gorenstein M. V., Li G. Z., Vissers J. P., Geromanos S. J. ( 2006) Absolute quantification of proteins by LCMSE: a virtue of parallel MS acquisition. Mol. Cell. Proteomics 5, 144– 156 [DOI] [PubMed] [Google Scholar]
- 6.Qian W. J., Jacobs J. M., Liu T., Camp D. G., 2nd, Smith R. D. ( 2006) Advances and challenges in liquid chromatography-mass spectrometry-based proteomics profiling for clinical applications. Mol. Cell. Proteomics 5, 1727– 1744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Washburn M. P., Wolters D., Yates J. R., 3rd ( 2001) Large-scale analysis of the yeast proteome by multidimensional protein identification technology. Nat. Biotechnol. 19, 242– 247 [DOI] [PubMed] [Google Scholar]
- 8.Constantopoulos T. L., Jackson G. S., Enke C. G. ( 1999) Effects of salt concentration on analyte response using electrospray ionization mass spectrometry. J. Am. Soc. Mass Spectrom. 10, 625– 634 [DOI] [PubMed] [Google Scholar]
- 9.Smith R. D., Tang K., Shen Y. ( 2006) Ultra-sensitive and quantitative characterization of proteomes. Mol. Biosyst. 2, 221– 230 [DOI] [PubMed] [Google Scholar]
- 10.Spahr C. S., Davis M. T., McGinley M. D., Robinson J. H., Bures E. J., Beierle J., Mort J., Courchesne P. L., Chen K., Wahl R. C., Yu W., Luethy R., Patterson S. D. ( 2001) Towards defining the urinary proteome using liquid chromatography-tandem mass spectrometry. I. Profiling an unfractionated tryptic digest. Proteomics 1, 93– 107 [DOI] [PubMed] [Google Scholar]
- 11.Cutillas P. R., Norden A. G., Cramer R., Burlingame A. L., Unwin R. J. ( 2003) Detection and analysis of urinary peptides by on-line liquid chromatography and mass spectrometry: application to patients with renal Fanconi syndrome. Clin. Sci. 104, 483– 490 [DOI] [PubMed] [Google Scholar]
- 12.Cutillas P. R., Norden A. G., Cramer R., Burlingame A. L., Unwin R. J. ( 2004) Urinary proteomics of renal Fanconi syndrome. Contrib. Nephrol. 141, 155– 169 [DOI] [PubMed] [Google Scholar]
- 13.Adachi J., Kumar C., Zhang Y., Olsen J. V., Mann M.The human urinary proteome contains more than 1500 proteins, including a large proportion of membrane proteins. Genome Biol. 20067R80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ong S. E., Mann M. ( 2005) Mass spectrometry-based proteomics turns quantitative. Nat. Chem. Biol. 1, 252– 262 [DOI] [PubMed] [Google Scholar]
- 15.Ow S. Y., Cardona T., Taton A., Magnuson A., Lindblad P., Stensjö K., Wright P. C. ( 2008) Quantitative shotgun proteomics of enriched heterocysts from Nostoc sp. PCC 7120 using 8-plex isobaric peptide tags. J. Proteome Res. 7, 1615– 1628 [DOI] [PubMed] [Google Scholar]
- 16.Cutillas P. R., Geering B., Waterfield M. D., Vanhaesebroeck B. ( 2005) Quantification of gel-separated proteins and their phosphorylation sites by LC-MS using unlabeled internal standards: analysis of phosphoprotein dynamics in a B cell lymphoma cell line. Mol. Cell. Proteomics 4, 1038– 1051 [DOI] [PubMed] [Google Scholar]
- 17.Turck C. W., Falick A. M., Kowalak J. A., Lane W. S., Lilley K. S., Phinney B. S., Weintraub S. T., Witkowska H. E., Yates N. A. ( 2007) The Association of Biomolecular Resource Facilities Proteomics Research Group Study: relative protein quantitation. Mol. Cell. Proteomics 6, 1291– 1298 [DOI] [PubMed] [Google Scholar]
- 18.Collins A. J., Kasiske B., Herzog C., Chen S. C., Everson S., Constantini E., Grimm R., McBean M., Xue J., Chavers B., Matas A., Manning W., Louis T., Pan W., Liu J., Li S., Roberts T., Dalleska F., Snyder J., Ebben J., Frazier E., Sheets D., Johnson R., Li S., Dunning S., Berrini D., Guo H., Solid C., Arko C., Daniels F., Wang X., Forrest B., Gilbertson D., St Peter W., Frederick P., Eggers P., Agodoa L. ( 2003) Excerpts from the United States Renal Data System 2003 Annual Data Report: atlas of end-stage renal disease in the United States. Am. J. Kidney Dis. 42,Suppl. 5, A5–7, S1–230 [PubMed] [Google Scholar]
- 19.Wolfe R. A., Ashby V. B., Milford E. L., Ojo A. O., Ettenger R. E., Agodoa L. Y., Held P. J., Port F. K. ( 1999) Comparison of mortality in all patients on dialysis, patients on dialysis awaiting transplantation, and recipients of a first cadaveric transplant. N. Engl. J. Med. 341, 1725– 1730 [DOI] [PubMed] [Google Scholar]
- 20.Paul L. C. ( 1999) Chronic allograft nephropathy: an update. Kidney Int. 56, 783– 793 [DOI] [PubMed] [Google Scholar]
- 21.Laupacis A., Keown P., Pus N., Krueger H., Ferguson B., Wong C., Muirhead N. ( 1996) A study of the quality of life and cost-utility of renal transplantation. Kidney Int. 50, 235– 242 [DOI] [PubMed] [Google Scholar]
- 22.Nankivell B. J., Borrows R. J., Fung C. L., O'Connell P. J., Allen R. D., Chapman J. R., Jr ( 2003) The natural history of chronic allograft nephropathy. N. Engl. J. Med. 349, 2326– 2333 [DOI] [PubMed] [Google Scholar]
- 23.Beckingham I. J., Nicholson M. L., Bell P. R. ( 1994) Analysis of factors associated complications following renal transplant needle core biopsy. Br. J. Urol. 73, 13– 15 [DOI] [PubMed] [Google Scholar]
- 24.Rush D. N., Nickerson P., Jeffery J. R., McKenna R. M., Grimm P. C., Gough J. ( 1998) Protocol biopsies in renal transplantation: research tool or clinically useful? Curr. Opin. Nephrol. Hypertens. 7, 691– 694 [DOI] [PubMed] [Google Scholar]
- 25.Nankivell B. J., Chapman J. R. ( 2006) Chronic allograft nephropathy: current concepts and future directions. Transplantation 81, 643– 654 [DOI] [PubMed] [Google Scholar]
- 26.Solez K., Colvin R. B., Racusen L. C., Sis B., Halloran P. F., Birk P. E., Campbell P. M., Cascalho M., Collins A. B., Demetris A. J., Drachenberg C. B., Gibson I. W., Grimm P. C., Haas M., Lerut E., Liapis H., Mannon R. B., Marcus P. B., Mengel M., Mihatsch M. J., Nankivell B. J., Nickeleit V., Papadimitriou J. C., Platt J. L., Randhawa P., Roberts I., Salinas-Madriga L., Salomon D. R., Seron D., Sheaff M., Weening J. J. ( 2007) Banff '05 Meeting Report: differential diagnosis of chronic allograft injury and elimination of chronic allograft nephropathy (‘CAN′). Am. J. Transplant. 7, 518– 526 [DOI] [PubMed] [Google Scholar]
- 27.Racusen L. C., Solez K., Colvin R. B., Bonsib S. M., Castro M. C., Cavallo T., Croker B. P., Demetris A. J., Drachenberg C. B., Fogo A. B., Furness P., Gaber L. W., Gibson I. W., Glotz D., Goldberg J. C., Grande J., Halloran P. F., Hansen H. E., Hartley B., Hayry P. J., Hill C. M., Hoffman E. O., Hunsicker L. G., Lindblad A. S., Marcussen N., Mihatsch M. J., Nadasdy T., Nickerson P., Olsen T. S., Papadimitriou J. C., Randhawa P. S., Rayner D. C., Roberts I., Rose S., Rush D., Salinas-Madrigal L., Salomon D. R., Sund S., Taskinen E., Trpkov K., Yamaguchi Y. ( 1999) The Banff 97 working classification for renal allograft pathology. Kidney Int. 55, 713– 723 [DOI] [PubMed] [Google Scholar]
- 28.Eisen M. B., Spellman P. T., Brown P. O., Botstein D.Cluster analysis and display of genome-wide expression patterns. Proc. Natl. Acad. Sci. U. S. A. (1998)95,14863– 14868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zweig M. H., Campbell G. ( 1993) Receiver-operating characteristic (ROC) plots: a fundamental evaluation tool in clinical medicine. Clin. Chem. 39, 561– 577 [PubMed] [Google Scholar]
- 30.Wilm M. S., Mann M. ( 1994) Electrospray and Taylor-Cone theory, Dole's beam of macromolecules at last?. Int J. Mass Spectrom. Ion Process. 136, 167– 180 [Google Scholar]
- 31.Gangl E. T., Annan M. M., Spooner N., Vouros P. ( 2001) Reduction of signal suppression effects in ESI-MS using a nanosplitting device. Anal. Chem. 73, 5635– 5644 [DOI] [PubMed] [Google Scholar]
- 32.Quintana L. F., Solé-Gonzalez A., Kalko S. G., Bañon-Maneus E., Solé M., Diekmann F., Gutierrez-Dalmau A., Abian J., Campistol J. M. ( 2009) Urine proteomics to detect biomarkers for chronic allograft dysfunction. J. Am. Soc. Nephrol. 20, 428– 435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Maurer H. H. ( 2006) Hyphenated mass spectrometric techniques-indispensable tools in clinical and forensic toxicology and in doping control. J. Mass Spectrom. 41, 1399– 1413 [DOI] [PubMed] [Google Scholar]
- 34.Teppo A. M., Honkanen E., Finne P., Törnroth T., Grönhagen-Riska C. ( 2004) Increased urinary excretion of alpha1-microglobulin at 6 months after transplantation is associated with urinary excretion of transforming growth factor-beta1 and indicates poor long-term renal outcome. Transplantation 78, 719– 724 [DOI] [PubMed] [Google Scholar]
- 35.Mas V., Maluf D., Archer K., Yanek K., Mas L., King A., Gibney E., Massey D., Cotterell A., Fisher R., Posner M. ( 2007) Establishing the molecular pathways involved in chronic allograft nephropathy for testing new noninvasive diagnostic markers. Transplantation 83, 448– 457 [DOI] [PubMed] [Google Scholar]
- 36.Cosio F. G., Grande J. P., Wadei H., Larson T. S., Griffin M. D., Stegall M. D. ( 2005) Predicting subsequent decline in kidney allograft function from early surveillance biopsies. Am. J. Transplant. 5, 2464– 2472 [DOI] [PubMed] [Google Scholar]
- 37.Yilmaz S., Isik I., Afrouzian M., Monroy M., Sar A., Benediktsson H., McLaughlin K. ( 2007) Evaluating the accuracy of functional biomarkers for detecting histological changes in chronic allograft nephropathy. Transpl. Int. 20, 608– 615 [DOI] [PubMed] [Google Scholar]
- 38.Hoyer J. R., Seiler M. W. ( 1979) Pathophysiology of Tamm-Horsfall protein. Kidney Int. 16, 279– 289 [DOI] [PubMed] [Google Scholar]
- 39.Kokot F., Duława J. ( 2000) Tamm-Horsfall protein updated. Nephron 85, 97– 102 [DOI] [PubMed] [Google Scholar]
- 40.Howie A. J., Brewer D. B. ( 1983) Extra-tubular deposits of Tamm-Horsfall protein in renal allografts. J. Pathol. 139, 193– 206 [PubMed] [Google Scholar]
- 41.Torffvit O., Kamper A. L., Strandgaard S. ( 1997) Tamm-Horsfall protein in urine after uninephrectomy/transplantation in kidney donors and their recipients. Scand. J. Urol. Nephrol. 31, 555– 559 [DOI] [PubMed] [Google Scholar]
- 42.Kaden J., Groth J., May G., Liedvogel B. ( 1994) Urinary Tamm-Horsfall protein as a marker of renal transplant function. Urol. Res. 22, 131– 136 [DOI] [PubMed] [Google Scholar]
- 43.Beshensky A. M., Wesson J. A., Worcester E. M., Sorokina E. J., Snyder C. J., Kleinman J. G. ( 2001) Effects of urinary macromolecules on hydroxyapatite crystal formation. J. Am. Soc. Nephrol. 12, 2108– 2116 [DOI] [PubMed] [Google Scholar]
- 44.Iñigo P., Campistol J. M., Lario S., Piera C., Campos B., Bescós M., Oppenheimer F., Rivera F. ( 2001) Effects of losartan and amlodipine on intrarenal hemodynamics and TGF-beta(1) plasma levels in a crossover trial in renal transplant recipients. J. Am. Soc. Nephrol. 12, 822– 827 [DOI] [PubMed] [Google Scholar]
- 45.Madeddu P., Emanueli C., El-Dahr S. ( 2007) Mechanisms of disease: the tissue kallikrein-kinin system in hypertension and vascular remodeling. Nat. Clin. Pract. Nephrol. 3, 208– 221 [DOI] [PubMed] [Google Scholar]
- 46.Phipps J. A., Feener E. P. ( 2008) The kallikrein-kinin system in diabetic retinopathy: lessons for the kidney. Kidney Int. 73, 1114– 1119 [DOI] [PubMed] [Google Scholar]
- 47.Bakris G. L. ( 2008) Slowing nephropathy progression: focus on proteinuria reduction. Clin. J. Am. Soc. Nephrol. 3,Suppl. 1, S3– 10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Taal M. W., Brenner B. M. ( 2008) Renal risk scores: progress and prospects. Kidney Int. 73, 1216– 1219 [DOI] [PubMed] [Google Scholar]
- 49.National Kidney Foundation ( 2002) K/DOQI™ clinical practice guidelines for chronic kidney disease: evaluation, classification and stratification. Am. J. Kidney Dis. 392Suppl. 1, S1– 266 [PubMed] [Google Scholar]