Abstract
Thioredoxin 1 (Trx1) is a key redox modulator that is functionally conserved across a wide range of species, including plants, bacteria, and mammals. Using a conserved CXXC motif, Trx1 catalyzes the reduction of cysteine disulfides and S-nitrosothiols. In contrast to small molecular reductants such as glutathione and cysteine that can reduce a wide range of oxidized proteins, Trx1 reduces only selected proteins via specific protein-protein interaction. Trx1 has been shown to regulate numerous signal transduction pathways, and its dysfunctions have been implicated in several diseases, including cancer, inflammation, and neurodegenerative and cardiovascular diseases. Identification of Trx1 target proteins may help to identify novel signaling mechanisms that are important for Trx1 antistress responses. In this study, we performed an ICAT proteomics study for the identification of Trx1 target proteins from the hearts of a cardiac specific Trx1-overexpressing transgenic mouse model (Tg-Trx1). Trx1-reduced proteins were distinguished from Trx1-induced proteins by comparison of the ICAT results with those obtained using a parallel iTRAQ (isobaric tags for relative and absolute quantitation) protein expression analysis. We were able to identify 78 putative Trx1 reductive sites in 55 proteins. Interestingly we identified a few protein functional networks that had not been shown previously to be regulated by Trx1, including the creatine-phosphocreatine shuttle, the mitochondrial permeability transition pore complex, and the cardiac contractile apparatus. The results presented here suggest that in addition to a general antioxidant function, Trx1 may be involved in the coordination of a wide array of cellular functions for maintaining proper cardiac energy dynamics and facilitating muscle contraction.
Thioredoxins (Trxs)1 are a class of antioxidant proteins that mediate the reduction of specific disulfide bonds and S-nitrosothiols within oxidized proteins. Trx and the NADPH-dependent thioredoxin reductase (TrxR) form a protein reductive system that plays essential roles in the clearance of elevated reactive oxygen species, the repair of oxidatively modified proteins, and the restoration of cellular redox homeostasis. Two isoforms of Trx have been widely studied: Trx1 is mostly found in the cytosol and nucleus, whereas Trx2 is mitochondrion-specific. Each Trx isoform is coupled with its own TrxR systems. Trx1 has been shown to promote cell growth and proliferation and to inhibit apoptosis by modulating both caspase-dependent and -independent pathways. Trx1 has also been demonstrated to be a potent regulator of a wide variety of transcription factors and other gene expression regulators by preserving the reductive states of specific cysteines within transiently oxidized proteins such as nuclear factor κB (1), hypoxia inducible factor-1α (2), histone deacetylase 4 (3), and glucocorticoid receptor (4). A wide range of bioanalytical techniques has been applied to identify Trx1 targets, including two-dimensional electrophoresis-based differential thiol labeling (5, 6), Trx affinity chromatography (7, 8), and ICAT (9). The ICAT technique is of particular interest to redox analysis because of its capability to aid the identification of potential reduction sites within substrate proteins as well as the quantification of the extent of reduction (10). This technique was pioneered by Aebersold and co-workers (11) for the quantification of proteins by isotope tagging of cysteines. The advantage of this technique resides within its ability for the enrichment of cysteine-containing peptides for the reduction of sample complexity, resulting in more depth in stable isotope-based quantitative proteome coverage than conventional shotgun proteomics methods (11). Cohen and co-workers (12, 13) have tailored this technique to probe the redox status of protein cysteines and identified redox-sensitive cysteines in cardiac sarcoplasmic reticulum proteins including ion channels that may be important for modulating cardiac functions. Furthermore they have also elegantly mapped differential cysteine thiol redox sensitivities of p21ras GTPase toward peroxynitrite and oxidized glutathione (14). More recently, Svensson and co-workers (9) have used a modified ICAT method to discover over 100 in vitro Trx reduction targets in plants in which Trx-induced changes in protein disulfides were quantified. This report has provided the largest number of Trx-targeting plant disulfides to date, demonstrating the feasibility of using this technique for uncovering Trx targets in mammalian systems. In this study, we adopted a redox ICAT strategy to detect potential Trx1 reduction targets in rodent tissues.
Trx1 has been associated with a wide variety of diseases with oxidative imbalance, including cancer (15), human immunodeficiency virus infection (16), neurodegenerative diseases (17), and cardiovascular diseases (18). We were interested in understanding the roles of Trx1 in the protection of heart function in a rodent model of cardiac hypertrophy, the adaptive enlargement of the heart when under stress. However, prolonged pathological hypertrophy has been associated with metabolic disorder, inadequate ATP supply, contractile dysfunction, and gradual development into heart failure (19). We have shown previously that overexpression of Trx1 (in Tg-Trx1 animals) plays a central role in the activation of cardioprotective signal transduction pathways within hypertrophic hearts (3, 20). Mechanistically Trx1 may also exert its function through the regulation of gene expression, translation, and post-translational modifications. In a previous RNA microarray study (21), we reported that a wide range of genes is significantly altered in the hearts of Tg-Trx1 mice, including the up-regulation of genes involved in oxidative phosphorylation and the tricarboxylic acid cycle and several stress-related transcriptional factors. Given its known function as a protein reductant, a significant aspect of Trx1 function is expected to be exerted through the selective reduction of target proteins. For example, recently we have reported that histone deacetylase 4 is an important target of Trx1 reduction in heart (3). Trx1 facilitates the formation of a histone deacetylase 4-containing multiprotein complex resulting in histone deacetylase 4 nuclear translocation and thus regulates the expression of antihypertrophic genes. It is likely that in addition to regulating the function of individual proteins Trx1 may also exert its cardiac protective function by coordinately regulating a series of protein networks. To identify such protein networks, we conducted an ICAT-based proteomics study to identify proteins whose cysteine thiols became more reduced in the hearts of Tg-Trx1 animals compared with the control animals. The ICAT results were compared with data obtained from the iTRAQ-based protein expression analysis to reveal genuine Trx1 reduction protein targets as opposed to Trx1-induced proteins. We discovered that protein networks associated with energy production and utilization processes, such as glycolysis, the tricarboxylic acid cycle, β-oxidation, the mitochondrial permeability transition pore (MPTP) complex, and the contractile apparatus of the myofibrils, were affected by Trx1 overexpression, suggesting a role for Trx1 in maintaining heart energetics.
MATERIALS AND METHODS
Chemicals and Reagents
HPLC grade ACN and water were purchased from J. T. Baker Inc. Sequencing grade modified trypsin was from Promega (Madison, WI). Tris, α-cyano-4-hydroxycinnamic acid, catalase, protease inhibitor mixture (P8340), and other chemicals were purchased from Sigma-Aldrich unless stated otherwise. MS calibration standard peptides, Glu-fibrinopeptide, and human adrenocorticotropic hormone 18–39 were bought from AnaSpec (San Jose, CA). Cleavable ICAT reagents and iTRAQ reagents were obtained from Applied Biosystems (ABI, Foster City, CA). Recombinant human DJ-1 (ab51198, Abcam, Cambridge, MA), human Trx1 (T8690, Sigma), rat TrxR1 (American Diagnostica, Greenwich, CT), anti-Trx1 antibody (ab16835, Abcam), anti-adenine-nucleotide translocase 1 (ANT1; sc-9299, Santa Cruz Biotechnology, Santa Cruz, CA), and anti-biotin M antibody (MB-9100, Vector Laboratories, Burlingame, CA) were used in this study.
Transgenic Mouse Generation and Transverse Aortic Constriction (TAC) Surgery
Mice with cardiac specific overexpression of Trx1 (Tg-Trx1) were generated on an FVB background using the α-myosin heavy chain promoter as described previously (21, 22). Induction of cardiac hypertrophy was accomplished by surgical constriction of the transverse thoracic aorta and was performed on both the control and Tg-Trx1 mice as reported earlier (23). Briefly mice were anesthetized with pentobarbital sodium solution (60 mg/kg, intraperitoneal) and ventilated using a rodent ventilator. The left chest was entered through the second intercostal space, and the aorta was isolated. A Prolene suture was placed around the aorta between the innominate artery and the left carotid artery. A 27-gauge needle was tied onto the aorta and later removed. The chest was then closed in layers. The animals were kept warm, and the rectal temperature was maintained at 37 °C. After weaning from the ventilator, the mice were kept in an incubator with humidified oxygen and returned to cages after recovering from anesthesia. All protocols regarding the use of animals were in compliance with the regulations of the Institutional Animal Care and Use Committee at the University of Medicine and Dentistry of New Jersey.
Protein Extraction
Protein extracts were prepared from ∼100 mg of diced left ventricular tissues after three cold PBS washes and were homogenized with a tissue homogenizer (Omni International, Marietta, GA) in 500 μl of lysis buffer. For the ICAT proteomics study, 6 m urea, 2% CHAPS, 1% Triton X-100, and 30 mm Tris-HCl at pH 7.5 was used. For the iTRAQ proteomics study, 25 mm triethylammonium bicarbonate and 20 mm Na2CO3 at pH 8.0 was used. Protease inhibitors (10 μl/ml), 2.5 mm sodium pyrophosphate, 1 mm β-glycerophosphate, and 1 mm Na3VO4 were added to both lysis buffers. Protein concentrations were measured with the BCA protein assay (Pierce). Protein extractions and sample preparation were quickly carried out on ice and with minimum exposure to air. Protein extracts were immediately precipitated with cold acetone (−20 °C) to remove unwanted interferences such as metal ions and other small antioxidant molecules that may induce non-biological redox variations of proteins. Furthermore both control and Tg-Trx1 samples were processed in parallel so the impact of artificial oxidation, if any, would be comparable across all the samples.
ICAT Labeling and Multidimensional Chromatography
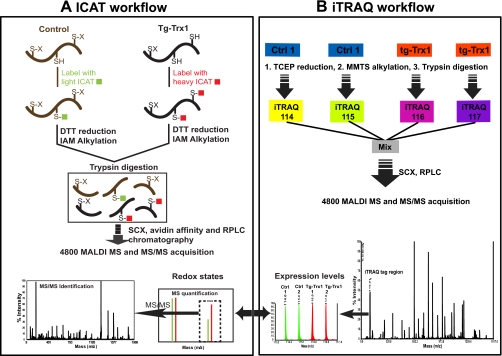
The ICAT labeling procedures used for this study have been described previously (10) and are illustrated in Fig. 1. In brief, 100 μg of each of the three independent control extracts was labeled with the light ICAT reagents, and the same amounts of Tg-Trx1 samples (three animals) were labeled with the heavy ICAT reagents at 37 °C for 2 h. No reduction and alkylation steps were performed prior to ICAT labeling to preserve the native protein thiol redox states. Excess ICAT reagents were removed with the addition of 50 mm DTT. Newly generated free thiols were alkylated with 50 mm iodoacetamide. The light (L) and heavy (H) ICAT labeled proteins were mixed and subjected to tryptic digestions at a 50:1 protein/enzyme ratio. Tryptic digested peptides were acidified and fractionated using strong cation exchange chromatography (SCX). SCX was run on an ABI Biocad Sprint System equipped with a PolySulfoethyl A column (200 mm × 4.6 mm; PolyLC Inc., Columbia, MD). The gradient profile of SCX consisted of 10 min of 100% mobile phase A (10 mm KH2PO4 and 20% ACN at pH 2.7) followed by 30 min of 0–25% mobile phase B (0.6 m KCl, 10 mm KH2PO4, and 20% ACN at pH 2.7) and 20 min of 25–100% B at 1 ml/min. Eluents were collected at a 2-min intervals and dried using a SpeedVac. Peptides in the SCX fractions were then enriched by biotin affinity chromatography using an avidin column provided in the cleavable ICAT kit according to the manufacturer's protocol. Removal of biotin moieties from the ICAT peptides was carried out with TFA cleavage at 37 °C for 2 h. After cleavage, peptides were dried and reconstituted in 5% ACN and 0.1% TFA (mobile phase A) for subsequent reversed phase LC (RPLC) separation using an LC Packings capillary HPLC system (Dionex, Sunnyvale, CA). Peptides were captured with an in-line trapping column (5 μm, 0.3 × 5 mm) and resolved on a PepMapTM C18 column (5 μm, 0.075 × 150 mm) with a 70-min linear gradient of 0–30% mobile phase B (95% ACN and 0.1% TFA) followed by 30 min of 30–90% mobile phase B at a flow rate of 400 nl/min. The RPLC eluents were mixed in line at a 1:2 ratio with a MALDI matrix (4 mg/ml α-cyano-4-hydroxycinnamic acid, 60% ACN, 0.1% TFA, 20 mm ammonium phosphate, and the internal mass calibrants) through a microtee and deposited onto the MALDI plates with a Probot (Dionex).
Fig. 1.
ICAT and iTRAQ work flow for Trx1 target discovery. IAM, iodoacetamide; Ctrl, control.
iTRAQ Labeling and Multidimensional Chromatography
Fifty micrograms of protein from each of the four samples (two controls and two Tg-Trx1 animals) was digested by trypsin and labeled with one of the four iTRAQ reagents according to the manufacturer's protocol. Peptides derived from the two control samples were labeled with iTRAQ tags 114 and 115, and the two Tg-Trx1 samples were labeled with iTRAQ tags 116 and 117 (see Fig. 1). The iTRAQ-labeled peptides were first combined; dried in a SpeedVac; then resuspended in 500 μl of Buffer A, which contained 10 mm KH2PO4 and 20% ACN, pH 2.7; and subjected to SCX fractionation using a PolySulfoethyl A column (4.6 × 200 mm, 5 μm, 300 Å; PolyLC Inc.) on an ABI Biocad Sprint System. A 60-min gradient program consisting of 10 min of mobile phase A and 40 min of linear gradient from 0 to 50% mobile phase B (10 mm KH2PO4, 20% ACN, and 600 mm KCl at pH 2.7) followed by a 10-min linear gradient from 50 to 100% B was performed at a constant flow rate of 1 ml/min. Thirty-five fractions were collected and desalted using PepCleanTM C18 spin columns (Pierce) according to manufacturer's protocols. RPLC separation of peptides within SCX fractions was identical to the procedures used for ICAT peptide analysis described above.
ICAT Analysis on Recombinant Human DJ-1
Recombinant human DJ-1 (0.5 μg/μl) was first oxidized with 200 μm H2O2 for 30 min in the dark. The reaction was quenched by addition of catalase (0.1 μg/ml). The oxidized DJ-1 solution (100 μl) was split in halves: one half was treated with the Trx1 reduction system, and the other half was treated with buffer only. The Trx1 reduction system was prepared as follows. Five micrograms of Trx1 was incubated in 2 μl of activation buffer (10 mm Tris and 2 mm EDTA, pH 7.5) for 15 min, and this solution was then mixed with 14 μl of reaction solution (10 mm Tris, 2 mm EDTA, 0.2 mm NADPH, and 1 μg of human Trx1 reductase, pH 7.5). Twenty-five micrograms of oxidized DJ-1 protein was added to the Trx1 reduction solution and then incubated at room temperature for 20 min. Trx1-reduced DJ-1 was then labeled with the ICAT H reagent, and the buffer-treated control was labeled with the ICAT L reagent. The remaining procedures followed the ICAT protocols described above except that the SCX step was carried out with an SCX cartridge provided in the ICAT labeling kit (ABI) according to the manufacturer's protocol.
MS and Database Search
ICAT
RPLC-purified peptides were analyzed on a 4800 MALDI-TOF/TOF mass spectrometer (ABI) operating in the reflectron mode. First 2,000 shots were accumulated for each MS spectrum with a mass range of m/z 800–3,800. Internal calibration standards were used for achieving a mass accuracy better than 50 ppm. Following MS analysis, the isotope ion cluster intensities of ICAT pair ions (heavy and light ions with a mass difference of 9.03 ± 0.03 Da) were extracted and quantified from the parent spectra with the GPS Explorer software (v3.5, ABI). Only ICAT pairs with at least 20% intensity differences and a signal-to-noise ratio over 50 were submitted for MS/MS analysis; 3,500 shots were accumulated for each MS/MS spectrum. The peak lists were generated with 4000 Series Explorer (v3.5, ABI). For peak detection, the signal-to-noise ratio threshold was set to 10, local noise window width was 250 m/z, and minimum peak width bin size was 2.9; resolution was set at 22,000 at m/z 2,400 for MS and 8,000 at m/z 2,000 for MS/MS. Peptide identification was performed with MASCOT (v1.9) integrated in the GPS Explorer software against a non-redundant Swiss-Prot database (release 54 containing 13,561 unique mouse sequences). The search parameters included one missed tryptic cleavage and 50-ppm MS and 0.3-Da MS/MS error tolerance. Variable modifications included ICAT L/H modifications, carbamidomethylation of cysteines, and methionine oxidation. Only peptides identified with confidence interval (C.I.) values at or above 95% were considered as confident identifications. Each MS/MS spectrum was subjected to manual inspection for the confirmation of positive identifications. False discovery rates were estimated to be less than 4% for all three experiments (calculation according to Peng et al. (24)). Typical ICAT quantification accuracy with this work flow on our instruments was evaluated by using BSA tryptic peptides of set mixing ratios. Analytical coefficient of variation values of less than 10% were observed (10). For the data presented in Table I, we decided to include only peptides with ICAT changes beyond 20% of the population median so that future biological validations are more likely to be successful.
Table I. Putative Trx1-mediated reduction protein network.
| Protein name (Swiss-Prot accession number) | Peptide sequencea | ICAT ratio(H/L) |
pvalueb | AvgcICATratio | ProteiniTRAQratiod | Reported Trxtargets (Refs.) | ||
|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | ||||||
| Metabolic enzymes | ||||||||
| Carbohydrate metabolism | ||||||||
| Acetyl-coenzyme A synthetase 2-like (Q99NB1) | 138ELLETTCR145 | 2.2 | 2.9 | 2.5 | 0.003 | 2.5 | 1.0 (12) | |
| Glyceraldehyde-3-phosphate dehydrogenase (P16858) | 233VPTPNVSVVDLTCR246 | 2.8 | 6.1 | 2.9 | 0.010 | 3.9 | 1.1 (18) | 29, 50–52 |
| l-Lactate dehydrogenase B chain (P16125) | 159VIGSGCNLDSAR170 | 1.5 | 2.3 | 1.5 | 0.027 | 1.8 | 1.0 (13) | |
| Phosphoglucomutase-1 (Q9D0F9) | 153TIEEYAICPDLK164 | 2.0 | 2.1 | 1.7 | 0.004 | 1.9 | 1.0 (15) | |
| Phosphoglycerate kinase 1 (P09411) | 49FCLDNGAK56 | 1.6 | 1.7 | 1.6 | 0.001 | 1.6 | 1.0 (13) | 51 |
| Pyruvate dehydrogenase E1 component subunit α (P35486) | 268FAAAYCR274 | 1.3 | 1.4 | 1.1 | 0.041 | 1.3 | 1.0 (16) | 28, 53 |
| 246LPC*IFICENNR226 | 2.0 | 2.2 | 1.9 | 0.002 | 2.0 | |||
| Pyruvate dehydrogenase E1 component subunit β (Q9D051) | 259EGIECEVINLR269 | 1.2 | 1.5 | 1.2 | 0.050 | 1.3 | 1.1 (14) | 28, 53 |
| Triose-phosphate isomerase (P17751) | 60IAVAAQNCYK69 | 1.7 | 1.8 | 1.8 | 0.001 | 1.8 | 1.1 (11) | 29, 51, 52 |
| Tricarboxylic acid cycle | ||||||||
| Citrate synthase (Q9CZU6) | 95GYSIPECQK103 | 1.5 | 1.7 | 1.6 | 0.003 | 1.6 | 1.0 (12) | |
| Isocitrate dehydrogenase (P54071) | 113CATITPDEAR122 | 1.6 | 2.3 | 1.6 | 0.017 | 1.8 | 1.1 (13) | 28, 52 |
| Malate dehydrogenase, mitochondrial (P08249) | 270EGVVECSFVQSK281 | 1.7 | 2.0 | 1.6 | 0.006 | 1.8 | 1.1 (20) | 28, 52 |
| 282ETECTYFSTPLLLGK296 | 1.8 | 2.5 | 1.9 | 0.008 | 2.1 | |||
| 92GCDVVVIPAGVPR104 | 1.4 | 1.5 | 1.4 | 0.002 | 1.4 | |||
| 79GYLGPEQLPDCLK91 | 1.7 | 2.9 | 1.9 | 0.018 | 2.2 | |||
| 204TIIPLISQCTPK215 | 1.9 | 2.5 | 1.9 | 0.006 | 2.1 | |||
| Fatty acid metabolism | ||||||||
| Carnitine O-palmitoyltransferase I (Q924X2) | 456SFTLISCK463 | 1.8 | 3.3 | 1.9 | 0.021 | 2.3 | 1.1 (18) | |
| Δ3,5-Δ2,4-Dienoyl-CoA isomerase (O35459) | 185YCTQDAFFQIK195 | 1.4 | 1.7 | 1.5 | 0.008 | 1.5 | 1.1 (11) | |
| Enoyl-CoA hydratase (Q8BH95) | 107TFQDCYSSK115 | 1.2 | 1.5 | 1.2 | 0.036 | 1.3 | 1.1 (11) | |
| Fatty acid-binding protein (P04117) | 114LVVECVMK121 | 2.3 | 3.4 | 2.5 | 0.005 | 2.7 | 1.1 (6) | |
| Long-chain-specific acyl-CoA dehydrogenase (P51174) | 346AFVDSCLQLHETK358 | 2.8 | 4.2 | 2.9 | 0.004 | 3.3 | 1.1 (20) | |
| 166CIGAIAMTEPGAGSDLQGVR185 | 1.5 | 1.5 | 1.5 | 0.000 | 1.5 | |||
| Long-chain-fatty-acid-CoA ligase 1 (P41216) | 100GIQVSNNGPCLGSR113 | 1.3 | 1.9 | 1.3 | 0.043 | 1.5 | 1.0 (26) | |
| 617GLQGSFEELCR627 | 1.7 | 2.3 | 1.8 | 0.008 | 1.9 | |||
| Very-long-chain-specific acyl-CoA dehydrogenase (P50544) | 231SSAIPSPCGK240 | 1.5 | 1.3 | 1.3 | 0.016 | 1.4 | 0.9 (23) | |
| Oxidative phosphorylation | ||||||||
| ATP synthase ϵ chain (P56382) | 15FSQICAK21 | 1.8 | 2.5 | 1.9 | 0.008 | 2.1 | 1.1 (3) | 28, 29 |
| Electron transfer flavoprotein-ubiquinone oxidoreductase (Q921G7) | 559FCPAGVYEFVPLEQGDGFR577 | 2.3 | 1.5 | 2.0 | 0.016 | 1.9 | 1.0 (25) | |
| NADH dehydrogenase α subcomplex subunit 10 (Q99LC3) | 59VITVDGNICSGK70 | 1.7 | 1.7 | 1.7 | 0.000 | 1.7 | 1.1 (13) | |
| NADH dehydrogenase protein 2 (Q91WD5) | 343IIEQCLNK350 | 2.1 | 2.4 | 1.7 | 0.008 | 2.1 | 1.1 (13) | |
| NADH dehydrogenase protein 6 (P52503) | 101TGTCGYCGLQFK112 | 2.0 | 1.7 | 1.6 | 0.006 | 1.8 | 1.0 (2) | |
| Succinate dehydrogenase flavoprotein subunit (Q8K2B3) | 76AAFGLSEAGFNTACLTK92 | 1.8 | 1.8 | 1.9 | 0.000 | 1.8 | 0.9 (17) | 28, 29 |
| 648TLNEADCATVPPAIR662 | 2.2 | 2.8 | 2.2 | 0.003 | 2.4 | |||
| 528VGSVLQEGCEK538 | 2.2 | 2.4 | 2.0 | 0.002 | 2.2 | |||
| Other metabolic proteins | ||||||||
| 2-Oxoglutarate dehydrogenase E1 component (Q60597) | 601SMTCPSTGLEEDVLFHIGK619 | 1.7 | 2.2 | 1.3 | 0.034 | 1.7 | 1.1 (32) | 53, 54 |
| Aldose reductase (P45376) | 42HIDCAQVYQNEK53 | 2.5 | 2.5 | 2.5 | 0.000 | 2.5 | 1.1 (11) | |
| Aspartate aminotransferase (P05202) | 186TCGFDFSGALEDISK200 | 3.6 | 3.0 | 3.6 | 0.001 | 3.4 | 1.0 (15) | 28 |
| 288VGAFTVVCK296 | 1.6 | 4.0 | 1.8 | 0.042 | 2.5 | |||
| Creatine kinase M-type (P07310) | 253FCVGLQK259 | 2.2 | 3.5 | 2.2 | 0.009 | 2.6 | 1.0 (19) | |
| Creatine kinase, sarcomeric mitochondrial (Q6P8J7) | 173GLSLPPACSR182 | 1.1 | 1.3 | 1.2 | 0.023 | 1.2 | 0.9 (19) | |
| 311LGYILTCPSNLGTGLR326 | 1.6 | 1.8 | 1.3 | 0.021 | 1.6 | |||
| 80MTPSGYTLDQCIQTGVDNPG HPFIK104 | 1.6 | 1.8 | 1.9 | 0.004 | 1.8 | |||
| Fumarylacetoacetate hydrolase (Q8R0F8) | 127SFTSSCPVSAFVPK140 | 1.7 | 1.6 | 1.4 | 0.008 | 1.6 | 1.1 (3) | |
| Malate dehydrogenase, cytoplasmic (P14152) | 150ENFSCLTR157 | 1.4 | 1.8 | 1.5 | 0.013 | 1.6 | 1.1 (16) | 28, 52, 55 |
| Nucleoside-diphosphate kinase B (Q01768) | 106GDFCIQVGR114 | 1.2 | 1.4 | 1.4 | 0.015 | 1.3 | 1.0 (11) | 28, 29 |
| Muscle and structural proteins | ||||||||
| Actin, α cardiac muscle 1 (P68033) | 1MCDDEETTALVCDNGSGLVKA GFAGDDAPR30 | 1.5 | 1.7 | 1.4 | 0.008 | 1.5 | 0.9 (16) | |
| Filamin-C (Q8VHX6) | 1090GAGTGGLGLTVEGPCEAK1107 | 1.8 | 1.7 | 1.4 | 0.011 | 1.6 | 1.0 (19) | |
| 617IECDDKGDGSC*DVR630 | 2.4 | 3.6 | 2.6 | 0.005 | 2.9 | |||
| Four and a half LIM domains protein 2 (O70433) | 132CQQPIGTK139 | 1.3 | 1.3 | 1.1 | 0.033 | 1.2 | 1.0 (6) | |
| 254CSLSLVGR261 | 1.3 | 1.2 | 1.6 | 0.034 | 1.4 | |||
| 84EEQLLC*TDCYSNEYSSK100 | 1.6 | 2.7 | 1.7 | 0.024 | 2.0 | |||
| 145ENQNFC*VPCYEK156 | 1.5 | 2.5 | 1.7 | 0.025 | 1.9 | |||
| 157QYALQCVQCK166 | 1.2 | 1.8 | 1.4 | 0.043 | 1.5 | |||
| Myosin light polypeptide 3 (P09542) | 181LMAGQEDSNGCINYEAFVK199 | 1.5 | 1.4 | 1.1 | 0.033 | 1.3 | 1.0 (13) | |
| Myosin-6 (Q02566) | 35TECFVPDDKEEYVK48 | 2.0 | 2.5 | 4.2 | 0.016 | 2.9 | 1.0 (83) | |
| Myosin-binding protein C (O70468) | 1251ATNLQGEAQCEC*R1263 | 1.4 | 1.8 | 1.8 | 0.013 | 1.7 | 0.8 (41) | |
| 562CEVSDENVR570 | 1.7 | 3.0 | 1.7 | 0.026 | 2.1 | |||
| 713LLCETEGR720 | 1.5 | 1.8 | 1.7 | 0.005 | 1.7 | |||
| Obscurin (A2AAJ9) | 928ADAGEYSCEAGGQK941 | 3.0 | 3.9 | 2.6 | 0.003 | 3.2 | 0.9 (10) | |
| 2972CQAGSAQSSAEVTVEAR2988 | 1.6 | 2.6 | 2.3 | 0.016 | 2.2 | |||
| Tropomyosin α-1 chain (P58771) | 190CAELEEELK198 | 1.5 | 2.8 | 1.5 | 0.043 | 1.9 | 0.9 (16) | |
| Transport and channel protein | ||||||||
| ADP/ATP translocase 1 (P48962) | 153EFNGLGDCLTK163 | 2.0 | 3.5 | 2.1 | 0.014 | 2.5 | 1.0 (14) | |
| Chloride intracellular channel protein 4 (Q9QYB1) | 228DEFTNTCPSDK238 | 1.7 | 1.7 | 2.3 | 0.011 | 1.9 | 1.0 (3) | |
| Sarcoplasmic/endoplasmic reticulum calcium ATPase 3 (Q64518) | 437VGEATETALTCLVEK451 | 1.6 | 1.6 | 1.5 | 0.001 | 1.6 | N/A | |
| Redox and chaperone proteins | ||||||||
| DnaJ homolog subfamily A member 2 (Q9QYJ0) | 303VIEPGCVR310 | 1.9 | 2.2 | 3.1 | 0.011 | 2.4 | 1.1 (3) | 56 |
| DnaJ homolog subfamily A member 3 (Q99M87) | 282GSIITNPCVVCR293 | 1.6 | 1.4 | 1.4 | 0.006 | 1.5 | 1.1 (4) | 56 |
| Peroxiredoxin-5 (P99029) | 83GVLFGVPGAFTPGCSK98 | 1.7 | 1.4 | 1.9 | 0.014 | 1.7 | 1.1 (8) | 9, 28, 52 |
| Peptidyl-prolyl cis-trans isomerase A (P17742) | 155KITISDCGQL164 | 1.3 | 1.3 | 1.5 | 0.011 | 1.4 | 1.1 (8) | 52, 57, 58 |
| Peptidyl-prolyl cis-trans isomerase (Q99KR7) | 196KIVITDCGQLS206 | 1.4 | 1.3 | 1.2 | 0.014 | 1.3 | 0.9 (4) | 52, 57, 58 |
| Protein DJ-1 (Q99LX0) | 49DVMICPDTSLEDAK62 | 1.4 | 1.4 | 1.9 | 0.023 | 1.6 | 0.9 (2) | |
| Others | ||||||||
| α2-HS-glycoprotein (P29699) | 323VGQPGAAGPVSPMCPGR339 | 1.7 | 1.2 | 1.4 | 0.036 | 1.4 | 1.3 (3) | |
| Apolipoprotein A-I-binding protein (Q8K4Z3) | 265YQLNLPSYPDTECVYR280 | 1.3 | 1.6 | 1.7 | 0.017 | 1.5 | 0.9 (2) | |
| PDZ and LIM domain protein 5 (Q8CI51) | 469FFAPECGR476 | 1.4 | 1.3 | 1.3 | 0.004 | 1.3 | 1.0 (4) | |
| Platelet glycoprotein 4 (Q08857) | 266FFSSDICR273 | 1.5 | 1.5 | 1.7 | 0.004 | 1.6 | 1.1 (6) | |
| 293FVLPANAFASPLQNPDNHCFC* TEK316 | 1.4 | 1.6 | 1.4 | 0.006 | 1.5 | |||
| Rab GDP dissociation inhibitor β (Q61598) | 194TDDYLDQPCC*ETINR208 | 1.3 | 1.4 | 1.7 | 0.020 | 1.5 | 1.1 (8) | |
| Serotransferrin (Q921I1) | 344NQQEGVCPEGSIDNSPVK361 | 1.7 | 1.6 | 1.8 | 0.002 | 1.7 | 1.0 (26) | |
a C represents an ICAT-labeled cysteine, C* indicates a carbamidomethylated cysteine; M indicates an oxidized methionine.
b One-tailed Student's t test was performed on the percentages of free thiol levels (converted from the ICAT ratios (R) with equations 1/(1 + R) × 100% for control animals and R/(1 + R) × 100% for Tg-Trx1 animals).
c Average.
d Numbers in parentheses indicate the number of unique peptides used for iTRAQ quantification calculation. N/A indicates that the protein was not found in the iTRAQ analysis.
iTRAQ
RPLC-separated iTRAQ-labeled peptides were analyzed on a 4800 MALDI-TOF/TOF MS instrument. MS spectra (m/z 800–3,500) were acquired in positive ion reflector mode, averaging 500 laser shots per spot. Data-dependent selection of the top 10 most intense ions in each MALDI spot were submitted for subsequent MS/MS analysis using 2-keV collision energy and 5 × 10−7-torr collision gas pressure with an accumulation of 2,000 shots per precursor mass. MS/MS ion peak lists were generated as described for the ICAT experiments. Protein database search by MASCOT (v1.9) was performed against the same Swiss-Prot database as that used for ICAT analysis. The following search parameters were used: trypsin with one missed cleavage; mass tolerance of 50 ppm for the precursor ions and 0.3 Da for the MS/MS fragment ions; fixed modifications including N-terminal iTRAQ labeling, iTRAQ-labeled lysines, and MMTS-modified cysteines; and variable modifications including methionine oxidation and iTRAQ-labeled tyrosines. Peptides identified with C.I. values no less than 95% were used for protein identification and quantitation. Protein expression ratios (Tg-Trx1/control) were calculated as described previously (25, 26). Briefly the iTRAQ reporter ion cluster areas were extracted using the GPS Explorer software (ABI), and only ion counts greater than 5,000 were used for quantification analysis. The individual reporter ion peak areas for each iTRAQ channel were normalized by the population median. For confident protein identification (average of at least two unique peptides per protein), the relative protein expression ratio distribution and the standard deviations between the animal groups were determined accordingly. In this study, two biological replicates of the iTRAQ-labeled sample were analyzed, and a Student's t test was performed to determine differentially expressed proteins. Proteins quantified with p values no greater than 0.05 and a protein expression ratio beyond 20% of the population median were considered as differentially expressed (26).
ANT1 Immunoprecipitation and Western Blotting
Heart proteins were extracted with a buffer containing 20 mm Tris-HCl, 150 mm NaCl, 1 mm EDTA, 1 mm EGTA, 1% Triton X-100, 1 mm sodium orthovanadate, 2.5 mm sodium pyrophosphate, and 1 mm β-glycerophosphate, pH 7.4, supplemented with protease inhibitor mixtures (10 μl/ml). Cell extracts (500 μg of proteins) were first treated with 200 μm H2O2 for 30 min followed by the addition of 0.1 μg/ml catalase to remove the excess H2O2. Different protein reduction conditions were applied separately to the oxidized proteins for 30 min at room temperature: 1) reaction buffer (2.0 mm EDTA and 10 mm Tris-HCl, pH 7.5) only; 2) 5 μg of Trx1 in the reaction buffer; 3) 5 μg of Trx1, 1 μg of TrxR, and 0.2 mm NADPH in the reaction buffer; and 4) 5 mm tris(2-carboxyethyl)phosphine (TCEP). The treated proteins were precipitated with cold acetone (−20 °C), and the pellets were washed twice with cold acetone. The pellets were resuspended in radioimmune precipitation assay buffer, and the protein cysteine thiols were alkylated with the addition of 0.2 mm (N-(6-(biotinamido)hexyl)-3′-(2′-pyridyldithio)-propionamide for 1 h. The biotinylated proteins were immunoprecipitated with an anti-ANT1 antibody. For Western blot analysis, the proteins were separated by 11% SDS-PAGE and transferred onto a nitrocellulose membrane. The membrane was probed with either an anti-biotin M antibody or an anti-ANT1 antibody, respectively. Horseradish peroxidase-conjugated secondary antibody (Santa Cruz Biotechnology) was used for detection and visualization by chemiluminescence.
Statistical Analyses
ICAT data are expressed as ICAT pair ratios (H/L: Tg-Trx1/control) from three independent experiments (Table I). Statistical significance was determined by performing a one-tailed Student's t test with Microsoft Excel, and a p value ≤0.05 was considered significant.
Trx1 Reductive Target Sequence Motif Search
An iterative statistical program (27), motif-x, was used to extract potential Trx1 reductive motifs among the identified target sequences. Cysteine was assigned as the centralized amino acid, and foreground sequences were extended to 15 amino acids on each side of the cysteine. The significance threshold was set at 0.000001.
RESULTS
Identification of Putative Trx1 Target Cysteines in Cardiac Proteins
Hypertrophy and increased oxidative stress in hearts were introduced with TAC surgery in both control and Tg-Trx1 mice. Western analysis confirmed the significant overexpression of Trx1 in transgenic animals (supplemental Fig. 1). TAC induced a significant increase in left ventricle weight/body weight (4.0 ± 0.2 mg/g), an index of cardiac hypertrophy in control mice, whereas the increases in left ventricle weight/body weight were significantly attenuated in Tg-Trx1 mice (3.5 ± 0.1 mg/g; p < 0.01), consistent with our previous observation (22). Both Tg-Trx1 and control animals, when subjected to TAC surgery, incurred elevated oxidation of many proteins (21, 22). However, the elevated Trx1 levels in the transgenic mice facilitate the reduction of specific cysteines in target proteins. We used a forward ICAT labeling strategy as described previously (10) to identify redox-sensitive cysteines that are putative targets of Trx1 in TAC-stressed mouse hearts (Fig. 1). ICAT H reagent was used to label Tg-Trx1 samples, whereas the ICAT L reagent was used to label the control samples. In such a scheme, Trx1-induced protein thiol reduction would be manifested as H/L ratios larger than 1.0. Within three independent ICAT experiments, we observed ∼2,000 ICAT ion pairs with a mean ICAT ratio of 1.15 (ICAT ratios presented in Table I; ratios were normalized by the population median of the individual experiment); ∼700 precursors were selected for MS/MS identifications. We identified 152, 166, and 195 unique peptides with C.I. value ≥95% in the three experiments (supplemental Table 1). According to our previous analysis of the ICAT redox proteomics work flow on our MS instrument (10), the typical quantification coefficient of variation is less than 10%. Therefore, a more stringent 20% ICAT quantification ratio was used as a cutoff value for determining significant reduction of protein thiol levels between control and Tg-Trx1 animals. Only peptides fulfilled the following criteria were included in Table I: (i) C.I. value ≥95%, (ii) identification in all three ICAT experiments, (iii) p value ≤0.05, and (iv) ICAT ratio changes beyond 20% of the population median.
We found that 78 cysteines within 55 proteins were significantly reduced by Trx1 overexpression in all three experiments (Table I; categorized by gene ontology functional groups), and an additional 70 peptides were found in at least two experiments (supplemental Table 1). Potential Trx1 target proteins demonstrated various degrees of sensitivity to Trx1 reduction, ranging from a 20 to ∼300% increase in free cysteine thiol levels. The majority of the Trx1-responsive proteins included structural proteins, ion channels, stress response proteins, and metabolic enzymes in the glycolysis pathway, the tricarboxylic acid cycle, oxidative phosphorylation, and β-oxidation of fatty acids.
We previously showed that TAC treatment resulted in increased oxidative stress in heart, likely resulting in elevated protein oxidation (22). A large number of proteins were found to contain cysteines highly sensitive to Trx1 reduction, including glyceraldehyde-3-phosphate dehydrogenase (3.8), pyruvate dehydrogenase E1 component subunit α (2.0), long-chain-specific acyl-CoA dehydrogenase (3.3), aldose reductase (2.5), ADP/ATP translocase 1 (2.5), and aspartate aminotransferase (3.4) (numbers in parentheses indicate -fold changes of free thiol levels, Tg-Trx1/control). Many proteins possessed more than one cysteine that was sensitive to Trx1 reduction but to different extents. For example, four different creatine kinase (Q6P8J7) peptides were found to have five cysteines: Cys63, Cys67, Cys90, Cys180, and Cys317. Cys63 and Cys67 were non-responsive to Trx1 as no significant differences were found between the light and heavy ICAT-labeled peptides (supplemental Table 1). Cys90, Cys180, and Cys317 responded positively to the Trx1-mediated reduction leading to an increase in heavy ICAT-labeled peptide ions by 20, 60, and 70%, respectively (Table I). Protein levels of creatine kinase were comparable between the control and Tg-Trx1 animals as evidenced by an iTRAQ ratio of 0.9.
To understand whether the observed increase of free thiol content was exclusively due to Trx1-mediated protein reduction or also due to alterations in protein expression, we compared the results of the ICAT analysis with those of the iTRAQ analysis of similar samples. Because our ICAT work flow did not contain the initial reduction step prior to ICAT labeling, increases in H/L ratios could be due to increases in protein amounts and/or their free thiol levels. The rationale for the comparison of ICAT and iTRAQ results was that if the protein levels were constant according to the iTRAQ method different ICAT ratios should be attributed to reversible thiol modifications. In the iTRAQ analysis, 10 proteins (including Trx1 with iTRAQ ratio of 1.5; p < 0.01) of 769 unique proteins (∼2%) were found to be significantly changed in the Tg-Trx1 animals. Changed proteins have been validated by Western blotting analysis with more animal numbers.2 Some proteins were identified with average iTRAQ ratios beyond 20% of the population median but were not deemed significant (p values >0.05), for example myosin-binding protein C. Myosin-binding protein C was found with an average iTRAQ ratio of 0.8 (p value of 0.26 from 41 peptides) and with average ICAT peptide ratios for three distinct peptides of 1.7, 2.1, and 1.7, respectively (Table I). It is likely that these peptides would have been detected as more reduced in Tg-Trx1 animals if this protein were not also down-regulated.
Validation of ANT1 as a Direct Trx1 Reduction Target
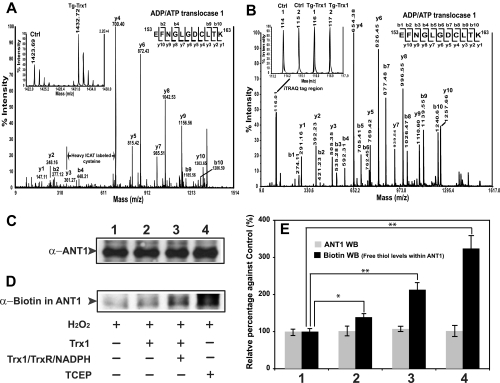
Following the ICAT proteomics study, we confirmed that one of the proteins, ANT1, contained an oxidized cysteine that can be directly reduced by Trx1 (Fig. 2). First the fragmentations of a heavy ICAT-labeled peptide (m/z 1432.72) generated an MS/MS ion profile that matched to ANT1 153EFNGLGDCLTK163 sequence with a rather complete y ion series (Fig. 2A). The MS quantification of the ICAT pair revealed a more than 2-fold increase in free thiols in this peptide when comparing Tg-Trx1 animals with wild type animals (Fig. 2A, inset). In a parallel iTRAQ experiment, the same peptide was identified with almost complete b and y ion series (Fig. 2B). iTRAQ reporter ions showed that comparable levels of ANT1 were present in both control and Tg-Trx1 samples (Fig. 2B, inset). By evaluating the ICAT and iTRAQ proteomics results, we concluded that overexpression of Trx1 was capable of reducing the oxidized Cys160 of ANT1. Next we sought to validate that ANT1 is a genuine target of Trx1 reduction using an alternative approach. Heart proteins were first subjected to H2O2 oxidation followed by treatment with different reductants including Trx1, Trx1/TrxR/NADPH, and TCEP. The free protein thiols in the treated proteins were labeled with (N-(6-(biotinamido)hexyl)-3′-(2′-pyridyldithio)-propionamide, immunoprecipitated with an anti-ANT1 antibody, and then blotted with different antibodies. We observed that ANT1 protein levels were constant across all four samples (Fig. 2, C and E). Western blot analysis with biotin showed that only trace levels of biotinylated ANT1 (a marker for free thiol levels) was detected in H2O2-treated samples, indicating that most of the ANT1 cysteines were oxidized (Fig. 2D, lane 1). Trx1 treatment led to a ∼30% increase in free thiols (Fig. 2D, lane 2). Treatment of the cell lysate with the Trx1/TrxR/NADPH reduction system increased protein thiols by ∼130% (Fig. 2D, lane 3), suggesting that Trx1 regeneration was important to its reduction of ANT1. Lastly a strong reductant, TCEP, restored ∼210% of the oxidized cysteines to the reduced states (Fig. 2D, lane 4). This analysis suggested that ANT1 contained redox-sensitive protein thiols that were sensitive to 200 μm H2O2 oxidation, and these oxidized cysteines could in turn be specifically reduced by the Trx1/TrxR/NADPH system (Fig. 2E). Furthermore TCEP nonspecifically reduced most of the oxidized cysteines, including those that were non-responsive to Trx1.
Fig. 2.
Trx1 reduction of ANT1. A, ICAT comparison of ANT1 peptide 153–163 between control and Tg-Trx1 animals. Heavy ICAT-labeled ion (m/z 1432.72) was selected for MS/MS fragmentation, and the fragment ion profile matched to 153EFNGLGDCLTK163 with a complete y ion series. A mass difference of 339.1 Da between y3 and y4 corresponded to a heavy ICAT-labeled Cys160. The MS quantification of the ICAT ion pair revealed that the peptide derived from Tg-Trx1 mouse was more reduced than that from the control animal (∼2.5-fold; see inset). B, in a parallel iTRAQ experiment, the same ANT1 peptide was identified with almost complete b and y ion series. Evaluation of the iTRAQ quantification region (see inset) confirmed that equivalent amounts of this ANT1 peptide were present in all four samples. Notably within the iTRAQ procedure, TCEP reduction and MMTS alkylation steps were performed; thus the mass difference between y3 and y4 of 149.0 Da corresponded to an MMTS-modified cysteine. By comparing both quantitative proteomics analyses, we concluded that Cys160 of ANT1 may be a Trx1 reduction target, and the protein levels were not affected by the overexpression of Trx1. C and D, oxidized ANT1 was specifically reduced by the Trx1/TrxR/NADPH system. Five hundred micrograms of mouse heart proteins was first treated with 200 μm H2O2 for 30 min in the dark. Excess H2O2 was removed with catalase (0.1 μg/ml). The oxidized proteins were subjected to different reduction treatments: 1) buffer, 2) 5 μg of Trx1, 3) 5 μg of Trx1 and TrxR/NADPH, and 4) 2 μl of 50 mm TCEP. Following the reduction treatments, proteins were precipitated with cold acetone (−20 °C) and then labeled with the biotinylation reagent for 1 h at room temperature. An anti-ANT1 antibody was used to precipitate ANT1 proteins. Five micrograms of the precipitated proteins was loaded onto an 11% SDS-PAGE gel, and the separated proteins were blotted and probed with either an anti-ANT1 antibody (C) or an anti-biotin antibody (D), respectively. ANT1 levels were constant across all treated samples (C). A trace amount of biotinylated ANT1 was detected in H2O2-treated samples (D, lane 1). Trx1 treatment increased cysteine thiols by 30% (D, lane 2). Application of the Trx1/TrxR/NADPH reduction system was able to restore over 100% more protein thiols (D, lane 3). Lastly a strong indiscriminate reductant, TCEP, reversed over 200% more oxidized cysteines to the reduced states (D, lane 4). E, statistical analysis of the blots suggested that ANT1 contained redox-sensitive cysteines thiols that were oxidized by H2O2, and selected cysteines (likely Cys160) could in turn be reduced by the Trx1/TrxR/NADPH system. Furthermore TCEP nonspecifically reduced most of the oxidized cysteines (ANT1 has four cysteines: Cys57, Cys129, Cys160, and Cys257; Error bars represent standard deviations. *, p < 0.01; **, p < 0.005; n = 3). WB, Western blot; Ctrl, control.
Trx1 System Reversibly Reduced Cys53 in DJ-1
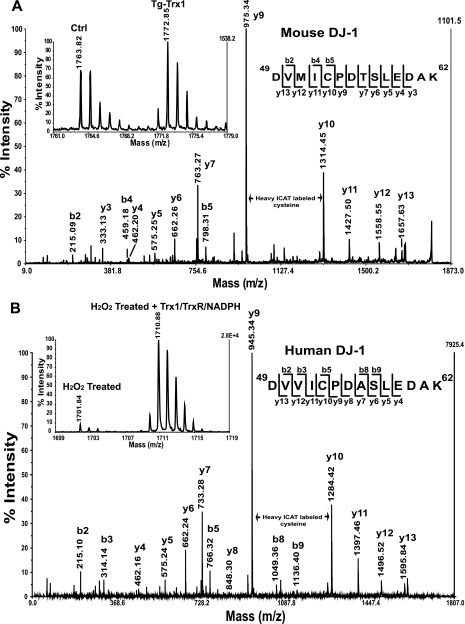
In Tg-Trx1 mice, Trx1 restored ∼60% of the free thiols in the DJ-1 peptide 49DVMICPDTSLEDAK62 compared with the control mouse heart (Fig. 3A, inset). An almost continuous series of y ions were observed in the MS/MS spectra (Fig. 3A). A mass difference of 339.1 Da between the y9 and y10 ions corresponded to the addition of a cleaved heavy ICAT label to Cys53. To confirm that the reduction of this specific cysteine was Trx1-dependent, we simulated oxidative stress by treating recombinant human DJ-1 protein with H2O2 (200 μm) for 30 min followed by removal of excess H2O2 with catalase (at a final concentration of 0.1 μg/ml). The oxidized DJ-1 samples were then incubated with either the activated Trx1/TrxR/NADPH system or blank reaction buffer for 20 min at room temperature and subjected to ICAT labeling. We observed only trace levels of available protein thiols labeled with the light ICAT reagent; however, the Trx1/TrxR/NADPH system reduced a significant amount of the oxidized Cys53 in peptide 49DVVICPDASLEDAK62 (Fig. 3B, inset). Although the tryptic peptide sequences are slightly different between mouse and human DJ-1, Cys53 is highly conserved in both species. In conclusion, both in vivo and in vitro experiments confirmed that Cys53 is a redox-sensitive cysteine, and its redox state can be regulated by the Trx1 reduction system.
Fig. 3.
Trx1 reduction of protein DJ-1. A, for Tg-Trx1 mice, 50% more free thiols were observed in DJ-1 peptide 49DVMICPDTSLEDAK62 compared with the control samples (see inset). Nearly complete series of y ions were observed in the MS/MS spectrum; a mass difference of 339.1 Da between y9 and y10 corresponded to the addition of a heavy ICAT label to Cys53. B, to confirm the direct reduction of Cys53 by Trx1, we evaluated the ability of Trx1 to reduce H2O2-oxidized human protein DJ-1 in an in vitro experiment. Following H2O2 oxidation of human DJ-1 for 30 min, catalase (0.1 μg/ml) was used to quench the remaining H2O2. The treated sample was divided into equal halves. One half was incubated with the Trx1/TrxR/NADPH system for 20 min at room temperature, and the other half was incubated with buffer. The ICAT labeling procedure was used to quantify the cysteine thiol levels. With H2O2 treatment, we observed only trace levels of protein thiols labeled with the light ICAT reagent; however, the Trx1 system was able to reduce a significant amount of oxidized Cys53 (see inset). Both in vivo and in vitro experiments confirmed that Cys53 within DJ-1 is redox-sensitive, and it can be reduced by Trx1. Ctrl, control.
Identification of Potential Trx1-interactive Sequence Motifs
We also examined all the putative Trx1 target sequences found in this study for the possible presence of consensus sequence motifs. Interestingly we discovered two potential CXXC motifs (where either one or both cysteines could be reduced by Trx1) resembling the catalytic site of Trx1 (supplemental Fig. 2). Interestingly three peptides from this study (NADH dehydrogenase protein 6 (101TGTCGYCGLQFK112 where C represents an ICAT-labeled cysteine), DnajA3 (282GSIITNPCVVCR293), and four and a half LIM domains protein 2 (157QYALQCVQCK166)) contained a CXXC motif where both cysteines were more reduced in Tg-Trx1 animals (Table I). Four and a half LIM domains protein 2 also had two additional peptides (84EEQLLC*TDCYSNEYSSK100 and 145ENQNFC*VPCYEK156 where C* indicates a carbamidomethylated cysteine) matched to the CXXC motif. However, because of the limited sample size reported here, future studies including proteomics and molecular modeling approaches may be needed to validate the predictive value of such motifs.
DISCUSSION
Heart Oxidative Stress and Trx1 Function
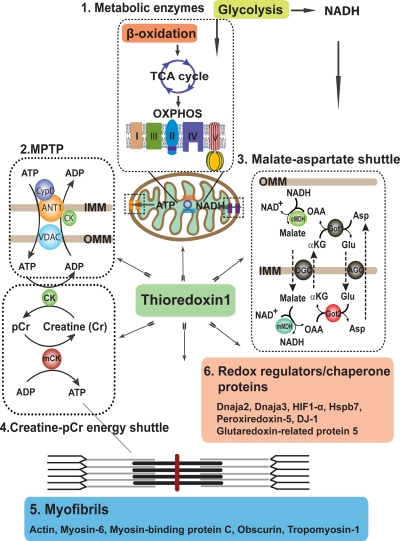
Understanding the components of cellular antistress pathways may provide clues on how to develop specific therapeutic agents for stress-related heart diseases. Trx1 and the affiliated redox regulatory system are important mechanism by which cardiac myocytes counter the detrimental effects of oxidative stress. Our earlier studies suggested that Trx1 is protective in cardiac myocytes in animal models of heart diseases by means of the reduction of specific protein targets (3). In addition, we also observed increased activities among several mitochondrial enzymes and elevated ATP content in Tg-Trx1 hearts (21). In this study, through a comparative ICAT proteomics study of Tg-Trx1 and control animals, we identified 55 potential Trx1 reduction targets in the heart, including several previously reported Trx1 targets (Table I), which validated our approach. In addition, we discovered novel Trx1 reduction protein networks for maintaining energy conductance from mitochondria to the cardiac contractile apparatus, which will be discussed below (Fig. 4).
Fig. 4.
Trx1-mediated energy production and utilization networks of cardiac proteins. Trx1 reduces metabolic proteins involved in glycolysis, β-oxidation, the tricarboxylic acid cycle, and oxidative phosphorylation, ensuring sustainable production of ATP for muscle contraction. Trx1 also regulates the MPTP, creatine-phosphocreatine energy shuttle, and malate-aspartate shuttle for both metabolite and ATP conductance among coordinated protein networks, ensuring the delivery of high energy phosphates to heart muscle. A large number of myofibril components are reduced by Trx1 supporting the contraction ability of the cardiac muscle. Trx1 also modulates a wide variety of redox regulator/chaperone proteins such as Dnaja2, Dnaja3, Hspb7, Prx5, DJ-1, and glutaredoxin-related protein 5, coordinating cellular antistress response. IMM, inner mitochondrial membrane; OMM, outer mitochondrial membrane; OAA, oxaloacetate; αKG, α-ketoglutarate; Glu, glutamate; Asp, aspartate; cMDH, cytosolic malate dehydrogenase; mMDH, mitochondrial malate dehydrogenase; Got1, cytosolic aspartate aminotransferase; Got2, mitochondrial aspartate aminotransferase; TCA, tricarboxylic acid cycle; CK, creatine kinase; mCK, muscular creatine kinase; pCr, phosphocreatine; CypD, cyclophilin-D; VDAC, voltage-dependent anion channel; OXPHOS, oxidative phosphorylation; AGC, aspartate-glutamate carrier; OGC, oxoglutarate carrier. Proteins colored in gray were not found in our ICAT results; they are depicted to enhance the clarity of the diagram.
Comparison of ICAT and iTRAQ Analysis Results
The importance of Trx1 has spurred enormous interest in identifying its targets, an endeavor that has been aided by the developments of innovative techniques such as fluorescent diagonal electrophoresis (5) and affinity chromatography (28, 29). However, these approaches have not been able to provide quantitative information regarding the degree or the specific sites of reduction; both are critical for relating protein redox status and the associated biological functions. ICAT is a cysteine-specific isotope labeling method for peptide quantification at the MS levels, whereas iTRAQ reagents label primary amines for isobaric peptide quantification at the MS/MS levels. Both methods have been widely used for large scale quantification of proteins (9, 10, 13, 25). To capture cysteine thiol redox status, the reduction/alkylation steps were omitted for this ICAT analysis. This strategy allowed the detection of the increase of specific protein thiols in Tg-Trx1 animals. The increase of the ICAT ratios could be due to Trx1-mediated gene/protein expression differences, protein reduction, or a combination of these elements. To differentiate these possibilities, we compared the ICAT proteomics results with those obtained from the iTRAQ expression analysis in which the TCEP reduction/MMTS alkylation steps were done to determine the relative protein expression levels of the same samples. Interestingly the expression levels of the 55 significant Trx1 reductive targets (Table I) were not significantly changed in the iTRAQ study, indicating that the increase in cysteine thiol levels in Trx1 mouse hearts was largely due to Trx1-mediated reduction of redox-sensitive thiols.
There were also indications that some proteins (peptides) are more oxidized in Tg-Trx1 animals (catalase (ICAT ratio of 0.48) and ubiquinol-cytochrome-c reductase complex core protein 1 (ICAT ratio of 0.65); supplemental Table 1). However, these proteins did not satisfy our stringent criteria for target selection (e.g. observed in all three independent experiments). The number of oxidized proteins in Tg-Trx1 animals was relatively small, and they are included in supplemental Table 1. We hypothesize that the reason why some proteins were oxidized more significantly in Tg-Trx1 animals may be due to Trx1-induced activation of oxidoreductases (Ero1α (30), for example) that can in turn oxidize downstream targets.
Validation of Trx1 Targets
Many of the proteins revealed from this study have been reported previously to be direct Trx1 reduction targets (Table I), validating the reliability of the current approach. However, Trx1 may also contribute to the reduction of oxidized cysteines indirectly, e.g. via the peroxiredoxins (31) or glutathione system (32). We conducted additional experiments to validate selected proteins as direct Trx1 reduction targets. One was ANT1, a key component of the mitochondrial permeability transition pore complex, which is a multiprotein complex containing ANT1, voltage-dependent anion channel, cyclophilin-D, creatine kinase, and Bcl/Bax family proteins (33). The MPTP exerts its function as a mitochondrial intermembrane conduit for ATP/ADP exchange to supply the high energy phosphates for muscle contraction. Dysregulation of MPTP is characterized by the depolarization of the mitochondrial inner membrane, disruption of the cross-membrane potential, mitochondrial swelling, and eventual release of mitochondrial apoptogens, including cytochrome c (34, 35). ANT1 has been reported to be vulnerable to oxidative modifications leading to aberrant MPTP activity that results in reduced ATP production and utilization and increased apoptosis (35). Mice deficient in ANT1 exhibit severe cardiomyopathy (36). There are four cysteines within mouse ANT1, several of which have been shown to be prone to oxidation (37, 38). Oxidation of Cys159 in human ANT1 (corresponding to the conserved Cys160 in mouse) within the binding site of adenine nucleotides has been shown to be detrimental to pore function and nucleotide transport (38). In this study, although ANT1 protein levels were comparable for all animals, Cys160 was found to be more reduced in Tg-Trx1 than wild type hearts (Tg-Trx1/control ratio of 2.5; Table I and Fig. 2, A and B). We also found that ANT1 was oxidized by H2O2 (Fig. 2D, lane 1), and plain Trx1 was able to partially reverse the oxidation of free thiols (Fig. 2, C and D, lane 2). The reduction effect was more pronounced when TrxR and NADPH were included for Trx1 regeneration (Fig. 2, C and D, lane 3), suggesting that ANT1 could be a direct target of Trx1 reduction. More importantly, Cys160 was likely to be a reactive cysteine within ANT1 that was reduced by Trx1 given that TCEP was more thorough at potentially reducing all oxidized cysteine thiols nonspecifically in ANT1 than was Trx1 (Fig. 2, C and D, lane 4). In this study, we also observed that other MPTP components were modulated by Trx1, including creatine kinase, voltage-dependent anion channel 1, and cyclophilin-D (supplemental Table 1), suggesting MPTP may be directly regulated by Trx1 in a redox-dependent manner to sustain the supply of ATP for the heart under TAC-induced oxidative stress.
In addition to validating Trx1 targets at the protein levels, we also performed experiments to confirm that such regulation is site-specific. DJ-1 is a redox-sensitive protein associated with the development of Parkinson disease (39). Although it is expressed in the heart, its cardiac function is largely unexplored. DJ-1 deficiency in neuronal cells has been shown to sensitize cells to oxidative stress and may result in cell death (40); overexpression of DJ-1 is cytoprotective (41). There are four cysteines in mouse DJ-1, several of which have been shown to be very sensitive to oxidative stress (39, 42). No reports have shown that Trx1 is able to repair DJ-1 oxidative damages. Our in vitro study showed that Cys53, a highly conserved residue among mammalian species, may be a Trx1 reductive target (Table I and Fig. 3A); this is significant given previous reports of cysteinylation on Cys53 upon oxidative stress (39). In a follow-up experiment, we were able to show that Cys53 in human DJ-1 is sensitive to H2O2 oxidation (Fig. 3B), and its oxidation can be almost completely reduced by direct interaction with Trx1 (Fig. 3B).
Trx1 Targets within Energy Pathways
Not much is known of the impact of Trx1 on metabolic protein functions in mammalian cells, especially in heart tissues, where the demand for metabolic fuel is high. Typically when diseased hearts become stressed, they become adaptively hypertrophied, exhibiting significant metabolic dysfunction (43). For example, the disruption of β-oxidation of fatty acids has been reported in hypertrophied hearts, resulting in both the reduction of acetyl-CoA supply for the tricarboxylic acid cycle and accumulation of toxic lipid derivatives (44). Deficiency of very-long-chain-acyl-CoA dehydrogenase is known to induce heart hypertrophy (45). Here we found that both long-chain- and very-long-chain-acyl-CoA dehydrogenases are potential Trx1 reduction targets (Table I), suggesting that within Tg-Trx1 animals the lipid metabolic profiles may be protected from oxidative stress to maintain proper heart functions (Fig. 4).
Creatine kinase plays an important role in shuttling ATP from the mitochondria to myofibrils, supporting robust heart muscle contraction. There are two forms of creatine kinases in cardiomyocytes, cytosolic creatine kinase and muscular creatine kinase, which work in tandem to transfer the high energy phosphates from ATP to the contractile apparatus (Fig. 4). Our ICAT analysis identified that both cytosolic creatine kinase and muscular creatine kinase are potential substrates for Trx1 reduction. Cys317 in the 311LGYILTCPSNLGTGLR326 peptide within the catalytic site of cytosolic creatine kinase has been shown to be sensitive to oxidation, resulting in the inhibition of its enzymatic activity (12, 46). About 60% more free thiols in this peptide were found in Tg-Trx1, suggesting that Trx1 may help to maintain the creatine kinase activity, facilitating energy conductance from mitochondria to the heart contractile machinery.
Oxidation of cardiac contractile proteins has been shown to impair the plasticity and contractility of muscles and could be reduced by antioxidants in vitro (47, 48). We observed that a wide spectrum of sarcomeric proteins were reduced by Trx1 overexpression (Table I), suggesting Trx1 may play a direct role in maintaining cardiac contractile apparatus functionality and energy utilization (Fig. 4).
Limitation of the Current Redox ICAT Method
Although the redox ICAT method is effective for finding potential Trx1 target proteins, many well characterized Trx1 targets such as peroxiredoxin 1 (Prx1), Prx2, and ribonucleotide reductase were missing from this study. There are several potential limitations of the current method for redox proteomics studies. 1) The peptides containing the Trx1-reactive cysteines may not be suitable for this analytical work flow. For example, the Prx1 tryptic peptide 38YVVFFFYPLDFTFVCPTEIIAFSDR62 (an underlined C represents a known Trx1 reduction target cysteine) contains a known Trx1 reductive site at Cys52 (49). This peptide is highly hydrophobic and may not be eluted from the RPLC column. Furthermore its mass of m/z 3376.5 may be too large to be fragmented efficiently by MALDI tandem MS. A similar issue may also be applicable to the Prx2 tryptic peptide 37YVVLFFYPLDFTFVCPTEIIAFSDHAEDFR66. On the other hand, the Trx1 reductive site in peroxiredoxin 5, 83GVLFGVPGAFTPGCSK98, was found in this study because this tryptic peptide was amenable to the LC/MS/MS analysis. 2) Tryptic peptides containing the Trx1 reductive sites from proteins including ribonucleotide reductase are amenable for LC/MS/MS analysis but may be relatively low in abundance to be detected. 3) Some peptides may show significant ICAT ratio changes in MS levels, but their MS/MS fragmentation may be poor due to their amino acid compositions or the attachment of a bulky ICAT tag. 4) Because the protein disulfide bond reduction step was omitted from this redox ICAT study, it is possible that some of the free cysteines buried inside non-linearized protein domains may not be labeled efficiently by the bulky ICAT reagents. 5) Redox-induced post-translational modifications on non-cysteine residues, such as phosphorylation, may occur non-uniformly in light and heavy ICAT-labeled peptides. Therefore quantification accuracy of ICAT may be affected. 6) Some proteins bound to Trx1 with high affinity may still be reduced in the control animals, leading to similar reduction levels compared with Tg-Trx1 animals regardless of the different Trx1 expression levels between the animal groups. Therefore, they may not be found in this ICAT difference-based study. Alternative approaches, including “hooking” to C35S Trx1 mutant, may be used to identify some of these Trx1 targets (7, 8).
Conclusions
In the present study, we performed a proteomics identification of Trx1 reduction target proteins from the hearts of cardiac specific Tg-Trx1 mice. Using the comparative analysis of both iTRAQ and ICAT results, we were able to reveal many putative Trx1 reduction substrates, several of which were previously unknown. We identified several protein networks whose functions may be regulated by Trx1, including the creatine-phosphocreatine shuttle, the MPTP complex, and the sarcomeric contractile apparatus. The results presented here suggest that in addition to its antioxidant function Trx1 may be involved in the coordination of a wide array of cellular signaling pathways to maintain cardiac function.
Footnotes
* This work was supported by National Institutes of Health Grant NS046593 (to H. L. for the NeuroProteomics Core Facility). This work was also supported by a grant from the foundation of the University of Medicine and Dentistry of New Jersey (to H. L. and J. S.).
 The on-line version of this article (available at http://www.mcponline.org) contains supplemental material.
The on-line version of this article (available at http://www.mcponline.org) contains supplemental material.
2 T. Liu, T. Ago, P. Zhai, J. Sadoshima, and H. Li, manuscript in preparation.
1 The abbreviations used are:
- Trx
- thioredoxin
- iTRAQ
- isobaric tags for relative and absolute quantitation
- RPLC
- reversed phase LC
- ANT1
- adenine-nucleotide translocase 1
- TAC
- transverse aortic constriction
- MPTP
- mitochondrial permeability transition pore
- TCEP
- tris(2-carboxyethyl)phosphine
- MMTS
- methyl methanethiosulfonate
- TrxR
- thioredoxin reductase
- L
- light
- H
- heavy
- SCX
- strong cation exchange chromatography
- C.I.
- confidence interval
- Prx
- peroxiredoxin
- GPS
- global proteomics server.
REFERENCES
- 1.Matthews J. R., Wakasugi N., Virelizier J. L., Yodoi J., Hay R. T.( 1992) Thioredoxin regulates the DNA binding activity of NF-kappa B by reduction of a disulphide bond involving cysteine 62. Nucleic Acids Res. 20, 3821– 3830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ema M., Hirota K., Mimura J., Abe H., Yodoi J., Sogawa K., Poellinger L., Fujii-Kuriyama Y.( 1999) Molecular mechanisms of transcription activation by HLF and HIF1alpha in response to hypoxia: their stabilization and redox signal-induced interaction with CBP/p300. EMBO J. 18, 1905– 1914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ago T., Liu T., Zhai P., Chen W., Li H., Molkentin J. D., Vatner S. F., Sadoshima J.( 2008) A redox-dependent pathway for regulating class II HDACs and cardiac hypertrophy. Cell 133, 978– 993 [DOI] [PubMed] [Google Scholar]
- 4.Makino Y., Yoshikawa N., Okamoto K., Hirota K., Yodoi J., Makino I., Tanaka H.( 1999) Direct association with thioredoxin allows redox regulation of glucocorticoid receptor function. J. Biol. Chem. 274, 3182– 3188 [DOI] [PubMed] [Google Scholar]
- 5.Yano H., Wong J. H., Lee Y. M., Cho M. J., Buchanan B. B.( 2001) A strategy for the identification of proteins targeted by thioredoxin. Proc. Natl. Acad. Sci. U.S.A. 98, 4794– 4799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Maeda K., Finnie C., Svensson B.( 2004) Cy5 maleimide labelling for sensitive detection of free thiols in native protein extracts: identification of seed proteins targeted by barley thioredoxin h isoforms. Biochem. J. 378, 497– 507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wong J. H., Cai N., Balmer Y., Tanaka C. K., Vensel W. H., Hurkman W. J., Buchanan B. B.( 2004) Thioredoxin targets of developing wheat seeds identified by complementary proteomic approaches. Phytochemistry 65, 1629– 1640 [DOI] [PubMed] [Google Scholar]
- 8.Motohashi K., Romano P. G., Hisabori T.( 2009) Identification of thioredoxin targeted proteins using thioredoxin single cysteine mutant-immobilized resin. Methods Mol. Biol. 479, 117– 131 [DOI] [PubMed] [Google Scholar]
- 9.Hägglund P., Bunkenborg J., Maeda K., Svensson B.( 2008) Identification of thioredoxin disulfide targets using a quantitative proteomics approach based on isotope-coded affinity tags. J. Proteome Res. 7, 5270– 5276 [DOI] [PubMed] [Google Scholar]
- 10.Fu C., Hu J., Liu T., Ago T., Sadoshima J., Li H.( 2008) Quantitative analysis of redox-sensitive proteome with DIGE and ICAT. J. Proteome Res. 7, 3789– 3802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gygi S. P., Rist B., Gerber S. A., Turecek F., Gelb M. H., Aebersold R.( 1999) Quantitative analysis of complex protein mixtures using isotope-coded affinity tags. Nat. Biotechnol. 17, 994– 999 [DOI] [PubMed] [Google Scholar]
- 12.Sethuraman M., McComb M. E., Heibeck T., Costello C. E., Cohen R. A.( 2004) Isotope-coded affinity tag approach to identify and quantify oxidant-sensitive protein thiols. Mol. Cell. Proteomics 3, 273– 278 [DOI] [PubMed] [Google Scholar]
- 13.Sethuraman M., McComb M. E., Huang H., Huang S., Heibeck T., Costello C. E., Cohen R. A.( 2004) Isotope-coded affinity tag (ICAT) approach to redox proteomics: identification and quantitation of oxidant-sensitive cysteine thiols in complex protein mixtures. J. Proteome Res. 3, 1228– 1233 [DOI] [PubMed] [Google Scholar]
- 14.Sethuraman M., Clavreul N., Huang H., McComb M. E., Costello C. E., Cohen R. A.( 2007) Quantification of oxidative posttranslational modifications of cysteine thiols of p21ras associated with redox modulation of activity using isotope-coded affinity tags and mass spectrometry. Free Radic. Biol. Med. 42, 823– 829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Powis G., Mustacich D., Coon A.( 2000) The role of the redox protein thioredoxin in cell growth and cancer. Free Radic. Biol. Med. 29, 312– 322 [DOI] [PubMed] [Google Scholar]
- 16.Bertini R., Howard O. M., Dong H. F., Oppenheim J. J., Bizzarri C., Sergi R., Caselli G., Pagliei S., Romines B., Wilshire J. A., Mengozzi M., Nakamura H., Yodoi J., Pekkari K., Gurunath R., Holmgren A., Herzenberg L. A., Herzenberg L. A., Ghezzi P.( 1999) Thioredoxin, a redox enzyme released in infection and inflammation, is a unique chemoattractant for neutrophils, monocytes, and T cells. J. Exp. Med. 189, 1783– 1789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lovell M. A., Xie C., Gabbita S. P., Markesbery W. R.( 2000) Decreased thioredoxin and increased thioredoxin reductase levels in Alzheimer's disease brain. Free Radic. Biol. Med. 28, 418– 427 [DOI] [PubMed] [Google Scholar]
- 18.Shioji K., Nakamura H., Masutani H., Yodoi J.( 2003) Redox regulation by thioredoxin in cardiovascular diseases. Antioxid. Redox Signal. 5, 795– 802 [DOI] [PubMed] [Google Scholar]
- 19.Ventura-Clapier R., Garnier A., Veksler V.( 2004) Energy metabolism in heart failure. J. Physiol. 555, 1– 13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ago T., Sadoshima J.( 2007) Thioredoxin1 as a negative regulator of cardiac hypertrophy. Antioxid. Redox Signal. 9, 679– 687 [DOI] [PubMed] [Google Scholar]
- 21.Ago T., Yeh I., Yamamoto M., Schinke-Braun M., Brown J. A., Tian B., Sadoshima J.( 2006) Thioredoxin1 upregulates mitochondrial proteins related to oxidative phosphorylation and TCA cycle in the heart. Antioxid. Redox Signal. 8, 1635– 1650 [DOI] [PubMed] [Google Scholar]
- 22.Yamamoto M., Yang G., Hong C., Liu J., Holle E., Yu X., Wagner T., Vatner S. F., Sadoshima J.( 2003) Inhibition of endogenous thioredoxin in the heart increases oxidative stress and cardiac hypertrophy. J. Clin. Investig. 112, 1395– 1406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sadoshima J., Montagne O., Wang Q., Yang G., Warden J., Liu J., Takagi G., Karoor V., Hong C., Johnson G. L., Vatner D. E., Vatner S. F.( 2002) The MEKK1-JNK pathway plays a protective role in pressure overload but does not mediate cardiac hypertrophy. J. Clin. Investig. 110, 271– 279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Peng J., Elias J. E., Thoreen C. C., Licklider L. J., Gygi S. P.( 2003) Evaluation of multidimensional chromatography coupled with tandem mass spectrometry (LC/LC-MS/MS) for large-scale protein analysis: the yeast proteome. J. Proteome Res. 2, 43– 50 [DOI] [PubMed] [Google Scholar]
- 25.Liu T., Donahue K. C., Hu J., Kurnellas M. P., Grant J. E., Li H., Elkabes S.( 2007) Identification of differentially expressed proteins in experimental autoimmune encephalomyelitis (EAE) by proteomic analysis of the spinal cord. J. Proteome Res. 6, 2565– 2575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hu J., Qian J., Borisov O., Pan S., Li Y., Liu T., Deng L., Wannemacher K., Kurnellas M., Patterson C., Elkabes S., Li H.( 2006) Optimized proteomic analysis of a mouse model of cerebellar dysfunction using amine-specific isobaric tags. Proteomics 6, 4321– 4334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schwartz D., Gygi S. P.( 2005) An iterative statistical approach to the identification of protein phosphorylation motifs from large-scale data sets. Nat. Biotechnol. 23, 1391– 1398 [DOI] [PubMed] [Google Scholar]
- 28.Balmer Y., Vensel W. H., Tanaka C. K., Hurkman W. J., Gelhaye E., Rouhier N., Jacquot J. P., Manieri W., Schürmann P., Droux M., Buchanan B. B.( 2004) Thioredoxin links redox to the regulation of fundamental processes of plant mitochondria. Proc. Natl. Acad. Sci. U.S.A. 101, 2642– 2647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kumar J. K., Tabor S., Richardson C. C.( 2004) Proteomic analysis of thioredoxin-targeted proteins in Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 101, 3759– 3764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Baker K. M., Chakravarthi S., Langton K. P., Sheppard A. M., Lu H., Bulleid N. J.( 2008) Low reduction potential of Ero1alpha regulatory disulphides ensures tight control of substrate oxidation. EMBO J. 27, 2988– 2997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kim J. A., Park S., Kim K., Rhee S. G., Kang S. W.( 2005) Activity assay of mammalian 2-cys peroxiredoxins using yeast thioredoxin reductase system. Anal. Biochem. 338, 216– 223 [DOI] [PubMed] [Google Scholar]
- 32.Kanzok S. M., Fechner A., Bauer H., Ulschmid J. K., Müller H. M., Botella-Munoz J., Schneuwly S., Schirmer R., Becker K.( 2001) Substitution of the thioredoxin system for glutathione reductase in Drosophila melanogaster. Science 291, 643– 646 [DOI] [PubMed] [Google Scholar]
- 33.Juhaszova M., Wang S., Zorov D. B., Nuss H. B., Gleichmann M., Mattson M. P., Sollott S. J.( 2008) The identity and regulation of the mitochondrial permeability transition pore: where the known meets the unknown. Ann. N.Y. Acad. Sci. 1123, 197– 212 [DOI] [PubMed] [Google Scholar]
- 34.Shanmuganathan S., Hausenloy D. J., Duchen M. R., Yellon D. M.( 2005) Mitochondrial permeability transition pore as a target for cardioprotection in the human heart. Am. J. Physiol. Heart Circ. Physiol. 289, H237– 242 [DOI] [PubMed] [Google Scholar]
- 35.Schwarz M., Andrade-Navarro M. A., Gross A.( 2007) Mitochondrial carriers and pores: key regulators of the mitochondrial apoptotic program? Apoptosis 12, 869– 876 [DOI] [PubMed] [Google Scholar]
- 36.Graham B. H., Waymire K. G., Cottrell B., Trounce I. A., MacGregor G. R., Wallace D. C.( 1997) A mouse model for mitochondrial myopathy and cardiomyopathy resulting from a deficiency in the heart/muscle isoform of the adenine nucleotide translocator. Nat. Genet. 16, 226– 234 [DOI] [PubMed] [Google Scholar]
- 37.Vieira H. L., Belzacq A. S., Haouzi D., Bernassola F., Cohen I., Jacotot E., Ferri K. F., El Hamel C., Bartle L. M., Melino G., Brenner C., Goldmacher V., Kroemer G.( 2001) The adenine nucleotide translocator: a target of nitric oxide, peroxynitrite, and 4-hydroxynonenal. Oncogene 20, 4305– 4316 [DOI] [PubMed] [Google Scholar]
- 38.Majima E., Koike H., Hong Y. M., Shinohara Y., Terada H.( 1993) Characterization of cysteine residues of mitochondrial ADP/ATP carrier with the SH-reagents eosin 5-maleimide and N-ethylmaleimide. J. Biol. Chem. 268, 22181– 22187 [PubMed] [Google Scholar]
- 39.Choi J., Sullards M. C., Olzmann J. A., Rees H. D., Weintraub S. T., Bostwick D. E., Gearing M., Levey A. I., Chin L. S., Li L.( 2006) Oxidative damage of DJ-1 is linked to sporadic Parkinson and Alzheimer diseases. J. Biol. Chem. 281, 10816– 10824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bretaud S., Allen C., Ingham P. W., Bandmann O.( 2007) p53-dependent neuronal cell death in a DJ-1-deficient zebrafish model of Parkinson's disease. J. Neurochem. 100, 1626– 1635 [DOI] [PubMed] [Google Scholar]
- 41.Lev N., Ickowicz D., Melamed E., Offen D.( 2008) Oxidative insults induce DJ-1 upregulation and redistribution: implications for neuroprotection. Neurotoxicology 29, 397– 405 [DOI] [PubMed] [Google Scholar]
- 42.Kinumi T., Kimata J., Taira T., Ariga H., Niki E.( 2004) Cysteine-106 of DJ-1 is the most sensitive cysteine residue to hydrogen peroxide-mediated oxidation in vivo in human umbilical vein endothelial cells. Biochem. Biophys. Res. Commun. 317, 722– 728 [DOI] [PubMed] [Google Scholar]
- 43.Ritchie R. H., Delbridge L. M.( 2006) Cardiac hypertrophy, substrate utilization and metabolic remodelling: cause or effect? Clin. Exp. Pharmacol. Physiol. 33, 159– 166 [DOI] [PubMed] [Google Scholar]
- 44.Barger P. M., Kelly D. P.( 1999) Fatty acid utilization in the hypertrophied and failing heart: molecular regulatory mechanisms. Am. J. Med. Sci. 318, 36– 42 [DOI] [PubMed] [Google Scholar]
- 45.Strauss A. W., Powell C. K., Hale D. E., Anderson M. M., Ahuja A., Brackett J. C., Sims H. F.( 1995) Molecular basis of human mitochondrial very-long-chain acyl-CoA dehydrogenase deficiency causing cardiomyopathy and sudden death in childhood. Proc. Natl. Acad. Sci. U.S.A. 92, 10496– 10500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Reddy S., Jones A. D., Cross C. E., Wong P. S., Van Der Vliet A.( 2000) Inactivation of creatine kinase by S-glutathionylation of the active-site cysteine residue. Biochem. J. 347, 821– 827 [PMC free article] [PubMed] [Google Scholar]
- 47.Ferreira L. F., Reid M. B.( 2008) Muscle-derived ROS and thiol regulation in muscle fatigue. J. Appl. Physiol. 104, 853– 860 [DOI] [PubMed] [Google Scholar]
- 48.Hertelendi Z., Tóth A., Borbély A., Galajda Z., van der Velden J., Stienen G. J., Edes I., Papp Z.( 2008) Oxidation of myofilament protein sulfhydryl groups reduces the contractile force and its Ca2+ sensitivity in human cardiomyocytes. Antioxid. Redox Signal. 10, 1175– 1184 [DOI] [PubMed] [Google Scholar]
- 49.Chae H. Z., Chung S. J., Rhee S. G.( 1994) Thioredoxin-dependent peroxide reductase from yeast. J. Biol. Chem. 269, 27670– 27678 [PubMed] [Google Scholar]
- 50.Fermani S., Sparla F., Falini G., Martelli P. L., Casadio R., Pupillo P., Ripamonti A., Trost P.( 2007) Molecular mechanism of thioredoxin regulation in photosynthetic A2B2-glyceraldehyde-3-phosphate dehydrogenase. Proc. Natl. Acad. Sci. U.S.A. 104, 11109– 11114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Alkhalfioui F., Renard M., Vensel W. H., Wong J., Tanaka C. K., Hurkman W. J., Buchanan B. B., Montrichard F.( 2007) Thioredoxin-linked proteins are reduced during germination of Medicago truncatula seeds. Plant Physiol. 144, 1559– 1579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Marchand C., Le Maréchal P., Meyer Y., Miginiac-Maslow M., Issakidis-Bourguet E., Decottignies P.( 2004) New targets of Arabidopsis thioredoxins revealed by proteomic analysis. Proteomics 4, 2696– 2706 [DOI] [PubMed] [Google Scholar]
- 53.Bunik V.( 2000) Increased catalytic performance of the 2-oxoacid dehydrogenase complexes in the presence of thioredoxin, a thiol-disulfide oxidoreductase. J. Mol. Catal. B Enzym. 8, 165– 174 [Google Scholar]
- 54.Bunik V., Raddatz G., Lemaire S., Meyer Y., Jacquot J. P., Bisswanger H.( 1999) Interaction of thioredoxins with target proteins: role of particular structural elements and electrostatic properties of thioredoxins in their interplay with 2-oxoacid dehydrogenase complexes. Protein Sci. 8, 65– 74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hara S., Motohashi K., Arisaka F., Romano P. G., Hosoya-Matsuda N., Kikuchi N., Fusada N., Hisabori T.( 2006) Thioredoxin-h1 reduces and reactivates the oxidized cytosolic malate dehydrogenase dimer in higher plants. J. Biol. Chem. 281, 32065– 32071 [DOI] [PubMed] [Google Scholar]
- 56.Choi H. I., Lee S. P., Kim K. S., Hwang C. Y., Lee Y. R., Chae S. K., Kim Y. S., Chae H. Z., Kwon K. S.( 2006) Redox-regulated cochaperone activity of the human DnaJ homolog Hdj2. Free Radic. Biol. Med. 40, 651– 659 [DOI] [PubMed] [Google Scholar]
- 57.Motohashi K., Koyama F., Nakanishi Y., Ueoka-Nakanishi H., Hisabori T.( 2003) Chloroplast cyclophilin is a target protein of thioredoxin. Thiol modulation of the peptidyl-prolyl cis-trans isomerase activity. J. Biol. Chem. 278, 31848– 31852 [DOI] [PubMed] [Google Scholar]
- 58.Motohashi K., Kondoh A., Stumpp M. T., Hisabori T.( 2001) Comprehensive survey of proteins targeted by chloroplast thioredoxin. Proc. Natl. Acad. Sci. U.S.A. 98, 11224– 11229 [DOI] [PMC free article] [PubMed] [Google Scholar]