Abstract
The ubiquitin-proteasome system is a central mechanism for controlled proteolysis that regulates numerous cellular processes in eukaryotes. As such, defects in this system can contribute to disease pathogenesis. In this pathway, E3 ubiquitin ligases provide platforms for binding specific substrates, thereby coordinating their ubiquitylation and subsequent degradation by the proteasome. Despite the identification of many E3 ubiquitin ligases, the identities of their specific substrates are still largely unresolved. The ankyrin repeat-containing protein with a suppressor of cytokine signaling box 2 (ASB2) gene that we initially identified as a retinoic acid-response gene in acute promyelocytic leukemia cells encodes the specificity subunit of an E3 ubiquitin ligase complex that is involved in hematopoietic cell differentiation. We have recently identified filamin A and filamin B as the first ASB2 targets and shown that ASB2 triggers ubiquitylation and proteasome-mediated degradation of these proteins. Here a global quantitative proteomics strategy is provided to identify substrates of E3 ubiquitin ligases targeted to proteasomal degradation. Indeed we used label-free methods for quantifying proteins identified by shotgun proteomics in extracts of cells expressing wild-type ASB2 or an E3 ubiquitin ligase-defective mutant of ASB2 under the control of an inducible promoter. Measurements of spectral count and mass spectrometric signal intensity demonstrated a drastic decrease of filamin A and filamin B in myeloid leukemia cells expressing wild-type ASB2 compared with cells expressing an E3 ubiquitin ligase-defective mutant of ASB2. Altogether we provide an original strategy that enables identification of E3 ubiquitin ligase substrates that have to be degraded.
The ubiquitin-proteasome system (UPS)1 plays an essential role in the regulation of protein stability in eukaryotic cells. Degradation of a protein by the UPS entails two successive steps: the covalent attachment of multiple ubiquitin molecules to the protein substrate and its degradation by the 26 S proteasome (1, 2). Ubiquitylation of protein substrates occurs through the sequential action of distinct enzymes: a ubiquitin-activating enzyme, E1; a ubiquitin-conjugating enzyme, E2; and a ubiquitin ligase, E3, responsible for the specific recognition of substrates. Increasing attention has been recently given to the UPS leading to the identification of hundreds of E3 ubiquitin ligases (E3s). Two major classes of E3s have been described: (i) E3s of the HECT (homologous to the E6-associated protein carboxyl terminus) domain family that function as ubiquitin carriers (3, 4) and (ii) E3s of the RING (really interesting new gene) or of the U box families that have no inherent catalytic activity but recruit an E2 enzyme toward substrates (5–7).
Classical approaches to identify substrates of E3s are based on the identification of interacting proteins. Although these have successfully led to the identification of a number of substrates of monomeric E3s, identification of substrates of multimeric E3s is very challenging because of the weak affinity of substrates for their requisite specificity subunit and because of the labile nature of the substrate complexed with the specificity subunit (8).
Acute promyelocytic leukemia (APL) is associated with six reciprocal translocations always involving the retinoic acid receptor α (RARα) gene (9–11). The RARα protein is a member of the nuclear receptor superfamily that stimulates myeloid differentiation in the presence of its ligand, all-trans-retinoic acid (RA). In more than 95% of APL, the t(15;17) translocation between the promyelocytic leukemia (PML) gene on chromosome 15 and the RARα gene on chromosome 17 produces the PML-RARα fusion protein (12). The PML-RARα protein enhances the repression of RARα target genes by increasing associations with corepressors (13–15) and by recruiting DNA methyltransferases (16). These complexes dissociate from the PML-RARα fusion protein in the presence of pharmacological concentrations of RA perhaps explaining why APL cells are sensitive to RA treatment. Indeed at pharmacological concentrations, RA induces complete remission in a high percentage of APL patients (17–19). By studying RA-induced differentiation of APL cells we have attempted to identify some of the genes that may be up-regulated during this process to further understand the control of growth and differentiation in leukemia (20). One gene identified in this manner, ASB2 (ankyrin repeat-containing protein with a suppressor of cytokine signaling box 2) is an RA-response gene involved in induced differentiation of myeloid leukemia cells (21–23).
The ASB2 protein is a subunit of a multimeric E3 ubiquitin ligase of the cullin-RING ligase family (24, 25). The ASB2 suppressor of cytokine signaling box can be divided into a BC box that defines a binding site for the Elongin BC complex and a Cul5 box that determines the binding specificity for Cullin5 (24, 26). Indeed the ASB2 protein, by interacting with the Elongin BC complex, can assemble with a Cullin5/Rbx1 or -2 module to reconstitute an active E3 ubiquitin ligase complex (23–25). Within this complex, the ASB2 protein is the specificity subunit involved in the recruitment of specific substrate(s). Furthermore endogenous ASB2 protein was copurified with ubiquitin ligase activity in RA-treated APL cells suggesting that, during induced differentiation of leukemia cells, the ASB2 protein may target proteins involved in blocking differentiation to destruction by the proteasome machinery (24). We recently identified actin-binding proteins filamin A (FLNa) and filamin B (FLNb) as ASB2 targets and showed that ASB2 triggers ubiquitylation and drives proteasome-mediated degradation of these proteins during RA-induced differentiation of myeloid leukemia cells (23).
With the aim to develop a strategy to identify E3 substrates that are degraded by the proteasome, we used an MS approach to identify ASB2 substrates in physiologically relevant settings. Indeed we used label-free quantitative proteomics to identify proteins that are absent or less abundant in cells that express wild-type ASB2 but that accumulate in cells expressing an ASB2 E3 ligase-defective mutant. Application of label-free MS methods that have the advantage to be simple, fast, and cheap enabled the identification of FLNa and FLNb as ASB2 substrates. This study provides a new strategy for the identification of E3 substrates that have to be degraded.
EXPERIMENTAL PROCEDURES
Cell Lines, Culture Conditions, and Cell Extracts
Clonal PLB985 cells stably transfected with ZnSO4-inducible vectors expressing wild-type ASB2 or an E3 ligase-defective mutant of ASB2 (PLB985/MT-FLAG-ASB2wt and PLB985/MT-FLAG-ASB2LA cells, respectively) were used as described previously (23). PLB985 cells transfected with the empty vector (PLB985/MT-FLAG) were used as controls. Exponentially growing cells were seeded at 1.5 × 105 cells/ml for 24 h and either induced by addition of 100 μm ZnSO4 for 8 h or left uninduced. Cell viability was evaluated using a standard trypan blue dye exclusion assay. Cells were maintained in a 5% CO2 incubator at 37 °C. Cell fractionations were carried out using the ProteoExtract Subcellular Proteome Extraction kit as recommended by the manufacturer (Calbiochem). All buffers were supplemented with 1 mm Na3VO4, 50 mm NaF, and 1% protease inhibitor mixture (Sigma). Proper subfractionation of cellular proteomes was verified using antibodies to Grb2 (cytosol and membrane/organelles), TAF6 (nucleus), and vimentin (cytoskeleton) (not shown). The cytosolic fraction was used for this study. Proteins were quantified using the BCA™ Protein Assay kit (Pierce).
Antibodies and Western Blots
The rabbit serum raised against ASB2 (1PNA) has been described previously (21). Antibodies to FLNa (PM6/317) and to FLNb were from Millipore. Anti-Talin 1 (H-300) and anti-Grb2 (C-23) were from Santa Cruz Biotechnology. Anti-non-muscle myosin II heavy chain A, anti-TAF6 (25TA-2G7), and anti-vimentin (V9) were from Covance, Euromedex, and Immunotech, respectively. Samples were separated by SDS-PAGE on a 7% acrylamide gel, transferred to nitrocellulose membranes, and analyzed by immunoblotting with the indicated antibodies.
Northern Blots
Total RNA extraction and hybridization were as described previously (21, 27). The ASB2 probe corresponded to the ASB2 open reading frame. The FLNa probe was an internal EcoRI/KpnI fragment of the FLNa cDNA. The Arbp probe was previously described as 36B4 probe (28).
In-gel Digestion
150 μg of each cytoplasmic extract of ZnSO4-treated cells were diluted in Laemmli buffer and boiled for 5 min before being separated on a 7% acrylamide SDS-PAGE gel (18-cm length). Proteins were visualized by colloidal Coomassie Blue staining. Each lane was cut into 40 slices that were washed in water followed by a second wash in 100 mm NH4HCO3 and a third wash in 100 mm NH4HCO3, ACN (1:1). All washing steps were performed for 5 min at 37 °C. Proteins were reduced and alkylated by successive incubations in 100 mm NH4HCO3, 10 mm DTT for 45 min at 56 °C and in 100 mm NH4HCO3, 55 mm iodoacetamide for 30 min at room temperature. An additional cycle of washes in NH4HCO3 and NH4HCO3/ACN was then performed. Proteins were digested by incubating each gel slice in a sufficient covering volume (50 μl) of modified porcine trypsin solution (12.5 ng/μl in 12.5 mm NH4HCO3; Promega). Trypsin digestion was performed overnight at 37 °C under shaking. Resulting peptides were extracted from the gel slices by successive incubations in 25 mm NH4HCO3, ACN (1:1) and 5% formic acid (FA), ACN (1:1) for 15 min at 37 °C under shaking. The two collected extractions were pooled, and the peptide mixture was dried under vacuum and stored at −20 °C.
Nano-LC-ESI-LTQ-Orbitrap MS/MS Analysis
For MS analysis, tryptic peptides were resuspended with 15 μl of 2% ACN, 0.05% TFA and submitted to nano-LC-MS/MS using an Ultimate3000 system (Dionex) coupled to an LTQ-Orbitrap mass spectrometer (Thermo Fisher Scientific) operating in positive mode with a spray voltage of 1.5 kV. 5 μl of each sample were loaded on a C18 precolumn (300-μm inner diameter × 5 mm; Dionex) at 20 μl/min in 5% ACN, 0.05% TFA. After 5-min desalting, the precolumn was switched on line with the analytical column (75-μm-inner diameter × 15-cm PepMap C18; Dionex) equilibrated in 95% solvent A (5% ACN, 0.2% FA) and 5% solvent B (80% ACN, 0.2% FA). Peptides were eluted using a 5–50% gradient of solvent B during 80 min at 300 nl/min flow rate. Data were acquired with Xcalibur (LTQ-Orbitrap Software version 2.2, Thermo Fisher Scientific). The mass spectrometer was operated in the data-dependent mode and was externally calibrated. Survey MS scans were acquired in the Orbitrap in the 300–2000 m/z range with the resolution set to a value of 60,000 at m/z 400. Up to five of the most intense multiply charged ions (2+, 3+, and 4+) per scan were CID fragmented in the linear ion trap. A dynamic exclusion window was applied within 60 s. All tandem mass spectra were collected using a normalized collision energy of 35%, an isolation window of 4 m/z, and one microscan. Other instrumental parameters included maximum injection times and automatic gain control targets of 250 ms and 500,000 ions for the FT MS and 100 ms and 10,000 ions for LTQ MS/MS, respectively.
Database Search and Data Analysis
Data were analyzed using Xcalibur software (version 2.0.6, Thermo Fisher Scientific), and MS/MS centroid peak lists were generated using the extract_msn.exe executable program (Thermo Fisher Scientific) integrated into the Mascot Daemon software (Mascot version 2.2.03, Matrix Science). The following parameters were set to create peak lists: parent ions in the mass range 400–4,500, no grouping of MS/MS scans, and threshold at 1,000. A peak list was created for each fraction (i.e. each gel slice) analyzed, and individual Mascot searches were performed for each fraction.
Data were searched using the Mascot server (Mascot version 2.2.03, Matrix Science) against Homo sapiens sequences in the Swiss-Prot TrEMBL database (69,287 sequences). This database consists of UniProtKB/Swiss-Prot Protein Knowledgebase Release 55.1 merged in house with UniProtKB/TrEMBL Protein Database Release 38.1. Mass tolerances in MS and MS/MS were set to 5 ppm and 0.8 Da, respectively, and the instrument setting was specified as “ESI Trap.” Trypsin was designated as the protease (specificity set for cleavage after Lys or Arg), and up to two missed cleavages were allowed. Oxidation of methionine, amino-terminal protein acetylation, and carbamidomethylation and propionamide on cysteine were searched as variable modifications; no fixed modification was set. Mascot results were parsed with the home-made and developed software MFPaQ version 4.0 (Mascot File Parsing and Quantification) (29), and protein hits were automatically validated if they were identified with at least either: (i) one top ranking peptide with a Mascot score of more than 42 (p value < 0.001), (ii) two top ranking peptides each with a Mascot score of more than 27 (p value < 0.0316), or (iii) three top ranking peptides each with a Mascot score of more than 22 (p value < 0.1). p values were determined by the Mascot Search program. Proteins identified with exactly the same set of peptides were grouped. Highly homologous protein hits, i.e. proteins identified with top ranking MS/MS queries also assigned to another protein hit of higher score, were detected by the MFPaQ software and were considered as individual hits and included in the final list only if they were additionally assigned a specific top ranking peptide of score higher than 30. Keratins were manually removed from the protein lists. Identification data of FLNa, FLNb, and Talin 1 are provided in supplemental Data 1 and 2. From all the validated result files corresponding to the fractions of a one-dimensional gel lane, MFPaQ was used to generate a unique non-redundant list of proteins (data not shown). The MFPaQ software was also used to compare these lists to determine whether the identified proteins were specific to wild-type ASB2 (ASB2wt)-expressing cells, to ASB2LA-expressing cells, or present in both cell lines.
To evaluate false positive rates, all the initial database searches were performed using the “decoy” option of Mascot, i.e. the data were searched against a combined database containing the real specified protein sequences (target database, Swiss-Prot TrEMBL human) and the corresponding reversed protein sequences (decoy database). MFPaQ used the same criteria to validate decoy and target hits, calculated the false discovery rate (FDR = number of validated decoy hits/(number of validated target hits + number of validated decoy hits) × 100) for each gel slice analyzed, and made the average of FDR for all slices belonging to the same gel lane (i.e. to the same sample). FDRs were below 1%.
Quantitative Analysis
Analysis of quantitative changes in protein abundance was measured by two label-free methods. To consider only the peptides that were unique for each filamin or talin, sequence alignments of FLNa, FLNb, and FLNc (provided as supplemental Data 3) and of Talin 1 and Talin 2 (supplemental Data 4) were performed using MultAlin.
For spectral counting, fragment spectra identifying peptides specific to proteins of interest and considered as top ranking peptides by the Mascot search algorithm (see supplemental Data 1 and 2) were summed up across all gel slices. This was performed manually using data obtained with the Mascot software. Two quantitative analyses were performed from two independent polyacrylamide gels.
For peptide chromatographic peak intensity measurements, the Xcalibur software (Thermo Fisher Scientific) was used to extract the ion current chromatogram of peptides with a mass tolerance of 5 ppm and to perform the area integration of the corresponding peaks using the Interactive Chemical Information System algorithm. Indeed five specific peptides of FLNa or Talin 1 were selected based on the following criteria: the Mascot peptide ion score was above 42 in the slice where the protein was identified with the best score; the peptide ion did not present several types of modification and was not present at multiple charge states. The data sampling across the MS channel was verified for the peptides of interest and was always above 10 points (supplemental Data 9 and 10). Therefore, for these peptides, ratios of the sum of the chromatographic peak areas measured in the slices where proteins were identified in ASB2LA-expressing cell extracts to the sum of the chromatographic peak area measured in the slices where proteins were identified in ASB2wt-expressing cell extracts were calculated. To estimate the relative abundance of FLNa and Talin 1, mean values and S.D. of these ratios were calculated in different biological and technical replicates (for details, see supplemental Data 5–8).
RESULTS
Model System: Cells Expressing a Functional or an E3 Ligase-defective ASB2 Protein
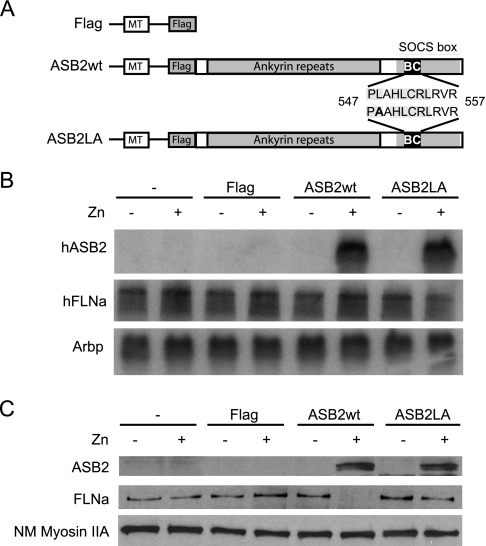
To identify novel substrates of E3s that are targeted for degradation, we set up a global strategy using label-free quantitative proteomics with cells induced to express either a functional or a defective E3 ligase (Fig. 1). To evaluate this method, expression vectors encoding FLAG-tagged ASB2wt or an ASB2 BC box mutant (ASB2LA) unable to interact with the Elongin BC complex and defective in ubiquitin ligase activity under the control of the zinc-inducible metallothionein (MT) promoter (MT-FLAG-ASB2wt and MT-FLAG-ASB2LA, respectively) were stably transfected into PLB985 myeloid leukemia cells (Fig. 2 A) (23). The MT-FLAG vector was used as an empty control (23). PLB985, PLB985/MT-FLAG, PLB985/MT-FLAG-ASB2wt, and PLB985/MT-FLAG-ASB2LA cells were incubated with or without ZnSO4 for 8 h. When zinc was added to the media, ASB2 mRNA (Fig. 2B) and protein (Fig. 2C) expression increased dramatically in PLB985/MT-FLAG-ASB2wt and PLB985/MT-FLAG-ASB2LA cells. ASB2wt and ASB2LA proteins were mainly detected in the cytosolic fractions (Fig. 2C and data not shown). Although equivalent amounts of ASB2wt and ASB2LA proteins were expressed, only wild-type ASB2 induced loss of FLNa (Fig. 2C) as shown previously (23). In contrast, the abundance of FLNa in untransfected cells or in cells transfected with the MT-FLAG vector was unaffected by ZnSO4 treatment (Fig. 2C). As expected, the abundance of FLNa mRNA in either cell line was not regulated in cells cultured without or with ZnSO4 (Fig. 2B).
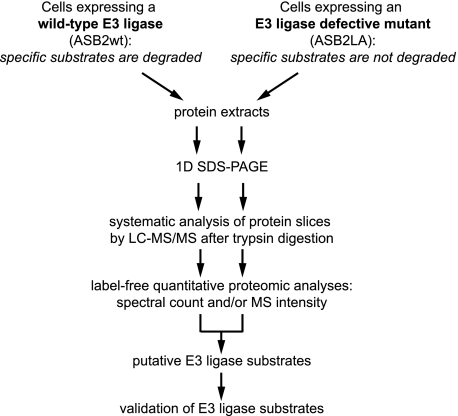
Fig. 1.
Experimental design for identification of E3 ubiquitin ligase substrates that are targeted for degradation. Our strategy uses label-free quantitative proteomics approaches to identify proteins that are absent or less abundant in cells that express a functional E3 ubiquitin ligase but that accumulate in cells that express an E3 ligase-defective mutant of this protein. This strategy was applied to identify substrates of ASB2 using myeloid leukemia cells expressing ASB2wt or an ASB2 BC box mutant (ASB2LA) that is unable to interact with the Elongin BC complex and defective in ubiquitin ligase activity under the control of an inducible promoter. 1D, one-dimensional.
Fig. 2.
ASB2-induced degradation of FLNa in myeloid leukemia cells. PLB985, PLB985/MT-FLAG, PLB985/MT-FLAG-ASB2wt, and PLB985/MT-FLAG-ASB2LA cells were left untreated (−) or were treated (+) with 100 μm ZnSO4 (Zn) for 8 h. A, schematic representation of ASB2wt and ASB2LA constructs under the control of the zinc-inducible MT promoter. The BC box mutation is indicated in bold. B, autoradiograms of mRNA of ASB2 (upper panel), FLNa (middle panel), and Arbp as an assessment of RNA quantities in each lane (lower panel). C, proteins of cytosolic extracts (15 μg) were separated by SDS-PAGE and immunoblotted for ASB2, FLNa, and non-muscle myosin heavy chain IIA (NM Myosin IIA). SOCS, suppressor of cytokine signaling.
Spectral Count Label-free Quantitative Proteomics Reveals ASB2 Substrate Candidates
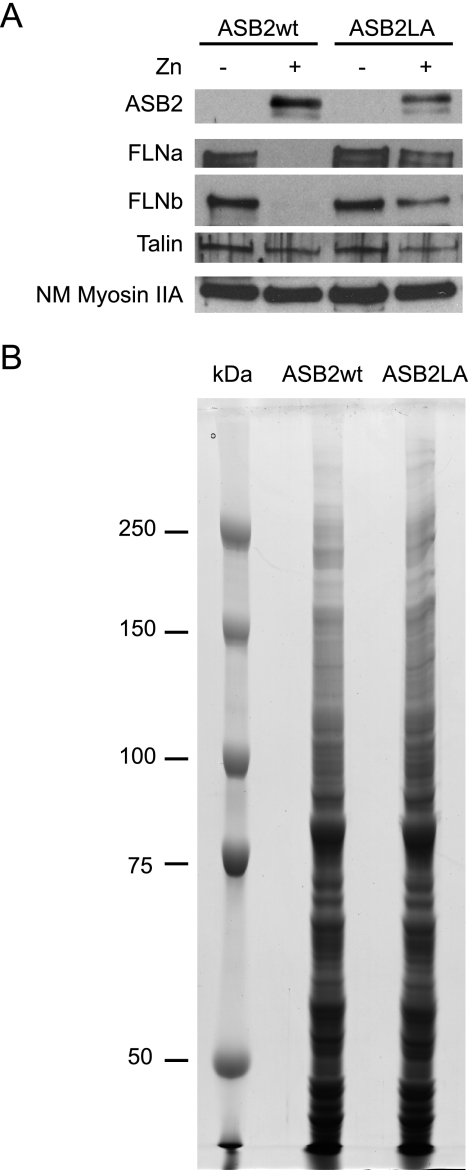
To identify proteins that are absent or less abundant in cells that express ASB2wt but that accumulate in cells expressing ASB2LA, PLB985/MT-FLAG-ASB2wt, and PLB985/MT-FLAG-ASB2LA, cells were incubated with or without ZnSO4 for 8 h. ASB2wt induced loss of both FLNa and -b (Fig. 3A). In contrast, no loss of another actin-binding protein, talin, or non-muscle myosin heavy chain IIA was observed in ASB2wt-expressing cells (Fig. 3A) showing ASB2 specificity for FLNa and -b. This is consistent with the fact that ASB2wt did not induce degradation of talin in transfected cells (23). Proteins from cytosolic extracts of ZnSO4-treated PLB985/MT-FLAG-ASB2wt and PLB985/MT-FLAG-ASB2LA cells (150 μg) were separated by SDS-PAGE and stained with colloidal Coomassie Blue (Fig. 3B). No obvious qualitative difference was observed between both lanes. Gel slices were cut for further trypsin-mediated protein digestion and LTQ-Orbitrap LC-MS/MS analysis. Proteins were identified using Mascot, and the automatic validation module of the MFPaQ software (29) was used to generate lists of proteins identified in both samples. Through the comparison of these two lists, the MFPaQ software produced three protein lists corresponding to (i) proteins found exclusively in ASB2wt-expressing cells, (ii) proteins found exclusively in ASB2LA-expressing cells, and (iii) proteins common to both samples (data not shown). These lists also contained the number of MS/MS spectra acquired that matched to all the peptides for each identified protein. When a protein was identified several times in consecutive gel slices, all the MS/MS spectra attributed to peptides belonging to the protein were taken into account. This information was added to the lists to have an indication of the respective abundance of the proteins in both samples. Because FLNa and FLNb belong to a protein family with high sequence homology that also includes FLNc (supplemental Data 3), they shared identical tryptic peptides. Thus, we considered only those spectrum-to-peptide matches that were unique to each FLN (supplemental Data 3). Although FLNa was detected in cells expressing either ASB2wt or ASB2LA, the spectral count number of peptides specific to FLNa was drastically lower in ASB2wt-expressing cells compared with ASB2LA-expressing cells (Table I). In contrast, no MS/MS spectrum corresponding to specific peptides of FLNb was acquired in the shotgun proteomics analysis performed with extracts of cells expressing ASB2wt, whereas some were detected in cells expressing ASB2LA (Table I). To estimate the relative abundance of FLNa in PLB985 cells induced to express ASB2LA and ASB2wt, the ratio of the number of MS/MS spectra of peptides specific to FLNa in ASB2LA-expressing cells to the number of MS/MS spectra of peptides specific to FLNa in ASB2wt-expressing cells was calculated. Indeed FLNa spectral count ratio was 11.3, whereas that of control Talin 1 was 1.2 (Table I). Furthermore a second experiment was performed leading to similar ratios (data not shown). Thus, the mean value of the FLNa spectral count ratio of these two analyses was 15.6 ± 6.1, whereas that of Talin 1 was 1.08 ± 0.02. Altogether spectral count ratios allowed us to identify FLNa and FLNb as potential substrates of ASB2 E3 ligase activity that are targeted for degradation.
Fig. 3.
ASB2 ubiquitin ligase activity triggers degradation of FLNa and -b in myeloid leukemia cells. PLB985/MT-FLAG-ASB2wt and PLB985/MT-FLAG-ASB2LA cells were left untreated (−) or were treated (+) with 100 μm ZnSO4 (Zn) for 8 h. A, cytosolic extracts (15 μg) were separated by SDS-PAGE and immunoblotted for ASB2, FLNa, FLNb, Talin 1, and non-muscle myosin heavy chain IIA (NM Myosin IIA). B, 150-μg aliquots of cytosolic extracts of PLB985/MT-FLAG-ASB2wt and PLB985/MT-FLAG-ASB2LA cells cultured with 100 μm ZnSO4 for 8 h were loaded on an SDS, 7% polyacrylamide gel. Proteins were revealed by colloidal Coomassie Blue staining. Molecular mass marker proteins were run in parallel.
Table I. Identification and label-free quantification of FLNa, FLNb, and Talin 1 in myeloid leukemia cells expressing wild-type or an E3 ubiquitin ligase-defective mutant of ASB2.
FLNa, FLNb, and Talin 1 were identified by shotgun proteomics analysis in cytosolic extracts of myeloid leukemia cells expressing wild-type ASB2 (wt) or the ASB2LA mutant (LA). Relative label-free quantification of these proteins was performed by spectral count. One representative experiment of two is shown. NA, not applicable.
| Protein | Swiss-Prot accession no. | Molecular mass | Identification |
Quantification |
|||||
|---|---|---|---|---|---|---|---|---|---|
| Mascot protein scorea |
Sequence coverageb |
Spectral countc |
Spectral count ratio (LA:wt) | ||||||
| wt | LA | wt | LA | wt | LA | ||||
| Da | % | ||||||||
| FLNa | P21333 | 280,564 | 1,270 | 5,941 | 28.8 | 66 | 79 | 893 | 11.3 |
| FLNb | O75369 | 278,021 | 107 | 417 | 1.8 | 13.9 | 0 | 43 | NA |
| Talin 1 | Q9Y490 | 269,599 | 8,232 | 6,908 | 69.9 | 61.8 | 1,146 | 1,265 | 1.1 |
a Mascot protein score obtained from the slice where the protein was identified with the best score.
b Corresponding sequence coverage.
c Number of total spectrum-to-peptide matches for all peptides specific to indicated proteins.
MS Intensity-based Label-free Quantification of FLNa Amount in ASB2-expressing Cells
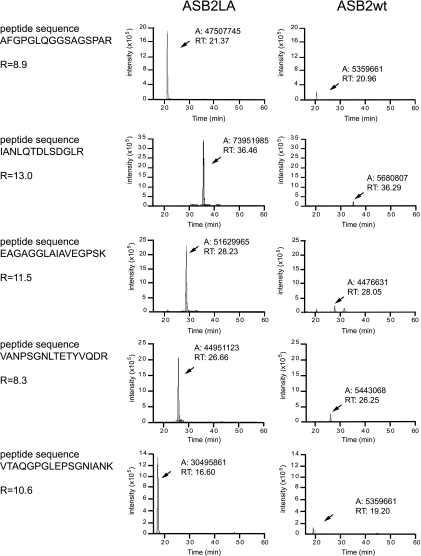
To further confirm the high ratio obtained with the spectral count method a label-free quantification was also performed by comparing the chromatographic peak area of specific peptides of FLNa and Talin 1 (Fig. 4 and supplemental Data 9 and 10). It was not possible to use this type of quantification for FLNb as no specific peptides could even be clearly detected in the MS spectra acquired during the shotgun analysis of extracts from ASB2wt-expressing cells. This quantification was performed with five selected specific peptides of each protein (see “Experimental Procedure” and supplemental Data 5, 6, 9, and 10). As shown in Fig. 4, the ratio of chromatographic peak area of peptides specific to FLNa in ASB2LA-expressing cells to chromatographic peak area of peptides specific to FLNa in ASB2wt-expressing cells indicated that the abundance of FLNa was drastically decreased in ASB2wt-expressing cells. To further quantify this ratio, biological and technical replicates were analyzed (Table II and supplemental Data 7 and 8). The mean ratio of chromatographic peak area of peptides specific to FLNa in ASB2LA-expressing cells to chromatographic peak area of peptides specific to FLNa in ASB2wt-expressing cells was 9.0 ± 1.9, whereas that of control Talin 1 was 1.0 ± 0.2 (Table II).
Fig. 4.
Quantification based on the chromatographic peak area of selected peptides specific to FLNa. Peak areas of the extracted ion chromatograms (XICs) of five selected peptides specific to FLNa were measured using the Interactive Chemical Information System peak detection method from the Xcalibur software. Peptide sequences are indicated on the left. For each FLNa peptide ion, XICs were extracted from the MS analysis of the slice where FLNa was identified with the best score (gel slice 6; see supplemental Data 5) from the cytosolic extracts of ASB2LA- (left) and ASB2wt (right)-expressing cells. The measured area (A) and the retention time (RT) of each chromatographic peak are specified. The ratio (R) indicated for each FLNa-specific peptide corresponds to the ratio of the chromatographic peak areas measured in the slice where FLNa was identified in ASB2LA-expressing cell extracts to the chromatographic peak area measured in the slice where FLNa was identified in ASB2wt-expressing cell extracts.
Table II . Relative amounts of FLNa and Talin 1 based on chromatographic peak areas of five selected specific peptides.
PLB985/MT-FLAG-ASB2wt and PLB985/MT-FLAG-ASB2LA cells were treated with 100 μm ZnSO4 for 8 h. Cytosolic cell extracts of two independent experiments were separated by SDS-PAGE. Filamin A and Talin 1 were identified by shotgun proteomics analysis, and their relative quantification was performed with the five specific peptides of each protein described in supplemental Data 7 and 8. For each peptide ion of FLNa and Talin 1, XICs were extracted from four MS analyses. Ratios indicated for each biological and technical replicate correspond to ratios of the sum of the chromatographic peak areas measured in slices where the protein was identified in ASB2LA-expressing cell extracts to the sum of the chromatographic peak area measured in slices where the protein was identified in ASB2wt-expressing cell extracts. a, b, c, and d are technical replicates from two biological replicates. Results are mean and S.D. from two independent experiments.
| Peptide | Experiment |
Average | |||
|---|---|---|---|---|---|
| a | b | c | d | ||
| FLNa | |||||
| EAGAGGLAIAVEGPSK | 4.6 | 8.4 | 8.0 | 8.0 | |
| IANLQTDLSDGLR | 4.8 | 9.5 | 9.2 | 8.9 | |
| VANPSGNLTETYVQDR | 8.2 | 8.8 | 12.1 | 12.5 | |
| AFGPGLQGGSAGSPAR | 7.1 | 11.2 | 11.9 | 11.9 | |
| VTAQGPGLEPSGNIANK | 6.6 | 10.1 | 9.3 | 9.3 | |
| 6.3 ± 1.5a | 9.6 ± 1.1a | 10.1 ± 1.8a | 10.1 ± 2.0a | 9.0 ± 1.9 | |
| Talin 1 | |||||
| LGAASLGAEDPETQVVLINAVK | 1.1 | 0.7 | 0.9 | 1.0 | |
| ERIPEAPAGPPSDFGLFLSDDDPK | 1.2 | 2.1 | 1.0 | 1.2 | |
| EADESLNFEEQILEAAK | 0.9 | 0.7 | 0.9 | 1.0 | |
| ILAQATSDLVNAIK | 0.5 | 1.0 | 0.8 | 1.3 | |
| LAQAAQSSVATITR | 0.6 | 0.8 | 0.9 | 0.9 | |
| 0.8 ± 0.3a | 1.1 ± 0.6a | 0.9 ± 0.1a | 1.1 ± 0.1a | 1.0 ± 0.2 | |
a Means and S.D. of the five different peptides of FLNa and Talin 1 in experiments a–d.
DISCUSSION
We have provided the development and testing of a strategy for the identification of potential substrates of E3s that are targeted to proteasome degradation. The robustness of this label-free quantitative proteomics approach was further highlighted by a drastic decrease of known ASB2 substrates, FLNa and FLNb, in cells that expressed wild-type ASB2.
In addition to genetics approaches and educated guesses, purification of interacting proteins is the classical way to identify substrates of E3s. However, in most cases, the covalent attachment of ubiquitin to proteins in the form of Lys-48-linked polyubiquitin chain leads rapidly to their degradation by the proteasome. Although proteasome inhibitors abolish substrate degradation, this treatment often leads to the formation of detergent-insoluble structures, named aggresomes, in which ubiquitylated proteins accumulate (30, 31). In this regard, it is noteworthy that our approach does not rely on the use of proteasome inhibitors and avoids difficulties linked to the extraction of proteins from insoluble structures such as aggresomes. Although two-hybrid screens have on occasion yielded E3-substrate interactions, this approach may be limited by the absence of post-translational modifications of the substrates that are required for their recognition by the E3. Our overall strategy is applicable to the proteomics analysis of cells expressing either a wild-type E3 or an E3 ligase-defective mutant in physiologically relevant settings and therefore represents a general strategy. In contrast to the purification of E3 ligase partners, this approach can be applied to low amounts of proteins. We note that we cannot exclude the possibility that decreased expression of a candidate protein following expression of a functional E3 is not due to its ubiquitylation. Because ubiquitin-mediated protein degradation is a highly dynamic process, induction of the E3 for a short period of time should reduce side effects due to expression of the E3. Nevertheless to conclusively demonstrate that a protein is a substrate of an E3, it is further necessary to demonstrate its ubiquitylation in vivo and/or in vitro. The proof of principle of this strategy is provided by our results highlighting FLNa and FLNb as potential substrates of ASB2. Indeed we have previously demonstrated that ASB2 can promote FLNa ubiquitylation using in vitro substrate ubiquitylation assays with highly purified proteins (23). Furthermore although our previous attempts to identify ASB2 substrates based on the identification of ASB2 partners such as yeast two-hybrid screens or immunoaffinity purification failed (data not shown), the global proteomics strategy developed here pointed to FLNa and FLNb as potential substrates of ASB2.
Mass spectrometry-based quantification methods have gained increasing popularity in proteomics over the past 5 years because of the gain in accuracy, but they are still technically challenging (32). One major approach is based on differential stable isotope labeling to create a specific tag that can be introduced into proteins or peptides in different ways. Alternatively label-free quantification methods that have recently emerged avoid the time-consuming and costly steps of introducing a label into proteins or peptides. Two major label-free protein quantification strategies are currently widely used: (i) spectral counting, counting the number of fragment spectra that identify peptides of a given protein (33), and (ii) peptide chromatographic peak intensity measurements, measuring the chromatographic peaks of peptide precursor ions belonging to a specific protein (34–36). The spectral counting approach was first used in this study because it is easy to implement in a shotgun proteomics strategy. Furthermore it can detect large changes between proteins (33). However, we are aware that this label-free approach is not the more accurate among the label-free mass spectrometric quantification techniques (37), but it clearly points to FLNa and FLNb as substrates of ASB2. Although the number of acquired spectra matching to FLNa is an indicator of its amount in ASB2wt- and ASB2LA-expressing cells, direct mass spectrometric signal intensity for several specific FLNa peptides further demonstrated the drastic decrease of FLNa in cells induced to express wild-type ASB2. Indeed the quantification based on chromatographic peak area of FLNa amount in myeloid leukemia cells expressing ASB2wt indicated that FLNa was dramatically reduced to 11.7 ± 2.4% of that observed in cells expressing an ASB2 mutant that is unable to stimulate FLNa degradation. This is in agreement with our previous results using qualitative and/or semiquantitative assays (23). It is noteworthy that our results identified FLNa, which is abundant, and FLNb, which is present at low levels, as ASB2 substrates in myeloid cells. Our approach will also distinguish spectrum-to-peptide matches that are unique for a particular protein from those common to different proteins. Indeed MS analysis has the advantage to allow the identification of peptides specific to a unique protein. Therefore, in contrast to approaches using antibodies for which the specificity is not always well known, MS approaches represent a tool to identify a particular protein among highly related proteins. Nevertheless the sorting of the specific peptides usually has to be performed manually because most of the database search algorithms are still limited in the exclusive assignments of these peptides to their corresponding proteins.
In summary, our study demonstrated the strength of label-free quantification using a highly sensitive fast MS instrument with high mass accuracy to identify ASB2 substrates. Our strategy will also aid future studies aimed to the identification of novel E3 substrates. Furthermore this study may result in a better understanding of ubiquitin-mediated protein degradation and its role in the onset of various human diseases.
Acknowledgments
We thank Prof. C. Moog-Lutz and D. Bouyssie for helpful discussions.
Footnotes
* This work was supported by the CNRS and Université Paul Sabatier and also by grants from the Agence Nationale de la Recherche (Programme Jeunes Chercheuses, Jeunes Chercheurs), the Comité Midi-Pyrénées de la Ligue contre le Cancer, the Association pour la Recherche sur le Cancer, and the Fondation pour la Recherche Médicale (to P. G. L.) and from the Agence Nationale de la Recherche (Plates-formes technologiques du vivant), the Fondation pour la Recherche Médicale (Programme Grands Equipements), and the Génopole Toulouse Midi-Pyrénées (to B. M.).
 The on-line version of this article (available at http://www.mcponline.org) contains supplemental material.
The on-line version of this article (available at http://www.mcponline.org) contains supplemental material.
1 The abbreviations used are:
- UPS
- ubiquitin-proteasome system
- APL
- acute promyelocytic leukemia
- ASB2
- ankyrin repeat-containing protein with a suppressor of cytokine signaling box 2
- E3
- E3 ubiquitin ligase
- FA
- formic acid
- FDR
- false discovery rate
- FLN
- filamin
- PML
- promyelocytic leukemia
- RA
- retinoic acid
- RARα
- retinoic acid receptor α
- RING
- really interesting new gene
- XIC
- extracted ion chromatogram
- E1
- ubiquitin-activating enzyme
- E2
- ubiquitin-conjugating enzyme
- LTQ
- linear trap quadrupole
- MFPaQ
- Mascot File Parsing and Quantification
- ASB2wt
- wild-type ASB2
- MT
- metallothionein.
REFERENCES
- 1.Glickman M. H., Ciechanover A. ( 2002) The ubiquitin-proteasome proteolytic pathway: destruction for the sake of construction. Physiol. Rev. 82, 373– 428 [DOI] [PubMed] [Google Scholar]
- 2.Pickart C. M. ( 2004) Back to the future with ubiquitin. Cell 116, 181– 190 [DOI] [PubMed] [Google Scholar]
- 3.Huibregtse J. M., Scheffner M., Beaudenon S., Howley P. M. ( 1995) A family of proteins structurally and functionally related to the E6-AP ubiquitin-protein ligase. Proc. Natl. Acad. Sci. U.S.A. 92, 2563– 2567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Scheffner M., Nuber U., Huibregtse J. M. ( 1995) Protein ubiquitination involving an E1-E2-E3 enzyme ubiquitin thioester cascade. Nature 373, 81– 83 [DOI] [PubMed] [Google Scholar]
- 5.Zheng N., Wang P., Jeffrey P. D., Pavletich N. P. ( 2000) Structure of a c-Cbl-UbcH7 complex: RING domain function in ubiquitin-protein ligases. Cell 102, 533– 539 [DOI] [PubMed] [Google Scholar]
- 6.Hatakeyama S., Nakayama K. I. ( 2003) U-box proteins as a new family of ubiquitin ligases. Biochem. Biophys. Res. Commun. 302, 635– 645 [DOI] [PubMed] [Google Scholar]
- 7.Petroski M. D., Deshaies R. J. ( 2005) Function and regulation of cullin-RING ubiquitin ligases. Nat. Rev. Mol. Cell Biol. 6, 9– 20 [DOI] [PubMed] [Google Scholar]
- 8.Jin J., Ang X. L., Shirogane T., Wade Harper J. ( 2005) Identification of substrates for F-box proteins. Methods Enzymol. 399, 287– 309 [DOI] [PubMed] [Google Scholar]
- 9.Melnick A., Licht J. D. ( 1999) Deconstructing a disease: RARalpha, its fusion partners, and their roles in the pathogenesis of acute promyelocytic leukemia. Blood 93, 3167– 3215 [PubMed] [Google Scholar]
- 10.Zelent A., Guidez F., Melnick A., Waxman S., Licht J. D. ( 2001) Translocations of the RARalpha gene in acute promyelocytic leukemia. Oncogene 20, 7186– 7203 [DOI] [PubMed] [Google Scholar]
- 11.Catalano A., Dawson M. A., Somana K., Opat S., Schwarer A., Campbell L. J., Iland H. ( 2007) The PRKAR1A gene is fused to RARA in a new variant acute promyelocytic leukemia. Blood 110, 4073– 4076 [DOI] [PubMed] [Google Scholar]
- 12.Rowley J. D., Golomb H. M., Dougherty C. ( 1977) 15/17 translocation, a consistent chromosomal change in acute promyelocytic leukaemia. Lancet 1, 549– 550 [DOI] [PubMed] [Google Scholar]
- 13.Grignani F., De Matteis S., Nervi C., Tomassoni L., Gelmetti V., Cioce M., Fanelli M., Ruthardt M., Ferrara F. F., Zamir I., Seiser C., Grignani F., Lazar M. A., Minucci S., Pelicci P. G. ( 1998) Fusion proteins of the retinoic acid receptor-alpha recruit histone deacetylase in promyelocytic leukaemia. Nature 391, 815– 818 [DOI] [PubMed] [Google Scholar]
- 14.Guidez F., Ivins S., Zhu J., Söderström M., Waxman S., Zelent A. ( 1998) Reduced retinoic acid-sensitivities of nuclear receptor corepressor binding to PML- and PLZF-RARalpha underlie molecular pathogenesis and treatment of acute promyelocytic leukemia. Blood 91, 2634– 2642 [PubMed] [Google Scholar]
- 15.Lin R. J., Nagy L., Inoue S., Shao W., Miller W. H., Jr, Evans R. M. ( 1998) Role of the histone deacetylase complex in acute promyelocytic leukaemia. Nature 391, 811– 814 [DOI] [PubMed] [Google Scholar]
- 16.Di Croce L., Raker V. A., Corsaro M., Fazi F., Fanelli M., Faretta M., Fuks F., Lo Coco F., Kouzarides T., Nervi C., Minucci S., Pelicci P. G. ( 2002) Methyltransferase recruitment and DNA hypermethylation of target promoters by an oncogenic transcription factor. Science 295, 1079– 1082 [DOI] [PubMed] [Google Scholar]
- 17.Warrell R. P., Jr, Frankel S. R., Miller W. H., Jr, Scheinberg D. A., Itri L. M., Hittelman W. N., Vyas R., Andreeff M., Tafuri A., Jakubowski A., Gabrilove J., Gordon M. S., Dmitrovsky E. N. ( 1991) Differentiation therapy of acute promyelocytic leukemia with tretinoin (all-trans-retinoic acid). N. Engl. J. Med. 324, 1385– 1393 [DOI] [PubMed] [Google Scholar]
- 18.Huang M. E., Ye Y. C., Chen S. R., Chai J. R., Lu J. X., Zhoa L., Gu L. J., Wang Z. Y. ( 1988) Use of all-trans retinoic acid in the treatment of acute promyelocytic leukemia. Blood 72, 567– 572 [PubMed] [Google Scholar]
- 19.Castaigne S., Chomienne C., Daniel M. T., Ballerini P., Berger R., Fenaux P., Degos L. ( 1990) All-trans retinoic acid as a differentiation therapy for acute promyelocytic leukemia. I. Clinical results. Blood 76, 1704– 1709 [PubMed] [Google Scholar]
- 20.Moog-Lutz C., Cavé-Riant F., Guibal F. C., Breau M. A., Di Gioia Y., Couraud P. O., Cayre Y. E., Bourdoulous S., Lutz P. G. ( 2003) JAML, a novel protein with characteristics of a junctional adhesion molecule, is induced during differentiation of myeloid leukemia cells. Blood 102, 3371– 3378 [DOI] [PubMed] [Google Scholar]
- 21.Guibal F. C., Moog-Lutz C., Smolewski P., Di Gioia Y., Darzynkiewicz Z., Lutz P. G., Cayre Y. E. ( 2002) ASB-2 inhibits growth and promotes commitment in myeloid leukemia cells. J. Biol. Chem. 277, 218– 224 [DOI] [PubMed] [Google Scholar]
- 22.Kohroki J., Fujita S., Itoh N., Yamada Y., Imai H., Yumoto N., Nakanishi T., Tanaka K. ( 2001) ATRA-regulated Asb-2 gene induced in differentiation of HL-60 leukemia cells. FEBS Lett. 505, 223– 228 [DOI] [PubMed] [Google Scholar]
- 23.Heuzé M. L., Lamsoul I., Baldassarre M., Lad Y., Lévêque S., Razinia Z., Moog-Lutz C., Calderwood D. A., Lutz P. G. ( 2008) ASB2 targets filamins A and B to proteasomal degradation. Blood 112, 5130– 5140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Heuzé M. L., Guibal F. C., Banks C. A., Conaway J. W., Conaway R. C., Cayre Y. E., Benecke A., Lutz P. G. ( 2005) ASB2 is an Elongin BC-interacting protein that can assemble with Cullin 5 and Rbx1 to reconstitute an E3 ubiquitin ligase complex. J. Biol. Chem. 280, 5468– 5474 [DOI] [PubMed] [Google Scholar]
- 25.Kohroki J., Nishiyama T., Nakamura T., Masuho Y. ( 2005) ASB proteins interact with Cullin5 and Rbx2 to form E3 ubiquitin ligase complexes. FEBS Lett. 579, 6796– 6802 [DOI] [PubMed] [Google Scholar]
- 26.Kamura T., Maenaka K., Kotoshiba S., Matsumoto M., Kohda D., Conaway R. C., Conaway J. W., Nakayama K. I. ( 2004) VHL-box and SOCS-box domains determine binding specificity for Cul2-Rbx1 and Cul5-Rbx2 modules of ubiquitin ligases. Genes Dev. 18, 3055– 3065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lutz P. G., Houzel-Charavel A., Moog-Lutz C., Cayre Y. E. ( 2001) Myeloblastin is an Myb target gene: mechanisms of regulation in myeloid leukemia cells growth-arrested by retinoic acid. Blood 97, 2449– 2456 [DOI] [PubMed] [Google Scholar]
- 28.Lutz P. G., Moog-Lutz C., Coumau-Gatbois E., Kobari L., Di Gioia Y., Cayre Y. E. ( 2000) Myeloblastin is a granulocyte colony-stimulating factor-responsive gene conferring factor-independent growth to hematopoietic cells. Proc. Natl. Acad. Sci. U.S.A. 97, 1601– 1606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bouyssié D., Gonzalez de Peredo A., Mouton E., Albigot R., Roussel L., Ortega N., Cayrol C., Burlet-Schiltz O., Girard J. P., Monsarrat B. ( 2007) Mascot file parsing and quantification (MFPaQ), a new software to parse, validate, and quantify proteomics data generated by ICAT and SILAC mass spectrometric analyses: application to the proteomics study of membrane proteins from primary human endothelial cells. Mol. Cell. Proteomics 6, 1621– 1637 [DOI] [PubMed] [Google Scholar]
- 30.Johnston J. A., Ward C. L., Kopito R. R. ( 1998) Aggresomes: a cellular response to misfolded proteins. J. Cell Biol. 143, 1883– 1898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.García-Mata R., Bebök Z., Sorscher E. J., Sztul E. S. ( 1999) Characterization and dynamics of aggresome formation by a cytosolic GFP-chimera. J. Cell Biol. 146, 1239– 1254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bantscheff M., Schirle M., Sweetman G., Rick J., Kuster B. ( 2007) Quantitative mass spectrometry in proteomics: a critical review. Anal. Bioanal. Chem. 389, 1017– 1031 [DOI] [PubMed] [Google Scholar]
- 33.Liu H., Sadygov R. G., Yates J. R., 3rd ( 2004) A model for random sampling and estimation of relative protein abundance in shotgun proteomics. Anal. Chem. 76, 4193– 4201 [DOI] [PubMed] [Google Scholar]
- 34.Bondarenko P. V., Chelius D., Shaler T. A. ( 2002) Identification and relative quantitation of protein mixtures by enzymatic digestion followed by capillary reversed-phase liquid chromatography-tandem mass spectrometry. Anal. Chem. 74, 4741– 4749 [DOI] [PubMed] [Google Scholar]
- 35.Wang W., Zhou H., Lin H., Roy S., Shaler T. A., Hill L. R., Norton S., Kumar P., Anderle M., Becker C. H. ( 2003) Quantification of proteins and metabolites by mass spectrometry without isotopic labeling or spiked standards. Anal. Chem. 75, 4818– 4826 [DOI] [PubMed] [Google Scholar]
- 36.Gao B. B., Stuart L., Feener E. P. ( 2008) Label-free quantitative analysis of 1D-PAGE LC/MS/MS proteome: application on angiotensin II stimulated smooth muscle cells secretome. Mol. Cell. Proteomics 7, 2399– 2409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Old W. M., Meyer-Arendt K., Aveline-Wolf L., Pierce K. G., Mendoza A., Sevinsky J. R., Resing K. A., Ahn N. G. ( 2005) Comparison of label-free methods for quantifying human proteins by shotgun proteomics. Mol. Cell. Proteomics 4, 1487– 1502 [DOI] [PubMed] [Google Scholar]