Abstract
Safe recombinant vaccines, based on a small number of antigenic proteins, are emerging as the most attractive, cost-effective solution against infectious diseases. In the present work, we confirmed previous data from our laboratory showing that whole viable bacterial cell treatment with proteases followed by the identification of released peptides by mass spectrometry is the method of choice for the rapid and reliable identification of vaccine candidates in Gram-positive bacteria. When applied to the Group B Streptococcus COH1 strain, 43 surface-associated proteins were identified, including all the protective antigens described in the literature as well as a new protective antigen, the cell wall-anchored protein SAN_1485 belonging to the serine-rich repeat protein family. This strategy overcomes the difficulties so far encountered in the identification of novel vaccine candidates and speeds up the entire vaccine discovery process by reducing the number of recombinant proteins to be tested in the animal model.
Vaccination is the safest, most attractive, and cost-effective solution to combat infectious diseases (1). Unfortunately vaccines against several pathogens are not yet available, and this is largely because of the difficulties encountered in the identification of the few pathogen components capable of eliciting protective immune responses.
Recently new genomics-based approaches have been described and shown to be very powerful for the discovery of vaccine candidates (2–4). However, these methods are labor-intensive and time-consuming in that the identification of the few protective antigens requires the screening of a large number of recombinant proteins in biological assays, which usually involve animal models. Therefore, the development of new strategies capable of substantially reducing the number of proteins to be tested would be highly desirable. Looking at the list of vaccines, either licensed or in advanced phase of development, that protect by eliciting antibody-mediated immunity, it appears that they include secreted toxins and/or highly expressed, surface-exposed molecules (5, 6). Hence the development of strategies capable of singling out this relatively small group of antigens from the plethora of pathogen components would substantially accelerate the vaccine discovery process.
We have recently proposed a novel proteomics-based approach, which has allowed the identification of Group A Streptococcus (GAS)1 proteins having domains protruding out of the bacterial surface (7). The approach is based on (i) the proteolytic treatment of bacteria under conditions that preserve cell viability and (ii) the analysis of the released peptides by mass spectrometry. The approach proved to be rapid and highly selective in that the large majority (>90%) of the identified proteins fell into the categories of cell wall proteins, lipoproteins, membrane proteins, and secreted proteins. Furthermore the method also allowed a semiquantitative evaluation of protein exposition and level of expression because, in general, the number of peptides identified from a given protein nicely correlates with the extent of its recognition by specific antibodies as judged by fluorescence-activated cell sorting analysis (7). Interestingly the list of surface-associated proteins included most of the published GAS protective antigens as well as new protective components such as the cell envelope proteinase Spy0416 (7), a protein attracting the interest of several laboratories for its important role in pathogenesis (8–10). To demonstrate that the proteomics-based approach represents a reliable and generally applicable strategy for the identification of vaccine components in Gram-positive bacteria, we have applied the same protocol to the Group B Streptococcus (GBS) for which a vaccine is not yet available on the market. GBS is a multiserotype Gram-positive opportunistic human pathogen that can lead to life-threatening infections in newborns and elderly adults (11–16).
Here we show that on the surface of the hypervirulent GBS COH1 strain there are 43 major proteins belonging to the families of cell wall proteins, lipoproteins, and membrane proteins. As in the case of GAS (7), the proteins identified comprise all protective antigens so far described in the literature (6, 17–26) as well as a new antigen, SAN_1485, which appears to be highly protective in the active maternal immunization mouse model. These data confirm the effectiveness of protease digestion coupled to mass spectrometry for the identification of surface-exposed antigens in Gram-positive bacteria and demonstrate the power of this technology for the rapid discovery of new vaccines.
EXPERIMENTAL PROCEDURES
Bacterial Surface Digestion
Streptococcus agalactiae serotype III COH1 strain was plated overnight in blood agar (Trypticase™ Soy Agar II with 5% sheep blood, BD Biosciences). Bacteria colonies were then grown at 37 °C in 200 ml of Todd-Hewitt broth (THB) in the presence of 5% CO2 until an A600 of 0.3 was reached. Bacteria were harvested by centrifugation at 3,500 × g for 10 min at 4 °C and washed twice with PBS. Cells were resuspended in 800 μl of PBS containing 40% sucrose (pH 7.4 for trypsin digestion and pH 6.0 for proteinase K digestion). Digestions were carried out with 10 μg of trypsin (Promega, Madison, WI) or 5 μg of proteinase K (Sigma) for 30 min at 37 °C. Bacterial cells were then spun down at 3,500 × g for 10 min at 4 °C, and the supernatants were filtered through 0.22-μm pore size filters (Millex, Millipore, Bedford, MA). Protease reactions were stopped with formic acid at 0.1% final concentration. Before analysis, PBS and sucrose were removed by an off-line desalting procedure using OASIS cartridges (Waters, Milford, MA) following the manufacturer's protocol. Desalted peptides were concentrated with a Centrivap Concentrator (Labconco, Kansas City, KS) and kept at −20 °C until further analysis.
Bacterial Surface Double Digestion
S. agalactiae serotype III COH1 strain was cultured and surface-digested as described above. Digestion supernatants were then denatured and reduced with Rapigest® (Waters) and 5 mm DTT at 100 °C, respectively, for 10 min. The pH was then adjusted to 8.0 using NH4HCO3, and an additional overnight proteolytic step with 2 μg of trypsin (Promega) at 37 °C was performed. The second digestion reaction was stopped with formic acid at 0.1% final concentration. The peptide mixtures were then desalted and concentrated as described above and stored at −20 °C until further analysis.
Protein Identification by Nano-LC/MS/MS
Peptides were separated by nano-LC on a NanoAcquity UPLC system (Waters) connected to a Q-ToF Premier ESI mass spectrometer equipped with a nanospray source (Waters). Samples were loaded onto a NanoAcquity 1.7-μm BEH130 C18 column (75 μm × 25 mm; Waters) through a NanoAcquity 5-μm Symmetry® C18 trap column (180 μm × 20 mm; Waters). Peptides were eluted with a 120-min gradient of 2–40% 98% acetonitrile, 0.1% formic acid solution at a flow rate of 250 nl/min. The eluted peptides were subjected to an automated data-dependent acquisition using the MassLynx software, version 4.1 (Waters) where an MS survey scan was used to automatically select multicharged peptides over the m/z ratio range of 300–2,000 for further MS/MS fragmentation. Up to eight different peptides were individually subjected to MS/MS fragmentation following each MS survey scan. For all samples, a second nano-LC/MS/MS analysis was carried out for the selective fragmentation of singly charged peptides. After data acquisition, individual MS/MS spectra were combined, smoothed, and centroided using ProteinLynx, version 3.5 (Waters) to obtain the peak list file. The Mascot Daemon application (Matrix Science Ltd., London, UK) was used for the automatic submission of data files to in-house licensed Mascot, version 2.2.1, running on a local server. Protein identification was achieved by searching in a locally curated database containing 52,399 sequences and 14,622,587 residues and obtained by combining protein sequence data derived from the eight completely sequenced GBS strains downloaded from the J. Craig Venter Institute and the S. agalactiae section of the NCBInr database. The Mascot search parameters were set to (i) 2 as the number of allowed missed cleavages (only for trypsin digestion), (ii) methionine oxidation and glutamine and asparagine deamidation as variable modifications, (iii) 0.3 Da as the peptide tolerance, and (iv) 0.3 Da as the MS/MS tolerance. Only significant hits were considered as defined by the Mascot scoring and probability system. The score thresholds for acceptance of peptide identification were ≥18 for trypsin digestion and ≥36 for proteinase K digestion.
Computational Analysis of Identified Proteins
A computational analysis of each identified protein sequence was performed with the PSORTb v.2.0 package (27) to predict the subcellular localization. For the detection of lipoprotein signal sequences and cleavage sites, LipPred (28) software was used.
Cloning, Expression, and Purification of GBS Protein SAN_1485
COH1 strain was used as the source of DNA for amplification of the identified surface antigen. The following PCR primers were designed to amplify the gene without predicted signal peptide coding sequences: 5′-GGAATTCCATATGGTCAAAAGTGTTATGACA-3′ (forward) and 5′-CTCTCTCAAGCTTTTCGGAGTTAGATACCG-3′ (reverse).
A PCR fragment from position 82 to 1739 downstream from the ATG codon of SAN_1485 was obtained. After digestion with NdeI and HindIII, the PCR product was introduced into the plasmid expression vector to generate His-tagged recombinant protein. His-tagged protein was obtained by cloning in pET21b+ vector (Novagen, Madison, WI), and Escherichia coli BL21DE3 cells (Novagen) were used as recipient. Procedures for protein expression and purification were as described previously (29).
Active Maternal Immunization
A maternal immunization/neonatal pup challenge model of GBS infection was used to verify the protective efficacy of SAN_1485 as described previously (6, 30). In brief, CD-1 female mice (6–8 weeks old) were immunized before breeding on days 1, 21, and 35. The mice received either PBS or 20 μg of protein/dose. Mice were bred 2–7 days after the last immunization. Within 48 h of birth, pups were injected intraperitoneally with 50 μl of GBS COH1 culture corresponding to LD90. Challenge inocula were prepared starting from frozen cultures diluted to the appropriate concentration with THB. Survival of pups was monitored for 2 days after challenge. Data were evaluated for statistical significance by Fisher's exact test (6).
RESULTS
Surface Proteome Analysis of GBS COH1 Revealed the Presence of 43 Surface-exposed Proteins
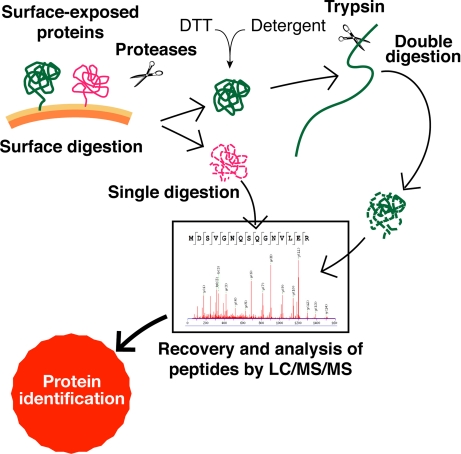
Fig. 1 illustrates the experimental approach used to identify the surface-exposed proteins of Group B Streptococcus COH1 strain. The method is an optimization of what has already been described for Group A Streptococcus (7). Whole bacteria were treated either with trypsin or proteinase K (bacterial “shaving” process), and the exposed protein domains and peptides released from the bacterial surface were collected by centrifugation and analyzed by MS/MS either directly or after a second protease treatment carried out under denaturing and reducing conditions. The second proteolytic digestion was added to the original protocol to allow the identification of those protein domains that, being particularly resistant to proteolysis, would otherwise remain undetected by LC/MS/MS. Protein identification was based on peptide matching between experimental MS/MS spectra and the theoretical spectra that were elaborated from a database containing the genome sequences of eight GBS strains and from the NCBInr database (S. agalactiae section; see “Experimental Procedures” for more details).
Fig. 1.
Representation of the proteomics strategy used to identify surface-exposed proteins. Peptides and polypeptides released into the supernatant by surface digestion were either directly analyzed by LC/MS/MS (single digestion) or subjected to a second step of digestion with trypsin after a denaturing and reducing treatment (double digestion). MS/MS spectra were then searched against a database containing protein sequence data derived from the eight completely sequenced GBS strains and from the S. agalactiae section of the NCBInr database for protein identification.
A total of 43 proteins were identified (Table I and supplemental Table 1S), 30 on the basis of two or more peptide matches and 13 with one peptide match only (peptide sequence details and Mascot scores are provided in supplemental Table 1S). Five of the 43 proteins identified were detected only after the second protease treatment.
Table I. Proteins identified on the surface of GBS COH1 strain.
The surface proteome of GBS COH1 strain is shown. Proteins are grouped according to their predicted subcellular localization. For each protein the following information is reported: NCBI gene ID, protein annotation and total number of identified peptides generated by either single or double digestion. See supplemental Tables 1S and 2S and supplemental material for details. ABC, ATP-binding cassette; RND, resistance-nodulation-cell division; MFP, membrane fusion proteins; CAMP, Christie, Atkins Munch-Peterson.
| Gene ID | Annotation | Single digestion | Double digestion | Total no. of identified peptides |
|---|---|---|---|---|
| Cell wall-anchoredproteins | ||||
| SAN_0509 | Reticulocyte-binding protein | ✓ | ✓ | 2 |
| SAN_0698 | Cell wall surface anchor family protein | ✓ | 1 | |
| SAN_1174 | Fibrinogen-binding protein | ✓ | 1 | |
| SAN_1369 | C5a peptidase precursor | ✓ | ✓ | 6 |
| SAN_1485 | Cell wall surface anchor family protein | ✓ | ✓ | 26 |
| SAN_1518 | Cell wall surface anchor family protein, putative | ✓ | 1 | |
| SAN_1577 | Cell wall surface anchor family protein | ✓ | 1 | |
| SAN_1578 | Amidase family protein | ✓ | 1 | |
| SAN_2207 | Pathogenicity protein, putative | ✓ | ✓ | 25 |
| Membrane proteins | ||||
| SAN_0172 | Conserved hypothetical protein | ✓ | 3 | |
| SAN_0480 | Serine protease (HtrA) | ✓ | 2 | |
| SAN_0545 | Unknown | ✓ | ✓ | 6 |
| SAN_0785 | Conserved hypothetical protein | ✓ | 5 | |
| SAN_1035 | Amino acid ABC transporter, binding protein | ✓ | 1 | |
| SAN_1460 | Efflux transporter, RND family, MFP subunit subfamily | ✓ | 2 | |
| SAN_1470 | Conserved hypothetical protein | ✓ | 3 | |
| SAN_1666 | Conserved hypothetical protein | ✓ | 1 | |
| Lipoproteins | ||||
| SAN_0273 | Protein of unknown function/lipoprotein, putative | ✓ | 2 | |
| SAN_0710 | Lipoprotein, putative | ✓ | 2 | |
| SAN_0850 | Protein of unknown function/lipoprotein, putative | ✓ | ✓ | 6 |
| SAN_1534 | Lipoprotein, putative | ✓ | 6 | |
| SAN_1636 | Peptidyl-prolyl cis-trans isomerase, cyclophilin-type | ✓ | ✓ | 2 |
| SAN_1898 | 5-Nucleotidase, lipoprotein e(P4) family | ✓ | 1 | |
| SAN_2137 | Lipoprotein, putative | ✓ | 1 | |
| SAN_2224 | Protein of unknown function/lipoprotein, putative | ✓ | ✓ | 2 |
| Extracellular proteins | ||||
| SAN_0024 | PcsB protein | ✓ | ✓ | 4 |
| SAN_0040 | Gro | ✓ | ✓ | 16 |
| SAN_0118 | N-Acetylmuramoyl-l-alanine amidase, family 4 protein | ✓ | 3 | |
| SAN_0317 | ABC transporter, substrate-binding protein | ✓ | ✓ | 18 |
| SAN_0872 | YaeC family protein | ✓ | ✓ | 4 |
| SAN_0970 | DNA entry nuclease | ✓ | 1 | |
| SAN_1132 | Iron compound ABC transporter, binding protein | ✓ | ✓ | 2 |
| SAN_1449 | Choline-binding protein D | ✓ | ✓ | 14 |
| SAN_1808 | N-Acetylmuramoyl-l-alanine amidase, family 4 | ✓ | ✓ | 9 |
| SAN_2140 | CAMP factor (CAMP-2) | ✓ | 1 | |
| Cytoplasmic proteins | ||||
| SAN_0083 | Ribosomal protein L2 (RplB) | ✓ | 5 | |
| SAN_0575 | Cell division protein DivIVA | ✓ | 2 | |
| SAN_0854 | Translation elongation factor Tu (Tuf) | ✓ | ✓ | 3 |
| SAN_1319 | Adherence and virulence protein A | ✓ | 1 | |
| SAN_1390 | Ribosomal protein L10 | ✓ | ✓ | 1 |
| Proteins with unpredictable subcellular localization | ||||
| SAN_1012 | Protein of unknown function/lipoprotein, putative | ✓ | 1 | |
| SAN_1360 | Protein of unknown function | ✓ | 1 | |
| SAN_2424 | Protein of unknown function | ✓ | 4 |
When the 43 proteins were subjected to computational analysis to establish their cellular compartmentalization as predicted by the PSORTb v2.0 (27) package in combination with the LipPred software (28) (see “Experimental Procedures” for details), eight proteins were classified as cell wall proteins (LPXTG motif-carrying proteins), nine were assigned to the transmembrane protein family carrying at least one transmembrane-spanning region, nine were lipoproteins, and eight were classified as secreted proteins (Table I). Of the remaining eight proteins, three had an unknown cellular compartmentalization, whereas the other five proteins were classified as cytoplasmic. However, these proteins have been found associated with the surface of many other bacterial species (31–35), and one of them, the adherence and virulence protein A, named SAN_1319, carries a fibronectin binding domain (the FbpA domain, PF05833) typically found in surface-exposed proteins (Table I and supplemental Table 1S). In conclusion, the proteins found associated with the GBS COH1 surface are fully consistent with their expected cellular location.
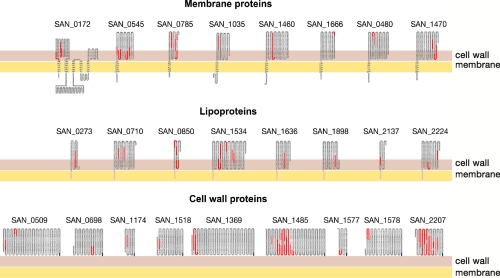
Also remarkably consistent are the positions of identified peptides with respect to the predicted topological organization of the proteins. Because proteases only have access to the protein regions protruding out of the cells, the peptides generated by proteolytic treatment are expected to reside within these regions. Indeed as shown in Fig. 2 , all peptides but one belong to domains that PSORTb predicts to be facing the external side of the bacterial membrane.
Fig. 2.
Schematic topological representation of each protein identified on the surface of COH1 GBS strain. Topological organization of the identified proteins was predicted using PSORTb information. The orange line represents the membrane, and the red line represents the cell wall. For lipoproteins, the lipoyl anchor is represented as a black segment embedded within the membrane, whereas for the cell wall-anchored proteins, the last C-terminal blue residues are the LPXTG signature anchored to the cell wall. The identified peptides are marked in red.
Similarly to what has been observed for Group A Streptococcus (7), the surface-exposed proteins experimentally found in COH1 strain represent only a fraction of the total number of membrane- and/or surface-associated proteins predicted from a computational analysis of the genome (Fig. 3). In particular, whereas the identified cell wall proteins and the lipoproteins constitute 40 and 20% of all predicted proteins, respectively, membrane proteins carrying one or more transmembrane-spanning regions (TMSRs) represent less than 2% of what PSORTb would predict.
Fig. 3.
COH1 GBS strain surface proteome. The 43 proteins belonging to the COH1 surface proteome were grouped into families based on their predicted cellular location. Red areas of each pie indicate the number of PSORTb-predicted proteins that were not found in the surface proteome, whereas the yellow areas represent the number of identified proteins belonging to each protein family.
Known Protective Antigens Are Part of the Surface Proteome
As already pointed out, vaccines known to elicit protective antibody responses are either secreted proteins or highly expressed, surface-exposed molecules. Therefore, we first asked whether the list of 43 proteins found on the surface of COH1 included the protective antigens so far described for GBS. Seven protective antigens have been reported so far (Table II): the C5a peptidase (SAN_1369) (17, 18), the Gro/surface immunogenic protein SIP (SAN_0040) (19, 20), the reticulocyte-binding protein/leucine-rich repeat protein (LrrG) (SAN_0509) (21), the protein Rib (22–24), and the three pilus variants PI-1, PI-2a, and PI-2b (6, 25, 26). The genes encoding these protective antigens are all present in the COH1 genome with the exception of the PI-2a pilus island; therefore six of the seven protective antigens were expected to be found on the surface of COH1. As shown in Table II, five of the six proteins, C5a peptidase, SIP, LrrG, and the backbone subunits of both PI-1 and PI-2a, were experimentally detected on the bacterial surface. The only missing protein was Rib, and this negative result was somehow unexpected because the protein is reported as being a conserved protective antigen (24). However, there is evidence that not all serotype III strains express Rib on their surface (22, 24), and this variability in surface expression might explain the absence of Rib in the COH1 surface proteome. To support this hypothesis, we measured Rib mRNA levels by RT-PCR in COH1 and in 2603V/R. The latter strain, belonging to serotype V, was chosen because the analysis of its surface proteins had revealed the presence of six peptides unequivocally belonging to the Rib protein.2 mRNA analysis demonstrated that Rib is differentially expressed in the two strains; the expression level was 10-fold higher in the 2603V/R strain with respect to COH1 (data not shown). This suggested that the inability to find Rib on the surface of COH1 is because of its poor expression in this strain.
Table II. Existing protective antigens for GBS.
The list shows reported Group B Streptococcus protective antigens. The presence or not on the COH1 genome and its identification on the surface proteome is reported for each antigen.
| Protein annotation (gene locusa) | Predicted protein localizationb | Presence of the gene in COH1 genome | Presence of the protein in COH1 surface proteome |
|---|---|---|---|
| C5a peptidase (SAN_1369) (17, 18) | Cell wall | Yes | Yes |
| SIP (SAN_0040) (19, 20) | Extracellular | Yes | Yes |
| LrrG (SAN_0509) (21) | Cell wall | Yes | Yes |
| PI-1 subunits (SAN_0698; SAN_0702) (5, 25, 26) | Cell wall | Yes | Yes (SAN_0698) |
| PI-2a subunits (5, 25, 26) | Cell wall | No | No |
| PI-2b subunits (SAN_1518; SAN_1519) (5, 25, 26) | Cell wall | Yes | Yes (SAN_1518) |
| Rib proteinc (22–24) | Cell wall | Yes | No |
aGene locus names according to NCBI and The Institute for Genomic Research.
btProtein localization as predicted by PSORTb v2.0.
ctThe COH1 genome is an unfinished sequence in “assembly” phase: 10 sequences are annotated as Rib proteins or surface protein Rib (SAN_0524, SAN_0528, SAN_2343, SAN_2398, SAN_2418, SAN_2435, SAN_2436, SAN_2437, SAN_2438, and SAN_2439). No peptide was identified from any of these sequences.
Surface Protein Analysis Revealed a Novel Protective Antigen
In addition to the protective antigens described above, the list of COH1 surface proteins includes a number of proteins that appeared to be abundantly expressed. For this reason, it was of interest to investigate whether some of these proteins might confer protection in the active maternal immunization model. In this context, one of these proteins, SAN_1485, appears to be particularly attractive. It is a 1085-amino acid-long protein with a cell wall-anchoring LPXTG motif and with a serine-rich repeat domain on its C-terminal part (Fig. 4). The protein was identified with as many as 25 peptide matches, indicating that it is highly expressed and exposed on the surface. All peptides fall in the N-terminal part of the molecule, the region that, in line with the topology of all cell wall-anchored proteins, is expected to be protruding out of the cell. No peptides belonging to the C-terminal serine-rich repeat domain were detected. On the basis of these data, SAN_1485 was expressed in E. coli as a C-terminal His tag fusion fragment starting from Val-28 up to Glu-580, and the purified protein was used for protection studies using the maternal immunization-neonatal pup challenge mouse model (see “Experimental Procedures” for details). As shown in Table III, over 80% of the offspring generated by immunized adult female mice survived an LD90 challenge dose of the two hypervirulent strains, COH1 and M781. Altogether these data confirm the power of surface proteome analysis in elucidating surface protein composition in Gram-positive bacteria and in identifying vaccine candidates.
Fig. 4.
Amino acid sequence of the novel protective antigen SAN_1485. The portions of the protein that were identified by surface digestion and mass spectrometry are marked in red. The gray background defines the region of the protein that was cloned and used for immunization. The LPXTG cell wall-anchoring motif is highlighted in the yellow box
Table III. Protective activity of the cell wall anchor family protein SAN_1485 compared with PBS as negative control and type III polysaccharide conjugated to CRM protein as positive control.
NT, not treated.
| Antigen | Challenge strains |
|||||
|---|---|---|---|---|---|---|
| COH1 |
M781 |
|||||
| No. of immunized mice | No. of mice that survived | Survival | No. of immunized mice | No. of mice that survived | Survival | |
| % | % | |||||
| SAN_1485 | 40 | 35 | 87 | 49 | 40 | 82 |
| CRM type III | NT | NT | NT | 45 | 43 | 95 |
| Adjuvant alone | 69 | 4 | 6 | 40 | 3 | 7 |
DISCUSSION
The main challenge for subunit-based vaccines is the identification of the microbial components capable of eliciting protective immune responses. Historically protective antigen identification has been carried out by fractionating microbial total extracts or culture supernatants using classical biochemical procedures and identifying those fractions containing specific components that cross-react with sera from convalescent patients. These components are then tested in appropriate in vitro/in vivo protection models to select the ones that finally reach human trials. This strategy for vaccine candidate selection has often proved to be laborious and time-consuming and relies on the assumption (not always true) that protective antigens are the ones that elicit immune responses during natural infections.
The advent of genomics and postgenomics, together with the development of high throughput technologies for gene cloning and expression and protein purification, have allowed researchers to take a brute force approach according to which libraries of recombinant proteins representing a large fraction of the entire proteome of any given pathogen can be produced and screened in appropriate surrogate of protection assays. This approach, first attempted by Ling Lissolo and co-workers (36), proved to be very effective for vaccine development against a number of bacterial pathogens, including Meningococcus B (5, 37), Group B Streptococcus (6), and Staphylococcus aureus (38), but also showed a few severe drawbacks. In particular, several hundred proteins for each pathogen have to be expressed, purified, and tested in appropriate animal models; this latter step was the real bottleneck of the entire process.
Because all subunit-based vaccines eliciting protective antibody responses fall in the category of secreted proteins and highly expressed, surface-exposed proteins (5, 6), a reliable experimental method capable of selectively identifying these two categories of antigens would substantially reduce the number of candidates to be tested in protection assays and ultimately speed up the whole vaccine discovery process. We have recently proposed a novel proteomics-based approach for the characterization of surface-exposed proteins in GAS that is based on the treatment of whole bacteria with proteases to selectively digest protein domains sticking out of the cell surface. The proteins identified in that study were all surface-associated and included most of the protective antigens described previously as well as a new potential vaccine candidate (7).
With the aim at demonstrating that this strategy can be effectively applied to other pathogens and become a general strategy for vaccine discovery in Gram-positive bacteria, we analyzed the surface proteome of the wild-type, hypervirulent, and highly encapsulated type III GBS strain COH1. The analysis led to the identification of 43 proteins, the large majority of which belong to the category of surface-associated and/or secreted proteins.
It is interesting to note that, for this analysis, we slightly modified the protocol previously described (7) for Group A Streptococcus to favor the generation of a sufficient amount of small peptides from highly stable surface proteins. The new protocol, which allowed the identification of five additional proteins, involves the addition of a reducing/denaturing step after protease treatment of the bacterial cells to further digest the large domains released after proteolysis that would otherwise be particularly recalcitrant to enzymatic hydrolysis. It is in fact known that surface proteins are by nature often structured to be resistant to harsh environmental conditions. This is, for example, the case of pilus proteins whose robustness and capacity to resist trypsin action was first observed more than 60 years ago by Lancefield and Dole (39). The mechanism that makes pilus subunits so stable has recently been elegantly elucidated by Baker and co-workers (40) who showed that these proteins form intramolecular isopeptidic bonds generated through an autocatalytic reaction. The authors have also proposed the formation of isopeptidic bonds as a general mechanism of stabilization of Gram-positive bacterial surface proteins in place of the disulfide bonds found in the Gram-negative counterparts.
Among the five proteins identified by double digestion only, SAN_0698 and SAN_1518 are known to be part of this highly organized, protease-resistant, pili structure (25, 26). This suggests that the other two proteins, i.e. SAN_1174 and SAN_1577, may also be highly structured proteins equally difficult to be cleaved in vivo. Remarkably the fifth protein identified by double digestion only is an in silico predicted cytoplasmic protein, the adherence and virulence protein A (SAN_1319). As reported above, this protein is probably involved in adhesion processes at the cell surface, and our data suggest a possible organization of this protein in a compact, protease-resistant folded structure.
As previously observed for Group A Streptococcus, the surface-exposed proteins experimentally identified in this study represent a minor fraction of what is expected to be associated with the membrane and cell wall. This is particularly true for proteins carrying one or more TMSRs: according to PSORTb, this family includes 440 proteins, but our experimental data indicate that only eight of them are surface-exposed under the growth conditions we used. At present, this apparent discrepancy between predictive analysis and experimental data is difficult to explain and requires further investigation. It is expected that a few of these proteins are not correctly assigned to the membrane compartment, particularly those carrying only one TMSR. Furthermore a non-negligible fraction of these proteins might not have been identified because of their poor expression and/or because their relatively small size does not allow them to cross the cell wall and the polysaccharidic capsule. However, poor expression and small size of external domains do not appear to be the only explanations for failing to find the proteins on the bacterial surface. In fact, we have recently accumulated data on the global transcription profile of GBS grown in THB or isolated from the blood of infected mice. Under both conditions, over 50% of the genes encoding predicted membrane proteins were either sufficiently well expressed (above the median value) or highly expressed.3 Furthermore the shortest of the eight experimentally identified membrane proteins is 188 amino acids long, indicating that proteins with extracellular domains above this size should be potentially capable of crossing both the cell wall and the capsule. A computational analysis indicates that more than half of the membrane proteins have domains longer than 200 amino acids, suggesting that structural constraints, in addition to protein size, determine the extent of protein exposition. We hypothesize that membrane proteins in Gram-positive bacteria are specifically designed to remain hidden under the thick coat of cell wall and capsule.
The list of identified proteins includes most of the protective antigens already described for GBS; the only protective antigen not present in the list is Rib. The explanation resides in the fact that although COH1 carries the rib gene (41) the gene is poorly expressed in this strain as demonstrated by RT-PCR. This is consistent with a previous report showing that Rib expression is variable among GBS strains belonging to serotype III. These data, in addition to explaining the reasons for not finding Rib on the COH1 surface, demonstrate the power of the surface protein analysis proposed here in providing a semiquantitative indication of the level of expression and surface exposition of each protein. This is particularly important for vaccine purposes because, as already pointed out, good vaccine candidates fall in the category of highly expressed and exposed proteins.
To the best of our knowledge, a number of proteins included in our list of surface-exposed antigens have never been tested in protection studies. It was therefore of interest to verify whether some of them could elicit protective immunity in the active maternal mouse model (6, 30). We focused our attention on one specific antigen, SAN_1485, which appeared particularly attractive because the presence of its gene has been recently associated with the hypervirulent genetic lineage restriction digest pattern III-3/ST-17 (42). This hypervirulent lineage is strongly associated with invasive infections in neonates (43, 44) and therefore of potential interest from a vaccine standpoint. Because the SAN_1485 is a 1085-amino acid protein with a molecular mass of 113 kDa, we decided to test the protective activity of only a fragment of the protein, the selection of which was guided by the result of the mass spectrometry analysis. The protein was in fact identified by the matching of 25 different peptides, all localized in the N-terminal portion of the protein. We therefore expressed a 550-amino acid-long fragment spanning the molecule from amino acid 28 to amino acid 580. The fragment turned out to protect over 80% of the pups challenged with either COH1 or M781; this is a protection level comparable to that obtained with the type III polysaccharide conjugated to CRM197 (Table III). Therefore, as was the case for Group A Streptococcus, the surface proteome analysis led to a rapid detection of a new, promising protective antigen against Group B Streptococcus infections.
Our method exploits the robustness of the Gram-positive bacterial cells provided by the thickness of the peptidoglycan layer. Therefore, although of general use for defining the composition of surface-exposed proteins in Gram-positive bacteria, the protocol might not be ideal for studying surface proteins in Gram-negative bacteria. Indeed cells lysis was observed when we treated Neisseria meningitidis and pathogenic E. coli strains with proteases under different experimental conditions.4 For the identification of outer membrane proteins in Gram-negative bacteria, we have recently proposed a new strategy that appears to be highly specific (45, 46). The strategy is based on the selection of mutants that spontaneously release to the growth medium high amounts of outer membrane vesicles (OMVs). The OMV production appears to be the result of membrane budding, and therefore OMVs are a bona fide representation of the protein composition of the bacterial membrane and periplasmic compartment that is embedded during the vesicle formation (47, 48). The proteomics characterization of OMVs produced by a N. meningitidis Δgna33 mutant strain and by a meningitis-associated E. coli IHE3034 ΔtolR strain revealed that all the identified proteins belong to the outer membrane and the periplasmic compartments (45, 46). Furthermore the most protective N. meningitidis antigens identified using a reverse vaccinology approach (5) and now in clinical trials (37) were also found to be part of the OMV proteomics analysis (45), highlighting the usefulness of OMV protein characterization as an effective and rapid approach to vaccine candidate discovery.
The advantage of using the proteomics-based strategy proposed here over genomics approaches such as “reverse vaccinology” (2, 49, 50) or “immunomics” (51, 52) for vaccine discovery deserves our last comment. The time required to complete the surface protein analysis of a single strain usually takes a very short time. Therefore, several strains can be processed so that a list of a few tens of surface-exposed proteins can be generated in a limited time frame. The identified proteins can also be ranked on the basis of their level of expression and conservation among the panel of strains analyzed, and therefore the most abundant and conserved proteins can be singled out and tested in protection studies in the appropriate in vitro/in vivo model. In practical terms, this implies that, in general, less than 30 proteins have to be cloned, expressed, and tested in vaccination experiments to select the best candidates to bring to development. If one takes into consideration that when reverse vaccinology was used to identify the protective antigens in N. meningitidis and Group B Streptococcus, 350 and 589 proteins, respectively, were screened using laborious and time-consuming assays (5, 6), the superiority of the surface proteome analysis over genomics approaches appears to be overwhelming.
Acknowledgments
CRM protein conjugated with GBS type III polysaccharide was a kind gift from Paolo Costantino and Francesco Berti from Novartis Vaccines and Diagnostics. We thank Antonietta Maiorino for expert secretarial assistance and Giorgio Corsi for artwork.
Footnotes
 The on-line version of this article (available at http://www.mcponline.org) contains supplemental material.
The on-line version of this article (available at http://www.mcponline.org) contains supplemental material.
2 E. Altindis, personal communication.
3 I. Margarit, personal communication
4 N. Norais, personal communication
1 The abbreviations used are:
- GAS
- Group A Streptococcus
- GBS
- Group B Streptococcus
- THB
- Todd-Hewitt broth
- NCBInr
- National Center for Biotechnology Information non-redundant
- TMSR
- transmembrane-spanning region
- SIP
- surface immunogenic protein
- Lrr
- leucine-rich repeat
- OMV
- outer membrane vesicle
- CRM
- cross-reactive material.
REFERENCES
- 1.Anonymous The value of vaccines. Nat. Rev. Microbiol. 200862 [Google Scholar]
- 2.Rappuoli R. ( 2000) Reverse vaccinology. Curr. Opin. Microbiol. 3, 445– 450 [DOI] [PubMed] [Google Scholar]
- 3.Grandi G. ( 2006) Genomics and proteomics in reverse vaccines. Methods Biochem. Anal. 49, 379– 393 [DOI] [PubMed] [Google Scholar]
- 4.Johri A. K., Paoletti L. C., Glaser P., Dua M., Sharma P. K., Grandi G., Rappuoli R. ( 2006) Group B Streptococcus: global incidence and vaccine development. Nat. Rev. Microbiol. 4, 932– 942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pizza M., Scarlato V., Masignani V., Giuliani M. M., Aricò B., Comanducci M., Jennings G. T., Baldi L., Bartolini E., Capecchi B., Galeotti C. L., Luzzi E., Manetti R., Marchetti E., Mora M., Nuti S., Ratti G., Santini L., Savino S., Scarselli M., Storni E., Zuo P., Broeker M., Hundt E., Knapp B., Blair E., Mason T., Tettelin H., Hood D. W., Jeffries A. C., Saunders N. J., Granoff D. M., Venter J. C., Moxon E. R., Grandi G., Rappuoli R. ( 2000) Identification of vaccine candidates against serogroup B meningococcus by whole-genome sequencing. Science 287, 1816– 1820 [DOI] [PubMed] [Google Scholar]
- 6.Maione D., Margarit I., Rinaudo C. D., Masignani V., Mora M., Scarselli M., Tettelin H., Brettoni C., Iacobini E. T., Rosini R., D'Agostino N., Miorin L., Buccato S., Mariani M., Galli G., Nogarotto R., Nardi Dei V., Vegni F., Fraser C., Mancuso G., Teti G., Madoff L. C., Paoletti L. C., Rappuoli R., Kasper D. L., Telford J. L., Grandi G. ( 2005) Identification of a universal Group B streptococcus vaccine by multiple genome screen. Science 309, 148– 150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rodríguez-Ortega M. J., Norais N., Bensi G., Liberatori S., Capo S., Mora M., Scarselli M., Doro F., Ferrari G., Garaguso I., Maggi T., Neumann A., Covre A., Telford J. L., Grandi G. ( 2006) Characterization and identification of vaccine candidate proteins through analysis of the group A Streptococcus surface proteome. Nat. Biotechnol. 24, 191– 197 [DOI] [PubMed] [Google Scholar]
- 8.Fernandez-Espla M. D., Garault P., Monnet V., Rul F. ( 2000) Streptococcus thermophilus cell wall-anchored proteinase: release, purification, and biochemical and genetic characterization. Appl. Environ. Microbiol. 66, 4772– 4778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Harris T. O., Shelver D. W., Bohnsack J. F., Rubens C. E. ( 2003) A novel streptococcal surface protease promotes virulence, resistance to opsonophagocytosis, and cleavage of human fibrinogen. J. Clin. Investig. 111, 61– 70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lei B., Mackie S., Lukomski S., Musser J. M. ( 2000) Identification and immunogenicity of group A Streptococcus culture supernatant proteins. Infect. Immun. 68, 6807– 6818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schuchat A., Wenger J. D. ( 1994) Epidemiology of group B streptococcal disease. Risk factors, prevention strategies, and vaccine development. Epidemiol. Rev. 16, 374– 402 [DOI] [PubMed] [Google Scholar]
- 12.Tyrrell G. J., Senzilet L. D., Spika J. S., Kertesz D. A., Alagaratnam M., Lovgren M., Talbot J. A. ( 2000) Invasive disease due to group B streptococcal infection in adults: results from a Canadian, population-based, active laboratory surveillance study—1996. Sentinel Health Unit Surveillance System Site Coordinators. J. Infect. Dis. 182, 168– 173 [DOI] [PubMed] [Google Scholar]
- 13.Harrison L. H., Elliott J. A., Dwyer D. M., Libonati J. P., Ferrieri P., Billmann L., Schuchat A. ( 1998) Serotype distribution of invasive group B streptococcal isolates in Maryland: implications for vaccine formulation. Maryland Emerging Infections Program. J. Infect. Dis. 177, 998– 1002 [DOI] [PubMed] [Google Scholar]
- 14.Hansen S. M., Uldbjerg N., Kilian M., Sørensen U. B. ( 2004) Dynamics of Streptococcus agalactiae colonization in women during and after pregnancy and in their infants. J. Clin. Microbiol. 42, 83– 89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dermer P., Lee C., Eggert J., Few B. ( 2004) A history of neonatal group B streptococcus with its related morbidity and mortality rates in the United States. J. Pediatr. Nurs. 19, 357– 363 [DOI] [PubMed] [Google Scholar]
- 16.Edwards M. S., Baker C. J. ( 2005) Group B streptococcal infections in elderly adults. Clin. Infect. Dis. 41, 839– 847 [DOI] [PubMed] [Google Scholar]
- 17.Cheng Q., Carlson B., Pillai S., Eby R., Edwards L., Olmsted S. B., Cleary P. ( 2001) Antibody against surface-bound C5a peptidase is opsonic and initiates macrophage killing of group B streptococci. Infect. Immun. 69, 2302– 2308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cheng Q., Debol S., Lam H., Eby R., Edwards L., Matsuka Y., Olmsted S. B., Cleary P. P. ( 2002) Immunization with C5a peptidase or peptidase-type III polysaccharide conjugate vaccines enhances clearance of group B Streptococci from lungs of infected mice. Infect. Immun. 70, 6409– 6415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Brodeur B. R., Boyer M., Charlebois I., Hamel J., Couture F., Rioux C. R., Martin D. ( 2000) Identification of group B streptococcal Sip protein, which elicits cross-protective immunity. Infect. Immun. 68, 5610– 5618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Martin D., Rioux S., Gagnon E., Boyer M., Hamel J., Charland N., Brodeur B. R. ( 2002) Protection from group B streptococcal infection in neonatal mice by maternal immunization with recombinant Sip protein. Infect. Immun. 70, 4897– 4901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Seepersaud R., Hanniffy S. B., Mayne P., Sizer P., Le Page R., Wells J. M. ( 2005) Characterization of a novel leucine-rich repeat protein antigen from group B streptococci that elicits protective immunity. Infect. Immun. 73, 1671– 1683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Larsson C., Stålhammar-Carlemalm M., Lindahl G. ( 1996) Experimental vaccination against group B streptococcus, an encapsulated bacterium, with highly purified preparations of cell surface proteins Rib and alpha. Infect. Immun. 64, 3518– 3523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Larsson C., Stålhammar-Carlemalm M., Lindahl G. ( 1999) Protection against experimental infection with group B streptococcus by immunization with a bivalent protein vaccine. Vaccine 17, 454– 458 [DOI] [PubMed] [Google Scholar]
- 24.Stålhammar-Carlemalm M., Stenberg L., Lindahl G. ( 1993) Protein rib: a novel group B streptococcal cell surface protein that confers protective immunity and is expressed by most strains causing invasive infections. J. Exp. Med. 177, 1593– 1603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lauer P., Rinaudo C. D., Soriani M., Margarit I., Maione D., Rosini R., Taddei A. R., Mora M., Rappuoli R., Grandi G., Telford J. L. ( 2005) Genome analysis reveals pili in Group B Streptococcus. Science 309105. [DOI] [PubMed] [Google Scholar]
- 26.Rosini R., Rinaudo C. D., Soriani M., Lauer P., Mora M., Maione D., Taddei A., Santi I., Ghezzo C., Brettoni C., Buccato S., Margarit I., Grandi G., Telford J. L. ( 2006) Identification of novel genomic islands coding for antigenic pilus-like structures in Streptococcus agalactiae. Mol. Microbiol. 61, 126– 141 [DOI] [PubMed] [Google Scholar]
- 27.Gardy J. L., Laird M. R., Chen F., Rey S., Walsh C. J., Ester M., Brinkman F. S. ( 2005) PSORTb v.2.0: expanded prediction of bacterial protein subcellular localization and insights gained from comparative proteome analysis. Bioinformatics 21, 617– 623 [DOI] [PubMed] [Google Scholar]
- 28.Taylor P. D., Toseland C. P., Attwood T. K., Flower D. R. ( 2006) LIPPRED: a web server for accurate prediction of lipoprotein signal sequences and cleavage sites. Bioinformation 1, 176– 179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Montigiani S., Falugi F., Scarselli M., Finco O., Petracca R., Galli G., Mariani M., Manetti R., Agnusdei M., Cevenini R., Donati M., Nogarotto R., Norais N., Garaguso I., Nuti S., Saletti G., Rosa D., Ratti G., Grandi G. ( 2002) Genomic approach for analysis of surface proteins in Chlamydia pneumoniae. Infect. Immun. 70, 368– 379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rodewald A. K., Onderdonk A. B., Warren H. B., Kasper D. L. ( 1992) Neonatal mouse model of group B streptococcal infection. J. Infect. Dis. 166, 635– 639 [DOI] [PubMed] [Google Scholar]
- 31.Marques M. A., Chitale S., Brennan P. J., Pessolani M. C. ( 1998) Mapping and identification of the major cell wall-associated components of Mycobacterium leprae. Infect. Immun. 66, 2625– 2631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dallo S. F., Kannan T. R., Blaylock M. W., Baseman J. B. ( 2002) Elongation factor Tu and E1 beta subunit of pyruvate dehydrogenase complex act as fibronectin binding proteins in Mycoplasma pneumoniae. Mol. Microbiol. 46, 1041– 1051 [DOI] [PubMed] [Google Scholar]
- 33.Spence J. M., Clark V. L. ( 2000) Role of ribosomal protein L12 in gonococcal invasion of Hec1B cells. Infect. Immun. 68, 5002– 5010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kurar E., Splitter G. A. ( 1997) Nucleic acid vaccination of Brucella abortus ribosomal L7/L12 gene elicits immune response. Vaccine 15, 1851– 1857 [DOI] [PubMed] [Google Scholar]
- 35.Padmalayam I., Anderson B., Kron M., Kelly T., Baumstark B. ( 1997) The 75-kilodalton antigen of Bartonella bacilliformis is a structural homolog of the cell division protein FtsZ. J. Bacteriol. 179, 4545– 4552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Grandi G. ( 2001) Antibacterial vaccine design using genomics and proteomics. Trends Biotechnol. 19, 181– 188 [DOI] [PubMed] [Google Scholar]
- 37.Giuliani M. M., Adu-Bobie J., Comanducci M., Aricò B., Savino S., Santini L., Brunelli B., Bambini S., Biolchi A., Capecchi B., Cartocci E., Ciucchi L., Di Marcello F., Ferlicca F., Galli B., Luzzi E., Masignani V., Serruto D., Veggi D., Contorni M., Morandi M., Bartalesi A., Cinotti V., Mannucci D., Titta F., Ovidi E., Welsch J. A., Granoff D., Rappuoli R., Pizza M. ( 2006) A universal vaccine for serogroup B meningococcus. Proc. Natl. Acad. Sci. U.S.A 103, 10834– 10839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Stranger-Jones Y. K., Bae T., Schneewind O. ( 2006) Vaccine assembly from surface proteins of Staphylococcus aureus. Proc. Natl. Acad. Sci. U.S.A 103, 16942– 16947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lancefield R. C., Dole V. P. ( 1946) The properties of T antigens extracted from group A hemolytic streptococci. J. Exp. Med. 84, 449– 471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kang H. J., Coulibaly F., Clow F., Proft T., Baker E. N. ( 2007) Stabilizing isopeptide bonds revealed in gram-positive bacterial pilus structure. Science 318, 1625– 1628 [DOI] [PubMed] [Google Scholar]
- 41.Tettelin H., Masignani V., Cieslewicz M. J., Donati C., Medini D., Ward N. L., Angiuoli S. V., Crabtree J., Jones A. L., Durkin A. S., Deboy R. T., Davidsen T. M., Mora M., Scarselli M., Margarit y Ros I., Peterson J. D., Hauser C. R., Sundaram J. P., Nelson W. C., Madupu R., Brinkac L. M., Dodson R. J., Rosovitz M. J., Sullivan S. A., Daugherty S. C., Haft D. H., Selengut J., Gwinn M. L., Zhou L., Zafar N., Khouri H., Radune D., Dimitrov G., Watkins K., O'Connor K. J., Smith S., Utterback T. R., White O., Rubens C. E., Grandi G., Madoff L. C., Kasper D. L., Telford J. L., Wessels M. R., Rappuoli R., Fraser C. M.Proc. Natl. Acad. Sci. U.S.A. (2005)102,13950– 13955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Seifert K. N., Adderson E. E., Whiting A. A., Bohnsack J. F., Crowley P. J., Brady L. J. ( 2006) A unique serine-rich repeat protein (Srr-2) and novel surface antigen (epsilon) associated with a virulent lineage of serotype III Streptococcus agalactiae. Microbiology 152, 1029– 1040 [DOI] [PubMed] [Google Scholar]
- 43.Jones N., Bohnsack J. F., Takahashi S., Oliver K. A., Chan M. S., Kunst F., Glaser P., Rusniok C., Crook D. W., Harding R. M., Bisharat N., Spratt B. G. ( 2003) Multilocus sequence typing system for group B streptococcus. J. Clin. Microbiol. 41, 2530– 2536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Luan S. L., Granlund M., Sellin M., Lagergård T., Spratt B. G., Norgren M. ( 2005) Multilocus sequence typing of Swedish invasive group B streptococcus isolates indicates a neonatally associated genetic lineage and capsule switching. J. Clin. Microbiol. 43, 3727– 3733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ferrari G., Garaguso I., Adu-Bobie J., Doro F., Taddei A. R., Biolchi A., Brunelli B., Giuliani M. M., Pizza M., Norais N., Grandi G. ( 2006) Outer membrane vesicles from group B Neisseria meningitidis delta gna33 mutant: proteomic and immunological comparison with detergent-derived outer membrane vesicles. Proteomics 6, 1856– 1866 [DOI] [PubMed] [Google Scholar]
- 46.Berlanda Scorza F., Doro F., Rodríguez-Ortega M. J., Stella M., Liberatori S., Taddei A. R., Serino L., Gomes Moriel D., Nesta B., Fontana M. R., Spagnuolo A., Pizza M., Norais N., Grandi G. ( 2008) Proteomics characterization of outer membrane vesicles from the extraintestinal pathogenic Escherichia coli DeltatolR IHE3034 mutant. Mol. Cell. Proteomics 7, 473– 485 [DOI] [PubMed] [Google Scholar]
- 47.Bernadac A., Gavioli M., Lazzaroni J. C., Raina S., Lloubès R. ( 1998) Escherichia coli tol-pal mutants form outer membrane vesicles. J. Bacteriol. 180, 4872– 4878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yem D. W., Wu H. C. ( 1978) Physiological characterization of an Escherichia coli mutant altered in the structure of murein lipoprotein. J. Bacteriol. 133, 1419– 1426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Masignani V., Rappuoli R., Pizza M. ( 2002) Reverse vaccinology: a genome-based approach for vaccine development. Expert Opin. Biol. Ther. 2, 895– 905 [DOI] [PubMed] [Google Scholar]
- 50.Mora M., Veggi D., Santini L., Pizza M., Rappuoli R. ( 2003) Reverse vaccinology. Drug. Discov. Today 8, 459– 464 [DOI] [PubMed] [Google Scholar]
- 51.Etz H., Minh D. B., Henics T., Dryla A., Winkler B., Triska C., Boyd A. P., Söllner J., Schmidt W., von Ahsen U., Buschle M., Gill S. R., Kolonay J., Khalak H., Fraser C. M., von Gabain A., Nagy E., Meinke A. ( 2002) Identification of in vivo expressed vaccine candidate antigens from Staphylococcus aureus. Proc. Natl. Acad. Sci. U.S.A 99, 6573– 6578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Henics T., Winkler B., Pfeifer U., Gill S. R., Buschle M., von Gabain A., Meinke A. L.( 2003) Small-fragment genomic libraries for the display of putative epitopes from clinically significant pathogens. BioTechniques 35, 196– 202204, 206 passim [DOI] [PubMed] [Google Scholar]