Abstract
Members of the human protein kinase superfamily are the major regulatory enzymes involved in the activity control of eukaryotic signal transduction pathways. As protein kinases reside at the nodes of phosphorylation-based signal transmission, comprehensive analysis of their cellular expression and site-specific phosphorylation can provide important insights into the architecture and functionality of signaling networks. However, in global proteome studies, low cellular abundance of protein kinases often results in rather minor peptide species that are occluded by a vast excess of peptides from other cellular proteins. These analytical limitations create a rationale for kinome-wide enrichment of protein kinases prior to mass spectrometry analysis. Here, we employed stable isotope labeling by amino acids in cell culture (SILAC) to compare the binding characteristics of three kinase-selective affinity resins by quantitative mass spectrometry. The evaluated pre-fractionation tools possessed pyrido[2,3-d]pyrimidine-based kinase inhibitors as immobilized capture ligands and retained considerable subsets of the human kinome. Based on these results, an affinity resin displaying the broadly selective kinase ligand VI16832 was employed to quantify the relative expression of more than 170 protein kinases across three different, SILAC-encoded cancer cell lines. These experiments demonstrated the feasibility of comparative kinome profiling in a compact experimental format. Interestingly, we found high levels of cytoplasmic and low levels of receptor tyrosine kinases in MV4–11 leukemia cells compared with the adherent cancer lines HCT116 and MDA-MB-435S. The VI16832 resin was further exploited to pre-fractionate kinases for targeted phosphoproteomics analysis, which revealed about 1200 distinct phosphorylation sites on more than 200 protein kinases. This hitherto largest survey of site-specific phosphorylation across the kinome significantly expands the basis for functional follow-up studies on protein kinase regulation. In conclusion, the straightforward experimental procedures described here enable different implementations of kinase-selective proteomics with considerable potential for future signal transduction and kinase drug target analysis.
Reversible protein phosphorylation represents the most common type of post-translational modification (PTM)1 in eukaryotic organisms. A plethora of studies on a large variety of proteins have established that site-specific phosphorylation events fulfill key functions in the activity control of signaling cascades and networks (1). Cellular protein phosphorylation is controlled by more than 500 members of the protein kinase superfamily, which comprises one of the largest enzyme families encoded by the human genome (2). Protein kinases represent the key elements in phosphorylation-based signal transmission. Aberrant protein kinase expression and/or activity, often because of gene amplification or mutational changes, is involved in pathological processes leading to malignant transformation and tumor development (3). Therefore, protein kinases have emerged as a major class of drug targets for therapeutic intervention (4–6). Given the diversity of molecular mechanisms related to de-regulated kinase function in human cancers, proteomic approaches could significantly enhance our understanding of disease-relevant kinase function and also help to optimize and adjust therapeutic strategies. In addition to assessing protein expression, the analysis of site-specific phosphorylations on protein kinases is of particular relevance, as these PTMs can be indicative of their cellular catalytic activities (7, 8). Protein kinases can not only modulate each other's functions and activities through site-specific phosphorylation events, but often also undergo site-specific autophosphorylation once they get activated (9). Thus, the comprehensive assessment of kinase-derived phosphopeptides can provide important insights into the regulation of these key players in phosphorylation-controlled signaling.
Regulatory enzymes such as protein kinases are often expressed at low cellular levels. This can impede their detection by LC-MS in highly complex peptide mixtures derived from total cell or tissue extracts. These analytical challenges are further aggravated in phosphoproteomic experiments due to the fact that many phosphopeptide species result from sub-stoichiometric phosphorylation events (10). Consequently, phosphopeptide isolation methods have proven to be essential. Among others, techniques such as immobilized metal affinity chromatography or enrichment by means of titanium dioxide (TiO2)-coated beads have found widespread use in MS-based phosphoproteomics (11–13). In addition, to reduce initial sample complexity, either protein fractionation by gel electrophoresis or peptide separation by strong cation exchange chromatography is typically included in contemporary phosphoproteomics workflows (14–16). These separation techniques in combination with LC-MS on state-of-the-art mass spectrometers enabled the identification of thousands of phosphorylation sites from total cellular extracts (15, 17, 18). Despite these impressive advances, such large-scale efforts require considerable instrument time, and the current methodology is still not comprehensive across the full dynamic range of the entire phosphoproteome. This creates the rationale for sub-proteome analyses to achieve high coverage and analytical sensitivity, which is particularly relevant for members of the protein kinase enzyme family.
To date, the only pre-fractionation techniques permitting the enrichment of more than a few protein kinases are affinity capture methods relying on immobilized and kinase-selective small molecule inhibitors (19–21). We and others have demonstrated that combinations of such kinase inhibitor resins efficiently pre-fractionate kinases for subsequent phosphorylation analysis (7, 22, 23). Ideally, capture molecules for kinase proteomics have two properties. First, they should exhibit high non-selectivity within the kinase superfamily. Second, they should efficiently discriminate between protein kinases and other classes of cellular proteins under the biochemical conditions of the pre-fractionation procedure.
In our efforts to characterize affinity reagents fulfilling these criteria, we quantitatively compared a selection of immobilized pyrido[2,3-d]pyrimidine-based inhibitors with respect to their proteome-wide kinase binding properties. Based on this assessment, an affinity matrix displaying the small molecule VI16832 was used as an enrichment tool for the comparative expression analysis of protein kinases in different cancer cell lines. The highly efficient VI16832 affinity resin further enabled a large-scale phosphoproteomics survey resulting in the identification and confident assignment of about 1200 phosphorylation sites on more than 200 distinct protein kinases.
EXPERIMENTAL PROCEDURES
Cell Culture
For quantitative kinase inhibitor studies MV4–11 (ATCC, CRL-9591) were grown in RPMI 1640 medium (Invitrogen) containing 20% fetal bovine serum (Invitrogen), MDA-MB-435S (435S, ATCC, HTB-129) (24), and HCT116 (ATCC, CCL-247) were grown in Dulbecco's modified Eagle's medium (Invitrogen) containing 10% fetal bovine serum. All media were supplemented with penicillin and streptomycin (Invitrogen). For quantitative MS-based studies, cells were labeled with either l-arginine and l-lysine (Arg0, Lys0), l-[U-13C6,14N4]arginine, and l-[2H4]lysine (Arg6, Lys4) or l-[U-13C6,15N4] and l-[U-13C6,15N2]lysine (Arg10, Lys8) (Cambridge Isotope Laboratories or Sigma) for six cell doublings to achieve complete labeling of cellular proteins. The adherent HCT116 and 435S cells were lysed directly on cell culture plates, MV4–11 cells were harvested by centrifugation before cell lysis.
Generation of Kinase Inhibitor Resins
The kinase inhibitor VI16832 was prepared as described (7). VI16741 and VI16743 were synthesized accordingly, except that 8-ethyl-2-methanesulfonyl-8H-pyrido[2,3-d]pyrimidine-7-one and 8-cyclopentyl-2-methanesulfonyl-8H-pyrido[2,3-d]pyrimidine-7-one were used as starting material instead of 8-bicyclo-[2.2.1]hept-2-yl-2-methanesulfonyl-8H-pyrido[2,3-d]pyrimidine-7-one in case of VI16832 (25). For preparation of the affinity resins used in the SILAC experiments, 2 volumes of a 3 mm inhibitor solution prepared in 50% dimethylformamide, 50% EtOH were mixed with 1 volume of drained ECH-Sepharose beads (GE Healthcare) and then subjected to carbodiimide-catalyzed immobilization according to described procedures (22). Coupling efficiencies were similar for all three inhibitors, resulting in a concentration of 1.5 mm immobilized ligand on Sepharose beads as determined by UV-Vis measurements (data not shown). Kinase enrichment in the phosphoproteomics experiments was performed with VI16832 covalently immobilized on epoxy-activated Sepharose (GE Healthcare) according to a reported coupling protocol (26). Here, 2 volumes of 1.5 mm VI16832 solution prepared in 50% DMSO, 50% 50 mm Na2CO3, pH 11 were subjected to 1 volume of drained epoxy-activated Sepharose to initiate the coupling reaction.
Cell Lysis and Affinity Enrichment
Cells were lysed in 50 mm Hepes-NaOH, pH 7.5, 150 mm NaCl, 0.5% Triton X-100, 1 mm EDTA, 1 mm EGTA, 1 mm phenylmethylsulfonyl fluoride, 10 mm NaF, 2.5 mm Na3VO4, 50 ng/ml calyculin A (Alexis Biochemicals, San Diego, CA), 10 μg/ml aprotinin, 10 μg/ml leupeptin, and 1% phosphatase inhibitor mixture 1 and 2 (Sigma) for 1 h at 4 °C. The cell debris was removed by centrifugation (20 min at 13,000 rpm) and by filtering through 0.22-μm mixed esters of cellulose membranes (Millipore). Protein concentration was measured using the BCA assay (Pierce). For comparative SILAC analysis of different inhibitor resins, 1.5 mg from each differentially labeled MV4–11 lysate was subjected to in vitro association with the respective kinase inhibitor resins. 30 μl of drained beads coupled with the respective kinase inhibitor were washed three times with lysis buffer and further three times with lysis buffer containing 1 m NaCl. Washed beads were incubated for 2.5 h at 4 °C in the dark with the lysates that had been adjusted to 1 m NaCl in a final volume of 650 μl. In each experiment, aliquots of the three differentially labeled lysates were pooled to determine the initial SILAC ratios and resulting correction factors for the quantification after affinity enrichment. Beads were washed twice with lysis buffer containing 1 m NaCl and twice with lysis buffer containing 150 mm NaCl. For elution, resin-bound proteins were incubated for 10 min with 50 μl 0.5% LDS buffer (Invitrogen) containing 50 mm dithiothreitol at 70 °C. Elution fractions were pooled and concentrated by a factor of three in a vacuum concentrator (Eppendorf). Moreover, aliquots of the different elution fractions were compared by immunoblotting with kinase-specific antibodies.
For SILAC-based comparison of protein kinases in MV4–11, HCT116, and 435S cells, total cell lysates were prepared as described above and all adjusted to 1.5-mg protein in a volume of 500 μl. This amount of protein was obtained upon lysis of 17 × 106 MV4–11, 7.3 × 106 HCT116, and 5.3 × 106 435S cells, respectively. The three lysates were pooled prior to incubation with 90 μl of drained VI16832 beads according to the same protocol as used for the inhibitor resin comparisons.
For immunoblotting of either different affinity-purified fractions from MV4–11 cells or of total cell lysates from MV4–11, HCT116, and 435S cells, the following antibodies were used: rabbit anti-CDC2, mouse anti-Met and rabbit anti-PAK4 (Cell Signaling Technology, Inc.), mouse anti-PLK1 (7), rabbit anti-Fer (27), rabbit anti-PYK2 (Millipore), goat anti-Axl, goat anti-CK1α, rabbit anti-DDR1 (C-20), rabbit anti-FAK (C-20), goat anti-Fes (C-19), rabbit anti-HCK (N-30), rabbit anti-JAK1 (HR-785), and rabbit anti-Syk (N-19) (all from Santa Cruz Biotechnology, Inc.).
Protein kinase enrichment for phosphorylation site mapping was performed using an ÄKTA explorer system and Tricorn 5/20 chromatography columns (GE Healthcare) packed with 500 μl of VI16832 resin. Cells were lysed in a volume of 35–40 ml per experiment. The protein amounts of the starting extracts used in the first and second experiments were: 435S, 85 and 120 mg; HCT116, 240 and 175 mg; MV4–11, 180 and 120 mg. Lysates were adjusted to 1 m NaCl prior to loading onto the VI16832 column at a flow rate of 0.07 ml/min. Subsequent washing and elution steps were performed as described previously (22). Protein-containing elution fractions were lyophilized, re-suspended in one tenth of the initial volume, and then desalted by protein precipitation prior to gel electrophoresis (28).
Sample Preparation and MS Analysis
For gel electrophoresis, ready-made 10% NuPAGE® Bis-Tris gels (Invitrogen) were used according to the manufacturer's instructions. Resolved proteins were stained using the Collodial Blue staining kit (Invitrogen). In all SILAC experiments, gels were cut into three slices followed by in-gel digestion with trypsin and peptide purification with StageTips as described (29, 30).
For phosphopeptide identifications, gels were cut in either three (experiment 1) or 6 (experiment 2) molecular weight regions prior to in-gel proteolysis with trypsin (29). Phosphopeptides were specifically enriched using titanium dioxide (TiO2) microspheres (31, 32). The TiO2 beads (GL Science, Tokyo, Japan) were first equilibrated by consecutive incubations with 20 mm NH4OH in 20% acetonitrile (ACN), pH 10.5, washing buffer (50% ACN, 0.1% trifluoroacetic acid) and loading buffer (5 g/liter 2,5-dihydrobenzoic acid in 55% ACN). Fractions of extracted peptides were adjusted to loading conditions and incubated for 30 min with 5 mg TiO2 beads at room temperature on a rotating wheel. Afterward, beads were washed once with 100 μl of loading buffer, three times with 1.5 ml of washing buffer, and phosphopeptides were eluted by incubating twice with 30 μl of 20 mm NH4OH in 20% ACN, pH 10.5. Eluates were combined and passed through C8 StageTips followed by a 30-μl rinse with 80% ACN, 0.5% acetic acid. After adjusting to a pH of 6, samples were concentrated to ∼3 μl and mixed with an equal volume of 4% ACN, 0.2% trifluoroacetic acid. MS analyses were done as described previously (7, 15). Briefly, peptide separations were done on 15-cm analytical columns (75-μm inner diameter) in-house packed with 3-μm C18 beads (Reprosil-AQ Pur, Dr. Maisch) using a nanoflow high pressure liquid chromatography system (Agilent Technologies 1100), which was coupled online to a LTQ-Orbitrap mass spectrometer (Thermo Fisher Scientific) via a nanoelectrospray ion source (Proxeon Biosystems). The LTQ-Orbitrap was operated in the data-dependent mode to automatically switch between full scan MS in the orbitrap analyzer (with resolution r = 60,000 at m/z 400) and the fragmentation of the five most intense peptide ions by either MS/MS or multi-stage activation in the LTQ part of the instrument, the latter being triggered on neutral loss species at 97.97, 48.99, or 32.66 m/z below the precursor ion for 30 ms (33). For all measurements with the orbitrap detector, a lock-mass strategy was used for internal calibration as described (34).
Peptide Identification, Quantitation, and Data Analysis
Raw MS files acquired from individual experiments were merged using the Raw2msm software (34), and the resulting msm files were searched against concatenated forward and reversed versions of the human IPI protein database version 3.13 (SILAC-based inhibitor comparison), version 3.19 (phosphorylation site mapping), or version 3.24 (SILAC-based kinome profiling) containing 57,032, 60,397, and 66,921 protein entries, using the MASCOT search engine (Matrix Science). All databases contained frequently occurring contaminants including human keratins, porcine trypsin, and endopeptidase Lys-C. Search parameters were set to up to three missed cleavages, mass tolerances of 25 ppm for MS, and 0.5 Da for MS/MS scans. Carbamidomethylation of cysteine was set as fixed modification; variable modifications included oxidized methionine, phosphorylation of serine, threonine and tyrosine, N-acetyl protein, N-pyroglutamine and in the SILAC experiments, the isotopic variants Lys4, Lys8, Arg6, and Arg10.
The html output files generated by MASCOT together with the raw data files were then further processed using the MSQuant software, version 1.4.0 used for SILAC-based inhibitor comparison and version 1.4.3 for SILAC-based kinome profiling and phosphorylation site identification). Prior to peptide quantification or computation of PTM scores, peptide datasets were filtered for a false-discovery rate (FDR) of less than 1% (p < 0.01) according to a target/decoy database searching strategy. To achieve a FDR of less than 1%, filtering criteria such as a peptide length ≥ 6 and a mass error < 5ppm were applied together with a minimal MASCOT score that ranged from 21 to 29 depending on the experiment.
MSQuant determines the average ratio over the peptide elution profile, and all precursor ion assignments used for quantitation were manually validated (15). Upon normalization for the initial SILAC pooling error, protein ratios were calculated as the mean of all ratios from uniquely assigned peptides.
To identify highly significant differences in relative protein abundance, the relative ratios of the protein quantifications from the two biological replicate experiments were analyzed for their normal distribution to account for the combined biological and technical variation in the quantitative MS analyses. Protein abundance was considered as significantly different (p < 0.01) in case ratios differed from the mean by 2.58 σ as determined from the “ratio of ratios” distributions of the biological replicate analyses.
The assignment of phosphorylation sites in identified phosphopeptides was done with the PTM scoring algorithm implemented in MSQuant as described previously by Olsen et al. (15). In our present study, phosphorylation sites were rated as class I in case of a localization probability of at least 0.95. The localization p values for all identified phosphopeptides as well as the corresponding annotated MS/MS spectra can be accessed online (35).For enrichment analysis of gene ontology (GO) categories, Cytoscape (36) together with the BinGO plugin (36, 37) was used to identify statistically over-represented GO molecular function terms compared with a reference dataset consisting of all IPI entries and their respective GO identifiers essentially as described (38).
RESULTS
Comparative Target Profiling for Kinase-selective Pre-fractionation Reagents
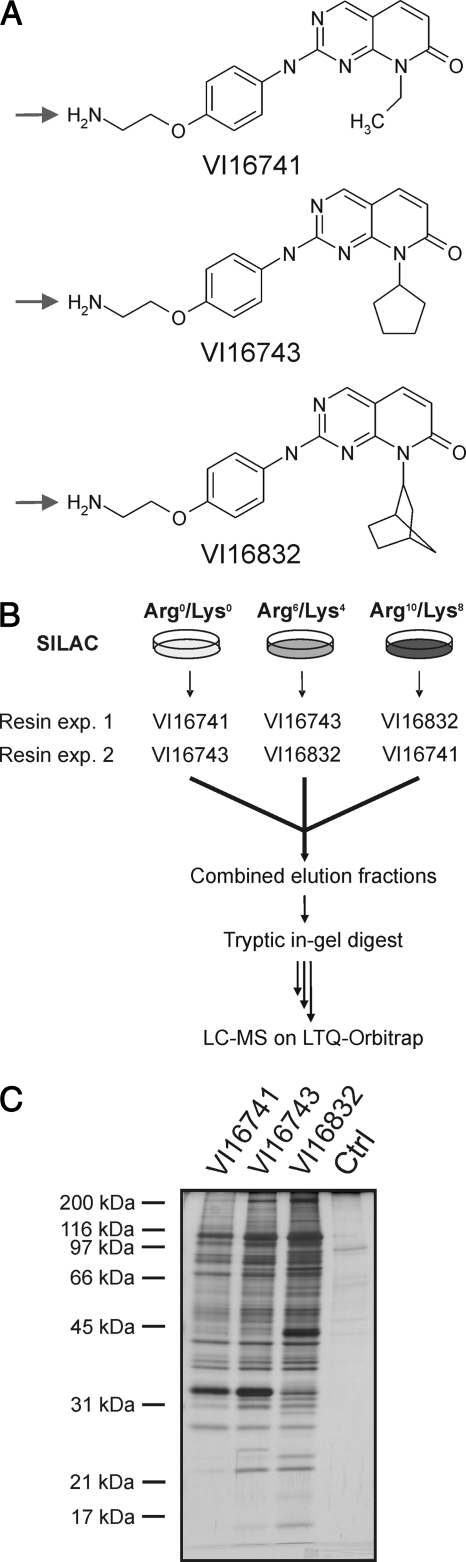
To enable broad kinase enrichment, immobilized kinase inhibitors should ideally exhibit considerable non-selectivity in conjunction with high affinity for many members of the protein kinase superfamily. The previously described pyrido[2,3-d]pyrimidine-based capture molecule PP58 exhibited high potency and non-selectivity for a subset of protein kinases comprising about 25% of the human kinome (39). These kinases possess a small amino acid (often a threonine residue) at a critical “gate-keeper” position, which does not interfere with the positioning of the inhibitor's dichlorophenyl moiety in a hydrophobic pocket located at their ATP binding sites (40). In contrast, this cavity is rather inaccessible in the majority of protein kinases with more spacious “gate-keeper” residues (40). We initially reasoned that this part of the kinome might be targeted by a PP58-related compound that lacks the dichlorophenyl group but is otherwise identical in structure. However, such a capture molecule exhibited fairly weak kinase affinity (data not shown). Therefore, based on previously described structure-activity relationship data for Cdk4 inhibition by pyrido[2,3-d]pyrimidine kinase antagonists (25), we have recently introduced compounds with larger cyclopentyl and norbornyl moieties (designated VI16743 and VI16832) at the N8 position to compensate for the observed drop in potency (7)2. As expected, these capture molecules retained considerable numbers of kinases. However, the actual effect of the N8 substituent on cellular target binding profiles has not been systematically analyzed. Such information would help to adjust pre-fractionation toward the cellular kinases that are of interest in individual projects. Therefore, we prepared pyrido[2,3-d]pyrimidine ligands with the cyclopentyl and norbornyl moieties (VI16743 and VI16832) as well as a derivative with a smaller ethyl moiety in the N8 position (VI16741) and immobilized all three compounds through their primary amino groups (Fig. 1A) (7).
Fig. 1.
Comparison of inhibitor resins by quantitative chemical proteomics. A, chemical structures of pyrido[2,3-d]pyrimidine-based inhibitors used as affinity capture reagents for protein kinase enrichment. The amino groups (marked by arrows) were used for immobilization on Sepharose beads. B, SILAC scheme and workflow used for comparative quantitative MS analysis of cellular proteins enriched by the different inhibitor resins from MV4–11 cell extracts. C, aliquots of the elution fractions from either kinase inhibitor resins or from a control resin (Ctrl) of ECH-Sepharose devoid of immobilized ligand were analyzed by SDS-PAGE and silver staining.
To compare the VI16741, VI16743, and VI16832 resins, we performed stable isotope labeling by amino acids in cell culture (SILAC) with the acute myelogenous leukemia (AML) cell line MV4–11 to enable quantitative MS analysis (41). These cells harbor an internal tandem duplication in the juxtamembrane domain of the FMS-like tyrosine kinase 3 (FLT3). This mutation is also present in a subset of AML patients and results in constitutive up-regulation of growth-promoting FLT3 tyrosine kinase activity (42). Upon quantitative incorporation of either normal arginine and lysine (Arg0/Lys0) or their isotopic variants (Arg6/Lys4 or Arg10/Lys8), lysates from three differentially SILAC-encoded MV4–11 cell populations were incubated with the affinity resins carrying covalently immobilized VI16741, VI16743, or VI16832 as capture ligands (Fig. 1, A and B). After in vitro association and elution of retained proteins, we first analyzed small aliquots of the elution fractions by gel electrophoresis and silver staining. Most proteins appeared to be present in all three fractions, albeit typically at somewhat higher levels in the VI16743 and VI16832 resin eluates (Fig. 1C). In addition, some protein bands showed specific resin binding, such as a prominent VI16741 and VI16743 resin-interacting 35 kDa protein identified as mitochondrial delta(3,5),delta(2,4)-dienoyl-CoA isomerase and a VI16832 resin-bound 50 kDa protein identified as the multifunctional protein ADE2 (data not shown), indicating that different hydrophobic moieties in the N8 position can confer selectivity as evident for these abundant purine-binding non-protein kinases.
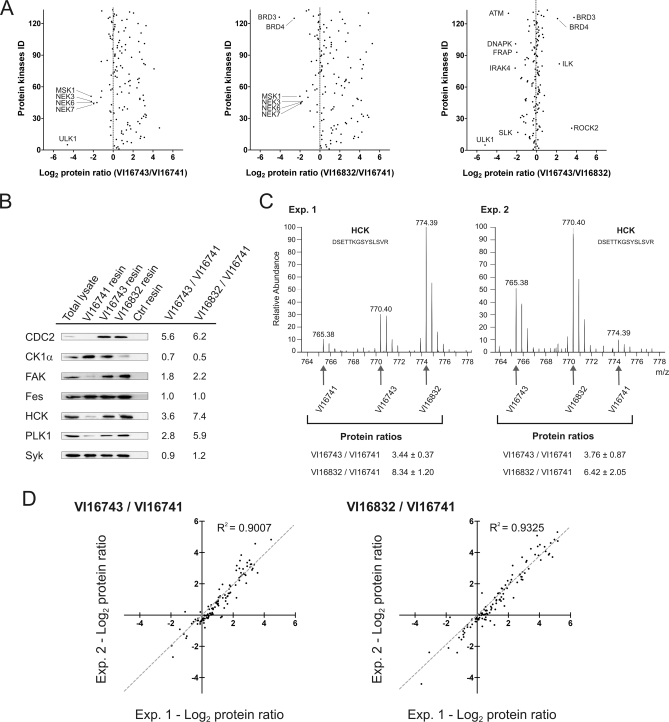
The large remainders of the elution fractions were combined prior to gel electrophoresis and tryptic digestion of proteins from three different molecular weight regions. The resulting mixtures of proteolytically derived peptides were analyzed by LC-MS on a LTQ-Orbitrap hybrid mass spectrometer. After database searching and filtering for an identification certainty greater than 99%, relative peptide abundances between VI16741-, VI16743-, or VI16832-enriched fractions were determined by SILAC-based quantification. Quantitative peptide data was then used to calculate the relative protein levels in the different resin eluates (supplemental Tables 1 and 2). In total, more than 130 distinct protein kinases could be identified and quantified in the inhibitor resin eluates. This demonstrates the capacity of the pyrido[2,3-d]pyrimidine-based affinity ligands to pre-fractionate and detect almost half of the expressed kinome, which is estimated to consist of up to 300 distinct protein kinases in a given mammalian cell (43). A considerable subset of protein kinases interacted more strongly with the VI16743 and V16832 resins than with the VI16741-containing beads, indicating that the space-filling cyclopentyl and norbonyl moieties in the N8 position resulted in an overall increase in potency with respect to kinase binding (Fig. 2A). In a few cases, bulky N8-substituents were not well tolerated, notably by various NIMA-related expressed kinase (NEK) family members (NEK3, 6, and 7) that were found in higher abundance in the VI16741 resin eluates. The differences between the VI16743 and V16832 resins were less pronounced, with relatively small subsets of protein kinases preferentially bound by either affinity matrix. Similar comparisons were also made for the more than 250 non-protein kinase proteins quantified from the affinity resin eluates (supplemental Fig. 1A). To verify the quantitative MS approach with a second assay, immunoblotting was done with a selection of kinase-specific antibodies. The outcome of this analysis was found in excellent agreement with the MS results (Fig. 2, B and C). To evaluate the reproducibility of the SILAC-based quantification, the protein kinase ratios obtained in biological replicate analysis were visualized in scatter blots (Fig. 2D). Notably, independent experimental ratios for VI16743 versus VI16741 as well as for VI16832 versus VI16741 resin binding were similar, demonstrating the accuracy and reliability of the quantitative MS approach. Comparable results were obtained for the identified non-protein kinases (supplemental Fig. 1B). Moreover, as determined from the distribution of the ratios of the replicate, log2-transformed protein quantifications, values of more than 2.65 or less than 0.38 indicated differential binding with high confidence (p < 0.01) (supplemental Fig. 2). Hence, the SILAC-enabled strategy applied here not only enabled the comparative profiling of pyrido[2,3-d]pyrimidine-derived kinase enrichment reagents, but further exemplifies the general utility of such approaches to quantitatively assess cellular target binding to distinct immobilized small molecule ligands in a compact experimental format (44).
Fig. 2.
Quantitative analysis of protein kinases enriched with three different pyrido[2,3-d]pyrimidine ligands. A, relative protein kinase binding to the affinity resins was determined by SILAC-based quantitative MS of unique peptides. For the three pair-wise resin comparisons, the averaged ratios of two independent experiments were plotted on a logarithmic scale (log2) for all protein kinases. B, aliquots of MV4–11 cell lysate and eluted protein from incubations with the indicated inhibitor resins or control (Ctrl) beads were separated by SDS-PAGE and immunoblotted with antibodies recognizing the kinases CDC2, CK1α, FAK, Fes, HCK, PLK1, and Syk. For comparison, ratios of relative kinase binding according to the quantitative MS analysis are shown. C, MS spectra of a HCK peptide detected as a triplet due to SILAC encoding. Upon swapped SILAC labeling, relative ion intensities changed accordingly in replicate experiments. Peptide species derived from the different affinity purifications are marked by arrows, and the resulting ratios are shown for both experiments. D, log2 transformed ratios of kinase binding to the different resins are shown in scatter blot comparisons of two independent experiments. Pearson correlation coefficients (R2) close to one indicated an overall high concordance of measured kinase ratios in biological replicates.
The pyrido[2,3-d]pyrimidine-based affinity resins are efficient and easy-to-use purification reagents for a considerable subset of the expressed kinome. Importantly, many of the isolated protein kinases have not been tractable biochemically by conventional immunoprecipitation approaches, due to the lack of high affinity antibodies for that purpose. In agreement with efficient protein kinase-selective enrichment, GO analysis revealed highly significant overrepresentation of corresponding GO molecular function terms in the resin-bound protein fraction (data not shown). Moreover, although protein kinases accounted for just one third of all protein identifications, they were on average identified and quantified with three times more peptides than non-protein kinases, indicating even higher purification efficiency as evident from GO analysis.
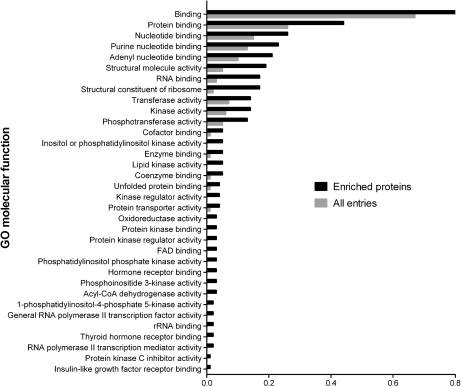
To further explore the binding characteristics of the pyrido[2,3-d]pyrimidine-based affinity ligands, we performed a GO analysis for over-represented molecular functions in the subset of resin-bound proteins that were not protein kinases (Fig. 3 and supplemental Table 3). This assessment revealed considerable enrichment for nucleotide-dependent enzymes such as oxidoreductases, dehydrogenases, and lipid kinases, which likely resulted from direct interactions with the purine-like pharmacophore provided by the immobilized inhibitors. Notably, important signaling factors were among these resin-bound enzymes such as phosphatidylinositol 3-kinase, phosphatidylinositol-4,5-bisphosphate 3-kinase and others. GO analysis of non-protein kinases further indicated over-representation of the molecular function “protein kinase binding”, which resulted from the specific co-purification of protein kinase interactors such as various cyclins as well as other regulatory kinase subunits.
Fig. 3.
Gene ontology analysis of non-protein kinases enriched with immobilized kinase inhibitors. Non-protein kinases that were identified with at least one unique peptide upon inhibitor affinity enrichment from MV4–11 cell extracts were compared with the entire list of IPI entries. Significantly over-represented GO molecular function terms (p < 0.001) are shown. Ratios represent the numbers of either inhibitor-enriched proteins or all IPI entries annotated to the listed GO molecular function terms divided by the respective numbers of all proteins with annotated GO molecular function terms.
Comparative Kinase Expression Analysis in Different Cancer Cell Lines
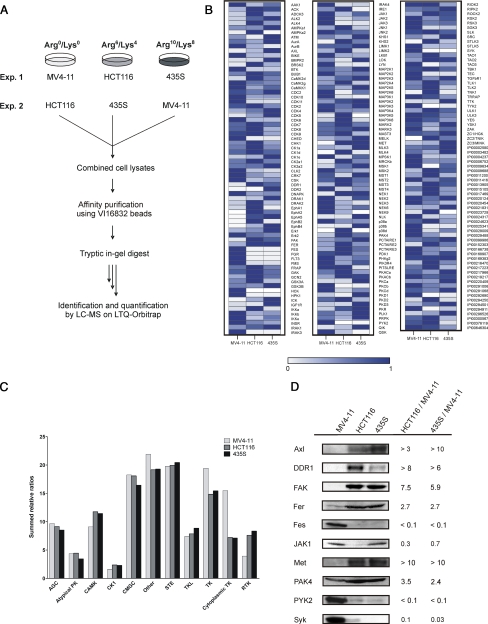
The quantitative comparison of pyrido[2,3-d]pyrimidine resins demonstrated efficient kinome fractionation by the VI16743 and VI16832 resins, and we chose VI16832-based affinity enrichment to monitor cell-type specific differences of kinase expression profiles. The identification of up-regulated kinases has considerable potential in tumor cell analysis, given that over-expression of protein kinases is frequently involved in carcinogenesis. To test this experimental strategy, three cancer cell lines (MV4–11 leukemia, HCT116 colon carcinoma, and 435S melanoma-derived cells) were differentially SILAC-encoded prior to cell lysis. Total cell extracts were combined and incubated with VI16832 inhibitor beads followed by sample processing and quantitative MS as described above (Fig. 4A). In total, more than 170 protein kinases and almost 40 other nucleotide-utilizing enzymes could be identified and quantified in their relative expression levels in the three cancer cell lines (supplemental Tables 4 and 5). Our affinity resin-based strategy covered more than twice as many protein kinases as a previously reported approach, which involved the use of reactive and biotinylated ATP conjugates for kinase expression profiling (45). Moreover, the VI16832 resin enabled the analysis of an almost similar number of protein kinases in a single sample as detected upon enrichment with up to seven distinct affinity ligands for protein kinases (7, 23). The comparison of replicate experiments indicated high reproducibility of the quantified protein ratios (supplemental Fig. 3). To generate expression profiles, protein levels detected in the cell line with the highest abundance were set to 100%. The quantified ratios were then used to determine percentage values for relative expression in the other two cell lines. In cases where no peptide ion signals were recorded in one or two of the analyzed cell lines, expression values were set to 0%. We then generated a heat map to visualize quantified protein kinases and other nucleotide-binding enzymes according to their relative expression patterns across the three cancer cell lines (Fig. 4B).
Fig. 4.
Comparative kinase expression analysis across different cancer cell lines. A, scheme illustrating the SILAC-based chemical proteomics workflow for quantitative comparison of VI16832-interacting sub-proteomes in three cancer cell lines. B, quantified ratios for protein kinases and other nucleotide-binding proteins were transformed into relative expression levels with the highest expression in any of the three cell lines set to 1. Based on these values, a heat map was generated to visualize relative expression levels across the analyzed cancer cell lines. C, summed relative expression values for the seven major kinase groups as well as the atypical and other kinases according to Manning et al. (2) were compared for MV4–11, HCT116, and 435S cells. In addition, members of the tyrosine kinase (TK) group were subdivided into cytoplasmic and receptor tyrosine kinase in the cell line comparison. D, total cell lysates of MV4–11, HCT116, and 435S cells were immunoblotted with antibodies recognizing the kinases Axl, DDR1, FAK, Fer, Fes, JAK1, Met, PAK4, PYK2, and Syk. For comparison, binding ratios according to the SILAC-based quantification of VI16832 resin-bound protein kinases are shown.
The summed relative ratios of protein kinases were rather similar for the seven major groups of the kinome as well as for the other and atypical kinases (Fig. 4C). Interestingly, when we divided the tyrosine kinase group into its cytoplasmic and receptor-type members, the overall relative expression of cytoplasmic PTKs was higher in the MV4–11 suspension cells compared with the adherent HCT116 and 435S cell lines. Syk, Tec family kinases (Btk, Tec) and several members of the Src family (Lyn, Fgr, HCK) were substantially higher expressed or exclusively detected in MV4–11 cells. Furthermore, we found two examples of an inverse correlation of closely related cytoplasmic PTKs; Fes and PYK2 were far more prominent in MV4–11, whereas their close relatives Fer and FAK were found in much higher levels in HCT116 and 435S cells. Unlike for cytoplasmic PTKs, HCT116 and 435S cells exhibited a higher overall level of receptor tyrosine kinases according to our analysis (Fig. 4C). However, FLT3 receptor tyrosine kinase expression was exclusively found in MV4–11 cells, demonstrating that our comparative analysis revealed this key oncogenic kinase in receptor tyrosine kinase pathogenesis.
To verify that differential resin binding measured by quantitative MS reflects cell-type specific kinase expression, we cultivated all three cell lines in normal growth media and prepared total cell extracts using denaturing lysis conditions. Subsequently, cell extracts were compared by immunoblotting against a selection of kinases, which exhibited significant cell-type specific differences according to the SILAC-based quantification of resin eluates. As shown in Fig. 4D, immunoblotting results were found in good agreement with the measured SILAC ratios. This indicated that other potential sources of variation that could affect the affinity purification approach, such as cell-type specific expression changes upon SILAC or differential solubilization of kinases due to non-denaturing cell lysis, apparently had no major influence on the quantitative cell line comparisons.
Large-scale Phosphoproteomics Analysis of Cancer Cell Lines upon Kinase Affinity Enrichment
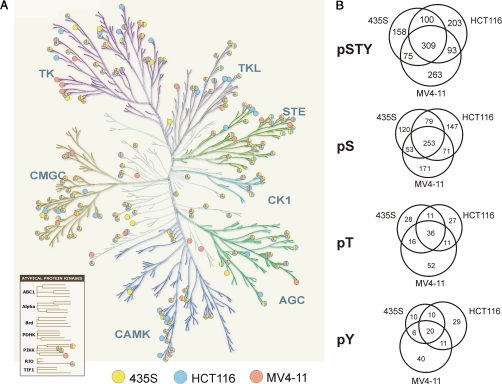
The efficient pre-fractionation of protein kinases from total cell extracts provides an experimental basis for the analysis of post-translational modifications with high analytical sensitivity. To exploit this potential for phosphoproteomics analysis, we employed the VI16832 resin for protein kinase enrichments from MV4–11, HCT116, and 435S cell lysates. In contrast to the batch purification protocol described above, a column chromatography set-up was employed to enable processing of larger amounts of starting material and thereby enhance the sensitivity of phosphopeptide detection. Affinity-purified proteins from the different cancer cell lysates were separated by gel electrophoresis prior to in-gel digestion with trypsin and subsequent phosphopeptide enrichment with TiO2 microspheres (31, 32). Raw data from the LC-MS analyses were filtered for phosphopeptide identifications within a false-discovery rate of less than 1% for each individual experiment. Phosphoproteomics analysis of VI16832-enriched fractions from MV4–11, HCT116, or 435S cells resulted in more than 8500 phosphopeptide identifications. These translated into almost 1700 distinct phosphopeptide species derived from 212 different members of the protein kinase superfamily. We further identified more than 1300 distinct phosphopeptides on 563 non-protein kinases (Table I). Notably, about 30% of the protein kinases and 50% of the other proteins were not detected in our previous analysis of kinase-enriched fractions from HeLa S3 cells. Using computational PTM scoring, more than 1200 phosphorylation sites on kinases and 900 on other proteins could be localized with high confidence (Table I and supplemental Table 6). All identified phosphopeptides can be accessed through the Phosida database, which also provides links to annotated fragmentation spectra harboring the identified phosphopeptides (35). The more than 200 identified protein kinases were rather evenly distributed in the dendrogram of the human kinome (Fig. 5A). This indicated that VI16832 did not select for sequence-related determinants restricted to certain subsets of the kinome and further highlighted the utility of this reagent as broadly kinase-selective enrichment tool for sensitive PTM analysis. The Venn diagrams show the cell line distribution of the identified phosphorylation sites on protein kinases. Importantly, the analysis of VI16832-retained proteins from the three cancer cell lines considerably increased the overall number of identified phosphorylation sites on protein kinases (Fig. 5B). However, as kinase-enriched fractions from the different cell extracts were subjected to individual, qualitative phosphopeptide mapping experiments, selective identifications did not necessarily indicate cell-type specific differences but could also be due to run-to-run variability inherent to LC-MS in the data-dependent acquisition mode. Phosphorylation sites were also identified on various nucleotide-binding enzymes as well as other proteins including regulatory subunits of protein kinases (Table I and supplemental Fig. 4A and supplemental Table 6). Although the retained non-kinase phosphoproteins accounted for 60–70% of all identifications, they were typically found with fewer phosphopeptides per protein. Moreover, on average, peptides from these proteins exhibited considerably lower signal intensities than protein kinase-derived peptides. Considering the sum of all phosphopeptide intensities as a measure for VI16832-enriched protein amount, more than 80% was derived from protein kinases. This value demonstrates the remarkable kinase selectivity of the phosphoproteomics workflow presented here (supplemental Fig. 4B).
Table I. Overview of results from phosphokinome analysis.
Numbers of identified phosphoproteins, phosphopeptide sequences, phosphopeptides, and confidently assigned phosphorylation sites (p > 0.95) are shown either for protein kinases or for all other proteins analyzed upon VI16832 affinity chromatography.
| Protein kinases | Other proteins | |
|---|---|---|
| Phosphoproteins | 212 | 563 |
| Phosphopeptide sequences | 1431 | 1224 |
| Phosphopeptides | 1695 | 1329 |
| Phosphosites (class I) | 1201 | 904 |
Fig. 5.
Phosphorylation site mapping across the human kinome. A, protein kinase-derived phosphopeptides identified upon enrichment by VI16832 affinity chromatography are marked in the dendrogram of the human kinome (2). Colors indicate in which of the analyzed cell lines phosphopeptides were found. The kinome tree illustration was adapted with permission from Cell Signaling Technology, Inc. B, cell line distribution of the identified phosphorylation sites on protein kinases, which could be confidently localized to specific serines, threonines, or tyrosines (class I sites; p > 0.95). Numbers are shown for all phosphorylations sites combined (pSTY) and separately for phosphoserine, -threonine, and -tyrosine (pS, pT, and pY).
When we analyzed the distribution of phosphoserine, -threonine, and -tyrosine sites, we found tyrosine phosphorylation to be more frequent on protein kinases than on other identified proteins (Table II). Even more intriguingly, tyrosine phosphorylation accounted for more than one third of the identified sites in the activation segment regions of protein kinases (Table II and Fig. 6). Activation segment phosphorylations often induce conformational changes that allow protein kinases to adopt and maintain their catalytically active states. Therefore, they are involved in the regulation of many members of the protein kinase superfamily (9). In this context, our results highlight the key role of tyrosine phosphorylation for the activity control of kinase-mediated cellular signaling. This is remarkable considering the low overall abundance of Tyr(P), which by comparison accounted for just 1.8% of all identified phosphorylation sites in the large-scale analysis of growth factor-induced signaling in total cell extracts by Olsen et al. (15). Moreover, as activation segment phosphorylation can serve as a direct biochemical read-out for cellular kinase activity; VI16832-mediated enrichment provides an experimental basis to monitor these key signaling events for more than 50 kinases per cell line (Fig. 6).
Table II. Distribution of Ser(P), Thr(P), and Tyr(P).
Distributions of phosphorylation sites by amino acid are shown for affinity purified fractions from 435S, HCT116, and MV4–11 cells. Percentages are shown for protein kinases, for the activation loop regions of protein kinases, and for all other phosphoproteins.
| Ser(P) | Thr(P) | Tyr(P) | |
|---|---|---|---|
| % | % | % | |
| Protein kinases | |||
| 435S | 78.5 | 14.1 | 7.4 |
| HCT116 | 77.7 | 12.1 | 10.2 |
| MV4–11 | 73.9 | 15.5 | 10.6 |
| PK activation loop | |||
| 435S | 32.2 | 28.8 | 39.0 |
| HCT116 | 28.6 | 27.0 | 44.4 |
| MV4–11 | 34.4 | 28.1 | 37.5 |
| Other phosphoproteins | |||
| 435S | 88.8 | 9.9 | 1.3 |
| HCT116 | 90.1 | 9.5 | 1.4 |
| MV4–11 | 86.3 | 9.2 | 4.5 |
Fig. 6.
Identified phosphorylation sites in the kinase activation loop region of protein kinases. Phosphopeptides with at least one confidently assigned phosphorylation site (class I, in bold red) in at least on cell line (marked by X) are shown and highlighted in yellow. Additional class III phosphorylation sites are shown in light red. Cell lines in which the same sequence and number of phosphorylations was found in the absence of site-determining information are indicated by X. The conserved tripeptide motifs DFG and APE, which define the borders of the activation segment, are highlighted in turquoise and green, respectively. In case activation segment phosphopeptides are shared among different members of the expressed kinome, alternative protein kinases are indicated in parentheses.
The comparison of the current dataset with the three largest previous phosphoproteomics studies on human proteins revealed both overlapping as well as complementary information about phosphorylation sites on protein kinases (supplemental Fig. 4C). Compared with a large-scale study of epidermal growth factor signaling by Olsen et al. (15) in which 254 out of 5674 confidently assigned phosphorylation sites mapped to protein kinases; about four times more sites were found on protein kinases. Moreover, we identified almost twice the number of phosphorylation sites on protein kinases as found in a recent large-scale study on mitotic phosphorylation (18). The total number of 1201 phosphosites that could be assigned in this study even surpassed the 1007 confidently localized phosphorylation events reported in our previous analysis of cell cycle-regulated changes in kinase-enriched fractions (7). Moreover, despite an overlap of 555 phosphosites between these two studies, we find almost 650 additional site-specific phosphorylations on protein kinases. In contrast to these earlier quantitative studies using more complex, SILAC-encoded samples, the analyses in our current study were done in a qualitative manner with the goal to promote comprehensive phosphorylation site mapping, which, to the best of our knowledge, resulted in the most extensive phosphoproteomics analysis of protein kinases reported to date.
DISCUSSION
In our present study, we immobilized kinase inhibitors from the pyrido[2,3-d]pyrimidine class of compounds to generate affinity resins for the pre-fractionation of protein kinase-enriched sub-proteomes (25). Cellular target capture was compared for three immobilized pyrido[2,3-d]pyrimidine derivatives using SILAC-based quantitative MS (41). The VI16743 and VI16832 resins were particularly efficient as purification tools, as these affinity resins were capable of retaining more than 130 distinct protein kinases from a single cell extract. Thus, they represent straightforward and easy-to-use purification reagents for a considerable subset of the expressed human kinome. Due to a lack of affine antibodies, many protein kinases are difficult to study by conventional immunoprecipitation approaches. In such cases, small molecule-based isolation can provide a straightforward alternative for targeted analysis; for example when sample processing and MS analysis is restricted to the molecular weight region comprising the kinase-of-interest. Thus, the datasets reported here specify a large number of protein kinases amenable for focused signal transduction analysis, which might involve quantitative MS to monitor PTM regulation upon different types of cell treatment.
The quantitative MS strategy for the comparison of pyrido[2,3-d]pyrimidine derivatives represents a generic approach. It can easily be extended to characterize other kinase-selective capture molecules, which retain subsets of the expressed kinome not efficiently purified by the VI16743 and VI16832 resins. Such comparative analyses should be useful to further improve on previously described multi-resin approaches, which combine immobilized kinase inhibitors with distinct target profiles for maximal coverage of the expressed kinome (7, 22, 23).
Kinase-selective proteomics focuses on a subset of the proteome, which is of high relevance for targeted therapeutic intervention in diseases such as human cancer. Our comparative analysis of three different cancer cell lines demonstrates quantitative profiling of kinase expression in a compact experimental format. Although only three cell lines can be compared by SILAC in a single experiment, further multiplexing is possible by merging data from parallel triple-labeling experiments through a shared reference sample (15). SILAC-based kinase profiling across larger collections of cancer cell lines could reveal subgroup-specific expression patterns, which might help to adjust targeted therapeutic interventions to the kinases involved in disease progression. Conceptually similar strategies could also be applied to the analysis of protein kinase-enriched fractions from primary tumor specimens; for example by employing chemical tagging with iTRAQ reagents as an alternative to SILAC-based quantification. In a recently published large-scale study by Rikova et al. (8), non-small cell lung cancer cell lines and tumors were comparatively analyzed for their phosphotyrosine-containing proteomes. Remarkably, this survey allowed clustering of the analyzed samples into different groups with distinct tyrosine kinase patterns. Compared with phosphotyrosine-directed approaches, proteomics of kinase-enriched sub-proteomes can be expected to provide both overlapping information (with respect to tyrosine kinases) as well as complementary data regarding serine/threonine kinases (and also other nucleotide-binding proteins). Protein expression data from the serine/threonine kinase branches of the human kinome likely reveal further insights into cancer cell biology, as exemplified by findings that overexpression of mitotic serine/threonine kinases, such as Aurora A and B, polo-like kinase1 and NEK2, can result in chromosomal instability and has been implicated in malignant transformation (46, 47). The pyrido[2,3-d]pyrimidine inhibitor resins described in this study are particularly useful for such sub-proteome surveys due to their ability to capture these key mitotic enzymes as well as many other additional serine/threonine kinases. Here, we have further exploited the enrichment abilities of the pyrido[2,3-d]pyrimidine-based capture ligand VI16832 to extensively map phosphorylation sites on protein kinases. Results from these large-scale analyses provide a multitude of new starting points for further functional studies on cellular kinase regulation. Combination of kinase-selective enrichment, quantitative MS, and phosphopeptide purification exhibits considerable potential for future studies, as kinome-wide comparisons on both the protein as well as the post-translational level across different cancer cell lines or tumor samples might significantly expand our knowledge about kinase drug targets and their oncogenic activities on a system-wide level.
Acknowledgments
We thank Axel Ullrich for his generous support of the present study with funding from the Department of Molecular Biology, Max Planck Institute of Biochemistry. The Fer-specific antibody was kindly provided by Peter A. Greer. The modified kinome dendrogram shown in Fig. 5A was reproduced with permission of Cell Signaling Technology, Inc.).
Footnotes
 The on-line version of this article (available at http://www.mcp.org) contains supplemental data.
The on-line version of this article (available at http://www.mcp.org) contains supplemental data.
2 T. Reinl, M. Nimtz, G. Kéri, J. Wehland, H. Daub, and L. Jänsch, submitted for publication.
1 The abbreviations used are:
- AML
- acute myelogenous leukemia
- FLT3
- FMS-like tyrosine kinase 3
- GO
- gene ontology
- IPI
- International Protein Index
- LC-MS
- liquid chromatography-mass spectrometry
- NEK
- NIMA-related expressed kinase
- PTK
- protein tyrosine kinase
- PTM
- post-translational modification
- SILAC
- stable isotope labeling by amino acids in cell culture
- ACN
- acetonitrile
- FDR
- false-discovery rate
REFERENCES
- 1.Ubersax J. A., Ferrell J. E., Jr.( 2007) Mechanisms of specificity in protein phosphorylation. Nat. Rev. Mol. Cell Biol. 8, 530– 541 [DOI] [PubMed] [Google Scholar]
- 2.Manning G., Whyte D. B., Martinez R., Hunter T., Sudarsanam S.( 2002) The protein kinase complement of the human genome. Science 298, 1912– 1934 [DOI] [PubMed] [Google Scholar]
- 3.Blume-Jensen P., Hunter T.( 2001) Oncogenic kinase signaling. Nature 411, 355– 365 [DOI] [PubMed] [Google Scholar]
- 4.Strebhardt K., Ullrich A.( 2008) Paul Ehrlich's magic bullet concept: 100 years of progress. Nat. Rev. Cancer 8, 473– 480 [DOI] [PubMed] [Google Scholar]
- 5.Krause D. S., Van Etten R. A.( 2005) Tyrosine kinases as targets for cancer therapy. N. Engl. J. Med. 353, 172– 187 [DOI] [PubMed] [Google Scholar]
- 6.Faivre S., Demetri G., Sargent W., Raymond E.( 2007) Molecular basis for sunitinib efficacy and future clinical development. Nat. Rev. Drug Discov. 6, 734– 745 [DOI] [PubMed] [Google Scholar]
- 7.Daub H., Olsen J. V., Bairlein M., Gnad F., Oppermann F. S., Körner R., Greff Z., Kéri G., Stemmann O., Mann M.( 2008) Kinase-selective enrichment enables quantitative phosphoproteomics of the kinome across the cell cycle. Mol. Cell 31, 438– 448 [DOI] [PubMed] [Google Scholar]
- 8.Rikova K., Guo A., Zeng Q., Possemato A., Yu J., Haack H., Nardone J., Lee K., Reeves C., Li Y., Hu Y., Tan Z., Stokes M., Sullivan L., Mitchell J., Wetzel R., Macneill J., Ren J. M., Yuan J., Bakalarski C. E., Villen J., Kornhauser J. M., Smith B., Li D., Zhou X., Gygi S. P., Gu T. L., Polakiewicz R. D., Rush J., Comb M. J.( 2007) Global survey of phosphotyrosine signaling identifies oncogenic kinases in lung cancer. Cell 131, 1190– 1203 [DOI] [PubMed] [Google Scholar]
- 9.Nolen B., Taylor S., Ghosh G.( 2004) Regulation of protein kinases; controlling activity through activation segment conformation. Mol. Cell 15, 661– 675 [DOI] [PubMed] [Google Scholar]
- 10.Steen H., Jebanathirajah J. A., Rush J., Morrice N., Kirschner M. W.( 2006) Phosphorylation analysis by mass spectrometry: myths, facts, and the consequences for qualitative and quantitative measurements. Mol. Cell. Proteomics 5, 172– 181 [DOI] [PubMed] [Google Scholar]
- 11.Collins M. O., Yu L., Choudhary J. S.( 2007) Analysis of protein phosphorylation on a proteome-scale. Proteomics 7, 2751– 2768 [DOI] [PubMed] [Google Scholar]
- 12.Schreiber T. B., Mausbacher N., Breitkopf S. B., Grundner-Culemann K., Daub H.( 2008) Quantitative phosphoproteomics - an emerging key technology in signal-transduction research. Proteomics 8, 4416– 4432 [DOI] [PubMed] [Google Scholar]
- 13.Macek B., Mann M., Olsen J. V.( 2009) Global and site-specific quantitative phosphoproteomics: principles and applications. Annu. Rev. Pharmacol. Toxicol. 49, 199– 221 [DOI] [PubMed] [Google Scholar]
- 14.Li X., Gerber S. A., Rudner A. D., Beausoleil S. A., Haas W., Villén J., Elias J. E., Gygi S. P.( 2007) Large-scale phosphorylation analysis of alpha-factor-arrested Saccharomyces cerevisiae. J. Proteome Res. 6, 1190– 1197 [DOI] [PubMed] [Google Scholar]
- 15.Olsen J. V., Blagoev B., Gnad F., Macek B., Kumar C., Mortensen P., Mann M.( 2006) Global, in vivo, and site-specific phosphorylation dynamics in signaling networks. Cell 127, 635– 648 [DOI] [PubMed] [Google Scholar]
- 16.Beausoleil S. A., Jedrychowski M., Schwartz D., Elias J. E., Villén J., Li J., Cohn M. A., Cantley L. C., Gygi S. P.( 2004) Large-scale characterization of HeLa cell nuclear phosphoproteins. Proc. Natl. Acad. Sci. U.S.A 101, 12130– 12135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Villén J., Beausoleil S. A., Gerber S. A., Gygi S. P.( 2007) Large-scale phosphorylation analysis of mouse liver. Proc. Natl. Acad. Sci. U.S.A. 104, 1488– 1493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dephoure N., Zhou C., Villén J., Beausoleil S. A., Bakalarski C. E., Elledge S. J., Gygi S. P.( 2008) A quantitative atlas of mitotic phosphorylation. Proc. Natl. Acad. Sci. U.S.A. 105, 10762– 10767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Daub H.( 2005) Characterization of kinase-selective inhibitors by chemical proteomics. Biochim. Biophys. Acta 1754, 183– 190 [DOI] [PubMed] [Google Scholar]
- 20.Godl K., Wissing J., Kurtenbach A., Habenberger P., Blencke S., Gutbrod H., Salassidis K., Stein-Gerlach M., Missio A., Cotten M., Daub H.( 2003) An efficient proteomics method to identify the cellular targets of protein kinase inhibitors. Proc. Natl. Acad. Sci. U.S.A. 100, 15434– 15439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rix U., Hantschel O., Dürnberger G., Remsing Rix L. L., Planyavsky M., Fernbach N. V., Kaupe I., Bennett K. L., Valent P., Colinge J., Köcher T., Superti-Furga G.( 2007) Chemical proteomic profiles of the BCR-ABL inhibitors imatinib, nilotinib, and dasatinib reveal novel kinase and nonkinase targets. Blood 110, 4055– 4063 [DOI] [PubMed] [Google Scholar]
- 22.Wissing J., Jänsch L., Nimtz M., Dieterich G., Hornberger R., Kéri G., Wehland J., Daub H.( 2007) Proteomics analysis of protein kinases by target class-selective prefractionation and tandem mass spectrometry. Mol. Cell. Proteomics 6, 537– 547 [DOI] [PubMed] [Google Scholar]
- 23.Bantscheff M., Eberhard D., Abraham Y., Bastuck S., Boesche M., Hobson S., Mathieson T., Perrin J., Raida M., Rau C., Reader V., Sweetman G., Bauer A., Bouwmeester T., Hopf C., Kruse U., Neubauer G., Ramsden N., Rick J., Kuster B., Drewes G.( 2007) Quantitative chemical proteomics reveals mechanisms of action of clinical ABL kinase inhibitors. Nat. Biotechnol. 25, 1035– 1044 [DOI] [PubMed] [Google Scholar]
- 24.Rae J. M., Creighton C. J., Meck J. M., Haddad B. R., Johnson M. D.( 2007) MDA-MB-435 cells are derived from M14 melanoma cells–a loss for breast cancer, but a boon for melanoma research. Breast Cancer Res. Treat. 104, 13– 19 [DOI] [PubMed] [Google Scholar]
- 25.Barvian M., Boschelli D. H., Cossrow J., Dobrusin E., Fattaey A., Fritsch A., Fry D., Harvey P., Keller P., Garrett M., La F., Leopold W., McNamara D., Quin M., Trumpp-Kallmeyer S., Toogood P., Wu Z., Zhang E.( 2000) Pyrido[2,3-d]pyrimidin-7-one inhibitors of cyclin-dependent kinases. J. Med. Chem. 43, 4606– 4616 [DOI] [PubMed] [Google Scholar]
- 26.Brehmer D., Greff Z., Godl K., Blencke S., Kurtenbach A., Weber M., Müller S., Klebl B., Cotten M., Kéri G., Wissing J., Daub H.( 2005) Cellular targets of gefitinib. Cancer Res. 65, 379– 382 [PubMed] [Google Scholar]
- 27.Senis Y. A., Craig A. W., Greer P. A.( 2003) Fps/Fes and Fer protein-tyrosinekinases play redundant roles in regulating hematopoiesis. Exp. Hematol. 31, 673– 681 [DOI] [PubMed] [Google Scholar]
- 28.Wessel D., Flügge U. I. ( 1984) A method for the quantitative recovery of protein in dilute solution in the presence of detergents and lipids. Anal. Biochem. 138, 141– 143 [DOI] [PubMed] [Google Scholar]
- 29.Shevchenko A., Tomas H., Havlis J., Olsen J. V., Mann M. ( 2006) In-gel digestion for mass spectrometric characterization of proteins and proteomes. Nat. Protoc. 1, 2856– 2860 [DOI] [PubMed] [Google Scholar]
- 30.Rappsilber J., Mann M., Ishihama Y. ( 2007) Protocol for micro-purification, enrichment, pre-fractionation and storage of peptides for proteomics using StageTips. Nat. Protoc. 2, 1896– 1906 [DOI] [PubMed] [Google Scholar]
- 31.Larsen M. R., Thingholm T. E., Jensen O. N., Roepstorff P., Jørgensen T. J. ( 2005) Highly selective enrichment of phosphorylated peptides from peptide mixtures using titanium dioxide microcolumns. Mol. Cell. Proteomics 4, 873– 886 [DOI] [PubMed] [Google Scholar]
- 32.Pinkse M. W., Uitto P. M., Hilhorst M. J., Ooms B., Heck A. J. ( 2004) Selective isolation at the femtomole level of phosphopeptides from proteolytic digests using 2D-NanoLC-ESI-MS/MS and titanium oxide precolumns. Anal. Chem. 76, 3935– 3943 [DOI] [PubMed] [Google Scholar]
- 33.Schroeder M. J., Shabanowitz J., Schwartz J. C., Hunt D. F., Coon J. J. ( 2004) A neutral loss activation method for improved phosphopeptide sequence analysis by quadrupole ion trap mass spectrometry. Anal. Chem. 76, 3590– 3598 [DOI] [PubMed] [Google Scholar]
- 34.Olsen J. V., de Godoy L. M., Li G., Macek B., Mortensen P., Pesch R., Makarov A., Lange O., Horning S., Mann M. ( 2005) Parts per million mass accuracy on an Orbitrap mass spectrometer via lock mass injection into a C-trap. Mol. Cell. Proteomics 4, 2010– 2021 [DOI] [PubMed] [Google Scholar]
- 35.Gnad F., Ren S., Cox J., Olsen J. V., Macek B., Oroshi M., Mann M.PHOSIDA (phosphorylation site database): management, structural and evolutionary investigation, and prediction of phosphosites. Genome Biol. 20078R250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Maere S., Heymans K., Kuiper M. ( 2005) BiNGO: a Cytoscape plugin to assess overrepresentation of gene ontology categories in biological networks. Bioinformatics 21, 3448– 3449 [DOI] [PubMed] [Google Scholar]
- 37.Shannon P., Markiel A., Ozier O., Baliga N. S., Wang J. T., Ramage D., Amin N., Schwikowski B., Ideker T. ( 2003) Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 13, 2498– 2504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Adachi J., Kumar C., Zhang Y., Olsen J. V., Mann M. The human urinary proteome contains more than 1500 proteins, including a large proportion of membrane proteins. Genome Biol. 2006;7:R80. doi: 10.1186/gb-2006-7-9-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wissing J., Godl K., Brehmer D., Blencke S., Weber M., Habenberger P., Stein-Gerlach M., Missio A., Cotten M., Müller S., Daub H. ( 2004) Chemical proteomic analysis reveals alternative modes of action for pyrido[2,3-d]pyrimidine kinase inhibitors. Mol. Cell. Proteomics 3, 1181– 1193 [DOI] [PubMed] [Google Scholar]
- 40.Blencke S., Zech B., Engkvist O., Greff Z., Orfi L., Horváth Z., Kéri G., Ullrich A., Daub H. ( 2004) Characterization of a conserved structural determinant controlling protein kinase sensitivity to selective inhibitors. Chem. Biol. 11, 691– 701 [DOI] [PubMed] [Google Scholar]
- 41.Mann M. ( 2006) Functional and quantitative proteomics using SILAC. Nat. Rev. Mol. Cell Biol. 7, 952– 958 [DOI] [PubMed] [Google Scholar]
- 42.Gilliland D. G., Griffin J. D. ( 2002) The roles of FLT3 in hematopoiesis and leukemia. Blood 100, 1532– 1542 [DOI] [PubMed] [Google Scholar]
- 43.Su A. I., Cooke M. P., Ching K. A., Hakak Y., Walker J. R., Wiltshire T., Orth A. P., Vega R. G., Sapinoso L. M., Moqrich A., Patapoutian A., Hampton G. M., Schultz P. G., Hogenesch J. B. ( 2002) Large-scale analysis of the human and mouse transcriptomes. Proc. Natl. Acad. Sci. U.S.A 99, 4465– 4470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang Y. X., Knyazev P. G., Cheburkin Y. V., Sharma K., Knyazev Y. P., Orfi L., Szabadkai I., Daub H., Kéri G., Ullrich A. ( 2008) AXL is a potential target for therapeutic intervention in breast cancer progression. Cancer Res. 68, 1905– 1915 [DOI] [PubMed] [Google Scholar]
- 45.Patricelli M. P., Szardenings A. K., Liyanage M., Nomanbhoy T. K., Wu M., Weissig H., Aban A., Chun D., Tanner S., Kozarich J. W. ( 2007) Functional interrogation of the kinome using nucleotide acyl phosphates. Biochemistry 46, 350– 358 [DOI] [PubMed] [Google Scholar]
- 46.Li J. J., Li S. A. ( 2006) Mitotic kinases: the key to duplication, segregation, and cytokinesis errors, chromosomal instability, and oncogenesis. Pharmacol. Ther. 111, 974– 984 [DOI] [PubMed] [Google Scholar]
- 47.Strebhardt K., Ullrich A.( 2006) Targeting polo-like kinase 1 for cancer therapy. Nat. Rev. Cancer 6, 321– 330 [DOI] [PubMed] [Google Scholar]