Abstract
There are relatively few protocols described for the in situ detection of microRNA (miRNA) and they often use cryostat sections, signal amplification and hybridization or washes of 50−60 °C. This protocol describes in situ miRNA detection that can be done in paraffin-embedded, formalin-fixed tissue. Detection of the miRNA precursors can be done by RT in situ PCR, which can theoretically detect one copy per cell. The key variable for the RT in situ PCR protocol is optimal protease digestion, which is then followed by overnight DNase digestion and target specific incorporation of the reported nucleotide into the amplified cDNA. Detection of mature miRNAs is achieved by in situ hybridization with locked nucleic acid probes. This part of the protocol involves a brief protease digestion, followed by an overnight hybridization, short low stringency wash and detection of the labeled probe. The key variables for this method include probe concentration and stringency conditions. Each miRNA in situ method takes 1 d. The final step of the protocol involves colabeling by immunohistochemistry for the putative target of the miRNA, which is done after the in situ hybridization step and takes a few hours.
INTRODUCTION
MicroRNAs are small, noncoding sequences of approximately 20−23 nucleotides that regulate gene expression by being able to inactivate specific mRNAs through annealing to their 3′-untranslated region. Evidence suggests that processing of the inactive precursor miRNA into the mature, active form by Dicer may be, in part, regulated by the sequestration of the precursor form, typically in the nucleus1,2. Therefore, precursor export may be one method that cells use to modulate miRNA expression and underscores the value of determining the specific cellular compartmentalization of the precursor and mature forms of a given miRNA.
MicroRNAs are involved in many biological processes such as cell differentiation, hypoxia, apoptosis, stem cell differentiation, proliferation, inflammation and response to infection. They are also central to oncogenesis where they have the potential to function as either tumor suppressors or oncogenes (oncomiRs) (reviews in refs. 3-7). miRNA profiles are highly correlated with cancer diagnosis, prognosis and response to therapy3-7. Thus, it is most likely that miRNA in situ analysis may play a major role in the clinical laboratories in terms of cancer diagnosis and therapeutics.
There is at present no agreed-upon criteria for the determination of miRNA targets8. Various bioinformatics algorithms (such as miRanda, TargetScan and PicTar) are one avenue that can be used to define possible miRNA target sites within a given mRNA. Such initial analyses can be supplemented by vehicles, including mFold8. Additionally, one may clone the 3′-untranslated region region of the target gene upstream of a reporter system, such as luciferase, and cotransfect this with the miRNA of interest and determine if gene expression is repressed8. Although useful, even the latter method does not simulate the exact conditions in vivo in the cells or tissue of interest. This can be better accomplished by studying the intact tissues of interest using in situ-based methodologies. A simple strategy for such analyses uses serial section testing by in situ hybridization for the miRNA and immunohistochemistry for the putative protein target. The serial sections are 4 μm apart. As most cells are much larger than 4 μm, one can study exactly the same cells for the expression of a given miRNA and its putative target. A more direct approach involves colabeling experiments on the same tissue using a protocol we developed for RNA and protein codetection9.
We describe a protocol that has three components: RT in situ PCR to detect the miRNA precursors, miRNA in situ hybridization with locked nucleic acid (LNA) probes and the analysis of putative miRNA targets by immunohistochemistry by colabeling for the protein of interest or, if this is not technically feasible, on serial sections. One could alternatively do RT in situ PCR without using direct incorporation of a reporter nucleotide into the amplicon but, rather, doing the RT in situ PCR and then doing an in situ hybridization step to detect the newly synthesized amplicon. This process is called RT PCR in situ hybridization and, in our experience, both methods work equally well assuming, of course, that the controls give the expected results10. The limitations of this protocol include the following: the primers used for RT in situ PCR cannot differentiate between the precursor and primary miRNA molecules; the LNA in situ hybridization method may not detect low-copy miRNAs, and certain proteins may be difficult to ‘unmask’ with the immunohistochemistry method; and, thus, although the automated system is ultrasensitive, protein expression can be scored as a false-negative result. The potential applications reflect the advantages of an in situ hybridization system over other miRNA detection systems where the tissue is destroyed before molecular analyses. These include identification of what specific cell type(s) contain the miRNA of interest, the determination of the subcellular compartment that contains the given miRNA and the ability to determine if the presence of a specific miRNA in a cell is associated with silencing of a given mRNA or protein.
Table 1 lists the key features of several recently published protocols for the in situ detection of miRNA using LNA probes11-15 and compares them with our protocol. Note that each of the other protocols uses a prehybridization step, routinely use temperatures of 50−60 °C for the hybridization and stringent washes, and commonly uses the tyramide signal amplification as well as cryostat sections. In comparison, our method has no prehybridization or tyramide signal amplification steps, uses paraffin-embedded, formalin-fixed tissue and, most importantly, addresses the key relationship between miRNA copy number, LNA probe concentration and the stringency of the wash for optimal results.
TABLE 1.
Comparison of different in situ hybridization protocols for miRNA detection.
| Reference | Protease/prehybrization | Probe label | Probe concentration | Hybridization time/temperature | Wash | TSA | Tissue | Colabel/RT in situ PCR |
|---|---|---|---|---|---|---|---|---|
| 13 | Yes/Yes | FITC | 1.2 pmol per 20 μl | 2 h/53 °C | 53 °C, 0.1× SCC | Yes | FFPE | No/No |
| 15 | No/Yes | FITC Digoxigenin | 1.0 pmol per 20 μl | 1 h/53 °C | 55 °C, 0.1× SCC | Yes | Frozen | No/No |
| 16 | No/Yes | Digoxigenin | 3.0 pmol per test | 1 h/55 °C | 55 °C, 0.1× SCC | Yes | Frozen | No/No |
| 12 | Yes/Yes | Digoxigenin | 0.1 pmol per 20 μl | 15 h/50 °C | 60 °C, 0.1× SCC | No | Frozen | Yes/No |
| 11 | No/Yes | FITC | Not listed | 3 h/37 °C | 37 °C, not listed | No | FFPE | No/No |
| This protocol | Yes/No | Digoxigenin | 2−40 pmol per 20 μl | 15 h/37 °C | 4 °C, 0.1× SCC | No | FFPE | Yes/Yes |
FITC, fluorescein isothiocyanate; FFPE, formalin-fixed, paraffin-embedded tissue; TSA, tyramide signal amplification.
There is relatively little data on in situ analysis of miRNA expression1,2,11-16. This reflects several issues, including the small size of the mature miRNA, which translates into a melting temperature (Tm) of the miRNA–DNA probe-hybridized complex, which may be too low to allow for its in situ detection. The Tm issue can be addressed with the LNA modification of the nucleotide bases. In the LNA modification, the furanose ring is chemically ‘locked’ through the use of a 2′-0, 4′-C methylene bridge, which limits the movement of the probe–target complex in three-dimensional space. The end result is an increase in Tm of approximately 20−30 °C if six nucleotides are LNA-modified1,12.
It is well documented that, for mRNA and DNA targets, the signal with in situ hybridization is dependent on the target copy number, probe concentration and conditions of stringency10. A major difference between our protocol and the other published protocols is that we demonstrate the need for less stringent conditions and higher probe concentrations if one is to detect low-copy miRNAs.
Experimental design
Overview of procedure steps
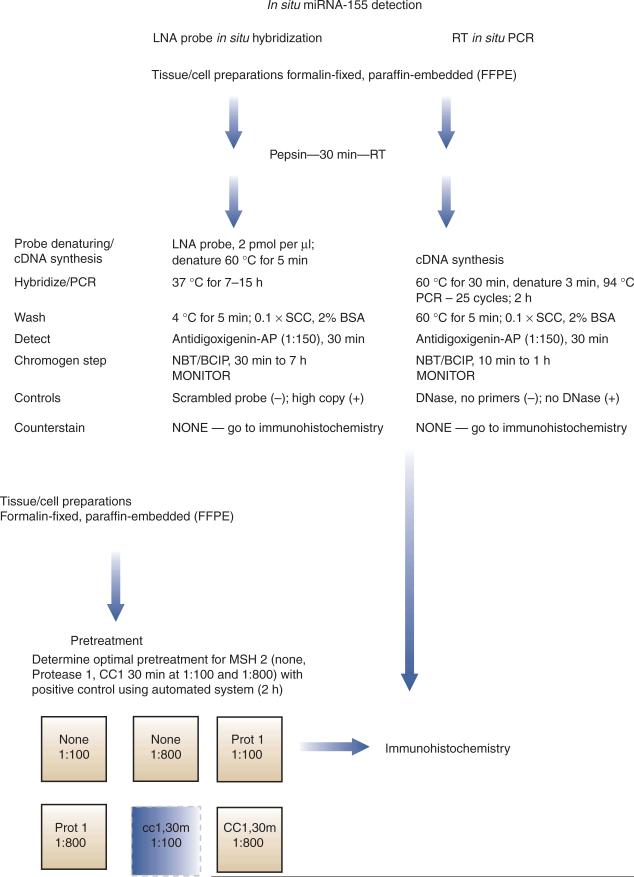
An overview of our protocol is provided in Figure 1. This flowchart diagram illustrates the detection of miRNAs by in situ hybridization using the LNA digoxigenin-labeled probe and of miRNA precursors using RT in situ PCR. It also shows the detection of possible protein targets by colabeling with immunohistochemistry. The diagram uses miR-155 and MSH2 in colon cancers as a specific example; actual data that were obtained from following these protocols are provided in Figure 2.
Figure 1.
A flowchart schematic for the detection of miRNA (mature and precursors) and putative protein targets by colabeling. A simplified protocol for miRNA detection by in situ hybridization with an LNA probe and RT in situ PCR is presented. The LNA probe most likely detects the mature miRNA, whereas the RT in situ PCR detects only the precursor molecules. After either in situ test, one can do colabeling with immunohistochemistry for the putative protein target, assuming that the optimal condition for the latter includes either protease digestion or cell conditioning. One needs to determine the optimal pretreatment for the antigen. The six boxes represent either no pretreatment (None) with dilute (1:800) or concentrated (1:100) primary antibody, protease 1 digestion (Prot 1) or cell conditioning for 30 min (cc1,30) at the two different concentrations. Figure 2c–e shows a representative example of results obtained using the protocol with the detection of miR-155 in colon cancer and colabeling with a possible target, MSH 2.
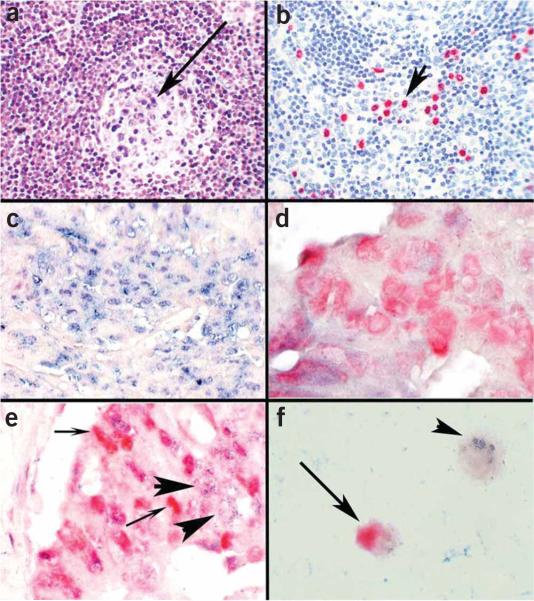
Figure 2.
Determination of putative targets of miRNAs by colocalization analysis with immunohistochemistry. The miRNA miR-155 is markedly upregulated in lymphoproliferative diseases, such as AIDS, and is also increased in colon cancer in which there is often dysregulation of the DNA repair gene MSH 2. (a) miR-155 is present in the atypical lymphocytes but not the endothelial cells as determined by in situ hybridization with an LNA probe; the arrow points to the germinal center where several blue-staining cells indicative of miR-155 expression are seen. Many more miR-155 cells are seen in the interfollicular zone. (b) By analyzing serial sections with immunohistochemistry using an antibody directed against HHV8 and a red chromogen, it was simple to demonstrate that there were over 25 times more miR-155-positive cells than HHV8-positive cells, showing that HHV-8 per se was not responsible for upregulating miR-155; the arrow points to several HHV-8 red staining cells. (c) We performed colocalization experiments with miR-155 and MSH 2, using cell conditioning for 30 min for the MSH2 protein. Note that in cancer cell groups strongly positive for miR-155 (blue, in situ hybridization with an LNA probe), MSH 2 (red, immunohistochemistry) is not evident. (d) Conversely, in colon cancer cell groups where the neoplastic cells expressed MSH 2 (red, immunohistochemistry), miR-155 (blue, in situ hybridization with an LNA probe) was not evident. (e) In some groups of cancer cells, both miR-155 and MSH 2 were present. The colocalization experiments in these groups showed that the cells making MSH 2 (small arrows, red—immunohistochemistry) did not contain miR-155 (large arrows, blue—in situ hybridization with an LNA probe) and vice versa. These data suggest that miR-155 may be downregulating MSH 2. (f) Colocalization experiments with cell lines can also be used to get useful information on whether a given miRNA may be regulating a specific protein. miR-16 downregulates the protein bcl-2, which is an important anti-apoptotic factor in several lymphomas. Colocalization experiments showed that miR-16-positive cells (small arrow, blue—in situ hybridization with an LNA probe) were bcl-2 negative, and the bcl-2-positive cells (large arrow, red—immunohistochemistry, ) were miR-16 negative, lending supporting evidence to a direct role for miR-16 in bcl-2 regulation. Panels a–c are at ×400 and panels d–f are at ×1,000 original magnification.
An outline of the main steps of PROCEDURE is given below:
Prepare silane-coated slides that each contain two to three 4-μm sections of paraffin-embedded, formalin-fixed tissue per slide.
For RT in situ PCR and LNA in situ hybridization, protease-digest the tissues for 30 min using pepsin (1.3 mg ml−1) at room temperature (20−22 °C).
For immunohistochemistry, determine the optimal conditions (proper dilution and pretreatment—no pretreatment, protease digestion or antigen retrieval) for the primary antibody using the automated immunohistochemistry system and a tissue or cell line known to have the target protein in abundance. This takes about 2 h. We use the Benchmark LT from Ventana Medical Systems and the Universal Ultraview Red or DAB kit.
For RT in situ PCR, digest tissue for 7−15 h in RNase-free DNase solution, then wash slides in DEPC-treated water to remove the DNase and in RNase-free 100% (vol/vol) ethanol, and air-dry.
For RT in situ PCR, apply miRNA precursor-amplifying solution, then do cDNA synthesis (30 min at 60 °C), denature at 94 °C for 3 min, then do PCR amplification for 25 cycles (60 °C for 1.5 min and 94 °C for 1 min—about 2 h) using digoxigenin as reporter nucleotide.
For LNA in situ hybridization, incubate probe (2 pmol μl−1) and tissue at 60 °C for 5 min, then hybridize for 7−15 h at 37 °C.
Wash RT in situ PCR slides in 0.2× SSC and 2% bovine serum albumin (BSA) at 60 °C for 5 min; wash LNA in situ hybridization slides in 0.2× SSC and 2% BSA at 4 °C for 5 min.
Incubate RT in situ PCR and LNA in situ hybridization slides in antidigoxigenin–alkaline phosphatase conjugate (1:150 dilution) for 30 min at 37 °C.
Incubate RT in situ PCR and LNA in situ hybridization slides in NBT/BCIP chromogen at 37 °C; monitor reaction under the microscope. Typically, RT in situ PCR requires 10−30 min incubation and LNA in situ hybridization requires 30 min to 7 h incubation.
After RT in situ PCR and LNA in situ hybridization, one can do immunohistochemistry for protein of interest. If so, do not counterstain the in situ-tested slides. Otherwise, one can do the RT in situ PCR and LNA in situ hybridization on serial sections (4 μm apart) from each other and from the immunohistochemistry test.
To interpret results, view results of controls.
Controls
For RT in situ PCR, the key positive control is the tissue not treated with DNase, whereas the negative control is the tissue section treated with DNase, but where the primers are omitted for the RT and PCR steps. Other positive and negative controls one may use for RT in situ PCR include the use of tissues known to be positive and negative, respectively, for the miRNA precursor as determined by quantitative real-time RT-PCR or northern blot analysis17,18. For LNA miRNA in situ hybridization, the standard negative control is the scrambled probe that has the same sequence as the miRNA probe, but the nucleotides are placed in random order. The positive control is tissue or cells known to have a high copy number of the target miRNA as documented by either northern blot or quantitative real-time RT-PCR. For immunohistochemistry, the positive control is a cell line or tissue known to contain the protein of interest; this information is often provided by the manufacturer and/or by a MEDLINE search of the literature. The negative controls include a cell line or tissue known to be negative for the protein, and the internal control, defined by the cell types in the positive control tissue that do not contain the target protein.
Optimization of reaction conditions—protease digestion
Protease digestion exposes the RNA target by removing the RNA–protein adducts that form during formalin fixation. We recommend the use of one protease, so the user can be familiar with its nuances. We prefer pepsin as it rarely causes tissue over digestion, as compared with proteinase K10. However, the latter is our preferred protease for immunohistochemistry, as the automated immunohistochemistry system from Ventana Medical Systems is geared to this protease. We also prefer proteinase K for in situ hybridization if using tissues that have been fixed for days to weeks in formalin, such as autopsy material. Proteinase K is the most commonly used protease with in situ hybridization. Its advantages are that it is much more stable than pepsin and can be stored at 4 °C for months and it is more active than pepsin, which is especially useful for tissues fixed for prolonged times, such as autopsy material. Its main disadvantage is that it tends to overdigest tissue, and thus if used, digestion times should be reduced to 5−10 min. With pepsin, most formalin-fixed tissues will allow detection of the miRNA target after 15−30 min of digestion with a concentration of 1.3 mg ml−1. However, with RT in situ PCR, the optimal protease digestion time can vary a great deal from tissue to tissue. One needs to determine the optimal protease digestion time for RT in situ PCR. This is accomplished by determining which protease digestion time yields an intense nuclear signal in most cell types with direct incorporation of the reporter nucleotide that is eliminated with pretreatment in the RNase-free DNase solution (review in ref. 10).
Primer design and specificity
The primers designed to anneal to the miRNA hairpin will amplify both the pri- and pre-miRNA, as the hairpin is contained within each. Thus, we shall refer to the amplification of miRNA precursors to represent both the pri- and pre-miRNA. One may use the same primers for RT in situ PCR for the miRNA precursors as used in the solution phase RT-PCR. We have tested side by side over 20 primer pairs in solution phase and RT in situ PCR for miRNA precursors, with excellent correlation1,2. Assuming the primers are about 20 nucleotides in size, we have noted that RT in situ PCR can amplify amplicons from 100 to around 600 nucleotides10.
Probe selection and labeling
Table 1 highlights other recently published protocols for the detection of miRNA in situ; each use LNA probes. We also recommend, as is done by these other protocols, LNA modification of at least 6 of the 17−21 nucleotides of the probe. If one wishes to label the probe (e.g., with a radioactive nucleotide for northern blot analysis), then we prefer the 3′-oligoprobe terminal transferase end-tailing kit by Enzo Life Sciences10. We recommend using LNA-modified probes that are already 5′-labeled with digoxigenin. The sequences of the mature miRNA are readily available on the Internet (http://microrna.sanger.ac.uk/sequences/).
Hybridization conditions
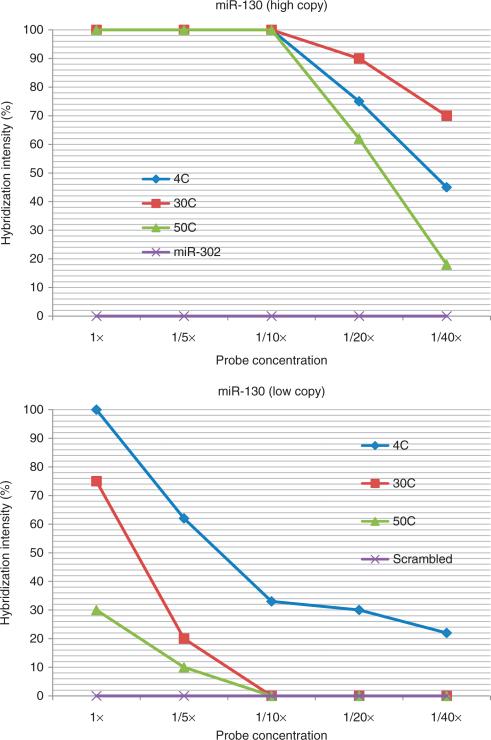
There is a critical relationship between miRNA copy number, probe concentration, stringency of the wash and intensity of signal when doing miRNA in situ hybridization with the LNA probe. This information is presented in Figures 3 and 4; the copy number data are provided by ref. 19. Note that for high-copy miRNAs, one can use 0.2 pmol μl−1 of the probe with a stringent wash of 50 °C and still get near maximum signal. However, if the mRNA copy number is reduced 94% relative to the high-copy miRNA, then maximum signal requires a concentration of 2 pmol μl−1 with a wash at 4 °C. Importantly, under these conditions, the scrambled probe shows no background. Thus, we recommend hybridization using 2 pmol μl−1 for 7−15 h with a wash at 4 °C for 5 min to be able to detect most miRNAs, even when present in relatively low copy number.
Figure 3.
Optimizing experiments for miRNA detection by in situ hybridization with an LNA probe: importance of miRNA copy number, probe concentration and temperature of the post-hybridization wash. The top panel depicts a series of experiments in which placenta were tested for miR-130 with an LNA probe. The concentration of the probe ranged from 2 pmol μl−1 (1×) to 0.05 pmol μl−1 (1/40×). The temperature of the post-hybridization wash was set at 4, 30 or 50 °C. We also tested miR-302 by in situ hybridization as a negative control, as this miRNA is not expressed in the placenta; the post-hybridization wash was at 4 °C. The data points represent the mean of duplicate analyses done blinded to the reaction conditions; the standard error of the mean was no more than 10% of the mean value. In comparison, the bottom panel depicts an equivalent series of experiments with miR-130 but with brain tissues. miR-130 is present in a copy number in brain that is about 93% less than in placenta19. The data with the scrambled probe (negative control) are included; the post-hybridization wash was done at 4 °C. Note the strong relationship between probe concentration, stringency of the post-hybridization wash and the relative miRNA copy number.
Figure 4.
LNA-based in situ detection of miRNAs: keys to successful hybridization. The placenta contains high-copy miR-13019. In situ hybridization for miR-130 shows an intense signal (brown, NBT/BCIP) with a probe concentration of 0.4 pmol μl−1 if the post-hybridization wash is at (a) 4 °C or (b) 50 °C. The inset shows no signal with the scrambled probe under the same conditions of either panel a or b. In comparison, the copy number of miR-130 is 93% less in the brain compared with the placenta19. Panel c shows a signal in the brain at a probe concentration of 0.05 pmol μl−1 if the post-hybridization wash is at 4 °C but no signal if the post-hybridization wash is at 50 °C, (d) even if the probe concentration is increased to 0.8 pmol ml−1. The inset in panel d shows that, under similar conditions, a strong signal is seen in mouse embryo if the stem cell marker miR-302 probe is used at a concentration of 0.4 pmol μl−1. By combining RT in situ PCR for the precursor with LNA in situ hybridization, one can determine that benign tissues (such as the leiomyoma in panel e) can show miR-221 precursors (blue signal) and not the mature form (inset—LNA probe where no signal was evident); note the nuclear localization of the miRNA precursor. (f) When the mature form of miR-221 is evident, as is typical in this invasive leiomyosarcoma, the miR-221 precursor (RTISPCR) and mature miR-221 signal from the LNA probe both tend to localize to the cytoplasm (each panel is at ×400 original magnification).
MATERIALS
REAGENTS
Formalin, 10%, neutral-buffered (Fisher Scientific, 23−245−684) ! CAUTION Allergenic; work in fume hood.
Diethylprocarbonate (DEPC) water (Fluka, 32490) ! CAUTION Carcinogenic; work in fume hood.
3′-End tailing oligonucleotide kit (Enzo Diagnostics, 42730)
Biotin-modified dUTP (Enzo Diagnostics, 42721)
Streptavidin—alkaline phosphatase detection kit (Enzo Diagnostics, 32895; contains washes, alkaline phosphatase conjugate, NBT, BCIP chromogen, counterstain, proteinase K, silane-coated glass slides) ▲ CRITICAL The most reliable source of the NBT/BCIP in our experience is from Enzo Diagnostics as part of their alkaline phosphatase-based detection systems.
20× SSC (Ventana Medical Systems, 950−110)
Antidigoxigenin alkaline phosphatase conjugate (Roche Diagnostics, 11 093 274 910) ▲ CRITICAL The most reliable source of the antidigoxigenin–alkaline phosphatase conjugate in our experience is Roche; do not freeze this conjugate, as it will lose much of its activity.
RNase-free DNase (Roche Diagnostics, 10 776 785 001)
RNase inhibitor (Roche Diagnostics, 3335 402)
Bovine albumin (MP Biomedicals, ICN820451)
In situ hybridization buffer (Enzo Diagnostics, 33808)
Pepsin powder (Dako, S3002)
Digoxigenin-11-dUTP (Roche, 11 558 706 910)
EZ rTth kit (Applied Biosystems, N8080179)
Ultraview universal alkaline phosphatase red detection kit for immunohistochemistry (Ventana Medical Systems, 760−501)
Proteinase K solution (Ventana Medical Systems, 760−2018)
Antibodies for immunohistochemistry: here is a partial list of antibodies that, from our experience, should allow for colabeling using our protocol: RANTES, IL-2, TNF-α, IFN-γ, IL-6, bcl-2, CBP, CD 133, CAB 39, FOXP1, PTEN (each from Abcam), CD3, CD20, CD45, CD68, CD56, MSH2, p27, GFAP, her 2 neu, Ki-67, S-100 (each from Ventana Medical Systems) and HMB-45 (from Enzo Clinical Labs)
EQUIPMENT
Silane-coated glass slides (Fisher Scientific, 12−550−15)
Coverslips (Fisher Scientific, 12−548−5M)
Protocol mounting media (Fisher Scientific, SP15−100)
Incubator (Fisher Scientific, 97−990E)
Hot plate (up to 100 °C) (Enzo Diagnostics)
Microscope and digital camera
Ventana Medical Systems Benchmark or Discovery-automated immunohistochemistry and in situ hybridization system
REAGENT SETUP
Pepsin solution
Prepare pepsin solution by diluting 13 mg of the pepsin into 9.5 ml of DEPC-treated water and 0.5 ml of 0.2 N HCl. The solution can be frozen at −20 °C in 1-ml aliquots and thawed immediately before use.
PROCEDURE
Tissue preparation
• TIMING 30 min
1 Obtain paraffin-embedded, formalin-fixed tissue. These samples are usually obtained from a surgical pathology laboratory. ▲ CRITICAL STEP Formalin fixation will inactivate infectious agents. However, it is recommended that one follows universal precautions when working with formalin-fixed tissues. One does not need to obtain formalin solution made in DEPC water, as the formalin probably inactivates any RNase10.
▲ CRITICAL STEP It should be stressed that fixatives that contain other ingredients, such as picric acid (Bouin's solution) or heavy metals (Zenker's solution), are contraindicated with any in situ-based method, as fixation with these molecules results in the rapid degradation of RNA and DNA (review in ref. 10). Furthermore, alcohol-based fixatives often yield poor morphology with in situ-based methods, especially in situ PCR10.
2 Examine representative hematoxylin-and-eosin (H&E) stain under the microscope (ref. 10 contains a section on H&E interpretation for those with minimal experience in histopathology) to confirm that the tissue has the appropriate histological features. For example, if one is doing a study on breast cancers, examine the H&E and confirm that the deeper levels of the block still contain the cancer.
3 Have a histology technician with a microtome place 2−3 sections that are 3−4 μm in thickness on sequentially labeled and numbered silane-coated slides. We float the sections on distilled water before placing on the slide and have not noted any improvement in signal using RNase-free water. Store slides in a box at room temperature. In our experience, the slides do not have to be treated with RNase inhibitor before adding the sections.
■ PAUSE POINT The slides can thus be stored for many years and still support in situ RNA analyses10.
Protease digestion
• TIMING 30 min
4 Place 50−200 μl of pepsin solution on each tissue section, depending on the tissue size. Digest the tissues for 30 min at room temperature.
▲ CRITICAL STEP Pepsin digestion (Steps 4−6) is necessary only for tissues that will be used for RT in situ PCR and LNA in situ hybridization (not for immunohistochemical analysis).
5 Stop the protease digestion by rinsing the slides for 30 s in DEPC-treated water, followed by 30 s in RNase-free 100% ethanol.
6 Air-dry slides.
▲ CRITICAL STEP Slides should be tested within 7 h.
▲ CRITICAL STEP Protein–RNA cross-links can also be degraded by heat. Thus, antigen retrieval can be used as an alternative method to expose the miRNA target.
In situ PCR/hybridization
7 Slides from Step 6 can be used for RT in situ PCR to allow precursor miRNA detection (option A) or for in situ hybridization with LNA probes to allow mature miRNA detection (option B).
(A) RT in situ PCR for miRNA precursor detection
• TIMING 1 d
DNase digestion. Place 10−30 μl of RNase-free DNase (dilute the stock solution 1:10 with the buffer from the EZ rTth kit) onto slides from Step 6 and incubate at 37 °C for 7−15 h.
Remove the DNase with a brief wash in DEPC-treated water and allow the slides to air-dry, which can be facilitated with a quick dip in RNase-free 100% ethanol.
Synthesize and amplify the miRNA-specific cDNA. Prepare the following solution as per the EZ r Tth kit: 10 μl of EZ buffer, 1.6 μl each of the four nucleotides, 1.6 μl of 2% BSA (to block adsorption of the enzyme on the glass), 0.6 μl of 1 nmol μl−1 digoxigenin dUTP, 3 μl of the primer set (the same primers used for detection of the miRNA precursors in solution phase can be used at a concentration of 20 μM; use irrelevant/scrambled or no primers for the negative control), 12.4 μl of 10 mM MnAcetate, 2 μl of rTth, 1.4 μl of RNase inhibitor and 12.6 μl of DEPC-treated water.
Apply 5−15 μl (depending on size) of the reaction mixture from Step A(iii) immediately to tissue sections from Step A(ii) and overlay with a polypropylene coverslip to prevent evaporation. This, in turn, can be anchored onto the slide with superglue or nail polish.
Place slides in an aluminum boat, which is then covered with mineral oil.
Perform cDNA synthesis by incubating slides for 30 min at 60 °C.
Amplify cDNA after denaturing at 94 °C for 3 min with 25 cycles at 60 °C annealing or extension (1.5 min) and 94 °C denaturation (1 min).
Rinse slides in xylene and 100% ethanol, respectively, for 3−5 min, and air-dry.
(B) In situ hybridization with LNA probes for mature miRNA detection
• TIMING 1 d
Dilute 125 pmol of the digoxigenin-labeled LNA probe with 62.5 μl of the Enzo in situ hybridization buffer.
Apply 5−10 μl of miRNA LNA probe cocktail (from Step B(i)) per tissue on slides from Step 6, cover with a sterile polypropylene coverslip, heat to 60 °C for 5 min and incubate for 7−15 h at 37 °C.
Stringency wash
• TIMING 5 min
8 Wash the RT in situ PCR slides (from Step 7A(viii)) in 0.2× SSC with 2% BSA at 60 °C for 5 min. Wash the LNA in situ slides (from Step 7B(ii)) in 0.2× SSC with 2% BSA at 4 °C for 5 min.
Detection of the digoxigenin-labeled miRNA precursor cDNA or LNA probe–miRNA complex
• TIMING 1 h
9 Incubate each slide with 100 μl of antidigoxigenin/alkaline phosphatase (1:150 dilution in PBS, pH 7) for 30 min at 37 °C.
10 Wash the slides with 500 μl of the pH 9.5 detection buffer for 1 min at room temperature.
11 Incubate the slides in the NBT/BCIP reagent at 37 °C (add 50 μl of each to 15 ml of the pH 9.5 detection buffer) for 5 min to several hours (see below).
12 Carefully monitor the colorimetric reaction under the microscope and stop by placing the slides in water when there is a strong signal in the positive control and no to minimal background in the negative control.
▲ CRITICAL STEP The incubation time in the chromogen for RT in situ PCR slides typically ranges from 5 to 20 min. For the LNA in situ slides, this typically ranges from 1 to 5 h.
13 Counterstain with nuclear fast red for 1 min, wash in water, rinse in 100% ethanol for 3−5 min, fresh xylene for 3−5 min, then coverslip with permount and view under the microscope.
▲ CRITICAL STEP Do not counterstain or coverslip if one wishes to do colabeling with immunohistochemistry after the in situ detection of the miRNA (see Fig. 1).
Immunohistochemical detection of miRNA targets or colocalization of an miRNA with its putative target
• TIMING 2 h
14 Optimize the pretreatment conditions and the concentration of the primary antibody. We use the automated immunohistochemistry system from Ventana Medical Systems called the Benchmark LT. As shown in Figure 1, take four slides from one tissue that is known to contain the target protein in high copy number. To determine the optimal conditions for that antigen, program the machine to incubate two of the slides in cell conditioning one solution (i.e., antigen retrieval) for 30 min using a concentrated primary antibody (1:100) and more dilute primary antibody (1:800), respectively. The other two slides should contain at least two sections per slide; treat one section with the Ventana Protease 1 for 4 min and leave the other section untreated. These two slides should be programmed under ‘no paraffin’ with the automated system and then also treated with the two disparate concentrations of the primary antibody as for the first two slides.
▲ CRITICAL STEP Both Ventana systems (the Benchmark and Discovery) allow one to determine the concentration of the primary antibody by manual titration. We recommend using the Ultraview Universal Red system, as we prefer the red signal for the protein and blue signal for miRNA colabeling.
15 Determine the optimal antibody dilution and pretreatment by checking for a strong signal in the cells known to have the target protein and no/minimal background in the cells that do not contain the protein.
▲ CRITICAL STEP In our experience, in over 80% of cases, this simple protocol will give the optimizing conditions. Most commonly, one will need to further dilute the primary antibody to eliminate background, as the Ultraview system is very sensitive. Rarely, optimal conditions require cell conditioning 2.
16 Colabel the protein and the miRNA. After one determines the optimal conditions (primary antibody concentration and pretreatment) with the automated immunohistochemistry system, the target protein can be labeled using this system and predetermined conditions (as described in Step 14) either on slides from Step 12 to colabel the protein and the miRNA of interest or using new slides that represent serial sections that are 4 μm apart from those on which in situ hybridization was carried out, and thus will contain the same cells.
▲ CRITICAL STEP Specifically, if the optimal conditions for antigen detection include protease digestion or antigen retrieval, one can often colabel using the miRNA in situ-tested slide. If the antigen requires cell conditioning for optimal conditions, then do the colabeling after miRNA detection using the same cell conditioning protocol; we have had good results under these conditions (Fig. 2). If the antigen requires no pretreatment, then one must do the miRNA detection and antigen detection on serial sections; direct colabeling will not work due to the protease digestion needed for miRNA detection and our observation that miRNA detection does not work well in colabeling if the immunohistochemistry is done first. If the antigen requires protease digestion, we recommend doing an additional protease digestion step with the Ventana Protease 1 for the immunohistochemistry colabeling, as the pepsin digestion for miRNA in situ is often too weak to expose the antigen for immunohistochemistry.
• TIMING
The time required for each step is detailed in the flowchart in Figure 1. We typically begin the LNA in situ and RT in situ PCR in the morning and finish the work by noon of the next day. The immunohistochemistry can be started in the morning and finished by mid-afternoon of the same day.
? TROUBLESHOOTING
RT in situ PCR
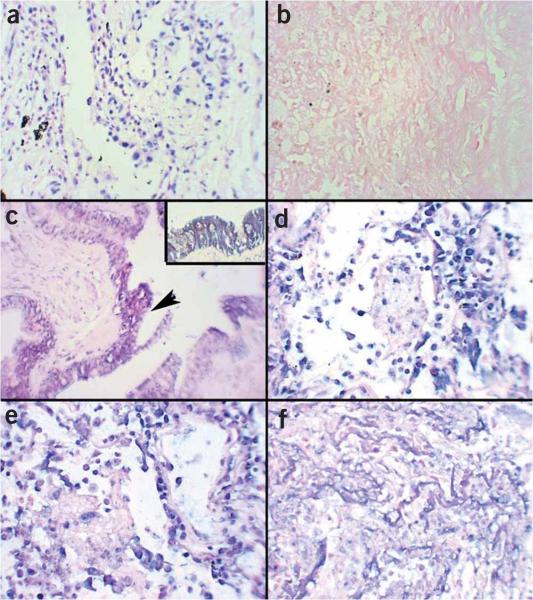
In our experience, the one essential variable with RT in situ PCR is protease digestion time: too little, and the DNase will not be able to degrade the host DNA, which will result in a false-positive nuclear-based signal in the negative control; too much, and the tissue morphology will be destroyed (Fig. 5).
Figure 5.
RT in situ PCR for precursor miRNA detection: keys to successful amplification. The key to troubleshooting with RT in situ PCR for miRNA precursor detection is to analyze the positive (no DNase) and negative (DNase, irrevelant or scrambled primers control) before examining the test section. (a) The PCR in situ positive control must show an intense nuclear-based signal that is completely eliminated in b the negative control for the results for c the miRNA precursors to be accurate. The test is for miRNA-126, which is present in the pulmonary epithelium; note how the signal is evident only in these cells (arrow) and not the surrounding tissue, which is an important histological-based internal control; the inset shows the cytoplasmic signal that localizes to the bronchiole epithelium. Note what happens if the protease digestion time is too short; a signal is seen in the negative control (DNase, no primers—panel d) and (e) many different cell types besides the bronchiole epithelium are positive in the test. Also note how the colorimetric reaction occurs in the nucleus as compared with the cytoplasmic pattern when the controls work properly (panel c). Finally, note that when the protease digestion time is too long, the cell morphology is no longer recognizable and only the more resistant basement membranes are seen; thus, no signal is evident (panel f). Each panel is at 400× except the inset, which is at 1,000× original magnification.
In sum, with RT in situ PCR10:
if a nuclear based signal is seen with the negative (DNase, irrelevant or no primers) control—increase protease digestion time and redo the experiment.
if tissue morphology is poor—decrease protease digestion time and/or dilute protease and redo the experiment.
if no signal is seen with the positive (no DNase) control, this most likely reflects either too short a protease digestion time or the use of a fixative that contains a heavy metal or picric acid or a detection system problem (inactive NBT/BCIP or the alkaline phosphatase/anti digoxigenin conjugate). The most likely reason is the protease, so the first step is to use a more concentrated protease for a longer time period. If this is not successful, test the detection system using a separate in situ assay (such as human papillomavirus 6/11 in situ and vulvar warts) that contains a very high copy number and invariably yields a strong signal. If no signal is evident, the most likely problem is inactive NBT/BCIP or the alkaline phosphatase conjugate.
LNA-based in situ hybridization
The most common problems are background (a signal in a tissue known to be negative for the miRNA) and no signal or too weak a signal in a tissue known to contain the miRNA in high-copy number. In our experience, the problem of no to weak signal is far more common than background. If background is noted, this is best solved by diluting the probe 1:1 with the in situ hybridization buffer and retesting the negative control tissue. The best solution for no signal is, as evident in Figure 3, to increase the concentration of the probe and to be certain that one is using a low-stringency post-hybridization wash done at 4 °C.
Immunohistochemistry
Background
The presence of a colorimetric reaction with a given primary antibody in the negative control tissue is usually solved by (i) diluting the antibody, (ii) determining which pretreatment is associated with background by testing the negative tissue initially with no pretreatment, protease digestion and antigen retrieval 1 solution and (iii) determining which condition(s) yields a colorimetric reaction. In our experience, it is very unusual not to be able to eliminate background with the automated system from Ventana Medical Systems by adjusting primary antibody concentration and pretreatment conditions.
No signal
The key here is to make certain that the tissue or cell line is known to contain the protein of interest and that the primary antibody is compatible with the Ventana system (it must be made in a mouse or a rabbit). Under these circumstances, the lack of signal is best solved by increasing the concentration of the primary antibody, increasing the hybridization time and scrupulously testing each possible pretreatment regime. Another trick of the trade is not to freeze and rethaw the primary antibody, as this will often result in a diminution of the signal. In our experience, it is very unusual not to be able to generate a signal with the Ultraview Universal systems in tissues known to contain the protein target. However, when doing colabeling experiments, we have found that the miRNA in situ hybridization must go first; the tissue handling during immunohistochemistry may render the miRNA otherwise undetectable. Thus, the antigen must be able to withstand protease digestion for detection by immunohistochemistry. In our experience, antigens that require cell conditioning pretreatment work fine with colabeling after miRNA in situ analysis; one uses the same cell conditioning regime.
ANTICIPATED RESULTS
The anticipated results for the different assays that make up this protocol are as follows:
RT in situ PCR for miRNA precursors
A strong nuclear-based signal is seen in the no-DNase control, and no signal is seen in the DNase, no/irrelevant primers control. This means that the RT in situ PCR is working properly and that any signal in the test slide denotes miRNA precursors. The expected result from the experimental sample is a signal, either nuclear or cytoplasmic, that localizes to specific cell types given the strong cell tropism exhibited by miRNAs (Fig. 5c).
LNA in situ hybridization for miRNA
A signal is seen in the known positive control and no signal is seen with the scrambled probe. This means that the LNA in situ hybridization reaction is working properly and that any signal in the test slide denotes miRNA, most likely the mature, active form (Figs. 2 and 4). The expected result from the experimental sample is a signal, either nuclear or cytoplasmic, that localizes to specific cell types given the strong cell tropism exhibited by miRNAs.
Immunohistochemistry
A signal is seen in the known positive control and no signal is seen with the known negative control under defined conditions of primary antibody concentration and pretreatment. This means that the immunohistochemistry reaction is working properly and that any signal in the test slide denotes the protein. The expected result from the experimental sample is a signal, either nuclear or cytoplasmic, that localizes to specific cell types given the strong cell tropism exhibited by proteins.
ACKNOWLEDGMENTS
We greatly appreciate the assistance and reagents from Ventana Medical Systems, and the direct assistance of Christopher Roberts, PhD, Kathleen Sergott and Margaret Nuovo, MD, who assisted with the photography and the figures. This work was supported by a grant from the Lewis Foundation (G.J.N.) and R21 CA114304 (T.D.S.).
Footnotes
Reprints and permissions information is available online at http://npg.nature.com/reprintsandpermissions/
References
- 1.Nuovo GJ. In situ detection of precursor and mature microRNAs in paraffin embedded, formalin fixed tissues and cell preparations. Methods. 2008;44:39–46. doi: 10.1016/j.ymeth.2007.10.008. [DOI] [PubMed] [Google Scholar]
- 2.Lee EJ, et al. Classification of microRNA processing patterns in tissues, cell lines, and tumors. RNA. 2008;14:35–42. doi: 10.1261/rna.804508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Calin GA, Croce CM. MicroRNA signatures in human cancers. Nat. Rev. Cancer. 2006;6:857–866. doi: 10.1038/nrc1997. [DOI] [PubMed] [Google Scholar]
- 4.Garzon R, Fabbri M, Cimmino A, Calin GA, Croce CM. MicroRNA expression and function in cancer. Trends Mol. Med. 2006;12:580–587. doi: 10.1016/j.molmed.2006.10.006. [DOI] [PubMed] [Google Scholar]
- 5.Yanaihara N, et al. Unique microRNA molecular profiles in lung cancer diagnosis and prognosis. Cancer Cell. 2006;9:189–198. doi: 10.1016/j.ccr.2006.01.025. [DOI] [PubMed] [Google Scholar]
- 6.Gramantieri L, et al. Cyclin G1 is a target of miR-122a, a microRNA frequently down-regulated in human hepatocellular carcinoma. Cancer Res. 2007;67:6092–6099. doi: 10.1158/0008-5472.CAN-06-4607. [DOI] [PubMed] [Google Scholar]
- 7.Cimmino A, et al. miR-15 and miR-16 induce apoptosis by targeting bcl-2. Proc. Natl. Acad. Sci. USA. 2005;102:13944–13949. doi: 10.1073/pnas.0506654102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kuhn DE, et al. Experimental validation of miR targets. Methods. 2008;44:47–54. doi: 10.1016/j.ymeth.2007.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nuovo GJ. Co labeling using RT in situ PCR: a review. J. Histochem. Cytochem. 2001;49:1329–1339. doi: 10.1177/002215540104901101. [DOI] [PubMed] [Google Scholar]
- 10.Nuovo GJ. PCR In Situ Hybridization: Protocols and Applications. 3rd edn Lippincott, Williams and Wilkins, Raven Press; New York: 1996. [Google Scholar]
- 11.Politz JC, Zhang F, Pederson T. MicroRNA-206 colocalizes with ribosome-rich regions in both the nucleolus and cytoplasm of rat myogenic cells. Proc. Natl. Acad. Sci. USA. 2006;103:18957–18962. doi: 10.1073/pnas.0609466103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Obernosterer G, Martinez J, Alenius M. Locked nucleic acid based in situ detection of microRNAs in mouse tissue sections. Nat. Protoc. 2007;2:1508–1514. doi: 10.1038/nprot.2007.153. [DOI] [PubMed] [Google Scholar]
- 13.Sempere LF, et al. Altered MicroRNA expression confined to specific epithelial cell subpopulations in breast cancer. Cancer Res. 2007;67:11612–11620. doi: 10.1158/0008-5472.CAN-07-5019. [DOI] [PubMed] [Google Scholar]
- 14.Schetter AJ, et al. MicroRNA expression profiles associated with prognosis and therapeutic outcome in colon adenocarcinoma. JAMA. 2008;299:425–436. doi: 10.1001/jama.299.4.425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Silahtaroglu AN, et al. Detection of microRNAs in frozen tissue sections by fluorescence in situ hybridization using locked nucleic acid probes and tyramide signal amplification. Nat. Protoc. 2007;2:2520–2528. doi: 10.1038/nprot.2007.313. [DOI] [PubMed] [Google Scholar]
- 16.Bak M, et al. MicroRNA expression in the adult mouse nervous system. RNA. 2008;14:432–444. doi: 10.1261/rna.783108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jiang J, Lee EJ, Gusev Y, Schmittgen TD. Real-time expression profiling of microRNA precursors in human cancer cell lines. Nucleic Acids Res. 2005;33:5394–5403. doi: 10.1093/nar/gki863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schmittgen TD, Jiang J, Liu Q, Yang L. A high-throughput method to monitor the expression of microRNA precursors. Nucleic Acids Res. 2004;32:43–47. doi: 10.1093/nar/gnh040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liang Y, Ridzon D, Wong L, Chen C. Characterization of microRNA expression profiles in normal human tissues. BMC Genomics. 2007;8:166. doi: 10.1186/1471-2164-8-166. [DOI] [PMC free article] [PubMed] [Google Scholar]