Abstract
Tumor necrosis factor (TNF) and lymphotoxin-α (LTA) are cytokines with a wide range of inflammatory and immunomodulatory activities. Type 1 diabetes is an autoimmune disease characterized by destruction of insulin-producing pancreatic β cells. The aim of the present study was to evaluate the association of polymorphisms in the TNF/LTA gene region with susceptibility to type 1 diabetes. We investigated 11 TNF/LTA tag polymorphisms, designed to capture the majority of common variation in the region, in 160 trio families from South Croatia. We observed overtransmission of alleles from parents to affected child at five variants: (rs909253, allele C, p = 1.2×10−4; rs1041981, allele A, p = 1.1×10−4; rs1800629 (G-308A), allele A, p = 1.2×10−4; rs361525(G-238A), allele G, p = 8.2×10−3 and rs3093668, allele G, p = 0.014). We also identified overtransmission of the rs 1800629(G-308A)-rs361525(G-238A) A-G haplotype, p = 2.384×10−5. The present study found an association of the TNF/LTA gene region with type 1 diabetes. A careful assessment of TNF/LTA variants adjusted for linkage disequilibrium with HLA loci is needed to further clarify the role of these genes in type 1 diabetes susceptibility in the population of South Croatia.
Keywords: Type 1 diabetes, Tumor necrosis factor, Lymphotoxin alpha, TDT, Tag polymorphism
1. Introduction
Type 1 diabetes mellitus (T1DM) results from a cellular-mediated autoimmune destruction of pancreatic β-cells [1]. It has a strong genetic component. The most important genetic factors for determining the risk of developing T1DM reside in the HLA class II loci but also, according to recent findings, in the HLA-B and HLA-A class I genes [2]. Several other gene regions have also been identified: a region 5′ to the INS gene, CTLA4, PTPN22, IL2RA, and the IFIH1 region [3,4]. A recent T1DM genome-wide association study identified another four regions at 12q24, 12q13, 16p13, and 18p11 [5].
The region between HLA class I and II, known as class III, contains a number of genes involved in inflammatory and immune response such as tumor necrosis factor (TNF) and lymphotoxin-α (LTA) [6]. Both genes mediate similar pleiotropic effects and both bind the same TNF receptors [7]. Several activated immune system cells (monocytes, macrophages, and natural killer cells) release TNF, which then induces the expression of HLA class II molecules on the surface of the immune system cells [8,9]. TNF is believed to be one of the main proinflammatory cytokines implicated in the destruction of pancreatic β-cells [10]. However, TNF has opposing effects on T1DM. It is shown in an animal model that TNF can be, supported by both interleukin-1 and interferon-γ, toxic for β-cells [11]. Also, nonobese diabetic (NOD) mice that overexpressed TNF-α in their β-cells are predisposed to diabetes [12]. Neonatal TNF treatment of NOD mice results in earlier onset and increased incidence of T1DM, however, TNF administration to adult NOD mice prevents them from developing T1DM [10]. Also, administration of recombinant TNF to fetal thymus organ culture has a significant impact on the development of T cells such that they no longer cause T1DM [13]. The LTA cytokine is produced by lymphocytes and mediates a large number of inflammatory, immunostimulatory and antiviral responses [6]. The role of LTA was demonstrated as essential for the normal development of peripheral lymphoid organs in mice models [14]. It was shown that administration of LTA in NOD mice and Bio-Breeding (BB) rats may have modulated autoimmunity and prevented development of T1DM [15]. Feugeas et al. observed lower LTA production in T1DM patients than in normal controls, implying that a defect of LTA production can be a T1DM risk factor [7].
A genetic correlation between TNF and LTA genes with T1DM is widely suggested, although some reports show that the association between alleles at the TNF/LTA locus and T1DM can be attributed to linkage disequilibrium (LD) with the susceptible DQB1-DRB1 haplotypes, instead of an independent effect [6,16]. However, several other studies found an independent effect of TNF/LTA with T1DM [17-20]. TNF involvement in the destruction of pancreatic β-cells [10,11] and multiple inflammatory activities of TNF and LTA cytokines make them plausible targets for revealing molecular susceptibility to T1DM. The joint analysis of TNF and LTA genes may be more informative in defining the risk/protection to T1DM susceptibility [7,18].
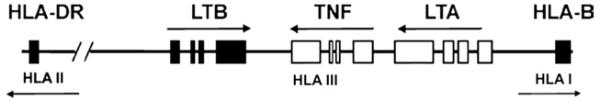
The TNF and LTA genes lie next to each other on chromosome 6p21.3. According to Makhatadze there is a localization of TNF and LTA genes within the HLA complex, as shown in Figure 1 [8]. Two TNF gene promoter single nucleotide polymorphisms (SNPs), G-238A (rs361525) and G-308A (rs1800629), have frequently been studied [6,16-22]. However, inconsistencies in SNP designation through various LTA gene studies makes difficult to distinguish and differentiate investigated SNPs [6,7,18,22]. Studies in North Indian, Hungarian, Bahraini, and Moroccan populations observed associations of TNF -308 SNP and TNF/LTA haplotypes with T1DM independently of HLA loci [17-20,22]. On the other hand, TNF/LTA T1DM associations in Chinese, Caucasian, and Polish populations were explained by dependence on HLA alleles [6,16,21].
Fig. 1.
Localization of TNF and LTA genes inside HLA complex, according to Makhatadze [8].
In this study we investigate 11 SNPs selected as tagging SNPs (tagSNP) for the TNF and LTA genes. These include the two extensively studied TNF promo ter SNPs (rs361525 and rs1800629), five other TNF SNPs (rs1800750, rs3093662, rs3093664, rs3093665, rs3093668), and four LTA SNPs (rs928815, rs909253, rs746868, rs1041981). These SNPs were selected to capture the majority of common variations in the TNF/LTA gene region.
The aim of the present study was to analyze the transmission of 11 TNF/LTA gene tag SNPs in case-parent trio samples and to evaluate their association with susceptibility to T1DM in the population of South Croatia.
2. Subjects and methods
2.1. Subjects
We typed 132 parent– offspring trios, 20 parent– offspring duos, and seven families with two and one family with three affected children from the population of South Croatia. Each proband was ascertained with T1DM according to the World Health Organization criteria. The sex distribution among affected children was 83 (49.11%) males and 86 (50.08%) females and the mean age at the onset of T1DM was 8.86 ± 5.36 (mean ± SD). Table 1 shows clinical, immunologic, and lifestyle characteristics of 88 of the patients who were involved in this study. Immunologic characteristics refer to all other immunologic diseases of patients beside T1DM, mostly Hashimoto thyreoiditis, asthma, and psoriasis. Clinical characteristics refer to history of other diseases of patients such as chicken pox, tonsillitis, scarlet fever, and various allergic reactions. We also show exposure to early introduction of milk formulas before 3 months of age and exposure to stress before the onset of diabetes such as the war in Croatia in the beginning of the 1990s, the loss of a dear person, and various family problems as lifestyle characteristics. We also show the area of living (urban/rural) of the patients. This study was approved by the ethics committee, and informed consent from patients and their parents was obtained prior to the blood sampling.
Table 1.
Clinical, immunological and some life style characteristics of 88 patients who were involved in the study of TNF/LTA genes
| Characteristic | No. of patients (%) N = 88 |
|---|---|
| Positive T1DM family history | 40 (45.4) |
| Other autoimmune diseases | 14 (15.9) |
| Introduction of milk formulas before 3 months of age | 50 (56.8) |
| Chicken pox | 34 (48.9) |
| Tonsillitis | 29 (32.9) |
| Scarlet fewer | 5 (5.7) |
| Allergic reactions | 13 (14.7) |
| Stress before T1DM onset | 33 (37.5) |
| Urban area of living | 55 (62.5) |
2.2. SNP selection and genotyping
Genomic DNA was extracted from peripheral blood leukocytes using the QIAamp DNA Blood Mini Kit (Qiagen, Hilden, Germany). Eleven haplotype-tagging SNPs (rs928815, rs909253, rs746868, rs1041981, rs1800750, rs1800629, rs361525, rs3093662, rs3093664, rs3093665, rs3093668) were selected across the TNF and LTA loci from SNP data generated in 32 Caucasian mother–father–child trios as previously described [23,24]. Genotypes were determined using a fluorescence-based competitive allele specific assay (Kaspar, Kbioscience, UK). Using the same methodology, and as a measure of quality control, all samples were checked for gender. This assay investigates the single base difference between a homologous exon of the zinc-finger genes ZFX and ZFY and is diagnostic for sex chromosome (assay and SNP selection details are available from the authors on request). Gender control genotyping identified six discrepancies when compared to database information. These six individuals and their families were excluded from further analysis.
2.3. Statistical analysis
Before association analysis, we performed quality control of the obtained trio genotypes. We tested for Mendelian inheritance and identified inconsistencies in two families, which were then excluded from the analysis. We evaluated the rate of missing genotypes using Plink version 1.00 (http://pngu.mgh.harvard.edu/purcell/plink/) [25]. The percentage of missing genotypes for SNPs rs746868 and rs3093664 was 5.6% and 2.3%, respectively, but were included in further analyses. For the remaining nine SNPs the percentage of missing genotypes remained less than 1%. Hardy-Weinberg equilibrium (HWE) in healthy parents was tested using Pedstats [26]. Two SNPs showed a slight deviation from HWE in healthy parents (rs928815, p = 0.039 and rs746868, p = 0.035) but were included in further analyses. Minor allele frequencies (MAF) were compared with the National Center for Biotechnology Information SNP database (NCBI dbSNP) MAF for the central European (CEU) population (www.ncbi.nlm.nih.gov/projects/SNP/). MAF in healthy parents was not concordant with NCBI dbSNP frequencies for the CEU population for rs3093664 SNP (MAF_healhy_parents = 0.503, MAF_NCBI = 0.065). Because of the two quality control discrepancies (significant MAF difference with CEU population and a small but noticeable genotype missingness), rs3093664 SNP was excluded from the association analysis.
The proportion of variation across the TNF/LTA region captured by tagSNPs from this study was calculated based on the HapMap [27], using Tagger (http://www.broad.mit.edu/mpg/tagger/server.html) [28]. After quality control, a total of 152 cleaned-up families and 10 tagSNPs were taken forward to further association analyses.
Single-point and multi-point association analyses were carried out using implementations of the transmission disequilibrium test (TDT) in Plink and Unphased [25,29]. Haplotype frequencies were estimated using the expectation-maximization algorithm implemented in Unphased [29]. The r2 and D′ measures of pairwise LD were calculated for all SNPs using Haploview [30]. Power calculations were performed using Quanto [31]. To obtain empirical p values, 10,000 permutations were run for each analysis, and permutation p values less than 0.05 were considered statistically significant.
To test whether our results would replicate in the largest genome-wide association analysis of T1DM, we applied for and received approval for Wellcome Trust Case Control Consortium (WTCCC) genotype data access [32]. We then conducted a case-control analysis in the WTCCC dataset of the same chromosomal region, as was investigated in our study, and compared the results.
3. Results
SNPs rs928815-rs746868, rs909253-rs1041981, and rs361525-rs3093668 were in tight LD with each other (D′ = 1, r2 =1;D′ = 1, r2 = 0.984 and D′ = 1, r2 = 0.797, respectively). Other SNPs showed some or no LD between each other. LD correlation between 10 investigated TNF/LTA SNPs in healthy parents can be seen in Supplement 1, Figures 1a and 1b. Seven of our tag SNPs were found in the HapMap (build 35) database (rs928815, rs909253, rs1041981, rs1800750, rs1800629, rs3093662, rs3093668). Based on the HapMap data, these seven SNPs capture 21 polymorphic sites (>61.9% of common variation) in the region (chr6:31634835-31655771) at an r2 threshold of greater than 0.8. The list of 21 TNF/LTA common variants and their 7 tagSNPs are shown in Supplement 2, Table 1.
The results of TDT single-point analysis, information about tagSNPs gene positions, allele changes and MAFs are shown in Table 2. We detected three statistically significant minor allele overtransmissions from parents to affected child for SNPs rs909253 (p = 1.2×10−4), rs1041981 (p = 1.1×10−4), and rs1800629 (G-308A) (p = 1.2×10−4) and two minor allele undertransmissions for SNPs rs361525 (G-238A) (p = 8.2×10−3) and rs3093668 (p = 0.014) after 10,000 permutations. Genotype data for affected children and their parents can be seen in Supplement 3, Table 2.
Table 2.
Transmission disequilibrium analysis of 10 TNF/LTA SNPs in 152 parent-offspring trio families
| SNP | Positiona | MAFb | M:Cc | T:Ud | ORe | L95e | U95e | χ 2 f | p Valueg |
|---|---|---|---|---|---|---|---|---|---|
| rs928815 | 5′ LTA | 0.406 | T:G | 48:69 | 0.6957 | 0.4813 | 1.005 | 3.769 | 0.052 |
| rs909253 | LTA-intron 1 | 0.396 | C:T | 81:39 | 2.077 | 1.417 | 3.043 | 14.7 | 0.00012 |
| rs746868 | LTA-intron 1 | 0.400 | C:G | 40:59 | 0.678 | 0.4538 | 1.013 | 3.646 | 0.056 |
| rs1041981 | LTA-exon 3 | 0.391 | A:C | 80:38 | 2.105 | 1.431 | 3.097 | 14.95 | 0.00011 |
| rs1800750 | 5′ of TNF | 0.003 | A:G | 0:1 | / | / | / | 1 | 0.317 |
| rs1800629 | 5′ of TNF | 0.269 | A:G | 65:28 | 2.321 | 1.491 | 3.616 | 14.72 | 0.00012 |
| rs361525 | 5′ of TNF | 0.006 | A:G | 0:7 | / | / | / | 7 | 0.00815 |
| rs3093662 | TNF-intron 1 | 0.038 | G:A | 9:16 | 0.5625 | 0.2486 | 1.273 | 1.96 | 0.162 |
| rs3093665 | TNF-3′UTR | 0.032 | C:A | 9:9 | 1 | 0.397 | 2.519 | 0 | 1 |
| rs3093668 | 3′ of TNF | 0.003 | C:G | 0:6 | / | / | / | 6 | 0.014 |
Position of the SNP within the gene.
MAF in T1DM children.
Minor allele (M) vs. common allele (C).
Copies of the minor allele transmitted (T) and nontransmitted (U).
Odds ratios (OR) with 95% lower and upper confidence intervals (L95,U95).
Chi square (χ2) test.
p Value after 10,000 permutations.
Haplotype-based TDT analysis showed T1DM association with rs1800629(G-308A) and rs361525(G-238A) haplotypes (p = 2.384×10−5), after 10,000 permutations. Specifically, the A-G rs1800629(G-308A)-rs361525(G-238A) haplotype was overtransmitted from parents to affected offspring (Table 3).
Table 3.
Transmission of rs1800629(G-308A)-rs361525(G-238A) haplotypes in152 parent–offspring trio families
| Haplotype | T | Freq T | NT | Freq NT | OR | p Valuea |
|---|---|---|---|---|---|---|
| 1-2 (A-G) | 72 | 0.2571 | 35 | 0.125 | 1 | 2.384×10−5 |
| 2-1 (G-A) | 1 | 0.0035 | 8 | 0.028 | 0.6 | |
| 2-2 (G-G) | 207 | 0.7393 | 237 | 0.8464 | 0.4 |
T, transmitted; freq T, frequency transmitted; NT, nontransmitted; freq NT, frequency nontransmitted; OR, odds ratio.
overall p-value.
None of our 10 investigated SNPs were analyzed in the WTCCC study of T1DM. This is because WTCCC genotyping platform, Affymetrix 500 K, did not have these SNPs incorporated within. There was only one SNP from TNF gene (rs1799964) that was part of Affymetrix 500 K chip. SNP rs1799964 showed significant association with T1DM in WTCCC case-control analysis (OR = 1.303, p = 3.948×10−8). However, based on the HapMap database rs1799964 is not in LD with any of our 10 TNF/LTA SNPs (Supplement 4, Figure 2), therefore we could not make any comparison of our association results with WTCCC results.
4. Discussion
In this study we analyzed 10 TNF/LTA gene SNPs with T1DM and found significant association in the families from South Croatia. SNPs from this study were designed to tag most of the common variation from the TNF/LTA gene region and were analyzed in T1DM for the first time. We detected three minor (rs909253, rs1041981, rs1800629 (G-308A)) and two major (rs361525 (G-238A), rs3093668) allele overtransmissions from parents to affected children. We also observed significant A-G rs1800629(G-308A)-rs361525(G-238A) haplotype overtransmission. This study had 90% statistical power to detect an effect (at α = 0.05) for SNPs rs909253, rs1041981, and rs1800629 and less than 80% power for SNPs rs361525(G-238A) and rs3093668 assuming an additive model.
Two SNPs that show overtransmission of the minor allele (rs909253, rs1041981) and two that overtransmit a major allele (rs361525(G-238A), rs3093668) are in LD with each other (Supplement 1). It is possible that only one SNP drives the association and the observed patterns are, consequently, caused by LD. Other SNP that shows very significant minor allele overtransmission, rs1800629(G-308A) is not in LD with any other investigated SNP. Interestingly, overtransmission of A-G rs1800629(G-308A)-rs361525(G-238A) haplotype shows stronger significance (2.384×10−5) than any of the single markers individually (rs1800629 (G-308A), p = 1.2×104; rs361525(G-238A), p = 8.15×0−3). As both promoter polymorphisms have been associated with transcriptional enhancement rate it is possible that when acting in cis these two markers show an even stronger interaction [33-35].
The HLA class III region, particularly around TNF, has been regarded as a susceptibility locus for T1DM [36]. The most frequently analyzed SNPs from this region are the two TNF promoter SNPs at positions −308 (rs1800629) and −238 (rs361525). LTA lies next to TNF, and several studies have combined TNF/LTA SNPs and analyzed them with respect to T1DM. A significant increase in TNF −308 allele A and G/A and A/A genotypes was found in North Indian T1DM cases [17,19]. Similarly, an increase in the prevalence of the TNF −308 A allele was reported in Hungarian diabetic patients [20]. Associations of the TNF/LTA haplotypes and LTA +249A/G SNP with T1DM, was suggested to be independent of HLA in the Moroccan and Bahraini populations [18,22]. On the other hand, Deng et al. excluded the −308 polymorphism as a marker forT1DM in Chinese and Caucasian datasets because of LD with the DR3-DQB1*0201 haplotype [21]. In addition, after adjusting the data for LD with DRB1-DQB1 and B18-DR3 haplotypes, Noble etal. observed a lack of TNF −308, −238, and LTA A1069G SNP association with T1DM [6]. The similar conclusions were drawn out from the TNF studies in southwestern Polish and in Japanese populations [16,33].
Several studies reported a small independent effect of TNF/LTA SNPs, particularly of rs1800629 (−308), on susceptibility to T1DM. In our study we also observed associations of several TNF/LTA SNPs, including of rs1800629 (−308), with T1DM. However, the South Croatian families analyzed in this study have not been HLA typed and therefore we can not determine whether the effects of the TNF/LTA variants are primary or secondary to HLA loci. A functional support for independent effects may be explained by transcription rate enhancement of TNF gene by TNF promoter −308 and −238 SNPs, functional control of TNF promoter SNPs on LTA expression and expression of other nearby genes [34,35,37-39]. An integrated role of TNF and other inflammatory cytokines in the destruction of pancreatic beta cells [19] provides further argument for a TNF/LTA T1DM effect.
In summary, the present study investigated the association of 10 TNF/LTA tag SNPs, specifically designed to capture the majority of common variation in the region, with T1DM in families from South Croatia. Selected SNPs were analyzed with T1DM for the first time. We observed associations of variants in the TNF/LTA gene region with T1DM. However, to fully understand the TNF/LTA involvement in T1DM susceptibility, a careful assessment of TNF/LTA genes adjusted for LD with HLA in a larger sample is needed.
Supplementary Material
Acknowledgments
The project is funded by the Croatian Ministry of Science, Education and Sports (number 216-1080315-0293). We thank The British Scholarship Trust for support for the VB study visit to Oxford. EZ is a Wellcome Trust Research Career Development Fellow.
Footnotes
Appendix. Supplementary data
Supplementary data associated with this article can be found, in the online version, at 10.1016/j.humimm.2008.12.010.
References
- 1.American Diabetes Association Diagnosis and Classification of Diabetes Mellitus. Diabetes Care. 2006;29(Suppl):S43–8. [PubMed] [Google Scholar]
- 2.Nejentsev S, Howson JM, Walker NM, Szeszko J, Field SF, Stevens HE, et al. Wellcome Trust Case Control Consortium. Localization of type 1 diabetes susceptibility to the MHC class I genes HLA-B and HLA-A. Nature. 2007;450:887–92. doi: 10.1038/nature06406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rich SS, Concannon P, Erlich H, Julier C, Morahan G, Nerup J, et al. The Type 1 Diabetes Genetics Consortium. Ann N Y Acad Sci. 2006;1079:1–8. doi: 10.1196/annals.1375.001. [DOI] [PubMed] [Google Scholar]
- 4.Smyth DJ, Cooper JD, Bailey R, Field S, Burren O, Smink LJ, et al. A genome-wide association study of nonsynonymous SNPs identifies a type 1 diabetes locus in the interferon-induced helicase (IFIH1) region. Nat Genet. 2006;38:617–9. doi: 10.1038/ng1800. [DOI] [PubMed] [Google Scholar]
- 5.Todd JA, Walker NM, Cooper JD, Smyth DJ, Downes K, Plagnol V, et al. Robust associations of four new chromosome regions from genome-wide analyses of type 1 diabetes. Nat Genet. 2006;39:857–64. doi: 10.1038/ng2068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Noble JA, Valdes AM, Lane JA, Green AE, Erlich HA. Linkage disequilibrium with predisposing DR3 haplotypes accounts for apparent effects of tumor necrosis factor and lymphotoxin-α polymorphisms on type 1 diabetes susceptibility. Hum Immunol. 2006;67:999–1002. doi: 10.1016/j.humimm.2006.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Feugeas JP, Caillens H, Poirier JC, Charron D, Marcelli-Barge A, Wautier JL. Influence of metabolic and genetic factors on tumor necrosis factor-α and lymphotoxin-α production in insulin-dependent diabetes mellitus. Diabetes Metab. 1997;23:295–301. [PubMed] [Google Scholar]
- 8.Makhatadze NJ. Tumor necrosis factor locus: Genetic organisation and biological implications. Hum Immunol. 1998;59:571–9. doi: 10.1016/s0198-8859(98)00056-1. [DOI] [PubMed] [Google Scholar]
- 9.Kim KA, Kim S, Chang I, Kim GS, Min YK, Lee MK, et al. IFN gamma/TNF alpha synergism in MHC class II induction: Effect of nicotinamide on MHC class II expression but not on islet-cell apoptosis. Diabetologia. 2002;45:385–93. doi: 10.1007/s00125-001-0755-8. [DOI] [PubMed] [Google Scholar]
- 10.Yang XD, Tisch R, Singer SM, Cao ZA, Liblau RS, Schreiber RD, et al. Effect of tumor necrosis factor alpha on insulin-dependent diabetes mellitus in NOD mice. I. The early development of autoimmunity and the diabetogenic process. J Exp Med. 1994;180:995–1004. doi: 10.1084/jem.180.3.995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rabinovitch A, Suarez-Pinzon WL. Cytokines and their roles in pancreatic islet beta-cell destruction and insulin-dependent diabetes mellitus. Biochem Pharmacol. 1998;55:1139–49. doi: 10.1016/s0006-2952(97)00492-9. [DOI] [PubMed] [Google Scholar]
- 12.Vaux DL, Flavell RA. Apoptosis genes and autoimmunity. Curr Opin Immunol. 2000;12:719–24. doi: 10.1016/s0952-7915(00)00168-0. [DOI] [PubMed] [Google Scholar]
- 13.Middlebrook AJ, Lebsack T, DeLuca D. TNF-alpha mediated modulation of T cell development and exacerbation of in vitro T1DM in fetal thymus organ culture. J Autoimmun. 2007;29:134–45. doi: 10.1016/j.jaut.2007.06.002. [DOI] [PubMed] [Google Scholar]
- 14.De Togni P, Goellner J, Ruddle NH, Streeter PR, Fick A, Mariathasan S, et al. Abnormal development of peripheral lymphoid organs in mice deficient in lymphotoxin. Science. 1994;264:703–7. doi: 10.1126/science.8171322. [DOI] [PubMed] [Google Scholar]
- 15.Takahashi K, Satoh J, Seino H, Zhu XP, Sagara M, Masuda T, et al. Prevention of type I diabetes with lymphotoxin in BB rats. Clin Immunol Immunopathol. 1993;69:318–23. doi: 10.1006/clin.1993.1187. [DOI] [PubMed] [Google Scholar]
- 16.Deja G, Jarosz-Chobot P, Polańska J, Siekiera U, Małecka-Tendera E. Is the association between TNF-alpha-308 A allele and DMT1 independent of HLA-DRB1, DQB1 alleles? Mediators Inflamm. 2006;2006:1–7. doi: 10.1155/MI/2006/19724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Das SN, Baniasadi V, Kapuria V. Association of -308 TNF-alpha promoter polymorphism with type 1 diabetes in North Indians. Int J Immunogenet. 2006;33:411–6. doi: 10.1111/j.1744-313X.2006.00632.x. [DOI] [PubMed] [Google Scholar]
- 18.Bouqbis L, Akhayat O, Garchon HJ, Calafell F, Izaabel H. TNFA-TNFB haplotypes modify susceptibility to type I diabetes mellitus independently of HLA class II in a Moroccan population. Tissue Antigens. 2003;61:72–9. doi: 10.1034/j.1399-0039.2003.610106.x. [DOI] [PubMed] [Google Scholar]
- 19.Kumar R, Goswami R, Agarwal S, Israni N, Singh SK, Rani R. Association and interaction of the TNF-alpha gene with other pro- and anti-inflammatory cytokine genes and HLA genes in patients with type 1 diabetes from North India. Tissue Antigens. 2007;69:557–67. doi: 10.1111/j.1399-0039.2007.00817.x. [DOI] [PubMed] [Google Scholar]
- 20.Krikovszky D, Vásárhelyi B, Tóth-Heyn P, Körner A, Tulassay T, Madácsy L. Association between G-308A polymorphism of the tumor necrosis factor-alpha gene and 24-hour ambulatory blood pressure values in type 1 diabetic adolescents. Clin Genet. 2002;62:474–7. doi: 10.1034/j.1399-0004.2002.620609.x. [DOI] [PubMed] [Google Scholar]
- 21.Deng GY, Maclaren NK, Huang HS, Zhang LP, She JX. No primary association between the 308 polymorphism in the tumor necrosis factor alpha promoter region and insulin-dependent diabetes mellitus. Hum Immunol. 1996;5:137–42. doi: 10.1016/0198-8859(95)00166-2. [DOI] [PubMed] [Google Scholar]
- 22.Stayoussef M, Al-Jenaidi FA, Al-Abbasi A, Al-Ola K, Khayyat H, Mahjoub T, et al. Modulation of type 1 diabetes susceptibility by TNFα −308G/A and lymphotoxin α +249A/G haplotypes in Bahraini patients: Lack of linkage disequilibrium with predisposing DQB1-DRB1 haplotypes. Clin Vaccine Immunol. 2008;15:379–81. doi: 10.1128/CVI.00410-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hanchard N, Rockett K, Udalova I, Wilson J, Keating B, Koch O, et al. An investigation of transmission ratio distortion in the central region of the human MHC. Genes Immun. 2006;7:51–8. doi: 10.1038/sj.gene.6364277. [DOI] [PubMed] [Google Scholar]
- 24.Hanchard N, Diakite M, Koch O, Keating B, Pinder M, Jallow M, et al. Implications of inter-population linkage disequilibrium patterns on the approach to a disease association study in the human MHC class III. Immunogenetics. 2006;58:465–70. doi: 10.1007/s00251-006-0118-1. [DOI] [PubMed] [Google Scholar]
- 25.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, et al. PLINK: A toolset for whole genome association and population-based linkage analysis. Am J Hum Genet. 2007;81:559–75. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wigginton JE, Abecasis GR. PEDSTATS: Descriptive statistics, graphics and quality assessment for gene mapping data. Bioinformatics. 2005;21:3445–7. doi: 10.1093/bioinformatics/bti529. [DOI] [PubMed] [Google Scholar]
- 27.International HapMap Consortium The International HapMap project. Nature. 2005;426:789–96. doi: 10.1038/nature02168. [DOI] [PubMed] [Google Scholar]
- 28.de Bakker PI, Yelensky R, Pe'er I, Gabriel SB, Daly MJ, Altshuler D. Efficiency and power in genetic association studies. Nat Genet. 2005;37:1217–23. doi: 10.1038/ng1669. [DOI] [PubMed] [Google Scholar]
- 29.Dudbridge F. Pedigree disequilibrium tests for multilocus haplotypes. Genet Epidemiol. 2003;25:115–21. doi: 10.1002/gepi.10252. [DOI] [PubMed] [Google Scholar]
- 30.Barrett JC, Fry B, Maller J, Daly MJ. Haploview: Analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–5. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- 31.Gauderman WJ. Candidate gene association studies for a quantitative trait, using parent-offspring trios. Genet Epidemiol. 2003;25:327–38. doi: 10.1002/gepi.10262. [DOI] [PubMed] [Google Scholar]
- 32.Wellcome Trust Case Control Consortium Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. 2007;447:661–78. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hamaguchi K, Kimura A, Seki N, Higuchi T, Yasunaga S, Takahashi M, et al. Analysis of tumor necrosis factor-alpha promoter polymorphism in type 1 diabetes: HLA-B and -DRB1 alleles are primarily associated with the disease in Japanese. Tissue Antigens. 2000;55:10–6. doi: 10.1034/j.1399-0039.2000.550102.x. [DOI] [PubMed] [Google Scholar]
- 34.Kroeger KM, Carville KS, Abraham LJ. The −308 tumor necrosis factor-alpha promoter polymorphism effects transcription. Mol Immunol. 1997;34:391–9. doi: 10.1016/s0161-5890(97)00052-7. [DOI] [PubMed] [Google Scholar]
- 35.Kaluza W, Reuss E, Grossmann S, Hug R, Schopf RE, Galle PR, et al. Different transcriptional activity and in vitro TNF-alpha production in psoriasis patients carrying the TNF-alpha 238A promoter polymorphism. J Invest Dermatol. 2000;114:1180–3. doi: 10.1046/j.1523-1747.2000.00001.x. [DOI] [PubMed] [Google Scholar]
- 36.Nishimura M, Obayashi H, Mizuta I, Hara H, Adachi T, Ohta M, et al. TNF, TNF receptor type 1, and allograft inflammatory factor-1 gene polymorphisms in Japanese patients with type 1 diabetes. Hum Immunol. 2003;64:302–9. doi: 10.1016/s0198-8859(02)00799-1. [DOI] [PubMed] [Google Scholar]
- 37.Wilson AG, Symons JA, McDowell TL, McDevitt HO, Duff GW. Effects of a polymorphism in the human tumor necrosis factor alpha promoter on transcriptional activation. Proc Natl Acad Sci U S A. 1997;94:3195–9. doi: 10.1073/pnas.94.7.3195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Knight JC, Keating BJ, Rockett KA, Kwiatkowski DP. In vivo characterization of regulatory polymorphisms by allele-specific quantification of RNA polymerase loading. Nat Genet. 2003;33:469–75. doi: 10.1038/ng1124. [DOI] [PubMed] [Google Scholar]
- 39.Zeggini E, Groves CJ, Parkinson JR, Halford S, Owen KR, Frayling TM, et al. Large-scale studies of the association between variation at the TNF/LTA locus and susceptibility to type 2 diabetes. Diabetologia. 2005;48:2013–7. doi: 10.1007/s00125-005-1902-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.