Abstract
Hedgehog (Hh) morphogens play fundamental roles in development whilst dysregulation of Hh signaling leads to disease. Multiple cell surface receptors are responsible for transducing and/or regulating Hh signals. Among these, the hedgehog-interacting protein (HIP) is a highly conserved, vertebrate-specific, inhibitor of Hh signaling. We have solved a series of crystal structures for the human HIP ectodomain and Desert Hh in isolation, as well as Sonic and Desert Hh-HIP complexes, with and without calcium. The interaction determinants, confirmed by biophysical studies and mutagenesis, reveal novel and distinct functions for Hh zinc- and calcium-binding sites; functions which appear common to all vertebrate Hhs. Zinc makes a key contribution to the Hh-HIP interface while calcium prevents electrostatic repulsion between the two proteins, thus playing a major modulatory role. This interplay of several metal-binding sites suggests a tuneable mechanism for regulation of Hh signaling.
A handful of secreted growth factors and morphogens are responsible for almost all developmental decisions during embryogenesis and, among these, Hedgehog (Hh) family proteins are ubiquitous players in tissue growth, patterning and morphogenesis1,2. Dysregulation of Hh signaling has pathological consequences, triggering developmental defects3, neurodegenerative diseases4 and cancer5.
The Hh signaling pathway is broadly conserved in vertebrates and invertebrates, but the number of Hh ligands has expanded during evolution (one Hh in Drosphila, and three in mammals: Sonic hedgehog (Shh), Desert hedgehog (Dhh) and Indian hedgehog (Ihh))1. Mature Hh proteins are derived from Hh protein precursors by auto-processing and lipid attachment to generate a dually lipid-modified amino-terminal signaling domain (Hh-N)6. These modifications facilitate the integration of Hh into multimeric lipoprotein particles7,8. Hh constructs lacking the lipid attachment sites have the same receptor binding properties, but their tissue distribution is impaired6,9.
The extracellular regulation of signaling is more complex in vertebrates than in invertebrates. Transduction of Hh signals into cells is mediated by the two transmembrane receptors Smoothened and Patched (PTC)10. The interference hedgehog protein family (Ihog in fly and CDO and BOC in human)11 and heparan sulphate proteoglycans (HSPG)12 act as crucial modulators in both vertebrates and invertebrates, while hedgehog-interacting protein (HIP)13 and growth arrest-specific gene 1 (GAS 1)14 are vertebrate-specific.
HIP is an inhibitor of Hh signaling. It is attached to the cell surface by a C-terminal helix and binds all three Hh ligands (Sonic, Indian and Desert Hh) with nanomolar affinity13. Cells exposed to Hh up-regulate both PTC and HIP expression13. A secreted version of HIP is also able to sequester Hh, thus interfering with Hh signaling15. HIP functions in cartilage development13, normal nerve development16 and axon guidance17 and its expression is altered in a variety of cancers18-21. Despite this wealth of functional data, the molecular mechanisms underlying HIP function are still unknown. We therefore undertook an analysis of the structural and binding characteristics of human HIP-Hh interactions.
RESULTS
Structure of the human HIP ectodomain
A series of HIP constructs were produced in mammalian HEK-293T cells (22, see methods for details). The full-length ectodomain of human HIP (eHIP, 56-670) fragmented during expression; mutation of the proteolytic recognition sequences (Arg189, Arg210 and Lys211 to alanine; eHIPS construct) or an N-terminal truncation of the first 213 amino acid residues (eHIP∆N construct) resulted in proteolytically stable and homogeneous proteins (Fig. 1a). The stabilized full-length ectodomain is dimeric whereas eHIP∆N is a monomer (Supplementary Fig.1) suggesting a function for the N-terminal cysteine-rich domain in mediating dimerisation, although we cannot rule out that dimerization might be a consequence of mutagenesis.
Figure 1.
Structure of the HIP ectodomain. (a) Schematic domain organization of the HIP receptor. SP: signal peptide; L1, L2: interdomain linker regions; EGF1, EGF2: epidermal growth factor repeat domains; Hx: membrane attachment helix. β-propeller blades are color-coded and numbered. Proteolytic cleavage site residues Arg189 and Arg210, identified by N-terminal sequencing, are highlighted. The crystallization construct (eHIP∆N) and the stabilised full-length ectodomain construct (eHIPS) are shown. (b) Selenomethionine SAD-phased and phase-extended electron density map (calculated to 2.8 Å, contoured at 1 σ) with rainbow-colored Cα trace of eHIP∆N. (c) Ribbon diagram of eHIP∆N with color coding as in a and b. The six blades of the β-propeller domain (each consisting of a 4-stranded β-sheet) are numbered as in a. The eleven disulphide bridges are shown in black stick representation and marked with Roman numerals. d, Electrostatic properties. eHIP∆N is shown as solvent accessible surface colored by electrostatic potential contoured at +/− 10 kT (red, acidic; blue, basic). The prominent negatively charged patch which interacts with Hh ligands is marked with a dotted circle.
Attempts to crystallize the stabilised ectodomain of HIP failed, but eHIP∆N yielded well-ordered crystals. The structure was solved using selenomethionine-labeled protein for phasing (Fig.1b). Refinement resulted in an R-factor of 20.1% (R-free: 23.9%) to 2.8 Å resolution with two eHIP∆N molecules in the crystallographic asymmetric unit (Supplementary Fig. 2). The eHIP∆N monomer is composed of an N-terminal 6-bladed β-propeller domain (residues 214-583) linked to two epidermal growth factor (EGF) repeat domains (residues 607-637 and 638-670, respectively) (Fig. 1c). The major atypical characteristic of the HIP β-propeller is a large protrusion of two loops from blade 3 which form an extensive negatively charged patch on the surface (Fig. 1d). Twenty eight residues connect the β-propeller to the EGF repeats. This well-ordered linker (colored orange in Fig. 1b and c) stretches across the β-propeller (over blades 6, 1 and 2). Disulphides (Fig. 1c) stabilising the linker (I, IV and VI) and the orientation of the first EGF repeat (II and VI) are conserved in all known HIP receptors (Supplementary Fig. 3). This rigid arrangement forms a structural unit; attempts to produce truncated forms of HIP lacking the first EGF repeat failed (unpublished data). The EGF repeats have a linear, elongated conformation with the C-terminus of the second EGF repeat pointing away from the β-propeller domain. The major difference between the two HIP molecules in the asymmetric unit is the relative orientation of the second EGF repeat (Supplementary Fig. 2) reflecting the flexibility between the EGF domains. This hinge may provide a necessary degree of freedom in the membrane-proximal region of the ectodomain (which is tethered to the cell surface only nine residues downstream of the second EGF repeat).
Structure of the HIP-Shh complex
We next produced (Supplementary Fig. 4) the complex between eHIP∆N and the N-terminal signaling domain of Shh (ShhN, a monomeric construct that lacks the lipid-modification sites) and determined its crystal structure to 3.2 Å resolution. The Hh ligand uses a surface oriented away from its termini (which are lipid-modified in vivo) to interact with the two distinctive loops (BL1 and BL2) protruding from blade 3 of the HIP β-propeller (Fig.1d, Fig. 2a and b). Two disulphide bridges at the beginning and the end of blade 3 (disulphide bridges II, III in Fig. 2a) stabilise the blade for Hh ligand reception whilst the C-terminal EGF repeats and membrane proximal region of HIP are oriented away from the ligand-binding site. The complex interface buries 840 Å2 of eHIP∆N and 759 Å2 of ShhN with a surface complementarity score (sc) of 0.66, and consists of 13 potential hydrogen bonds, 157 non-bonded van-der-Waals contacts and one zinc ion, involving a total of 14 residues of the HIP receptor and 20 residues of ShhN (Supplementary Fig. 3 and 5a).
Figure 2.
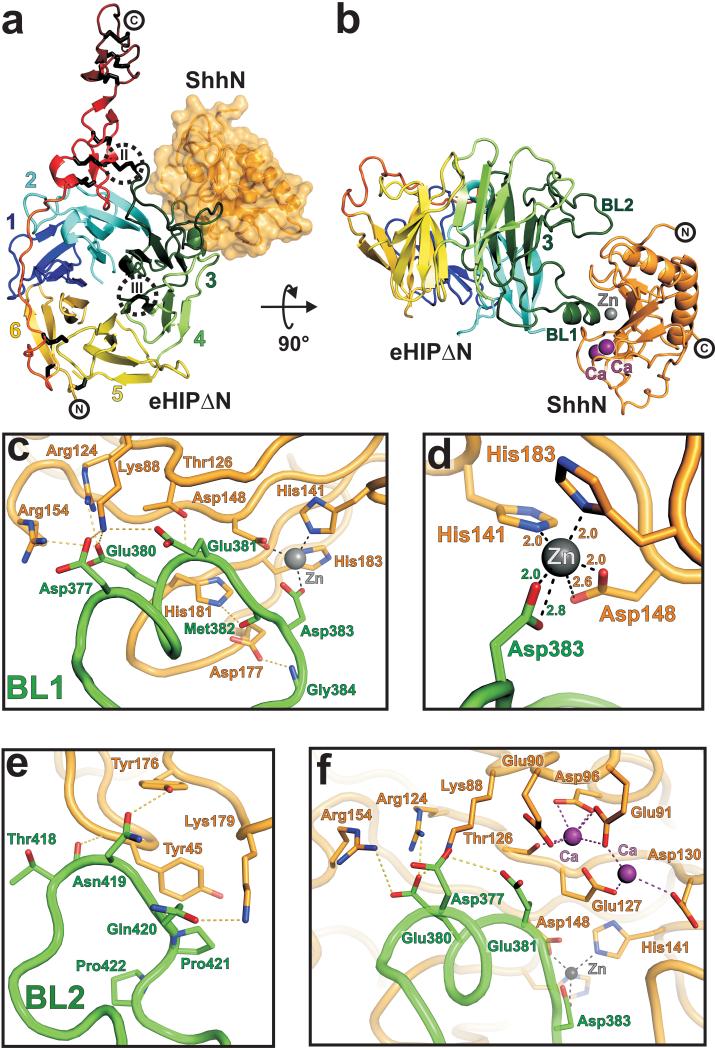
Structure of the Shh-HIP complex. (a) Cartoon representation of eHIP∆N (orientation, β-propeller blade numbering, and color coding as in Fig. 1c) in complex with ShhN depicted as solvent accessible surface (in orange). Disulphide bridges (II and III), stabilizing HIP β-propeller blade 3, are highlighted (dotted circles). (b) Cartoon representation of the eHIP∆N-ShhN complex. The two binding loops (BL1 and BL2) of HIP are labeled. The zinc ion is shown in grey and the two calcium ions in violet. (c-f) Close up views of the Shh-HIP interactions. eHIP∆N (green) and ShhN (orange) main chains are shown as coils. Residues involved in complex interactions are drawn in stick representation (oxygen: red, nitrogen: blue). The zinc ion is shown in grey and the calcium ions in violet. Potential hydrogen bonds are marked as yellow dotted lines, interactions with the zinc ion as grey dotted lines and with calcium as violet dotted lines. Contacts of HIP-BL1 with ShhN are shown in c, the zinc-binding site is detailed in d, HIP-BL2 interactions are shown in e and the properties of the calcium-binding site in f.
Detailed interactions are depicted in Fig. 2c-f. In eHIP∆N, BL1 (residues 370-391, harbouring a small α-helical segment) forms the major interaction interface inserting into the Shh zinc-binding cavity (Fig. 2b and c). The carboxylate group of HIP-Asp383, located at the tip of BL1, completes the coordination of the zinc-binding site, which is formed by Shh residues His141, Asp148 and His183 (Fig. 2d). The presence of the zinc was verified by coordination geometry23, X-ray absorption- and fluorescence scans and anomalous scattering (Supplementary Fig. 6). Two calcium ions, coordinated by Shh residues Glu90, Glu91, Asp96, Thr126, Glu127 and Asp130 (Fig. 2f), are also clearly visible in the electron density maps (Supplementary Fig. 6d) and appear to play an important role in the stabilisation of a Shh loop (Lys88-Gly94; discussed below). The second HIP loop contributing to binding (BL2) interacts with a Shh surface patch adjacent to the zinc-binding cavity forming three potential hydrogen bonds and several van-der-Waals contacts (Fig. 2e).
Importance of metal-binding sites for HIP-Hh interactions
Although the eHIP∆N-ShhN interface is relatively small and the surface complementarity score is at the lower end of the scale for physiological protein-protein interactions, binding constants in the nanomolar range were observed using both cell-based 13 and surface plasmon resonance (SPR) experiments (Fig. 3a and b, Supplementary Fig. 7). HIP ectodomain (including the N-terminal cysteine-rich dimerisation domain; Fig. 1a) and eHIP∆N show similar Hh-binding behaviour, although the full-length form has somewhat enhanced binding which may reflect its dimeric state (Fig. 3). For both HIP constructs the presence of zinc in the Hh metal-binding site is crucial (Fig. 3). Addition of 10 mM EDTA (a chelator for divalent cations) completely abolished Hh-binding (Fig. 3d) or, in the presence of calcium, mutation of the zinc-coordinating HIP-Asp383 (Fig. 2c and d) to arginine or alanine abolished or reduced Hh-binding by greater than 25-fold, respectively (Fig. 3a). Thus the tight Hh-HIP interaction can be explained by the contribution of HIP to the coordination of zinc in the Hh ligand metal-binding site (Fig. 2d). Hall et al. 24 first identified zinc in the crystal structure of ShhN, and pointed out a similarity to the zinc-containing active sites of some metalloproteases. Zinc appears to play a crucial structural role, binding ShhN with very high affinity (Kd ≤ 100 pM)25, however, the HIP-Shh structure reveals a second role. Zinc-binding is the key mediator of complex formation; indeed, the zinc-binding site is not conserved in Drosophila (Supplementary Fig. 5a), which correlates with the absence of HIP in flies.
Figure 3.
Binding properties of Hh-HIP interactions. (a) Table of binding constants (Kd) measured by SPR between different HIP constructs and ShhN or DhhN, respectively. Data are expressed as mean ± standard error. ND: not determinable. (b-d) Binding of the stabilized HIP ectodomain (eHIPS) to ShhN. The left panels are representative sets of experimental sensorgrams from typical equilibrium-based binding experiments, with reference substraction. Different concentrations of ShhN were injected over surfaces coupled with eHIP in the presence (b) or the absence (c) of calcium and in the presence of 10 mM EDTA (d), respectively. For all injections the primary experimental traces reached equilibrium and returned to baseline after the injection. The right panels are plots of the equilibrium binding response (RU) from the sensorgrams as a function of ShhN concentration (0.5 to 5000 nM). Each plot is derived from a different independent experiment, but within each plot, the three curves are derived from a single series of injections of ShhN over a biosensor chip in which three experimental surfaces have been coated with different amounts of eHIPS. This allowed the use of global fitting to improve data averaging. Best-fit binding curves with a global fit of the experimental data to a uniform Kd value are shown as colored lines.
The low affinity (Kd ≥ 100 μM,26) calcium-binding site (Fig. 2f) adopts the same conformation as described by McLellan et al.26 and found to be important for Shh binding to various cell-surface receptors. We confirmed this finding in SPR experiments where addition of 2 mM calcium caused a marked increase of the Hh-HIP binding affinity (Kd from 100 nM to 6 nM; Fig. 3a-c, Supplementary Fig. 7). However, our structure reveals that neither the two calcium ions nor the loops coordinating them contribute specific interactions to the complex interface suggesting a secondary role for the calcium ions in HIP-binding. We therefore investigated the effect of calcium absence on the structure of Hh ligands and, subsequently, on Hh-HIP structures. In the previously published uncomplexed ShhN structure24, in the absence of calcium, the side chains of Glu90, Glu91 are oriented towards the solvent (Fig. 4a). To verify whether this applies to other Hh ligands, we solved crystal structures of the N-terminal signaling domain of human Desert Hh (DhhN, equivalent to the ShhN construct) with and without bound calcium. The overall architecture of DhhN, including the zinc-binding site, closely matches ShhN (r.m.s.d 0.5 Å for 150 Cα atoms; Fig. 4a) as expected from the high degree of sequence identity (77%, Supplementary Fig. 5). The conformations of the Glu90 and Glu91 side chains in the calcium-free forms of ShhN24 and DhhN match, while for DhhN with calcium bound, these residues contribute to metal-coordination and adopt the conformation observed in the ShhN-eHIP∆N complex (Fig. 4a, Supplementary Fig. 8).
Figure 4.
Molecular determinants of Hh-receptor interactions. (a) Structural superpositions. DhhN with calcium (slate) and without (magenta), and ShhN with calcium (orange) and without (yellow; PDB 1VHH) are shown as coils. Zinc and calcium ions are depicted as spheres. The close-up highlights loop Lys88-Gly94, Glu90 and Glu91 (stick representation) change conformation upon calcium-binding. (b) ShhN solvent-accessible surface colored by residue conservation (Supplementary Fig. 5). Hh-binding loops of HIP are shown in cyan. (c) Comparison of ShhN- and DhhN-HIP complexes. ShhN surface is colored orange. Shh loop Lys88-Gly94, ordered only in the calcium-bound ShhN-HIP, is highlighted in red. Hh-binding loops of HIP are shown as coils (ShhN-eHIP∆N with calcium: green; ShhN-eHIP∆N without calcium: blue; DhhN-eHIP∆N without calcium - molecule 1: yellow, molecule 2: cyan). A third ligand-binding loop of HIP (BL3) is observed in only one copy of the DhhN-eHIP∆N asymmetric unit (dotted ellipse, see also Supplementary Fig. 9). (d) Effects of calcium-binding. ShhN-eHIP∆N complexes are shown as coils (with calcium: orange/green, without calcium: violet/blue). Spheres highlight HIP-Glu381 and Shh-Glu90. Shh-loop Lys88-Gly94 is depicted as a dotted line. (e) Comparison of the Shh-HIP and Shh-CDO (PDB 3D1M) complexes. Shh ligands are superimposed (HIP: green, CDO: blue, HIP-complexed ShhN: orange, CDO-complexed ShhN: red). Metal ions are depicted as spheres.
We produced, crystallized and solved the structures of ShhN-eHIP∆N and DhhN-eHIP∆N complexes in the absence of calcium (Supplementary Fig. 4). The structures show that the Hh residues contributing to the Hh-HIP interface are highly similar across family members and vertebrate species (Fig. 4b, Supplementary Fig. 5), conserving the architecture of the ligand-receptor interfaces (Fig. 4c, Supplementary Fig. 9). In the absence of calcium the Hh loop Lys88-Gly94 becomes disordered, suggesting that calcium-binding is important to prevent electrostatic repulsion between the negatively charged residues of Hh loop Lys88-Gly94 and a negatively charged patch (formed by HIP residues Asp377, Glu380, Glu381, Asp383) on the apposing HIP surface (Fig. 2f). In the presence of calcium, stabilisation of the loop allows the side chains of HIP-Glu381 and Shh-Glu90 to contribute to the complex by forming additional van-der-Waals interactions (Fig. 4d), consistent with the observed 20- to 60-fold increase in affinity between Hh ligands and HIP under these conditions (Fig. 3a).
A previously reported structure of ShhN in complex with the third fibronectin type III domain of CDO showed calcium to be essential for binding26 and the conformation of the calcium-binding site is essentially identical to the ones we observed in the DhhN and ShhN-eHIP∆N structures (Supplementary Fig. 8). A comparison of the Shh-HIP and Shh-CDO complexes shows a partial overlap of the receptor-binding interfaces which includes residues of the Shh loop Lys88-Gly94; however, HIP and CDO have very different orientations relative to the Hh ligand (Fig. 4e). The Shh-CDO interface does not include direct interactions with the zinc-binding site and is less extensive, in agreement with solution binding assays: KdShh-Cdo = 1300 nM26 and KdShh-HIP = 6 nM (Fig. 3a). In our Shh-HIP and Dhh-HIP complexes the zinc-binding site is crucial for complex formation and even in the absence of calcium the Hh-HIP affinities are 10-times higher than that reported for the Shh-CDO (Fig. 3a).
DISCUSSION
The structural similarity between the Hh zinc-binding site and the active site of zinc-hydrolases strongly suggests that these classes of proteins are evolutionarily related. However, alteration of Shh residues, which by analogy should be essential for hydrolytic activity, did not impair signaling via PTC, thus ruling out a functional requirement for enzymatic catalysis27. The fact that this site is not conserved in any Drosophila species sequenced to date, although it is maintained in all other closely related Hh orthologues from insects and other invertebrates (Supplementary Fig. 5), further suggests that it is not required for the canonical signaling pathways such as PTC. Nevertheless, vertebrates appear to have exploited the presence of this zinc-binding site to diversify the repertoire of Hh interacting proteins to include the HIP receptor, thus introducing an additional level of regulation in this signaling pathway.
Mutations of Shh-His140 and -Cys183 (residues directly or indirectly involved in zinc-chelation) cause holoprosencephaly (HPE) and further facial anomalies as well as cancer in human patients28,29. The ShhN-CDO interaction appears to be zinc independent26 and, as discussed above, Drosphila Hh lacks the zinc binding site. Although site-directed mutagenesis of residues forming the zinc-binding site led to a marked decrease of Shh potency in a cellular differentiation assay, it was not clear whether this effect was due solely to a decrease in protein stability25. However, since functional activity was not completely abolished, interactions with other vertebrate Hh receptors (e.g. PTC) may not be entirely dependent on the presence of zinc. In contrast, zinc clearly plays a central role in Hh ligand-binding to HIP, thus the zinc- and calcium-binding sites have distinct functions in defining the specificities of Hh interactions with cell-surface receptors.
For the Hh-HIP interaction our results reveal a secondary role for the calcium-binding sites. The local concentration of extracellular calcium is highly variable30, thus the occupancy of the low affinity-binding site in Hh may vary. The juxtaposition of one metal-binding site, which plays the lead role in protein-protein interaction, with additional sites, which modulate the binding affinity, is strongly reminiscent of ligand-binding by the integrin family of cell adhesion molecules. For example, the structure of the integrin αIIbβ3 – fibrinogen γC peptide interface is centred on the integrin metal ion-dependent adhesion site (MIDAS; a high affinity magnesium-binding site) but has direct contributions from the adjacent to MIDAS (ADMIDAS) calcium-binding site31. In general, for the integrin system, the MIDAS metal ion has been shown to have a direct role in ligand-binding while the ADMIDAS metal ion has a regulatory function. Similarly, our results suggest that the interplay between the zinc- and calcium-binding sites in vertebrate Hh ligands may allow extracellular calcium concentration to play an important modulatory role.
Accession codes
Protein Data Bank: Atomic coordinates and structure factors of ShhN-eHHIP∆N (with and without Ca2+), DHHN-eHHIP∆N, eHHIP∆N and DHHN (with and without Ca2+) have been deposited with accession numbers 2WFX, 2WG4, 2WG3, 2WFT, 2WFR and 2WFQ, respectively.
METHODS
Production of the human hedgehog-interacting protein HIP ectodomain
We cloned human HIP ectodomain constructs (GenBank NP_071920; eHIP: residues 56-670; eHIP∆N: residues 214-670) into the pHLsec vector22 and expressed these by transient transfection in HEK-293T cells in the presence of kifunensine32. Proteins were purified from dialysed conditioned medium by immobilised metal-affinity chromatography and treated with endoglycosidase F132 to cleave glycosidic bonds of N-linked sugars. Proteins were concentrated and further purified by size-exclusion chromatography in 10mM HEPES, pH 7.5, 150 mM NaCl. Selenomethionine-labelled eHIP∆N (SeMet-eHIP∆N) was produced as described previously22.
Production of ShhN and DhhN, and complex formation with HIP
The N-terminal signaling domain of mouse sonic hedgehog (residues 40-194, GenBank NP_033196) was produced as described above although no kifunensine was used since ShhN is not glycosylated. Mouse ShhN differs in only one residue to human ShhN (a structurally conserved exchange of Thr68 to Ser) and most likely the structure of human ShhN is essentially identical to that of the murine orthologue. DhhN (residues 39-194, GenBank NP_066382) was cloned into the pET22b expression plasmid (Novagen) and expressed in E. coli Rosetta(DE3)pLysS cells. Cultures were grown at 37 °C to an A600 of 0.8, cooled to 20 °C, induced with 0.25 mM isopropyl-beta-D-thiogalactopyranoside (IPTG) and then grown for ~15h before harvesting. Bacterial pellets were resuspended in 25 mM sodium phosphate, pH 8.0, 500 mM NaCl, 1 mM β-mercaptoethanol and EDTA-free protease inhibitor cocktail (Roche). Cells were broken using a Basic Z model cell disruptor (Constant Systems) and fractionated by centrifugation (30,000 xg, 5 °C, 30 min). DhhN was purified from the supernatant by immobilised metal-affinity chromatography and size-exclusion chromatography in 10 mM HEPES, pH 7.5, 150 mM NaCl.
The eHIP∆N-ShhN and eHIP∆N-DhhN complexes were formed by mixing a molar ratio of 1.5:1 purified hedgehog ligand to eHIP∆N receptor, incubated for 1 h at room temperature and purified by size-exclusion chromatography in 10 mM HEPES, pH 7.5, 150 mM NaCl, with or without 2 mM CaCl2.
Crystallization and data collection
Prior to crystallization proteins were concentrated by ultrafiltration (eHIP∆N: 4 mg ml-1, eHIP∆N-ShhN: 8 mg ml-1, eHIP∆N-DhhN: 10 mg ml-1; DhhN: 7 mg ml-1). Nano-litre crystallization trials were set-up using a Cartesian Technologies robot (100 nl protein solution plus 100 nl reservoir solution) in 96-well Greiner plates33, placed in a TAP Homebase storage vault maintained at 295 K and imaged via a Veeco visualization system. Native and selenomethionine-labeled eHIP∆N crystallized in 0.1 M Tris-HCl, pH 8.5, 3.5 M potassium formate, the eHIP∆N-ShhN complex in 0.1 M sodium acetate, pH 4.6, 0.5 M potassium thiocyanate, the eHIP∆N-DhhN in 1.4 M sodium/potassium phosphate, pH 5.0 and DhhN in 0.1 M Tris-HCl, pH 8.5, 0.2 M lithium sulfate, 0.2 M sodium thiocyanate, 30% (w/v) PEG4000. For the DhhN-calcium complex, crystals were grown as above and subsequently soaked for 5 h with 50 mM CaCl2 dissolved in reservoir solution.
Diffraction data were collected at 100 K, crystals being flash-cooled in a cryo N2 gas stream. Prior to flash-freezing, crystals were treated with the appropriate cryo protectant solutions (eHIP∆N: 0.1 M Tris-HCl, 4.5 M potassium formate; eHIP∆N-ShhN: perfluoropolyether oil PFO-X125/03 (Lancaster Synthesis); eHIP∆N-DhhN and DhhN: 25% (v/v) glycerol in mother liquor). Data were collected at beamline ID29 (native and selenomethionine-labeled eHIP∆N (λ=0.9792), eHIP∆N-ShhN with calcium (λ=0.9762), eHIP∆N-DhhN (λ=0.9793) and DhhN with calcium (λ=0.9762)) at the European Synchrotron Radiation Facility, France, and at beamlines I02 (DhhN without calcium (λ=0.9507)) and I03 (eHIP∆N-ShhN without calcium (λ=0.8900)) at the Diamond Light Source, UK. X-ray data were processed and scaled with the HKL suite34. The program XPREP (http://www.bruker-nonius.com) was used to calculate quality indicators and to merge data. Data collection statistics are shown in Table 1.
Table 1.
Data collection and refinement statistics
| SeMet- eHIP∆N |
Native eHIP∆N |
eHIP∆N-ShhN with calcium |
eHIP∆N-ShhN without calcium |
eHIP∆N- DhhN without calcium |
DhhN- without calcium |
DhhN with calcium |
|
|---|---|---|---|---|---|---|---|
| Data collection | |||||||
| Space group | P3121 | P3121 | P3221 | P3221 | P212121 | P212121 | P212121 |
| Cell dimensions | |||||||
| a, b, c (Å) | 101, 101, 309 | 101, 101, 306 | 89, 89, 171 | 88, 88, 172 | 102, 110, 144 | 41, 42, 84 | 40, 42, 84 |
| α, β, γ (°) | 90, 90, 120 | 90, 90, 120 | 90, 90, 120 | 90, 90, 120 | 90, 90, 90 | 90, 90, 90 | 90, 90, 90 |
| Resolution (Å) | 20.0-3.5 (3.6-3.5) |
20.0-2.8 (2.9-2.8) |
20.0-3.2 (3.3-3.2) |
20.0-3.15 (3.25-3.15) |
20.0-2.6 (2.7-2.6) |
30.0-1.85 (1.95-1.85) |
40.0-1.95 (2.05-1.95) |
| Rsym (%) | 12.9 (62.3) | 11.5 (73.7) | 18.7 (72.8) | 20.9 (88.5) | 18.4 (81.0) | 12.9 (68.0) | 11.8 (74.9) |
| I / σI | 15.7 (3.4) | 14.7 (3.4) | 10.6 (2.5) | 9.9 (2.8) | 15.3 (2.7) | 14.6 (4.5) | 10.5 (2.6) |
| Completeness (%) | 99.5 (99.7) | 99.5 (99.9) | 100 (100) | 99.2 (99.8) | 100.0 (100.0) | 99.6 (99.7) | 93.6 (91.8) |
| Redundancy | 11.3 (8.1) | 9.1 (8.9) | 6.5 (6.7) | 7.5 (7.8) | 10.0 (10.1) | 12.7 (12.0) | 6.8 (6.5) |
| Refinement | |||||||
| Resolution (Å) | 20.0-2.8 (2.86-2.80) |
20.0-3.2 (3.35-3.20) |
20.0-3.15 (3.35-3.15) |
20.0-2.6 (2.67-2.60) |
30.0-1.85 (1.90-1.85) |
40.0-1.95 (2.00-1.95) |
|
| No. reflections | 45399 (2611)a | 16254 (1640)b | 13191 (2559)b | 48634 (3477)c | 12205 (876)d | 10413 (718)d | |
| Rwork /Rfree | 20.1 (29.7)/ 23.9 (34.2) |
24.1 (32.7)/ 29.9 (38.5) |
23.4 (28.9)/ 29.2 (34.1) |
22.7 (33.0)/ 28.1 (40.2) |
18.9 (23.8)/ 22.9 (27.9) |
18.2 (25.8)/ 22.5 (30.0) |
|
| No. atoms | |||||||
| Protein | 6578 | 4465 | 4455 | 17348 | 2599 | 2609 | |
| Zinc | 0 | 1 | 1 | 2 | 1 | 1 | |
| Calcium | 0 | 3 | 0 | 0 | 0 | 2 | |
| Water | 38 | 0 | 0 | 135 | 104 | 96 | |
| B-factors | |||||||
| Protein | 81 | 75 | 63 | 38 | 32 | 32 | |
| Zinc | - | 60 | 44 | 28 | 30 | 30 | |
| Calcium | - | 73 | - | - | - | 46 | |
| Water | 60 | - | - | 34 | 25 | 22 | |
| R.m.s. deviations | |||||||
| Bond lengths (Å) | 0.014 | 0.004 | 0.004 | 0.011 | 0.013 | 0.014 | |
| Bond angles (°) | 1.794 | 0.524 | 0.557 | 1.329 | 1.382 | 1.455 |
Values in parentheses are for highest-resolution shell.
The asymmetric unit contains two eHIP∆N molecules.
The asymmetric unit contains one eHIP∆N-ShhN complex.
The asymmetric unit contains two eHIP∆N-DhhN complexes.
The asymmetric unit contains two DhhN molecules.
Structure determination and refinement
The eHIP∆N crystal structure was determined by single anomalous dispersion (SAD) analysis. The positions of 12 selenium atoms were determined by using SHELXD35. This solution was input into AUTOSHARP36 for phase calculation, improvement and phase extension using the high-resolution native data to 2.8 Å resolution. The resulting map was of high quality and allowed tracing of the whole polypeptide chain. An initial model was built automatically using RESOLVE37 and completed by hand using COOT38. Non-crystallographic symmetry restraints were used to refine the model using the programs BUSTER39, REFMAC40 and Phenix41.
The DhhN structures were solved by molecular replacement using PHASER42 with the structure of mouse ShhN (PDB 1VHH,24) as search model. This solution was input into ARP/WARP43 for automated model building and manually adjusted and refined using COOT38 and REFMAC40.
The eHIP∆N-ShhN complexes with and without calcium and the eHIP∆N-DhhN complex were solved by molecular replacement using Phaser with the eHIP∆N structure and the ShhN (PDB 1VHH,24) or DhhN as search models, respectively. The resulting electron density maps were of high quality and allowed manual adjustment and refinement of the complex structures using COOT38, BUSTER39, REFMAC40 and Phenix41.
Refinement statistics are given in Table 1, all data within the indicated resolution range were included. Stereochemical properties were assessed by MOLPROBITY44 and PROCHECK45. Ramachandran statistics are as follows (favoured/disallowed in %): ShhN-eHIP∆N with calcium: 92.4/0, ShhN-eHIP∆N without calcium: 92.7/0, DhhN-eHIP∆N: 95.7/0.1, eHIP∆N: 93.0/0, DhhN with calcium: 96.3/0, DhhN without calcium: 96.3/0. Superpositions were calculated using SHP46 and electrostatic potentials were generated using APBS47. SC48 was used to calculate the surface complementarity scores. Buried surface areas of protein-protein interactions were calculated using the PISA webserver (http://www.ebi.ac.uk/msd- srv/prot_int/pistart.html).
Site directed mutagenesis
Mutations were generated by a two-step overlapping PCR using Pyrobest Polymerase (Takara). PCR products were cloned into the pHLsec mammalian expression vector resulting in protein constructs with a C-terminal hexa-histidine or with a C-terminal BirA recognition sequence22. Mutant proteins were secreted at similar levels to the wildtype eHIP∆N and ShhN (unpublished data). The stringent quality control mechanisms specific to the mammalian cell secretory pathway ensure that secreted proteins are correctly folded49.
Multiangle Light Scattering (MALS)
MALS experiments were carried out using an analytical Superdex S200 10/30 column (GE Heathcare) with online static light scattering (DAWN HELEOS II, Wyatt Technology, Santa Barbara, CA), differential refractive index (Optilab rEX, Wyatt Technology) and Agilent 1200 UV (Agilent Techologies) detectors. Proteins used for MALS were fully glycosylated and had previously been purified by size-exclusion chromatography and concentrated to approximately 2 mg/ml. Data were analysed using the ASTRA software package (Wyatt Technology).
Surface Plasmon resonance (SPR) binding studies
SPR experiments were performed using a Biacore T100 machine (GE Healthcare, Uppsala, Sweden) at 25 °C in 10 mM HEPES, pH 7.5, 150 mM NaCl, 0.05% polysorbate 20 (v/v), supplemented with 2 mM calcium chloride where indicated. All proteins were homogeneous with full biological activity and underwent gel filtration in running buffer immediately before use. Protein concentrations were determined from the absorbance at 280 nm using calculated molar extinction coefficients. Proteins for surface attachment were enzymatically biotinylated within an engineered C-terminal tag. The native membrane insertion topology was recapitulated by attachment of 200-2000 response units (RU) of biotinylated protein to surfaces on which 3000 RU of streptavidin were coupled via primary amines50. Experiments with wild type proteins were performed in both orientations and with mutant proteins in one orientation. The signal from experimental flow cells was corrected by subtraction of a blank and reference signal from a mock or irrelevant protein coupled flow cell. In all experiments analyzed, the experimental trace returned to baseline after each injection (Fig. 3b-d, Supplementary Fig. 7) and data fit to a simple 1:1 Langmuir model of binding. Kd values were obtained by nonlinear curve fitting of the Langmuir binding isotherm (bound = C*max/(Kd + C), where C is analyte concentration and max is the maximum analyte binding) to the data using the Levenberg-Marquardt algorithm implemented in the program Origin (Microcal Software, Northampton, MA).
Illustrations
Figures were produced using the programs PYMOL (www.pymol.org), Adobe Photoshop (Adobe Systems) and Corel Draw (Corel Corporation).
Supplementary Material
ACKNOWLEDGEMENTS
We thank the staff of ESRF beamline ID 29 and Diamond beamlines I02 and I03 for assistance with data collection, T. Walter for help with crystallization, G. Sutton for help with MALS experiments, S. Graham and D. Stuart for discussions. We acknowledge the use of the crystallization facilities provided by the MRC-funded Oxford Protein Production Facility (OPPF). The work was funded by the Wellcome Trust. A.R.A. is an MRC Career Development Fellow. C.A.O’C. is an MRC Senior Fellow. E.Y.J. is a CR-UK Principal Research Fellow. C.S. is a Wellcome Trust Career Development Fellow.
Footnotes
Note: Supplementary information is available on the Nature Structural & Molecular Biology website.
REFERENCES
- 1.Ingham PW, McMahon AP. Hedgehog signaling in animal development: paradigms and principles. Genes Dev. 2001;15:3059–87. doi: 10.1101/gad.938601. [DOI] [PubMed] [Google Scholar]
- 2.Varjosalo M, Taipale J. Hedgehog: functions and mechanisms. Genes Dev. 2008;22:2454–72. doi: 10.1101/gad.1693608. [DOI] [PubMed] [Google Scholar]
- 3.Roessler E, et al. Mutations in the human Sonic Hedgehog gene cause holoprosencephaly. Nat Genet. 1996;14:357–60. doi: 10.1038/ng1196-357. [DOI] [PubMed] [Google Scholar]
- 4.Dellovade T, Romer JT, Curran T, Rubin LL. The hedgehog pathway and neurological disorders. Annu Rev Neurosci. 2006;29:539–63. doi: 10.1146/annurev.neuro.29.051605.112858. [DOI] [PubMed] [Google Scholar]
- 5.Yauch RL, et al. A paracrine requirement for hedgehog signalling in cancer. Nature. 2008;455:406–10. doi: 10.1038/nature07275. [DOI] [PubMed] [Google Scholar]
- 6.Mann RK, Beachy PA. Novel lipid modifications of secreted protein signals. Annu Rev Biochem. 2004;73:891–923. doi: 10.1146/annurev.biochem.73.011303.073933. [DOI] [PubMed] [Google Scholar]
- 7.Zeng X, et al. A freely diffusible form of Sonic hedgehog mediates long-range signalling. Nature. 2001;411:716–20. doi: 10.1038/35079648. [DOI] [PubMed] [Google Scholar]
- 8.Panakova D, Sprong H, Marois E, Thiele C, Eaton S. Lipoprotein particles are required for Hedgehog and Wingless signalling. Nature. 2005;435:58–65. doi: 10.1038/nature03504. [DOI] [PubMed] [Google Scholar]
- 9.Pepinsky RB, et al. Identification of a palmitic acid-modified form of human Sonic hedgehog. J Biol Chem. 1998;273:14037–45. doi: 10.1074/jbc.273.22.14037. [DOI] [PubMed] [Google Scholar]
- 10.Lum L, Beachy PA. The Hedgehog response network: sensors, switches, and routers. Science. 2004;304:1755–9. doi: 10.1126/science.1098020. [DOI] [PubMed] [Google Scholar]
- 11.Wilson CW, Chuang PT. New “hogs” in Hedgehog transport and signal reception. Cell. 2006;125:435–8. doi: 10.1016/j.cell.2006.04.016. [DOI] [PubMed] [Google Scholar]
- 12.Beckett K, Franch-Marro X, Vincent JP. Glypican-mediated endocytosis of Hedgehog has opposite effects in flies and mice. Trends Cell Biol. 2008;18:360–3. doi: 10.1016/j.tcb.2008.06.001. [DOI] [PubMed] [Google Scholar]
- 13.Chuang PT, McMahon AP. Vertebrate Hedgehog signalling modulated by induction of a Hedgehog-binding protein. Nature. 1999;397:617–21. doi: 10.1038/17611. [DOI] [PubMed] [Google Scholar]
- 14.Kang JS, Zhang W, Krauss RS. Hedgehog signaling: cooking with Gas1. Sci STKE. 2007;2007:pe50. doi: 10.1126/stke.4032007pe50. [DOI] [PubMed] [Google Scholar]
- 15.Coulombe J, Traiffort E, Loulier K, Faure H, Ruat M. Hedgehog interacting protein in the mature brain: membrane-associated and soluble forms. Mol Cell Neurosci. 2004;25:323–33. doi: 10.1016/j.mcn.2003.10.024. [DOI] [PubMed] [Google Scholar]
- 16.Parmantier E, et al. Schwann cell-derived Desert hedgehog controls the development of peripheral nerve sheaths. Neuron. 1999;23:713–24. doi: 10.1016/s0896-6273(01)80030-1. [DOI] [PubMed] [Google Scholar]
- 17.Bourikas D, et al. Sonic hedgehog guides commissural axons along the longitudinal axis of the spinal cord. Nat Neurosci. 2005;8:297–304. doi: 10.1038/nn1396. [DOI] [PubMed] [Google Scholar]
- 18.Olsen CL, Hsu PP, Glienke J, Rubanyi GM, Brooks AR. Hedgehog-interacting protein is highly expressed in endothelial cells but down-regulated during angiogenesis and in several human tumors. BMC Cancer. 2004;4:43. doi: 10.1186/1471-2407-4-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tada M, et al. Down-regulation of hedgehog-interacting protein through genetic and epigenetic alterations in human hepatocellular carcinoma. Clin Cancer Res. 2008;14:3768–76. doi: 10.1158/1078-0432.CCR-07-1181. [DOI] [PubMed] [Google Scholar]
- 20.Taniguchi H, et al. Intrahepatic mRNA levels of type I interferon receptor and interferon-stimulated genes in genotype 1b chronic hepatitis C. Association between IFNAR1 mRNA level and sustained response to interferon therapy. Intervirology. 2007;50:32–9. doi: 10.1159/000096310. [DOI] [PubMed] [Google Scholar]
- 21.Tojo M, Kiyosawa H, Iwatsuki K, Kaneko F. Expression of a sonic hedgehog signal transducer, hedgehog-interacting protein, by human basal cell carcinoma. Br J Dermatol. 2002;146:69–73. doi: 10.1046/j.1365-2133.2002.04583.x. [DOI] [PubMed] [Google Scholar]
- 22.Aricescu AR, Lu W, Jones EY. A time- and cost-efficient system for high-level protein production in mammalian cells. Acta Crystallogr D Biol Crystallogr. 2006;62:1243–50. doi: 10.1107/S0907444906029799. [DOI] [PubMed] [Google Scholar]
- 23.Harding MM. Geometry of metal-ligand interactions in proteins. Acta Crystallogr D Biol Crystallogr. 2001;57:401–11. doi: 10.1107/s0907444900019168. [DOI] [PubMed] [Google Scholar]
- 24.Hall TM, Porter JA, Beachy PA, Leahy DJ. A potential catalytic site revealed by the 1.7-A crystal structure of the amino-terminal signalling domain of Sonic hedgehog. Nature. 1995;378:212–6. doi: 10.1038/378212a0. [DOI] [PubMed] [Google Scholar]
- 25.Day ES, et al. Zinc-dependent structural stability of human Sonic hedgehog. Biochemistry. 1999;38:14868–80. doi: 10.1021/bi9910068. [DOI] [PubMed] [Google Scholar]
- 26.McLellan JS, et al. The mode of Hedgehog binding to Ihog homologues is not conserved across different phyla. Nature. 2008;455:979–83. doi: 10.1038/nature07358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fuse N, et al. Sonic hedgehog protein signals not as a hydrolytic enzyme but as an apparent ligand for patched. Proc Natl Acad Sci U S A. 1999;96:10992–9. doi: 10.1073/pnas.96.20.10992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Orioli IM, et al. Identification of novel mutations in SHH and ZIC2 in a South American (ECLAMC) population with holoprosencephaly. Hum Genet. 2001;109:1–6. doi: 10.1007/s004390100537. [DOI] [PubMed] [Google Scholar]
- 29.Ribeiro LA, Richieri-Costa A. Single median maxillary central incisor, hypophyseal tumor, and SHH mutation. Am J Med Genet A. 2005;136A:346–7. doi: 10.1002/ajmg.a.30625. [DOI] [PubMed] [Google Scholar]
- 30.Hofer AM, Brown EM. Extracellular calcium sensing and signalling. Nat Rev Mol Cell Biol. 2003;4:530–8. doi: 10.1038/nrm1154. [DOI] [PubMed] [Google Scholar]
- 31.Springer TA, Zhu J, Xiao T. Structural basis for distinctive recognition of fibrinogen gammaC peptide by the platelet integrin alphaIIbbeta3. J Cell Biol. 2008;182:791–800. doi: 10.1083/jcb.200801146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chang VT, et al. Glycoprotein structural genomics: solving the glycosylation problem. Structure. 2007;15:267–73. doi: 10.1016/j.str.2007.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Walter TS, et al. A procedure for setting up high-throughput nanolitre crystallization experiments. Crystallization workflow for initial screening, automated storage, imaging and optimization. Acta Crystallogr D Biol Crystallogr. 2005;61:651–7. doi: 10.1107/S0907444905007808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 35.Schneider TR, Sheldrick GM. Substructure solution with SHELXD. Acta Crystallogr D Biol Crystallogr. 2002;58:1772–9. doi: 10.1107/s0907444902011678. [DOI] [PubMed] [Google Scholar]
- 36.Vonrhein C, Blanc E, Roversi P, Bricogne G. Automated structure solution with autoSHARP. Methods Mol Biol. 2007;364:215–30. doi: 10.1385/1-59745-266-1:215. [DOI] [PubMed] [Google Scholar]
- 37.Terwilliger TC. Automated side-chain model building and sequence assignment by template matching. Acta Crystallogr D Biol Crystallogr. 2003;59:45–9. doi: 10.1107/S0907444902018048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–32. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 39.Blanc E, et al. Refinement of severely incomplete structures with maximum likelihood in BUSTER-TNT. Acta Crystallogr D Biol Crystallogr. 2004;60:2210–21. doi: 10.1107/S0907444904016427. [DOI] [PubMed] [Google Scholar]
- 40.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D Biol Crystallogr. 1997;53:240–55. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 41.Adams PD, et al. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr D Biol Crystallogr. 2002;58:1948–54. doi: 10.1107/s0907444902016657. [DOI] [PubMed] [Google Scholar]
- 42.McCoy AJ, Grosse-Kunstleve RW, Storoni LC, Read RJ. Likelihood-enhanced fast translation functions. Acta Crystallogr D Biol Crystallogr. 2005;61:458–64. doi: 10.1107/S0907444905001617. [DOI] [PubMed] [Google Scholar]
- 43.Perrakis A, Morris R, Lamzin VS. Automated protein model building combined with iterative structure refinement. Nat Struct Biol. 1999;6:458–63. doi: 10.1038/8263. [DOI] [PubMed] [Google Scholar]
- 44.Davis IW, et al. MolProbity: all-atom contacts and structure validation for proteins and nucleic acids. Nucleic Acids Res. 2007;35:W375–83. doi: 10.1093/nar/gkm216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hooft RW, Vriend G, Sander C, Abola EE. Errors in protein structures. Nature. 1996;381:272. doi: 10.1038/381272a0. [DOI] [PubMed] [Google Scholar]
- 46.Stuart DI, Levine M, Muirhead H, Stammers DK. Crystal structure of cat muscle pyruvate kinase at a resolution of 2.6 A. J Mol Biol. 1979;134:109–42. doi: 10.1016/0022-2836(79)90416-9. [DOI] [PubMed] [Google Scholar]
- 47.Baker NA, Sept D, Joseph S, Holst MJ, McCammon JA. Electrostatics of nanosystems: application to microtubules and the ribosome. Proc Natl Acad Sci U S A. 2001;98:10037–41. doi: 10.1073/pnas.181342398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lawrence MC, Colman PM. Shape complementarity at protein/protein interfaces. J Mol Biol. 1993;234:946–50. doi: 10.1006/jmbi.1993.1648. [DOI] [PubMed] [Google Scholar]
- 49.Trombetta ES, Parodi AJ. Quality control and protein folding in the secretory pathway. Annu Rev Cell Dev Biol. 2003;19:649–76. doi: 10.1146/annurev.cellbio.19.110701.153949. [DOI] [PubMed] [Google Scholar]
- 50.O’Callaghan CA, et al. BirA enzyme: production and application in the study of membrane receptor-ligand interactions by site-specific biotinylation. Anal Biochem. 1999;266:9–15. doi: 10.1006/abio.1998.2930. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.





