Summary
Linkage-specific polyubiquitin recognition is thought to make possible the diverse set of functional outcomes associated with ubiquitination. Thus far, mechanistic insight into this selectivity has been largely limited to single domains that preferentially bind to lysine 48-linked polyubiquitin (K48-polyUb) in isolation. Here we propose a mechanism, linkage-specific avidity, in which multiple ubiquitin binding domains are arranged in space so that simultaneous, high-affinity interactions are optimum with one polyUb linkage, but unfavorable or impossible with other polyUb topologies and monoUb. Our model is human Rap80, which contains tandem ubiquitin interacting motifs (UIMs) that bind to K63-polyUb at DNA double-strand breaks. We show how the sequence between the Rap80 UIMs positions the domains for efficient, avid binding across a single K63 linkage, thus defining selectivity. We also demonstrate K48-specific avidity in a different protein, ataxin-3. Using tandem UIMs, we establish the general principles governing polyUb linkage selectivity and affinity in multivalent ubiquitin receptors.
Introduction
The covalent attachment of the small protein ubiquitin (Ub) to other proteins is an essential step in an enormous variety of cellular processes (Pickart and Eddins, 2004). Substrates can be modified with a single Ub unit or polymeric Ub chains assembled by the linkage of one Ub C-terminus to any of seven lysines on another Ub molecule (Peng et al., 2003). Whereas ubiquitination with chains linked though lysine 48 (K48-polyUb) leads to degradation of the substrate protein at the 26S proteasome (Pickart and Cohen, 2004), lysine 63-linked polyUb and monoUb function as distinct but non-proteolytic signaling elements (Sun and Chen, 2004) in pathways such as endocytosis and DNA repair. The prevailing model holds that this functional diversity is possible because downstream receptors can distinguish the Ub forms by selective binding (Pickart and Fushman, 2004).
We suspected that some multivalent Ub binding proteins may achieve linkage selectivity by exploiting the distinct orientation and spacing of Ub units that result from a particular polyUb linkage. Multiple ubiquitin binding domains (UBDs) could be arrayed in space to optimize simultaneous interactions with both Ub units in a configuration characteristic of one type of Ub-Ub linkage, but not another. For the target polyUb linkage, binding by the first of multiple UBDs to one Ub would position the second UBD favorably for interaction with a nearby Ub in the chain. Because the second binding event occurs between binding partners at high local concentrations, it is potentially much more favorable. This could form the basis of linkage-specific polyUb recognition, as well as polyUb vs monoUb specificity. This type of cooperative binding is termed avidity; hence, we call this mechanism ‘linkage-specific avidity’.
We examined the human protein Rap80 as a potential model of linkage-specific avidity. At the site of DNA double-strand breaks in human cells, an early signaling cascade leads to the recruitment of the Ub E2 enzyme Ubc13 and the E3 RNF8 which then ubiquitinate one or more substrates at the site of the damage with K63-linked polyUb (Bennett and Harper, 2008; Kolaset et al., 2007; Mailand et al., 2007; Wang and Elledge, 2007; Huen et al., 2007). Rap80 links these early events to proteins critical for repair by binding to the K63-polyUb signal with N-terminal tandem UIMs (tUIMs), while a central Rap80 domain binds an associated complex that includes Abraxas, the tumor suppressor BRCA1, and the deubiquitinating enzyme BRCC36 (Kim et al., 2007; Sobhian et al., 2007; Wang et al., 2007; Wang and Elledge, 2007; Yan et al., 2007). The importance of the localization activity is evident from clinical BRCA1 mutants that do not assemble into the complex with Rap80 and thus fail to reach the sites of DNA damage (Sobhian et al., 2007).
Although UIM domains are not known to have a large linkage preference in isolation, Rap80 bound more K63-polyUb than K48-polyUb in GST pull-down experiments (Kim et al, 2007; Sobhian et al., 2007). UIMs are 18 - 21 amino acids in length and are ideal for structure prediction and molecular modeling because of their simple, α-helical structure (Fisher et al., 2003; Hofmann and Falquet, 2001). We performed modeling based on the structure of a UIM bound to monoUb and found that the 7-residue linker between the Rap80 tUIMs could position the domains ideally for simultaneous interactions with two K63-linked Ub units. In contrast, modeling based on a K48-diUb structure predicted that avid interaction with K48-diUb would be impossible or inefficient, requiring longer K48 chains for simultaneous contacts. Here we show that, consistent with these models, the Rap80 UIM linker defines selectivity by optimizing avid binding across a single K63 linkage. We demonstrate that linkage-specific avidity also underlies the selective preference for K48-polyUb in a tUIM protein with a shorter linker, human ataxin-3. Using tUIMs as a model, we establish the general principles that underlie linkage-specific avidity and polyUb affinity in multi-UBD proteins.
Results
Rap80 UIMs bind monoUb weakly
To examine Ub and polyUb binding by Rap80, we produced a His6-tagged version of the Rap80 tUIM peptide (Figure 1A) and fluorescently labeled it on a single cysteine that naturally occurs in the C-terminal UIM sequence (Figure 1B). To resolve the contributions of the individual UIM domains to binding, we created single-UIM peptides with either of the 8-residue consensus Ub binding sites (UIM1 and UIM2, Figure 1A) deleted. Because these peptides are small and relatively mobile in solution, binding to Ub or polyUb was detected as an increase in the peptide fluorescence anisotropy. Control experiments showed that the fluorescent label did not affect Ub binding (Supplementary Data Figure S1). Typical of UIM domains (Fisher et al., 2003), UIM1 and UIM2 bound monoUb very weakly (KdUIM1 = 510 μM, KdUIM2 = 520 μM; Figure 1C). The tUIM peptide bound monoUb with the same low affinity (KdtUIM = 520 μM). These results indicate that both UIM domains can interact with Ub, and that binding of monoUb by the tUIMs is not cooperative.
Figure 1.
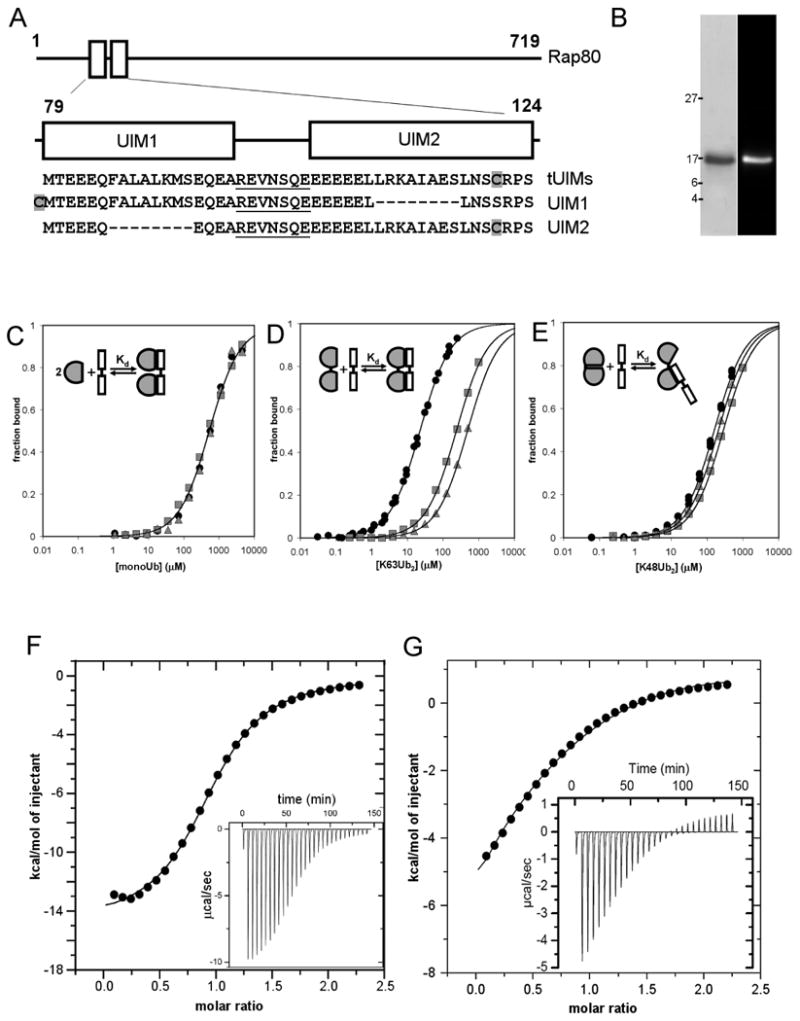
Binding studies with Rap80 UIM peptides reveal that differences in avid binding underlie K63-linked polyUb binding selectivity. (A) The Rap80 protein is shown schematically with the UIMs indicated in blocks (top). The sequences of the tUIM and single UIM peptides are shown, each with the linker sequence underlined and the cysteine residue used for fluorescent labeling shaded grey (bottom). (B) Coomassie staining (left) and fluorescence image (right) of purified Rap80 tUIM peptide after SDS-PAGE. (C-E) Fluorescence anisotropy binding data for the Rap80 tUIM peptide (filled circles), UIM1 (grey squares), and UIM2 (grey triangles) titrated with monoUb (C), K63-Ub2 (D), or K48Ub2 (E). Schematic binding models for the interaction of each ubiquitin species with the tUIM peptide are inset in each plot. (F, G) Isothermal titration calorimetry measurements for the Rap80 tUIM peptide with K63-Ub2 (F) or K48-Ub2 (G). Raw data traces are inset in the integrated, fit data plots.
Rap80 binds avidly across a single K63-linked diUb
Next we measured binding of the Rap80 UIM peptides to K63-linked diUb (K63-Ub2). The single UIM peptides bound K63-Ub2 with roughly the same affinity as for monoUb (KdUIM1 = 230 μM and KdUIM2 = 470 μM for K63-Ub2; Figure 1D). In contrast, the affinity of the tUIM peptide was much tighter (KdtUIM = 22 ± 1 μM for K63-Ub2). Binding data that included higher ligand (K63-Ub2) concentrations fit a complex model of divalent-divalent interactions well, with no change in the Kd value of interest (Figure S2).
To verify the binding constant measured by fluorescence anisotropy, we performed isothermal titration calorimetry (ITC) of the Rap80 tUIM peptide with K63-Ub2. The ITC data neared complete saturation and were fit by a single-site model (KdITC = 17.6 ± 0.7 μM, n = 0.97 sites; Figure 1F), in excellent agreement with the anisotropy binding constant and supporting our interpretation of those measurements. These data demonstrate a relatively high-affinity interaction between the Rap80 tUIMs and K63-Ub2 that depends on both UIM domains, indicating that the interaction is avid, with simultaneous binding to both Ub units in the K63-Ub2. As a result of this avidity, Rap80 tUIMs show a 24-fold preference for K63-Ub2 over monoUb.
Rap80 cannot bind avidly across a single K48-linked diUb
We next compared the affinities of the Rap80 UIM peptides for K48-linked diUb (K48-Ub2). UIM1 and UIM2 bound K48-Ub2 with roughly the same affinity as the tUIM peptide (KdUIM1 = 280 μM, KdUIM2 = 200 μM, KdtUIM = 157 ± 8 μM; Figure 1E). This indicates that the interaction between the tUIM peptide and K48-Ub2 employs only one UIM domain and therefore is not avid.
To confirm this result, we performed an ITC titration of K48-Ub2 with the tUIM peptide (Figure 1G). The ITC data reveal two distinct binding sites on K48-Ub2; both a negative and a positive enthalpy interaction are evident. These data cannot be fit by a single-site model as used for the avid K63-Ub2 interaction. Rather, the ITC data are fit well by a sequential binding model, with the first-site affinity in good agreement with the anisotropy value (Kd1,ITC = 171 ± 40 μM), and the second-site affinity in good agreement with the interaction of a single UIM with monoUb (Kd2,ITC = 470 ± 140 μM). It is likely that the weaker interaction was not observed in the fluorescence anisotropy titration because the ligand concentrations were lower than those used in the ITC measurement. Overall, these data indicate that the Rap80 tUIMs bind avidly to K63-Ub2, but not across a single K48 linkage. This linkage-specific avidity underlies a 7-fold preference for K63-Ub2 over K48-Ub2.
Closely linked domains promote affinity and selectivity in avid polyUb interactions
To test the possibility that longer K48-linked chains could allow non-adjacent Ub units to interact simultaneously with a single set of tUIMs, we measured the affinity of the tUIM peptide for K48-linked tetraUb (K48-Ub4). The tUIM peptide bound K48-Ub4 more than eleven-times tighter than K48-Ub2 (KdK48Ub4 = 14 μM), an increase in affinity indicative of avid binding to Ub4. In spite of avid interactions with longer K48 chains, the tUIM peptide still bound K63-Ub4 with a four-fold preference over K48-Ub4 (KdK63Ub4 = 3.6 μM). The decrease in Kd for K63-Ub4 over K63-Ub2 can be understood because Ub4 contains more binding units in molar terms, and because the longer chain presents a linear array of binding sites that may favor re-binding to neighboring sites. These measurements indicate that avid binding to non-adjacent Ub units is less efficient than avid binding across a single isopeptide linkage.
By the same reasoning, longer linkers between UIM domains should lower affinity and thus lower linkage selectivity. To test this, we extended the 7-amino acid sequence between the Rap80 UIMs to 14 residues (REVNSQEREVNSQE). This peptide bound K63-Ub2 substantially more weakly, and with very little selectivity (Table 1). Next we examined the yeast tUIM protein Vps27, which has an unstructured 25-residue linker (Swanson et al., 2003). As expected, the Vps27 tUIMs bound K63-Ub4 and K48-Ub4 indistinguishably. In contrast, the Vps27 UIMs connected with a 7-residue linker bound K63-linked chains with high affinity and specificity (Table 1). These results demonstrate that, for both polyUb chains and linked UBDs, binding units tightly coupled in space provide the maximum opportunity for high affinity and highly linkage-selective interactions.
Homology models of Rap80 bound to diUb reveal the structural basis of avid K63-polyUb recognition
Multiple secondary structure prediction programs indicate that the Rap80 UIM domains are strongly α-helical whereas the linker sequence is weakly α-helical. Circular dichroism (CD) measurements support this (Figure S6 and S7). Figure 2B is a structural model based on these predictions. We noticed that if the linking sequence acquired helical structure when bound to K63-Ub2, the tUIM peptide would comprise one continuous α-helix with an integer number of helical turns between the UIM domains. This would orient the Ub interaction surfaces of both UIMs towards the same face of the peptide in a configuration ideal for simultaneous interactions with both hydrophobic patches of the linear (Varadan et al., 2004) K63-Ub2 molecule. Next, we examined the five structures of UIM domains bound to monoUb (Hirano et al., 2006; Swanson et al., 2003; Wang, et al., 2005). UIMs in all of these structures shared a common orientation on the ubiquitin surface, along the axis created by the C-terminus and K63 (Figure 2A). This would allow two UIMs on a single helix to simultaneously bind two K63-linked ubiquitins. To model this Rap80·K63Ub2 interaction, we used the coordinates from the NMR structure of Vps27 UIM1 bound to monoUb (Swanson et al., 2003) to place each of the Rap80 UIMs on a Ub, and then connected the UIMs with an α-helical segment of appropriate length. Remarkably, the resulting configuration of the bound ubiquitins easily allows a K63 linkage and is a good match for the known K63-Ub2 structure (Figure S4). Figure 2D presents this model of Rap80 bound to K63-Ub2. Consistent with our binding data, we could not model simultaneous UIM interactions to an open form of K48-Ub2, even with an unstructured UIM linker (Figure S5).
Figure 2.
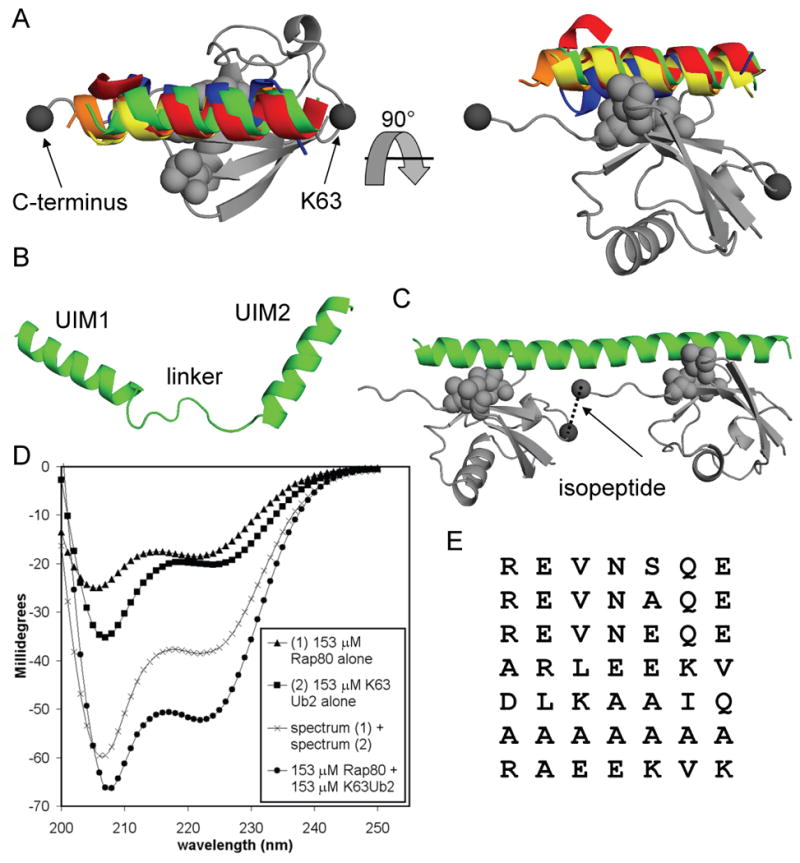
Modeling and structural measurements reveal the molecular basis of linkage-specific avidity for the Rap80 tUIM peptide. (A) The structures of 5 UIM domains [S5a UIM1 from 1YX5.pdb (red), S5a UIM2 from 1YX6.pdb (blue), Vps27 UIM1 from 1Q0W.pdb (green), and the double-sided UIM from Hrs, 2D3G.pdb (orange and yellow)] bound to monoUb (grey) were aligned using the Ub coordinates from each complex. The Ub coordinates from 1QOW.pdb are shown. The ubiquitin hydrophobic patch is shown in light grey spheres for all models. Two perspectives show the common UIM orientation on the surface of ubiquitin with respect to the C-terminus and K63 (dark grey spheres). (B) The homology model of unbound Rap80 tUIMs shows an unstructured linker. (C) The model for Rap80 (green) bound to K63-linked diUb (grey) shows how a helical linker orients the UIM domains for optimum simultaneous interactions. The isopeptide bond is shown between the C-terminus and K63 (dark grey spheres). (D) CD spectra for 153 μM Rap80 alone (triangles), 153 μM K63-Ub2 alone (squares), and a mixture of 153 μM Rap80 with 153 μM K63-Ub2 (circles). The sum of the unbound Rap80 and K63-Ub2 spectra (crosses) is shown for comparison to the bound complex. (E) An alignment of all tested 7-residue tUIM linkers that support K63-specific binding shows no sequence conservation.
To test directly the structural transition predicted for the Rap80 linker, we produced a version of the Rap80 tUIM peptide without additional vector-derived residues and measured the CD spectra of the peptide in the unbound and the K63-Ub2 bound state. Figure 2D shows the sum of the unbound Rap80 and unbound K63-Ub2 spectra compared to the spectrum of Rap80 and K63-Ub2 mixed at nearly saturating concentrations. Because large shifts in ubiquitin secondary structure are typically not seen upon binding, the difference in the two spectra at 222 nm indicates that Rap80 acquires significant helical structure when bound to K63-Ub2. Whole-spectrum interpretations of these data indicate that the unbound form of Rap80 is 30-35% α-helix, while the K63-Ub2 bound form is nearly 100% α-helix. Simpler estimates of helical content predict less helicity (Figure S6). These data support a model for the Rap80·K63-Ub2 interaction in which the 7-residue linker becomes helical in the bound state and precisely positions the UIM domains for avid binding across a single K63 linkage.
In our model of the Rap80·K63-Ub2 interaction, direct side-chain contacts between the Rap80 linker and ubiquitin are not predicted to be important. We tested 7 different 7 residue tUIM linkers (Figure 2E) and found that they all supported high affinity, K63-specific polyUb interactions, even though a wide variety of side chains were represented at each position within the linker. This excludes a major role for direct linker side-chain·ubiquitin contacts in defining affinity and linkage selectivity. Details on each of these constructs appear elsewhere in the text. The common property of all these linkers is some degree of predicted helical content, supporting our model of the Rap80·K63-Ub2 interaction.
Factors governing polyUb linkage specificity and affinity for tUIMs
We expected that tUIM binding properties would be affected by both the length and structure of the linking sequence, and by the intrinsic affinities of the individual UIM domains for ubiquitin. We constructed a set of peptides in which the Rap80 UIMs were linked by 1 to 9 alanine residues in order to examine linker length systematically in a way that was roughly independent from the linker structure and UIM affinity. K63-Ub2 binding data for the alanine linker set are presented in Figure 3A. The plot of Kd vs. linker length is well fit by a sine wave that repeats every 3.5 residues, or approximately 1 α-helical turn (Figure 3B). This is consistent with our model in which a single, continuous helix of tUIMs only engages the K63-Ub2 efficiently when the UIM binding sites are separated by a near-integer number of helical turns, and thus face the same side of the helix. The optimum linker length for K63-polyUb binding, 7 residues, also most satisfies this rule (i.e., 6.9 turns between binding sites), whereas the weakest binders (2A and 9A are >25-fold weaker than 7A) have linkers that keep the UIM binding sites almost totally out of helical phase with each other (5.6 turns and 7.5 turns between binding sites, respectively). This strong correlation demonstrates that α-helical linkers can dramatically modulate the affinity for K63-polyUb by controlling the orientation of the UIM domains with respect to each other, thus reducing or eliminating avidity.
Figure 3.
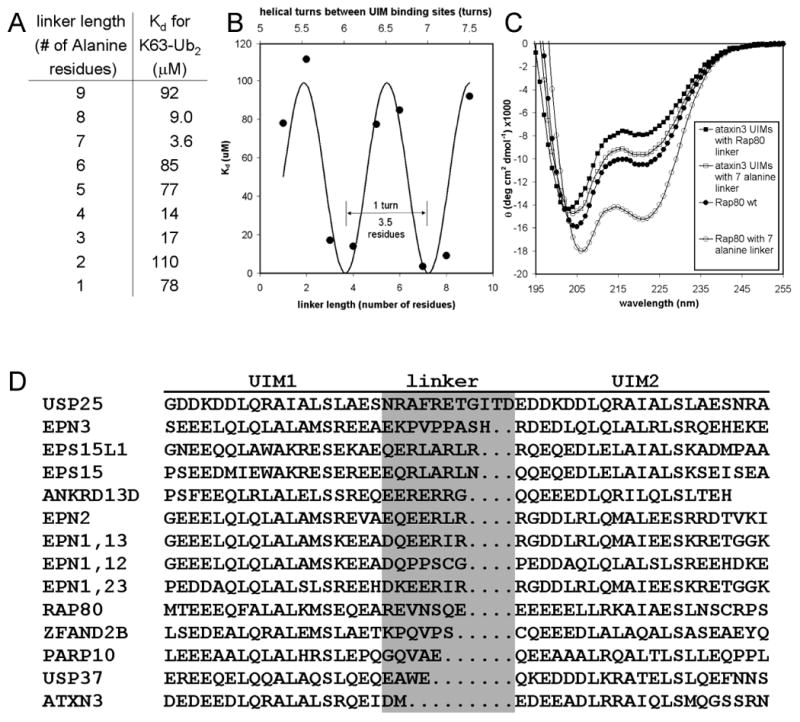
Binding and structural measurements reveal the factors governing polyUb linkage-specificity and affinity for tUIMs. (A) Summary of fluorescence anisotropy binding data collected for Rap80 tUIMs with various all-alanine linkers interacting with K63-Ub2. (B) The pattern of K63-Ub2 affinities demonstrates that linker length can dramatically modulate linkage-specificity by controlling the orientation of the UIM domains with respect to each other, thus affecting avidity. (C) CD spectra for various tandem UIM constructs establishes the link between structure and polyUb binding for tandem UIMs. (D) An alignment of tUIMs from humans shows diverse linker lengths. Linkers that can easily adopt helical conformations are expected to follow the K63-polyUb binding pattern presented in (B).
We observed the same pattern of affinities when shorter linkers derived from the Rap80 sequence were tested (compare REVNSQ and REVAAQ to wild type in Table 1), indicating further support for our model of K63-Ub2 bound Rap80. Even removing the linking sequence entirely to position the binding sites in phase reduced K63-Ub2 affinity only 2-fold compared to wildtype, whereas the 2-residue linker ‘DM’ binds K63-Ub2 with the weakest affinity of all (Table 1). This may indicate some tolerance for variation in the lateral distance between UIMs, but strict requirements for domain orientations. We note that affinity for K48-Ub2 was largely independent of the linker length or composition in this data set and for the alanine linkers tested, consistent with the finding that K48-Ub2 recognition is not avid (Table 1).
Intrinsic helical propensity in the linking sequence should correlate with tighter avid binding, because more ordered linkers more effectively pre-organize the second receptor site for binding after the first site is bound. In a striking example of this, Rap80 with the strongly helical 7-alanine linker binds K63-Ub2 6-fold more tightly than Rap80 with the weakly helical wild-type linker (Table 1; note no change in K48-Ub2 affinity). CD measurements confirm the difference in helicity; in Figure 3C, note the marked difference in the extent of the trough at 222 nm, which is one measure of helical content.
When the UIMs from ataxin-3 were spliced together with the Rap80 linker, the result was a much less helical peptide that was only weakly selective for K63-Ub2, and showed no K63 linkage preference for longer chains. When the same UIMs were joined by the more helical 7-alanine linker, helicity and K63 selectivity were improved (Figure 3C, Table 1). This suggests that there are minimal structural requirements for a linker in the context of its UIMs to achieve linkage specificity through avidity. This also supports our model in which the Rap80 linker defines selectivity through domain positioning, and not through specific contacts with Ub.
Using secondary structure prediction tools, we found several other linker variants predicted to have more helical content than the wild-type Rap80 linker (Figure S7). Indeed, all of the linker variants bound K63-Ub2 more tightly than wild-type Rap80, and the trends in affinity and selectivity closely matched the trend in predicted linker helical content (see Table 1 entries for S101A, S101E, DLKAAIQ and ARLEEKV linkers). CD measurements confirmed the underlying difference in helical propensity for one set of these peptides, though the changes were subtle (Figure S7). These results support our structural model of Rap80 bound to K63-Ub2, and indicate that linker sequences that reduce the flexibility between UIMs promote high affinity and linkage-selective interactions.
The 2-amino acid linker of ataxin-3 tUIMs defines K48 selectivity
Since the 7-residue linker alone accounts for most of the linkage preference of Rap80, we wondered if different-length linkers in other tUIM proteins could define different polyUb specificities by linkage-specific avidity. At least 12 human proteins contain closely spaced tUIMs (Figure 3D). Because ataxin-3 UIMs have a conserved 2-residue linker (Figure S8), and because previous studies indicated that ataxin-3 binds with high affinity to K48-polyUb (Chai et al., 2004), we tested whether the 2-residue linker conferred K48 selectivity. Indeed, ataxin-3 tUIMs bound K48-Ub4 with a 5-fold preference over K63-Ub4 (Table 1). Remarkably, the ataxin-3 linker alone was sufficient to transfer a 6-fold K48 preference to the Rap80 UIM domains (Table 1). The effect was apparently not due to linker-specific side-chain contacts with ubiquitin, as a 2-alanine linker between the Rap80 UIMs conferred K48-Ub4 specificity nearly as well (Table 1).
With 7-residue linkers, differences in avidity and thus linkage-specificity were apparent for differently-linked diubiquitins. In contrast, for 2-residue linkers, interactions with Ub2 were apparently not avid for either linkage, and were of low affinity and negligible selectivity (Table 1). Instead, high-affinity, avid binding was seen only for longer chains, where K48 linkage-specificity was evident. This indicates that receptors can distinguish polyUb linkages by avidity when the unit of avid recognition is longer polyUb chains. We note that the 2-alanine linker form of the Rap80 tUIM bound K63-Ub2 the most weakly of all the tUIMs in the alanine-linker series. Further binding and structural studies will be required to determine if 2-residue linkers position UIM domains for truly optimum K48-polyUb binding, or whether this arrangement simply excludes K63-polyUb most efficiently.
Discussion
Linkage-specific polyUb binding
The paradigm for linkage-selective polyUb recognition has been shaped mostly by examples from the Ub associated (UBA) domains. The molecular basis for K48-selective binding has been described for UBA2 from human hHR23A (Varadan et al., 2005), and the UBA in Mud1 from fission yeast (Trempe et al., 2005). In these cases, all of the elements that confer selectivity are contained within a single UBA domain, and specific recognition of K48-polyUb is achieved through binding at an interface centered on the Ub-Ub isopeptide bond. While this manuscript was in preparation, a study by Lo et al. revealed how the CC2-LZ domain of NEMO binds linear and K63-linked diubiquitin. Both halves of the dimeric CC2-LZ domain engage linear and K63-linked diubiquitin along slightly different extended surfaces that include the Ub-Ub junction.
While selective recognition of polyUb chains by single domains is likely an important mechanism, multiple UBDs are commonly found in Ub-binding proteins and protein complexes (Hicke, et al., 2005; Reyes-Turcu et al., 2008). In these cases, large cooperative binding effects may make the individual domain interactions less relevant. For example, multivalency is thought to underly the physiologically relevant affinities achieved at the endosome when Ub receptors bind oligomerized, multi-monoubiquitinated, or polyubiquitinated cargoes with a series of tandem UBDs that, in isolation, each bind Ub poorly (Barriere et al., 2006; Haglund et al., 2003; Hawryluk et al., 2006; Hicke, and Dunn, 2003). Here we have demonstrated that non-selective or weakly linkage-selective single UIMs can also achieve considerable linkage specificity when the arrangement of the domains makes avid binding to one polyUb topology more favorable than others. Our model differs from the isopeptide-centered recognition in the examples above because linkage-specific avidity requires no specific contacts at or near the isopeptide bond.
It remains to be determined whether UBDs other than UIMs can exert a linkage preference through avidity. A role for linkage-specific avidity in specific binding for other polyUb topologies or for Ub-like protein polymers such as polySUMO (Kerscher, Felberbaum, and Hochstrasser, 2006) is also speculative. Nonetheless, using tUIMs as a model, we have established some of the general principles that should govern these multi-valent interactions. Avid binding is highly sensitive to the orientation of the binding units with respect to each other and the flexibility between the units. Systems with less flexibility are expected to have higher affinities and specificities because entropic costs are higher when more flexible linkers need to bring binding units together in space, and because flexible linkers do not restrict binding units to a small set of potentially distinguishing conformations. Also, the intrinsic affinities of individual domains contribute to specificity and affinity; because avidity at best only multiplies receptor affinities, weak-binding receptors will gain less from a similar multivalent arrangement than tight ones (Bobrovnik, 2007; Schleif, and Wolberger, 2004). Potentially, UBDs distant in primary sequence, or even on separate polypeptides, could achieve linkage selectivity through avid interactions so long as flexibility between properly positioned domains is minimized.
Protein function and linkage-specific avidity
We have shown that the sequence linking UIM domains can be the major determinant of K63 or K48 linkage preference. Among human proteins with close tUIMs, the largest group have linkers similar to the Rap80 length (i.e., 7 or 8 residues), which in our studies support K63-specific binding (Figure 3D). It will be interesting to see if future studies bear out this predicted relationship to K63-polyUb.
Though we measured a K48 preference for ataxin-3, there is evidence for a preference to depolymerize mixed K63/K48-linked polymers (Winborn et al., 2008). Mixed-linkage chains have only recently been studied functionally, and their structures and basis for their recognition are unknown (Ikeda and Dikic, 2008; Kim et al., 2007). We note that, because mixed chains contain more than one type of isopeptide bond, specific recognition could not be achieved by a single isopeptide-directed interaction; instead, multi-valent interactions across more than one Ub unit may be required. Careful binding studies are needed to understand mixed-chain recognition by, for example, the ataxin-3 tUIMs.
Our solution-state measurements with a soluble fragment of Rap80 (residues 1 - 233) longer than the minimal tUIMs indicate only a 7-fold preference for K63-Ub4 over K48-Ub4 (Figure S3). Although published results indicate an apparently large selectivity for long K63-linked chains over K48-linked chains by Rap80 fragments, (Kim et al., 2007; Sobhian et al., 2007), it is important to recognize that these studies used immobilized, GST-fused versions of Rap80 to pull-down polyUb chains, and qualitative immunoblotting to assess the binding. Because avidity defines Rap80's linkage preference, artificial valencies introduced though GST dimerization and immobilization could have a profound impact on the results. One pull-down result also suggested that tight binding to K6-linked polyUb is possible (Sobhian et al., 2007). Because BRCA1, an indirect binding partner of Rap80, is an E3 ligase that can make K6-linked chains in vitro, the possibility of an interaction with K6 is provocative. However, several lines of evidence suggest that the in vivo binding partner for the Rap80 UIMs is a K63-linked polyUb chain (Huen et al., 2007; Kolas et al., 2007; Mailand et al., 2007; Plans et al., 2006; Wang and Elledge, 2007; Yan and Jetten, 2008), and in vivo roles for K6 chains have not been established.
Rap80 is phosphorylated on multiple serine residues by the ATM kinase in response to DNA damage. Serine 101 in the tUIM linker is one of these sites, although its phosphorylation is not required for efficient localization to DNA damage foci (Kim et al., 2007; Sobhian et al., 2007). Nonetheless, in light of our results that the nature of the linker can affect polyUb binding, we wondered if this modification could drive a structural transition in the linking sequence that could activate K63-polyUb binding. Although we failed to produce a phosphopeptide suitable for binding studies, the series of linker variants that we tested suggests that S101 phosphorylation is unlikely to promote the relatively large structural transition required to dramatically affect affinity and selectivity (see S101E in Table 1). However, other modifications may indeed be used to regulate tUIM affinity. A recent study shows that residues in and around the tUIMs of USP25 (Figure 3D) are SUMOylated, and that this modification regulates polyUb binding and enzymatic activity of the enzyme (Meulmeester et al., 2008).
To date, the functional significance of linkage-specificity has not been demonstrated for any polyUb receptor (Kim, and Rao, 2006). While the Rap80 UIMs achieve specificity that is comparable in magnitude to other linkage-selective proteins (Raasi et al., 2005), the somewhat modest preferences of these proteins (typically less than 10-fold) may call into question the functional role of linkage preference. In vivo studies will be required to determine the physiological significance of linkage-specificity for Rap80 and other polyUb-binding proteins.
Experimental Procedures
Plasmids and Proteins
The Supplemental Materials contain a list of the peptides used and their sequences (Table S1), along with details of their source DNAs, cloning, expression, purification, and fluorescent labeling.
Fluorescence anisotropy binding assays
Fluorescence anisotropy measurements were made using a Fluoromax 4 fluorometer in L format, thermostatted at 25 °C. Excitation and emission monochromators were set at 492 nm and 520 nm, respectively. Slit widths were 3 nm for diUb binding assays (1 μM fluorophore) or 6 nm for tetraUb binding assays (0.1 μM fluorophore). Anisotropies were calculated by the instrument software, and the overall intensity of the fluorescence signal was monitored at each point with the polarizers oriented at the magic-angle. All measurements were made in fluorescence buffer (25 mM Na phosphate pH 7.4, 150 mM NaCl, 5 mM β-mercaptoethanol, 1 mM EDTA, and either 0.005% surfactant P20 (BIACORE) or 0.05% Brij35). Concentrations of the fluorescent proteins were calculated using the fluorescein extinction coefficient at 494 nm, 68000 M-1cm-1. (poly)Ub concentrations were assessed by absorbance at 280 nm using the extinction coefficient 0.16 (mg/ml)-1(Pickart, and Raasi, 2005). Fluorescence anisotropy data were fit with a single-site binding model as described (Wilkinson, 2004). The equilibrium constants for wild-type Vps27 peptide binding to K63 and K48 Ub4 were determined by competition with the fluorescent Vps27 7aa linker peptide as described (Wilkinson, 2004).
ITC measurements
ITC titrations were performed on a Microcal VP-ITC at 30 °C in ITC buffer (25 mM phosphate pH 7.4, 150 mM NaCl, 10 mM β-mercaptoethanol, 1 mM EDTA). Each titration used 29 × 10 ul injections. For the titration of Rap80 tUIM peptide with K63-Ub2, the cell contained Rap80 peptide at 250 μM and the syringe contained 2.50 mM K63-Ub2. For the titration of K48-Ub2, the cell contained 450 μM K48-Ub2 and the syringe contained 4.50 mM Rap80 peptide. A version of the Rap80 tUIM peptide with a C-terminal 3-amino acid extension (DWS) was used for the ITC experiments to allow accurate peptide concentration determinations from absorbance at 280 nm.
CD measurements
CD measurements were performed on a Jasco J-810 spectropolarimeter using a 0.2 mm pathlength cuvette. Samples were first dialysed in 10 mM Na phosphate buffer, pH 7.4, with 100 mM NaCl. Peptide concentrations were independently determined before each CD reading from absorbance measurements (fluorescein) of samples in the CD cuvette.
Supplementary Material
Acknowledgments
We thank David Fushman for providing the K63-Ub2 structure coordinates. Work in this study was supported in part by NIH Roadmap grant RR020839.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Barriere H, Nemes C, Lechardeur D, Khan-Mohammad M, Fruh K, Lukacs GL. Molecular basis of oligoubiquitin-dependent internalization of membrane proteins in Mammalian cells. Traffic. 2006;3:282–297. doi: 10.1111/j.1600-0854.2006.00384.x. [DOI] [PubMed] [Google Scholar]
- Bennett EJ, Harper JW. DNA damage: ubiquitin marks the spot. Nat Struct Mol Biol. 2008;1:20–22. doi: 10.1038/nsmb0108-20. [DOI] [PubMed] [Google Scholar]
- Bobrovnik SA. The influence of rigid or flexible linkage between two ligands on the effective affinity and avidity for reversible interactions with bivalent receptors. J Mol Recognit. 2007;4:253–262. doi: 10.1002/jmr.836. [DOI] [PubMed] [Google Scholar]
- Chai Y, Berke SS, Cohen RE, Paulson HL. Poly-ubiquitin binding by the polyglutamine disease protein ataxin-3 links its normal function to protein surveillance pathways. J Biol Chem. 2004;5:3605–3611. doi: 10.1074/jbc.M310939200. [DOI] [PubMed] [Google Scholar]
- Di Francesco V, Garnier J, Munson PJ. Improving protein secondary structure prediction with aligned homologous sequences. Protein Sci. 1996;1:106–113. doi: 10.1002/pro.5560050113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncan LM, Piper S, Dodd RB, Saville MK, Sanderson CM, Luzio JP, Lehner PJ. Lysine-63-linked ubiquitination is required for endolysosomal degradation of class I molecules. EMBO J. 2006;8:1635–1645. doi: 10.1038/sj.emboj.7601056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher RD, Wang B, Alam SL, Higginson DS, Robinson H, Sundquist WI, Hill CP. Structure and ubiquitin binding of the ubiquitin-interacting motif. J Biol Chem. 2003;31:28976–28984. doi: 10.1074/jbc.M302596200. [DOI] [PubMed] [Google Scholar]
- Haglund K, Sigismund S, Polo S, Szymkiewicz I, Di Fiore PP, Dikic I. Multiple monoubiquitination of RTKs is sufficient for their endocytosis and degradation. Nat Cell Biol. 2003;5:461–466. doi: 10.1038/ncb983. [DOI] [PubMed] [Google Scholar]
- Hawryluk MJ, Keyel PA, Mishra SK, Watkins SC, Heuser JE, Traub LM. Epsin 1 is a polyubiquitin-selective clathrin-associated sorting protein. Traffic. 2006;3:262–281. doi: 10.1111/j.1600-0854.2006.00383.x. [DOI] [PubMed] [Google Scholar]
- Hicke L, Dunn R. Regulation of membrane protein transport by ubiquitin and ubiquitin-binding proteins. Annu Rev Cell Dev Biol. 2003:141–172. doi: 10.1146/annurev.cellbio.19.110701.154617. [DOI] [PubMed] [Google Scholar]
- Hicke L, Schubert HL, Hill CP. Ubiquitin-binding domains. Nat Rev Mol Cell Biol. 2005;8:610–621. doi: 10.1038/nrm1701. [DOI] [PubMed] [Google Scholar]
- Hirano S, Kawasaki M, Ura H, Kato R, Raiborg C, Stenmark H, Wakatsuki S. Double-sided ubiquitin binding of Hrs-UIM in endosomal protein sorting. Nat Struct Mol Biol. 2006;3:272–277. doi: 10.1038/nsmb1051. [DOI] [PubMed] [Google Scholar]
- Hofmann K, Falquet L. A ubiquitin-interacting motif conserved in components of the proteasomal and lysosomal protein degradation systems. Trends Biochem Sci. 2001;6:347–350. doi: 10.1016/s0968-0004(01)01835-7. [DOI] [PubMed] [Google Scholar]
- Huen MS, Grant R, Manke I, Minn K, Yu X, Yaffe MB, Chen J. RNF8 transduces the DNA-damage signal via histone ubiquitylation and checkpoint protein assembly. Cell. 2007;5:901–914. doi: 10.1016/j.cell.2007.09.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeda F, Dikic I. Atypical ubiquitin chains: new molecular signals. ‘Protein Modifications: Beyond the Usual Suspects’ review series. EMBO Rep. 2008;6:536–542. doi: 10.1038/embor.2008.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanayama A, Seth RB, Sun L, Ea CK, Hong M, Shaito A, Chiu YH, Deng L, Chen ZJ. TAB2 and TAB3 activate the NF-kappaB pathway through binding to polyubiquitin chains. Mol Cell. 2004;4:535–548. doi: 10.1016/j.molcel.2004.08.008. [DOI] [PubMed] [Google Scholar]
- Kerscher O, Felberbaum R, Hochstrasser M. Modification of proteins by ubiquitin and ubiquitin-like proteins. Annu Rev Cell Dev Biol. 2006:159–180. doi: 10.1146/annurev.cellbio.22.010605.093503. [DOI] [PubMed] [Google Scholar]
- Kim H, Chen J, Yu X. Ubiquitin-binding protein RAP80 mediates BRCA1-dependent DNA damage response. Science. 2007;5828:1202–1205. doi: 10.1126/science.1139621. [DOI] [PubMed] [Google Scholar]
- Kim HT, Kim KP, Lledias F, Kisselev AF, Scaglione KM, Skowyra D, Gygi SP, Goldberg AL. Certain pairs of ubiquitin-conjugating enzymes (E2s) and ubiquitin-protein ligases (E3s) synthesize nondegradable forked ubiquitin chains containing all possible isopeptide linkages. J Biol Chem. 2007;24:17375–17386. doi: 10.1074/jbc.M609659200. [DOI] [PubMed] [Google Scholar]
- Kim I, Rao H. What's Ub chain linkage got to do with it? Sci STKE. 2006;330:18. doi: 10.1126/stke.3302006pe18. [DOI] [PubMed] [Google Scholar]
- Kolas NK, Chapman JR, Nakada S, Ylanko J, Chahwan R, Sweeney FD, Panier S, Mendez M, Wildenhain J, Thomson TM, et al. Orchestration of the DNA-damage response by the RNF8 ubiquitin ligase. Science. 2007;5856:1637–1640. doi: 10.1126/science.1150034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo Y, Lin S, Rospigliosi CC, Conze DB, Wu C, Ashwell JD, Eliezer D, Wu H. Structural Basis for Recognition of Diubiquitins by NEMO. Molecular Cell. 2009 doi: 10.1016/j.molcel.2009.01.012. in press. doi:10.1016/j.molcel. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mailand N, Bekker-Jensen S, Faustrup H, Melander F, Bartek J, Lukas C, Lukas J. RNF8 ubiquitylates histones at DNA double-strand breaks and promotes assembly of repair proteins. Cell. 2007;5:887–900. doi: 10.1016/j.cell.2007.09.040. [DOI] [PubMed] [Google Scholar]
- Meiler J, Baker D. Coupled prediction of protein secondary and tertiary structure. Proc Natl Acad Sci U S A. 2003;21:12105–12110. doi: 10.1073/pnas.1831973100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meulmeester E, Kunze M, Hsiao HH, Urlaub H, Melchior F. Mechanism and consequences for paralog-specific sumoylation of ubiquitin-specific protease 25. Mol Cell. 2008;5:610–619. doi: 10.1016/j.molcel.2008.03.021. [DOI] [PubMed] [Google Scholar]
- Peng J, Schwartz D, Elias JE, Thoreen CC, Cheng D, Marsischky G, Roelofs J, Finley D, Gygi SP. A proteomics approach to understanding protein ubiquitination. Nat Biotechnol. 2003;8:921–926. doi: 10.1038/nbt849. [DOI] [PubMed] [Google Scholar]
- Pickart CM, Cohen RE. Proteasomes and their kin: proteases in the machine age. Nat Rev Mol Cell Biol. 2004;3:177–187. doi: 10.1038/nrm1336. [DOI] [PubMed] [Google Scholar]
- Pickart CM, Eddins MJ. Ubiquitin: structures, functions, mechanisms. Biochim Biophys Acta. 2004;1-3:55–72. doi: 10.1016/j.bbamcr.2004.09.019. [DOI] [PubMed] [Google Scholar]
- Pickart CM, Fushman D. Polyubiquitin chains: polymeric protein signals. Curr Opin Chem Biol. 2004;6:610–616. doi: 10.1016/j.cbpa.2004.09.009. [DOI] [PubMed] [Google Scholar]
- Pickart CM, Raasi S. Controlled synthesis of polyubiquitin chains. Methods Enzymol. 2005:21–36. doi: 10.1016/S0076-6879(05)99002-2. [DOI] [PubMed] [Google Scholar]
- Plans V, Scheper J, Soler M, Loukili N, Okano Y, Thomson TM. The RING finger protein RNF8 recruits UBC13 for lysine 63-based self polyubiquitylation. J Cell Biochem. 2006;3:572–582. doi: 10.1002/jcb.20587. [DOI] [PubMed] [Google Scholar]
- Raasi S, Pickart CM. Ubiquitin chain synthesis. Methods Mol Biol. 2005:47–55. doi: 10.1385/1-59259-895-1:047. [DOI] [PubMed] [Google Scholar]
- Raasi S, Pickart CM. Rad23 ubiquitin-associated domains (UBA) inhibit 26 S proteasome-catalyzed proteolysis by sequestering lysine 48-linked polyubiquitin chains. J Biol Chem. 2003;11:8951–8959. doi: 10.1074/jbc.m212841200. [DOI] [PubMed] [Google Scholar]
- Raasi S, Varadan R, Fushman D, Pickart CM. Diverse polyubiquitin interaction properties of ubiquitin-associated domains. Nat Struct Mol Biol. 2005;8:708–714. doi: 10.1038/nsmb962. [DOI] [PubMed] [Google Scholar]
- Reyes-Turcu FE, Shanks JR, Komander D, Wilkinson KD. Recognition of polyubiquitin isoforms by the multiple ubiquitin binding modules of isopeptidase T. J Biol Chem. 2008;28:19581–19592. doi: 10.1074/jbc.M800947200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rost B, Yachdav G, Liu J. The PredictProtein server. Nucleic Acids Res. 2004;(Web Server):W321–6. doi: 10.1093/nar/gkh377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato Y, Yoshikawa A, Yamagata A, Mimura H, Yamashita M, Ookata K, Nureki O, Iwai K, Komada M, Fukai S. Structural basis for specific cleavage of Lys 63-linked polyubiquitin chains. Nature. 2008;7211:358–362. doi: 10.1038/nature07254. [DOI] [PubMed] [Google Scholar]
- Schleif R, Wolberger C. Arm-domain interactions can provide high binding cooperativity. Protein Sci. 2004;10:2829–2831. doi: 10.1110/ps.04908404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sloper-Mould KE, Jemc JC, Pickart CM, Hicke L. Distinct functional surface regions on ubiquitin. J Biol Chem. 2001;32:30483–30489. doi: 10.1074/jbc.M103248200. [DOI] [PubMed] [Google Scholar]
- Sobhian B, Shao G, Lilli DR, Culhane AC, Moreau LA, Xia B, Livingston DM, Greenberg RA. RAP80 targets BRCA1 to specific ubiquitin structures at DNA damage sites. Science. 2007;5828:1198–1202. doi: 10.1126/science.1139516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun L, Chen ZJ. The novel functions of ubiquitination in signaling. Curr Opin Cell Biol. 2004;2:119–126. doi: 10.1016/j.ceb.2004.02.005. [DOI] [PubMed] [Google Scholar]
- Swanson KA, Kang RS, Stamenova SD, Hicke L, Radhakrishnan I. Solution structure of Vps27 UIM-ubiquitin complex important for endosomal sorting and receptor downregulation. EMBO J. 2003;18:4597–4606. doi: 10.1093/emboj/cdg471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trempe JF, Brown NR, Lowe ED, Gordon C, Campbell ID, Noble ME, Endicott JA. Mechanism of Lys48-linked polyubiquitin chain recognition by the Mud1 UBA domain. EMBO J. 2005;18:3178–3189. doi: 10.1038/sj.emboj.7600797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varadan R, Assfalg M, Haririnia A, Raasi S, Pickart C, Fushman D. Solution conformation of Lys63-linked di-ubiquitin chain provides clues to functional diversity of polyubiquitin signaling. J Biol Chem. 2004;8:7055–7063. doi: 10.1074/jbc.M309184200. [DOI] [PubMed] [Google Scholar]
- Varadan R, Assfalg M, Raasi S, Pickart C, Fushman D. Structural determinants for selective recognition of a Lys48-linked polyubiquitin chain by a UBA domain. Mol Cell. 2005;6:687–698. doi: 10.1016/j.molcel.2005.05.013. [DOI] [PubMed] [Google Scholar]
- Varadan R, Walker O, Pickart C, Fushman D. Structural properties of polyubiquitin chains in solution. J Mol Biol. 2002;4:637–647. doi: 10.1016/s0022-2836(02)01198-1. [DOI] [PubMed] [Google Scholar]
- Wagner S, Carpentier I, Rogov V, Kreike M, Ikeda F, Lohr F, Wu CJ, Ashwell JD, Dotsch V, Dikic I, Beyaert R. Ubiquitin binding mediates the NF-kappaB inhibitory potential of ABIN proteins. Oncogene. 2008;26:3739–3745. doi: 10.1038/sj.onc.1211042. [DOI] [PubMed] [Google Scholar]
- Wang B, Elledge SJ. Ubc13/Rnf8 ubiquitin ligases control foci formation of the Rap80/Abraxas/Brca1/Brcc36 complex in response to DNA damage. Proc Natl Acad Sci U S A. 2007;52:20759–20763. doi: 10.1073/pnas.0710061104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang B, Matsuoka S, Ballif BA, Zhang D, Smogorzewska A, Gygi SP, Elledge SJ. Abraxas and RAP80 form a BRCA1 protein complex required for the DNA damage response. Science. 2007;5828:1194–1198. doi: 10.1126/science.1139476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q, Young P, Walters KJ. Structure of S5a bound to monoubiquitin provides a model for polyubiquitin recognition. J Mol Biol. 2005;3:727–739. doi: 10.1016/j.jmb.2005.03.007. [DOI] [PubMed] [Google Scholar]
- Whitty A. Cooperativity and biological complexity. Nat Chem Biol. 2008;8:435–439. doi: 10.1038/nchembio0808-435. [DOI] [PubMed] [Google Scholar]
- Wilkinson KD. Quantitative analysis of protein-protein interactions. Methods Mol Biol. 2004:15–32. doi: 10.1385/1-59259-762-9:015. [DOI] [PubMed] [Google Scholar]
- Winborn BJ, Travis SM, Todi SV, Scaglione KM, Xu P, Williams AJ, Cohen RE, Peng J, Paulson HL. The deubiquitinating enzyme ataxin-3, a polyglutamine disease protein, edits lys63 linkages in mixed linkage ubiquitin chains. J Biol Chem. 2008;39:26436–26443. doi: 10.1074/jbc.M803692200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan J, Jetten AM. RAP80 and RNF8, key players in the recruitment of repair proteins to DNA damage sites. Cancer Lett. 2008 doi: 10.1016/j.canlet.2008.04.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan J, Kim YS, Yang XP, Li LP, Liao G, Xia F, Jetten AM. The ubiquitin-interacting motif containing protein RAP80 interacts with BRCA1 and functions in DNA damage repair response. Cancer Res. 2007;14:6647–6656. doi: 10.1158/0008-5472.CAN-07-0924. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


