Abstract
Epithelial cell behavior is coordinated by the composition of the surrounding extracellular matrix (ECM); thus ECM protein identification is critical for understanding normal biology and disease states. Proteomic analyses of ECM proteins have been hindered by the insoluble and digestion-resistant nature of ECM. Here we explore the utility of combining rapid ultrasonication- and surfactant-assisted digestion for the detailed proteomics analysis of ECM samples. When compared with traditional overnight digestion, this optimized method dramatically improved the sequence coverage for collagen I, revealed the presence of hundreds of previously unidentified proteins in Matrigel, and identified a protein profile for ECM isolated from rat mammary glands that was substantially different from that found in Matrigel. In a three-dimensional culture assay to investigate epithelial cell-ECM interactions, mammary epithelial cells were found to undergo extensive branching morphogenesis when plated with mammary gland-derived matrix in comparison with Matrigel. Cumulatively these data highlight the tissue-specific nature of ECM composition and function and underscore the need for optimized techniques, such as those described here, for the proteomics characterization of ECM samples.
Extracellular matrix (ECM)1 is a critical component of the tissue microenvironment. ECM plays a pivotal role in embryonic stem cell development and differentiation (1, 2) as well as many physiological (3) and pathological processes, including cancer progression (4, 5). Cell regulation by ECM has been studied with high frequency in recent years (7, 8). However, our ability to globally characterize ECM composition both in vitro and in vivo has been severely limited because of several unique attributes of ECM proteins such as high molecular weight glycans and the presence of covalent protein cross-links (6, 9, 10). Traditional proteomics approaches have proven to be ineffective for the identification of ECM proteins as demonstrated by the fact that collagens, despite being the most abundant protein in mammals, are significantly underrepresented in tissue-based proteomics data sets.
Ultrasonication has long been used for the digestion of bioorganic materials to allow for maximal and reproducible extraction and hence the accurate identification of small molecule and inorganic analytes (11). More recently, Capelo et al. (12) have used ultrasonic energy to catalyze tryptic digestion of proteins for subsequent mass spectrometry-based identification. Here we sought to determine whether this method could be optimized to prepare ECM samples for mass spectrometry-based analysis. For method development, we used rat tail collagen as a representative ECM protein for which current proteomics approaches have proven relatively unsuccessful. Type I collagen is defined as a right-handed triple helix heterotrimer comprising two identical α1 chains and one α2 chain that form a fibrillar network (6). The physical properties of the triple helical structure render the protein resistant to proteasch as trypsin (9). In this work, we focused our efforts on developing a digestion approach that improves our ability to perform proteomics analysis on a type I collagen preparation and then used this method to identify the protein composition of EHS murine chondrosarcoma matrix (10), herein referred to as Matrigel, and a matrix preparation from rat mammary tissue.
In this study, we developed a digestion approach suitable for a two-dimensional liquid chromatography-tandem mass spectrometry-based analysis of ECM proteins. Our digestion approach involves three cycles of ultrasonication for rapid initial trypsin digestion followed by overnight digestion using an acid-labile surfactant. This approach resulted in significant improvement in collagen peptide identification and the identification of numerous ECM proteins previously uncharacterized in Matrigel and in mammary tissue. The application of our ECM-optimized ultrasonic assisted trypsin digestion method is anticipated to significantly advance the identification of tissue- and disease state-specific ECM proteins.
EXPERIMENTAL PROCEDURES
Materials
Rat tail type I collagen and Matrigel were purchased from BD Bioscience. BSA, ammonium bicarbonate, DTT, tris(2-carboxyethyl)phosphine, and iodoacetamide were all purchased from Sigma-Aldrich. Formic acid (FA), TFA, and potassium chloride were obtained from Fluka (Buchs, Switzerland), and ACN was from Burdick and Jackson (Morristown, NJ). Trypsin (sequencing grade, l-1-tosylamido-2-phenylethyl chloromethyl ketone-treated) was from Promega (Madison, WI).
Reduction, Alkylation, and Standard Overnight Digestion
Reduction of disulfide bonds was achieved by addition of DTT to a final concentration of 5 mm (pH 7.4 using 25 mm ammonium bicarbonate) and incubation for 30 min at 70 °C. After cooling to room temperature, iodoacetamide was added (15 mm), and the samples were incubated in the dark at room temperature for 45 min. Standard overnight digestion was carried out by adding 1.2 μg of trypsin (3 μl)/100 μg of collagen or 100 μg of Matrigel (in 100 μl) and incubated overnight at 37 °C.
Surfactant-assisted Trypsin Digestion (SATD)
Digestion was performed as above with the addition of Rapigest SF Surfactant (Waters, Milford, MA) according to the manufacture's protocol with surfactant added to a final concentration of 0.1%. After overnight digestion at 37 °C, the solution was acidified with TFA to a final concentration of 0.5%. The sample was incubated at 37 °C for 30 min and then spun at 15,000 RCF for 10 min. The resulting supernatant was utilized for proteomics analyses.
Ultrasonication-assisted Tryptic Digestion (UATD)
Double deionized water was added to 100 μg of sample to a total volume of 100 μl in a 600-μl Eppendorf tube. The tube was secured using a ring stand and positioned such that the sonicator tip was 0.5 cm from the bottom of the tube with the bottom third of the tube submerged in a wet ice bath. Trypsin (1.2 μg) was added to the sample. A 450-watt Branson Sonifier (Branson, Danbury, CT) was used at power level 1 (∼10 watts), 50% duty cycle, and the sonicator was cycled on for 60 s, off for 30 s, and back on again for 60 s. Additional trypsin (1.2 μg) was added to the sample, and the same sonication cycle was applied for one or two additional cycles.
Mammary ECM Isolation
Adult female Sprague-Dawley nulliparous rats (Harlan-Teklad, Indianapolis, IN) were housed and euthanized at 18 weeks of age in compliance with the University of Colorado Health Sciences Center Animal Care and Use Committees and National Institutes of Health Policy on Humane Care and Use of Laboratory Animals. Lymph node-free inguinal mammary glands were harvested, snap frozen, and stored at −80 °C. Mammary ECM isolation was performed based on a previously described protocol (3) using pooled inguinal mammary gland tissue from six rats. Briefly frozen glands were pulverized and extracted in a high salt/N-ethylmaleimide solution (3.4 m NaCl, 50 mm Tris-HCl, pH 7.4, 4 mm EDTA, 2 mm N-ethylmaleimide) containing protease inhibitor mixture (Sigma). Homogenates were enriched for ECM by two cycles of centrifugation (RCFmax 110,000 × g, 30 min, 4 °C), resuspended in medium salt/urea solution and extracted overnight at 4 °C. Samples were pelleted at RCFmax 110,000 × g, and the ECM-enriched supernatants were extensively dialyzed (molecular mass cutoff, 12,000–14,000 kDa; Spectrum, Rancho Dominguez, CA) against low salt buffer at 4 °C.
Three-dimensional (3D) Cell Culture Assay
The protocol was modified from Krause et al. (13). Briefly a 1:1 volume ratio of 600 μg/ml virgin rat mammary ECM or 600 μg/ml Matrigel and 10.0 mg/ml Matrigel was prepared. Log phase MCF12A cells (40,000/well) were suspended in 500 μl of the ECM/Matrigel mixture and seeded into 10.5-mm well inserts (0.4-μm pores) of a 12-well plate (BD Biosciences). The gels were allowed to solidify for 60 min at 37 °C before adding 600 μl of culture medium onto each gel and 2 ml into each well. Culture medium consisted of Dulbecco's modified Eagle's medium/F-12 (Hyclone, Logan, UT), 5% horse serum (Hyclone), 10 μg/ml insulin (Intergen, Burlington, MA), 500 ng/ml hydrocortisone (Sigma), 20 ng/ml recombinant human epidermal growth factor (BD Biosciences), and 100 ng/ml cholera toxin (List Biological Laboratories, Campbell, CA). Images (100×) were taken on a Zeiss Axiovert 25 microscope at day 13 of culture.
Strong Cation Exchange
Strong cation exchange (SCX) fractionation was performed on the Matrigel digest using 10 × 2.1-mm and 250 × 2.1-mm polysulfylethyl A columns in series. A gradient of KCl (300 mm) in 20% ACN, 0.1% FA was carried out over 45 min with a flow rate of 100 μl/min using a multidimensional chromatography system (Ettan MDLC, GE Healthcare). Because of lower peptide abundance, SCX fractionation for in vivo mammary ECM was performed using two 10 × 2.1-mm polysulfylethyl A columns in series. For this sample the KCl gradient was carried out over 25 min with a flow rate of 80 μl/min. Each 100-μl fraction was concentrated to ∼40 μl to remove the ACN. A total of 8 μl of each fraction was subjected to nLC-MS/MS analysis.
Electrospray Ionization Mass Spectrometry
Samples were analyzed on an LTQ-FT Ultra hybrid mass spectrometer. Peptide desalting and separation were achieved using a dual capillary/nanopump HPLC system (Agilent 1200, Agilent, Palo Alto, CA). On this system 8 μl of sample was loaded onto a trapping column (ZORBAX 300SB-C18; dimensions, 5 × 0.3 mm; 5 μm) and washed with 5% ACN, 0.1% FA at a flow rate of 15 μl/min for 5 min. At this time the trapping column was put on line with the nanopump at a flow rate of 350 nl/min. An 85-min gradient from 8% ACN to 40% ACN was used to separate the peptides. The column was made from an in-house pulled 360-nm-outer diameter/100-nm-inner diameter fused silica capillary packed with Jupiter C18 resin (Phenomenex, Torrance, CA). The column was kept at a constant 40 °C using an in-house built column heater (14). Data acquisition was performed using the instrument-supplied Xcalibur (version 2.0.6) software. The LC runs were monitored in positive ion mode by sequentially recording survey MS scans (m/z 400–2000) in the ICR cell, whereas three MS2 scans were obtained in the ion trap via CID for the most intense ions. After two acquisitions of a given ion within 45 s, the ion was excluded for 150 s.
MS Data Analysis
The Raw Distiller (University of California San Francisco) program was used to create deisotoped centroided peak lists from the raw spectra into the Mascot “.mgf” format using the default settings. These peak lists were searched against the IPI.Mouse (v3.53.fasta) database (55,200 protein sequences) for Matrigel or IPI.Rat (v3.53.fasta) database (39,924 protein sequences) for rat mammary ECM samples using Protein Prospector, LC Batch-Tag Web (Version 5.0, University of California San Francisco), and MascotTM server (Version 2.2, Matrix Science; only in supplemental data). Precursor mass tolerance was set to ±6 ppm and ±0.6 Da for fragment ion tolerances. Trypsin specificity was used allowing for one missed cleavage. The modifications of Met oxidation, protein N-terminal acetylation, and peptide N-terminal pyroglutamic acid formation were allowed, and Cys carbamidomethylation was set as a fixed modification. For the type I collagen (COL1) experiments hydroxylation of proline and lysine was added to the variable modifications. In addition, for the COL1 experiments Mascot reporting was filtered to only allow for peptide matches with an ion score of >6 and a protein hit requiring a bold red match. In Protein Prospector searches were performed allowing for up to eight variable modifications per peptide to allow for the high level of Pro hydroxylation that is present in many collagen tryptic peptides. Experiments using COL1 with ultrasonication but without trypsin were searched with no enzyme specificity (i.e. Fig. 1, lane 5). For all searches, false discovery rates were determined by searching against decoy databases (reversed and randomized) using the default parameters used by the two search engines. The expect cutoff used for both programs was E < 0.005 and having two or more unique peptide hits. The remaining peptide identifications that mapped to unique protein sequences and met the above expect cutoff are give in supplemental spreadsheets. Secondary collagen database searches were performed using Protein Prospector for the two in vivo derived sample sets. The same search parameters were used with the following exceptions: a total of six variable modifications were allowed, hydroxylation/oxidation of proline was allowed, and more stringent identification criteria were used. The identifications had to achieve an expect value of less than 0.001 and individual peptide score of greater than 10, and the identification had to arise from spectra that had not been identified in the original search. A match was considered unique only once for each peptide sequence despite the number of and type of modifications (often a peptide was identified with two, three, or four hydroxylated/oxidized prolines, and these matches were used to report one unique peptide selected by the highest score). The collagen rat database consisted of unique collagen entries in IPI.Rat (40 proteins), and likewise the collagen mouse database consisted of all unique collagen entries in IPI.Mouse (53 proteins).
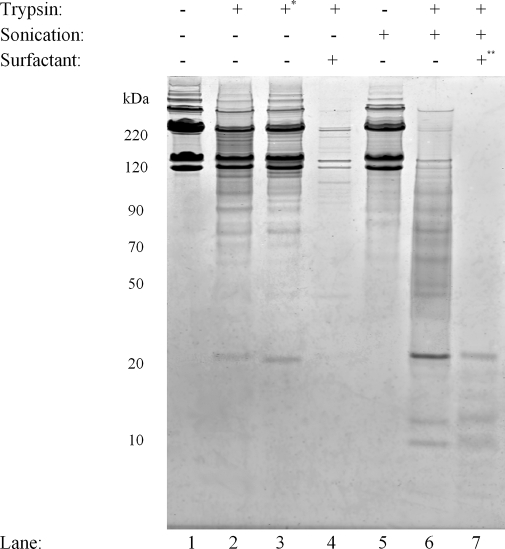
Fig. 1.
Collagen type I digestion efficiency. A 1D SDS-PAGE image of COL1 digestion conditions is shown. 21 μl of COL1 at 1 mg/ml was loaded into each lane. Lane 1, undigested COL1; lane 2, COL1 reduced and alkylated followed by overnight digestion with trypsin; lane 3, same as lane 2 but without reduction and alkylation (*); lane 4, reduction, alkylation, and tryptic digestion overnight with Rapigest; lane 5, COL1 reduced and treated with three cycles of ultrasonication; lane 6, COL1 reduced and treated with three cycles of ultrasonication with trypsin present; lane 7, COL1 treated with three cycles of ultrasonication-assisted trypsin digestion followed by (**) tryptic digestion overnight in the presence of Rapigest.
RESULTS
Rat Tail Type I Collagen Digestion
Collagen was chosen for method optimization to determine the parameters necessary for efficient tryptic digestion of fibrillar ECM proteins. Two distinct rat tail collagen preparations were prepared by various digestion methods (triplicate for preparation 1 and quadruplicate for preparation 2). The efficiency of the various digestion methods was evaluated based on gel band intensity and peptide identification by mass spectrometry. Overnight tryptic digestion at 37 °C resulted in limited proteolysis with an average of seven and five unique α1 and α2 chain peptides identified, respectively, by nLC-MS/MS analysis to give 5% coverage for each of the chains (Fig. 1 , lanes 3, and Table I , row 2). Table I, row 1 shows that with addition of reduction and alkylation, 12 and 27 unique peptides were identified with 7 and 12% coverage, respectively. There appears to be little difference in the banding pattern and intensity between the sample that was first reduced and alkylated (lane 2) and the sample that was not (lane 3). However, the peptide identification results indicate a measurable difference in digestion efficiency.
Table I. MS/MS identification of collagen I digests.
Results are averages from all runs. The top results are in bold. o/n, overnight.
| Conditionsa | Ultrasonication | COL1 subunit | Score | Unique peptides | Coverage |
|---|---|---|---|---|---|
| % | |||||
| o/n digestion (Fig. 1,lane 2) | α1 | 397 | 12 | 7 | |
| α2 | 817 | 27 | 12 | ||
| o/n digestion (Fig. 1, lane 3) | α1 | 242 | 7 | 5 | |
| α2 | 167 | 5 | 5 | ||
| Surfactant-assisted o/n trypsin digestion (Fig. 1, lane 4) | α1 | 1471 | 47 | 28 | |
| α2 | 1304 | 42 | 31 | ||
| No trypsin (Fig. 1, lane 5)b | v | α1 | 0 | 0 | 0 |
| α2 | 0 | 0 | 0 | ||
| UATD (Fig. 1, lane 6) | v | α1 | 1053 | 33 | 18 |
| α2 | 1380 | 42 | 22 | ||
| UATD then surfactant-assisted o/n digestion (Fig. 1, lane 7) | v | α1 | 2779 | 85 | 35 |
| α2 | 2026 | 60 | 39 |
a n = 7 unless otherwise noted.
b n = 4.
Next we applied a digestion approach that has proven very successful for other difficult to digest samples, such as membrane proteins. Collagen I was mixed with Rapigest, an acid-labile surfactant that does not interfere with tryptic activity (15). The sample was then reduced, alkylated, and allowed to digest overnight with trypsin. The 1D gel banding (Fig. 1, lane 4) and peptide identification results showed marked improvement over the standard digestion methods with an average of 47 unique collagen α1 (28% coverage) and 42 unique α2 peptides identified (31% coverage; Table I). We then applied an ultrasonication cycle that consisted of sonication for 60 s, a 30-s break, and a 60-s sonication with this cycle repeated two times. The banding pattern indicates that without protease ultrasonication exposure alone does not significantly reduce collagen 1 band size (Fig. 1, lane 5). Consistent with this observation, quadruplicate nLC-MS/MS analysis (three from the first collagen preparation and one from the second) resulted in no peptide identifications even with no enzyme specificity searches (Table I, row 4). Using the ultrasonication treatment with trypsin added before the cycle resulted in a dramatic reduction of the high molecular weight bands observed (lane 6) and significant peptide identification results with 33 and 42 unique peptides to give 18 and 22% coverage (Table I, row 6), respectively, despite the very short digestion time totaling 6 min of ultrasonication exposure. Finally we applied the ultrasonication method followed by surfactant-assisted overnight digestion, which further increased digestion (lane 7). This combined approach showed a substantial improvement in the number of peptides identified by nLC-MS/MS analysis with an average of 85 and 60 unique peptides and 35 and 39% coverage for α1 and α2 chains, respectively. Cumulatively the combined approach of ultrasonication followed with surfactant-assisted digestion had an additive effect on tryptic digestion efficiency of collagen I with a significant increase in the protein identification scores and protein sequence coverage compared with overnight digestion.
Matrigel Digestion and Proteomics Analysis
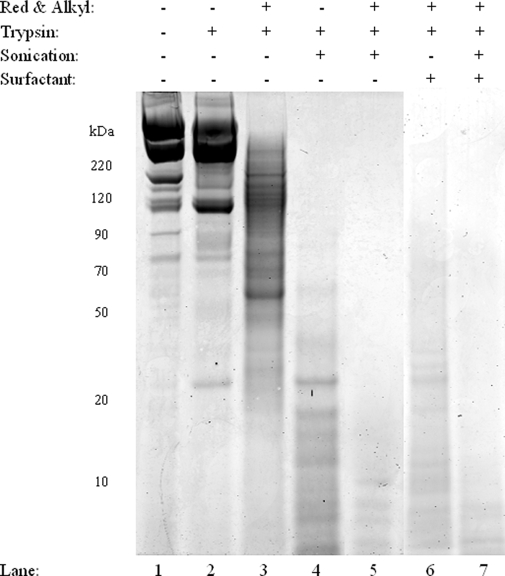
To evaluate the ability of the optimized digestion protocol to increase the number of proteins identified in basement membrane derived from the EHS mouse chondrosarcoma, Matrigel was digested via classic overnight digestion (both with and without surfactant) with ultrasonication-assisted trypsin digestion and with the approaches combined. The results are shown in Fig. 2. Similar to the 1D gel results obtained for the collagen I sample, when Matrigel was digested overnight using the standard tryptic digestion protocol, very little digestion occurred (Fig. 2, lane 2). Matrigel subjected to reduction and alkylation followed by overnight tryptic digestion showed an improvement in digestion (Fig. 2, lane 3). However, even under these conditions, high molecular weight bands were observed, demonstrating incomplete digestion. Improved digestion was achieved with ultrasonication-assisted digestion (Fig. 2, lane 4) with reduction before and then again after sonication-assisted digestion followed by alkylation (Fig. 2, lane 5). We confirmed that the decrease in banding observed on the gels resulted in increased tryptic peptides by 2-h nLC-MS/MS runs. The ultrasonication-assisted digestion approach resulted in the identification of ∼50% more proteins and a 5-fold increase in protein scores for the top five protein identifications (results not shown). Protein staining from the sample exposed to surfactant with trypsin indicated extensive digestion (Fig. 2, lane 6). However, the sequential treatment of ultrasonication- and surfactant-assisted digestion approaches resulted in the least amount of observable protein staining, suggesting that this approach would increase protein identification (Fig. 2, lane 7). With these promising results obtained using our combined digestion approach, we pursued an in-depth proteomics analysis of Matrigel.
Fig. 2.
Digestion of Matrigel. A 1D SDS-PAGE image shows the effect of disulfide status on the efficiency of Matrigel digestion using the SATD, UATD, and combined methods (lanes 3–7) versus overnight trypsin digestion (lane 2). All lanes were loaded with 21 μg of Matrigel. Without reduction of the disulfides, higher molecular weight bands remained after the digestion procedure (lanes 2 and 4). Samples digested using UATD showed few higher molecular weight bands (lane 5). The combination of UATD and SATD (lane 7) showed the least amount of protein staining. Red & Alkyl, reduction and alkylation.
Having digested Matrigel with the combined digestion method we identified proteins using a two-dimensional chromatographic fractionation approach followed by tandem MS analysis on an LTQ-ICR mass spectrometer. A total of 64 fractions were collected during a SCX run, and 36 were analyzed by nLC-MS/MS based on highest UV signal intensity. By searching the data against the International Protein Index database (IPI.Mouse) we identified numerous basement membrane and traditional ECM proteins. The vast majority of protein as determined by the number of peptides identified, protein sequence coverage, and best expect value were the basement membrane components laminin α, β, and γ; nidogen (entactin); and perlecan (HSPG2). We identified additional classes of proteins including metabolic enzymes, transcription factors, soluble extracellular proteins, and proteases. In all, 310 protein entries were identified with high confidence using the criteria of an expect value of less than 0.005 and a minimum of two unique peptides. The resulting false discovery rate (FDR) was calculated to be 0.26%. The top 25 ranked protein identifications followed by additional ECM proteins identified are shown in Table II. A complete list of identifications is provided in the supplemental data including additional peptide identifications that map to unique protein sequences (total FDR of 0.71%).
Table II. Partial list of proteins identified from Matrigel.
The number of unique peptides identifying a protein with shared tryptic peptides is in parentheses (homologous identifications). Classic ECM proteins in the top 25 identifications are in bold. SPARC, secreted protein acidic and rich in cysteine.
| Protein name | Rank | Accession no. | Unique peptides | Protein score | Best expect value |
|---|---|---|---|---|---|
| Top 25 proteins | |||||
| Laminin subunit α1 | 1 | IPI00113726.3 | 150 | 4845 | 48.7 |
| Laminin subunit β1 | 2 | IPI00338785.3 | 113 | 3699 | 62.8 |
| Perlecan (HSPG2) | 3 | IPI00515360.8 | 104 | 3326 | 37.6 |
| Laminin subunit γ1 | 4 | IPI00400016.1 | 80 | 2789 | 55.3 |
| Nidogen-1 | 5 | IPI00111793.1 | 61 | 1982 | 58.5 |
| Laminin subunit α5 | 6 | IPI00116913.3 | 23 | 823 | 10.6 |
| Vimentin | 7 | IPI00227299.6 | 29 | 815 | 53.6 |
| Fibronectin 1a | 8 | IPI00352163.3 | 28 | 842 | 21 |
| Serum albumin | 9 | IPI00131695.3 | 24 | 740 | 39.8 |
| Peroxidasin homolog | 10 | IPI00461384.5 | 21 | 664 | 21.3 |
| 78-kDa glucose-regulated protein | 11 | IPI00319992.1 | 22 | 729 | 33.9 |
| Lamin-B1a | 12 | IPI00230394.5 | 15 | 443 | 30.1 |
| ATP synthase subunit β, mitochondrial | 13 | IPI00468481.2 | 13 | 459 | 38.8 |
| Fibrinogen β chain | 14 | IPI00279079.1 | 15 | 438 | 32.4 |
| Low density lipoprotein receptor-related protein 2 | 15 | IPI00349520.5 | 15 | 473 | 4.7 |
| 60-kDa heat shock protein | 16 | IPI00308885.6 | 13 | 423 | 22 |
| Myosin-9 | 17 | IPI00123181.4 | 13 | 444 | 9.6 |
| Heat shock 70-kDa protein 9a | 18 | IPI00880839.1 | 12 | 416 | 21.1 |
| Prolyl 4-hydroxylase, β polypeptidea | 19 | IPI00122815.3 | 11 | 321 | 27.1 |
| SPARC | 20 | IPI00126343.1 | 11 | 349 | 44.4 |
| Transitional endoplasmic reticulum ATPase | 21 | IPI00622235.5 | 12 | 356 | 26.2 |
| Serotransferrin | 22 | IPI00139788.2 | 12 | 356 | 20.8 |
| UDP-glucose ceramide glucosyltransferase-like 1 | 23 | IPI00762897.2 | 12 | 356 | 11 |
| Isoform 2 of spectrin α chain, brain | 24 | IPI00753793.2 | 10 | 331 | 6.6 |
| Complement component factor H | 25 | IPI00130010.4 | 9 | 290 | 12.8 |
| Other ECM proteins | |||||
| Laminin β2 | 2-1 | IPI00109612.2 | 9(7) | 260 | 6.5 |
| Perlecan isoform (HSPG2) | 3-1 | IPI00113824.1 | 88 (3) | 2813 | 35.5 |
| Nidogen-2 | 5-1 | IPI00719919.2 | 10 (9) | 317 | 12.6 |
| Agrn; isoform 2 of agrin | 36 | IPI00378698.6 | 8 | 247 | 7.5 |
| Collagen α2(IV) | 43 | IPI00338452.3 | 8/10a | 234/332a | 7 |
| Isoform 2 of collagen α1(XVIII) | 118 | IPI00131476.2 | 4/1a | 112/11a | 4.1 |
| Collagen α1(IV) | 120 | IPI00109588.4 | 3/8a | 92/166a | 3.1 |
| Isoform C of fibulin-1 | 170 | IPI00230432.2 | 3 | 74 | 5.4 |
| Laminin subunit α4 | 215 | IPI00223446.5 | 4 | 98 | 3.4 |
| Collagen α1(XV) | 394 | IPI00409035.5 | 2 | 43 | 2.9 |
| von Willebrand factor A domain-containing protein 1 | 409 | IPI00331609.7 | 1 | 32 | 3.9 |
| Sparcl1; SPARC-like protein 1 | 615 | IPI00308484.3 | 1 | 26 | 2 |
| Collagen α1(I)a | IPI00329872.1 | 11a | 239a | 21.6 | |
| Collagen α2(I)a | IPI00222188.4 | 3a | 78a | 7.7 | |
| Collagen α1(VII)a | IPI00134652.3 | 2a | 28a | 5.1 |
a Identified in collagen-only database search with hydroxylated/oxidized proline as a variable modification.
In addition to searching the data against the complete protein sequence database, we performed a Protein Prospector search against a database that only contained mouse collagen entries. This allowed for the identification of hydroxyproline residues, which are a common collagen modification. This variable modification was not allowed in the original search as it drastically increases the false discovery rate. Resulting peptide identifications that originated from spectra that had not been assigned in the original search are listed in the supplemental data summarized in Table II. We observed additional peptides that increased the sequence coverage for already identified collagens XVIII α1 (from four to five unique peptides) and IV α1 (from three to 11 peptides) and α2 (from eight to 18 peptides) entries (Table II). In addition, three new protein identifications for collagens I α1 (11 peptides) and α2 (three peptides) and VII α1 (two peptides) were made.
Tissue-based Mammary ECM Functional Characterization, Digestion, and Proteomics Analysis
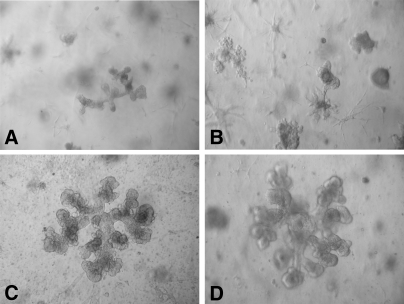
It is anticipated that ECM composition will be tissue-specific because of the unique functions of each organ. To this end, ECM was isolated from the mammary glands of adult rats, and its function was compared with Matrigel in a 3D cell culture model developed by Sonnenschein and co-workers (13). Human immortalized mammary MCF 12A epithelial cells underwent extensive branching morphogenesis when the cells were cultured with mammary matrix, whereas little branching morphogenesis occurred when these cells were cultured with Matrigel (Fig. 3). This observation indicates that epithelial cell-ECM interactions are, as predicted, optimized when the cells and ECM are derived from the same tissue. To make progress toward understanding these tissue-specific cell-ECM interactions, we applied our digestion approach to this in vivo derived mammary matrix. For nLC-MS/MS analysis, the mammary matrix sample was prepared identically to Matrigel with the exception that less starting material was available, and thus fewer cation exchange fractions (20) were collected and analyzed.
Fig. 3.
3D cell culture model of mammary gland branching morphogenesis. Human mammary epithelial cells undergo extensive branching morphogenesis in the presence of matrix derived from virgin rat mammary glands. A and B, MCF12A cells cultured in 3D with Matrigel for 14 days (100\x). C and D, MCF12A cells cultured in 3D with rat mammary matrix for 13 days (100\x).
Using our optimized digestion method, we identified peptides from 248 proteins in rat mammary matrix (FDR, 0.07%). The top identification was to the ECM protein perlecan with over 600 peptides identified; 96 of these were unique to give 37.5% sequence coverage (). The second highest scoring unique ECM protein was collagen α3 (VI). Decorin, fibrillin 1, and fibronectin were also identified in the top five ranked proteins. Several collagens were identified in the mammary matrix but not in Matrigel, including the fibular collagens type I and II. When the data were searched against a rat collagen database several additional unique peptides were identified. Most notably, collagen I α1 and α2 chain identifications gained an additional 52 and 40 unique peptides, respectively (Table III). Based on these additional identifications, collagens I α1 and α2, III α1, and VI α1 move up to the top 25 identifications list. As with Matrigel, there were a large number of cellular contaminant proteins in addition to known extracellular matrix proteins.
Table III. Partial list of proteins identified from mammary ECM preparation.
The number of unique peptides identifying a protein with shared tryptic peptides is in parenthesis (homologous identifications). Classic ECM proteins in the top 25 identifications are in bold. SPARC, secreted protein acidic and rich in cysteine; EGF, epidermal growth factor.
| Protein name | Rank | Accession no. | Unique peptides | Protein score | Best expect value |
|---|---|---|---|---|---|
| Top 25 proteins | |||||
| Perlecan (HSPG2) | 1 | IPI00388323.5 | 96 | 3125 | 37.5 |
| Collagen α3(VI) | 2 | IPI00565677.2 | 74 | 2421 | 33.9 |
| Fibrillin-1 | 3 | IPI00204006.1 | 75 | 2471 | 32.5 |
| Collagen α1(XIV) | 4 | IPI00360766.4 | 53/14a | 1641/262a | 36.9 |
| Fibronectin | 5 | IPI00200757.1 | 51 | 1684 | 32.8 |
| Filamin-A | 6 | IPI00409539.3 | 50 | 1646 | 31.7 |
| Laminin β2 | 7 | IPI00212868.1 | 45 | 1429 | 35.8 |
| Plectin 6 | 8 | IPI00209000.2 | 39 | 1177 | 11.8 |
| AHNAK nucleoprotein | 9 | IPI00769072.2 | 45 | 1471 | 12.3 |
| Spectrin α chain | 10 | IPI00209258.4 | 39 | 1325 | 23.1 |
| Periostin, osteoblast-specific factor | 11 | IPI00193018.2 | 38 | 1300 | 52.1 |
| Tenascin-X | 12 | IPI00394165.2 | 45 | 1292 | 15.4 |
| Vimentin | 13 | IPI00230941.5 | 45 | 1417 | 69.1 |
| Serum albumin | 14 | IPI00191737.6 | 38 | 1274 | 68.8 |
| Laminin γ1 | 15 | IPI00363849.2 | 32 | 1117 | 33.2 |
| Lamin-A | 16 | IPI00201060.4 | 36 | 1229 | 45.3 |
| Myosin-11 | 17 | IPI00764167.1 | 31 | 1059 | 20.6 |
| Nidogen-1 | 18 | IPI00231136.5 | 29 | 930 | 28.1 |
| Laminin α4 | 19 | IPI00361106.4 | 26 | 884 | 19.6 |
| Spectrin β | 20 | IPI00555287.2 | 22 | 755 | 14.2 |
| Type II cytoskeletal 5 | 21 | IPI00382153.4 | 25 | 746 | 44.1 |
| Decorin | 22 | IPI00199861.1 | 23 | 699 | 55.9 |
| Tubulin β-5 chain | 23 | IPI00197579.2 | 17 | 576 | 45.9 |
| Complement inhibitory factor H | 24 | IPI00208659.1 | 20 | 638 | 23.5 |
| Lamin-B1 | 25 | IPI00231418.5 | 20 | 665 | 42.8 |
| Other ECM proteins | |||||
| Perlecan (HSPG2) 284-kDa protein | 1-1 | IPI00778528.1 | 56 (7) | 1854 | 32.3 |
| Nidogen-2 | 18-1 | IPI00372786.6 | 23 (22) | 742 | 24.9 |
| Osteoglycin | 26 | IPI00362931.1 | 19 | 558 | 41.9 |
| Collagen α1(VI) | 27 | IPI00371853.3 | 19/5a | 626/122a | 25 |
| Laminin β1 | 28 | IPI00365542.3 | 23 | 682 | 20.4 |
| Laminin α2 chain | 30 | IPI00361301.3 | 17 | 519 | 8.1 |
| Prolargin | 31 | IPI00190287.1 | 16 | 492 | 34.2 |
| Collagen α2(VI) | 36 | IPI00372839.3 | 14/3a | 490/63a | 20.6 |
| Lumican | 46 | IPI00206403.1 | 14 | 423 | 37 |
| Biglycan | 52 | IPI00191090.1 | 11 | 349 | 34.7 |
| Collagen α1(I) | 57 | IPI00188909.2 | 8/52a | 259/1891a | 4.7 |
| Collagen α1(XVIII) | 61 | IPI00214859.5 | 6/2a | 186/30a | 5.9 |
| Collagen α2(I) | 64 | IPI00188921.1 | 6/40a | 179/1473a | 5.5 |
| Collagen α1(XV) | 68 | IPI00364868.3 | 6/2a | 193/43a | 5.4 |
| Collagen α2(IV) | 82 | IPI00365380.3 | 6/2a | 182/44a | 6.3 |
| Fibulin-5 | 86 | IPI00326179.3 | 5 | 139 | 14.5 |
| Laminin α5 | 91 | IPI00190577.4 | 5 | 162 | 2.5 |
| Collagen α1(VII) | 102 | IPI00767686.2 | 4/1a | 127/13a | 2.3 |
| Bone marrow proteoglycan | 114 | IPI00206023.1 | 4 | 111 | 22.9 |
| Agrin | 121 | IPI00562438.1 | 4 | 105 | 4 |
| Papilin, proteoglycan-like sulfated glycoprotein | 144 | IPI00367768.3 | 4 | 103 | 6.1 |
| Collagen α1(IV) | 154 | IPI00362887.3 | 2/1a | 80/19a | 2.2 |
| Sulfated glycoprotein 1 | 240 | IPI00195160.1 | 2 | 48 | 3.6 |
| Versican core protein | 261 | IPI00190776.1 | 2 | 49 | 1.2 |
| von Willebrand factor | 291 | IPI00210224.5 | 1 | 35 | 0.6 |
| SPARC | 295 | IPI00189424.2 | 1 | 34 | 4.7 |
| Laminin α1 | 319 | IPI00363534.4 | 4 | 68 | 1.9 |
| von Willebrand factor A domain-containing protein 1 | 337 | IPI00471665.1 | 1 | 32 | 4.8 |
| Similar to collagen α3(VI) | 380 | IPI00370868.3 | 1 | 27 | 0.8 |
| EGF-containing fibulin-like extracellular matrix protein 1 | 388 | IPI00365221.4 | 1 | 26 | 1.8 |
| Collagen α1(III)a | IPI00366944.2 | 35a | 1304a | 46 | |
| Collagen α1(II)a | IPI00394380.4 | 5a | 143a | 14.6 | |
| Collagen α1(V)a | IPI00365380.3 | 2a | 77a | 8.2 | |
| Collagen α2(V)a | IPI00366945.3 | 2a | 68a | 5 |
a Identified in collagen-only database search with hydroxylated/oxidized proline as a variable modification.
DISCUSSION
In the present study, we investigated a novel method for the in-solution digestion of ECM proteins, which are resistant to standard trypsin-based digestion methods. Part of the rationale for optimizing digestion protocols for ECM is based on the recent appreciation of ECM proteins as critical regulators of epithelial, mesenchymal, and stem cell function. ECM influences cell behavior through multiple mechanisms including cell shape, polarity, gene expression, migration, invasion, proliferation, tissue-specific differentiation, and death pathways (16–21, 30). In fact, in cancer cells, ECM has been identified as being as important as epithelial cell mutations with respect to expression of the transformed phenotype (4, 22, 23). Despite these important roles, the protein composition of ECM in tissues is largely unknown. This is because, to date, proteomics methods for the detailed biochemical characterization of ECM have been severely limited because of challenges with sample digestion. For example, using the current methods to digest collagen-enriched samples, only up to four peptides were identified for collagen I obtained from fibroblast-derived matrices (24). Consistent with these published data, after tryptic digestion of rat tail collagen I, we identified an average of only seven and five peptides for the two subunits. By reducing and alkylating the sample before digestion, significantly more peptides were identified bringing the average unique peptides identified to 12 and 27 for the two subunits. This difference is likely due to disulfide bonds that conserve tertiary structure, resulting in protection from proteolysis. In contrast, using our combined digestion approach, we identified an average of 85 unique peptides for the α1 chain and 60 unique peptides for the α2 chain. Differences in identification results for the two distinct collagen preparations were minimal, indicating the versatility of this approach.
Ultrasonication has been reported to involve gas bubble nucleation, growth, and then rapid collapse, a process known as cavitation. The energy released during the last step has been reported to produce elevated temperatures, pressures, and free radical formation (.H and .OH) (25). Using our digestion method we observed minimal global sample heating from the ultrasonication cycles with an observed range of 38–48 °C. In preliminary experiments exploring the effects of ultrasonication without trypsin, we observed a loss of protein banding on 1D gels when high power settings were used (data not shown). These results are similar to the findings of Davies (26) and Davies et al. (27) in which protein banding was diminished when protein samples were treated with radiation-generated .OH in the presence of O2. It appears that with sufficient energy input extensive amide backbone cleavage occurs possibly by a similar radical-based mechanism. Guided by these preliminary experiments we developed our current method that uses low power input and cycling of the ultrasonication exposure to minimize temperature-related effects.
To determine the effect of our ultrasonication conditions on trypsin activity we tested the ability of ultrasonication-treated versus control trypsin to digest reduced and alkylated BSA. We observed an ∼60% decrease in peptides identified with ultrasonication-treated trypsin, indicating a partial loss of enzymatic activity (data not shown). However, despite this partial loss in trypsin activity, the combined ultrasonication- and surfactant-assisted tryptic digestion method allowed us to characterize the protein composition of two tissue ECM preparations with unprecedented resolution. It is likely that some or all of the loss in trypsin activity with ultrasonication is a result of autolysis similar to that observed in standard overnight digestions. In addition, protein banding is retained using the ultrasonication method without trypsin present, demonstrating the requirement for trypsin proteolysis (Fig. 1, lane 5).
Identification of the protein composition of Matrigel is of interest because since its discovery in the 1970s, Matrigel has served as a basement membrane substitute for cell culture work where it has been shown to support differentiation (28), growth (29), and organotypic morphogenesis (30) of various cell lines. In addition, Matrigel is the ECM preparation of choice in tumor xenograft studies where it facilitates tumor cell survival, growth, and metastasis in immunocompromised mice (31, 32). Although Matrigel has been widely accepted as the standard ECM matrix for the majority of 3D cell culture work and xenograft studies around the world, proteomics characterization of this matrix has not been reported. In this study, we found that Matrigel includes, but is not limited to, traditional ECM proteins such as laminin 1, nidogen, perlecan, collagen IV, and fibronectin (10). Additional extracellular and cellular proteins of several classes were also identified. Several of these were matrix-modifying enzymes such as lysyl oxidases, prolyl 3-hydroxylases, and lysyl hydroxylases, which are essential for the stability and intermolecular cross-linking of collagen.
Several cellular proteins identified in Matrigel are known members of focal adhesions or cell motility complexes such as integrin β1, talin, α-actinin, filamin, vinculin, kinectin, Arp2/3, and focal adhesion kinase. Their identification suggests that some cell-ECM protein interactions may be conserved throughout the ECM isolation procedure. In addition, several cytoskeletal and intermediate filament proteins associated with focal adhesions were identified. The question of whether these non-ECM proteins are being isolated through stable protein-protein interactions with cell surface ECM receptors or are nonspecific artifacts of the purification procedure is of interest and remains to be determined. Surprisingly the stress response proteins hypoxia up-regulated protein 1, peroxidasin, heat shock proteins 70 (1A, 1B, 5, 8, and 9), HSP 60, and HSP 90 α and β were relatively abundant in Matrigel. HSP can mediate “domestic” danger signals, which represent an important initiator of immune response to cellular stress (33). Moreover many HSPs were suggested as endogenous ligands for Toll-like receptors (34). In summary, the optimized digestion method allowed us to detect proteins that may have functional consequences for the numerous in vitro and in vivo experiments in which Matrigel is used.
Tissue specificity of ECM matrix was evident from the 3D culture assay where glandular morphogenesis of human mammary epithelial cells was promoted to a much greater extent by ECM proteins extracted from rat mammary glands than by Matrigel. Several differences in protein composition were observed between rat mammary matrix and Matrigel that may contribute to these differences. Although the current data do not provide direct protein quantification, the relative protein rank, sequence coverage, and number of unique peptides can be used in a semiquantitative manner. First relative to Matrigel, mammary matrix was found to be high in fibronectin (FN) and low in laminin 1 (α1/β1/γ1). Fibronectin expression at both the mRNA and protein levels has been shown to be dependent on the developmental state of the mammary gland (3). Specifically FN is up-regulated during periods of glandular expansion that occur during puberty and pregnancy and is down-regulated with glandular regression that occurs after lactation (3). Consistent with these observations, mammary FN expression has been shown to be up-regulated by ovarian hormones (35). This dynamic expression pattern of FN in the mammary gland is in contrast to reported laminin 1 mRNA and protein levels, which are maintained at a relatively constant level across pregnancy, lactation, and weaning-induced gland regression (3). Further in comparison with laminin, the addition of fibronectin in a 3D culture model increased proliferation of MCF10A cells, a human immortalized but non-transformed line, resulting in larger acinar size. However, branching morphogenesis, as observed with our tissue-specific mammary matrix, was not observed in that study (36). More recently, it has been reported that the addition of collagen I is sufficient to induce branching morphogenesis in 3D cultured mammary epithelial cells (13). Of potential relevance, collagen II, III, and V peptides were identified only in rat mammary matrix, and significantly more peptides were identified for type I collagen when compared with Matrigel. Thus fibrillar collagens in concert with fibronectin may facilitate the branching morphogenesis in the 3D culture assay that was induced by the rat mammary matrix preparation.
In the rat mammary matrix, collagen VI and fibrillin, which form microfibrillar assemblies that are important for tissue elasticity, cell attachment, and motility, were identified as the second and third top protein hits with 74 and 75 unique peptide identifications, respectively. The fourth protein hit, collagen XIV (undulin) (37), which belongs to the FACIT (fibril-associated collagen with interrupted triple helices) group of collagens, is able to bind collagen type VI and decorin and is thought to serve as a cross-linker between other collagens (38, 39). Despite having low or no binding to fibrillar collagens I-V, the non-helical region of type XIV collagen mediates the contraction of collagen I gels by fibroblasts, thus potentially influencing the mechanical properties of collagen fibrils (40). Consequently collagen type XIV/undulin may be involved in stabilizing collagen fibrils and influencing their three-dimensional arrangement in the mammary gland. Additionally tenascin-X was identified and has been reported to contribute to matrix stability and is possibly involved in collagen fibril formation (41).
Matrigel and rat mammary matrix also differed in composition of secreted proteoglycans, ECM components with high negative charge that bind to a plethora of proteins including growth factors, cytokines, and signaling molecules (42). Perlecan was abundant in both matrices, whereas decorin was prevalent in rat mammary matrix. Several proteoglycans such as periostin, prolagrin, versican, and the small leucine-rich proteoglycan osteoglycin were only found in the rat mammary matrix. Perlecan was first isolated from the murine EHS tumor and is known to be an integral component of basement membranes with binding sites for laminin-1, collagen IV, and fibronectin. Further perlecan is essential for the functioning of signaling molecules implicated in branching morphogenesis such as Wnt and hedgehog as well as serving as a buffer for inflammatory cytokines (42, 43). Decorin, which also binds growth factors and cytokines, mediates collagen fiber assembly and thus tissue tensional properties. As 3D culture modeling is increasingly refined to reflect the in vivo microenvironment, it will be important to address the effects of these various proteoglycans on cell behavior in the context of the major ECM adhesion proteins. Finally new method development to minimize the level of cellular contamination of ECM preparations will further facilitate functional assays and proteomics analysis.
It has become increasingly clear that the ECM plays a significant role in determining cell fate and behavior. To make progress toward quantitative proteomics analysis of ECM, improvements in sample preparation are necessary. Our ultrasonication followed by overnight surfactant-assisted tryptic digestion approach significantly enhanced the analysis of ECM proteins and is flexible enough to work with samples from a wide range of in vitro and in vivo systems. This valuable method will allow more comprehensive characterization of complex ECM mixtures with the intent of understanding the role of ECM in tissue- and disease-specific processes.
Acknowledgments
We thank Pavel Strop for critical reading and comments on the manuscript and Lindsay Hosford and Joanna Marcinkiewicz for early method development contributions. We also thank Al Burlingame, Robert Chalkley, Aenoch Lynn, Shenheng Guan, and Peter Baker for support and access to Protein Prospector and Raw Distiller.
Footnotes
* This work was supported, in whole or in part, by National Institutes of Health Grants P30 CA046934-17 through the University of Colorado Comprehensive Cancer Center Core Support and S10RR023015 from the National Center for Research Resources. This work was also supported by Department of Defense Grant BC051532 and the Butcher Foundation (to P. S.) and the Cancer League of Colorado (to K. H.).
The on-line version of this article (available at http://www.mcponline.org) contains supplemental material.
- ECM
- extracellular matrix
- COL1
- type I collagen
- EHS
- Engelbreth-Holm-Swarm
- FN
- fibronectin
- nLC
- nanoflow LC
- LTQ
- linear ion trap
- FA
- formic acid
- SATD
- surfactant-assisted trypsin digestion
- RCF
- relative centrifugal force
- UATD
- ultrasonication-assisted tryptic digestion
- 3D
- three-dimensional
- SCX
- strong cation exchange
- IPI
- International Protein Index
- 1D
- one-dimensional
- FDR
- false discovery rate
References
- 1.Ingber D. E. ( 2008) Can cancer be reversed by engineering the tumor microenvironment? Semin. Biol. 18, 356– 364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Engler A. J., Sen S., Sweeney H. L., Discher D. E. ( 2006) Matrix elasticity directs stem cell lineage specification. Cell 126, 677– 689 [DOI] [PubMed] [Google Scholar]
- 3.Schedin P., Mitrenga T., McDaniel S., Kaeck M. ( 2004) Mammary ECM composition and function are altered by reproductive state. Mol. Carcinog. 41, 207– 220 [DOI] [PubMed] [Google Scholar]
- 4.Schedin P., Elias A. ( 2004) Multistep tumorigenesis and the microenvironment. Breast Cancer Res. 6, 93– 101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Radisky E. S., Radisky D. C. ( 2007) Stromal induction of breast cancer: inflammation and invasion. Rev. Endocr. Metab. Disord. 8, 279– 287 [DOI] [PubMed] [Google Scholar]
- 6.Gelse K., Pöschl E., Aigner T. ( 2003) Collagens—structure, function, and biosynthesis. Adv. Drug Deliv. Rev. 55, 1531– 1546 [DOI] [PubMed] [Google Scholar]
- 7.Schedin P., O'Brien J., Rudolph M., Stein T., Borges V. ( 2007) Microenvironment of the involuting mammary gland mediates mammary cancer progression. J. Mammary Gland Biol. Neoplasia 12, 71– 82 [DOI] [PubMed] [Google Scholar]
- 8.Fata J. E., Werb Z., Bissell M. J. ( 2004) Regulation of mammary gland branching morphogenesis by the extracellular matrix and its remodeling enzymes. Breast Cancer Res. 6, 1– 11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Peyrol S., Raccurt M., Gerard F., Gleyzal C., Grimaud J. A., Sommer P. ( 1997) Lysyl oxidase gene expression in the stromal reaction to in situ and invasive ductal breast carcinoma. Am. J. Pathol. 150, 497– 507 [PMC free article] [PubMed] [Google Scholar]
- 10.Yuan L. S., Siegel M., Choi K., Khosla C., Miller C. R., Jackson E. N., Piwnica-Worms D., Rich K. M. ( 2007) Transglutaminase 2 inhibitor, KCC009, disrupts fibronectin assembly in the extracellular matrix and sensitizes orthotopic glioblastomas to chemotherapy. Oncogene 26, 2563– 2573 [DOI] [PubMed] [Google Scholar]
- 11.Bermejo P., Capelo J. L., Mota A., Madrid Y., Camara C. ( 2004) Enzymatic digestion and ultrasonication: a powerful combination in analytical chemistry. Trends Anal. Chem. 23, 654– 663 [Google Scholar]
- 12.Capelo J. L., Maduro C., Vilhena C. ( 2005) Discussion of parameters associated with the ultrasonic solid-liquid extraction for elemental analysis (total content) by electrothermal atomic absorption spectrometry. An overview. Ultrason. Sonochem. 12, 225– 232 [DOI] [PubMed] [Google Scholar]
- 13.Krause S., Maffini M. V., Soto A. M., Sonnenschein C. ( 2008) A novel 3D in vitro culture model to study stromal-epithelial interactions in the mammary gland. Tissue Eng. Part C Methods 14, 261– 271 [DOI] [PubMed] [Google Scholar]
- 14.Speers A. E., Blackler A. R., Wu C. C. ( 2007) Shotgun analysis of integral membrane proteins facilitated by elevated temperature. Anal. Chem. 79, 4613– 4620 [DOI] [PubMed] [Google Scholar]
- 15.Umar A., Dalebout J. C., Timmermans A. M., Foekens J. A., Luider T. M. ( 2005) Method optimisation for peptide profiling of microdissected breast carcinoma tissue by matrix-assisted laser desorption/ionisation-time of flight and matrix-assisted laser desorption/ionisation-time of flight/time of flight-mass spectrometry. Proteomics 5, 2680– 2688 [DOI] [PubMed] [Google Scholar]
- 16.Egeblad M., Littlepage L. E., Werb Z. ( 2005) The fibroblastic coconspirator in cancer progression. Cold Spring Harb. Symp. Quant. Biol. 70, 383– 388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Werb Z. ( 1997) ECM and cell surface proteolysis: regulating cellular ecology. Cell 91, 439– 442 [DOI] [PubMed] [Google Scholar]
- 18.Schenk S., Quaranta V. ( 2003) Tales from the crypt[ic] sites of the extracellular matrix. Trends Cell Biol. 13, 366– 375 [DOI] [PubMed] [Google Scholar]
- 19.Barcellos-Hoff M. H. ( 2001) It takes a tissue to make a tumor: epigenetics, cancer and the microenvironment. J. Mammary Gland Biol. Neoplasia 6, 213– 221 [DOI] [PubMed] [Google Scholar]
- 20.Tlsty T. D. ( 2001) Stromal cells can contribute oncogenic signals. Semin. Cancer Biol. 11, 97– 104 [DOI] [PubMed] [Google Scholar]
- 21.Mueller M. M., Fusenig N. E. ( 2004) Friends or foes—bipolar effects of the tumour stroma in cancer. Nat. Rev. Cancer. 4, 839– 849 [DOI] [PubMed] [Google Scholar]
- 22.Hendrix M. J., Seftor E. A., Seftor R. E., Kasemeier-Kulesa J., Kulesa P. M., Postovit L. M. ( 2007) Reprogramming metastatic tumour cells with embryonic microenvironments. Nat. Rev. Cancer 7, 246– 255 [DOI] [PubMed] [Google Scholar]
- 23.Paszek M. J., Zahir N., Johnson K. R., Lakins J. N., Rozenberg G. I., Gefen A., Reinhart-King C. A., Margulies S. S., Dembo M., Boettiger D., Hammer D. A., Weaver V. M. ( 2005) Tensional homeostasis and the malignant phenotype. Cancer Cell 8, 241– 254 [DOI] [PubMed] [Google Scholar]
- 24.Pflieger D., Chabane S., Gaillard O., Bernard B. A., Ducoroy P., Rossier J., Vinh J. ( 2006) Comparative proteomic analysis of extracellular matrix proteins secreted by two types of skin fibroblasts. Proteomics 6, 5868– 5879 [DOI] [PubMed] [Google Scholar]
- 25.Riesz P., Berdahl D., Christman C. L. ( 1985) Free radical generation by ultrasound in aqueous and nonaqueous solutions. Environ. Health Perspect. 64, 233– 252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Davies K. J. ( 1987) Protein damage and degradation by oxygen radicals. I. General aspects. J. Biol. Chem. 262, 9895– 9901 [PubMed] [Google Scholar]
- 27.Davies K. J., Delsignore M. E., Lin S. W. ( 1987) Protein damage and degradation by oxygen radicals. II. Modification of amino acids. J. Biol. Chem. 262, 9902– 9907 [PubMed] [Google Scholar]
- 28.Kim H. S., Lee B. L., Bae S. I., Kim Y. I., Park J. G., Kleinman H. K., Kim W. H. ( 1998) Differentiation of a colon cancer cell line on a reconstituted basement membrane in vitro. Int. J. Exp. Pathol. 79, 443– 451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xu C., Inokuma M. S., Denham J., Golds K., Kundu P., Gold J. D., Carpenter M. K. ( 2001) Feeder-free growth of undifferentiated human embryonic stem cells. Nat. Biotechnol. 19, 971– 974 [DOI] [PubMed] [Google Scholar]
- 30.Schedin P., Strange R., Mitrenga T., Wolfe P., Kaeck M. ( 2000) Fibronectin fragments induce MMP activity in mouse mammary epithelial cells: evidence for a role in mammary tissue remodeling. J. Cell Sci. 113, 795– 806 [DOI] [PubMed] [Google Scholar]
- 31.Fridman R., Kibbey M. C., Royce L. S., Zain M., Sweeney M., Jicha D. L., Yannelli J. R., Martin G. R., Kleinman H. K. ( 1991) Enhanced tumor growth of both primary and established human and murine tumor cells in athymic mice after coinjection with Matrigel. J. Natl. Cancer Inst. 83, 769– 774 [DOI] [PubMed] [Google Scholar]
- 32.Mehta R. R., Graves J. M., Hart G. D., Shilkaitis A., Das Gupta T. K. ( 1993) Growth and metastasis of human breast carcinomas with Matrigel in athymic mice. Breast Cancer Res. Treat. 25, 65– 71 [DOI] [PubMed] [Google Scholar]
- 33.Zhang X., Mosser D. M. ( 2008) Macrophage activation by endogenous danger signals. J. Pathol. 214, 161– 178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tsan M. F., Gao B. ( 2004) Heat shock protein and innate immunity. Cell. Mol. Immunol. 1, 274– 279 [PubMed] [Google Scholar]
- 35.Woodward T. L., Mienaltowski A. S., Modi R. R., Bennett J. M., Haslam S. Z. ( 2001) Fibronectin and the alpha(5)beta(1) integrin are under developmental and ovarian steroid regulation in the normal mouse mammary gland. Endocrinology 142, 3214– 3222 [DOI] [PubMed] [Google Scholar]
- 36.Williams C. M., Engler A. J., Slone R. D., Galante L. L., Schwarzbauer J. E. ( 2008) Fibronectin expression modulates mammary epithelial cell proliferation during acinar differentiation. Cancer Res. 68, 3185– 3192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ehnis T., Dieterich W., Bauer M., Schuppan D. ( 1998) Localization of a cell adhesion site on collagen XIV (undulin). Exp. Cell Res. 239, 477– 480 [DOI] [PubMed] [Google Scholar]
- 38.Olsen B. R. ( 1997) Collagen IX. Int. J. Biochem. Cell Biol. 29, 555– 558 [DOI] [PubMed] [Google Scholar]
- 39.Brown J. C., Mann K., Wiedemann H., Timpl R. ( 1993) Structure and binding properties of collagen type XIV isolated from human placenta. J. Cell Biol. 120, 557– 567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nishiyama T., McDonough A. M., Bruns R. R., Burgeson R. E. ( 1994) Type XII and XIV collagens mediate interactions between banded collagen fibers in vitro and may modulate extracellular matrix deformability. J. Biol. Chem. 269, 28193– 28199 [PubMed] [Google Scholar]
- 41.Egging D., van den Berkmortel F., Taylor G., Bristow J., Schalkwijk J. ( 2007) Interactions of human tenascin-X domains with dermal extracellular matrix molecules. Arch. Dermatol. Res. 298, 389– 396 [DOI] [PubMed] [Google Scholar]
- 42.Bishop J. R., Schuksz M., Esko J. D. ( 2007) Heparan sulphate proteoglycans fine-tune mammalian physiology. Nature 446, 1030– 1037 [DOI] [PubMed] [Google Scholar]
- 43.Perrimon N., Bernfield M. ( 2001) Cellular functions of proteoglycans—an overview. Semin. Cell Dev. Biol. 12, 65– 67 [DOI] [PubMed] [Google Scholar]