Summary
Deciphering the epigenetic “code” remains a central issue in transcriptional regulation. Here, we report the identification of a MPN+/JAMM domain-containing histone H2A deubiquitinase (2A-DUB, or KIAA1915/MYSM1) specific for monoubiquitinated H2A (uH2A), that has permitted delineation of a strategy for specific regulatory pathways of gene activation. 2A-DUB regulates transcription by coordinating histone acetylation and deubiquitination, and destabilizing the association of linker histone H1 with nucleosomes. 2A-DUB interacts with p/CAF in a co-regulatory protein complex, with its deubiquitinase activity modulated by the status of acetylation of nucleosomal histones. Consistent with this mechanistic role, 2A-DUB participates in transcriptional regulation events in androgen receptor-dependent gene activation, and the levels of uH2A are dramatically decreased in prostate tumors, serving as a cancer-related mark. We suggest that H2A ubiquitination represents a widely-used mechanism for many regulatory transcriptional programs, and predict that various H2A ubiquitin ligases/deubiquitinases will be identified for specific cohorts of regulated transcription units.
Introduction
Histone modifications are believed to play central roles in chromatin remodeling and transcriptional regulation (reviewed in Jenuwein and Allis, 2001, Li et al., 2007). In contrast to other relatively well-characterized histone modifications, e.g. acetylation and methylation, the role of histone ubiquitination in gene expression and chromatin remodeling remains the least understood despite a long history of its discovery (reviewed in Osley, 2004, Zhang, 2003). This is due, in part, to the fact that histones are largely monoubiquitinated, which is not linked to degradation, and a lack of information regarding the histone ubiquitination/deubiquitination enzymes. While earlier studies suggest that ubiquitination of histones can participate in both gene activation and repression (Baarends et al. 1999, Huang et al. 1986, Levinger and Varshavsky, 1982, Nickel et al., 1989), recent studies, by identifying specific E2/E3 ubiquitin ligases and deubiquitinases of H2A/H2B, revealed that H2A ubiquitination is crucial for X-inactivation and Polycomb group (PcG) complex-dependent gene silencing (de Napoles et al., 2004, Fang et al., 2004, Wang et al., 2004), and both ubiquitination and deubiquitination of H2B are at least partially required for transcriptional activation/elongation (Daniel et al., 2004, Henry et al., 2003, Kao et al., 2004, Pavri et al., 2006, Wood et al., 2003). Intriguingly, H2B ubiquitination was found to be a prerequisite for methylation of H3K4 and H3K79, which might be important for the regulatory functions in gene expression (Dover et al., 2002, Ng, et al., 2002, Shahbazian et al., 2005, Sun and Allis, 2002).
Histone H2A was the first protein shown to be ubiquitinated (Goldknopf et al., 1975) and the site was mapped to the highly conserved C-terminal Lys 119 residue (Goldknopf and Busch, 1977, Nickel and Davie, 1989). In mammalian cells, a remarkable portion (about 10%) of total H2A is monoubiquitinated. Similarly, a ubiquitination site has also been mapped to lysine residues in the C-terminus of H2B (Lys 120 in human and Lys 123 in yeast) (Thorne et al., 1987). Recently, ubiquitination events of H1, H3 and H4 have also been reported in mammalian cells (Chen et al., 1998, Pham and Sauer, 2000, Wang et al., 2006). Several E2/E3 enzymes involved in histone ubiquitination have been recently identified, e.g. yeast Rad6/Bre1 and human RNF20/40 for H2B, polycomb group component Ring1B for H2A, Cul4-DDB-Roc1 complex for H3/H4, and TAFII250 for linker histone H1 (Pham and Sauer, 2000, Robzyk et al., 2000, Wang et al., 2004, Wang et al., 2006, Zhu et al., 2005). On the other hand, while deubiquitinases (Ubp8, Ubp10 and Usp7) have been reported for H2B in transcriptional regulation (Emre et al., 2005, Gardner et al., 2005, Henry et al., 2003, van der Knaap et al., 2005), corresponding enzymes for H2A have not been identified, and the roles/mechanisms of histone H2A ubiquitination in gene expression remain largely unknown. Here we report that a MPN+/JAMM domain-containing histone H2A deubiquitinase (2A-DUB, or KIAA1915/MYSM1) is required for full activation of several transcriptional events including androgen receptor (AR)-regulated target genes in prostate cancer cells. By forming a regulatory protein complex with the histone acetyltransferase (HAT) p300/CBP-associated factor (p/CAF), 2A-DUB regulates transcriptional initiation and likely also elongation by stepwise coordination of histone acetylation, H2A deubiquitination and linker histone dissociation.
Results
Identification of a histone H2A deubiquitinase
In a screen of candidate proteins for potential transcriptional coregulators, we identified KIAA1915/MYSM1 (hereafter referred to as 2A-DUB) as a positive regulator of androgen receptor (AR) activity on an AR responsive element (ARE)-containing reporter gene (Fig. S1). Interestingly, 2A-DUB contains a MPN+/JAMM domain that has been shown to possess an intrinsic metalloprotease-like activity that is able to hydrolyze the isopeptide bonds of nedd8 and/or ubiquitin chains (Cope et al., 2002, Maytal-Kivity et al., 2002, Verma et al., 2002, Yao and Cohen, 2002); hence we reasoned that it might serve specific enzymatic functions in transcriptional regulation. In addition to the putative enzymatic function, 2A-DUB contains SANT and SWIRM domains (Fig. 1A). The SANT domain is a well-documented motif capable of binding to histones and DNA, existing in many transcriptional regulators e.g. N-CoR, SMRT, CoREST and Myb (Boyer et al., 2004). Similarly, SWIRM domains appear frequently in chromatin-associated proteins i.e. the recently identified histone demethylase LSD1, and favor interaction with linker DNA and/or N-terminal tails of histone H3 (Da et al., 2006, Qian et al., 2005, Tochio et al., 2006). The C-terminus of 2A-DUB also contains a generic LXXLL motif, existing in many coactivators for interaction with agonist-bound nuclear receptors (Glass and Rosenfeld, 2000). Taken together, the combination of functional domains of 2A-DUB implicates it as a putative deubiquitinase involved in transcriptional activation by nuclear receptors. 2A-DUB proved to be predominately localized in the nucleus where it interacts with core histones (Fig. S2), implying the possibility of functioning as a histone modifier.
Figure 1. Identification of a deubiquitinase of histone H2A.
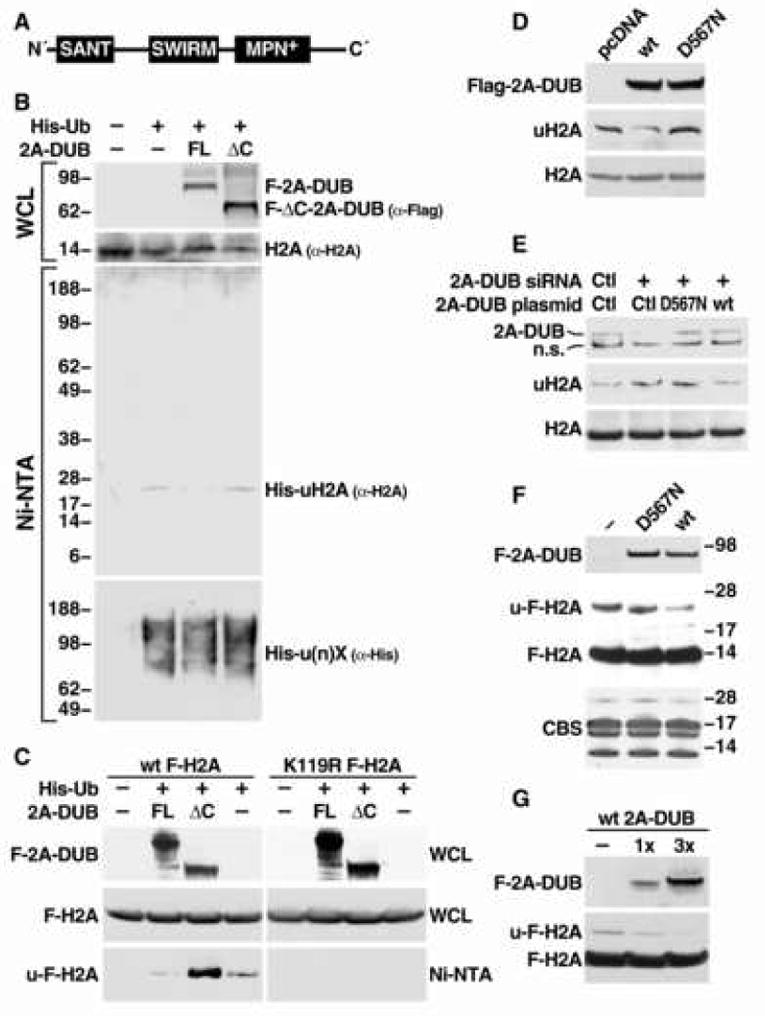
(A) Schematic representation of the domain organization of 2A-DUB/KIAA1915/MYSM1 protein. (B) In vivo ubiquitination assay for H2A after overexpression of human full-length (FL) and C-terminal enzymatic JAMM/MPN+ domain-deleted mutant (ΔC) 2A-DUB (aa 17-531) in HEK293T cells. Molecular weight (kDa) is shown on the left. WCL represents whole cell lysates. (C) In vivo ubiquitination assay in HEK293 cells stably expressing wild-type (wt) and K119R Flag-tagged H2A. Anti-Flag antibody was used to detect signals from Flag-H2A. (D) Detection of endogenous uH2A and H2A levels by Western blot analysis after overexpression of Flag-tagged full-length wt and D567N mutant 2A-DUB in HEK293T cells. (E) Detection of endogenous uH2A and H2A levels by Western blotting upon siRNA-mediated knock-down of endogenous 2A-DUB or co-expression of wt or D567N 2A-DUB with siRNA against 2A-DUB. (F) In vitro deubiquitination assay for H2A. Affinity-purified wt and D567N 2A-DUB (transfected into HEK293T cells) were used as enzymatic source (about 2 μg), and acid-extracted histones from wt Flag-H2A-expressing HEK293 cells was used as the substrate (about 1.5 μg). Western analysis of 2A-DUB and Flag-H2A showed the signals of enzymes and substrates, and the Commassie Blue staining (CBS) displayed the loading of core histone substrates. (G) Using 1 μg (1×) or 3 μg (3×) of 2A-DUB in the same assays as (F).
We then investigated its putative deubiquitinase functions in HEK293T cells. Full-length or MPN+/JAMM enzymatic domain-deficient ΔC mutant protein was co-expressed with a His6-ubiquitin expression plasmid, and His6-ubiquitinated proteins in cell lysates were pulled down by Ni+-NTA beads under denaturing conditions. Ubiquitinated histones were detected by Western blot analysis using specific antibodies against core histones H2A, H2B, H3 and H4. Intriguingly, only the monoubiquitinated form of H2A (uH2A) was found to be reduced by overexpression of full-length 2A-DUB, while monoubiquitinated H2B (uH2B) was not altered, and H3/H4 ubiquitination could not be detected under these assay conditions (Fig. 1B, Fig S3 and data not shown). Given the fact that uH2A and uH2B are the main monoubiquitinated histones and both are involved in regulation of gene expression, determining the specificity of 2A-DUB to uH2A was of particular interest. We further used Flag-tagged wild-type and mutant (K119R) H2A harboring a mutation in the major monoubiquitinated residue in these assays, showing that the K119 in wild-type H2A is the site deubiquitinated by 2A-DUB whereas ubiquitination of the mutant H2A could not be observed (Fig 1C). To investigate whether the endogenous uH2A status is regulated by 2A-DUB, soluble chromatin extracts were subject to Western blot analysis using a specific monoclonal antibody directed against uH2A (Vassilev et al., 1995, Fig. S4A) in HEK293T cells. Wild-type 2A-DUB substantially decreased uH2A levels, whereas the D567N mutant, in which the key aspartic acid residue required for the metal-binding-dependent deubiquitinase activity was substituted to a nonfunctional asparagine residue (Ambroggio et al., 2004, Cope et al., 2002, Verma et al., 2002, Yao and Cohen, 2002), lost the ability to deubiquitinate uH2A (Fig. 1D). In cell lines stably overexpressing 2A-DUB, the levels of uH2A inversely correlated with the expression levels of transfected 2A-DUB (Fig. S4A). To further test its role by a loss-of-function approach, we checked the uH2A levels after knocking down 2A-DUB expression in HEK293T cells using specific siRNAs. Upon efficient knockdown (>80%) of 2A-DUB, global uH2A levels increased substantially (Fig. 1E). Simultaneous expression of wild-type 2A-DUB, but not the D567N mutant, neutralized the induced uH2A levels (Fig. 1E). Finally we evaluated whether 2A-DUB is a direct histone H2A deubiquitinase by in vitro assays using core histones purified from HEK293 cells stably expressing Flag-tagged wild-type H2A (Fig. S5A). Compared to their mutant counterparts (K119R H2A), wild-type histones displayed significant ∼24 kDa bands (the expected size of uH2A) recognized by anti-Flag, anti-H2A and anti-ubiquitin antibodies, indicating these bands indeed represented ubiquitinated Flag-H2A (Fig. S5A and Fig. 3A). Upon incubating with wild-type and D567N 2A-DUB proteins affinity-purified from stable ectopic-expressing HEK293 cells, the Flag-tagged uH2A band (∼24 kDa) was remarkably reduced by the wild-type, but not by the mutant enzyme (Fig. 1F). By increasing the amount of wild-type 2A-DUB in the in vitro assay, ubiquitin group was released from u-Flag-H2A in a dose-dependent manner (Fig. 1G). Together, these observations reveal that 2A-DUB, dependent upon the MPN+/JAMM metalloprotease activity, is a histone H2A deubiquitinase, suggesting that 2A-DUB could exert its transcriptional regulatory function by this specific histone-modifying enzymatic activity.
Figure 3. Coordination of histone acetylation, H2A deubiquitination and H1 phosphorylation/dissociation in nucleosomes.
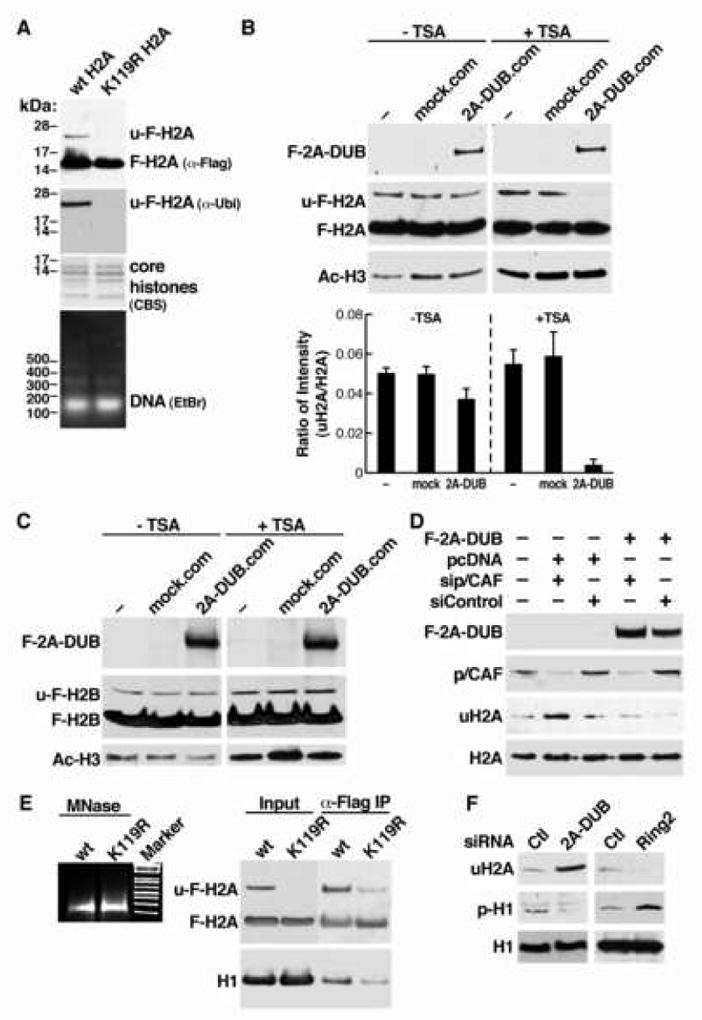
(A) Characterization of Flag-H2A-containing mono/oligonucleosomes. Purified nucleosomes from HEK293T cells stably expressing wt and K119R Flag-tagged H2A were evaluated for specificity of the 24-kD ubiquitinated Flag-H2A band (u-F-H2A) by Western blot using anti-Flag and anti-ubiquitin antibodies. Composition of core histones was checked by Coomassie blue staining (CBS), and DNA size of MNase-digested nucleosomes was detected by ethidium bromide (EtBr) staining after gel separation. (B) In vitro deubiquitination assay using affinity-purified 2A-DUB protein complex (2A-DUB.com) and mock purified materials from control cells (mock.com) as the enzymatic source, and using Flag-tagged H2A-containing mono/oligonucleosomes purified from TSA-treated or untreated cells as the substrate. Levels of 2A-DUB, u-F-H2A, F-H2A and acetylation of histone H3 (Ac-H3) were detected by Western blot analysis. Lower panel: quantification of three independent assays. (C) Similar assays as (B), using Flag-tagged H2B-containing mono/oligonucleosomes as the substrate. (D) Flag-2A-DUB and siRNA against p/CAF were transfected into HEK293T cells as indicated. Levels of corresponding proteins were detected by Western blots. (E) Detection of composition of associated linker histone H1 in immuno-purified wt or K119R Flag-H2A-containing mononucleosomes. Mononucleosomal DNA was stained by EtBr (left). (F) Evaluation of phosphorylation of H1 after manipulating uH2A levels by knocking down 2A-DUB or H2A E3 ligase Ring2 in HEK293T cells.
A functional 2A-DUB/HAT protein complex
To gain biochemical insights to functions of 2A-DUB, we generated HEK293 cells stably expressing human 2A-DUB tagged with three Flag epitopes at the amino-terminus (Fig. 2A). Nuclear extracts of a pool of cells stably expressing Flag-2A-DUB or control HEK293 cells were subject to M2 anti-Flag resin chromatography. Then the 3xFlag peptide eluates were separated by SDS-PAGE, and the specific protein bands in Flag-2A-DUB-containing lysates were analyzed by mass spectrometry after digestion by a standard protocol (see Experimental Procedures). In total we identified about 10 polypeptides specifically co-purified with Flag-tagged 2A-DUB in HEK293 cells (Fig. 2B and C). Intriguingly, a histone acetyltransferase (HAT) p300/CBP-associated factor (p/CAF) was co-purified with 2A-DUB, suggesting a potential functional link between histone deubiquitination and acetylation, in agreement with the general ideas that histone acetylation is mostly a mark of gene activation while histone H2A ubiquitination is involved in repression/silencing as proposed by recent reports (de Napoles et al., 2004, Wang et al., 2004). Of note, a portion of 2A-DUB was found to be ubiquitinated (mono- or di-) at a specific lysine residue close to the “core” of the metalloprotease domain, implying that the deubiquitinase activity might be regulated by ubiquitination. Co-immunoprecipitation assays further confirmed a specific interaction of 2A-DUB with p/CAF, as well as interactions with RNA-binding motif protein 10 (RBM10), thyroid hormone receptor (T3R)-interacting protein 5 (Trip5) that are co-purified components in the 2A-DUB protein complex (Fig. 2D, left panel). Reciprocal immunoprecipitation using anti-p/CAF also pulled down endogenous 2A-DUB and Trip5, suggesting they indeed form a coregulator complex (Fig. 2D, right panel). Using free core histones and nucleosomal histones as the substrate, the 2A-DUB complex in the in vitro HAT assay also possesses enzymatic activity that acetylates core histones in a similar pattern to that observed for recombinant p/CAF, preferentially targeting histones H3 and H4 (Fig. 2E and data not shown), revealing that p/CAF in the 2A-DUB complex is enzymatically active, and the function of the complex may require both histone H2A deubiquitination and histone acetylation activities.
Figure 2. Affinity purification of the 2A-DUB protein complex.
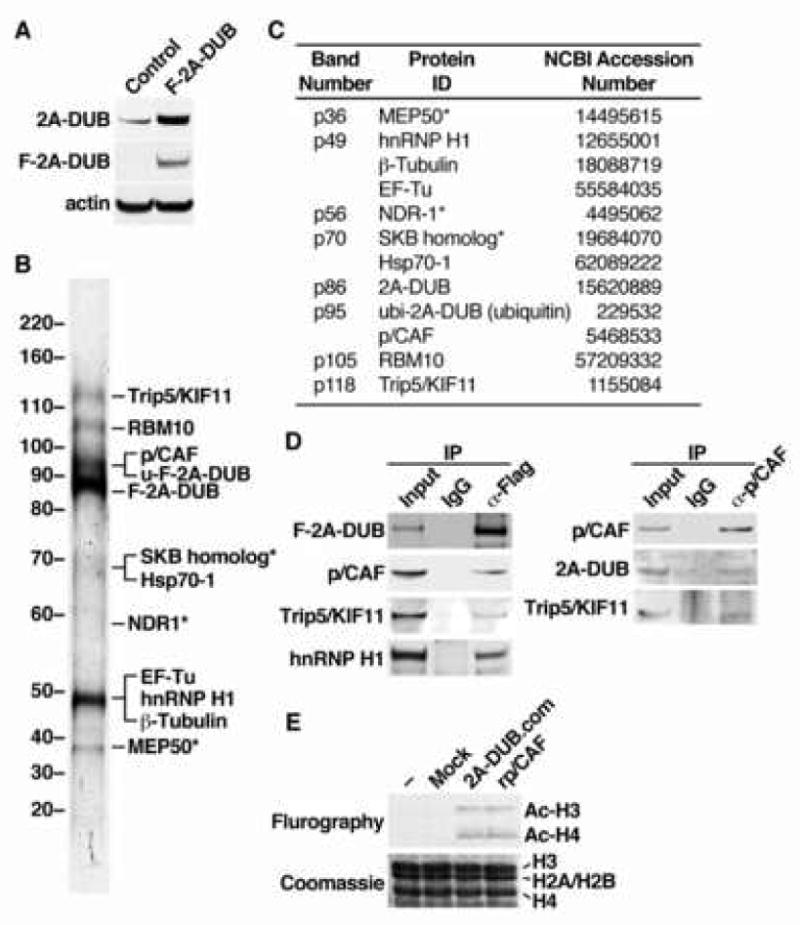
(A) Western blot analysis of HEK293 cell lines used for affinity purification. Control and F-2A-DUB cells were stably transfected with an empty vector and a vector encoding Flag-tagged human 2A-DUB, respectively. (B) Silver staining of co-purified peptides in the affinity-purified 2A-DUB protein complex from HEK293 cells after separation by SDS-PAGE. The molecular weight was shown on the left and the identity of the peptide band, analyzed by mass spectrometry, was shown on the right. Asterisks represent bands that were also observed in the materials from the parallel affinity purification of the control cell line, or known to interact with M2 anti-Flag resin. (C) Detailed information of mass spectrometric analysis of the co-purified peptides in the 2A-DUB protein complex. (D) Left panel: co-immunoprecipitation (Co-IP) of 2A-DUB and proteins co-purified with 2A-DUB. Flag-2A-DUB was transiently transfected into HEK293T cells, and immunoprecipitated from the nuclear extracts. Co-immunoprecipitated proteins were detected by Western blot analysis. Right panel: IP using anti-p/CAF to test the co-immunoprecipitated endogenous proteins in HEK293T cells. Inputs: 2%. (E) In vitro histone acetyltransferase (HAT) assay for free core histones using affinity-purified 2A-DUB complex (2A-DUB.com), recombinant HAT domain of p/CAF (rp/CAF) and materials from affinity purification of control cells (mock).
Coordination of histone acetylation, H2A deubiquitination and H1 dissociation
Co-existence of 2A-DUB and p/CAF in a protein complex suggests the intriguing possibility that H2A ubiquitination and histone acetylation act stepwise to coordinate regulation of gene expression, which may underlie the contributions of H2A ubiquitination to transcriptional regulation. To test whether the status of histone acetylation influence the deubiquitinase activity of 2A-DUB, we designed an in vitro deubiquitination assay using acetylation-manipulated mono/oligonucleosomes as the substrate. We used a histone deacetylase (HDAC) inhibitor trichostatin A (TSA) to accumulate hyperacetylated nucleosomal histones in stable HEK293 cells expressing Flag-H2A or H2B, which is efficiently incorporated into nucleosomes (Fig. S5 and S6). The mono/oligonucleosomes were prepared by micrococcal nuclease (MNase) digestion from TSA-treated or non-treated cells. DNA gel analysis confirmed that >90% were mononucleosomes (Fig. 3A). Flag-tagged histone-containing nucleosomes were next partially purified by M2 anti-Flag resin chromatography, with minimal incorporation of endogenous histones H2A/H2B (Fig. 3A, Fig. S5B, C). As shown before, the ∼24-kDa bands, not present in Flag-H2A/H2B in which the main lysine residues for monoubiquitination were substituted, represent monoubiquitinated Flag-H2A and Flag-H2B in the Western blot analysis (Fig. 3A and S5). Using Flag-H2A-containing nucleosomes, we found that the deubiquitinase activity of the 2A-DUB complex (Fig. 2) was very minimal for substrates purified from cells that were not treated with TSA, suggesting hypoacetylated nucleosomes harbor a “barrier” for the catalytic activity of 2A-DUB (Fig 3B). On the other hand, 2A-DUB activity was remarkably augmented in TSA-treated hyperacetylated nucleosomes, as shown by the substantially decreased level of the Flag-uH2A band (Fig. 3B). These data reveal that 2A-DUB preferentially deubiquitinates uH2A in hyperacetylated nucleosomes, suggesting that the enzymatic complex may first utilize p/CAF or other HATs to generate “optimized” substrates for 2A-DUB, which then removes the ubiquitin from H2A to yield a favorable histone modification status for transcriptional regulation/chromatin remodeling events. In contrast, we did not find any evidence that decreased uH2A levels in mononucleosomes augment p/CAF HAT activity by in vitro HAT assay using ubiquitinated wt and non-ubiquitinated K119R H2A-containing nucleosomes (data not shown). This suggests coordination of the histone modifications in accord with the general strategy suggested for sequential epigenetic regulation (Shi et al., 2005, Sun and Allis, 2002). Consistent with the in vivo results (Fig. S3) the 2A-DUB complex demonstrated no effect on Flag-H2B in nucleosome-based in vitro deubiquitinase assay, regardless acetylation status (Fig. 3C). Independently evaluating the role of p/CAF in 2A-DUB-dependent H2A deubiquitination, by specific siRNA-dependent knock-down p/CAF, revealed that reduction of p/CAF expression increased uH2A levels (Fig. 3D, lane 2 and 3). Moreover, 2A-DUB overexpression-induced decrease of uH2A was partially antagonized by p/CAF siRNA but not control siRNA (Fig. 3D, lane 4 and 5), suggesting a pivotal role of p/CAF in augmenting the deubiquitinase activity of 2A-DUB. Interestingly, siRNAs against CBP showed no effect on the activity of 2A-DUB, suggesting that site-specific regulation of histones H3/H4 acetylation modulates 2A-DUB function (data not shown).
We next attempted to identify downstream effectors of uH2A to elucidate the repressive role of uH2A in transcriptional regulation. Based on the “trans-tail” cross-talk between uH2B and H3K4/K36/K79 methylation (Dover et al., 2002, Henry et al., 2003, Ng, et al., 2002, Shahbazian et al., 2005, Sun and Allis, 2002), we first examined whether uH2A regulates histone methylation, focusing on repressive marks including H3K9 and H3K27, but found no alterations of these histone methylation marks upon increasing uH2A by siRNA knockdown of 2A-DUB (Fig. S7). We therefore isolated Flag-H2A-containing mononucleosomes from stable HEK293 cells expressing Flag-tagged wt or K119R H2A by affinity purification (Fig. 3E), comparing the composition of candidate histone marks in mononucleosomes. Because histone acetylation significantly increases the exchange dynamics of linker histone H1 to nucleosomes, and because the C-terminus of H2A, in which the ubiquitinated K119 is located, is the only histone tail that is capable of reaching the linker DNA and histone (Luger et al., 1997), we checked the composition of Ac-H3K9, Ac-H3K14 and linker histone H1 in wt (ubiquitinated) and K119R (non-ubiquitinated) H2A mononucleosomes by Western blot analysis. Strikingly, we found that the associated linker histone H1 was largely diminished in K119R H2A-containing mononucleosomes (Fig. 3E), indicating that deubiquitination of uH2A correlates with the dissociation of linker histones from core nucleosomes. This finding is consistent with recent suggestions that uH2A enhanced the binding of histone H1 to reconstituted nucleosomes in vitro (Jason et al., 2005). In contrast, H1 associated equally well with both ubiquitinated wt and non-ubiquitinated K120/125R Flag-H2B-containing mononucleosomes in a similar experiment (Fig. S8). We further found that H1 phosphorylation is remarkably decreased when uH2A levels were increased in response to siRNA-mediated knockdown of 2A-DUB in HEK293T cells, while reduction of uH2A levels by knocking down Ring2/Ring1B, a H2A E3 ligase (de Napoles et al., 2004, Wang et al., 2004), led to a clear increase of the phosphorylation of H1 (Fig. 3F). Given the fact that H1 phosphorylation is well established to augment the dissociation of H1 from nucleosomes (Contreras et al., 2003, Dou et al., 2002), it is possible that in cells the displacement of H1 could be triggered by raised phosphorylation in response to lower uH2A levels. In line with our observations, it has also been proposed that histone acetylation may facilitate H1 dissociation (Ju et al., 2004, Misteli et al., 2000, Nightingale et al., 1998). Displacement of linker histones has been shown generally, but not invariably, to be involved in gene activation (and in some cases gene repression), which may explain the dual roles of uH2A in transcriptional regulation. Of course, it is likely that uH2A also has other downstream effectors that function either synergistically or independently, and at different steps of transcriptional regulation (W.Z., P.Z., M.G.R., unpublished data).
2A-DUB and H2A ubiquitination in transcriptional initiation and elongation
Based on the actions of uH2A and our preliminary data (Fig. S1), we focused on androgen receptor (AR) to study the role of 2A-DUB and uH2A in regulated transcriptional activation. Using an androgen responsive element (ARE)-driven luciferase reporter, we found that DHT-induced activation of ARE-reporter was enhanced by wt 2A-DUB but antagonized by enzymatically inactive D567N mutant (Fig. 4A), indicating that 2A-DUB functions as a coactivator of AR in a manner depending on its deubiquitinase activity. Further, upon knockdown of 2A-DUB by two independent specific siRNAs or on reduction of p/CAF by siRNA (Fig. 1E and Fig. S9), DHT-dependent activation was significantly reduced (Fig. 4A). To test whether p/CAF, a HAT that interacts with 2A-DUB and augments the H2A deubiquitinase activity of 2A-DUB (Fig. 2 and 3), plays a role in 2A-DUB-dependent activation, we compared 2A-DUB-induced activation in the presence and absence of siRNA against p/CAF. Upon transfection of control siRNA, 2A-DUB further augmented DHT-induced ARE-dependent reporter activation by over 2.5 fold, whereas knockdown of p/CAF fully abolished the activity (Fig. 4B), consistent with the observation that histone acetylation and p/CAF help to optimize the H2A deubiquitinase activity of 2A-DUB that is crucial for transcriptional activation. 2A-DUB and p/CAF had no effect on basal reporter activity, suggesting that their activity is AR-dependent (Fig. 4A). Furthermore, we evaluated the role of 2A-DUB in regulation of endogenous target genes of AR. DHT-induced activation of AR target genes PSA, Nkx3.1 (Fig. 4C) and KLK2 (data not shown) were substantially impaired by efficient siRNA-mediated knockdown of 2A-DUB in LNCaP prostate cancer cells (Fig. 4C). Interestingly, 2A-DUB itself is also moderately upregulated by DHT, suggesting a positive regulatory feedback between AR and 2A-DUB (Fig. 4C). As expected, chromatin immunoprecipitation (ChIP) demonstrated that AR and p/CAF are recruited to the promoter of PSA in response to DHT (Fig. 5A). Using validated antibodies against 2A-DUB and uH2A (Fig. S4B, C), ChIP assays revealed that 2A-DUB is enriched on the promoter region of PSA in LNCaP cells after DHT treatment (Fig. 5A). Correlating with the recruitment of 2A-DUB and p/CAF, a clear decrease of uH2A levels and a coincidently increased level of acetylated H3 (Ac-H3) were observed upon DHT-treatment in LNCaP cells (Fig. 5A), suggesting that 2A-DUB and p/CAF co-activate AR target genes by coordinated modulation of uH2A and histone acetylation status. To test the impact of 2A-DUB/uH2A on linker histone H1 displacement, we evaluated the amount of H1 on the PSA promoter, finding a remarkable portion of H1 was dismissed from the promoter region upon DHT-treatment (Fig. 5B). The ligand-induced dissociation of H1 was largely diminished when 2A-DUB was knocked down by specific siRNA (Fig. 5B), consistent with the observation that loss of ubiquitination of H2A augments the dismissal of H1 (Fig. 3E). To directly test the functional role of p/CAF and 2A-DUB in the regulatory complex, we knocked down p/CAF by specific siRNA, and then checked the occupancy of 2A-DUB and uH2A on the PSA promoter. Interestingly, while the recruitment of 2A-DUB was not affected (Fig. 5C, left panel), DHT-dependent decrease of uH2A levels was substantially impaired by siRNA against p/CAF (Fig. 5C, right panel), suggesting that presence of p/CAF in the protein complex described here is required for the H2A deubiquitinating activity of 2A-DUB during PSA gene induction. Because androgen/AR signaling exerts key roles in prostate cancer (Chen et al., 2004, Heinlein and Chang, 2004, Zhu et al., 2006), regulation of AR-dependent transcription by 2A-DUB implies that uH2A could be a disease-related mark, analogous to acetylation and methylation marks found to be useful in prognosis of prostate cancer (Seligson et al., 2005). Strikingly, anti-uH2A immunostaining of patient-matched prostate cancer tissue microarrays revealed that uH2A levels are remarkably lower in prostate tumors compared to those in histologically normal prostate tissues from the same patients (Fig. 5D). Out of 35 prostate cancer patients, a 4-grade scoring method of uH2A staining revealed 18 with significant decrease (≥2 grades) of uH2A in tumors, 15 with moderate decrease (<2 grades), while only 2 with no difference compared to corresponding normal tissues. The average grade of uH2A levels is significantly lower in prostate tumors (Fig. 5E), thus correlating low uH2A levels with amplified androgen signaling in prostate cancer.
Figure 4. 2A-DUB in activation of androgen/AR-dependent transcription.
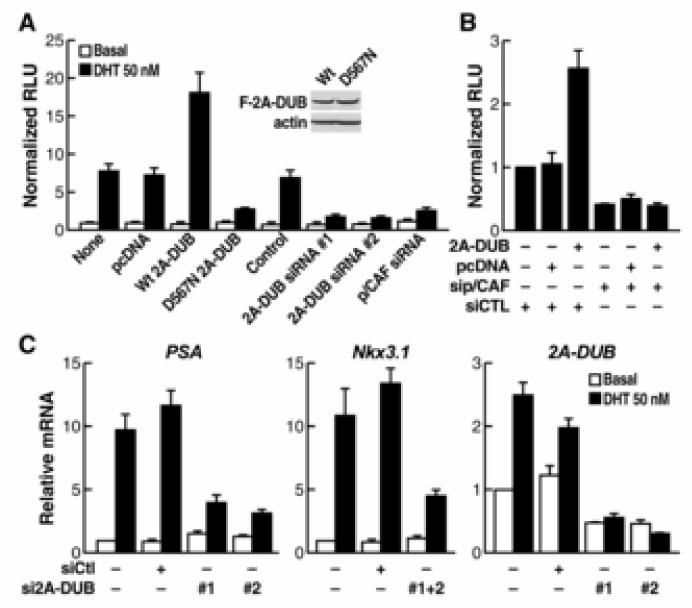
(A) Reporter assays using an ARE-dependent luciferase reporter in LNCaP prostate cancer cells. Cells were transfected with reporter and different plasmids or siRNA as indicated and then treated with or without AR agonist DHT. The activity of specific luciferase was normalized with that of a co-transfected unregulated renilla luciferase reporter. Values are the relative values to normalized activity from basal, reporter-only transfected cells (bar 1, set as 1), and represent means±SEM of three independent experiments. The similar expression levels of wt and D567N 2A-DUB are shown by Western blot analysis. (B) Assays of ARE-driven luciferase reporter in LNCaP cells. Cells were transfected with indicated plasmids or siRNA in combination with reporters, and then treated with DHT. Normalized relative values are calculated as described in (A). (C) Expression of endogenous AR target genes and 2A-DUB analyzed by quantitative RT-PCR. LNCaP cells were transfected with indicated siRNA, and then treated with DHT if needed. Values (normalized to corresponding values of internal control gene HPRT) are means±SEM of three independent experiments.
Fig. 5. The role of 2A-DUB and uH2A in androgen signaling in prostate cancer cells.
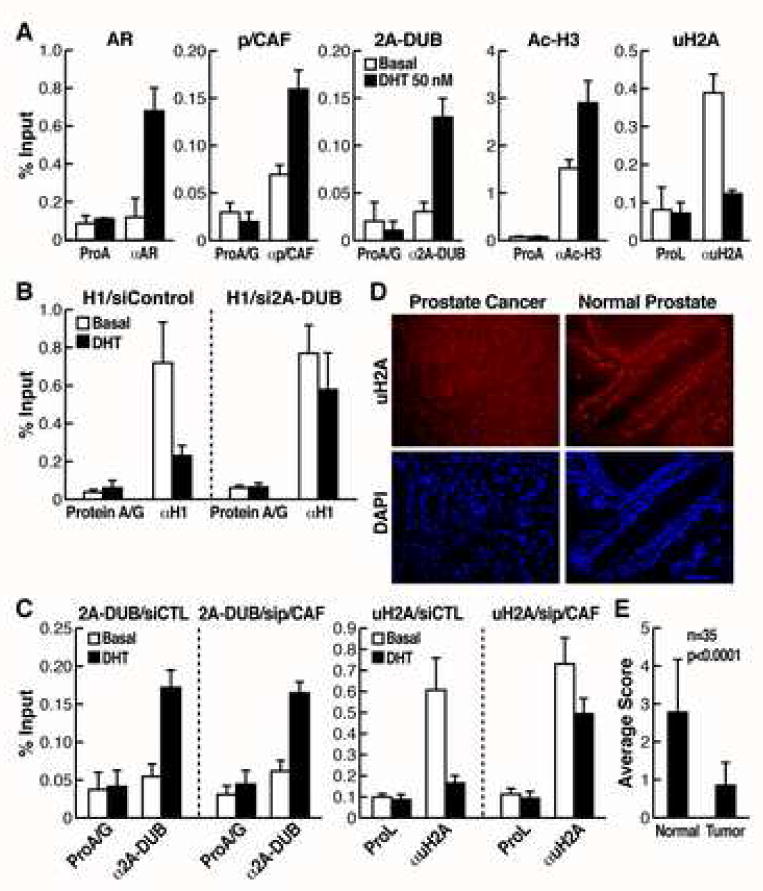
(A) ChIP assays of the promoter of PSA, a target gene of AR. LNCaP cells were cultured in the charcoal-stripped medium and then challenged with DHT for 1 hr. Soluble chromatin was immunoprecipitated by the antibodies indicated, and the bound DNA was analyzed by quantitative PCR using a primer pair flanking the AREs of the PSA promoter. Values (ratios of ChIP to corresponding inputs) are means±SEM of at least two independent experiments. (B) The occupancy of linker histone H1 was detected by quantitative ChIP assays on the promoter of PSA in LNCaP cells, transfected with control or 2A-DUB siRNA. (C) The occupancy of 2A-DUB and uH2A was detected by quantitative ChIP assays on the promoter of PSA in LNCaP cells, transfected with control or p/CAF siRNA. (D) Immunofluorescent staining of uH2A for a tissue microarray of prostate cancer. DAPI staining was performed to visualize the nuclear DNA. One representative staining is shown. (E) Semi-quantification of the uH2A staining by a four-grade scoring system (see Experimental Procedure). Values are mean±SD of grades scored for benign prostate tissues or corresponding prostate tumors. Paired Student's t test was used to calculate the p value.
We further investigated the role of 2A-DUB in other regulated transcriptional events. 2A-DUB deubiquitinase activity was required for full activation of AP-1-, ISGF-3- and estrogen receptor (ER)-dependent transcription as well as for AR-regulated gene expression (Fig. S1), as revealed by reporter assays or mRNA expression of endogenous gene targets (Fig. S10A-C). In contrast, RARβ2, a known target gene of retinoic acid receptor α (RARα), was not detectibly regulated by 2A-DUB, and thyroid hormone receptor (T3R)-mediated transcription was repressed by 2A-DUB apparently independent of the deubiquitinase activity (Fig. S10D, E). These observations suggest that 2A-DUB and uH2A could be specific for the regulation of a cohort of genes, while other yet undiscovered ligases/deubiquitinases may participate in regulating a different cohort of uH2A-dependent transcriptional events, exemplified by PcG- (de Napoles et al., 2004, Wang et al., 2004) and LPS-regulated gene expression in macrophages (W.Z., P.Z., M.G.R., unpublished).
On testing the binding of 2A-DUB across the PSA gene locus, we found that it is also present on exonic regions. Given the fact that histone acetylation and p/CAF are suggested to participate in transcriptional elongation (Cho et al., 1998, Daniel et al., 2004, Henry et al., 2003), this finding raises an intriguing issue whether 2A-DUB and uH2A are involved in both transcriptional initiation and elongation events. We therefore performed ChIP assays at the enhancer, promoter and exon 4 regions of PSA (Fig. 6A) for the phospho-serine 5 of Pol II CTD (pS5-Pol II) initiation mark, the phospho-serine 2 of Pol II CTD (pS2-Pol II) elongation mark (Komarnitsky et al., 2000) and the elongation-related histone mark trimethyl-H3K36 (H3K36M3) (Bannister et al., 2005, Krogan et al., 2003, Li et al., 2003, Vakoc et al., 2006, Xiao et al., 2003). As expected, DHT treatment causes the recruitment of AR to both enhancer and promoter regions, but not exon 4 of PSA in LNCaP cells (Fig. 6B left). Consistent with its role in gene activation, 2A-DUB was enriched at both the promoter and exon 4 in response to DHT, suggesting that it could participate in both initiation and elongation (Fig. 6C left). We normalized uH2A levels to the corresponding H2A levels to ascertain that the alteration was due to histone modification rather than histone displacement, finding a clear reduction at the promoter and exon 4 by DHT treatment (Fig. 6D left), which correlates with the recruitment of 2A-DUB. pS5-polII was enhanced mostly at the promoter region while pS2-polII and H3K36M3 were mainly enriched in exonic regions, consistent with their respective roles in gene expression (Fig. 6E-G left). Upon transfection of 2A-DUB siRNA in LNCaP cells, 2A-DUB occupancy in the presence of DHT was clearly diminished at both the promoter and exon 4, indicating that the knockdown was efficient (Fig. 6C right). Nonetheless, the recruitment of AR was unaffected by 2A-DUB siRNA (Fig 6B right), implying that loss of 2A-DUB does not influence the binding of major components of the transcriptional complex. Correlating with the dismissal of 2A-DUB, uH2A (normalized to H2A) was maintained at substantially high levels even after treatment with DHT (Fig. 6D right), revealing that 2A-DUB is crucial in removing ubiquitin from H2A in both promoter and exonic regions during PSA activation. Significantly, although cells were treated with DHT as usual, the amount of pS5-polII on the promoter was reduced upon loss of 2A-DUB and increased levels of uH2A (Fig. 6E right), which argues that 2A-DUB and resultant deubiquitination of uH2A participate in AR-mediated transcriptional initiation for the PSA gene. More importantly, both pS2-polII and H3K36M3 were remarkably diminished by siRNA of 2A-DUB even in the presence of DHT, mainly on the exon 4 region (Fig. 6F and G, right). We also noted that, upon treating cells with 2A-DUB siRNA, pS2-polII occupancy on the exon 4 reduced by >80% (Fig. 6F, bar 6 vs. 12), while on the promoter pS5-polII occupancy diminished by about 60% (Fig. 6E, bar 4 vs. 10), suggesting that the impaired elongation is due mainly to inefficient initiation, but with additional specific defects in elongation. Taken together, the ChIP data indicate that 2A-DUB, with the consequent alteration of uH2A status, participates in the initiation and likely the elongation steps of androgen/AR-induced gene activation, in agreement with our expression data (Fig. 4 and 5).
Figure 6. 2A-DUB in regulation of transcriptional initiation and elongation.
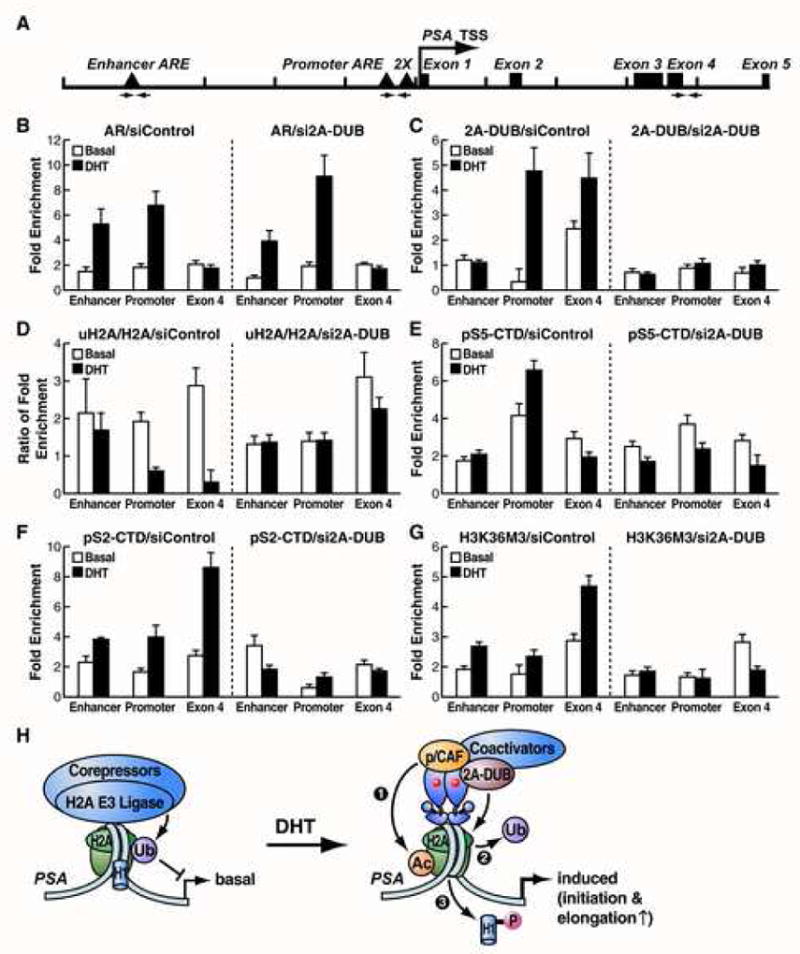
(A) Schematic representation of the genomic locus (10 kb) of the PSA gene. Arrows represent the primer sets used for ChIP analysis of the enhancer, promoter and exon 4 region of PSA. TSS: transcription start site. Quantitative ChIP assays were performed for AR (B), 2A-DUB (C), uH2A and H2A (D), PolII (phospho-serine 5 of CTD, pS5-CTD) (E), PolII (phospho-serine 2 of CTD, pS2-CTD) (F) and H3K36M3 (G). LNCaP cells were cultured in the charcoal-stripped medium. Following transfection of control (left panels) or 2A-DUB (right panels) siRNA, cells were treated with DHT for 1 hr, then ChIP was performed as described in Experimental Procedures. Enrichment folds compared to the corresponding control (bead alone) ChIP were calculated and shown. For (D), the ratios of enrichment folds of uH2A to H2A were shown. Values are mean±SD of three biological repeats. One representative of at least two independent experiments is shown. (H) A proposed model of the role of 2A-DUB and uH2A in transcriptional regulation. See the text for details.
Discussion
H2A ubiquitination coordinates histone acetylation and H1 dissociation
The central finding in our study is that deubiquitination of H2A, which is augmented by hyperacetylation of nucleosomal histones, enhances the phosphorylation and dissociation of linker histone H1. This stepwise coordination of histone modifications, mediated by a co-regulatory functional complex containing 2A-DUB and p/CAF, is critically involved in transcriptional regulation of a specific cohort of target genes. Identification of the histone H2A deubiquitinase 2A-DUB as a transcriptional coactivator has revealed insights into the linkage between histone H2A ubiquitination and transcriptional regulation, with 2A-DUB specificity for uH2A providing a valuable tool to delineate roles of H2A monoubiquitination. While it is currently unclear why 2A-DUB is H2A-specific, the structural domains within 2A-DUB suggest that this reflects the ability of the SANT domain to interact with DNA and histones (Boyer et al., 2004), and the SWIRM domain to bind to linker DNA between nucleosomes (Da et al., 2006, Qian et al., 2005). In concert with the fact that the C-terminus of H2A, where lysine 119 is located, is the histone tail that is able to touch linker DNA/histones (Luger et al., 1997), 2A-DUB may utilize the SWIRM or SANT domain to facilitate the accessibility to H2A for deubiquitination.
Together with other effectors of uH2A, e.g. FACT (W.Z., P.Z. and M.G.R., unpublished), dissociation of linker histone H1 from nucleosomes by deubiquitination of uH2A could be important in uH2A-related transcriptional regulation. Our findings are consistent with the in vitro observation that assembly of uH2A into nucleosomes facilitates the association of H1 (Jason et al., 2005), and the nucleosomal structural observations that the C-terminus of H2A is the fragment of core histones that is able to interact with linker histones (Luger et al., 1997). Interestingly, manipulating uH2A status by knocking down H2A-specific E3 ligase or deubiquitinase led to inverse alteration of H1 phosphrylation (Fig. 3F), which could be due to exchange of still unidentified kinases/phosphatases for H1 following alteration of uH2A levels. Consequently, hyperphosphorylation of H1 caused by H2A deubiquitination may trigger the higher dissociation rate of H1 from core nucleosomes (Dou et al., 2002). Displacement of linker histones, which is facilitated by their phosphorylation, is generally linked to gene activation but has also been linked to repression in a few instances, coincident with the contradictory, dual roles for uH2A in both gene activation and repression.
Unexpectedly, specific interactions between p/CAF and 2A-DUB, as components of a protein complex, seem to coordinate histone ubiquitination and acetylation. In yeast, the transcriptional activator SAGA complex also contains a HAT Gcn5, the yeast homolog of p/CAF, and a histone H2B deubiquitinase Ubp8 (Daniel et al., 2004, Henry et al., 2003). Both HAT and deubiquitination activities are required in SAGA-activated transcriptional initiation and elongation (Daniel et al., 2004, Henry et al., 2003). Since uH2A is undetectable in budding yeast (Swerdlow et al., 1990), the 2A-DUB-p/CAF complex could be a functional analogue of the yeast SAGA complex by specifically targeting uH2A in a cohort of transcription events in vertebrates. Notably, there are RNA-binding proteins co-purified in the 2A-DUB complex, similar to the yeast SAGA complex and the human STAGA complex (Martinez et al., 2001), suggesting a common feature of these complexes in coupling transcription and RNA processing or RNA-dependent regulatory function in transcription. Because 2A-DUB was not detected in the biochemically-purified p/CAF core complex (Ogryzko et al., 1998), we can suggest that the two core complexes are distinct, although sharing some functional similarities. The regulatory 2A-DUB-p/CAF complex is of functional importance because the activity of 2A-DUB is augmented by hyperacetylated nucleosomes and p/CAF. Because both the SANT and SWIRM domains serve as the modules for binding to histone H3 tail, which is a target for acetylation by p/CAF and other HATs, we are tempted to speculate that this acetylation may enhance the accessibility of uH2A to 2A-DUB.
The role of 2A-DUB in transcriptional regulation
We have identified several regulated transcriptional events that are modulated by 2A-DUB, including inflammation- and hormone-related gene expression. Remarkably, 2A-DUB acts as a coactivator of AR, based on its intrinsic deubiquitinase activity. AR requires a number of histone-modifying cofactors for its transcriptional activity, e.g. HAT CBP/p300, histone demethylases LSD1 and JHDM2 (Metzger et al., 2005, Yamane et al., 2006). Here we show that the 2A-DUB-p/CAF complex is also required for the full activation of AR by stepwise coordination of histone acetylation and deubiquitination. Collectively, a series of distinct histone-modifying coactivators are utilized by AR to remove repressive marks (Me-H3K9/K27, uH2A, etc.) and add active marks (Ac-H3K9/K14, Me-H3K4, etc.) to achieve the “optimal” modulation of nucleosomal architecture for transcriptional activation. Importantly, we also provide evidence that 2A-DUB and uH2A participate in transcriptional initiation and possibly also in elongation. At the initiation step, the 2A-DUB-p/CAF complex is recruited to the promoter region of PSA in response to ligand, removing the repressive uH2A mark from the acetylated nucleosomes and dissociating linker histones in a stepwise manner (Fig. 6H). As 2A-DUB may interact with AR via its SWIRM domain (Metzger et al., 2005), recruitment of the complex might be AR-dependent. By creating a chromatin structure favorable for transcription by the coordinated histone modification/displacement, the initiating Pol II complex may be recruited more efficiently to the promoter to launch transcription. During elongation, we attempt to suggest that the 2A-DUB-p/CAF complex is transferred to the elongation complex, probably via p/CAF, performing the stepwise coordination of histone modifications to add/remove active/repressive marks to facilitate elongating Pol II activity. In this regard, transient histone H2A/H2B displacement may also play a role because removing ubiquitin from H2A substantially increases the interaction between H2A and FACT (W.Z., P.Z. and M.G.R., unpublished), a positive factor for elongation that acts as a histone chaperone to exert displacement of H2A/H2B dimer (Belotserkovskaya et al., 2003).
Interestingly, the levels of monoubiquitination of H2A are significantly diminished in prostate cancer compared to benign prostate tissues (Fig. 5D, E), suggesting that the activation of a cohort of AR target genes in such cancer is likely to refer to, in part, low uH2A levels. Because a PcG complex contains an E3 ligase of H2A (Ring2/Ring1B) that is required for its gene silencing function (de Napoles et al., 2004, Wang et al., 2004), and the PcG protein EZH2 is overexpressed in hormone resistant and metastatic prostate cancer (Varambally et al., 2002), a Me-H3K27/uH2A-based repression could be critically involved in prostate cancer biology. Thus, 2A-DUB-dependent activation and EZH2-dependent repression mechanisms may be utilized by prostate cancer cells to modulate distinct cohorts of genes using a similar epigenetic strategy, whereas the globally-increased uH2A levels may reflect a predominant role of H2A deubiquitination in a number of chromatin-related regulatory events (e.g. mitosis) in prostate cancer biology in addition to transcriptional regulation. uH2A may thus serve as a highly useful cancer-related marker, in addition to other important histone modification marks that provide insights into prognosis (Seligson et al., 2005). Our data, together with those in the literature, suggest an important role of histone-modifying enzymes and consequent histone modifications in regulating androgen signaling in normal prostate biology and in pathogenesis of prostate cancer.
In summary, our data have uncovered monoubiquitination of histone H2A as a widely-used mechanism for modulating regulatory programs of gene expression, which is likely to be accomplished a series of dedicated ubiquitin E3 ligases and deubiquitinases responsible for specific cohorts of regulated transcription units.
Experimental Procedures
Cells, antibodies, siRNAs and other reagents
Human embryonic kidney cell lines HEK293 and HEK293T, human prostate cancer cell line LNCaP, human breast cancer cell line MCF-7, human cervical cancer cell line HeLa, and mouse embryocarcinoma cell line P19 were originally purchased from ATCC and were cultured under standard conditions. The details of other cell lines, antibodies, siRNA and reagents are described in Supplemental Data.
Affinity purification of 2A-DUB protein complex
The detailed purification procedure has been described previously (Ju et al., 2004). Details are available in Supplemental Data.
In vitro deubiquitination assay
The in vitro deubiquitination assay was modified from a method described previously (Cope et al., 2002, Verma, et al., 2002). Details are described in Supplemental Data.
Other experimental procedures are described in Supplemental Data.
Supplementary Material
Acknowledgments
We thank T. Nagase and H. Koga for cDNA of human and mouse KIAA1915, M. Oren for H2A/H2B constructs, R.J. Matusik for ARR2PB reporter, J. Köhrle for Dio-luciferase reporter, D. Monroe and T. Spelsberg for U2OS-ERα cells, C. Nelson for technical assistance, J. Hightower and M. Fisher for figure and manuscript preparation, and E. Bourk for critical reading of the manuscript. P.Z. is supported by a postdoctoral fellowship from the Human Frontier Science Program (HFSP). M.G.R. is an investigator with the Howard Hughes Medical Institute. This work was supported by grants from NIH to M.G.R. and C.K.G., NCI to P.T. and the PCRP to M.G.R.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Ambroggio XI, Rees DC, Deshaies RJ. JAMM: a metalloprotease-like zinc site in the proteasome and signalosome. PLoS Biol. 2004;2(1):E2. doi: 10.1371/journal.pbio.0020002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baarends WM, Hoogerbrugge JW, Roest HP, Ooms M, Vreeburg J, Hoeijmakers JH, Grootegoed JA. Histone ubiquitination and chromatin remodeling in mouse spermatogenesis. Dev Biol. 1999;207:322–333. doi: 10.1006/dbio.1998.9155. [DOI] [PubMed] [Google Scholar]
- Bannister AJ, Schneider R, Myers FA, Thorne AW, Crane-Robinson C, Kouzarides T. Spatial distribution of di- and tri-methyl lysine 36 of histone H3 at active genes. J Biol Chem. 2005;280(18):17732–17736. doi: 10.1074/jbc.M500796200. [DOI] [PubMed] [Google Scholar]
- Belotserkovskaya R, Oh S, Bondarenko VA, Orphanides G, Studitsky VM, Reinberg D. FACT facilitates transcription-dependent nucleosome alteration. Science. 2003;301(5636):1090–1093. doi: 10.1126/science.1085703. [DOI] [PubMed] [Google Scholar]
- Boyer LA, Latek RR, Peterson CL. The SANT domain: a unique histone-tail-binding module? Nat Rev Mol Cell Biol. 2004;5(2):158–163. doi: 10.1038/nrm1314. [DOI] [PubMed] [Google Scholar]
- Chen CD, Welsbie DS, Tran C, Baek SH, Chen R, Vessella R, Rosenfeld MG, Sawyers CL. Molecular determinants of resistance to antiandrogen therapy. Nat Med. 2004;10(1):33–39. doi: 10.1038/nm972. [DOI] [PubMed] [Google Scholar]
- Chen HY, Sun JM, Zhang Y, Davie JR, Meistrich ML. Ubiquitination of histone H3 in elongating spermatids of rat testes. J Biol Chem. 1998;273:13165–13169. doi: 10.1074/jbc.273.21.13165. [DOI] [PubMed] [Google Scholar]
- Cho H, Orphanides G, Sun X, Yang XJ, Ogryzko V, Lees E, Nakatani Y, Reinberg D. A human RNA polymerase II complex containing factors that modify chromatin structure. Mol Cell Biol. 1998;18(9):5355–5363. doi: 10.1128/mcb.18.9.5355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Contreras A, Hale TK, Stenoien DL, Rosen JM, Mancini MA, Herrera RE. The Dynamic Mobility of Histone H1 Is Regulated by Cyclin/CDK Phosphorylation. Mol Cell Biol. 2003;23:8626–8636. doi: 10.1128/MCB.23.23.8626-8636.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cope GA, Suh GS, Aravind L, Schwarz SE, Zipursky SL, Koonin EV, Deshaies RJ. Role of predicted metalloprotease motif of Jab1/Csn5 in cleavage of Nedd8 from Cul1. Science. 2002;298(5593):608–611. doi: 10.1126/science.1075901. [DOI] [PubMed] [Google Scholar]
- Da G, Lenkart J, Zhao K, Shiekhattar R, Cairns BR, Marmorstein R. Structure and function of the SWIRM domain, a conserved protein module found in chromatin regulatory complexes. Proc Natl Acad Sci U S A. 2006;103(7):2057–2062. doi: 10.1073/pnas.0510949103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniel JA, Torok MS, Sun ZW, Schieltz D, Allis CD, Yates JR, 3rd, Grant PA. Deubiquitination of histone H2B by a yeast acetyltransferase complex regulates transcription. J Biol Chem. 2004;279(3):1867–1871. doi: 10.1074/jbc.C300494200. [DOI] [PubMed] [Google Scholar]
- de Napoles M, Mermoud JE, Wakao R, Tang YA, Endoh M, Appanah R, Nesterova TB, Silva J, Otte AP, Vidal M, Koseki H, Brockdorff N. Polycomb group proteins Ring1A/B link ubiquitylation of histone H2A to heritable gene silencing and X inactivation. Dev Cell. 2004;7(5):663–76. doi: 10.1016/j.devcel.2004.10.005. [DOI] [PubMed] [Google Scholar]
- Dou Y, Bowen J, Liu Y, Gorovsky MA. Phosphorylation and an ATP-dependent process increase the dynamic exchange of H1 in chromatin. J Cell Biol. 2002;158(7):1161–1170. doi: 10.1083/jcb.200202131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dover J, Schneider J, Tawiah-Boateng MA, Wood A, Dean K, Johnston M, Shilatifard A. Methylation of histone H3 by COMPASS requires ubiquitination of histone H2B by Rad6. J Biol Chem. 2002;277(32):28368–28371. doi: 10.1074/jbc.C200348200. [DOI] [PubMed] [Google Scholar]
- Emre NC, Ingvarsdottir K, Wyce A, Wood A, Krogan NJ, Henry KW, Li K, Marmorstein R, Greenblatt JF, Shilatifard A, Berger SL. Maintenance of low histone ubiquitylation by Ubp10 correlates with telomere-proximal Sir2 association and gene silencing. Mol Cell. 2005;17(4):585–594. doi: 10.1016/j.molcel.2005.01.007. [DOI] [PubMed] [Google Scholar]
- Fang J, Chen T, Chadwick B, Li E, Zhang Y. Ring1b-mediated H2A ubiquitination associates with inactive X chromosomes and is involved in initiation of X inactivation. J Biol Chem. 2004;279(51):52812–52815. doi: 10.1074/jbc.C400493200. [DOI] [PubMed] [Google Scholar]
- Gardner RG, Nelson ZW, Gottschling DE. Ubp10/Dot4p regulates the persistence of ubiquitinated histone H2B: distinct roles in telomeric silencing and general chromatin. Mol Cell Biol. 2005;25(14):6123–6139. doi: 10.1128/MCB.25.14.6123-6139.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glass CK, Rosenfeld MG. The coregulator exchange in transcriptional functions of nuclear receptors. Genes Dev. 2000;14(2):121–141. [PubMed] [Google Scholar]
- Goldknopf IL, Busch H. Isopeptide linkage between nonhistone and histone 2A polypeptides of chromosomal conjugate-protein A24. Proc Natl Acad Sci U S A. 1977;74(3):864–868. doi: 10.1073/pnas.74.3.864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldknopf IL, Taylor CW, Baum RM, Yeoman LC, Olson MO, Prestayko AW, Busch H. Isolation and characterization of protein A24, a “histone-like” nonhistone chromosomal protein. J Biol Chem. 1975;250:7182–7187. [PubMed] [Google Scholar]
- Heinlein CA, Chang C. Androgen receptor in prostate cancer. Endocr Rev. 2004;25(2):276–308. doi: 10.1210/er.2002-0032. [DOI] [PubMed] [Google Scholar]
- Henry KW, Wyce A, Lo WS, Duggan LJ, Emre NC, Kao CF, Pillus L, Shilatifard A, Osley MA, Berger SL. Transcriptional activation via sequential histone H2B ubiquitylation and deubiquitylation, mediated by SAGA-associated Ubp8. Genes Dev. 2003;17(21):2648–2663. doi: 10.1101/gad.1144003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang SY, Barnard MB, Xu M, Matsui S, Rose SM, Garrard WT. The active immunoglobulin κ chain gene is packaged by non-ubiquitin-conjugated nucleosomes. Proc Natl Acad Sci U S A. 1986;83:3738–3742. doi: 10.1073/pnas.83.11.3738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jason LJ, Finn RM, Lindsey G, Ausio J. Histone H2A ubiquitination does not preclude histone H1 binding, but it facilitates its association with the nucleosome. J Biol Chem. 2005;280(6):4975–4982. doi: 10.1074/jbc.M410203200. [DOI] [PubMed] [Google Scholar]
- Jenuwein T, Allis CD. Translating the histone code. Science. 2001;293(5532):1074–1080. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- Ju BG, Solum D, Song EJ, Lee KJ, Rose DW, Glass CK, Rosenfeld MG. Activating the PARP-1 sensor component of the groucho/TLE1 corepressor complex mediates a CaMKinase IIdelta-dependent neurogenic gene activation pathway. Cell. 2004;119(6):815–829. doi: 10.1016/j.cell.2004.11.017. [DOI] [PubMed] [Google Scholar]
- Kao CF, Hillyer C, Tsukuda T, Henry K, Berger S, Osley MA. Rad6 plays a role in transcriptional activation through ubiquitylation of histone H2B. Genes Dev. 2004;18(2):184–195. doi: 10.1101/gad.1149604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komarnitsky P, Cho EJ, Buratowski S. Different phosphorylated forms of RNA polymerase II and associated mRNA processing factors during transcription. Genes Dev. 2000;14(19):2452–2460. doi: 10.1101/gad.824700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krogan NJ, Kim, Tong A, Golshani A, Cagney G, Canadien V, Richards DP, Beattie BK, Emili A, Boone C, Greenblatt J. Methylation of histone H3 by Set2 in Saccharomyces cerevisiae is linked to transcriptional elongation by RNA polymerase II. Mol Cell Biol. 2003;23(12):4207–4218. doi: 10.1128/MCB.23.12.4207-4218.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levinger L, Varshavsky A. Selective arrangement of ubiquitinated and D1 protein-containing nucleosomes within the Drosophila genome. Cell. 1982;28:375–385. doi: 10.1016/0092-8674(82)90355-5. [DOI] [PubMed] [Google Scholar]
- Li B, Carey M, Workman JL. The role of chromatin during transcription. Cell. 2007;128(4):707–719. doi: 10.1016/j.cell.2007.01.015. [DOI] [PubMed] [Google Scholar]
- Li B, Howe L, Anderson S, Yates JR, III, Workman JL. The Set2 histone methyltransferase functions through the phosphorylated carboxyl-terminal domain of RNA polymerase II. J Biol Chem. 2003;278(11):8897–8903. doi: 10.1074/jbc.M212134200. [DOI] [PubMed] [Google Scholar]
- Luger K, Mader AW, Richmond RK, Sargent DF, Richmond TJ. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature. 1997;389(6648):251–260. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- Martinez E, Palhan VB, Tjernberg A, Lymar ES, Gamper AM, Kundu TK, Chait BT, Roeder RG. Human STAGA complex is a chromatin-acetylating transcription coactivator that interacts with pre-mRNA splicing and DNA damage-binding factors in vivo. Mol Cell Biol. 2001;21(20):6782–6795. doi: 10.1128/MCB.21.20.6782-6795.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maytal-Kivity V, Reis N, Hofmann K, Glickman MH. MPN+, a putative catalytic motif found in a subset of MPN domain proteins from eukaryotes and prokaryotes, is critical for Rpn11 function. BMC Biochem. 2002;3:28. doi: 10.1186/1471-2091-3-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metzger E, Wissmann M, Yin N, Muller JM, Schneider R, Peters AH, Gunther T, Buettner R, Schule R. LSD1 demethylates repressive histone marks to promote androgen-receptor-dependent transcription. Nature. 2005;437(7057):436–439. doi: 10.1038/nature04020. [DOI] [PubMed] [Google Scholar]
- Misteli T, Gunjan A, Hock R, Bustin M, Brown DT. Dynamic binding of histone H1 to chromatin in living cells. Nature. 2000;408:877–881. doi: 10.1038/35048610. [DOI] [PubMed] [Google Scholar]
- Ng HH, Xu RM, Zhang Y, Struhl K. Ubiquitination of histone H2B by Rad6 is required for efficient Dot1-mediated methylation of histone H3 lysine 79. J Biol Chem. 2002;277(38):34655–34657. doi: 10.1074/jbc.C200433200. [DOI] [PubMed] [Google Scholar]
- Nickel BE, Allis CD, Davie JR. Ubiquitinated histone H2B is preferentially located in transcriptionally active chromatin. Biochemistry. 1989;28:958–963. doi: 10.1021/bi00429a006. [DOI] [PubMed] [Google Scholar]
- Nickel BE, Davie JR. Structure of polyubiquitinated histone H2A. Biochemistry. 1989;28:964–968. doi: 10.1021/bi00429a007. [DOI] [PubMed] [Google Scholar]
- Nightingale KP, Wellinger RE, Sogo JM, Becker PB. Histone acetylation facilitates RNA polymerase II transcription of the Drosophila hsp26 gene in chromatin. EMBO J. 1998;17(10):2865–2876. doi: 10.1093/emboj/17.10.2865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogryzko VV, Kotani T, Zhang X, Schiltz RL, Howard T, Yang XJ, Howard BH, Qin J, Nakatani Y. Histone-like TAFs within the PCAF histone acetylase complex. Cell. 1998;94(1):35–44. doi: 10.1016/s0092-8674(00)81219-2. [DOI] [PubMed] [Google Scholar]
- Osley MA. H2B ubiquitylation: the end is in sight. Biochim Biophys Acta. 2004;1677(13):74–78. doi: 10.1016/j.bbaexp.2003.10.013. [DOI] [PubMed] [Google Scholar]
- Pavri R, Zhu B, Li G, Trojer P, Mandal S, Shilatifard A, Reinberg D. Histone H2B monoubiquitination functions cooperatively with FACT to regulate elongation by RNA polymerase II. Cell. 2006;125(4):703–717. doi: 10.1016/j.cell.2006.04.029. [DOI] [PubMed] [Google Scholar]
- Pham AD, Sauer F. Ubiquitin-activating/conjugating activity of TAFII250, a mediator of activation of gene expression in Drosophila. Science. 2000;289:2357–2360. doi: 10.1126/science.289.5488.2357. [DOI] [PubMed] [Google Scholar]
- Qian C, Zhang Q, Li S, Zeng L, Walsh MJ, Zhou MM. Structure and chromosomal DNA binding of the SWIRM domain. Nat Struct Mol Biol. 2005;12(12):1078–1085. doi: 10.1038/nsmb1022. [DOI] [PubMed] [Google Scholar]
- Robzyk K, Recht J, Osley MA. Rad6-dependent ubiquitination of histone H2B in yeast. Science. 2000;287(5452):501–504. doi: 10.1126/science.287.5452.501. [DOI] [PubMed] [Google Scholar]
- Seligson DB, Horvath S, Shi T, Yu H, Tze S, Grunstein M, Kurdistani SK. Global histone modification patterns predict risk of prostate cancer recurrence. Nature. 2005;435(7046):1262–1266. doi: 10.1038/nature03672. [DOI] [PubMed] [Google Scholar]
- Shahbazian MD, Zhang K, Grunstein M. Histone H2B ubiquitylation controls processive methylation but not monomethylation by Dot1 and Set1. Mol Cell. 2005;19(2):271–277. doi: 10.1016/j.molcel.2005.06.010. [DOI] [PubMed] [Google Scholar]
- Shi YJ, Matson C, Lan F, Iwase S, Baba T, Shi Y. Regulation of LSD1 histone demethylase activity by its associated factors. Mol Cell. 2005;19(6):857–864. doi: 10.1016/j.molcel.2005.08.027. [DOI] [PubMed] [Google Scholar]
- Sun ZW, Allis CD. Ubiquitination of histone H2B regulates H3 methylation and gene silencing in yeast. Nature. 2002;418(6893):104–108. doi: 10.1038/nature00883. [DOI] [PubMed] [Google Scholar]
- Swerdlow PS, Schuster T, Finley D. A conserved sequence in histone H2A which is a ubiquitination site in higher eucaryotes is not required for growth in Saccharomyces cerevisiae. Mol Cell Biol. 1990;10(9):4905–4911. doi: 10.1128/mcb.10.9.4905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorne AW, Sautiere P, Briand G, Crane-Robinson C. The structure of ubiquitinated histone H2B. EMBO J. 1987;6:1005–1010. doi: 10.1002/j.1460-2075.1987.tb04852.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tochio N, Umehara T, Koshiba S, Inoue M, Yabuki T, Aoki M, Seki E, Watanabe S, Tomo Y, Hanada M, Ikari M, Sato M, Terada T, Nagase T, Ohara O, Shirouzu M, Tanaka A, Kigawa T, Yokoyama S. Solution structure of the SWIRM domain of human histone demethylase LSD1. Structure. 2006;14(3):457–468. doi: 10.1016/j.str.2005.12.004. [DOI] [PubMed] [Google Scholar]
- Vakoc CR, Sachdeva MM, Wang H, Blobel GA. Profile of histone lysine methylation across transcribed mammalian chromatin. Mol Cell Biol. 2006;26(24):9185–9195. doi: 10.1128/MCB.01529-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Knaap JA, Kumar BR, Moshkin YM, Langenberg K, Krijgsveld J, Heck AJ, Karch F, Verrijzer CP. GMP synthetase stimulates histone H2B deubiquitylation by the epigenetic silencer USP7. Mol Cell. 2005;17(5):695–707. doi: 10.1016/j.molcel.2005.02.013. [DOI] [PubMed] [Google Scholar]
- Varambally S, Dhanasekaran SM, Zhou M, Barrette TR, Kumar-Sinha C, Sanda MG, Ghosh D, Pienta KJ, Sewalt RG, Otte AP, Rubin MA, Chinnaiyan AM. The polycomb group protein EZH2 is involved in progression of prostate cancer. Nature. 2002;419(6907):624–629. doi: 10.1038/nature01075. [DOI] [PubMed] [Google Scholar]
- Vassilev AP, Rasmussen HH, Christensen EI, Nielsen S, Celis JE. The levels of ubiquitinated histone H2A are highly upregulated in transformed human cells: partial colocalization of uH2A clusters and PCNA/cyclin foci in a fraction of cells in S-phase. J Cell Sci. 1995;108:1205–1215. doi: 10.1242/jcs.108.3.1205. [DOI] [PubMed] [Google Scholar]
- Verma R, Aravind L, Oania R, McDonald WH, Yates JR, 3rd, Koonin EV, Deshaies RJ. Role of Rpn11 metalloprotease in deubiquitination and degradation by the 26S proteasome. Science. 2002;298(5593):611–615. doi: 10.1126/science.1075898. [DOI] [PubMed] [Google Scholar]
- Wang H, Wang L, Erdjument-Bromage H, Vidal M, Tempst P, Jones RS, Zhang Y. Role of histone H2A ubiquitination in Polycomb silencing. Nature. 2004;431(7010):873–878. doi: 10.1038/nature02985. [DOI] [PubMed] [Google Scholar]
- Wang H, Zhai L, Xu J, Joo HY, Jackson S, Erdjument-Bromage H, Tempst P, Xiong Y, Zhang Y. Histone H3 and H4 ubiquitylation by the CUL4-DDB-ROC1 ubiquitin ligase facilitates cellular response to DNA damage. Mol Cell. 2006;22(3):383–394. doi: 10.1016/j.molcel.2006.03.035. [DOI] [PubMed] [Google Scholar]
- Wood A, Krogan NJ, Dover J, Schneider J, Heidt J, Boateng MA, Dean K, Golshani A, Zhang Y, Greenblatt JF, Johnston M, Shilatifard A. Bre1, an E3 ubiquitin ligase required for recruitment and substrate selection of Rad6 at a promoter. Mol Cell. 2003;11(1):267–274. doi: 10.1016/s1097-2765(02)00802-x. [DOI] [PubMed] [Google Scholar]
- Xiao T, Hall H, Kizer KO, Shibata Y, Hall MC, Borchers CH, Strahl BD. Phosphorylation of RNA polymerase II CTD regulates H3 methylation in yeast. Genes Dev. 2003;17(5):654–663. doi: 10.1101/gad.1055503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamane K, Toumazou C, Tsukada Y, Erdjument-Bromage H, Tempst P, Wong J, Zhang Y. JHDM2A, a JmjC-containing H3K9 demethylase, facilitates transcription activation by androgen receptor. Cell. 2006;125(3):483–495. doi: 10.1016/j.cell.2006.03.027. [DOI] [PubMed] [Google Scholar]
- Yao T, Cohen RE. A cryptic protease couples deubiquitination and degradation by the proteasome. Nature. 2002;419(6905):403–407. doi: 10.1038/nature01071. [DOI] [PubMed] [Google Scholar]
- Zhang Y. Transcriptional regulation by histone ubiquitination and deubiquitination. Genes Dev. 2003;17(22):2733–2740. doi: 10.1101/gad.1156403. [DOI] [PubMed] [Google Scholar]
- Zhu B, Zheng Y, Pham AD, Mandal SS, Erdjument-Bromage H, Tempst P, Reinberg D. Monoubiquitination of human histone H2B: the factors involved and their roles in HOX gene regulation. Mol Cell. 2005;20(4):601–611. doi: 10.1016/j.molcel.2005.09.025. [DOI] [PubMed] [Google Scholar]
- Zhu P, Baek SH, Bourk EM, Ohgi KA, Garcia-Bassets I, Sanjo H, Akira S, Kotol PF, Glass CK, Rosenfeld MG, Rose DW. Macrophage/cancer cell interactions mediate hormone resistance by a nuclear receptor derepression pathway. Cell. 2006;124(3):615–629. doi: 10.1016/j.cell.2005.12.032. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


