Abstract
Unsaturated glucuronyl hydrolase (UGL) categorized into the glycoside hydrolase family 88 catalyzes the hydrolytic release of an unsaturated glucuronic acid from glycosaminoglycan disaccharides, which are produced from mammalian extracellular matrices through the β-elimination reaction of polysaccharide lyases. Here, we show enzyme characteristics of pathogenic streptococcal UGLs and structural determinants for the enzyme substrate specificity. The putative genes for UGL and phosphotransferase system for amino sugar, a component of glycosaminoglycans, are assembled into a cluster in the genome of pyogenic and hemolytic streptococci such as Streptococcus agalactiae, Streptococcus pneumoniae, and Streptococcus pyogenes, which produce extracellular hyaluronate lyase as a virulent factor. The UGLs of these three streptococci were overexpressed in Escherichia coli cells, purified, and characterized. Streptococcal UGLs degraded unsaturated hyaluronate and chondroitin disaccharides most efficiently at approximately pH 5.5 and 37 °C. Distinct from Bacillus sp. GL1 UGL, streptococcal UGLs preferred sulfated substrates. DNA microarray and Western blotting indicated that the enzyme was constitutively expressed in S. agalactiae cells, although the expression level increased in the presence of glycosaminoglycan. The crystal structure of S. agalactiae UGL (SagUGL) was determined at 1.75 Å resolution by x-ray crystallography. SagUGL adopts α6/α6-barrel structure as a basic scaffold similar to Bacillus UGL, but the arrangement of amino acid residues in the active site differs between the two. SagUGL Arg-236 was found to be one of the residues involved in its activity for the sulfated substrate through structural comparison and site-directed mutagenesis. This is the first report on the structure and function of streptococcal UGLs.
Cell surface polysaccharides play an important role in linking neighboring cells and protecting cells against physicochemical stress such as osmotic pressure or invasion by pathogens. Glycosaminoglycans such as chondroitin, hyaluronan, and heparin are highly negatively charged polysaccharides with a repeating disaccharide unit consisting of an uronic acid residue (glucuronic or iduronic acid) and an amino sugar residue (glucosamine or galactosamine) (1), and they are widely present in mammalian cells as an extracellular matrix responsible for cell-to-cell association, cell signaling, and cell growth and differentiation (2). For example, in humans, glycosaminoglycans exist in tissues such as the eye, brain, liver, skin, and blood (3). Except for hyaluronan, glycosaminoglycans such as chondroitin sulfate, dermatan sulfate, keratan sulfate, heparin sulfate, and heparan sulfate are often sulfated. Chondroitin consists of d-glucuronic acid (GlcA)2 and N-acetyl-d-galactosamine (GalNAc) with a sulfate group(s) at position 4 or 6 or both (4). Hyaluronan, is composed of GlcA and N-acetyl-d-glucosamine (GlcNAc) (5).
The adhesion of pathogenic bacteria to mammalian cells is regarded as a primary mechanism of bacterial infection, followed by secondary effects of the infectious process. Polysaccharides, including the glycosaminoglycans that form part of the cell surface matrix, are typical targets for microbial pathogens that invade host cells, and many specific interactions between pathogens and these polysaccharides have been described (6). Glycosaminoglycans in the extracellular matrix are also degraded enzymatically by hydrolases and lyases (1). Generally, hydrolases cleave the glycoside bonds between the glycosyl oxygen and the anomeric carbon atom through the addition of water and play an important role in glycosaminoglycan metabolism in mammals (7). On the other hand, bacterial pathogens invading host cells degrade glycosaminoglycans through the action of lyases. Bacterial polysaccharide lyases recognize the uronic acid residue in polysaccharides, cleave the glycoside bonds through the β-elimination reaction without water addition, and produce unsaturated saccharides with the unsaturated uronic acid residue having a C C double bond at the nonreducing terminus (8).
C double bond at the nonreducing terminus (8).
Streptococci such as group B Streptococcus agalactiae, group nonassigned Streptococcus pneumoniae, and group A Streptococcus pyogenes are typical pyogenic and hemolytic pathogens causing severe infections (e.g. pneumonia, bacteremia, sinusitis, or meningitis) (9–11). In S. pneumoniae, hyaluronate lyase, neuraminidases, autolysin, choline-binding protein A, and pneumococcal surface protein A are suggested to function as cell surface virulent factors (12). Hyaluronate lyase degrades the extracellular matrix component hyaluronan in mammalian cells through the β-elimination reaction and releases unsaturated disaccharide, indicating that the enzyme produced by pathogenic bacteria functions as a spreading factor (13). Because hyaluronate lyase is commonly produced by the three pyogenic and hemolytic streptococci (14–16), the structure and function of their enzymes have been intensively studied (17, 18). Groups A, B, C, and G streptococci also produce hyaluronate lyase (19), suggesting that the enzyme is ubiquitously present in pathogenic streptococci. Streptococcal hyaluronate lyase can also act on sulfated and nonsulfated chondroitin (20). The metabolism of the resultant unsaturated disaccharides in streptococci, however, remains to be clarified.
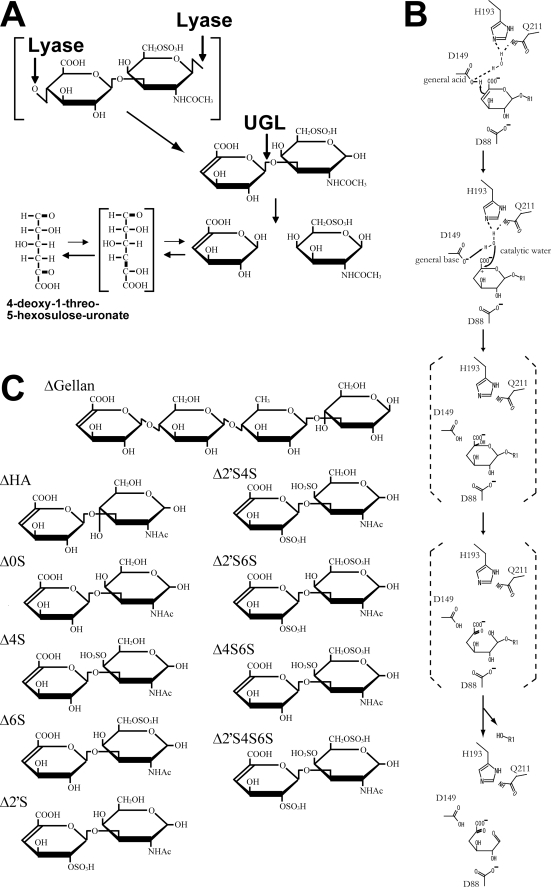
Unsaturated glucuronyl hydrolase (UGL), a member of the glycoside hydrolase family 88 in the CAZY data base (21), acts on unsaturated oligosaccharides having an unsaturated GlcA (ΔGlcA) with β-glycoside bond, such as ΔGlcA-GalNAc produced by chondroitin lyase and ΔGlcA-GlcNAc produced by hyaluronate lyase (22) (Fig. 1A). We have first identified the UGL-coding gene in Bacillus sp. GL1 (23) and clarified the structure and function of the enzyme by x-ray crystallography (24–27). The enzyme reaction generates ΔGlcA and the leaving saccharide. ΔGlcA is spontaneously converted to 4-deoxy-1-threo-5-hexosulose-uronate (Fig. 1A) because the ringed form of ΔGlcA has not been obtained because of keto-enole equilibrium (23, 28). In contrast with general glycoside hydrolases with retention or inversion catalytic mechanism of an anomeric configuration, UGL uniquely triggers hydrolysis of vinyl ether groups in unsaturated saccharides but not of the glycoside bond (26) (Fig. 1B). This article deals with the characteristics of streptococcal UGLs by using recombinant enzymes, gene expression in S. agalactiae cells by DNA microarray, and structural determinants of S. agalactiae UGL for substrate specificity by x-ray crystallography and site-directed mutagenesis.
FIGURE 1.
UGL reaction. A, degradation scheme of Δ6S by UGL. B, catalytic reaction mechanism of UGL. C, structures of unsaturated oligosaccharides. ΔGellan, unsaturated gellan tetrasaccharide; ΔHA, unsaturated hyaluronan disaccharide; Δ0S, unsaturated chondroitin disaccharide; Δ2′S, unsaturated chondroitin disaccharide sulfated at C-2 position of ΔGlcA residue; Δ2′S4S, unsaturated chondroitin disaccharide sulfated at C-2 position of ΔGlcA residue and C-4 position of GalNAc residue; Δ2′S6S, unsaturated chondroitin disaccharide sulfated at C-2 position of ΔGlcA residue and C-6 position of GalNAc residue; Δ4S6S, unsaturated chondroitin disaccharide sulfated at C-4 and C-6 positions of GalNAc residue; Δ2′S4S6S, unsaturated chondroitin disaccharide sulfated at C-2 position of ΔGlcA residue and C-4 and C-6 positions of GalNAc residue.
EXPERIMENTAL PROCEDURES
Homology and Sequence Alignment Analysis
To find novel proteins homologous to Bacillus sp. GL1 UGL (BacillusUGL), the BLAST and CLUSTALW programs were used to search for a sequence similarity and for multiple sequence alignment, respectively. Both programs are available on the DDBJ server (http://www.ddbj.nig.ac.jp/).
Microorganisms and Culture Conditions
S. agalactiae strain NEM316 (CIP 82.45) was from the Institut Pasteur. S. pneumoniae strain R6 (ATCC BAA-255) and S. pyogenes strain M1 GAS SF370 (ATCC 700294) were from the American Type Culture Collection. For genome DNA isolation, the three streptococci were statically grown at 30 °C under 5% CO2 in tryptic soy broth (Difco) containing 5% defibrinated sheep blood (Nippon Bio-Test Laboratories). To investigate gene expression, S. agalactiae cells were statically grown at 30 °C in tryptic soy broth with or without hyaluronan (Fluka). As a host for plasmid amplification, Escherichia coli strain DH5α (Toyobo) cells were cultured at 37 °C in LB medium (29) containing sodium ampicillin (0.1 mg/ml). E. coli strain HMS174(DE3)pLysS (Novagen) was used for expressing the three streptococcal UGLs. For expression in E. coli, the cells were aerobically precultured at 30 °C in LB medium supplemented with sodium ampicillin (0.1 mg/ml). At ∼0.5 turbidity at 600 nm, isopropyl-β-d-thiogalactopyranoside was added to the culture at 0.1 mm final concentration, and the cells were further cultured at 16 °C for 44 h.
DNA Manipulations
Genomic DNA isolation, subcloning, transformation, and gel electrophoresis were performed as described previously (29). The nucleotide sequences of streptococcal UGL genes were determined by the dideoxy chain termination method using automated DNA sequencer model 377 (Applied Biosystems) (30). Restriction endonucleases and DNA modifying enzymes were from Takara Bio and Toyobo.
Assays for Enzymes and Proteins
Streptococcal UGLs were assayed by monitoring the decrease in absorbance at 235 nm arising from the double bond (molar extinction coefficient ϵ235 = 4,800 m−1 cm−1) in the substrate. Standard assay was conducted at 30 °C in 20 mm Tris-HCl (pH 7.5), 0.15 m NaCl, and 0.2 mm substrate. For kinetic assay, the enzyme assay was performed three times using various concentrations (0.1–0.8 mm) of unsaturated chondroitin disaccharide sulfated at C-6 position of GalNAc residue (Δ6S) (Sigma) as a substrate. One unit of enzyme activity was defined as the amount of enzyme required to degrade 1.0 μmol of substrate/min by using a cuvette with a 1-cm light path. TLC was performed to investigate the enzyme activity using a high concentration of substrate and enzyme as described previously (23). Protein content was determined by the Bradford method (31), with bovine serum albumin as the standard. For the purified enzyme, protein concentration was estimated by measuring absorbance at 280 nm. Absorbance coefficients of S. agalactiae UGL (SagUGL), S. pneumoniae UGL (SpnUGL), and S. pyogenes UGL (SpyUGL) are 2.30, 2.15, and 2.21 for 1 mg/ml protein, respectively.
Construction of the Overexpression System
An overexpression system for SagUGL, SpnUGL, and SpyUGL was constructed in E. coli cells as follows. To clone the streptococcal UGL genes, PCR was conducted in a reaction mixture (0.1 ml) consisting of 5 units KOD polymerase (Toyobo), 0.25 μg of genomic DNA, 40 pmol of forward and reverse primers, 20 nmol of dNTPs, 100 nmol of MgCl2, 5 μl of dimethyl sulfoxide, and the commercial reaction buffer supplied with KOD polymerase. Forward and reverse primers for the SagUGL gene were 5′-GGCATATGATGAAAATAAAACCGGTCAAGG-3′ and 5′-CCCTCGAGTTACCAATAAAGTTCCCAGTCT-3′; for the SpnUGL gene, 5′-GGCATATGATAAAAAAGGTTACGATTGAAA-3′ and 5′-GGCTCGAGCTACCAATATAGGTTCCAGTCT-3′; and for the SpyUGL gene, 5′-GGCCATGGCACGACCTTTAAAAACCATTGC-3′ and 5′-GGCTCGAGTTACCAGTAAGGGTTCCAGTCT-3′, with a restriction site (NdeI, NcoI, or XhoI, underlined) added to each of their 5′ regions. PCR conditions were as follows: 98 °C for 30 s, 50 °C for 2 s, and 74 °C for 2 min with a total of 30 cycles. Final extension was performed by one cycle at 72 °C for 10 min. Nucleotide sequences of the PCR products were confirmed to completely match those of streptococcal UGL genes by DNA sequencing. The PCR products were ligated with HincII-digested pUC119 (Takara Bio), and the resultant plasmids were digested with NdeI or NcoI and XhoI to isolate streptococcal UGL genes. DNA fragments for streptococcal UGL genes were ligated with NdeI or NcoI and XhoI-digested pET21b or pET21d (Novagen). The resultant plasmids were designated pET21b-SagUGL, pET21b-SpnUGL, and pET21d-SpyUGL.
Purification
Unless otherwise specified, all operations were carried out at 0–4 °C. E. coli cells harboring pET21b-SagUGL, pET21b-SpnUGL, or pET21d-SpyUGL were grown in 3.0 liters of LB medium, collected by centrifugation at 6,000 × g and 4 °C for 5 min, washed with 20 mm potassium phosphate (pH 7.0), and then resuspended in the same buffer. The cells were ultrasonically disrupted (Insonator model 201M; Kubota) at 0 °C and 9 kHz for 20 min, and the clear solution obtained by centrifugation at 20,000 × g and 4 °C for 20 min was used as the cell extract. SagUGL was purified from the cell extract by anion exchange chromatography (DEAE-Toyopearl 650M; Tosoh, 2.6 × 9.5 cm), followed by anion exchange chromatography (MonoQ; GE Healthcare, 1.0 × 10 cm), and finally by gel filtration chromatography (GFC) (HiLoad 16/60 Superdex 75 pg; GE Healthcare, 1.6 × 60 cm). SpnUGL was purified from the cell extract by anion exchange chromatography (DEAE-Toyopearl 650M), followed by ammonium sulfate precipitation (50–70% saturation), and finally by GFC (HiLoad 16/60 Superdex 75 pg). SpyUGL was purified from the cell extract by ammonium sulfate precipitation (50–70% saturation) and by GFC (Sephacryl S-200HR; GE Healthcare, 2.7 × 58 cm). The active fractions were combined and dialyzed against 20 mm Tris-HCl (pH 7.5). The dialysate was concentrated to ∼8 mg/ml by ultrafiltration using a Centriprep (molecular mass cut-off, 10 kDa) (Millipore). The concentrate was used as the purified enzyme source.
Determination of Molecular Mass
To determine the molecular weight of streptococcal UGLs, SDS-PAGE (32) and GFC (HiLoad 16/60 Superdex 75 pg) were performed.
Optimal pH and Temperature and Thermal Stability
The experiments were conducted at 30 °C by using Δ6S as the substrate and purified streptococcal UGL. To determine the optimal pH, reactions were performed at 30 °C in the following 20 mm buffers: sodium acetate, potassium phosphate, and Tris-HCl. To determine the optimal temperature, reactions were performed at various temperatures in 20 mm Tris-HCl (pH 7.5). To determine the thermal stability, after preincubation of the enzyme at various temperatures for 5 min, residual activity was measured at 30 °C in 20 mm Tris-HCl (pH 7.5).
Substrate Specificity
Substrate specificity of streptococcal UGLs was determined using various unsaturated saccharides (Fig. 1C) at 0.2 mm, including Δ6S, unsaturated gellan tetrasaccharide (33), unsaturated hyaluronan disaccharide, unsaturated chondroitin disaccharide, unsaturated chondroitin disaccharide sulfated at the C-4 position of GalNAc residue (Δ4S), unsaturated chondroitin disaccharide sulfated at the C-2 position of ΔGlcA residue, unsaturated chondroitin disaccharide sulfated at the C-2 position of ΔGlcA residue and the C-4 position of GalNAc residue, unsaturated chondroitin disaccharide sulfated at the C-2 position of ΔGlcA residue and the C-6 position of GalNAc residue, unsaturated chondroitin disaccharide sulfated at the C-4 and C-6 positions of GalNAc residue, and unsaturated chondroitin disaccharide sulfated at the C-2 position of ΔGlcA residue and the C-4 and C-6 positions of GalNAc residue. These unsaturated disaccharides were from Seikagaku Kogyo.
DNA Microarray
Total RNAs for DNA microarray were prepared as follows. S. agalactiae cells grown at exponential growth phase (turbidity at 600 nm, 0.7) were collected at 6,000 × g and 4 °C for 5 min and treated with RNAprotect bacteria reagent (Qiagen) for stabilization of RNA. Based on the manufacturer's instructions, total RNAs were extracted with an RNeasy kit (Qiagen) and treated with DNase (RNase-free DNase set; Qiagen) supplied with the kit. The genome sequence and open reading frame prediction for S. agalactiae were provided by the Genome Sequence Project (9) (gib.genes.nig.ac.jp/single/index.php?spid=Saga_NEM316). DNA microarray from NimbleGen Systems includes 2,094 target genes fixed on the glass slide by fixing at least 20 unique probes consisting of 60-mer synthetic oligonucleotides for each gene. Biotin labeling, fragmentation, and hybridization were done by NimbleGen Systems. The arrays were scanned with an Axon Genepix 4000B scanner at 532- and 5-mm resolution and analyzed by quantile normalization and robust multiarray averaging (34). These normalized data were processed with NANDEMO analysis 1.0.0 software (Roche Applied Science). Student's t test for analyzing the mean log ratios of two samples and subsequent Bonferroni adjustment for multiple testing (2,094 open reading frames on arrays) were applied as a rigorous criterion for significantly changed signal intensity. Changes with p < 0.05 were considered statistically significant.
Western Blotting
Antibodies to the purified SpyUGL were raised in a rabbit, and the serum was used as polyclonal antibodies against streptococcal UGLs. To investigate SagUGL expression in S. agalactiae, the cell extracts were subjected to SDS-PAGE, followed by Western blotting using anti-SpyUGL antibodies as described previously (35). Anti-IgG conjugated with horseradish peroxidase (GE Healthcare) and a POD immunostaining kit (Wako Pure Chemical Industries) were used to visualize the protein band.
Crystallization and Structure Determination
To determine the three-dimensional structure of SagUGL, the purified enzyme was crystallized by the hanging drop vapor diffusion method. The 3 μl of proteins (8 mg/ml in 20 mm Tris-HCl, pH 7.5) was mixed with equal volume of a reservoir solution (100 mm MES-NaOH, pH 6.0, 1.9 m ammonium sulfate). The protein solution was then incubated at 20 °C, and single crystals grew up for ∼1 month. The crystal was flash frozen under a cold nitrogen gas stream at −173 °C after soaking in mother liquor containing 20% glycerol as a cryoprotectant. X-ray diffraction data were collected at λ = 0.7 Å using a Jupiter 210 CCD detector (Rigaku) at BL38B1 station of SPring-8 (Hyogo, Japan). The data were processed and scaled up to 1.75 Å with the HKL2000 program (36). The structure was determined by molecular replacement with the Molrep program (37) supplied in a CCP4 program package (38) by using coordinates of BacillusUGL (Protein Data Bank code 1VD5) as an initial model. Structure refinement was conducted with the Refmac5 program (39). Randomly selected 5% reflections were excluded from refinement and used to calculate Rfree. After each refinement cycle, the model was adjusted manually using the winCoot program (40). Final model quality was checked with the PROCHECK program (41). Model structure of SagUGL binding to Δ4S was built using coordinates of chondroitin-4 sulfate (Protein Data Bank code 1C4S), followed by energy minimization with the Refmac5 program (39). Figures for the protein structure were prepared using the PyMol program (42). Coordinates used in this work were taken from the RCSB Protein Data Bank (43).
Site-directed Mutagenesis
To substitute Asp-115, Asp-175, Arg-236, and Arg-236 of SagUGL with Asn, Asn, His, and Ala, respectively, eight oligonucleotides were synthesized as follows: D115N sense primer, 5′-GCATTAGATCACCACAACTTAGGATTTCTTTACACACC-3′; D115N antisense primer, 5′-GGTGTGTAAAGAAATCCTAAGTTGTGGTGATCTAATGC-3′; D175N sense primer, 5′-CTATCGCTTAATTATCAATTGCTTACTTAATATCC-3′; D175N antisense primer, 5′-GGATATTAAGTAAGCAATTGATAATTAAGCGATAG-3′; R236H sense primer, 5′-CCGTTAAAAGGTGTCACACATCAGGGTTATAGTGATG-3′; R236H antisense primer, 5′-CATCACTATAACCCTGATGTGTGACACCTTTTAACGG-3′; R236A sense primer, 5′-CCGTTAAAAGGTGTCACAGCACAGGGTTATAGTGATG-3′; and R236A antisense primer, 5′-CATCACTATAACCCTGTGCTGTGACACCTTTTAACGG-3′. Underlining in the oligonucleotide sequences indicates the mutation positions. Site-directed mutagenesis was performed using the plasmid pET21b-SagUGL as a template and synthetic oligonucleotides as sense and antisense primers by employing methods of a QuikChange site-directed mutagenesis kit (Stratagene), except that KODplus polymerase (Toyobo) was used for PCRs. The resultant plasmids with a mutation were designated pD115N, pD175N, pR236H, and pR236A, respectively. The mutations were confirmed by DNA sequencing. E. coli host strain (HMS174(DE3)) cells were transformed with the plasmids, respectively.
RESULTS AND DISCUSSION
Streptococcal Proteins Similar to Glycoside Hydrolase Family 88 UGL
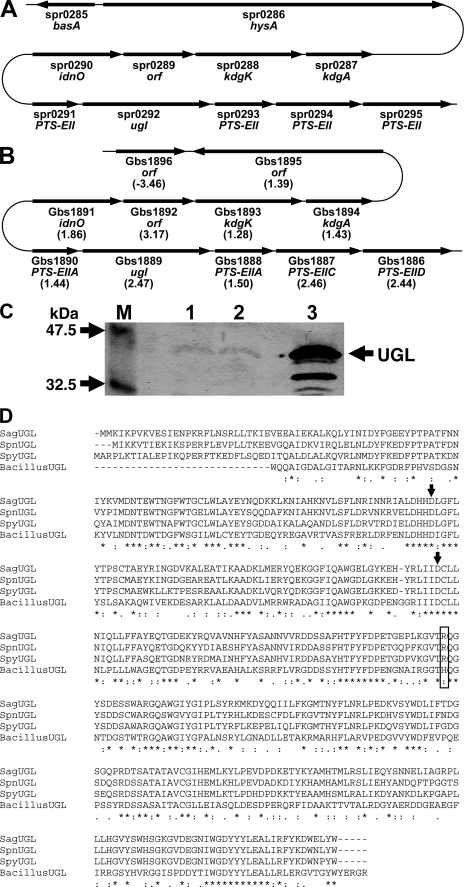
To find streptococcal proteins homologous to BacillusUGL (GenBankTM accession number AB019619), the BLAST program was used to search for sequence similarity against the streptococcal genome databases. As described previously (22), S. pneumoniae and S. pyogenes have an open reading frame similar to BacillusUGL: S. pneumoniae spr0292 (Uniprot ID, Q8DR77; 396 residues, 46.0 kDa) and S. pyogenes Spy0632 (Uniprot ID, Q9A0T3; 399 residues, 46.1 kDa). In S. pneumoniae, the gene for UGL homolog (spr0292) is located near the hyaluronate lyase (spr0286) gene (hysA) in the genome (10) (Fig. 2A), suggesting that the two proteins are cooperatively involved in degrading glycosaminoglycans. A UGL homologous protein is also encoded in the genome of S. agalactiae: S. agalactiae Gbs1889 (Uniprot ID, Q8E372; 398 residues, 46.6 kDa). Gbs1889 showed 40.3% identity with BacillusUGL in a 352-amino acid overlap, and the three streptococcal proteins are mutually similar (identity, over 69.1%) (Fig. 2D). Based on high identity, the three proteins are assigned to unsaturated glucuronyl hydrolase on each genome data base. In the three pathogenic streptococci, hyaluronate lyases are believed to function as a virulent factor (14–16), suggesting that UGL and hyaluronate lyase play an important role in the bacterial infection process. Because SpnUGL was predicted to be localized in the cytoplasm by the PSORT program (www.psort.org/) and BacillusUGL is demonstrated to be expressed in the intracellular fraction (23), streptococcal UGLs are probably cytoplasmic enzymes. Interestingly, the UGL gene of the three streptococci is located in a gene cluster for the putative phosphotransferase system to incorporate amino sugar (GalNAc), a component of glycosaminoglycans (Fig. 2, A and B). The phosphotransferase system consists of several domains such as enzymes IIA (E-IIA), IIB (E-IIB), IIC (E-IIC), and IID (E-IID) and incorporate solutes across the cytoplasmic membrane through phosphate addition to the substrates (44). The streptococcal genetic organization around the UGL gene strongly suggests that UGL could universally function in pathogenic bacteria as a virulent factor responsible for the complete degradation of glycosaminoglycans after depolymerization by polysaccharide lyase, followed by import of the resultant unsaturated disaccharides through phosphotransferase system. Furthermore, in the genome of the three streptococci, the putative genes coding for 5-keto-d-gluconate reductase, 2-keto-3-deoxygluconate kinase, and 2-keto-3-deoxy-6-phosphogluconate aldolase involved in the metabolism of 2-keto-3-deoxygluconate are located upstream of the UGL gene (Fig. 2, A and B). 5-Keto-d-gluconate reductase catalyzes a reversible reduction of 5-keto-d-gluconate to form d-gluconate (45). When 5-keto-d-gluconate reductase can also catalyze the reduction reaction of 4-deoxy-1-threo-5-hexosulose-uronate, a reaction product by UGL (Fig. 1A), the resultant product probably becomes a substrate for 2-keto-3-deoxygluconate kinase and is assimilated in the 2-keto-3-deoxygluconate metabolism pathway. The experimental data are required for elucidating the predicted pathway.
FIGURE 2.
UGL gene and protein in streptococci. A, S. pneumoniae gene cluster. B, S. agalactiae gene cluster. Except for Gbs1896, the values in parentheses indicate the increased level of each gene expression in S. agalactiae cells grown in the presence of hyaluronan compared with that in the absence of the polysaccharide based on the DNA microarray. The gene expression level except for Gbs1895 between the two bacterial cells was significantly different (p < 0.05). Details regarding the gene assignment have been described in the text. The open reading frame indicates a hypothetical protein. C, expression of the UGL protein in S. agalactiae cells. The cells grown in the presence and absence of hyaluronan were subjected to Western blotting using anti-SpyUGL antibodies. Lane M, prestained protein markers with molecular masses of 47.5 and 32.5 kDa; lane 1, S. agalactiae cells grown in the absence of hyaluronan; lane 2, S. agalactiae cells grown in the presence of hyaluronan; lane 3, purified SagUGL. Purified SagUGL in lane 3 included faint degraded products. D, amino acid sequence alignment of streptococcal and bacillus UGLs. The arrows indicate the catalytically important Asp residues. Possible streptococcal UGL arginine residues (SagUGL, Arg-236) involved in binding to sulfate group of Δ4S are boxed. Identical and similar amino acid residues among the four proteins are denoted by asterisks and dots, respectively.
Enzyme Characteristics
To examine whether streptococcal UGL-homologous proteins show unsaturated glucuronyl hydrolase activity, the proteins expressed in E. coli were purified (supplemental Table S1 and supplemental Fig. S1) and subjected to the enzyme assay. In the presence of each protein, the absorbance at 235 nm derived from the substrate in the reaction mixture decreased, indicating that the three proteins exhibit enzyme activity. Gbs1889, spr0292, and Spy0632 are designated SagUGL, SpnUGL, and SpyUGL, respectively. The properties of the purified enzymes are as follows.
Molecular Weight
The molecular masses of SagUGL, SpnUGL, and SpyUGL as determined by SDS-PAGE were 43 kDa (supplemental Fig. S1). These values were almost comparable with the theoretical one (46 kDa) deduced from the predicted amino acid sequences of the enzymes. In GFC on HiLoad 16/60 Superdex 75 pg, all of the enzymes were eluted between bovine serum albumin (67 kDa) and ovalbumin (43 kDa), indicating that the enzymes are monomeric.
pH and Temperature
SagUGL, SpnUGL, and SpyUGL were most active at approximately pH 5.5 in 20 mm potassium phosphate and at 37 °C (supplemental Fig. S2). ∼50% of enzyme activity was observed above 45 °C for 5 min in 20 mm Tris-HCl (pH 7.5) (supplemental Fig. S2).
Effect of Reducing Agents, Monosaccharides, and Metals
Table 1 shows the effects of different chemicals on enzyme. Streptococcal UGLs were significantly inhibited by uronic acid such as GlcA or galacturonic acid, suggesting that these saccharides were accommodated in the active site of the enzymes. Metal ions and thiol reagents at 1 mm had no significant effects on enzyme activity.
TABLE 1.
Effects of reducing agents, monosaccharides, and metals on streptococcal UGL activity
| Chemical | Concentration | Relative activity |
||
|---|---|---|---|---|
| SagUGL | SpnUGL | SpyUGL | ||
| mm | % | |||
| None | 100 | 100 | 100 | |
| Reducing agents | ||||
| Dithiothreitol | 1 | 120 | 98 | 100 |
| Glutathione (reduced) | 1 | 140 | 130 | 94 |
| 2-Mercaptoethanol | 1 | 120 | 90 | 80 |
| Sugars | ||||
| d-GlcA | 5 | 1.6 | 3.3 | 2.2 |
| d-Glucose | 5 | 120 | 100 | 60 |
| d-Mannose | 5 | 100 | 100 | 78 |
| d-Galactose | 5 | 120 | 110 | 79 |
| l-Rhamnose | 5 | 110 | 110 | 65 |
| d-Glucosamine | 5 | 100 | 130 | 98 |
| d-Sucrose | 5 | 110 | 100 | 81 |
| d-Xylose | 5 | 110 | 110 | 67 |
| d-Galacturonic acid | 5 | 0.37 | 1.1 | NDa |
| 2-Deoxyglucose | 5 | 97 | 100 | 89 |
| l-Fucose | 5 | 95 | 100 | 50 |
| GalNAc | 5 | 90 | 97 | 95 |
| GlcNAc | 5 | 83 | 98 | 96 |
| Metals | ||||
| CoCl2 | 1 | 45 | ND | ND |
| CaCl2 | 1 | 92 | 101 | 97 |
| MnCl2 | 1 | 73 | 95 | 23 |
| MgCl2 | 1 | 91 | 101 | 110 |
| ZnCl2 | 1 | ND | ND | 6.1 |
| AlCl3 | 1 | 35 | 140 | ND |
| LiCl | 1 | 76 | 94 | 130 |
| NaCl | 1 | 78 | 96 | 130 |
| KCl | 1 | 78 | 97 | 130 |
a ND, not detected; 0.2% < of the control (none).
Substrate Specificity
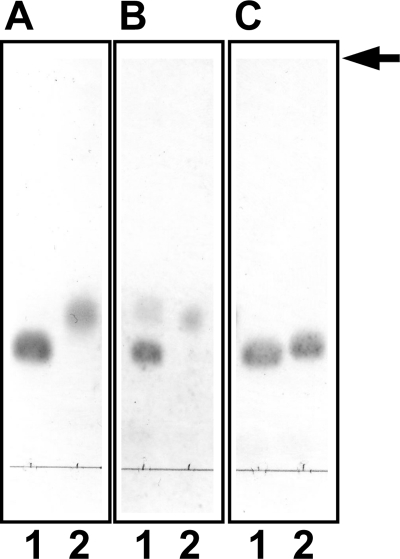
Table 2 shows substrate specificity of SagUGL, SpnUGL, and SpyUGL. Δ6S was degraded most efficiently by all the streptococcal UGLs. Unlike BacillusUGL (22), the unsaturated gellan tetrasaccharide produced from bacterial biofilm was almost inert on the streptococcal UGLs. Although the degradation level was extremely low, SagUGL and SpyUGL can act on unsaturated chondroitin disaccharide with sulfate group(s). To confirm no activity of BacillusUGL and SpnUGL on Δ4S, TLC was performed using a large amount of the enzymes. Δ4S was degraded by SpnUGL but not by BacillusUGL, whereas both enzymes acted on Δ6S (Fig. 3). Thus, there is a significant difference in the substrate specificity between streptococcal and bacillus UGLs. Streptococcal UGLs seem to have a tendency to act on sulfated substrates.
TABLE 2.
Substrate specificity of streptococcal and bacillus UGLs
The abbreviations used are as follows: ΔGellan, unsaturated gellan tetrasaccharide; ΔHA, unsaturated hyaluronan disaccharide; Δ0S, unsaturated chondroitin disaccharide; Δ2′S, unsaturated chondroitin disaccharide sulfated at C-2 position of ΔGlcA residue; Δ2′S4S, unsaturated chondroitin disaccharide sulfated at C-2 position of ΔGlcA residue and C-4 position of GalNAc residue; Δ2′S6S, unsaturated chondroitin disaccharide sulfated at C-2 position of ΔGlcA residue and C-6 position of GalNAc residue; Δ4S6S, unsaturated chondroitin disaccharide sulfated at C-4 and C-6 positions of GalNAc residue; Δ2′S4S6S, unsaturated chondroitin disaccharide sulfated at C-2 position of ΔGlcA residue and C-4 and C-6 positions of GalNAc residue.
| Substrate | Relative activity |
|||
|---|---|---|---|---|
| SagUGL | SpnUGL | SpyUGL | BacillusUGLa | |
| % | ||||
| Δ6S | 100 | 100 | 100 | 21 |
| Δ0S | 3.4 | 16 | 10 | 100 |
| ΔHA | 0.61 | 2.0 | 1.1 | 62 |
| Δ2′S | 0.011 | 0.31 | 0.34 | 17 |
| ΔGellan | 0.050 | NDb | 0.11 | 78 |
| Δ4S | 0.0090 | ND | 0.076 | ND |
| Δ2′S6S | 0.0044 | ND | 0.051 | ND |
| Δ4S6S | 0.019 | ND | 0.014 | ND |
| Δ2′S4S | 0.0075 | ND | ND | ND |
| Δ2′S4S6S | ND | ND | ND | ND |
a Cited from Ref. 22.
b ND, not detected; 0.002% < toward Δ6S.
FIGURE 3.
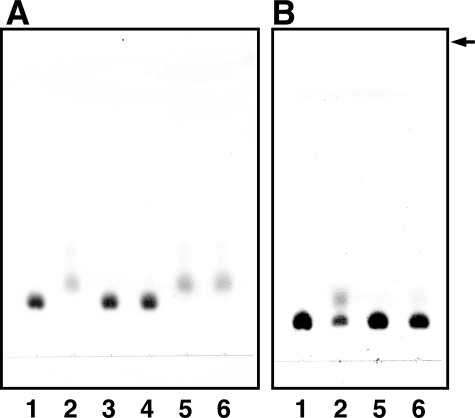
Degradation of Δ4S by SpnUGL but not by BacillusUGL. The 10 mm substrate Δ4S or Δ6S incubated at 30 °C with 2 mg/ml BacillusUGL (A) or SpnUGL (B) or without enzyme (C) was subjected to TLC analysis. Lane 1, Δ4S; lane 2, Δ6S. The arrow indicates the position to which the developing solvent reached.
Kinetics
The Km values of SagUGL, SpnUGL, and SpyUGL for Δ6S were 0.54 ± 0.19, 0.18 ± 0.039, and 0.39 ± 0.10 mm, respectively, suggesting that streptococcal UGLs mutually show similar affinity with the substrate. On the other hand, the kcat value of SagUGL is the highest (24 ± 4.3 s−1) in comparison with those of SpnUGL (1.3 ± 0.12 s−1) and SpyUGL (4.0 ± 0.65 s−1), indicating that SagUGL is relatively active on the substrate rather than the other two enzymes.
To the best of our knowledge, two UGLs from Bacillus sp. GL1 (23) and Flavobacterium heparinum (46) have been characterized, although F. heparinum UGL is specific to unsaturated disaccharides from heparin/heparan sulfate. The molecular weight, optimal reaction conditions, and thermal stability of streptococcal UGLs were comparable with those of Bacillus and F. heparinum UGLs. Except for the reaction turnover (kcat), there is no significant difference in enzyme characteristics among streptococcal UGLs. Distinct from BacillusUGL, the streptococcal UGLs were capable of degrading unsaturated disaccharides derived from mammalian sulfated glycosaminoglycans. This substrate specificity probably enables streptococci to readily invade mammalian cells through the degradation of extracellular sulfated glycosaminoglycans. Because SagUGL showed the highest activity, the expression and structural analysis of SagUGL is focused on hereafter.
Expression of UGL in Streptococcus
To investigate UGL gene expression in S. agalactiae cells, DNA microarray was conducted using the streptococcal cells grown with or without hyaluronan (supplemental Table S2). Hyaluronan induced the expression of the genetic cluster for UGL and the phosphotransferase system (Fig. 2B), although the increased level was not very high (less than 3-fold). Hyaluronan also slightly up-regulated the transcriptional level of putative genes involved in 2-keto-3-deoxygluconate metabolism. The SagUGL protein expression in both S. agalactiae cells grown with or without hyaluronan was confirmed by Western blotting with anti-SpyUGL antibodies (Fig. 2C). These results suggest that SagUGL is constitutively expressed, but the slight increase in its expression level occurs in the presence of hyaluronan.
Crystal Structure of SagUGL
To investigate structure determinants for different substrate specificity between streptococcal and bacillus UGLs, x-ray crystallography of SagUGL was performed. SagUGL yielded single crystals suitable for x-ray analysis (supplemental Fig. S1). The crystal structure of SagUGL was determined at 1.75 Å resolution via molecular replacement using BacillusUGL structure (Protein Data Bank code 1VD5) as an initial model. Data collection and model refinement statistics are summarized in Table 3. The final model contains one monomer enzyme starting from Met-2 to Trp-398, three sulfate ions, three glycerol molecules, and 407 water molecules. The N-terminal amino acid (Met-1) could not be assigned because of the thin electron density for the corresponding residue in the 2Fo − Fc map. Most (89.6%) nonglycine residues lie within the most favored regions of the Ramachandran plot as defined in the PROCHECK program (41), and no residues lie within the disallowed regions.
TABLE 3.
Statistics of SagUGL for x-ray diffraction data and structure refinement
| Data collection | |
| Wavelength (Å) | 0.70 |
| Resolution range (Å) | 50.00–1.75 (1.81–1.75)a |
| Space group | P3221 |
| Unit cell parameters (Å) | a = b = 116.08, c = 78.13 |
| Total observations | 774,026 |
| Independent reflections | 61,457 |
| Completeness (%) | 100 (100) |
| I/σ(I) | 33.2 (3.89) |
| Rmerge | 0.056 (0.38) |
| Refinement | |
| R factor (%) | 18.2 |
| Free R factor (%) | 21.4 |
| No. of molecules/asymmetric unit | 1 |
| No. of nonhydrogen atoms | |
| Protein | 3,329 |
| Heterogen | 33 |
| Solvent | 407 |
| Average B factor (Å2) | 21.1 |
| Root mean square deviation from ideal | |
| Bond length (Å) | 0.007 |
| Bond angles (degree) | 1.001 |
a The values for the outer resolution shell are given in parentheses.
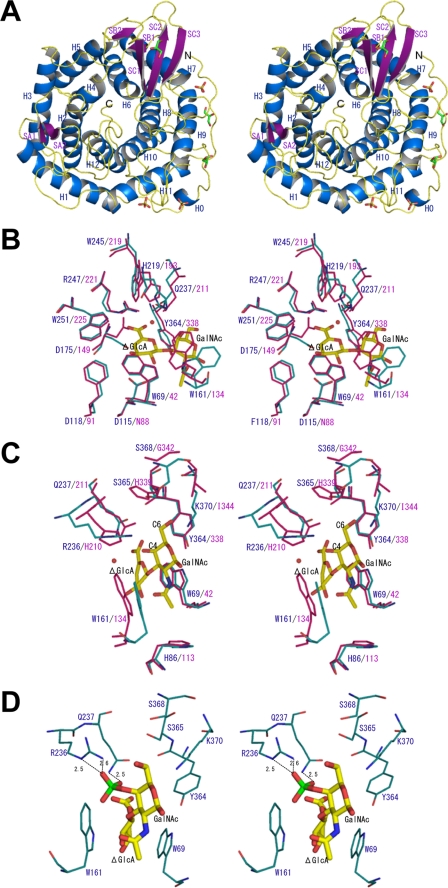
A ribbon diagram of SagUGL is shown in Fig. 4A. The enzyme is ∼45 × 45 × 40 Å in size. SagUGL consists of the α6/α6-barrel structure seen in the six-hairpin glycosylases superfamily of the SCOP data base (scop.mrc-lmb.cam.ac.uk/scop/), which contains glucoamylases (47), cellulase catalytic domains (48), N-acyl-d-glucosamine epimerase (49), and unsaturated galacturonyl hydrolase (50). The root mean square deviation value for overall 370 Cα atoms between SagUGL and BacillusUGL is 1.5 Å, indicating that their structures are essentially the same. According to the designation of the secondary structure element of BacillusUGL (24), SagUGL contains 12 α helices (H1–H12) and 7 β strands (SA1-SA2, SB1-SB2, and SC1-SC3). Helices H1, H3, H5, H7, H9, and H11 form the outer barrel, and the other helices, H2, H4, H6, H8, H10, and H12, form the inner barrel. The N-terminal 27 residues are peculiar to SagUGL. This N-terminal region was observed obviously in the crystal structure and lies along with outer barrel (Fig. 4A). One short α helix (H0) is additionally formed in the N-terminal loop region of SagUGL.
FIGURE 4.
Crystal structure of SagUGL (stereo diagram). A, the colors blue, purple, and yellow denote the secondary structure elements α-helices, β-sheets, and loops, respectively. Bound sulfate ions and glycerol molecules are represented as stick models. B and C, cyan represents SagUGL, and pink denotes BacillusUGL D88N in complex with unsaturated chondroitin disaccharide. Amino acid residues at subsite −1 (B) and +1 (C) are represented in the stick models. Substrate bound in D88N is also shown in the stick models. Carbon, oxygen, and nitrogen atoms of bound saccharides are colored by yellow, pink, and blue, respectively. D, energy-minimized model of SagUGL binding to Δ4S. Arg-236 and Gln-237 bind to the sulfate group of Δ4S through the formation of hydrogen bonds (broken lines). Hydrogen bond lengths are indicated. Carbon, oxygen, nitrogen, and sulfur atoms of bound saccharides are colored by yellow, pink, blue, and green, respectively.
Active Site Structure
To determine the structure/function relationship, x-ray crystallography of substrate-bound SagUGL was attempted but failed. Thus, structural comparison between SagUGL and ligand-bound BacillusUGL was conducted to identify the structural determinants for different substrate specificity between streptococcal and bacillus UGLs. In BacillusUGL, the central cleft surrounded by aromatic and positively charged residues functions as the active site (26). Crystal structures of BacillusUGL in complex with unsaturated chondroitin disaccharide, unsaturated hyaluronan disaccharide, and unsaturated gellan tetrasaccharide reveal that UGL is unusual in that no atom forms hydrogen bonds to the substrate glycosidic oxygen to be hydrolyzed (25, 26). Based on the complex structures and reaction product analysis using 18O water, the novel catalytic mechanism to hydrolyze the glycoside bond in which Asp-149 attacks the C-4 C-5 double bond of ΔGlcA was proposed (26) (Fig. 1B). Asp-149 acts as a general acid and base catalyst to protonate the ΔGlcA C-4 atom and to deprotonate the water molecule. The deprotonated water molecule attacks the ΔGlcA C-5 atom to yield unstable hemiketal; this is followed by spontaneous conversion to an aldehyde (4-deoxy-l-threo-5-hexosulose-uronate) and saccharide through hemiacetal formation and cleavage of the glycosidic bond. BacillusUGL strictly recognizes ΔGlcA at subsite −1 rather than the saccharide at subsite +1 (25). It is reasonable in that the enzyme acts on various unsaturated oligosaccharides but not on saturated oligosaccharides.
C-5 double bond of ΔGlcA was proposed (26) (Fig. 1B). Asp-149 acts as a general acid and base catalyst to protonate the ΔGlcA C-4 atom and to deprotonate the water molecule. The deprotonated water molecule attacks the ΔGlcA C-5 atom to yield unstable hemiketal; this is followed by spontaneous conversion to an aldehyde (4-deoxy-l-threo-5-hexosulose-uronate) and saccharide through hemiacetal formation and cleavage of the glycosidic bond. BacillusUGL strictly recognizes ΔGlcA at subsite −1 rather than the saccharide at subsite +1 (25). It is reasonable in that the enzyme acts on various unsaturated oligosaccharides but not on saturated oligosaccharides.
Most residues in the active site cleft of SagUGL and BacillusUGL are well conserved in both primary and tertiary structures (Figs. 2D and 4, B and C). In Fig. 4B, side chain conformation of the catalytic residue (Asp-149 in BacillusUGL and Asp-175 in SagUGL) differs between the two. The side chain of Asp-149 moves upon substrate binding (26), and conformation of SagUGL Asp-175 is similar to that of Asp-149 of substrate-free BacillusUGL. Side chain conformation of SagUGL Trp-161 and BacillusUGL Trp-134 is also different. This side chain stacks with sugar ring of various substrates at subsite +1 in BacillusUGL by altering its conformation. Therefore, the conformational differences in the two amino acid residues seen in Fig. 4B may not be effective for the difference in enzyme characteristics of SagUGL and BacillusUGL. On the other hand, amino acid residues at subsite +1 are somewhat different between the two (Fig. 4C). His-210 forming a hydrogen bond to O-4 of GalNAc residue in BacillusUGL in complex with unsaturated chondroitin disaccharide (26) is substituted with Arg-236 in SagUGL (Figs. 2D and 4C). His-339 interacting with C-6 of GalNAc residue in BacillusUGL is also replaced by Ser-365 in SagUGL. In addition to these substitutions of residues interacting with the substrate, other differences are observed at subsite +1. BacillusUGL Gly-342 and Ile-344 have no interaction with the substrate, but substitution of these residues with Ser-368 and Lys-370 residues in SagUGL may affect substrate binding, because their side chains are directed to C-6 atom of GalNAc and can interact with the substrate (Δ6S) when C-6 is sulfated (Fig. 4C). Although the overall electron density of SagUGL was good enough to determine the precise position of amino acid residues, that for a loop containing Arg-236 and Gln-237, was slightly poor to determine the exact side chain structure of some residues in the loop. This feature was the same in the structure determined using another crystal belonging to the space group of P21 (data not shown). These observations suggest conformational disorder of the loop. Because the mutant enzyme of BacillusUGL that substituted Gln-211, a neighbor residue of His-210 in BacillusUGL (Arg-236 of SagUGL), with alanine shows lower kcat and higher Km (26), the loop disorder is considered to regulate SagUGL enzymatic activity. SagUGL Arg-236, Ser-365, Ser-368, and Lys-370 are common to the other two SpnUGL and SpyUGL (Fig. 2D). The loop disorder and/or differences in residues (Arg-236 versus His-210, Ser-365 versus His-339, Ser-368 versus Gly-342, and Lys-370 versus Ile-344) are therefore attributed to the difference seen in substrate specificity between streptococcal and bacillus UGLs, i.e. BacillusUGL prefers unsulfated substrate and cannot degrade Δ4S, although SagUGL showed a high specificity for Δ6S and acted on Δ4S.
Amino Acid Residue Binding to Sulfate Group
Asp-149 of BacillusUGL is important for catalysis, and substitution of this residue with Asn reduced kcat by 1,000-fold compared with that of the wild-type enzyme (26). Mutant enzyme replaced Asp-88 by Asn also yields a decreased kcat value (1/10,000) and an increased Km value (10-fold), indicating the importance of this residue for catalytic reaction. To investigate roles of corresponding residues (Asp-175 and Asp-115) in SagUGL, mutants SagUGL D175N and D115N were constructed by site-directed mutagenesis. Compared with the wild-type enzyme with kcat of 24 ± 4.3 s−1 and Km of 0.54 ± 0.19 mm, substitution of Asp-175 with Asn in SagUGL decreased kcat 0.038 ± 0.067 s−1 but had no effect on Km (0.53 ± 0.18 mm). On the other hand, it was very difficult to determine kcat and Km exactly by kinetic study of SagUGL D115N because Km of SagUGL D115N is much higher than 1 mm. Enzyme assay with higher concentration of unsaturated substrate is difficult because absorbance at 235 nm derived from the C-4 C-5 double bond of the substrate is very high. SagUGL D115N and D175N were incapable of acting on Δ6S even when 10 mm substrate and 0.1 mg/ml enzyme were used (Fig. 5A). The increase in Km value in SagUGL D115N but not in D175N and the drastic decrease in kcat in both are similar to those seen in BacillusUGL D88N and D149N.
C-5 double bond of the substrate is very high. SagUGL D115N and D175N were incapable of acting on Δ6S even when 10 mm substrate and 0.1 mg/ml enzyme were used (Fig. 5A). The increase in Km value in SagUGL D115N but not in D175N and the drastic decrease in kcat in both are similar to those seen in BacillusUGL D88N and D149N.
FIGURE 5.
SagUGL mutant enzyme activity by TLC analysis. Lane 1, no enzyme; lane 2, wild-type enzyme; lane 3, D115N; lane 4, D175N; lane 5, R236H; lane 6, R236A. A, degradation of Δ6S. Reactions were performed at 30 °C for 30 min using 10 mm substrate and 0.1 mg/ml enzyme. B, degradation of Δ4S. Reactions were carried out at 30 °C for 60 min using 10 mm substrate and 2 mg/ml enzyme. The arrow indicates the position to which the developing solvent reached.
As indicated by comparison of the active site structure, some amino acid residues interacting with substrate at subsite +1 are different between SagUGL and BacillusUGL. This difference is probably crucial for determining substrate specificity. To clarify the effects of difference in residues at subsite +1 on substrate specificity of UGLs, SagUGL Arg-236 was replaced by His or Ala. Similar to the case of SagUGL D115N, kinetics studies of SagUGL R236H and R236A were difficult because of their higher Km values. Unlike SagUGL D115N and D175N, both SagUGL R236H and R236A effectively degraded Δ6S (Fig. 5A). As shown in TLC using high concentration of substrate and enzyme (Fig. 5B), SagUGL R236H and R236A were almost inactive on Δ4S, although the substrate was degraded by the wild-type enzyme. These results indicate that SagUGL Arg-236 is one of the residues involved in its activity for the sulfated substrate. This is supported by the energy-minimized model of SagUGL binding to Δ4S (Fig. 4D). Arg-236 and Gln-237 can bind to the sulfate group of Δ4S through the formation of hydrogen bonds.
In conclusion, pathogenic streptococci produce UGL together with polysaccharide lyase responsible for completely degrading glycosaminoglycans, and the streptococcal UGLs prefer sulfated substrates. This suggests that the substrate specificity is feasible for bacterial infection through degradation of mammalian extracellular matrices with sulfate groups. The amino acid sequence in the active site specific to streptococcal UGLs is probably involved in interacting with the substrate sulfate group.
Supplementary Material
Acknowledgments
We thank Drs. K. Hasegawa and S. Baba of the Japan Synchrotron Radiation Research Institute for kind help in data collection. Diffraction data for crystals were collected at the BL-38B1 station of SPring-8 (Hyogo, Japan) with the approval of the Japan Synchrotron Radiation Research Institute. We also thank Kayo Yumoto for excellent technical assistance.
This work was supported in part by grants-in-aid from the Japan Society for the Promotion of Science (to K. M. and W. H.) and by Targeted Proteins Research Program (to W. H.) from the Ministry of Education, Culture, Sports, Science, and Technology of Japan. This work was also supported in part by Research Fellowships from the Japan Society for the Promotion of Science for Young Scientists (to Y. M.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Tables S1 and S2 and Figs. S1 and S2.
- GlcA
- d-glucuronic acid
- GalNAc
- N-acetyl-d-galactosamine
- UGL
- unsaturated glucuronyl hydrolase
- ΔGlcA
- unsaturated GlcA
- BacillusUGL
- Bacillus sp. GL1 UGL
- Δ6S
- unsaturated chondroitin disaccharide sulfated at C-6 position of GalNAc residue
- SagUGL
- S. agalactiae UGL
- SpnUGL
- S. pneumoniae UGL
- SpyUGL
- S. pyogenes UGL
- GFC
- gel filtration chromatography
- Δ4S
- unsaturated chondroitin disaccharide sulfated at C-4 position of GalNAc residue
- MES
- 4-morpholineethanesulfonic acid.
REFERENCES
- 1.Ernst S., Langer R., Cooney C. L., Sasisekharan R. ( 1995) Crit. Rev. Biochem. Mol. Biol. 30, 387– 444 [DOI] [PubMed] [Google Scholar]
- 2.Gandhi N. S., Mancera R. L. ( 2008) Chem. Biol. Drug Des. 72, 455– 482 [DOI] [PubMed] [Google Scholar]
- 3.Lindahl U., Höök M. ( 1978) Annu. Rev. Biochem. 47, 385– 417 [DOI] [PubMed] [Google Scholar]
- 4.Laurent T. C., Fraser J. R. ( 1992) FASEB J. 6, 2397– 2404 [PubMed] [Google Scholar]
- 5.Iozzo R. V. ( 1998) Annu. Rev. Biochem. 67, 609– 652 [DOI] [PubMed] [Google Scholar]
- 6.Sawitzky D. ( 1996) Med. Microbiol. Immunol. 184, 155– 161 [DOI] [PubMed] [Google Scholar]
- 7.Davies G., Henrissat B. ( 1995) Structure 3, 853– 859 [DOI] [PubMed] [Google Scholar]
- 8.Linhardt R. J., Avci F. Y., Toida T., Kim Y. S., Cygler M. ( 2006) Adv. Pharmacol. 53, 187– 215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Glaser P., Rusniok C., Buchrieser C., Chevalier F., Frangeul L., Msadek T., Zouine M., Couvé E., Lalioui L., Poyart C., Trieu-Cuot P., Kunst F. ( 2002) Mol. Microbiol. 45, 1499– 1513 [DOI] [PubMed] [Google Scholar]
- 10.Hoskins J., Alborn W. E., Jr., Arnold J., Blaszczak L. C., Burgett S., DeHoff B. S., Estrem S. T., Fritz L., Fu D. J., Fuller W., Geringer C., Gilmour R., Glass J. S., Khoja H., Kraft A. R., Lagace R. E., LeBlanc D. J., Lee L. N., Lefkowitz E. J., Lu J., Matsushima P., McAhren S. M., McHenney M., McLeaster K., Mundy C. W., Nicas T. I., Norris F. H., O'Gara M., Peery R. B., Robertson G. T., Rockey P., Sun P. M., Winkler M. E., Yang Y., Young-Bellido M., Zhao G., Zook C. A., Baltz R. H., Jaskunas S. R., Rosteck P. R., Jr., Skatrud P. L., Glass J. I. ( 2001) J. Bacteriol. 183, 5709– 5717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ferretti J. J., McShan W. M., Ajdic D., Savic D. J., Savic G., Lyon K., Primeaux C., Sezate S., Suvorov A. N., Kenton S., Lai H. S., Lin S. P., Qian Y., Jia H. G., Najar F. Z., Ren Q., Zhu H., Song L., White J., Yuan X., Clifton S. W., Roe B. A., McLaughlin R. ( 2001) Proc. Natl. Acad. Sci. U.S.A. 98, 4658– 4663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jedrzejas M. J. ( 2001) Microbiol. Mol. Biol. Rev. 65, 187– 207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Paton J. C., Andrew P. W., Boulnois G. J., Mitchell T. J. ( 1993) Annu. Rev. Microbiol. 47, 89– 115 [DOI] [PubMed] [Google Scholar]
- 14.Gase K., Ozegowski J., Malke H. ( 1998) Biochim. Biophys. Acta 1398, 86– 98 [DOI] [PubMed] [Google Scholar]
- 15.Berry A. M., Lock R. A., Thomas S. M., Rajan D. P., Hansman D., Paton J. C. ( 1994) Infect. Immun. 62, 1101– 1118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hynes W. L., Dixon A. R., Walton S. L., Aridgides L. J. ( 2000) FEMS Microbiol. Lett. 184, 109– 112 [DOI] [PubMed] [Google Scholar]
- 17.Li S., Kelly S. J., Lamani E., Ferraroni M., Jedrzejas M. J. ( 2000) EMBO J. 19, 1228– 1240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mello L. V., De Groot B. L., Li S., Jedrzejas M. J. ( 2002) J. Biol. Chem. 277, 36678– 36688 [DOI] [PubMed] [Google Scholar]
- 19.Günther E., Ozegowski J. H., Köhler W. ( 1996) Zentralbl. Bakteriol. 285, 64– 73 [DOI] [PubMed] [Google Scholar]
- 20.Pritchard D. G., Lin B., Willingham T. R., Baker J. R. ( 1994) Arch. Biochem. Biophys. 315, 431– 437 [DOI] [PubMed] [Google Scholar]
- 21.Cantarel B. L., Coutinho P. M., Rancurel C., Bernard T., Lombard V., Henrissat B. ( 2009) Nucleic Acids Res. 37, D233– 238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mori S., Akao S., Nankai H., Hashimoto W., Mikami B., Murata K. ( 2003) Protein Expr. Purif. 29, 77– 84 [DOI] [PubMed] [Google Scholar]
- 23.Hashimoto W., Kobayashi E., Nankai H., Sato N., Miya T., Kawai S., Murata K. ( 1999) Arch. Biochem. Biophys. 368, 367– 374 [DOI] [PubMed] [Google Scholar]
- 24.Itoh T., Akao S., Hashimoto W., Mikami B., Murata K. ( 2004) J. Biol. Chem. 279, 31804– 31812 [DOI] [PubMed] [Google Scholar]
- 25.Itoh T., Hashimoto W., Mikami B., Murata K. ( 2006) Biochem. Biophys. Res. Commun. 344, 253– 262 [DOI] [PubMed] [Google Scholar]
- 26.Itoh T., Hashimoto W., Mikami B., Murata K. ( 2006) J. Biol. Chem. 281, 29807– 29816 [DOI] [PubMed] [Google Scholar]
- 27.Hashimoto W., Itoh T., Maruyama Y., Mikami B., Murata K. ( 2007) Int. Microbiol. 10, 233– 243 [PubMed] [Google Scholar]
- 28.Linker A., Meyer K., Weissmann B. ( 1955) J. Biol. Chem. 213, 237– 248 [PubMed] [Google Scholar]
- 29.Sambrook J., Fritsch E. F., Maniatis T. ( 1989) Molecular Cloning: A Laboratory Manual, 2nd Ed., Cold Spring Harbor Laboratory, Cold Spring Harbor, NY [Google Scholar]
- 30.Sanger F., Nicklen S., Coulson A. R. ( 1977) Proc. Natl. Acad. Sci. U.S.A. 74, 5463– 5467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bradford M. M. ( 1976) Anal. Biochem. 72, 248– 254 [DOI] [PubMed] [Google Scholar]
- 32.Laemmli U. K. ( 1970) Nature 227, 680– 685 [DOI] [PubMed] [Google Scholar]
- 33.Hashimoto W., Maesaka K., Sato N., Kimura S., Yamamoto K., Kumagai H., Murata K. ( 1997) Arch. Biochem. Biophys. 339, 17– 23 [DOI] [PubMed] [Google Scholar]
- 34.Irizarry R. A., Hobbs B., Collin F., Beazer-Barclay Y. D., Antonellis K. J., Scherf U., Speed T. P. ( 2003) Biostatistics 4, 249– 264 [DOI] [PubMed] [Google Scholar]
- 35.Hashimoto W., Suzuki H., Yamamoto K., Kumagai H. ( 1995) J. Biochem. 118, 75– 80 [DOI] [PubMed] [Google Scholar]
- 36.Otwinowski Z., Minor W. ( 1997) Methods Enzymol. 276, 307– 326 [DOI] [PubMed] [Google Scholar]
- 37.Vagin A., Teplyakov A. ( 1997) J. Appl. Crystallogr. 30, 1022– 1025 [Google Scholar]
- 38.Collaborative Computational Project ( 1994) Acta Crystallogr. D. Biol. Crystallogr. 50, 760– 76315299374 [Google Scholar]
- 39.Murshudov G. N., Vagin A. A., Dodson E. J. ( 1997) Acta Crystallogr. D. Biol. Crystallogr. 53, 240– 255 [DOI] [PubMed] [Google Scholar]
- 40.Emsley P., Cowtan K. ( 2004) Acta Crystallogr. D. Biol. Crystallogr. 60, 2126– 2132 [DOI] [PubMed] [Google Scholar]
- 41.Laskowski R. A., MacArthur M. W., Moss D. S., Thornton J. M. ( 1993) J. Appl. Crystallogr. 26, 283– 291 [Google Scholar]
- 42.DeLano W. L. ( 2004) The PyMOL Molecular Graphics System, DeLano Scientific LLC, San Carlos, CA [Google Scholar]
- 43.Berman H. M., Westbrook J., Feng Z., Gilliland G., Bhat T. N., Weissig H., Shindyalov I. N., Bourne P. E. ( 2000) Nucleic Acids Res. 28, 235– 242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Brinkkötter A., Klöss H., Alpert C., Lengeler J. W. ( 2000) Mol. Microbiol. 37, 125– 135 [DOI] [PubMed] [Google Scholar]
- 45.Yum D. Y., Lee B. Y., Pan J. G. ( 1999) Appl. Environ. Microbiol. 65, 3341– 3346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Myette J. R., Shriver Z., Kiziltepe T., McLean M. W., Venkataraman G., Sasisekharan R. ( 2002) Biochemistry 41, 7424– 7434 [DOI] [PubMed] [Google Scholar]
- 47.Murzin A. G., Brenner S. E., Hubbard T., Chothia C. ( 1995) J. Mol. Biol. 247, 536– 540 [DOI] [PubMed] [Google Scholar]
- 48.Juy M., Amrt A. G., Alzari P. M., Poljak R. J., Claeyssens M., Béguin P., Aubert J. P. ( 1992) Nature 357, 89– 91 [Google Scholar]
- 49.Itoh T., Mikami B., Maru I., Ohta Y., Hashimoto W., Murata K. ( 2000) J. Mol. Biol. 303, 733– 744 [DOI] [PubMed] [Google Scholar]
- 50.Itoh T., Ochiai A., Mikami B., Hashimoto W., Murata K. ( 2006) J. Mol. Biol. 360, 573– 585 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.