Abstract
In this study, we investigated the metabolic phenotype of the NF-κB p50 knock-out (p50-KO) mice. Compared with wild type mice, the p50-KO mice had an increase in food intake, but a decrease in body fat content. On chow diet, their blood glucose dropped much more than the wild type (WT) mice in the insulin tolerance test. Their glucose infusion rate was 30% higher than that of the WT mice in the hyperinsulinemic-euglycemic clamp. Their hepatic glucose production was suppressed more actively by insulin, and their insulin-induced glucose uptake was not altered in skeletal muscle or adipose tissue. In the liver, their p70S6K (S6K1) protein was significantly lower, and tumor necrosis factor-α (TNF-α) expression was much higher. Their S6K1 protein was reduced by TNF-α treatment in the primary culture of hepatocytes. S6K1 reduction was blocked by the proteasome inhibitor MG132. In their livers, IKK2 (IKKβ) activity was reduced together with IKKγ. Their S6K1 degradation was dependent on IKK2 deficiency. Reconstitution of the S6K1 protein in their liver blocked the increase in insulin sensitivity. S6K1 degradation was not observed in hepatocytes of the WT mice. The data suggest that inactivation of NF-κB p50 leads to suppression of IKK2 activity in the liver. IKK2 deficiency leads to S6K1 inhibition through TNF-induced protein degradation. The S6K1 reduction may contribute to insulin sensitivity in p50-KO mice. This study suggests that hepatic S6K1 may be a drug target in the treatment of insulin resistance.
Chronic inflammation occurs in adipose tissue and liver in obesity (1, 2). It is generally believed that inflammation contributes to pathogenesis of insulin resistance through activation of IKK/NF-κB and JNK signaling pathways. Serine kinase IKK2 (IKKβ) contributes to insulin resistance by activation of transcription factor NF-κB (3–6) or induction of IRS-1 serine phosphorylation (7, 8). Data from Ikk2 transgenic mice suggest that NF-κB may be involved in the pathogenesis of insulin resistance (4). However, the role of NF-κB has not been directly tested by gene knock-out in mice.
NF-κB contains two subunits, p50 (NF-κB1) and p65 (RelA), in most cases. The p50 knock-out (p50-KO) mice were first reported in 1995, in which the p50 gene was globally inactivated by gene deletion (9). The mice are normal in growth, but deficient in B lymphocyte function. The p50-KO mice have been used in the study of many biological systems including the immune system (10, 11), muscle dystrophy (12), liver regeneration (13, 14), aging (15), and brain function (16). However, the metabolic phenotype remains unclear in this line of KO mice. Whole body knock-out of p65 (p65-KO) was reported in 1995 for embryonic lethality (17, 18).
S6K1 (p70 ribosomal protein S6 kinase) is a serine kinase in the PI3K/Akt signaling pathway. It mediates nutrient (amino acid and glucose) and insulin signals in the regulation of insulin sensitivity (19, 20). Global inactivation of S6K1 in knock-out mice protects the mice against diet-induced insulin resistance (21). An increase in S6K1 activity leads to insulin resistance in mice (22). In humans, inhibition of the S6K activity by rapamycin leads to protection against insulin resistance (23). S6K1 contributes to insulin resistance by phosphorylation of IRS-1 at several serine residues (24–28). However, it is largely unknown how S6K1 protein abundance is regulated in cells.
To collect direct evidence for regulation of insulin resistance by NF-κB, we conducted a hyperinsulinemic-euglycemic clamp assay in the p50-KO mice. The data suggest that global inactivation of NF-κB p50 enhances insulin sensitivity in mice primarily through the liver. The molecular mechanism is related to inhibition of the S6K1 activity through a proteasome-dependent mechanism, which is a consequence of increased TNF-α activity and reduced IKK2 activity in the liver of p50-KO mice.
EXPERIMENTAL PROCEDURES
Animal Models
Male p50-KO (Nf-κb1 KO) mice with the B6X129PF2 gene background were purchased at 4 weeks of age from The Jackson Laboratory (Bar Harbor, ME). The age- and gender-matched wild type mice in the same gene background were purchased and used as the control. The mice were subject to a 1-week quarantine after arrival. All of the mice were housed at 4 mice/box in the animal facility with a 12:12-h light-dark cycle and constant temperature (22–24 °C). The mice had free to access water and chow diet. All procedures were performed in accordance with the National Institutes of Health guidelines for the care and use of animals and approved by the Institutional Animal Care and Use Committee at the Pennington Biomedical Research Center.
High Fat Diet (HFD)
HFD (D12331, Research Diets, New Brunswick, NJ) contains 58% of kcal as fat. The mice were fed the HFD beginning at 5 weeks of age. Their body weight and composition were examined every 2 weeks.
Cell Culture and Reagents
Mouse embryonic fibroblast (MEF) cell lines including WT and Ikk2-KO (Ikk2−/−) were described elsewhere (7). The cells were maintained in 10% fetal bovine serum, Dulbecco's modified Eagle's medium in a 5% CO2 incubator. The cells were starved in Dulbecco's modified Eagle's medium containing 0.2% fatty acid-free bovine serum albumin overnight before treatment with 10 ng/ml TNF-α (T-6674, Sigma). Rapamycin (A-275) was purchased from Biomol International (Plymouth Meeting, PA). 15PdGJ2 (catalog number 538927) was from Calbiochem (EMD, Gibbstown, NJ).
Nuclear Magnetic Resonance
Body composition was measured using quantitative nuclear magnetic resonance (NMR) as described elsewhere (29). In the test, a 6-week-old conscious and unrestrained mouse was placed into a small tube, and then the tube was inserted into a Brucker model mq10 NMR analyzer (Brucker, Milton, ON, Canada). The fat and lean mass were recorded within 1 min. Measurements were made in triplicate.
Intraperitoneal Glucose Tolerance Test (GTT) and Insulin Tolerance Test (ITT)
GTT was conducted by intraperitoneal injection of glucose at 250 mg/kg of body weight in mice after an overnight fast. ITT was conducted by intraperitoneal injection of insulin (I9278, Sigma) at 0.75 unit/kg of body weight in mice after a 4-h fast as described elsewhere (29). Blood glucose was monitored in the tail vein blood using the FreeStyle blood glucose monitoring system (TheraSense, Phoenix, AZ).
Hyperinsulinemic-Euglycemic Clamp
The p50-KO mice and wild type control mice were purchased from The Jackson Laboratory at 5 weeks of age and shipped to the Mouse Metabolic Phenotype Center of Vanderbilt University (Nashville, TN). The mice were fed a chow diet at the animal facility center of Vanderbilt University. The clamp was conducted in mice at 10 weeks of age. The clamp was performed in chronically catheterized (jugular vein and carotid artery) conscious mice after a 5-h fast (29). Catheters were inserted 4–5 days prior to performing the clamp. Only mice returning to within ∼10% of presurgical body weight were studied. The arterial catheter was used for blood sampling, and the venous catheter was used for infusing glucose, blood, tracers, and insulin. Catheters were attached to a swivel. Mice were unrestrained and not handled thereafter to minimize stress. The clamp period (t = 0 min) began at 1300 h. A primed continuous [3H]glucose infusion (5 μCi of bolus and 0.05 μCi/min) was given at t = −120 min to measure glucose turnover. An insulin infusion was started at t = 0 min (2 milliunits/kg/min; Humulin R, Eli Lilly). Saline-washed erythrocytes were infused (5–6 μl/min) during the experimental period to prevent an ∼5% fall in hematocrit. The [3-3H]glucose was increased to 0.1 μCi/min to minimize changes in specific activity. Glucose (5 μl) was measured every 10 min, and euglycemia was maintained using a variable glucose infusion rate. Samples (10 μl) to determine glucose-specific activity were taken at t = −5 min and every 10 min from t = 80–120 min. At t = 120 min a bolus of 2-[14C]deoxyglucose was given, and samples were taken at t = 122, 125, 130, 135, and 145 min. At t = 145 min animals were anesthetized, and tissues were collected and rapidly frozen for subsequent analysis. Samples (50 μl) were taken to measure plasma insulin at t = 0, 120, and 145 min.
Western Blot
Fresh fat and muscles were collected and frozen in liquid nitrogen. The whole cell lysate protein was extracted in lysis buffer with sonication and analyzed in Western blots as described elsewhere (29). Antibodies to IKK1 (sc-7182), IκBα (sc-371), IRS-1 (sc-7200), pIRS-1 Tyr-632 (Tyr-628 in mouse) (sc-17196), IRS-2 (sc-8299), IKKγ (sc-8330), and p65 (sc-8008x) were purchased from Santa Cruz. Antibodies to tubulin (ab7291), β-actin (ab6276), p50 (ab7971), pIRS-1 Ser-636 (Ser-632 in mouse) (ab53038), and S6K (ab9366) were from Abcam (Cambridge, UK). IKK2 antibody (05-535) was from Upstate, and pAkt Ser-473 (KP24001) was from Calbiochem. pAkt Thr-308 (number 9275), pS6K (Thr-389, number 9205), and pIKK (IKK1 Ser-180/IKK2 Ser-181, number 2681) were from Cell Signaling Technology (Danvers, MA). To detect multiple signals from one membrane, the membrane was stripped with a stripping buffer.
S6K Reconstitution
The S6K adenovirus was constructed by subcloning mouse S6K cDNA into the adenoviral vector in the ViraPower Adenoviral Expression System (K4930-00, Invitrogen). The S6K cDNA was amplified in PCR from a plasmid vector of hemogglutinin-S6K1 (8984, Addgene, Cambridge, MA). Then, it was inserted into adenovirus plasmid vector pAd/CMV/V5-DEST through the TOPO pENTR vector (K2400-20). The viral vector was transfected into 293A cells for virus production after digestion with the PacI endonuclease. The adenovirus was purified and titrated according to the manufacturer's instructions. The p50−/− mice were injected with S6K adenovirus or GFP adenovirus at 1 × 109 virus/mouse at 8 weeks of age through the tail vein. Three days later, ITT was performed to evaluate insulin sensitivity after a 4-h fast.
Primary Hepatocytes
The primary hepatocytes were isolated from mice at 6 weeks of age according to the procedure described elsewhere (30). Briefly, the mice were anesthetized by intraperitoneal injection of ketamine at a dose of 100 mg/kg of body weight. The liver was first perfused with Ca2+-free Hanks' balanced solution (Invitrogen) at a rate of 5 ml/min for 8 min through the vena cava, followed by continuous perfusion for 12 min with serum-free Williams' Medium E (Invitrogen) containing collagenase (Worthington, Type II, 50 units/ml) and 10 mm HEPES. Hepatocytes were harvested after centrifugation on Percoll (Sigma). The purified hepatocytes were placed into collagen-coated plates in Williams' Medium E supplemented with 10% fetal bovine serum for 24 h before the experiment. The cytokine treatment was conducted in serum-free medium after overnight serum starvation.
Quantitative Real-time RT-PCR
mRNA was examined in total RNA that was extracted from frozen tissues (kept at −80 °C) or cultured cells using Tri-Reagent (T9424, Sigma) as described elsewhere (31). TaqMan RT-PCR primers and probes were used to determine mRNA for Tnf-α (Mm00443258_m1), Il-6 (Mm00446190_m1), and S6k (Mm00659517_m1). The reagents were purchased from Applied Biosystems (Foster City, CA). Mouse ribosome 18S rRNA_s1 (without intron-exon junction) was used as an internal control. The mRNA result for each gene was normalized with the 18S signal. The reaction was conducted with a 7900 HT Fast real-time PCR system (Applied Biosystems, Foster City, CA).
Cytokine Assay
Proteins for TNF-α and IL-6 were measured in the hepatocyte lysate using ELISA Kits (MTA00 and M6000B, R&D Systems, Inc., Minneapolis, MN). The assay was conducted according to the manufacturer's instructions.
Hematoxylin and Eosin Staining
The fresh liver was collected immediately after sacrifice of the mouse and fixed in 10% neutral buffered formalin solution (HT50-1-2, Sigma). Tissue slides were obtained through serial cross-section cutting at 8-μm thickness and processed with a standard procedure. Briefly, the slides were deparaffinated and stained in hematoxylin (number 101542, Surgipath) for 15 min and rinsed in water until sections were blue. Then, slides were stained in eosin (E4009, Sigma), dehydrated quickly in 95% ethanol, and treated with phenazine methosulfate. The sections were mounted, and the image was taken with a Nikon microscope (Eclipse TS100).
Statistical Analysis
Data are presented as mean ± S.E. All in vitro experiments were repeated independently at least three times with consistent results. Signals in Western blots were quantified by densitometry using the NIH ImageJ software. Student's t test was used in statistical analysis of the data. p < 0.05 was considered as statistically significant.
RESULTS
The p50-KO Mice Are Lean and Resistant to Dietary Obesity
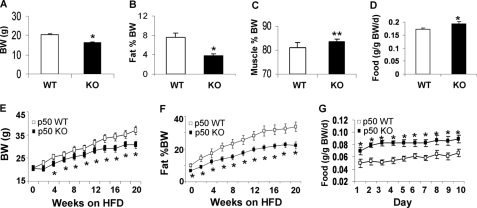
The p50-KO mice are normal in development, growth, and reproduction (9). To understand their metabolism, we monitored body weight, fat mass, and food intake. Compared with the WT mice, p50-KO mice had 10% less body weight at 6 weeks of age (Fig. 1A). Their fat mass was 50% less (Fig. 1B). Their lean body mass was 3% higher (Fig. 1C). Their food intake was higher after normalization with body weight (Fig. 1D). On the HFD, these differences remained between the KO and WT mice. The KO mice were lower in their body weight and fat content (Fig. 1, E and F). Their food intake was higher than that of the WT mice (Fig. 1G). The data suggest that the p50-KO mice have a lean phenotype and are resistant to diet-induced obesity.
FIGURE 1.
Decreased body weight and fat content in p50-KO mice. The study was conducted in mice at 6 weeks of age. A, body weight (BW). B, body fat content. Body composition was determined in p50-KO mice at 6 weeks of age using NMR. C, body muscle content. D, food intake normalized with body weight. E, body weight on the HFD. F, fat content on the HFD. G, food intake at 10 weeks on the HFD and normalized with body weight. Each value represents the mean ± S.E. (n = 8). *, p < 0.05; **, p < 0.001 by Student's t test.
Insulin Sensitivity Is Enhanced in the p50-KO Mice
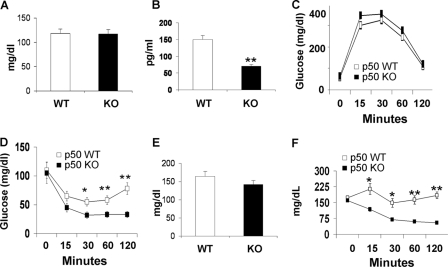
The p50-KO mice were normal in fasting glucose, but their fasting insulin was lower compared with that of the WT mice (Fig. 2, A and B). Their glucose metabolism was evaluated in a GTT and a ITT. Their glucose tolerance was not altered (Fig. 2C), but their insulin tolerance was enhanced significantly. The blood glucose dropped much faster in the KO mice as indicated by the ITT result (Fig. 2D), suggesting a higher level of insulin sensitivity. On the HFD, the KO mice had a similar phenotype regarding glucose metabolism, as they exhibited an identical fasting glucose as the WT mice (Fig. 2E). Their insulin tolerance remained higher (Fig. 2F). In GTT, no difference was observed between the KO and WT mice (data not shown). The result suggests that the p50-KO mice are more sensitive to insulin, and they are protected from diet-induced insulin resistance.
FIGURE 2.
Insulin sensitivity in mice. In panels A–D, the mice were on chow diet, and the assays were conducted in mice at 8–9 weeks of age. In panels E and F, the mice were on the HFD. A, fasting glucose. B, fasting insulin. C, GTT. D, ITT. E, fasting glucose at 10 weeks on HFD. F, ITT at 12 weeks on the HFD. Data are presented as the mean ± S.E. (n = 10). *, p < 0.05; **, p < 0.001 by Student's t test.
The Role of Liver in the Enhanced Insulin Sensitivity
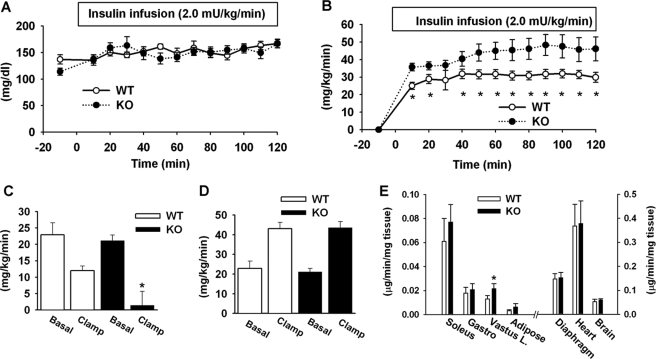
To identify the tissues/organs that are responsible for increased insulin sensitivity, we performed the hyperinsulinemic-euglycemic clamp test in p50-KO mice after a 5-h fast. During the clamp, the blood glucose was maintained at the same levels in KO and WT mice (Fig. 3A). Arterial insulin concentration was increased from 0.5 ± 0.1 to 1.3 ± 0.2 ng/ml in the KO mice and 0.9 ± 0.1 to 1.7 ± 0.2 ng/ml in the WT mice during the clamp. In the p50-KO mice, the glucose infusion rate was 30% higher (Fig. 3B), and the endogenous glucose production was suppressed by insulin more significantly. In the KO mice, the hepatic glucose production was almost not detectable during the clamp (Fig. 3C). In WT mice, hepatic glucose production was only suppressed by 40% of the basal level. The whole body glucose utilization rate was not different between KO and WT mice (Fig. 3D). The tissue-specific glucose uptake was examined in the major insulin-sensitive tissues including soleus, vastus lateralis and gastrocnemius muscles, epididymal fat, diaphragm, and heart. The brain was examined as a negative control. No difference was observed between the KO and WT mice in most of the tissues, although glucose uptake was modestly increased in the vastus lateralis muscle of KO mice (Fig. 3E). These results exclude the role of peripheral tissues and highlight the role of liver in the insulin sensitivity in the p50-KO mice.
FIGURE 3.
Hyperinsulinemic-euglycemic clamp test. The test was conducted in mice at 8 weeks of age. A, blood glucose level in clamp. B, glucose infusion rate during the course of the clamp. C, hepatic glucose production rate. D, glucose clearance rate. E, glucose uptake in peripheral tissues during the clamp. Skeletal muscles include soleus, gastrocnemius (Gastro), and superficial vastus lateralis (Vastus L.). Epididymal fat was used. Each data point represents the mean ± S.E. (n = 10). *, p < 0.05.
Increased IRS Activities and Reduced S6K Protein in the Liver
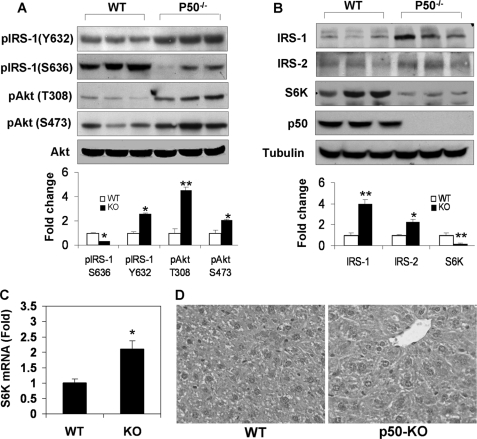
To understand the molecular mechanism of insulin sensitivity in the liver, we examined the insulin signaling pathway in liver tissue after the mice were challenged with a bolus of insulin. The pathway activity was determined by the phosphorylation status and protein abundance of the signaling proteins. Tyr phosphorylation of IRS-1 is catalyzed by the insulin receptor and is responsible for activation of PI3K/Akt kinases, which are required for Glut4 translocation and glucose uptake. Tyrosine phosphorylation was monitored with a phospho-specific antibody to Tyr-632 (Tyr-628 in the mouse IRS-1), a parameter of the insulin receptor activity (32). Tyrosine phosphorylation was enhanced in p50-KO mice (Fig. 4A). Consistently, Akt phosphorylation at Thr-308 and Ser-473 was increased in KO mice (Fig. 4A), suggesting an enhancement in insulin signaling activity. Serine phosphorylation of IRS-1 generally leads to inhibition of insulin signaling and often serves as an insulin resistance marker. We examined the IRS-1 serine phosphorylation with a phospho-specific antibody to Ser-636, which is a target of S6K1 serine kinase in the human IRS-1 (Ser-632 in the mouse IRS-1) (21, 28). In p50-KO mice, IRS-1 serine phosphorylation was reduced significantly (Fig. 4A). The reduction was associated with a 2-fold elevation in IRS-1 protein (Fig. 4B), suggesting less degradation of the IRS-1 protein. Serine phosphorylation usually promotes degradation of the IRS-1 protein. IRS-2 protein abundance was also increased in KO mice (Fig. 4B).
FIGURE 4.
Reduction of S6K protein in the liver of p50-KO mice. A, insulin signaling in the liver. The WT and p50-KO mice were fasted for 4 h before the insulin challenge (0.75 units/kg, intraperitoneal). IRS-1 and Akt phosphorylation was examined in the liver lysate 15 min after insulin injection. B, protein abundance for IRS-1/2 and S6K in the liver of p50-KO mice. In panels A and B, quantification results are presented in the bar figures for each signal in the blots. C, S6K mRNA determined by quantitative RT-PCR in the liver. D, microscope image of liver tissue. The image of liver tissue with hematoxylin and eosin staining was taken under a microscope with a ×40 objective lens. In this figure, Western blot experiments were repeated three times with consistent results. The representative blots are presented. In the bar figure, each data point represents the mean ± S.E. (n = 6). *, p < 0.05; **, p < 0.001.
Interestingly, the protein abundance of S6K was dramatically reduced in KO mice (Fig. 4B, S6K). As a major serine kinase in the negative feedback loop in the insulin signaling pathway, S6K may contribute to the increased insulin activity through the reduction in activity. S6k mRNA was examined, and its level was increased by 100% in the KO mouse liver (Fig. 4C). Association of the increased mRNA with the reduced protein suggests a quick turnover of S6K protein in the liver of p50-KO mice. No change was observed in protein abundance or phosphorylation in the insulin receptor (data not shown). Absence of NF-κB p50 protein was confirmed in the liver of KO mice (Fig. 4B). Morphology of the liver was examined under a microscope. With hematoxylin and eosin staining, no difference was observed in the structure or cell morphology in the livers of control and KO mice (Fig. 4D).
Reduced Insulin Sensitivity after Reconstitution of S6K in Liver
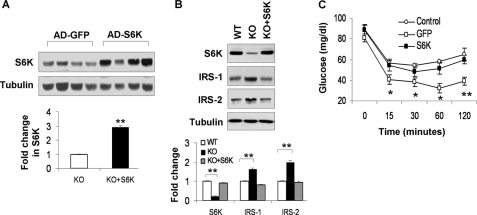
Given the role of S6K in the regulation of IRS-1 function (21, 27, 28, 33), we hypothesized that loss of S6K might be responsible for the increased insulin sensitivity in the liver of p50-KO mice. To test this possibility, we restored S6K protein abundance in the liver of p50-KO mice by expression of an exogenous S6k gene. The restoration was completed by injection of S6k adenovirus into the tail vein. The control mice were injected with a Gfp adenovirus. In this model, the S6K protein was increased by 2-fold in the liver of KO mice (Fig. 5A). The restored level is comparable with that in the WT liver (Fig. 5B). After S6K restoration, IRS-1 and IRS-2 proteins were reduced in the liver of KO mice, reaching levels comparable with those in the WT mice (Fig. 5B). ITT was conducted after S6K reconstitution to determine insulin sensitivity. The difference in insulin sensitivity between the p50-KO and WT mice became undetectable following restoration of S6K (Fig. 5C). These data suggest that the decrease in S6K activity may contribute to the insulin sensitization in the p50-KO mice.
FIGURE 5.
Reconstitution of S6K in the liver of p50-KO mice. p50−/− mice were injected with S6K adenovirus or GFP adenovirus at 1 × 109 virus/mouse at the age of 8 weeks. Three days later, ITT was performed to evaluate insulin sensitivity after 4 h of starvation. The liver was collected after the ITT test and used in examination of the S6K protein. A, Western blot of S6K in the liver (upper panel). Each lane represents one mouse. The mean value is presented in the bar figure in the lower panel. B, S6K reconstitution in the liver of p50-KO mice. The S6K protein was determined in the liver of reconstituted mice in a Western blot. Quantification of each band is presented in the lower panel. C, ITT after S6K reconstitution. Age- and gender-matched wild type mice were used in the control. GFP adenovirus (open square) was used as a virus control for S6K adenovirus (filled square). Data are presented as the mean ± S.E. (n = 7). *, p < 0.05; **, p < 0.001.
S6K Degradation in the Hepatocytes of p50-KO Mice
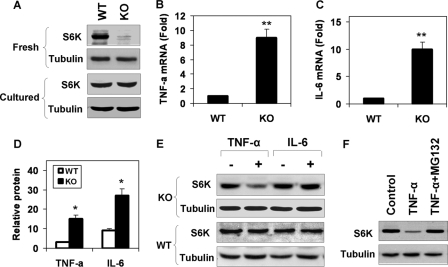
To investigate the mechanism of S6K reduction, we examined S6K protein in the primary hepatocytes of p50-KO mice. In freshly isolated hepatocytes, the S6K protein was significantly lower in p50-KO mice (Fig. 6A). However, after 24 h in cell culture, the difference was no longer detectable, suggesting that S6K degradation was dependent on the microenvironment in the liver. In search of the liver-associated factors, we examined gene expressions and observed an up-regulation of Tnf-α and Il-6 in mRNA in the p50-KO mice (Fig. 6, B and C). In the p50-KO mice, expression of the two genes was 7-fold (Tnf-α) and 9-fold (Il-6) higher than that in WT mice, respectively. The increase was detected in protein as well in the liver lysates (Fig. 6D).
FIGURE 6.
S6K degradation induced by TNF-α. In this study, 6-week-old mice were used. A, S6K protein in primary hepatocytes. The S6K was examined in fresh and cultured (24 h) hepatocytes of WT and p50-KO mice in a Western blot. B, TNF-α mRNA in liver. C, IL-6 mRNA in liver. D, cytokine proteins in the liver. The proteins were determined in the whole cell lysate of liver tissue by enzyme-linked immunosorbent assay. E, S6K degradation in the hepatocytes. The S6K protein was examined in primary hepatocytes after TNF-α (10 ng/ml) or IL-6 (10 ng/ml) treatments for 4 h. F, inhibition of S6K degradation by the proteasome inhibitor. S6K degradation was induced by TNF-α in the hepatocytes of p50-KO mice. The cells were pretreated with MG132 (15 μm) for 30 min to inhibit the proteasome. The Western blot experiments were repeated three times with consistent results. Representative blots are presented. In the bar figure, data are presented as the mean ± S.E. (n = 6). *, p < 0.05.
The increase in S6k mRNA in the liver of p50-KO mice (Fig. 4C) suggests that S6K protein reduction may not be a result of inhibition at the mRNA level. In an early study, we observed that TNF-α was able to induce serine phosphorylation of S6K (34). Protein degradation is commonly observed after serine phosphorylation. It is possible that serine modification is involved in control of S6K1 protein stability. To test this possibility, we examined S6K1 degradation in primary hepatocytes after TNF-α treatment. A reduction in the S6K protein was observed in the cells of p50-KO mice, but not in those of the WT mice (Fig. 6E). When IL-6 was tested in the same system, no change was observed in the S6K protein. These data suggest that TNF-α is a key factor in the liver for the S6K reduction. The TNF-induced S6K degradation was blocked by the proteasome inhibitor MG132 (Fig. 6F), suggesting that proteasome activity is required for S6K degradation. This group of data suggests that in the absence p50, TNF-α may induce degradation of the S6K protein in a proteasome-dependent manner.
Decreased IKK2 Activity in p50-KO Cells
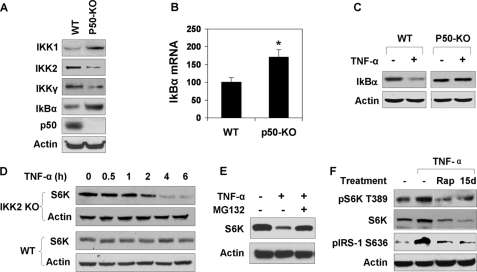
The above data suggest that TNF-induced degradation is specific to p50-KO hepatocytes. In search of the molecular mechanism, we examined IKK2 activity because IKK2 was reported to induce S6K phosphorylation through mTOR (35). In the p50-KO liver, IKK2 protein was reduced by 70% in the whole cell lysate as shown by a Western blot (Fig. 7A). In the IKK complex, the IKK2 activity is usually controlled by IKKγ (NEMO). IKKγ was also reduced in p50-KO cells. Interestingly, IKK1 (IKKα) and IκBα protein abundance was increased more than 50% in p50-KO cells (Fig. 7A). The increase in the IκBα protein was consistent with elevated mRNA levels of IκBα (Fig. 7B), suggesting a constant elevation of transcriptional activity of NF-κB p65. To determine the signaling activity of IKK2 in p50-KO cells, IκBα degradation was examined in primary hepatocytes. IκBα is a substrate of IKK2, and its degradation depends on IKK2-mediated phosphorylation. The degradation was induced by TNF-α in WT cells, but not in p50-KO cells (Fig. 7C). These data suggest that IKK2 activity is deficient in the KO cells.
FIGURE 7.
IKK2 controls S6K degradation in response to TNF-α. A, signaling proteins in the NF-κB pathway. The Western blot was conducted with liver lysates of five p50-KO mice and five control mice. The results are consistent among mice in each group. A representative blot with one mouse in each group is shown. B, Iκbα mRNA in liver. The result represents the mean ± S.E. (n = 5). C, IKK2 deficiency. IKK2 phosphorylation and IκBα degradation were examined in primary hepatocytes treated with TNF-α (10 ng/ml) for 15 min. D, S6K degradation in Ikk2−/− cells. The MEF cells were treated with TNF-α (10 ng/ml) in serum-free medium for the times indicated. E, inhibition of S6K degradation by the proteasome inhibitor in Ikk2-KO cells. MG132 (15 μm) was added into the medium at 30 min before the 4-h treatment with TNF-α. F, mTOR in S6K activation by TNF-α. HepG2 cells were pretreated with the mTOR inhibitor rapamycin (Rap, 200 nm) or the IKK2 inhibitor 15dPGJ2 (15d, 5 μm) followed by TNF-α treatment. pS6K (Thr-389) and pIRS-1 (Ser-636) were examined in a Western blot. The Western blot experiments were repeated three times with consistent results. Representative blots are presented. *, p < 0.05 by Student's t test (n = 5).
Association of IKK2 deficiency with S6K reduction led us to test the role of IKK2 in the control of S6K degradation. Ikk2−/− MEF cells were used in this study. In the WT MEFs, S6K protein was not changed by TNF-α treatment. In Ikk2 null cells, S6K degradation was induced by TNF-α (Fig. 7D). The degradation was time-dependent. At 4 h into the TNF-α treatment, 90% of S6K protein disappeared in the Ikk2−/− MEFs. The degradation was completely inhibited by the proteasome inhibitor MG132 (Fig. 7E), but not influenced by inhibitors to JNK (SP600125), mTOR (rapamycin), or IKK2 (15dPGJ2) (data not shown). This group of data suggests a role of IKK2 in control of the stability of the S6K protein.
mTOR Controls S6K Degradation
It was reported that both mTOR and IKK2 are required for S6K serine phosphorylation in response to TNF-α (35). If this pathway is deficient in p50-KO mice, the phosphorylation of S6K by mTOR will be reduced. Such reduction may lead to S6K protein degradation. To test this possibility, we examined S6K degradation after inhibition of mTOR or IKK2. Rapamycin was used to block mTOR activity in HepG2 cells. As expected, rapamycin reduced S6K phosphorylation at Thr-389 and IRS-1 phosphorylation at Ser-636 (Fig. 7F). IRS-1 is a substrate of S6K1. When IKK2 was inhibited by 15PdGJ2, a similar inhibition was observed in S6K activity (Fig. 7F). In these conditions, S6K protein was reduced in TNF-treated cells (Fig. 7F). In the absence of mTOR or IKK2 inhibition, S6K protein was increased by TNF-α. These data suggest that TNF-α induces S6K activation through the IKK2/mTOR pathway. This pathway may protect S6K from degradation.
DISCUSSION
The metabolic phenotype of p50-KO mice is characterized by an increase in hepatic insulin sensitivity. The increase in insulin sensitivity is supported by the reduction in fasting insulin and enhancement in insulin tolerance. The results from the hyperinsulinemic-euglycemic clamp suggest that the liver contributes to an increase in the glucose infusion rate in p50-KO mice. The clamp study suggests that the peripheral tissues may not contribute to insulin sensitivity because glucose uptake was not significantly changed in skeletal muscles and fat tissues. Additionally, p50-KO mice are lean and protected against diet-induced obesity. The mechanism of the lean phenotype may be related to the increase in energy expenditure in the KO mice.
The cellular and molecular mechanisms were investigated for insulin sensitivity in the liver. The result suggests that IKK2 activity is reduced in the p50-KO liver. This was observed with elevated expression of NF-κB target genes (Tnf-α, IκBα, and Il-6), suggesting an elevation of the transcriptional activity of NF-κB p65. This is not surprising as p50 inhibits p65 activity in transcriptional activation of target genes (36, 37). IKK2 inhibition may be a result of negative feedback in the NF-κB pathway. Although p50-KO mice have been used in a variety of studies (10–16), the IKK2 activity was not examined in those studies. We believe that expression of NF-κB target genes, such as A20 (38, 39) and Cyld (40), whose products are two cytoplasmic proteins (40, 41), may contribute to IKK2 inhibition. A20 and CYLD act through modification of ubiquitination. A20 is able to inactivate IKKγ in the IKK complex through deubiquitination (42). We observed that the IKKγ protein was reduced together with IKK2 in the liver of p50-KO mice.
The current study enriches the concept about inflammation in regulation of insulin sensitivity. In the inflammation signaling pathway, IKK2 activity contributes to the development of insulin resistance in the liver (4, 43). In transgenic mice, overexpression of IKK2 decreased insulin sensitivity (4), and knock-out of IKK2 protected mice from insulin resistance (43). IKK2 deficiency from Ikkγ knock-out also enhanced insulin sensitivity in mice (44). These studies suggest that in the absence of IKK2 activity, inflammation will not be able to induce insulin resistance. However, it was not tested if insulin sensitivity is enhanced over the control when IKK2 is inhibited. In the current study, our data suggest that inflammation enhances insulin sensitivity when IKK2 is deficient in the liver.
The current study suggests a new role of IKK2 in regulation of S6K activity. IKK2 was reported to induce S6K phosphorylation through mTOR (35). The report provides a link between IKK2 and S6K in the TNF-α signaling pathway. However, it is not clear if IKK2-induced phosphorylation controls stability of the S6K protein. Our data suggest that IKK2-mediated S6K phosphorylation acts to stabilize the protein in the physiological condition. In the absence of IKK2 activity, the S6K protein will be degraded in response to TNF-α. This was observed in p50-KO hepatocytes, Ikk2 null cells, and cells treated with the IKK2 inhibitor. With normal activity, IKK2 should protect S6K from degradation in wild type cells.
Hepatic S6K represents a new target for insulin sensitization. In the molecular mechanism of insulin signaling, TNF-α activates multiple serine kinases including IKK2, JNK, and S6K (1, 34). Signal integration of these serine kinases remains to be investigated. The current study suggests that signals of IKK2 and S6K are integrated in the TNF-α signaling pathway. S6K inhibits insulin sensitivity by phosphorylation of IRS-1 proteins at several serine residues (21, 27, 28, 33). In the liver of p50-KO mice, IRS-1 phosphorylation was enhanced at Tyr-632 and reduced at Ser-636. These changes were associated with the increase in IRS-1/IRS-2 proteins and elevated phosphorylation of Akt (Thr-308 and Ser-473). This pattern of changes provides a molecular mechanism of insulin sensitization in the liver of KO mice. Restoration of S6K activity by the S6K adenovirus reversed insulin sensitivity in KO mice. The data suggest a critical role of S6K in the liver. It is expected that inhibition of the hepatic S6K by a pharmacological agent should improve insulin sensitivity.
The proteasome-mediated degradation of S6K may represent a new level of S6K regulation. In the PI3K/Akt/mTOR pathway, S6K activity is regulated by serine phosphorylation from the mTOR kinase. In p50-KO mice, phosphorylation catalyzed by the IKK2/mTOR pathway may not lead to S6K degradation. The association of deficiency in this pathway with S6K degradation suggests that mTOR may protect S6K from degradation. Regulation of S6K protein by degradation is largely unknown. Two recent studies suggest that a recombinant S6K protein can be ubiquitinated and degraded in a proteasome-dependent manner (45, 46). However, the full-length S6K protein was not used in the studies. It is not clear if the full-length S6K protein is subject to the degradation. Additionally, it is not clear under what biological conditions the S6K degradation occurs. We found that full-length S6K was degraded in response to TNF-α in the p50-KO hepatocytes.
In the current study, TNF-α was shown to induce degradation of endogenous S6K in the p50-KO cells. Absence of IKK2 activation is required for TNF activity, suggesting that mTOR activation by IKK2 may protect S6K protein from degradation induced by TNF-α. The S6K reduction was detected in the freshly isolated hepatocytes, but not in cultured primary hepatocytes. This conditional degradation might be related to dilution of TNF-α by the culture medium or loss of Kupffer cells, which are macrophage-like cells in the liver with a strong potential in TNF-α production. In the cultured primary hepatocytes, addition of exogenous TNF-α led to degradation of the S6K protein in p50-KO cells, but not in WT cells.
The current study was based on comparison of p50-KO mice with wild type mice. Compensation of gene deficiency is a common biological response to gene knock-out in animal models. In some cases, the compensation may lead to an unpredictable phenotype. The current study presents a unique phenotype in p50-KO mice. Compensation for absence of the p50 gene may contribute to the phenotype. In the human body, the complete loss of p50 function may not occur. Such a phenotype may not apply to humans. However, the study suggests that hepatic S6K may be a drug target in the treatment of insulin resistance.
In summary, the metabolic phenotypes and molecular mechanisms have been studied in the p50-KO mice with a focus on insulin sensitivity. Based on the findings, we proposed a molecular mechanism of insulin sensitivity in the p50-KO mice. In the absence of NF-κB p50, S6K inhibition leads to insulin sensitization by increasing the IRS-1 and IRS-2 activities in the liver. The S6K inhibition is from the S6K protein degradation that is induced by TNF-α, whose expression is increased in the liver through the elevated transcriptional activity of NF-κB p65. TNF-induced S6K degradation is dependent on reduction in the IKK2 activity, which is decreased in the liver probably through a negative feedback from the enhanced activity of NF-κB p65. The data suggest that hepatic S6K is a potential target in the treatment of insulin resistance.
Acknowledgments
We highly appreciate support from Dr. Jianhua Shao at the University of Kentucky in purification of adenovirus and conduction of S6K reconstitution in the p50-KO mice. We highly appreciate the outstanding editorial support from Dr. Michael Keenan, Department of Human Ecology, Louisiana State University. The quantitative RT-PCR study was conducted in the genomic core that is supported by the Clinical Nutrition Research Unit Grant 1P30 DK072476 from the National Institutes of Health, NIDDK. Hyperinsulinemic-euglycemic clamp studies were performed at the Vanderbilt Mouse Metabolic Phenotyping Center, which is supported by Grant U24-DK59637.
This work was supported, in whole or in part, by National Institutes of Health Grant DK68036 (to J. Y.). This work was also supported by the American Diabetes Association Research Award 7-07-RA-189 (to J. Y.).
- NF-κB
- nuclear factor κB
- HFD
- high fat diet, 58% calorie in fat
- IκBα
- inhibitor κBα
- IKK2
- IκBαkinase 2 (IKKβ)
- JNK
- c-Jun NH2-terminal kinase
- p50 (NF-κB1)
- NF-κB p50 subunit
- p65 (RelA)
- NF-κB p65 subunit
- S6K
- p70 ribosomal protein S6 kinase
- TNF-α
- tumor necrosis factor-α
- mTOR
- mammalian target of rapamycin
- KO
- knock-out
- MEF
- mouse embryonic fibroblast
- GTT
- glucose tolerance test
- ITT
- insulin tolerance test
- GFP
- green fluorescent protein
- RT
- reverse transcriptase
- IL
- interleukin
- WT
- wild type
- PI3K
- phosphatidylinositol 3-kinase
- IRS
- insulin receptor substrate.
REFERENCES
- 1.Hotamisligil G. S. ( 2006) Nature 444, 860– 867 [DOI] [PubMed] [Google Scholar]
- 2.Shoelson S. E., Lee J., Goldfine A. B. ( 2006) J. Clin. Invest. 116, 1793– 1801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yuan M., Konstantopoulos N., Lee J., Hansen L., Li Z. W., Karin M., Shoelson S. E. ( 2001) Science 293, 1673– 1677 [DOI] [PubMed] [Google Scholar]
- 4.Cai D., Yuan M., Frantz D. F., Melendez P. A., Hansen L., Lee J., Shoelson S. E. ( 2005) Nat. Med. 11, 183– 190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Greten F. R., Eckmann L., Greten T. F., Park J. M., Li Z. W., Egan L. J., Kagnoff M. F., Karin M. ( 2004) Cell 118, 285– 296 [DOI] [PubMed] [Google Scholar]
- 6.Gao Z., He Q., Peng B., Chiao P. J., Ye J. ( 2006) J. Biol. Chem. 281, 4540– 4547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gao Z., Hwang D., Bataille F., Lefevre M., York D., Quon M. J., Ye J. ( 2002) J. Biol. Chem. 277, 48115– 48121 [DOI] [PubMed] [Google Scholar]
- 8.Herschkovitz A., Liu Y. F., Ilan E., Ronen D., Boura-Halfon S., Zick Y. ( 2007) J. Biol. Chem. 282, 18018– 18027 [DOI] [PubMed] [Google Scholar]
- 9.Sha W. C., Liou H. C., Tuomanen E. I., Baltimore D. ( 1995) Cell 80, 321– 330 [DOI] [PubMed] [Google Scholar]
- 10.Zhong H., Voll R. E., Ghosh S. ( 1998) Mol. Cell 1, 661– 671 [DOI] [PubMed] [Google Scholar]
- 11.Bohuslav J., Kravchenko V. V., Parry G. C., Erlich J. H., Gerondakis S., Mackman N., Ulevitch R. J. ( 1998) J. Clin. Invest. 102, 1645– 1652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hunter R. B., Kandarian S. C. ( 2004) J. Clin. Invest. 114, 1504– 1511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tharappel J. C., Nalca A., Owens A. B., Ghabrial L., Konz E. C., Glauert H. P., Spear B. T. ( 2003) Toxicol. Sci. 75, 300– 308 [DOI] [PubMed] [Google Scholar]
- 14.Tharappel J. C., Spear B. T., Glauert H. P. ( 2008) Toxicol. Appl. Pharmacol. 226, 338– 344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lu Z. Y., Yu S. P., Wei J. F., Wei L. ( 2006) Neuroscience 139, 965– 978 [DOI] [PubMed] [Google Scholar]
- 16.Jhaveri K. A., Ramkumar V., Trammell R. A., Toth L. A. ( 2006) Am. J. Physiol. Regul. Integr. Comp. Physiol. 291, R1516– R1526 [DOI] [PubMed] [Google Scholar]
- 17.Beg A. A., Sha W. C., Bronson R. T., Ghosh S., Baltimore D. ( 1995) Nature 376, 167– 170 [DOI] [PubMed] [Google Scholar]
- 18.Doi T. S., Takahashi T., Taguchi O., Azuma T., Obata Y. ( 1997) J. Exp. Med. 185, 953– 961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Patti M. E., Kahn B. B. ( 2004) Nat. Med. 10, 1049– 1050 [DOI] [PubMed] [Google Scholar]
- 20.Um S. H., D'Alessio D., Thomas G. ( 2006) Cell Metab. 3, 393– 402 [DOI] [PubMed] [Google Scholar]
- 21.Um S. H., Frigerio F., Watanabe M., Picard F., Joaquin M., Sticker M., Fumagalli S., Allegrini P. R., Kozma S. C., Auwerx J., Thomas G. ( 2004) Nature 431, 200– 205 [DOI] [PubMed] [Google Scholar]
- 22.Le Bacquer O., Petroulakis E., Paglialunga S., Poulin F., Richard D., Cianflone K., Sonenberg N. ( 2007) J. Clin. Invest. 117, 387– 396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Krebs M., Brunmair B., Brehm A., Artwohl M., Szendroedi J., Nowotny P., Roth E., Fürnsinn C., Promintzer M., Anderwald C., Bischof M., Roden M. ( 2007) Diabetes 56, 1600– 1607 [DOI] [PubMed] [Google Scholar]
- 24.Ozes O. N., Akca H., Mayo L. D., Gustin J. A., Maehama T., Dixon J. E., Donner D. B. ( 2001) Proc. Natl. Acad. Sci. U.S.A. 98, 4640– 4645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Harrington L. S., Findlay G. M., Gray A., Tolkacheva T., Wigfield S., Rebholz H., Barnett J., Leslie N. R., Cheng S., Shepherd P. R., Gout I., Downes C. P., Lamb R. F. ( 2004) J. Cell Biol. 166, 213– 223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Carlson C. J., White M. F., Rondinone C. M. ( 2004) Biochem. Biophys. Res. Commun. 316, 533– 539 [DOI] [PubMed] [Google Scholar]
- 27.Tremblay F., Brûlé S., Hee Um S., Li Y., Masuda K., Roden M., Sun X. J., Krebs M., Polakiewicz R. D., Thomas G., Marette A. ( 2007) Proc. Natl. Acad. Sci. U.S.A. 104, 14056– 14061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang J., Gao Z., Yin J., Quon M. J., Ye J. ( 2008) J. Biol. Chem. 283, 35375– 35382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gao Z., Wang Z., Zhang X., Butler A. A., Zuberi A., Gawronska-Kozak B., Lefevre M., York D., Ravussin E., Berthoud H. R., McGuinness O., Cefalu W. T., Ye J. ( 2007) Am. J. Physiol. Endocrinol. Metab. 292, E84– E91 [DOI] [PubMed] [Google Scholar]
- 30.Liu H. Y., Collins Q. F., Xiong Y., Moukdar F., Lupo E. G., Jr., Liu Z., Cao W. ( 2007) J. Biol. Chem. 282, 14205– 14212 [DOI] [PubMed] [Google Scholar]
- 31.Ye J., Gao Z., Yin J., He Q. ( 2007) Am. J. Physiol. Endocrinol. Metab. 293, E1118– E1128 [DOI] [PubMed] [Google Scholar]
- 32.Esposito D. L., Li Y., Cama A., Quon M. J. ( 2001) Endocrinology 142, 2833– 2840 [DOI] [PubMed] [Google Scholar]
- 33.Khamzina L., Veilleux A., Bergeron S., Marette A. ( 2005) Endocrinology 146, 1473– 1481 [DOI] [PubMed] [Google Scholar]
- 34.Gao Z., Zuberi A., Quon M. J., Dong Z., Ye J. ( 2003) J. Biol. Chem. 278, 24944– 24950 [DOI] [PubMed] [Google Scholar]
- 35.Lee D. F., Kuo H. P., Chen C. T., Hsu J. M., Chou C. K., Wei Y., Sun H. L., Li L. Y., Ping B., Huang W. C., He X., Hung J. Y., Lai C. C., Ding Q., Su J. L., Yang J. Y., Sahin A. A., Hortobagyi G. N., Tsai F. J., Tsai C. H., Hung M. C. ( 2007) Cell 130, 440– 455 [DOI] [PubMed] [Google Scholar]
- 36.Ballard D. W., Dixon E. P., Peffer N. J., Bogerd H., Doerre S., Stein B., Greene W. C. ( 1992) Proc. Natl. Acad. Sci. U.S.A. 89, 1875– 1879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ruben S. M., Narayanan R., Klement J. F., Chen C. H., Rosen C. A. ( 1992) Mol. Cell. Biol. 12, 444– 454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Opipari A. W., Jr., Hu H. M., Yabkowitz R., Dixit V. M. ( 1992) J. Biol. Chem. 267, 12424– 12427 [PubMed] [Google Scholar]
- 39.Zhang S. Q., Kovalenko A., Cantarella G., Wallach D. ( 2000) Immunity 12, 301– 311 [DOI] [PubMed] [Google Scholar]
- 40.Kovalenko A., Chable-Bessia C., Cantarella G., Israël A., Wallach D., Courtois G. ( 2003) Nature 424, 801– 805 [DOI] [PubMed] [Google Scholar]
- 41.Song H. Y., Rothe M., Goeddel D. V. ( 1996) Proc. Natl. Acad. Sci. U.S.A. 93, 6721– 6725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mauro C., Pacifico F., Lavorgna A., Mellone S., Iannetti A., Acquaviva R., Formisano S., Vito P., Leonardi A. ( 2006) J. Biol. Chem. 281, 18482– 18488 [DOI] [PubMed] [Google Scholar]
- 43.Arkan M. C., Hevener A. L., Greten F. R., Maeda S., Li Z. W., Long J. M., Wynshaw-Boris A., Poli G., Olefsky J., Karin M. ( 2005) Nat. Med. 11, 191– 198 [DOI] [PubMed] [Google Scholar]
- 44.Wunderlich F. T., Luedde T., Singer S., Schmidt-Supprian M., Baumgartl J., Schirmacher P., Pasparakis M., Brüning J. C. ( 2008) Proc. Natl. Acad. Sci. U.S.A. 105, 1297– 1302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Panasyuk G., Nemazanyy I., Filonenko V., Gout I. ( 2008) Biochem. Biophys. Res. Commun. 369, 339– 343 [DOI] [PubMed] [Google Scholar]
- 46.Wang M. L., Panasyuk G., Gwalter J., Nemazanyy I., Fenton T., Filonenko V., Gout I. ( 2008) Biochem. Biophys. Res. Commun. 369, 382– 387 [DOI] [PubMed] [Google Scholar]