Abstract
ARR19 (androgen receptor corepressor of 19 kDa), which encodes for a leucine-rich protein, is expressed abundantly in the testis. Further analyses revealed that ARR19 was expressed in Leydig cells, and its expression was differentially regulated during Leydig cell development. Adenovirus-mediated overexpression of ARR19 in Leydig cells inhibited testicular steroidogenesis, down-regulating the expression of steroidogenic enzymes, which suggests that ARR19 is an antisteroidogenic factor. Interestingly, cAMP/luteinizing hormone attenuated ARR19 expression in a fashion similar to that of GATA-1, which was previously reported to be down-regulated by cAMP. Sequence analysis of the Arr19 promoter revealed the presence of two putative GATA-1 binding motifs. Further analyses with 5′ deletion and point mutants of putative GATA-1 binding motifs showed that these GATA-1 binding sites were critical for high promoter activity. CREB-binding protein coactivated GATA-1 and markedly increased the activity of the Arr19 promoter. Both GATA-1 and CREB-binding proteins occupied the GATA-1 motifs within the Arr19 promoter, which was repressed by cAMP treatment. Altogether, these findings demonstrate that ARR19 is the target gene of GATA-1 and suggest that ARR19 gene expression in testicular Leydig cells is regulated by luteinizing hormone/cAMP signaling via the control of GATA-1 expression, resulting in the control of testicular steroidogenesis.
Arr19 (androgen receptor corepressor of 19 kDa), also referred to as Cklfsf2a, is a member of the chemokine-like factor superfamily (CKLFSF), a group of novel proteins that operate as a functional bridge between chemokines and members of the transmembrane 4 superfamily. Members of the CKLFSF family have been suggested to perform an important role across a broad range of physiological and pathological processes. In humans, the CKLFSF family harbors nine genes, CKLF and CKLFSF1 to -8, which exhibit high degrees of similarity at both the amino acid and structural levels. Among these, CKLF, CKLKSF1, and CKLFSF2 are closely related to each other and are tightly linked on chromosome 16q22.1 (1, 2). Human CKLFSF2 is expressed abundantly in the testis and has two counterparts in the mouse, Cklfsf2a and Cklfsf2b. Cklfsf2a and Cklfsf2b evidence a similar expression pattern and amino acid identities of 47.6 and 45.5%, respectively, with human CKLFSF2 (3). Mouse Cklfsf2a/Arr19 was originally cloned as a potential androgen response gene in the testis and was further characterized as a novel androgen receptor corepressor (4). The Arr19 gene is located on chromosome 8 and encodes for a leucine-rich and highly hydrophobic 19-kDa protein. ARR19 is expressed abundantly in the testis and moderately in other male reproductive organs, such as the prostate (3, 4).
Testicular gene expression and function are tightly regulated by the pituitary tropic hormones, luteinizing hormone (LH) and FSH. In response to hormonal signals, gene expression is modulated via the cAMP-dependent intracellular pathway, resulting in the activation of protein kinase A. The best studied target of cAMP/protein kinase A signaling is cAMP-response element (CRE)2-binding protein (CREB) transcription factor, which binds as a dimer to the 8-bp palindromic sequence CRE detected in the regulatory regions of some cAMP-regulated genes (5). In the testis, however, the promoter of several cAMP-regulated genes, such as StAR (steroidogenic acute regulatory protein), inhibin α, P450c17, p450scc, and aromatase, lacks the “classical” CRE elements. This suggests that other transcription factors, besides CREB, must also function as downstream effectors of cAMP signaling. GATA-1, a transcription factor, is expressed in murine testicular cells and in the tumor cell lines derived from them (6–14). Interestingly, it has been reported that the mRNA and protein levels of GATA-1 are negatively regulated by cAMP/gonadotropin in testicular cell lines, thereby suggesting its involvement in the gene regulation of cAMP-regulated genes in testicular cells (15).
GATA-1 belongs to the GATA-binding protein family, members of which share conserved zinc finger motifs in their DNA-binding domains and recognize a consensus sequence (T/A)GATA(A/G) (16–20). GATA-1 was originally identified as a transcription factor that was exclusively required for the cell-specific expression of globin genes and other erythroid lineage-specific genes (20–24, 25). The results of previous studies have suggested that GATA-1 is capable of regulating target gene expression via its interaction with other factors as well as through its own activation domain (26, 27). Some proteins, including Friend of GATA-1 (FOG, FOG-1, or ZFPM1), tumor suppressor protein retinoblastoma (RB), p300, and CREB-binding protein (CBP), have been shown to interact with GATA-1 and to modulate its transcriptional activity (26, 28–33).
Previous studies point to CBP as a potential co-activator of GATA-1. CBP, a protein with histone acetyltransferase property (34–36, 37), binds to GATA-1 in vitro and in vivo (38). Several mechanisms by which CBP intervenes in transcription regulation have been suggested. The binding of CBP to transcription factors may constitute a means by which histone acetyltransferase can be recruited into the vicinity of the nucleosomes. CBP can also serve as a bridging molecule between components of general transcription machinery and enhanceosome complexes (39). Furthermore, CBP has been shown to acetylate transcription factors, including GATA-1, which may exert a direct effect on their function (40). It was recently reported that the acetylation of GATA-1 promotes its association with other proteins (41).
In the present study, we show that Arr19, a control factor for testicular steroidogenesis, is a novel target gene of GATA-1 in testicular Leydig cells. The expression and down-regulation pattern of ARR19 by cAMP/LH were determined to be very similar to those of GATA-1, in both a time- and dose-dependent fashion. Analysis of the Arr19 promoter revealed that two putative GATA-1 binding sites are functional and necessary for the gene regulation of Arr19 by cAMP/LH. Moreover, CBP coupled with GATA-1 markedly stimulated the Arr19 promoter activity. In addition, we show that both GATA-1 and CBP occupy the GATA-1 binding sites in the testis as well as cells, which were repressed by cAMP treatment. These results indicate that the expression of Arr19 is important for testicular steroidogenesis and is regulated by cAMP/LH via the control of GATA-1 expression in testicular Leydig cells.
EXPERIMENTAL PROCEDURES
Animals and Treatment
FVB mice (postnatal days 14, 21, 28, and 56) were purchased from a commercial supplier (Daehan Laboratories, Daejeon, Korea). The selection of mouse ages was based on previous reports of the development of adult Leydig cells (42). Animals were sacrificed by CO2 asphyxiation, and testes were extracted for Western blot analysis. For ChIP assays and Western blot analyses, 14-day-old mice were injected intraperitoneally with 10 IU of human chorionic gonadotrophin (hCG; Sigma) for 6 h. Immunohistochemical analyses were performed using 14-day-old young mice and 56-day-old adult mice. Ethical treatment of the animals was carried out according to National Institutes of Health standards.
Plasmids
Mammalian expression vectors of GATA-1 (pXM-GATA-1) and CBP (pcDNA3-CBP) have been described previously (35, 43). In order to clone the promoter region of the Arr19 gene, a PCR fragment was amplified using primers harboring the KpnI and SacI restriction enzyme sites in their forward and reverse primers, respectively. The primer sequences (sense, 5′-GATGTCTCAGGGGTACTTGC-3′; antisense, 5′-GTTGTTAGCAGGTAGTTCTC-3′) were derived from DNA sequences of a mouse genomic data base, which is publicly available. PCR was conducted using a Pfu PCR kit in accordance with the manufacturer's recommendations (Elpis Biotech, Inc.). The fragments were purified using a gel extraction kit (ConcertTM rapid gel extraction system; Life Technology) and cloned into the pBluescript® II KS+/− phagemid vector. The insert was isolated via double digestion of positive clones with KpnI and SacI and was then subcloned directionally upstream of the luciferase reporter gene of the pGL3-basic vector (Promega). This construct was designated ARR19-P1. Deletion constructs of the Arr19 promoter were derived from the ARR19-P1 plasmid with restriction digestion via the following enzymes: ARR19-P2 (KpnI/AvaI), ARR19-P3 (NsiI/NheI), ARR19-P4 (SacI/SacI), and ARR19-P5 (BstXI). ΔCRE-ARR19-P1 was constructed from ARR19-P1, which was digested with AatII, blunt-ended, and self-ligated. Mutations of GATA-1 motifs in the Arr19 promoter were created using two sets of the following mutagenic primers (sense (5′-TAAGTAAGTTTCGTTTTTCTTCATT-3′) and antisense (5′-ATGAAGAAAAACAGAGAAACTTACTTA-3′); sense (5′-CATTCCTATTCCTCTGCCACGA GGGCAG-3′) and antisense (5′-CTGCCCTCGTGGCAGAGGAATAGGAATG-3′)), using a QuikChange site-directed mutagenesis kit (Stratagene) in accordance with the manufacturer's recommendations. The mutations of the GATA-1 motifs were confirmed by DNA sequencing.
Cell Culture and Transient Transfection Assay
MA-10 cells were maintained in RPMI 1640 medium supplemented with 25 mm HEPES, 2 mm l-glutamine, 15% horse serum, and antibiotics, and R2C cells were maintained in F10 medium supplemented with 15% horse serum and 2.5% fetal bovine serum. HEK 293T cells were maintained in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum and antibiotics. All cells were cultured at 37 °C in 5% CO2. For hormone treatments, MA-10 cells were plated at a density of 3.0 × 106 cells/100-mm Petri dish in medium containing 10% charcoal-stripped serum. Forty hours later, 8-bromo-cyclic AMP (Sigma) or LH was added to the medium as indicated.
Transfections were conducted using Superfect (Qiagen) for HEK 293T cells and Lipofectamine Plus (Invitrogen) for MA-10 cells, in accordance with the manufacturer's recommendations. The cells were plated in 24-well plates and were transfected with expression vectors, a reporter gene, and the LacZ expression plasmid, pCMVβ (Clontech), as a control for transfection efficiency. The total amount of DNA was kept constant by the addition of appropriate amounts of pcDNA3 empty vector. After 24 h of incubation, the cells were harvested in lysis buffer containing 0.1% Triton X-100 and 0.2 m Tris-HCl (pH 8.0). Luciferase and β-galactosidase activities were assayed as described previously (44). The levels of luciferase activity were normalized to β-galactosidase activity.
Primary Leydig Cell Purification
Purification of mouse Leydig cells was carried out as described previously (45), with some modification. Animals were first anesthetized and then killed by decapitation. Six testes per set (14, 21, 28, and 56 days) were removed, and testicular cells were dispersed by treating the decapsulated testes with collagenase (0.25 mg/ml; Sigma) in M199 media (Invitrogen) at room temperature for 20 min with gentle shaking. After incubation, the dispersed tissues were diluted with M199, and the solution was filtered. Interstitial cells were precipitated by centrifugation of the filtrate and were washed once with M199 and twice with phosphate-buffered saline.
Preparation of Recombinant Adenovirus
For the ectopic expression of the mouse Arr19, an adenoviral delivery system was used (46). Briefly, the Arr19 cDNA was cloned into pAdTrack-CMV shuttle vector. Homologous recombination was performed by transformation of adEasy-BJ5138 competent cells with pAdTrack-CMV-ARR19 together with adenoviral gene carrier vector. The recombinant viruses were selected, amplified in HEK 293 cells, and purified by cesium chloride density centrifugation. Viral titers were measured using Adeno-X rapid titer (BD Biosciences) according to the manufacturer's instructions.
Western Blot Analysis
Cell lysates were prepared in radioimmune precipitation cell lysis buffer (50 mm Tris-HCl, pH 8.0, 100 mm NaCl, 1% Nonidet P-40, 5 mm EDTA, pH 8.0, 1 mm Na3VO4, 1 mm Na4P2O7, 1 g/ml aprotinin, 0.1 g/ml leupeptin, 1 g/ml pepstatin A, 0.1 mm phenylmethylsulfonyl fluoride) and were separated via SDS-PAGE. Proteins were transferred onto nitrocellulose transfer membranes and were subsequently subjected to Western blot analysis with anti-ARR19, anti-Atp8b3 (produced in our laboratory), anti-GATA-1, anti-CBP, anti-DAX-1, anti-3β HSD, anti-P450scc, anti-StAR, and anti-actin (Santa Cruz Biotechnology, Inc., Santa Cruz, CA) antibodies. Signals were then visualized using an ECL kit (Amersham Biosciences). Actin signals were used as a loading control.
Northern Blot Analysis
Total RNA was prepared from MA-10 cells using Tri Reagent (Molecular Research Center, Inc.). Twenty micrograms of total RNA was separated on a 1.2% denaturing agarose gel, transferred onto Zeta-Probe nylon membrane (Bio-Rad), and immobilized by UV cross-linking. The membrane was hybridized with a random-primed 32P-labeled GATA-1 and Arr19 cDNA probe, as described previously (4, 15). The membrane was reprobed for glyceraldehydes-3-phosphate dehydrogenase as a loading control.
Radioimmunoassay
Testosterone concentrations were measured by radioimmunoassay. The exponentially growing R2C cells were cultured in F10 medium supplemented with 15% charcoal-stripped fetal bovine serum. Culture medium was collected for radioimmunoassay at several time points (0–36 h) of infection with Ad-GFP or Ad-ARR19. The experiment was repeated three times, and assay procedures were followed as described previously (44).
Immunohistochemistry
The testes were fixed in 4% paraformaldehyde and embedded in paraffin. Tissue sections (4–6 μm) were subjected to immunohistochemistry in accordance with the standard procedure (47) using rabbit anti-ARR19 or rat monoclonal anti-GATA-1 antibody (Santa Cruz Biotechnology). A horseradish peroxidase-streptavidin Histostain-Plus (Zymed, S. San Francisco, CA) system was employed to visualize the signals. The samples were analyzed via light and phase-contrast microscopy (Leica DMRXA microscope; Leica AG, Heerbrugg, Switzerland).
5′-Rapid Amplification of cDNA Ends (5′-RACE)
In order to map the transcription start site of the Arr19 gene, we utilized a 5′-RACE SMART RACE cDNA amplification kit (48, 49). Total RNA was isolated from MA-10 cells, and first strand cDNA was synthesized using a modified lock-docking oligo(dT) primer and SMART II A oligonucleotide, in accordance with the manufacturer's suggestions. The first round of the RACE PCR using UPM 10× universal Primer A mix and gene-specific MR1 primers (5′-GAACAGGTCAGGAGACACCAG-3′) yielded no distinct amplification products, except for a faint streak (data not shown). However, the nested PCR of the first PCR products using nested universal Primer A and a second set of gene-specific MR2 primers (5′-CTGTTCAGCGACTGCAGACAA-3′) yielded a band in the range of ∼190 bp. This band was gel-purified and cloned into the T/A PCR cloning vector. Four clones were sequenced and analyzed.
Immunoprecipitation
Nuclear extracts of MA-10 cells were prepared by lysing cells in hypotonic buffer containing 10 mm Hepes-KOH (pH 8.0), 1.5 mm MgCl2, 10 mm KCl, 0.1 mm ZnCl2, 0.5 mm dithiothreitol, 0.5 mm phenylmethylsulfonyl fluoride, 0.5 μg/ml leupeptin, and 2 μg/ml aprotinin. After 20 min of swelling on ice, cells were vortexed and spun at 1500 rpm for 5 min. Nuclei were extracted with high salt buffer containing 20 mm Hepes-KOH (pH 8.0), 25% glycerol, 420 mm NaCl, 1.5 mm MgCl2, 0.1 mm ZnCl2, 0.5 mm dithiothreitol, 0.5 mm phenylmethylsulfonyl fluoride, 0.5 μg/ml leupeptin, and 2 μg/ml aprotinin for 20 min on ice. After centrifugation, supernatant was diluted to reduce the NaCl concentration to 150 mm. Immunoprecipitations were performed with anti-CBP antibody (Santa Cruz Biotechnology) or nonimmune serum. GATA-1 was immunoprecipitated with anti-GATA-1 antibody (Santa Cruz Biotechnology) or isotype-matched irrelevant antibody as a control (anti-PECAM; Santa Cruz Biotechnology). Immune complexes were recovered with protein G-Sepharose (for IgG; Amersham Biosciences) and separated by SDS-PAGE, followed by Western blot analysis. We observed a strong propensity of GATA-1, together with anti-GATA-1 monoclonal antibodies, to bind to protein A- or G-Sepharose beads, nonspecifically, which was reduced after multiple washes with 350 mm NaCl.
Electrophoretic Mobility Shift Assay
An electrophoretic mobility shift assay was performed as described previously (9). In brief, nuclear cell extract (1 μg) prepared from MA-10 cells was incubated with radiolabeled double-stranded oligonucleotides containing either GATA-1A (5′-GGGGAAGAAAAAGATAGAAACTTA-3′) or GATA-1B (5′-GGGGCCCTCGTGGGATAGGAATAG-3′) in the presence of 1 μg of poly(dI/dC). The binding reactions were performed at room temperature for 30 min and on ice for another 30 min. For competition analysis, a 200-fold excess of specific nonradiolabeled double-stranded oligonucleotides containing each GATA motif was added together with radiolabeled oligonucleotides to the reaction mixtures. NurRE double-stranded oligomers (5′-GGGGTGATATTTACCTCCAAATGCAAATGCCA-3′) were used as nonspecific competitors. Anti-GATA-1 or anti-PECAM (as a nonspecific control) was added prior to incubation on ice for 30 min. The binding reactions were analyzed on a 5% polyacrylamide gel, as described previously (9).
Chromatin Immunoprecipitation (ChIP) Assay
Testicular cells of decapsulated testis (50) and MA-10 cells were cross-linked with 1% formaldehyde and incubated for 20 min with TSE I buffer (100 mm Tris-HCl, pH 9.4, and 10 mm dithiothreitol) at 30 °C, and the cells were washed and processed for ChIP assays as described previously (44). Anti-GATA-1 and anti-CBP antibodies were utilized for immunoprecipitation. The immunoprecipitated DNA was then subjected to PCRs using two sets of primers: sense 5′-GATGTCTCAGGGGTACTTGCCACTA-3′ and antisense 5′-CACTGGCCTGAACTTGACTAGCTATTC-3′ to amplify a 447-bp fragment spanning the region from −1123 to −676, and sense 5′-AGCCCTTTCCCCTGAGAAACTCACACA-3′ and antisense 5′-GTTGGGTGGCAGAGAGCTTAGTTTCC-3′ to amplify a 473-bp fragment spanning the region from −856 to −383, which harbors the putative GATA-1 binding sites of the Arr19 promoter. As negative controls, no antibody or normal rabbit serum was added for immunoprecipitation, and PCRs were done using β-actin primer pairs (sense, 5′-GAGACCTTCAACACCCCAGCC-3′; antisense, 5′-CCGTCAGGCAGCTCATAGCTC-3′), which amplify a 362-bp region spanning exon 4 of the β-actin gene.
Real Time PCR
Various transcripts in proper medium-treated cells were analyzed by real time PCR using a LightCycler, as described previously (51).
RESULTS
Expression of ARR19 in Leydig Cells during the Development of Mouse Testis
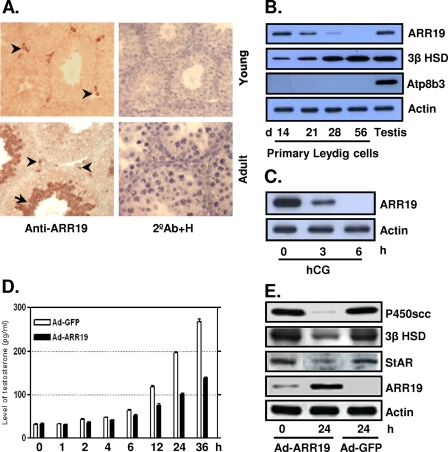
Our previous study showed that ARR19 is abundantly expressed in the testis, and its expression appears to be developmentally regulated (4). In order to evaluate the function and gene regulation of Arr19 in mouse testis, we initially attempted to identify the cell types that express ARR19. Immunohistochemical analyses were conducted using testis sections prepared from both young mice at day 14 and adult mice at day 56 (Fig. 1A). In young testis, the expression of ARR19 was detected only in interstitial regions between seminiferous tubules, whereas in adult testis, ARR19 was highly expressed in the majority of the haploid germ cells and in a small population of cells in the interstitial area.
FIGURE 1.
Expression of ARR19 in testicular Leydig cells and its function as a steroidogenic control factor. A, ARR19 expression was detected in the interstitial compartment in 14-day-old mouse testis (Young) and in a subset population of the interstitial cells in 56-day-old testis (Adult). ARR19 was also expressed within seminiferous tubules (haploid spermatids) in 56-day-old testis (Adult). The arrowheads indicate cells that express ARR19 within the interstitial region, and an arrow indicates ARR19-expressing germ cells. 2°Ab+H, immunocytochemistry with the secondary antibody (2°Ab) only for a negative control along with nuclear staining with hematoxylene (H). B, ARR19 expression was down-regulated during the development of Leydig cells. Primary Leydig cells were isolated from 14-, 21-, 28-, and 56-day-old mouse testes, and Western blot analyses were subsequently performed. Atp8b3 is a germ cell marker that is used to check for cross-contamination. C, ARR19 expression was down-regulated by LH (hCG) signaling. 14-day-old mice were injected with hCG for 0–6 h, followed by Western blot analyses of whole testis extracts. D, testosterone biosynthesis was inhibited by adenovirus-mediated overexpression of ARR19 (Ad-ARR19). R2C cells, which were grown in media supplemented with 15% charcoal-stripped serum, were infected with Ad-ARR19 or control Ad-GFP virus, and media were collected at different time points (0–36 h) for radioimmune assays. E, the expression of steroidogenic enzyme genes was inhibited by ARR19 overexpression in R2C cells. R2C cells were infected with Ad-ARR19 and control Ad-GFP for 24 h. The whole cell proteins were extracted, followed by Western blot analysis with anti-P450scc, anti-3β HSD, and anti-StAR antibody.
Since the expression of ARR19 was located in interstitial regions and the interstitial regions are mainly populated by Leydig cells in mouse testis, we suspected that ARR19 is expressed in Leydig cells. To test this hypothesis, we performed Western blot analysis by using primary Leydig cells isolated from postnatal days 14, 21, 28, and 56 (Fig. 1B), based on the development of adult Leydig cells in the testis (42). As expected, ARR19 expression was high at day 14 and gradually decreased to moderate levels at days 21 and 28. At day 56, ARR19 expression was almost totally abrogated. Taken together, these results suggest that the ARR19 is expressed in early stages of adult Leydig cells and also imply that its expression is developmentally regulated during the differentiation of adult Leydig cells.
Previously, it was well documented that LH/hCG controls the differentiation of adult Leydig cells through intracellular secondary messenger cAMP during development. To evaluate the possibility of LH/cAMP involvement in ARR19 expression, 14-day-old mice were injected intraperitoneally with hCG (human chorionic gonadotropin) for 0–6 h, and ARR19 expression was analyzed by Western blot analysis using whole testis extracts (Fig. 1C). Interestingly, the expression of ARR19 was completely abrogated within 6 h of treatment, whereas it was moderately inhibited at 3 h of hCG treatment. Taken together, these findings strongly suggest that ARR19 is expressed in the early stages of adult Leydig cell development, and its expression is controlled by LH/cAMP signaling.
Effect of Overexpression of ARR19 on Steroidogenesis in Leydig Cells
Since steroidogenesis in adult Leydig cells is controlled by steroidogenic enzyme genes, such as StAR, 3β HSD, P450scc, etc., and the expression of these genes is regulated by pituitary gonadotropin luteinizing hormone, LH, we attempted to determine the effect of adenovirus-mediated overexpression of Arr19 (Ad-ARR19) in the R2C rat Leydig cell line, which is constitutively steroidogenic (44). The testosterone level of cultured medium became lower than that of the control (Ad-GFP) after 2 h of infection with Ad-ARR19 and reached only ∼50% of the control level after 36 h of infection (Fig. 1D). This inhibition of testosterone biosynthesis was accompanied by decreased protein levels of steroidogenic enzymes, such as StAR, 3β HSD, and P450scc (Fig. 1E). The protein levels began to decrease after just 1 or 2 h of infection (data not shown) and were significantly reduced after 24 h of infection. These results suggest that ARR19 plays a crucial role in the control of testosterone production in Leydig cells, which occurs through the regulation of steroidogenic enzyme gene expressions.
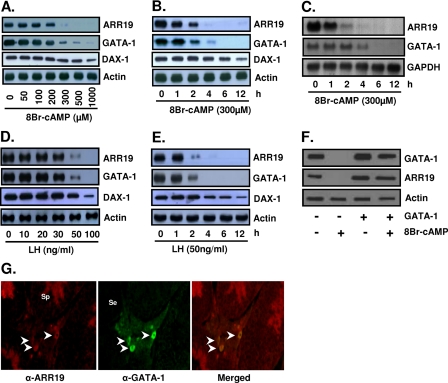
cAMP/LH Suppresses ARR19 Expression via GATA-1 Regulation in Leydig Cells
Promoter analysis of the Arr19 gene revealed the existence of putative GATA-1 binding sites. This suggests that the Arr19 gene may be regulated by the GATA-1 transcription factor. Since GATA-1 is expressed in MA-10 mouse Leydig cells and its expression is regulated by cAMP (15), we first assessed the expression of ARR19 and the effect of cAMP on ARR19 expression in MA-10 cells. As shown in Fig. 2A, the ARR19 protein was expressed in MA-10 cells, and its expression was inhibited by cAMP in a dose-dependent manner, as was GATA-1 protein expression. The administration of cAMP at 500 μm for 4 h completely abolished ARR19 protein expression. The inhibitory effect of cAMP on ARR19 protein expression was also demonstrated to be time-dependent (Fig. 2B). The reduction in the ARR19 protein level could be detected 1 h after treatment of MA-10 cells with 300 μm cAMP. Less than 5–10% of the ARR19 protein remained in the cells treated for 4 h with 300 μm cAMP, and treatment for longer periods of time resulted in the total abolishment of ARR19 protein expression. A similar inhibitory effect of cAMP on mRNA levels of Arr19 and GATA-1 was also observed, although the mRNA level of GATA-1 was less rapidly reduced than its protein level (Fig. 2C). These inhibitory effects of cAMP on ARR19 expression evidenced a pattern quite similar to those observed with GATA-1 expression, thus suggesting a correlation between the expressions of these two genes in MA-10 cells.
FIGURE 2.
Reduction of ARR19 expression by cAMP/LH treatments in MA-10 Leydig cells. A, MA-10 cells were treated with different doses of cAMP for 4 h prior to harvesting. B and C, MA-10 cells were treated with 300 μm cAMP and harvested at different time points. D, MA-10 cells were treated with different doses of LH for 2 h prior to harvesting. E, MA-10 cells were treated with 50 ng/ml LH and harvested at different time points. F, MA-10 cells transfected with (+) or without (−) GATA-1 expression plasmids were treated with or without 300 μm cAMP for 6 h prior to harvesting. A, B, D, E, and F, total cell extracts (50 μg of protein) were subjected to Western blot analysis for ARR19 and GATA-1 protein levels. DAX-1 was used as a positive control for the LH/cAMP response. C, total RNAs (20 μg) were subjected to Northern blot analysis for Arr19 and GATA-1 mRNA levels. G, coexpression of ARR19 and GATA-1 in the interstitial area of mouse testis. ARR19 expression was detected in a small number of cells in the interstitial compartment and haploid spermatids within seminiferous tubules (left column). GATA-1 expression was detected in some cells within the interstitial region (middle column). Overlapping of ARR19 and GATA-1 signals is shown in the right column. Arrowheads, cells that express ARR19 and GATA-1 within the interstitial region; Se, Sertoli cell; Sp, spermatid. 8Br-cAMP, 8-bromo-cyclic AMP.
Since cAMP is a secondary signaling molecule of the LH signaling pathway in Leydig cells, we anticipated a similar inhibitory effect of LH on the expressions of the ARR19 and GATA-1 proteins. In order to verify this expectation, we conducted Western blot analyses with MA-10 cells, which were treated with different concentrations of LH for different amounts of time. As shown in Fig. 2D, 2 h of treatment with 50 ng/ml LH clearly reduced the expression of ARR19 as well as GATA-1 protein, and 2 h of LH treatment at 100 ng/ml completely abolished both ARR19 and GATA-1 expression. The inhibitory effect of LH on the Arr19 and GATA-1 genes was also shown to occur in a time-dependent manner (Fig. 2E). LH treatment at a concentration of 50 ng/ml for 2 h significantly reduced the expressions of the ARR19 and GATA-1 proteins, and treatment for 4 h or longer resulted in the total abolishment of ARR19 expression. The expression of DAX-1, a positive control that was previously reported to be down-regulated by cAMP/LH (52), was partially inhibited at a particular amount and duration of treatment with cAMP/LH used for complete down-regulation of ARR19 and GATA-1. Furthermore, to confirm the direct relationship between GATA-1 and ARR19 expression in MA-10 cells, GATA-1 was expressed ectopically, and the pattern of ARR19 and GATA-1 down-regulation was observed with or without cAMP treatment (Fig. 2F). The results clearly indicate that the ectopic expression of GATA-1 rescues its complete down-regulation of ARR19 protein upon treatment with 300 μm cAMP for 6 h. Furthermore, immunohistochemical analysis of mouse testis revealed the coexpression of ARR19 with GATA-1 proteins in certain cells within the interstitial area of the testis (Fig. 2G). Taken together, these results demonstrate that the patterns of the inhibitory effect of LH on GATA-1 and ARR19 protein expression are extremely similar and also show that the down-regulation of ARR19 is rescued by GATA-1, which strongly suggests the possibly of Arr19 gene regulation by GATA-1 in MA-10 cells.
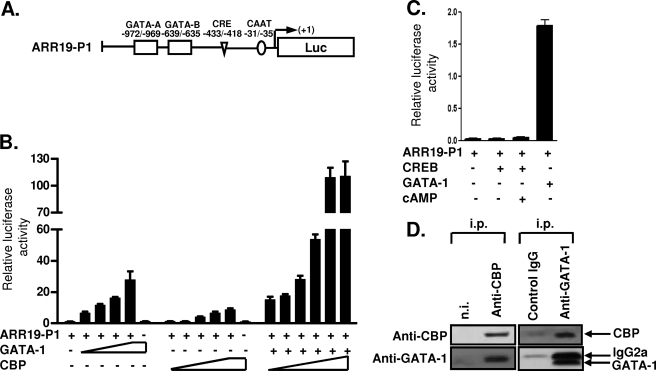
Activation of the ARR19 Promoter by the GATA-1 Transcription Factor
To analyze the promoter region of Arr19, we cloned the 1.3-kb upstream region of the Arr19 open reading frame by PCR with upstream specific primers and bacterial artificial chromosome harboring genomic sequences upstream of the mouse Arr19 gene as a template. The transcription start site of the Arr19 gene was then determined using a 5′-RACE SMART RACE cDNA amplification kit (48, 49) (data not shown) and was verified via comparison with the sequences of expressed sequence tag clones (GenBankTM accession number BY706860), which were acquired from a mouse testis cDNA library that was constructed using the cap trapper method (53).
For the study of the Arr19 promoter, the reporter plasmid pARR19-P1 was constructed by cloning a 1-kb upstream fragment from the transcription initiation site of the Arr19 gene into the pGL3basic promoterless luciferase construct (Fig. 3A). The 1-kb upstream region harbored the consensus sequence of CAAT, CRE, and GATA-1 motifs. Transient transfection assays of HEK 293T cells with the reporter and the GATA-1 expression plasmid revealed that GATA-1 induced the expression of the reporter luciferase gene in a dose-dependent manner (Fig. 3B), whereas other members of the GATA-binding protein family, GATA-2, -3, and -4, did not (data not shown). Meanwhile, the putative CRE was probably nonfunctional, because the Arr19 promoter activity was only slightly affected by the expression of the CREB either with or without cAMP treatment (Fig. 3C). CREB has been known to activate the transcription of CRE-containing promoters following an elevation of intracellular cAMP levels (25).
FIGURE 3.
Activation of the Arr19 promoter by GATA-1 transcription factor. A, schematic representation of the 1-kb Arr19 promoter (ARR19-P1) marked with putative GATA-1 and CRE motifs, which were identified using the MatInspector Professional Program. B, HEK 293T cells were transiently transfected with the ARR19-P1 reporter construct along with increasing amounts of GATA-1 or CBP expression plasmid or increasing amounts of CBP with a constant amount of GATA-1 expression plasmid, as indicated. C, MA-10 cells were transiently transfected with the ARR19-P1 reporter construct along with CREB or GATA-1 expression plasmid. After 36 h of transfection, cells were incubated for another 6 h with or without cAMP (300 μm). Values represent the means ± S.E. of at least three independent experiments. D, CBP was precipitated with rabbit anti-CBP serum or, as a control, rabbit nonimmune serum (n.i.) containing equal amounts of total IgG. GATA-1 protein was precipitated with rat anti-GATA-1 monoclonal antibody or an isotype-matched irrelevant control rat monoclonal antibody. The secondary anti-rat IgG antibody used for Western blot analysis recognized the rat IgG2a used for the GATA-1 immunoprecipitation. i.p., antibodies used for immunoprecipitations.
Coactivation of the Transcriptional Activity of GATA-1 by CBP on the ARR19 Promoter
The CBP also induced the reporter expression from the Arr19 promoter but to a much lesser extent than was observed with GATA-1 (Fig. 3B). Interestingly, CBP together with GATA-1 synergistically activated the Arr19 promoter, thereby resulting in a magnificent induction of the reporter expression as compared with what was observed with each factor alone (Fig. 3B). It was previously reported that CBP functions as a coactivator for the basic transcription factor and also for GATA-1 in MEL cells, thus enhancing their transcriptional activity (32, 38).
Since the putative CRE in the Arr19 promoter was nonfunctional (Fig. 3C) and thus CREB probably did not bind to it, we thought that CBP protein might work with GATA-1, rather than with CREB, in the Arr19 promoter. To determine whether CBP functions as a coactivator for GATA-1 via physical interaction, as previously reported in erythroid cells (38), we investigated the association of GATA-1 with CBP in MA-10 cells. Nuclear extracts of MA-10 cells were subjected to immunoprecipitation followed by Western blot analysis. As shown in Fig. 3D, anti-CBP antibody, but not control rabbit serum, coprecipitated GATA-1 protein with CBP, even after extensive washings in a buffer containing 350 mm NaCl. The inverse experiment shows that monoclonal anti-GATA-1 antibody, but not an isotype-matched control antibody, coprecipitated CBP protein with GATA-1. These findings suggest that in MA-10 cells, CBP interacts with GATA-1 and coregulates the expression of ARR19.
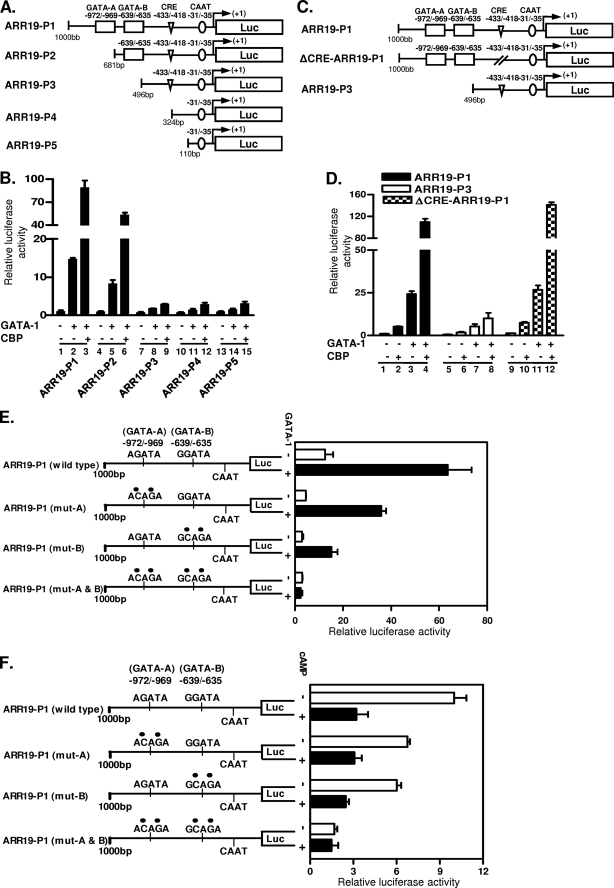
Mapping of GATA-1 Binding Sites in the ARR19 Promoter
In order to define the region(s) necessary for the GATA-1 activation of the Arr19 promoter and its cooperation with CBP, a series of deletion mutants of the Arr19 promoter were constructed (Fig. 4A). Reporter assays indicated that the deletion of the region from −1000 to −682 (ARR19-P2), which removes the first putative GATA-1 binding site (GATA-A), reduced the GATA-1-enhanced 1-kb Arr19 promoter activity by ∼40% (Fig. 4B, compare column 2 with column 5). In contrast, the deletion of the region from −1000 to −497 (ARR19-P3) and further deletion up to −325 (ARR19-P4) and to −111 (ARR19-P5), which removes both of the putative GATA-1 binding sites (GATA-A and GATA-B), reduced the GATA-1-enhanced 1-kb Arr19 promoter activity by ∼80% (Fig. 4B, compare column 2 with columns 8, 11, and 14).
FIGURE 4.
Mapping and mutational analyses of GATA-1 binding sites in the ARR19 promoter. A, schematic representation of deletion mutants of the Arr19 promoter. B, HEK 293T cells were transiently transfected with each deletion mutant construct of the Arr19 promoter and GATA-1 expression plasmid along with (+) or without (−) the CBP expression construct. C, schematic representation of Arr19 promoter mutants. ΔCRE-ARR19-P1 is a mutated Arr19 promoter at the putative CRE with a 4-bp deletion. D, HEK 293T cells were transiently transfected with each promoter construct along with GATA-1 and CBP expression plasmid. E, Arr19 promoter constructs are schematically depicted with the indication of wild-type or mutated sequences (●) of GATA-1 binding motifs. MA-10 cells were transiently transfected with each promoter construct along with (+) or without (−) the GATA-1 expression plasmid. F, MA-10 cells were transiently transfected with each promoter construct and were treated with (+) or without (−) cAMP for 6 h prior to harvesting. Values represent the means ± S.E. of at least three independent experiments.
Meanwhile, the coexpression of CBP with GATA-1 showed that their synergistic action for the activation of the Arr19 promoter was dramatically reduced with deletion beyond the point of −497 (Fig. 4B, compare columns 3 and 6 with columns 9, 12, and 15), which removes both of the putative GATA-1 binding sites, and caused only minimal activation of the Arr19 promoter by GATA-1. These results indicate that the region between −1000 and −497, which harbors two putative GATA-1 binding sites, is crucial for the profound activity of the Arr19 promoter, which is achieved via the promoter activation by GATA-1 protein and its augmentation by the CBP protein.
Within the 1-kb Arr19 promoter region, one putative CRE was identified between −433 and −418. In order to determine the role of the putative CRE in the promoter activity, although it seemed to be nonfunctional (Fig. 3C), we constructed ΔCRE-ARR19-P1, in which 4 bp (−425 to −422) of the CRE was deleted (Fig. 4C). Transient transfection analyses showed that the deletion of the putative CRE did not significantly affect the activation of the Arr19 promoter by CBP via its synergistic action with GATA-1 (Fig. 4D, compare column 4 with column 12). These results confirm that the putative CRE is nonfunctional and suggest that CBP functions via protein interaction with GATA-1 (Fig. 3D) as well as with the basic transcription factor, as reported previously (31, 33), rather than interacting with CREB on the ARR19 promoter.
Mutation Analysis of GATA-1 Binding Motifs in the Arr19 Promoter
The results of deletion analyses of the 1-kb Arr19 promoter indicated that both putative GATA-1 binding motifs (Fig. 4, GATA-A and GATA-B) were probably functional. In order to confirm this, we prepared point mutants of each GATA-1 binding site by site-directed mutagenesis and conducted transient transfection analyses in MA-10 cells, which endogenously express both the GATA-1 and Arr19 genes. The cells were transfected with each GATA mutant of the Arr19 promoter with or without the GATA-1 expression plasmid. As shown in Fig. 4E, the mutation of the GATA-A site significantly reduced the Arr19 promoter activity as compared with the wild type in either the presence or absence of exogenous GATA-1 protein expression. The mutation of GATA-B attenuated the promoter activity to a greater extent than was seen with the mutation of GATA-A site, thereby suggesting that GATA-B is a major GATA-1 functional site. The mutation of both GATA motifs markedly reduced Arr19 promoter activity and completely abolished the ability of the Arr19 promoter to be transactivated by GATA-1, which was manifested by the minimal effect of exogenous GATA-1 protein expression on the promoter activity.
The importance of both GATA-1 motifs for the profound activity of the Arr19 promoter was further verified by the effects of cAMP on the GATA-1 mutated constructs (Fig. 4F). Mutation of either of the GATA motifs resulted in a lower response to cAMP treatment, which inhibits GATA-1 expression (Fig. 2), as compared with the wild type. Mutation of both GATA-1 motifs completely abolished the ability of the Arr19 promoter to respond to cAMP signaling. Collectively, these results show that both of the GATA-1 binding motifs in the Arr19 promoter are functional, and they appear to be the only GATA-1 binding sites within the 1-kb promoter region that we have analyzed thus far.
GATA-1 Protein Binds to the Regulatory Elements in the ARR19 Promoter
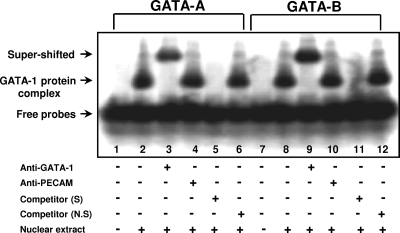
The binding of GATA-1 protein on the GATA motifs in the Arr19 promoter at −972/−969 (GATA-A) and −639/−635 (GATA-B) was confirmed by electrophoretic mobility shift assays (Fig. 5). Radiolabeled oligomers for each GATA motif were incubated with nuclear extracts of MA-10 cells. One major shift band was produced with both GATA-A and GATA-B oligomers. When the antibody against GATA-1 protein was added to the reactions, a supershifted band was observed, indicating the binding of GATA-1 protein on both GATA-A and GATA-B oligomers. No supershifted band was detected with isotype-matched anti-PECAM antibody.
FIGURE 5.
Binding of GATA-1 protein to the GATA-1 motifs in the Arr19 promoter. An electrophoretic mobility shift assay was performed with nuclear extracts (1 μg of total protein each) prepared from MA-10 cells and radiolabeled oligomers spanning −987/−960 (GATA-A) or −650/−628 (GATA-B) sequences of the Arr19 promoter. A 200-fold excess of specific or nonspecific nonradiolabeled oligomers was used for binding competition, and 1 μl (200 ng) of specific or nonspecific antibody (anti-PECAM) was used for supershift of the DNA·GATA-1 complex.
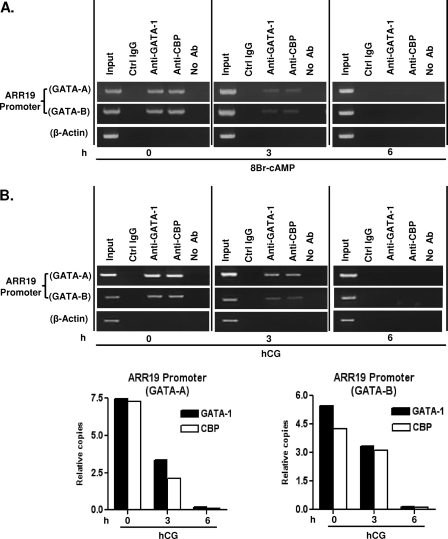
In an effort to further evaluate the association of GATA-1 and CBP protein with the GATA-1 motifs in the Arr19 promoter, we conducted ChIP assays with MA-10 cells that were treated with 300 μm cAMP at several time points (0, 3, and 6 h) during treatment. GATA-1, CBP, and Arr19 genes are endogenously expressed in MA-10 cells. Chromatins from the MA-10 cells were immunoprecipitated with anti-GATA-1 and anti-CBP antibody, and PCRs were conducted with a specific set of primers spanning each GATA-1 binding site in the Arr19 promoter. As shown in Fig. 6A, both GATA-1 regulatory elements (GATA-A and GATA-B) were occupied by GATA-1 and CBP protein, and the occupancies were reduced at 3 h of treatment, probably due to the reduced expression of GATA-1 protein with cAMP treatment (Fig. 2B). As expected, at 6 h of treatment, the occupancies of both GATA-1 and CBP protein were totally abrogated. No amplification of these elements was observed in the absence of specific antiserum (Fig. 6, No Ab) or with normal rabbit serum (Ctrl IgG).
FIGURE 6.
Association of GATA-1 and CBP protein with the GATA-1 motifs in the Arr19 promoter in MA-10 cells and in the testis. A, ChIP assays were conducted with MA-10 cells treated with 300 μm cAMP for 0, 3, and 6 h, and the chromatin was immunoprecipitated with anti-GATA-1 antibody and anti-CBP antibody. B, ChIP assays were conducted with the extract of 14-day-old mouse testis treated with hCG (10 IU, intraperitoneally) for 0, 3, and 6 h, and the chromatin was immunoprecipitated with anti-GATA-1 antibody and anti-CBP antibody. Immunoprecipitations with normal rabbit serum (Ctrl IgG) and without antibody (No Ab) or PCRs for β-actin DNA were employed as negative controls for nonspecific immunoprecipitations. PCRs were conducted with two sets of primer pairs in order to amplify the GATA-1 binding elements within the Arr19 promoter. Additional real time PCR was performed to quantify the relative amount of GATA-1 and CBP proteins associated with the Arr19 promoter under the treatment of hCG relative to the start point (0 h).
To confirm whether the regulation of Arr19 by GATA-1 and CBP in MA-10 cells reflects the situation in vivo, 14-day-old mice were injected intraperitoneally with hCG, and ChIP assays were then conducted. Chromatins from mouse testis were immunoprecipitated with anti-GATA-1 and anti-CBP antibody, and quantitative PCRs were conducted with the specific set of primers, which were used for MA-10 cells, spanning each GATA-1 binding site (Fig. 6B). Interestingly, the results were very similar to those found in MA-10 cells; both GATA-1 regulatory elements were occupied by GATA-1 and CBP proteins, and the occupancies by both proteins were gradually reduced with hCG treatment, resulting in the complete abolishment at 6 h of treatment. Together, these results suggest that GATA-1 protein binds directly to the GATA-1 binding motifs within the Arr19 promoter in vivo and in vitro together with CBP and regulates Arr19 gene expression in testicular Leydig cells.
DISCUSSION
In this study, we demonstrate that ARR19 is expressed in testicular Leydig cells, and its expression is a direct target of GATA-1, the gene expression of which is down-regulated by LH signaling. Arr19 was originally isolated as a putative androgen-induced gene from murine testis, and its expression was detected in male reproductive organs, including the testis and prostate (4). GATA-1 and GATA-4 have been suggested to be important in the regulation of gonadal development and function. GATA-1 is expressed in mouse testicular Sertoli cells from postnatal day 7 onward (54). Later in development, it is expressed in adult Sertoli cells of stage VII–IX seminiferous tubules. However, no study has yet been conducted regarding the differential expressions of GATA-1 and ARR19 in the interstitial compartment during the development of the testis.
The interstitium is populated by androgen-producing Leydig cells, which are physiologically and structurally heterogeneous. Leydig cells are the primary source of testosterone in males, and their differentiation in the testis during puberty is a signature event in the development of the male body. It has been hypothesized but remains far from proven that adult Leydig cells first arise from undifferentiated stem cells (stem Leydig cells; SLCs) (44–46). In rats, the putative SLCs are present in the testis at birth, and some of their progeny express Leydig cell-specific genes by 11 days postpartum, thus becoming committed to the Leydig cell lineage (57, 58). The committed cells subsequently undergo phase transitions through the progenitor (PLC) and immature stages and ultimately to the terminally differentiated adult Leydig cell stage (59). The PLCs proliferate and also exhibit certain aspects of differentiated function (60). LH receptors first appear as the PLCs differentiate (61), and the development of the steroidogenic capacity of PLCs requires stimulation by LH (62).
Our immunohistochemical analyses of 14-day-old and adult mouse testis demonstrated that ARR19 protein was highly expressed in the majority of interstitial cells during prepuberty, but its expression was restricted to a small population of interstitial cells during adulthood (Fig. 1A). Since SLCs and PLCs are present in the 14-day-old mouse testis prior to the onset of steroidogenic activity, and their numbers are very low in the adult testis, we speculate that the ARR19-expressing cells are putative precursors of Leydig cells and/or PLCs. In addition, Western blot analyses revealed that ARR19 was expressed in primary Leydig cells isolated from developing mouse testis, and its expression was gradually decreased with the age of the mouse testis (Fig. 1B). This suggests the involvement of LH/cAMP, an important mediator of Leydig cell differentiation, in the regulation of Arr19 gene expression, which we confirmed using 14-day-old testis injected with hCG and MA-10 Leydig cells treated with LH/cAMP (Figs. 1C and 2). Similar gene regulation by LH was previously reported with undifferentiated SLC markers, such as c-Kit and platelet-derived growth factor receptor (42). However, further studies are required to correctly identify the ARR19-positive cells with regard to the differentiation of Leydig cells.
With regard to the function of ARR19, we hypothesized that ARR19 would inhibit adult Leydig cell differentiation and/or testicular steroidogenesis, because ARR19 was highly expressed in earlier stages of Leydig cell development. In keeping constant with the hypothesis, the overexpression of ARR19 in R2C Leydig cells, which are constitutive steroidogenic and ARR19-negative, inhibited testosterone biosynthesis. Interestingly, the expression of steroidogenic enzyme genes, such as 3β HSD, StAR, and P450scc, was markedly reduced, suggesting that Arr19 may act as a corepressor of transcription factor(s). Arr19 has been previously reported to function as a corepressor of androgen receptor (4). However, whether Arr19 directly or indirectly down-regulates the expression of steroidogenic enzyme genes is yet unknown, and we are presently investigating the matter and the mechanism involved.
Previous studies have demonstrated that GATA-1 is expressed in Sertoli and Leydig cells of murine testis as well as the tumor cell lines derived from these testicular cells (15, 54–58, 60, 61). GATA-1 transactivates the inhibin/activin α gene in Sertoli and Leydig tumor cell lines (56, 61), and this is achieved through the interaction with two GATA motifs in the promoter region (56). The mutation of either or both of the GATA motifs markedly reduced the basal promoter activity of the α-subunit gene in testicular cells, but this did not influence the stimulatory effects of cAMP on the transcription of the α-subunit gene (56). Thus, cAMP positively regulated the inhibin α-subunit gene, a target of GATA-1, although it negatively regulated the GATA-1 mRNA and protein levels. The mechanism by which the inhibin α-subunit gene is regulated by cAMP seems to differ considerably from that of Arr19. The elevation of inhibin α-subunit mRNA by cAMP occurred via the CRE motif in the promoter region of the α-subunit (56), which also appeared to compensate for the decreased promoter activity resulting from the cAMP-mediated reduction of GATA-1 protein levels.
Several studies have shown that GATA-1 activity is regulated by acetylation and phosphorylation (32, 64, 65). The acetylation of GATA-1 increases DNA binding activity and stimulates the transcription of the GATA-1-dependent promoter (32, 37), whereas MAPK phosphorylation of GATA-1, in cooperation with acetylation, is involved in its degradation through the ubiquitination (64). The phosphatidylinositol 3-kinase/AKT signaling pathway is identified as a mediator of Epo-induced phosphorylation of GATA-1 in erythroid cells, which enhances GATA-1 transcriptional activity (65). Our data showed that the levels of ARR19 and GATA-1 protein were rapidly decreased by cAMP/LH treatment, both leaving less than 50% within 2 h (Figs. 2, B and E). In addition, the decrease of Arr19 mRNA level by cAMP was faster than that of GATA-1 (Fig. 2C), in contrast to the simultaneous decrease of their protein levels. These results raise the possibility of additional layers of regulation of GATA-1 activity, such as protein degradation or dissociation from the Arr19 promoter, by cAMP/LH. A previous study has shown that GATA-4 protein, another key regulator of gonadal gene expression, is phosphorylated by protein kinase A in response to cAMP in Leydig cells (63). It will be worthwhile to investigate whether the cAMP/protein kinase A signaling pathway can also modify GATA-1 protein, resulting in the modulation of GATA-1 activity.
In MA-10 cells, the expression of ARR19 was negatively regulated by LH/cAMP, and, moreover, the pattern of its negative regulation was similar to that of the GATA-1. In addition, the cloning and characterization of the Arr19 promoter showed that there were two functional GATA motifs, thereby indicating that Arr19 is a novel target gene of GATA-1. Sequence analysis of the Arr19 promoter also revealed a putative CRE motif, which resides proximally to the two GATA motifs. However, the CRE appears to be nonfunctional, since the expression of exogenous CREB protein exerted no effects on the Arr19 promoter (Fig. 3D). In accordance with this observation, the deletion of the CRE had no effect on the promoter activity of the Arr19 gene (Fig. 4D). Thus, CBP appears to function as a coactivator for GATA-1 on the Arr19 promoter.
In summary, we have demonstrated 1) the differential expression of ARR19 in Leydig cells during the development of mouse testis, 2) the inhibition of steroidogenesis by overexpression of ARR19 via the regulation of steroidogenic enzyme gene expressions, 3) the effects of LH/cAMP signaling on ARR19 and GATA-1 expression in MA-10 cells, and 4) the promoter regulation of Arr19 by GATA-1, which was augmented by CBP in vivo and in vitro. Together, our results suggest that Arr19 gene expression is controlled by LH/cAMP signaling via GATA-1 gene regulation in testicular Leydig cells. Further, LH/cAMP-mediated regulation of Arr19 gene expression suggests that ARR19 may play an important role in the differentiation of Leydig cells, which is controlled by LH.
This work was supported by Chonnam National University and by a Korea Research Foundation grant funded by the Korean Government (KRF-2006-005-J03002).
- CRE
- cAMP-response element
- CREB
- CRE-binding protein
- CBP
- CREB-binding protein
- LH
- luteinizing hormone
- SLC
- stem Leydig cell
- PLC
- progenitor Leydig cell
- RACE
- rapid amplification of cDNA ends
- ChIP
- chromatin immunoprecipitation
- hCG
- human chorionic gonadotrophin.
REFERENCES
- 1.Han W., Ding P., Xu M., Wang L., Rui M., Shi S., Liu Y., Zheng Y., Chen Y., Yang T., Ma D. ( 2003) Genomics 81, 609– 617 [DOI] [PubMed] [Google Scholar]
- 2.Han W., Lou Y., Tang J., Zhang Y., Chen Y., Li Y., Gu W., Huang J., Gui L., Tang Y., Li F., Song Q., Di C., Wang L., Shi Q., Sun R., Xia D., Rui M., Tang J., Ma D. ( 2001) Biochem. J. 357, 127– 135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Li T., Han W., Yang T., Ding P., Rui M., Liu D., Wang Y., Ma D. ( 2006) Int. J. Biochem. Cell Biol. 38, 420– 429 [DOI] [PubMed] [Google Scholar]
- 4.Jeong B. C., Hong C. Y., Chattopadhyay S., Park J. H., Gong E. Y., Kim H. J., Chun S. Y., Lee K. ( 2004) Mol. Endocrinol. 18, 13– 25 [DOI] [PubMed] [Google Scholar]
- 5.Mayr B., Montminy M. ( 2001) Nat. Rev. Mol. Cell Biol. 2, 599– 609 [DOI] [PubMed] [Google Scholar]
- 6.Ito E., Toki T., Ishihara H., Ohtani H., Gu L., Yokoyama M., Engel J. D., Yamamoto M. ( 1993) Nature 362, 466– 468 [DOI] [PubMed] [Google Scholar]
- 7.Yomogida K., Ohtani H., Harigae H., Ito E., Nishimune Y., Engel J. D., Yamamoto M. ( 1994) Development 120, 1759– 1766 [DOI] [PubMed] [Google Scholar]
- 8.Onodera K., Yomogida K., Suwabe N., Takahashi S., Muraosa Y., Hayashi N., Ito E., Gu L., Rassoulzadegan M., Engel J. D., Yamamoto M. ( 1997) J. Biochem. 121, 251– 263 [DOI] [PubMed] [Google Scholar]
- 9.Feng Z. M., Wu A. Z., Chen C. L. C. ( 1998) Mol. Endocrinol. 12, 378– 390 [DOI] [PubMed] [Google Scholar]
- 10.Viger R. S., Mertineit C., Trasler J. M., Nemer M. ( 1998) Development 125, 2665– 2675 [DOI] [PubMed] [Google Scholar]
- 11.Ketola I., Rahman N., Toppari J., Bielinska M., Porter-Tinge S. B., Tapanainen J. S., Huhtaniemi I. T., Wilson D. B., Heikinheimo M. ( 1999) Endocrinology 140, 1470– 1480 [DOI] [PubMed] [Google Scholar]
- 12.Chen C. L. C., Feng Z. M., Chung K., Boitani C., Bardin C. W. ( 1996) Program of the 78th Annual Meeting of the Endocrine Society, San Francisco 1996, Abstr. P2, p. 564, Endocrine Society, Chevy Chase, MD [Google Scholar]
- 13.Feng Z. M., Wu A. Z., Zhang Z., Chen C. L. C. ( 2000) Mol. Endocrinol. 14, 1820– 1835 [DOI] [PubMed] [Google Scholar]
- 14.Ketola I., Anttonen M., Vaskivuo T., Tapanainen J. S., Toppari J., Heikinheimo M. ( 2002) Eur. J. Endocrinol. 147, 397– 406 [DOI] [PubMed] [Google Scholar]
- 15.Zhang Z., Wu A. Z., Feng Z. M., Mruk D., Cheng C. Y., Chen C. L. ( 2002) Endocrinology 143, 829– 836 [DOI] [PubMed] [Google Scholar]
- 16.Ko L. J., Engel J. D. ( 1993) Mol. Cell. Biol. 13, 4011– 4022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Merika M., Orkin S. H. ( 1993) Mol. Cell. Biol. 13, 3999– 4010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Whyatt D. J., deBoer E., Grosveld F. ( 1993) EMBO J. 12, 4993– 5005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yamamoto M., Ko L. J., Leonard M. W., Beug H., Orkin S. H., Engel J. D. ( 1990) Genes Dev. 4, 1650– 1662 [DOI] [PubMed] [Google Scholar]
- 20.Evans T., Reitman M., Felsenfeld G. ( 1988) Proc. Natl. Acad. Sci. U.S.A. 85, 5976– 5980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wall L., deBoer E., Grosveld F. ( 1988) Genes Dev. 2, 1089– 1100 [DOI] [PubMed] [Google Scholar]
- 22.Martin D. I., Orkin S. H. ( 1990) Genes Dev. 4, 1886– 1898 [DOI] [PubMed] [Google Scholar]
- 23.Tsai S. F., Martin D. I., Zon L. I., D'Andrea A. D., Wong G. G., Orkin S. H. ( 1989) Nature 339, 446– 451 [DOI] [PubMed] [Google Scholar]
- 24.Tsai S. F., Strauss E., Orkin S. H. ( 1991) Genes Dev. 5, 919– 931 [DOI] [PubMed] [Google Scholar]
- 25.Manna P. R., Dyson M. T., Eubank D. W., Clark B. J., Lalli E., Sassone-Corsi P., Zeleznik A. J., Stocco D. M. ( 2002) Mol. Endocrinol. 16, 184– 199 [DOI] [PubMed] [Google Scholar]
- 26.Tsang A. P., Visvader J. E., Turner C. A., Fujiwara Y., Yu C., Weiss M. J., Crossley M., Orkin S. H. ( 1997) Cell 90, 109– 119 [DOI] [PubMed] [Google Scholar]
- 27.Nishikawa K., Kobayashi M., Masumi A., Lyons S. E., Weinstein B. M., Liu P. P., Yamamoto M. ( 2003) Mol. Cell. Biol. 23, 8295– 8305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fox A. H., Kowalski K., King G. F., Mackay J. P., Crossley M. ( 1998) J. Biol. Chem. 273, 33595– 33603 [DOI] [PubMed] [Google Scholar]
- 29.Fox A. H., Liew C., Holmes M., Kowalski K., Mackay J., Crossley M. ( 1999) EMBO J. 18, 2812– 2822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lipinski M. M., Jacks T. ( 1999) Oncogene 18, 7873– 7882 [DOI] [PubMed] [Google Scholar]
- 31.Kwok R. P., Lundblad J. R., Chrivia J. C., Richards J. P., Bächinger H. P., Brennan R. G., Roberts S. G., Green M. R., Goodman R. H. ( 1994) Nature 370, 223– 226 [DOI] [PubMed] [Google Scholar]
- 32.Boyes J., Byfield P., Nakatani Y., Ogryzko V. ( 1998) Nature 396, 594– 598 [DOI] [PubMed] [Google Scholar]
- 33.Chrivia J. C., Kwok R. P., Lamb N., Hagiwara M., Montminy M. R., Goodman R. H. ( 1993) Nature 365, 855– 859 [DOI] [PubMed] [Google Scholar]
- 34.Bannister A. J., Kouzarides T. ( 1996) Nature 384, 641– 643 [DOI] [PubMed] [Google Scholar]
- 35.Chen H., Lin R. J., Schiltz R. L., Chakravarti D., Nash A., Nagy L., Privalsky M. L., Nakatani Y., Evans R. M. ( 1997) Cell 90, 569– 580 [DOI] [PubMed] [Google Scholar]
- 36.Ogryzko V. V., Schiltz R. L., Russanova V., Howard B. H., Nakatani Y. ( 1996) Cell 87, 953– 959 [DOI] [PubMed] [Google Scholar]
- 37.Hung H. L., Lau J., Kim A. Y., Weiss M. J., Blobel G. A. ( 1999) Mol. Cell. Biol. 19, 3496– 3505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Blobel G. A., Nakajima T., Eckner R., Montminy M., Orkin S. H. ( 1998) Proc. Natl. Acad. Sci. U.S.A. 95, 2061– 2066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Barrett D. M., Gustafson K. S., Wang J., Wang S. Z., Ginder G. D. ( 2004) Mol. Cell. Biol. 24, 6194– 6204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Vo N., Goodman R. H. ( 2001) J. Biol. Chem. 276, 13505– 13508 [DOI] [PubMed] [Google Scholar]
- 41.Lamonica J. M., Vakoc C. R., Blobel G. A. ( 2006) Blood 108, 3736– 3738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ge R. S., Dong Q., Sottas C. M., Papadopoulos V., Zirkin B. R., Hardy M. P. ( 2006) Proc. Natl. Acad. Sci. U.S.A. 103, 2719– 2724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kamei Y., Xu L., Heinzel T., Torchia J., Kurokawa R., Gloss B., Lin S. C., Heyman R. A., Rose D. W., Glass C. K., Rosenfeld M. G. ( 1996) Cell 85, 403– 414 [DOI] [PubMed] [Google Scholar]
- 44.Hong C. Y., Park J. H., Ahn R. S., Im S. Y., Choi H. S., Soh J., Mellon S. H., Lee K. ( 2004) Mol. Cell. Biol. 24, 2593– 2604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Moraga P. F., Llanos M. N., Ronco A. M. ( 1997) J. Endocrinol. 154, 201– 209 [DOI] [PubMed] [Google Scholar]
- 46.Kim J. Y., Kim H. J., Kim K. T., Park Y. Y., Seong H. A., Park K. C., Lee I. K., Ha H., Shong M., Park S. C., Choi H. S. ( 2004) Mol. Endocrinol. 18, 2880– 2894 [DOI] [PubMed] [Google Scholar]
- 47.Shi S. R., Cote R. J., Liu C., Yu M. C., Castelao J. E., Ross R. K., Taylor C. R. ( 2002) Appl. Immunohistochem. Mol. Morphol. 10, 368– 373 [DOI] [PubMed] [Google Scholar]
- 48.Campbell S. E., Sood A., Argyle D. J., Nasir L., Argyle S. A., Bennet D. ( 2002) Gene 286, 233– 240 [DOI] [PubMed] [Google Scholar]
- 49.Song F., Goodman R. M. ( 2002) Gene 290, 115– 124 [DOI] [PubMed] [Google Scholar]
- 50.Eacker S. M., Agrawal N., Qian K., Dichek H. L., Gong E. Y., Lee K., Braun R. E. ( 2008) Mol. Endocrinol. 22, 623– 635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Fan W., Yanase T., Morinaga H., Okabe T., Nomura M., Daitoku H., Fukamizu A., Kato S., Takayanagi R., Nawata H. ( 2007) J. Biol. Chem. 282, 7329– 7338 [DOI] [PubMed] [Google Scholar]
- 52.Song K. H., Park Y. Y., Park K. C., Hong C. Y., Park J. H., Shong M., Lee K., Choi H. S. ( 2004) Mol. Endocrinol. 18, 1929– 1940 [DOI] [PubMed] [Google Scholar]
- 53.Carninci P., Westover A., Nishiyama Y., Ohsumi T., Itoh M., Nagaoka S., Sasaki N., Okazaki Y., Muramatsu M., Schneider C., Hayashizaki Y. ( 1997) DNA Res. 4, 61– 66 [DOI] [PubMed] [Google Scholar]
- 54.Ge R. S., Shan L. X., Hardy M. P. ( 1996) in The Leydig Cell ( Payne A. H., Hardy M. P., Russell L. D. eds) 1st Ed., pp. 159– 174, Cache River Press, Vienna, IL [Google Scholar]
- 55.Haider S. G. ( 2004) Int. Rev. Cytol. 233, 181– 241 [DOI] [PubMed] [Google Scholar]
- 56.Davidoff M. S., Middendorff R., Enikolopov G., Riethmacher D., Holstein A. F., Müller D. ( 2004) J. Cell Biol. 167, 935– 944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mendis-Handagama S. M., Ariyaratne H. B. ( 2001) Biol. Reprod. 65, 660– 671 [DOI] [PubMed] [Google Scholar]
- 58.Lo K. C., Lei Z., Rao Ch. V., Beck J., Lamb D. J. ( 2004) Endocrinology 145, 4011– 4015 [DOI] [PubMed] [Google Scholar]
- 59.Ge R. S., Dong Q., Sottas C. M., Chen H., Zirkin B. R., Hardy M. P. ( 2005) Biol. Reprod. 72, 1405– 1415 [DOI] [PubMed] [Google Scholar]
- 60.Haider S., Rai U. ( 1986) Gen. Comp. Endocrinol. 64, 321– 329 [DOI] [PubMed] [Google Scholar]
- 61.Wing T. Y., Ewing L. L., Zegeye B., Zirkin B. R. ( 1985) Endocrinology 117, 1779– 1787 [DOI] [PubMed] [Google Scholar]
- 62.Cooke B. A. ( 1996) in The Leydig Cell ( Payne A. H., Hardy M. P., Russell L. D. eds) 1st Ed., pp. 352– 363, Cache River Press, Vienna, IL [Google Scholar]
- 63.Tremblay J. J., Viger R. S. ( 2003) J. Biol. Chem. 278, 22128– 22135 [DOI] [PubMed] [Google Scholar]
- 64.Hernandez-Hernandez A., Ray P., Litos G., Ciro M., Ottolenghi S., Beug H., Boyes J. ( 2006) EMBO J. 25, 3264– 3274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zhao W., Kitidis C., Fleming M. D., Lodish H. F., Ghaffari S. ( 2006) Blood 107, 907– 915 [DOI] [PMC free article] [PubMed] [Google Scholar]