Abstract
The low density lipoprotein receptor-related protein (LRP) is the principal clearance receptor for serpins and serpin-proteinase complexes. The ligand binding regions of LRP consist of clusters of cysteine-rich ∼40-residue complement-like repeats (CR), with cluster II being the principal ligand-binding region. To better understand the specificity of binding at different sites within the cluster and the ability of LRP to discriminate in vivo between uncomplexed and proteinase-complexed serpins, we have systematically examined the affinities of plasminogen activator inhibitor-1 (PAI-1) and proteinase nexin-1 (PN-1) in their native, cleaved, and proteinase-complexed states to (CR)2 and (CR)3 fragments of LRP cluster II. A consistent blue shift of the CR domain tryptophan fluorescence suggested a common mode of serpin binding, involving lysines on the serpin engaging the acidic region around the calcium binding site of the CR domain. High affinity binding of non-proteinase-complexed PAI-1 and PN-1 occurred to all fragments containing three CR domains (3–59 nm) and most that contain only two CR domains, although binding energies to different (CR)3 fragments differed by up to 18% for PAI-1 and 9% for PN-1. No detectable difference in affinity was seen between native and cleaved serpin. However, the presence of proteinase in complex with the serpin enhanced affinity modestly and presumably nonspecifically. This may be sufficient to give preferential binding of such complexes in vivo at the relevant physiological concentrations.
The low density lipoprotein receptor-related protein (LRP)2 is a member of the LDL receptor family of mosaic-like receptors (1). Ligand binding occurs to regions composed of multiple copies of a ∼40-residue cysteine-rich, calcium-binding domain, termed variously CR (for complement-like repeat) or Ldl-A. In the case of LDLR, there is a single cluster of seven CR domains, whereas in LRP, there are four clusters (designated I–IV) composed of 2, 8, 10, and 11 CR domains, respectively. Unlike LDLR, which has a very limited range of protein ligands, LRP is known to bind and internalize a very wide range of structurally unrelated proteins, including serpins and their proteinase complexes, and activated forms of the panproteinase inhibitor α2-macroglobulin (α2M) (2). Cluster II is the principal ligand-binding region, although many of the ligands to this cluster have also been reported to bind to cluster IV (2). That the wider range of ligands for LRP is not solely related to the much greater number of CR domains compared with LDLR is shown by the quite wide range of ligands for VLDLR, which, like LDLR, has only a single cluster of CR domains, albeit of eight rather than seven domains.
Given that serpins are able to undergo a number of conformational transitions, most notably as a result of formation of complexes during proteinase inhibition, many early studies on the in vivo receptor-mediated clearance of serpins focused on the relative rates of clearance of the different conformational forms (3–5). It was shown for several serpins that their complexes with proteinase were cleared much more rapidly than native, cleaved, or latent forms. This led to the idea that a neoepitope is formed in the complex (6). From comparison of the internalization properties of PAI-1 complexes with different proteinases, it was further suggested that the neoepitope is localized to the serpin moiety, thus implying that the conformation of the serpin in the serpin-proteinase complex is sufficiently different to permit discrimination between complexed and uncomplexed serpin by the clearance receptor (7). However, the determination of x-ray structures of the serpin α1PI with two different proteinases showed that the serpin moiety is almost identical in conformation to cleaved α1PI (8, 9). Studies on other serpin-proteinase complexes, including those of PAI-1 with four different proteinases, all suggested equivalent structures for the complexes and hence no major conformational difference for the serpin moiety compared with the cleaved form (10, 11). Although the consequence of binding of serpins in their various conformational forms appears to result only in internalization and degradation, the consequences for binding of activated α2Ms are more complex. In addition to internalization, there is also good evidence for signal transduction, resulting in a range of intracellular changes, such as increase in Ca2+ and phosphorylation (12–14).
Given these variations in ligand-receptor interaction (namely the wide versus narrow specificity of LRP and LDLR, the apparent discrimination by LRP between different serpin conformational states, and the different cellular consequences of binding serpins and activated α2Ms to LRP), it is of great interest to understand the molecular level basis for these behaviors. We have already shown that the receptor binding domain of α2M, which contains the full binding region of the intact protein, shows a 30-fold preference for binding to one region of cluster II of LRP compared with an adjacent region (15). With the goal of determining whether there is comparable selectivity for serpins, we have now examined the binding of two closely related serpins, PAI-1 and PN-1, in different conformational states, to overlapping fragments of cluster II from LRP. We found that the serpin ligands bound tightly to many regions of cluster II, although with up to 12-fold difference in Kd for binding of PAI-1 to different (CR)3 fragments and up to 5-fold difference for PN-1. Most importantly, we found that, for both PAI-1 and PN-1, native and cleaved conformations bound with similar affinities, and, for PAI-1, the higher affinity of proteinase-complexed versus non-complexed serpin arose solely from a small additional, most likely nonspecific, binding contribution from the proteinase.
EXPERIMENTAL PROCEDURES
Expression, Purification, and Refolding of CR Constructs
All CR constructs were expressed in 2YT medium. CR34, CR45, CR56, CR78, and CR567 were cloned in pGEX-2T, modified to contain a TEV cleavage site, and expressed as GST fusion proteins in Escherichia coli BL21 cells. Cells were grown to A600 = 0.6–1.0 before induction with 1 mm isopropyl 1-thio-β-d-galactopyranoside. The cells were harvested after 5–6 h at 37 °C. GST fusion proteins were purified from cleared cell lysate by GSH-Sepharose chromatography, and the GST tag was removed by TEV cleavage during overnight dialysis against 4 liters of 20 mm Tris-HCl, pH 8.0, 50 mm NaCl, 4 mm EDTA, containing 14 mm β-ME. GST and uncleaved GST-CR fusion proteins were removed by passage through the GSH column.
CR89, CR345, CR456, and CR678 were cloned in pQE-30, modified to contain a GB1 fusion partner and a TEV cleavage site, and CR45 was cloned in pQE-30, modified to contain a NusA fusion partner and a TEV cleavage site. These His6-tagged fusion proteins were expressed in E. coli SG13009 cells containing the plasmid pRARE. The CR fusion proteins were purified from cell lysate by Ni2+ or TALON chromatography, and the fusion partners were removed by TEV cleavage during overnight dialysis against phosphate-buffered saline containing either 14 or 7 mm β-ME.
For these small domains, refolding represents only the correct formation of the three disulfides, since stable tertiary structure is then reversibly induced by Ca2+ binding. Prior to such refolding, all (CR)x species were further purified by Q-Sepharose HP chromatography, using a gradient of 0–1000 mm NaCl in 20 mm Tris-HCl, pH 8.0, and 6 m urea. The denatured (CR)x species were diluted to 0.1 mg/ml with 50 mm Tris-HCl, pH 8.5, 50 mm NaCl, 10 mm CaCl2 and refolded by dialysis against buffer containing 14 mm β-ME and 8 mm 2-hydroxyethyl disulfide. For increased folding efficiency, all (CR)x species except CR78 were mixed with the chaperone GST-receptor-associated protein (RAP) as a GST fusion protein (iodoacetamide-treated GST-RAP) at a 1:1.25 (CR/RAP) ratio (15). After 24 h at room temperature with N2 bubbling, the dialysis was continued for 24 h at 4 °C without N2. Finally, the mixture was dialyzed against 2 × 4 liters of 20 mm Tris-HCl, pH 7.8, 50 mm NaCl, 1 mm CaCl2 at 4 °C. The refolding mixture was loaded on GSH-Sepharose, and folded CR constructs (i.e. those capable of binding to RAP and therefore retained on the column) were eluted with 40 mm Tris-HCl, pH 8.0, 100 mm NaCl, 8 mm EDTA. Folded (CR)x species were further purified by Q-Sepharose HP chromatography (50–1000 mm NaCl in 20 mm Tris-HCl, pH 8.0, and 0.1 mm CaCl2). If additional purification was needed, calcium was removed by EDTA, and the (CR)x species passed through a Superdex 75 size exclusion column equilibrated in 20 mm Tris-HCl, pH 7.8, 150 mm NaCl, and 4 mm EDTA. CR3 was expressed and refolded as described previously (16).
As a quality control between each step of purification, samples were analyzed by reduced and non-reduced SDS-PAGE and non-denaturing PAGE. The presence of a single band on SDS-PAGE with a reduction in mobility under reducing conditions indicated homogeneous samples with formed disulfide bridges. A single band on non-denaturing PAGE indicated the presence of a single folding product. The final products were judged to be more than 95% pure by SDS-PAGE.
Expression and Purification of PAI-1
A pQE-30 vector (Qiagen) with the cDNA sequence corresponding to residues 1–379 from human PAI-1 with the four stabilizing mutations N150H/K154T/Q319L/M354I (variant 14-1B) (17) was constructed by standard methods and verified by sequencing. The final expressed construct includes an N-terminal extension (MRGSH6GSAV), consisting of an His6 tag and a GSA linker followed by the PAI-1 sequence with the numbering Val1-His2-His3-Pro4, etc. PAI-1 was expressed in E. coli SG13009 cells (Qiagen) grown to A600 = 0.8–1.0 and induced for 2 h at 37 °C with 1 mm isopropyl 1-thio-β-d-galactopyranoside before harvesting. PAI-1 was purified by Ni2+ affinity chromatography, followed by size exclusion chromatography on Superdex 75 (GE Healthcare) in 5 mm Tris, pH 6.6, 1000 mm NaCl. The full-length protein, including the N-terminal extension, was confirmed by mass spectroscopy to give the correct theoretical mass of 44.22 kDa. Sample purity better than 95% was estimated by SDS-PAGE analysis. PAI-1 concentrations were determined spectrophotometrically, using an extinction coefficient of 18,100 m−1 cm−1 at 280 nm. PAI-1 used throughout this work is the stable 14-1B variant containing the four mutations.
Expression of PN-1
A pRSETc vector (Invitrogen), modified with an N-terminal His6 tag, TEV cleavage site, and PN-1 cDNA was a kind gift of Dr. Peter Andreasen (Aarhus University). PN-1 was expressed in BL21(DE3) cells, grown to A600 = 0.6–0.8, and induced with 1 mm isopropyl 1-thio-β-d-galactopyranoside. The cells were grown overnight at 20 °C before harvesting. PN-1 was purified from the cell lysate by TALON chromatography. The His6 tag was removed by TEV cleavage during overnight dialysis against phosphate-buffered saline buffer containing 7 mm β-ME. PN-1 was further purified by heparin-Sepharose chromatography (200–2000 mm NaCl in 20 mm Tris-HCl, pH 7.4) and Q-Sepharose HP chromatography (50–1000 mm NaCl in 20 mm Tris-HCl, pH 8.0). PN-1 concentrations were determined spectrophotometrically, using an extinction coefficient of 18,800 m−1 cm−1 at 280 nm.
Preparation of Cleaved and Complexed Serpins
Prior to PAI-1 cleavage with proteinase, inhibitory activity of the protein was determined from the band shift upon complex formation with urokinase, monitored by SDS-PAGE, and found to be close to 100%. Reactive center loop-cleaved PAI-1 (rccPAI-1) was made by incubation with a 1.1–1.5-fold excess of trypsin (Sigma) for 10–15 min at 37 °C in 20 mm Tris, pH 7.4, 500 mm NaCl. The proteolytic reaction was quenched by adding a molar excess of the irreversible trypsin inhibitor Nα-p-tosyl-l-lysine chloromethyl ketone (TLCK) (Sigma). Unlike urokinase, trypsin produces mostly cleaved PAI-1 rather than stable covalent complex. rccPAI-1 was separated from trypsin·PAI-1 covalent complex and excess inactivated trypsin by two rounds of size exclusion chromatography through a 200-ml Superdex 75 column in 20 mm Tris, pH 7.4, 300 mm NaCl, and 1 mm CaCl2. Purity of the resulting sample was estimated to be better than 95% by SDS-PAGE.
rccPN-1 was generated by cleavage of the reactive center loop with porcine pancreatic elastase (PPE). PN-1 was incubated at room temperature for 1 h with PPE at a 1:100 (w/w) PPE/PN-1 ratio. The reaction was stopped by the addition of 100 μm 3,4-dichloroisocoumarin, and rccPN-1 was purified by heparin-Sepharose chromatography, as described above for purification of PN-1.
The covalent complex of PAI-1·trypsin was prepared by incubating a 4:1 molar ratio of PAI-1 to trypsin in 10 mm Tris, pH 7.4, 500 mm NaCl for 10 min at 37 °C before termination of the reaction with a molar excess of TLCK. The PAI-1 complex with lmw-uPA was prepared by incubation of the proteinase with a 1.3-fold molar excess of PAI-1 in 20 mm Tris, pH 7.4, 440 mm NaCl for 20 min at 37 °C. lmw-uPA was a kind gift from Dr. Grant Blouse. In both cases, complexes were base line-separated from uncomplexed PAI-1 by size exclusion chromatography on a Superdex 75 column in 20 mm MES, pH 5.5, 1000 mm NaCl. The stability and purity of the complexes were confirmed by SDS-PAGE analysis.
Fluorescence Spectroscopy
Fluorescence measurements were made on a PTI Quantamaster instrument, equipped with double monochromators on both the excitation and emission sides. All experiments were performed in quartz cuvettes using 1200 μl of sample in 20 mm Tris-HCl, pH 7.4, 150 mm NaCl, 1 mm CaCl2, supplemented with 0.1% (w/v) polyethylene glycol 8000. Fluorescence emission spectra were recorded from 300 to 450 nm in 4-nm steps, with an excitation wavelength of 295 nm. Since all proteins examined contain one or more tryptophans, reference spectra of the individual components were recorded and subtracted from spectra of complexes to obtain the difference perturbation spectrum for the binding of each CR. Protein concentrations of 50–1000 nm were used. To examine the calcium dependence of (CR)x ligand binding, experiments were performed in the presence of 10 mm EDTA.
Binding experiments with serpin or serpin-protease complexes to (CR)x species were performed with a fixed amount of the (CR)x species of between 50 and 1000 nm, depending on the affinity. Two-μl additions up to 40–60 μl of ligand were made, reaching a final ligand concentration between 350 and 2500 nm. For experiments involving lysine analogues, 500 nm CR3 was titrated with 1–2 m stock solutions of the lysine species to final concentrations between 5 and 30 mm.
The fluorescence change at the wavelength of maximum perturbation (328 nm) was measured as an average of 60 s at each titration step, and the subsequent titration curve was corrected for added fluorophore by subtracting a corresponding titration curve for ligand without CR. The resulting curve was then fitted to a standard 1:1 binding isotherm with floating Fmax, KD, and CR concentration. The fitted CR concentrations provided an independent estimate of the individual CR folding efficiencies (i.e. the concentration of high affinity species), which were routinely in the range of 50–100%.
Size Exclusion Chromatography
Size exclusion chromatography was carried out at room temperature on a 47-ml (10 × 600 mm) analytical Superdex 75 column equilibrated in 20 mm Tris-HCl, pH 7.4, 150 mm NaCl, 1 mm CaCl2. Samples were injected from a 100-μl loop at a flow rate of 0.5 ml/min. Mixtures of 9 μm (40 μg/injection) cleaved PAI-1 with the indicated amounts of CR56 or CR456 were preincubated for 10 min at room temperature prior to loading. The concentration of the protein applied to the column was chosen to be high enough (micromolar range) to ensure saturation of the complex, even after dilution on the column. Estimates of the stoichiometry of the complexes were obtained by integrating the absorbance at 280 nm of each peak and using the individual protein extinction coefficients, the initial molar ratio of proteins in the mixture, and the assumption that all cleaved PAI-1 was present in complex and that the late eluting peak contained only CR protein. These last two assumptions were justified given the absence of any peak at the position of cleaved PAI-1 and the position of the late eluting peak being the same as for free CR56 or CR456. The elution positions of cleaved PAI-1, CR56, and CR456 as well as additional molecular weight standards of bovine serum albumin (66 kDa) and carbonic anhydrase (58/29 kDa, dimer/monomer) were obtained in separate runs.
SDS and Non-denaturing PAGE
SDS-PAGE was performed using Novex 10–20% gels (Invitrogen) with standard Laemmli buffers, with or without reducing agent (β-ME) in the loading buffer. When loading trypsin·PAI-1 complex, a small amount of TLCK was added to the samples prior to loading buffer. All samples were burst-heated to 95 °C before loading. Non-denaturing PAGE was performed using Novex 8% gels, prerun for 3.5 h at 15 mA in 375 mm Tris-HCl, pH 8.8, 5 mm CaCl2 to introduce Ca2+ to the gel, which is essential for binding of CR fragments to their protein ligands. Gels were run in standard Tris-glycine buffer system, supplemented with 5 mm CaCl2 at 15 mA for 2 h. For gel shift experiments, samples of equimolar concentration of rccPAI-1 and CR, adjusted for activity, were incubated for 10 min at room temperature prior to loading.
RESULTS
Integrity of the CR Fragments
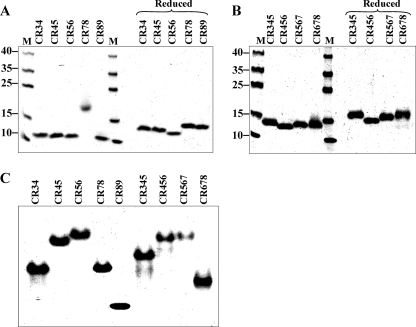
Even when mammalian cells are used for their overexpression, LRP and its fragments require the chaperone RAP to promote correct folding of the CR domains, each of which contains three disulfide bonds (18). Despite this, homogeneous, correctly folded species are not always produced (19). When CR fragments are expressed as inclusion bodies in E. coli, the problem of obtaining correctly folded material is exacerbated by the need to form the correct disulfides in each domain from denatured material. Carrying out the refolding in the presence of RAP and Ca2+ helps but still results in poor efficiency of correct refolding (5–25% depending on the fragment). Nevertheless, the higher level of expression in E. coli still results in greater overall yields. Refolded CR fragments used in the present study have therefore been subjected to additional affinity purification steps to ensure that they are homogeneous and correctly folded, as described previously (15).3 Sharp single bands were observed on SDS-PAGE under non-reducing conditions, which shifted upon reduction (Fig. 1, A and B), consistent with a unique set of disulfide bonds being present in each fragment. Native PAGE also gave well defined single bands, consistent with homogeneity of the various species (Fig. 1C).
FIGURE 1.
Homogeneity of CR fragments, judged by PAGE. A, (CR)2 fragments run under SDS-denaturing non-reducing (left) and reducing (right) conditions. B, (CR)3 fragments run under SDS-denaturing non-reducing (left) and reducing (right) conditions. C, (CR)2 and (CR)3 fragments run under non-denaturing conditions. Note that, although CR78 migrated anomalously under non-reducing conditions (A), mass spectrometry and analytical ultracentrifugation confirmed that it was a monomeric species. Lane M, molecular mass standards.
Mode of Binding of CR Fragments to Serpins
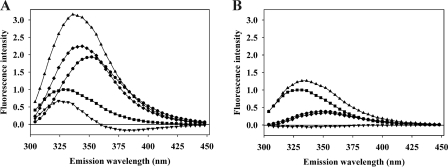
We have previously shown for the binding both of the receptor binding domain of α2M and of RAP and its domains to fragments CR345 and CR567 of LRP that the tryptophan fluorescence of the CR domains undergoes a 5–6-nm blue shift, albeit without a large change in intensity (15).3 The blue shift is understandable from the x-ray structures of several complexes of ligands with CR domains from LDLR and VLDLR, which show that a lysine side chain from the protein ligand fits into a pocket on the CR domain that contains the Ca2+-binding site and that is lined on one side by the single tryptophan present in most of these domains (an exception is phenylalanine in domain 4 of LDLR) (21–23). The occurrence of such a blue shift for the tryptophan emission spectrum of a CR domain upon ligand binding is thus likely to be diagnostic for such a binding mode. For all of the (CR)2 and (CR)3 fragments examined here for binding to PAI-1 and PN-1 species, an analogous blue shift, with relatively small overall change in fluorescence intensity, was seen (see Fig. 2A for an example), suggesting that the same kind of engagement of lysine side chains from the serpin is used in binding to the CR domains. This common blue shift was then exploited to follow binding between CR fragments and serpin ligands by monitoring the change in intensity at the wavelength of greatest signal perturbation.
FIGURE 2.
Representative tryptophan fluorescence spectra of a (CR)3 fragment binding to cleaved PAI-1. A, 200 nm rccPAI-1 (■), 200 nm CR456 (●), 1:1 mixture of rccPAI-1 and CR456 (▴), difference spectrum between a 1:1 mixture of rccPAI-1 and CR456 and rccPAI-1 (♦), and difference spectrum between a 1:1 mixture of rccPAI-1 and CR456 and the sum of spectra of rccPAI-1 and CR456 (▾). B, effect of 10 mm EDTA added to buffer. 200 nm rccPAI-1 (■), 200 nm CR456 (●), 1:1 mixture of rccPAI-1 and CR456 (▴), difference spectrum between 1:1 mixture of rccPAI-1 and CR456 and rccPAI-1 (♦), and difference spectrum between 1:1 mixture of rccPAI-1 and CR456 and the sum of spectra of rccPAI-1 and CR456 (▾). Note that the much lower intensity of the spectrum of CR456 in the presence of EDTA compared with the Ca2+ complex in A is as expected from the typical large increase in tryptophan fluorescence of CR domains upon binding Ca2+ (20).
Both to confirm earlier studies that ligand binding to CR domains depends on the presence of Ca2+, which is known to fix the structure of each domain (24), and that the fluorescence perturbation seen reports on binding of the ligand to the structured domain, we examined binding of CR456 to cleaved PAI-1 in the presence of EDTA. As expected, no perturbation of fluorescence was observed (Fig. 2B).
Contribution of Lysine to Binding to CR Domains
Mutagenesis studies on several LRP ligands have shown that lysine residues play a crucial role in ligand binding to LRP (25–28). In addition, the x-ray structure of the tandem CR domain pair LA34 from the LDL receptor to the third domain (D3) from RAP has provided an atomic level explanation for why this might be so (23). In that structure, two well separated lysine residues on D3 each engage one CR domain, with the ϵ-amino group of each lysine side chain coordinating to acidic residues that both form and surround the Ca2+-binding site of each domain. The hydrophobic portion of the lysine side chain interacts with the indole ring of the single tryptophan present in each CR domain. Even where mutagenesis has provided quantitation of the effect of mutating specific lysine residues, the replacement residue (methionine) was still capable of interacting with the trytophan, so that the full contribution of the lysine side chain could not be evaluated (15). To better estimate the contribution to binding solely from a lysine residue, we used our fluorescence approach to examine binding of lysine and analogues to the single CR domain CR3. In keeping with the expectation that it would bind to the Ca2+ site and so also interact with the single tryptophan in CR3, the endogenous tryptophan fluorescence was perturbed in a saturable manner when lysine was added (data not shown). From analysis of the binding titrations, a Kd for lysine of 2.2 mm was obtained, which corresponds to a binding energy of −14.9 kJ mol−1 (Table 1). Since lysine in a peptide linkage would have neither free α-amino or carboxyl groups, we also examined binding of l-lysine methyl ester and N-acetyl-l-lysine methyl ester. Blocking of the carboxyl enhanced the binding energy by 2.6 kJ mol−1, whereas blocking the α-amino reduced the affinity by 2.0 kJ mol−1. Consistent with this, ϵ-aminocaproic acid bound with 2.4 kJ mol−1 lower affinity than l-lysine. Finally, we examined binding of l-arginine and found a preference for lysine over arginine by 3.7 kJ mol−1. These results suggest that the engagement of a single lysine residue by the Ca2+-binding region of a CR domain is worth a minimum of −15.5 kJ mol−1. This can be enhanced by an additional positive charge close enough to the lysine to engage some of the acidic residues surrounding the Ca2+-binding site, perhaps accounting for why certain lysines are preferred over others as potential high affinity binding sites for CR domains.
TABLE 1.
Affinity of lysine and related species for CR3
| Species | Kd | ΔG |
|---|---|---|
| mm | kJ mol−1 | |
| l-Lysine | 2.2 ± 0.1 | −14.9 |
| l-Lysine methyl ester | 1.3 ± 0.3 | −17.5 |
| N-Acetyl l-lysine methyl ester | 1.7 ± 0.2 | −15.5 |
| ϵ-Aminocaproic acid | 6.4 ± 0.2 | −12.5 |
| l-Arginine | 10.2 ± 0.4 | −11.2 |
Gel Shift Assay
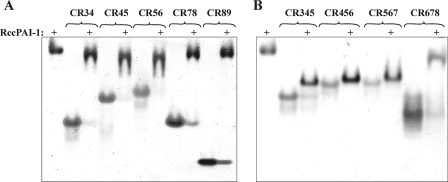
As an initial, qualitative way of examining binding between cleaved PAI-1 and different CR fragments, a gel shift assay was employed. For the three-domain constructs (CR345, CR456, CR567, and CR678), binding was tight enough to cause a band shift, although the effect was relatively small for CR678 (Fig. 3). With the exception of CR89, all of the two-domain constructs also caused a band shift (Fig. 3). This showed that cleaved PAI-1 could bind tightly to all of the (CR)3 fragments, spanning most of cluster II, and even to smaller fragments composed of only two domains.
FIGURE 3.
Gel shift assay of binding of all (CR)x fragments to cleaved PAI-1. Shown are 8% non-denaturing polyacrylamide gels for binding of (CR)2 species (A) and (CR)3 species (B) to rccPAI-1. Each (CR)x species is shown alone and in a 1:1 complex with rccPAI-1. Cleaved PAI-1 is shown in the leftmost lane of each gel.
Binding of Cleaved PAI-1 to Different LRP Fragments
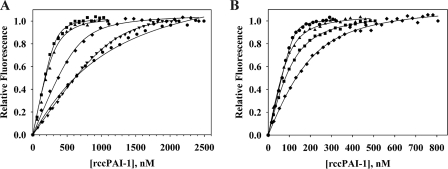
Using the common blue shift in the tryptophan emission spectrum of the CR domain of the various CR fragments upon binding to cleaved PAI-1, binding curves were obtained for cleaved PAI-1 binding to the five (CR)2 and four (CR)3 fragments. All titrations gave saturation of the change in tryptophan fluorescence and could be fitted to a single site binding process (Fig. 4). It should be noted that, because of the need to use at least 50 nm CR fragment to ensure adequate sensitivity, the tightest dissociation constants (i.e. those with values in the single digit nm range) should be considered approximate, since the titration data showed relatively little curvature. Nevertheless, it is clear that cleaved PAI-1 can bind with high affinity even to most (CR)2 fragments and that such binding can occur at quite distinct sites within cluster II, although there is a preference for CR456 over the overlapping species CR345, CR567, and CR678, which represents an up to 18% difference in binding energy. With the single exception of CR89, the dissociation constants, obtained from non-linear least squares fitting, were in the range of 18–90 nm for (CR)2 fragments and 3–59 nm for (CR)3 fragments (Table 2). The outlier is CR89, which gave a reproducible Kd of 1.2 μm. This much weaker Kd value is in keeping with the absence of a gel shift for its complex with cleaved PAI-1 (Fig. 2).
FIGURE 4.
Titration curves for binding of (CR)x fragments to cleaved PAI-1. A, titration curves for all two-domain fragments with fits. ♦, 500 nm CR34; ■, 300 nm CR45; ▴, 300 nm CR56; ▾, 1000 nm CR78; ●, 500 nm CR89 titrated with rccPAI-1. B, titration curves for all three domain fragments. ▴, 200 nm CR345; ●, 200 nm CR456; ■, 200 nm CR567; ♦, 500 nm CR678 titrated with rccPAI-1. The curves are fits to a 1:1 binding isotherm model and normalized to a relative fluorescence of 1. Kd determinations are the averages of a minimum of three independent titrations.
TABLE 2.
Affinity of cleaved PAI-1 for different (CR)2 and (CR)3 fragments
| CR fragmenta |
|||||||||
|---|---|---|---|---|---|---|---|---|---|
| CR34 | CR45 | CR56 | CR78 | CR89 | CR345 | CR456 | CR567 | CR678 | |
| Kd (nm) | 62 ± 15 | 18 ± 6 | 48 ± 18 | 90 ± 13 | 1200 ± 200 | 22 ± 4 | 2.7 ± 0.5 | 32 ± 10 | 59 ± 5 |
| ΔG0 (kJ mol−1) | −41.1 ± 0.6 | −44.1 ± 0.9 | −41.7 ± 1 | −40.1 ± 0.4 | −33.7 ± 0.5 | −43.6 ± 0.5 | −48.8 ± 0.5 | −42.7 ± 0.8 | −41.2 ± 0.2 |
| ΔFmax/F0b | 0.53 | 0.53 | 0.22 | 0.50 | 0.24 | 0.48 | 0.55 | 0.28 | 0.26 |
aKd values are the average and spread of best fits to multiple independent determinations.
bΔFmax/F0 is the change in fluorescence intensity at saturation relative to the starting fluorescence intensity of the (CR)x species, measured at the wavelength of observation.
Binding of Different PAI-1 Species
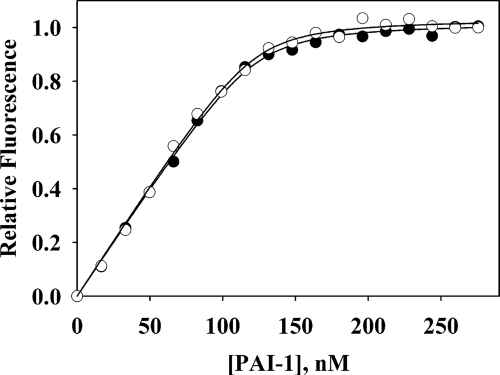
Much has been made of the discrimination shown by LRP in binding native, cleaved, and complexed forms of serpins in general and PAI-1 in particular (3, 7, 29). We therefore wanted to make a quantitative comparison between the binding of such different PAI-1 species to defined high affinity fragments of LRP. For this comparison, additional binding titrations were carried out for native 14-1B PAI-1 and for covalent complexes of PAI-1 with the physiologically relevant proteinase uPA and the non-target proteinase trypsin. Titrations of cleaved and native PAI-1 with CR456 were visually indistinguishable (Fig. 5). The Kd values for binding of native or cleaved PAI-1 to CR345, CR456, and CR567, although showing variation between the CR species, were indistinguishable between the native and cleaved forms (Table 3). For covalent complexes between PAI-1 and trypsin or lmw-uPA, the presence of proteinase increased the affinity for both CR345 and CR567, with a greater increase for uPA than for the non-target trypsin (Table 3). However, the increases were small in the context of the additional binding energy each represents (6–11% for the uPA complexes).
FIGURE 5.
Titration curves for binding of cleaved and native PAI-1 to CR456. Native (●) or cleaved (○) PAI-1 were titrated into 200 nm CR456. Raw data have been normalized to a relative fluorescence of 1 and show fits to a 1:1 binding isotherm model.
TABLE 3.
Affinity of various PAI-1 species for CR fragments
Kd values are the average and spread of best fits to multiple independent determinations. ND, not determined.
| CR45 | CR345 | CR456 | CR567 | |
|---|---|---|---|---|
| Native | ||||
| Kd (nm) | ND | 16 ± 5 | 3 ± 1 | 38 ± 19 |
| ΔG0 (kJ mol−1) | −44.4 ± 0.8 | −48.5 ± 0.8 | −42.3 ± 1.3 | |
| Cleaved | ||||
| Kd (nm) | 18 ± 6 | 22 ± 4 | 2.7 ± 0.5 | 32 ± 10 |
| ΔG0 (kJ mol−1) | −44.1 ± 0.9 | −43.6 ± 0.5 | −48.8 ± 0.5 | −42.7 ± 0.8 |
| lmw-uPA | ||||
| Kd (nm) | 8 ± 3 | 8 ± 1 | ND | 5 ± 2 |
| ΔG0 (kJ mol−1) | −47.3 ± 1.0 | −46.1 ± 0.3 | −47.3 ± 1 | |
| Trypsin | ||||
| Kd (nm) | ND | 18 ± 2 | ND | 14 ± 1 |
| ΔG0 (kJ mol−1) | −44.1 ± 0.3 | −44.7 ± 0.2 | ||
Stoichiometry of Complex Formation
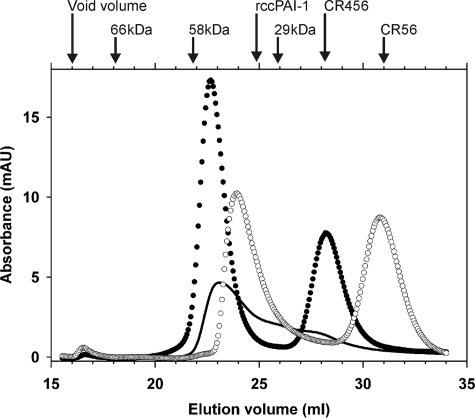
Although all of the titrations presented above gave good fits to a binding model of one molecule of (CR)2 or (CR)3 binding to one molecule of PAI-1, we wanted to obtain independent confirmation of this 1:1 stoichiometry that was not affected by the presence of any incorrectly folded CR domains. Accordingly, complexes of cleaved PAI-1 with either (CR)2 or (CR)3 species were fractionated by size exclusion chromatography, and the eluting species were characterized by elution position and composition. When cleaved PAI-1 was incubated with a substoichiometric amount of CR456, the earliest eluting peak appeared at a position earlier than PAI-1 alone and suggestive of a total mass of ∼56 kDa, which is only consistent with the presence of a single molecule of PAI-1 (44 kDa) in the complex (Fig. 6). When cleaved PAI-1 was incubated with 3 eq of CR456 and fractionated, the earliest eluting peak appeared at a position consistent with a mass of ∼57 kDa, suggesting again that only one PAI-1 molecule was present in the complex but also that, even with a large excess of CR456, only one molecule of the latter (∼14 kDa) was present. This was independently confirmed by quantitation of the integrated absorbance of the complex peak and the peak for unbound CR456 (elution position ∼28 ml). This gave a stoichiometry for the PAI-1·CR456 complex of 1:1.26. A similar experiment in which excess CR56 was incubated with cleaved PAI-1 gave an early eluting peak at a slightly later position than for the PAI-1·CR456 complex, reflecting the smaller size of CR56 (∼9 kDa) and thus also consistent with a 1:1 complex. Again, this was confirmed by analysis of the integrated absorbances of the peaks corresponding to complexed and free CR45, which gave a stoichiometry of PAI-1/CR56 for the early eluting peak of 1:0.94.
FIGURE 6.
Fractionation of PAI-1·(CR)x complexes by size exclusion chromatography. Elution profiles on a 47-ml Superdex 75 column of 9 μm cleaved PAI-1 preincubated with 0.5 eq of CR456 (solid line), with 3 eq of CR456 (●), or with 4 eq of CR56 (○). The elution positions of the individual protein components, run separately, together with those of bovine serum albumin (66 kDa) and the monomeric (29 kDa) and dimeric (58 kDa) forms of carbonic anhydrase are indicated at the top.
Comparison of PAI-1 with PN-1
PN-1 is the second member of the serpin clade E to which PAI-1 also belongs (30). PN-1 and PAI-1 consequently have the same gene structure and also have similar proteinase target specificity (31). However, they are located on different chromosomes and have quite different sequences (42% identity) and different tissue distributions, with PAI-1 being present in blood plasma, whereas PN-1 is secreted from various cell types, including glial cells (32). It was therefore of interest to compare the ability of cleaved and native PN-1 to bind to the LRP fragments examined above for binding to PAI-1. For the four triple-domain species CR345, CR456, CR567, and CR678 and the two-domain species CR34, CR45, CR56, and CR78, cleaved PN-1 bound with an affinity either similar or higher than that for cleaved PAI-1 to the same species (Table 4). CR89 bound much less tightly, but this was also the case for binding to cleaved PAI-1. A difference was that although cleaved PAI-1 showed a preference for binding to CR456, with binding energy 11–18% higher than for other (CR)3 species, PN-1 showed a preference for binding to CR567, with binding energy 5–9% higher than for other (CR)3 species. A comparison of native PN-1 with cleaved PN-1 was made for a single three-domain species, CR456, with no difference in affinity found (Table 4).
TABLE 4.
Dissociation constants (nm) of PN-1 versus PAI-1 species for selected CR fragments
Kd values are the average and spread of best fits to multiple independent determinations. ND, not determined.
| CR34 | CR45 | CR56 | CR78 | CR89 | CR345 | CR456 | CR567 | CR678 | |
|---|---|---|---|---|---|---|---|---|---|
| PN-1N | ND | ND | ND | ND | ND | ND | 7 ± 1 | ND | ND |
| PN-1C | 7 ± 3 | 18 ± 2 | 10 ± 1 | 13 ± 7 | 130 ± 36 | 7.7 ± 0.2 | 7 ± 2 | 2.8 ± 0.4 | 14 ± 9 |
| PAI-1N | ND | ND | ND | ND | ND | 16 ± 5 | 3 ± 1 | 38 ± 19 | ND |
| PAI-1C | 62 ± 15 | 18 ± 6 | 48 ± 18 | 90 ± 13 | 1200 ± 200 | 22 ± 4 | 2.7 ± 0.5 | 32 ± 10 | 59 ± 5 |
DISCUSSION
We have presented a systematic analysis of the binding of native, cleaved and proteinase-complexed PAI-1, as well as of native and cleaved PN-1, to a range of two-domain and three-domain fragments from the principal ligand-binding region of LRP, cluster II. With the exception of CR89, cleaved PAI-1 could bind with high affinity (Kd = 18–90 nm) to all tandem fragments, and with even higher affinity (Kd = 3–59 nm) to triple-domain fragments that cover the region from CR3 to CR8. Nevertheless, there was some preference for the site of binding, such that cleaved PAI-1 bound to CR456 more tightly than to other (CR)3 species, whereas cleaved PN-1 bound more tightly to CR567. The stoichiometries of these complexes was 1:1, whether the LRP fragment was (CR)2 or (CR)3. Such a combination of high affinity and relatively low specificity, either for a given serpin to different sites or different serpins to the same site, is similar to what we found recently in a detailed examination of the binding of RAP and RAP fragments to (CR)2 and (CR)3 fragments from the same LRP cluster II.3
The basis for this may be that on the one hand lysine residues appear to make a dominant contribution to binding to a CR domain and on the other that there are relatively modest requirements to be such a lysine. Thus, we have shown above that a lysine side chain contributes about 15.5 kJ mol−1 to binding a single CR domain. If the lysine has an additional positive charge close to the backbone, an additional ∼2 kJ mol−1 of binding energy results. Although such a single binding interaction may appear to be small (Kd of 1–6 mm), it should be realized that the affinities reported in Tables 2 and 3 for binding of (CR)2 or (CR)3 species, which are nearly all in the range of −41 to −49 kJ mol−1, could be nearly completely accounted for by each of two CR domains binding to a single such lysine to give a total binding interaction of ∼−35 kJ mol−1. Binding of a third CR domain by the same mechanism would actually give too strong an interaction and thus suggests that, although both CR domains of (CR)2 species engage a lysine on the ligand, the third domain of (CR)3 fragments binds in a less specific mode and contributes only a small additional amount. As examples from Table 2, cleaved PAI-1 binds to CR34 with ΔG of −41.1 kJ mol−1 but to CR345 with only an additional 2.5 kJ mol−1, whereas it binds to CR56 with ΔG of −41.7 kJ mol−1 but to CR567 with only an additional 1 kJ mol−1. Interestingly, in the case of the binding of the receptor binding domain of α2M to (CR)x fragments, it was found that mutagenesis of two very close lysine residues (1370 and 1374), which had been shown to be critical for receptor binding (26), reduced the affinity of the receptor binding domain by ∼13 kJ mol−1 (15), consistent with loss of a single CR-lysine interaction (here it should be noted that the residues were mutated to methionine so that the hydrophobic interaction with tryptophan could still occur, whereas the close juxtapositioning of Lys1370 and Lys1374 would probably make it impossible for each to separately engage a CR domain). This conclusion with respect to the number of CR domains engaged by a serpin ligand of LRP also parallels what was seen in our recent study on RAP, where each of the three domains bound tightly to (CR)2 species but gave only modestly higher affinity when a third CR domain was present.3 The minimal requirements placed on the lysine may thus be (i) that it is exposed in such a way that it can fit into the binding cleft that is part of the calcium binding site present in each of the domains CR3-CR9 (33), which would probably favor lysines that occur on the outer face of α-helices or in exposed loops rather than on the surface of β-sheets; (ii) that it (preferably) has an additional positive charge close to it to modestly enhance the binding interaction; and (iii) that two such lysines are at an appropriate separation such that each can engage a CR domain without interference but not so far apart that both CR domains cannot be simultaneously engaged. In regard to the latter, it should be noted that the separation of the α-carbons of the two lysines that separately engage a CR domain in the LA34·RAP-D3 complex is about 21 Å. Although the four-residue linker between LA3 and LA4 might allow a slightly larger separation between the lysines, a shorter one would result in clashes between the two CR domains.
Such modest requirements for coordinating lysines, together with the ability of a given pair of CR domains to modestly vary their separation and orientation by virtue of their short flexible linkers would help to explain why mutagenesis of many lysine and arginine residues in PAI-1, either singly or in combinations of up to three residues, had effects on binding to LRP of only 3–5-fold in Kd, representing no more than a 7% reduction in binding energy (28, 34), since removal of one lysine might readily be compensated by engagement of a different lysine by the CR domain, made possible by the flexible linkers of 3–10 residues that connect the CR domains (35). Although this might seem to imply that most proteins should therefore be able to bind to LRP or that a given protein should be able to bind multiple (CR)x species at different locations on the surface, it should be realized that the restrictions listed above on the properties of lysines that are suitable for coordination to CR domains, although modest, are nevertheless stringent enough to severely limit the number of potential high affinity binding regions. In this regard, it is a possibly relevant observation that all LRP-binding proteins are also heparin-binding proteins, implying that they all have a region of high density of exposed basic residues. This is also in keeping with our findings for PN-1, the second member of the E clade of human serpins, which binds to cluster II fragments with similar high affinity and low specificity as PAI-1 and with comparable lack of discrimination between the native and cleaved conformations. Given the large differences in sequence between PN-1 and PAI-1, it would be unlikely that structurally equivalent lysines would be used for binding in each case. Nevertheless, both have heparin binding regions that may involve the same general region of the protein and so are likely to have the necessary concentration of exposed “hot spot” lysines that would provide two lysines at an appropriate separation to engage two CR domains. The binding of kringle domains to exposed lysine side chains, which forms the basis for exosite interactions of many kringle-containing proteins is another example of a seemingly low requirement for binding (an exposed lysine with a carboxyl close by) that nevertheless occurs in only a limited number of target proteins (36).
Although this conclusion of degenerate binding sites on both LRP and the ligand is at variance with some literature reports that failed to find binding of some fragment combinations for which we here find tight binding, it should be borne in mind (i) that the affinities reported here are close to those reported for intact ligand to intact LRP and (ii) that our cluster II fragments are free from any flanking linkers or fusion partners, such as have been present in previous studies, that might modify the binding.
Also in keeping with the high affinity but low specificity of binding of cleaved serpin to different regions of cluster II is that both native PAI-1 and PN-1 bind as tightly as the cleaved serpin, with Kd for binding to CR456 of ∼3 nm for each PAI-1 species and ∼7 nm for each PN-1 species and indistinguishable binding of native and cleaved PAI-1 to other (CR)3 species. Although this observation has also been made by one other group for PAI-1 (34), it is at variance with an assertion in the same study and elsewhere (7) that there is a cryptic binding site on PAI-1 for LRP that only becomes exposed in complex with proteinase. In the former study, lysine 69 was identified as the epitope responsible in PAI-1, based on mutagenesis data that gave a 5-fold increase in Kd for a variant PAI-1 with mutation at this site and in complex with proteinase but no effect for the uncomplexed variant. However, x-ray structures of native and cleaved PAI-1 show this residue to be in essentially identical environments (37, 38), whereas the expected structure of the proteinase complex would also most likely leave this residue unaffected by the presence of proteinase (10). Indeed, the possibility that there might be another epitope that is only exposed in a proteinase-complexed serpin, but not the cleaved serpin, is at variance with all structural information on the serpin moiety in serpin-proteinase complexes compared with cleaved serpin (8–10, 39).
A second important conclusion of the present study is that, although proteinase complex formation does increase the affinity of the serpin for a given LRP fragment, the increase represents only a few kJ mol−1, which also represents only a few percent of the total binding energy. Thus, the lmw-uPA complex with PAI-1 binds to CR567 with a Kd of 5 nm compared with 32 nm for cleaved PAI-1. This should be viewed in the context of specific binding of uPA alone to LRP having a dissociation constant of ∼50 nm (40). Thus, the affinity of the uPA·PAI-1 complex is clearly not the sum of the individual binding interactions of uPA and PAI-1 by many kJ mol−1. Given this small, presumably nonspecific contribution of the proteinase in complex, it is not surprising that a non-cognate proteinase, such as trypsin, also increases the affinity, in our study to a Kd of 14 nm. Interestingly, this minor role for the nature of the proteinase was also pointed out earlier, although with the assertion that the proteinase played no role (7). Although it was not possible to measure the affinity of the trypsin·PAI-1 and lmw-uPA·PAI-1 complexes with the higher affinity CR456, because of experimental limitations of our fluorescence approach, it is likely that the affinity would be enhanced in a similar manner and thus make this site the somewhat preferred binding site for such complexes in intact LRP. Consistent with this, the uPA·PAI-1 complex binds to LRP with Kd of 0.2–1 nm (28, 40, 41). The possible significance of a modest increase in affinity for PAI-1 to LRP upon complex formation with proteinase is that it could tip the Kd from a value that is well above the usual physiological concentrations of PAI-1 (20 ng/ml, ∼0.5 nm) (42) to one that is close to those encountered in vivo. In this way, it could be ensured that the PAI-1·proteinase complex would bind efficiently to LRP, whereas uncomplexed PAI-1, whether native, cleaved, or even latent, would bind very poorly. It is therefore not the absolute affinity of a serpin or serpin-proteinase complex for LRP that is important but rather the affinity relative to the concentrations that would be encountered in vivo. By extension, one would expect lower affinities for more abundant serpins and higher affinities for rarer ones. In keeping with this idea, antithrombin, whose physiological concentration is very much higher (∼2.3 μm) (43) binds very much less tightly to LRP, even when complexed to proteinase (40, 41).
Acknowledgments
We thank Dr. Peter Andreasen for the kind gift of PN-1 cDNA, Dr. Grant Blouse for the gift of lmw-uPA, and Dr. Steven Olson for helpful comments on the manuscript.
This work was supported, in whole or in part, by National Institutes of Health Grants R01 GM54414 and R01HL79430. This work was also supported by fellowships from the Danish Research Council and the Carlsberg Foundation (to J. K. J.).
3 Jensen, J. K., Dolmer, K., Schar, C., and Gettins, P. G. W. (2009) Biochem. J., in press.
- LRP
- low density lipoprotein receptor-related protein
- α2M
- α2-macroglobulin
- β-ME
- β-mercaptoethanol
- CR
- complement-like repeat
- (CR)x
- LRP fragment containing x CR domains
- CRxy
- LRP fragment containing domains CRx and CRy
- CRxyz
- LRP fragment containing domains CRx, CRy and CRz
- LDLR
- low density lipoprotein receptor
- PAI-1
- plasminogen activator inhibitor 1
- PN-1
- proteinase nexin 1
- RAP
- receptor-associated protein
- rccPAI-1
- reactive center-cleaved PAI-1
- rccPN-1
- reactive center-cleaved PN-1
- TEV
- tobacco etch virus
- GST
- glutathione S-transferase
- TLCK
- Nα-p-tosyl-l-lysine chloromethyl ketone
- PPE
- porcine pancreatic elastase
- uPA
- urokinase-type plasminogen activator
- lmw-uPA
- low molecular weight uPA
- MES
- 4-morpholineethanesulfonic acid.
REFERENCES
- 1.Krieger M., Herz J. ( 1994) Annu. Rev. Biochem. 63, 601– 637 [DOI] [PubMed] [Google Scholar]
- 2.Herz J., Strickland D. K. ( 2001) J. Clin. Invest. 108, 779– 784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mast A. E., Enghild J. J., Pizzo S. V., Salvesen G. ( 1991) Biochemistry 30, 1723– 1730 [DOI] [PubMed] [Google Scholar]
- 4.de Smet B. J., de Boer J. P., Agterberg J., Rigter G., Bleeker W. K., Hack C. E. ( 1993) Blood 81, 56– 61 [PubMed] [Google Scholar]
- 5.Malek R., Aulak K. S., Davis A. E., 3rd ( 1996) Clin. Exp. Immunol. 105, 191– 197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Joslin G., Fallon R. J., Bullock J., Adams S. P., Perlmutter D. H. ( 1991) J. Biol. Chem. 266, 11282– 11288 [PubMed] [Google Scholar]
- 7.Stefansson S., Muhammad S., Cheng X. F., Battey F. D., Strickland D. K., Lawrence D. A. ( 1998) J. Biol. Chem. 273, 6358– 6366 [DOI] [PubMed] [Google Scholar]
- 8.Huntington J. A., Read R. J., Carrell R. W. ( 2000) Nature 407, 923– 926 [DOI] [PubMed] [Google Scholar]
- 9.Dementiev A., Dobó J., Gettins P. G. ( 2006) J. Biol. Chem. 281, 3452– 3457 [DOI] [PubMed] [Google Scholar]
- 10.Backovic M., Stratikos E., Lawrence D. A., Gettins P. G. ( 2002) Protein Sci. 11, 1182– 1191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tesch L. D., Raghavendra M. P., Bedsted-Faarvang T., Gettins P. G., Olson S. T. ( 2005) Protein Sci. 14, 533– 542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bacskai B. J., Xia M. Q., Strickland D. K., Rebeck G. W., Hyman B. T. ( 2000) Proc. Natl. Acad. Sci. U.S.A. 97, 11551– 11556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Qiu Z., Strickland D. K., Hyman B. T., Rebeck G. W. ( 2002) J. Biol. Chem. 277, 14458– 14466 [DOI] [PubMed] [Google Scholar]
- 14.Lutz C., Nimpf J., Jenny M., Boecklinger K., Enzinger C., Utermann G., Baier-Bitterlich G., Baier G. ( 2002) J. Biol. Chem. 277, 43143– 43151 [DOI] [PubMed] [Google Scholar]
- 15.Dolmer K., Gettins P. G. ( 2006) J. Biol. Chem. 281, 34189– 34196 [DOI] [PubMed] [Google Scholar]
- 16.Dolmer K., Huang W., Gettins P. G. ( 2000) J. Biol. Chem. 275, 3264– 3269 [DOI] [PubMed] [Google Scholar]
- 17.Lawrence D. A., Olson S. T., Palaniappan S., Ginsburg D. ( 1994) Biochemistry 33, 3643– 3648 [DOI] [PubMed] [Google Scholar]
- 18.Li Y., Lu W., Schwartz A. L., Bu G. ( 2002) Biochemistry 41, 4921– 4928 [DOI] [PubMed] [Google Scholar]
- 19.Meijer A. B., Rohlena J., van der Zwaan C., van Zonneveld A. J., Boertjes R. C., Lenting P. J., Mertens K. ( 2007) Biochim. Biophys. Acta 1774, 714– 722 [DOI] [PubMed] [Google Scholar]
- 20.Dolmer K., Huang W., Gettins P. G. W. ( 1998) Biochemistry 37, 17016– 17023 [DOI] [PubMed] [Google Scholar]
- 21.Rudenko G., Henry L., Henderson K., Ichtchenko K., Brown M. S., Goldstein J. L., Deisenhofer J. ( 2002) Science 298, 2353– 2358 [DOI] [PubMed] [Google Scholar]
- 22.Verdaguer N., Fita I., Reithmayer M., Moser R., Blaas D. ( 2004) Nat. Struct. Biol. 11, 429– 434 [DOI] [PubMed] [Google Scholar]
- 23.Fisher C., Beglova N., Blacklow S. C. ( 2006) Mol. Cell 22, 277– 283 [DOI] [PubMed] [Google Scholar]
- 24.Huang W., Dolmer K., Gettins P. G. W. ( 1999) J. Biol. Chem. 274, 14130– 14136 [DOI] [PubMed] [Google Scholar]
- 25.Lalazar A., Weisgraber K. H., Rall S. C., Jr., Giladi H., Innerarity T. L., Levanon A. Z., Boyles J. K., Amit B., Gorecki M., Mahley R. W. ( 1988) J. Biol. Chem. 263, 3542– 3545 [PubMed] [Google Scholar]
- 26.Arandjelovic S., Hall B. D., Gonias S. L. ( 2005) Arch. Biochem. Biophys. 438, 29– 35 [DOI] [PubMed] [Google Scholar]
- 27.Lee D., Walsh J. D., Mikhailenko I., Yu P., Migliorini M., Wu Y., Krueger S., Curtis J. E., Harris B., Lockett S., Blacklow S. C., Strickland D. K., Wang Y. X. ( 2006) Mol. Cell 22, 423– 430 [DOI] [PubMed] [Google Scholar]
- 28.Skeldal S., Larsen J. V., Pedersen K. E., Petersen H. H., Egelund R., Christensen A., Jensen J. K., Gliemann J., Andreasen P. A. ( 2006) FEBS J. 273, 5143– 5159 [DOI] [PubMed] [Google Scholar]
- 29.Picard V., Marque P. E., Paolucci F., Aiach M., Le Bonniec B. F. ( 1999) J. Biol. Chem. 274, 4586– 4593 [DOI] [PubMed] [Google Scholar]
- 30.Silverman G. A., Bird P. I., Carrell R. W., Church F. C., Coughlin P. B., Gettins P. G., Irving J. A., Lomas D. A., Luke C. J., Moyer R. W., Pemberton P. A., Remold-O'Donnell E., Salvesen G. S., Travis J., Whisstock J. C. ( 2001) J. Biol. Chem. 276, 33293– 33296 [DOI] [PubMed] [Google Scholar]
- 31.Gettins P. G. ( 2002) Chem. Rev. 102, 4751– 4804 [DOI] [PubMed] [Google Scholar]
- 32.Guenther J., Nick H., Monard D. ( 1985) EMBO J. 4, 1963– 1966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jensen G. A., Andersen O. M., Bonvin A. M., Bjerrum-Bohr I., Etzerodt M., Thøgersen H. C., O'Shea C., Poulsen F. M., Kragelund B. B. ( 2006) J. Mol. Biol. 362, 700– 716 [DOI] [PubMed] [Google Scholar]
- 34.Horn I. R., van den Berg B. M., Moestrup S. K., Pannekoek H., van Zonneveld A. J. ( 1998) Thromb. Haemost. 80, 822– 828 [PubMed] [Google Scholar]
- 35.Beglova N., North C. L., Blacklow S. C. ( 2001) Biochemistry 40, 2808– 2815 [DOI] [PubMed] [Google Scholar]
- 36.Panizzi P., Boxrud P. D., Verhamme I. M., Bock P. E. ( 2006) J. Biol. Chem. 281, 26774– 26778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stout T. J., Graham H., Buckley D. I., Matthews D. J. ( 2000) Biochemistry 39, 8460– 8469 [DOI] [PubMed] [Google Scholar]
- 38.Jensen J. K., Gettins P. G. ( 2008) Protein Sci. 17, 1844– 1849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dobó J., Gettins P. G. ( 2004) J. Biol. Chem. 274, 9264– 9269 [DOI] [PubMed] [Google Scholar]
- 40.Nykjaer A., Kjøller L., Cohen R. L., Lawrence D. A., Garni-Wagner B. A., Todd R. F., 3rd, van Zonneveld A. J., Gliemann J., Andreasen P. A. ( 1994) J. Biol. Chem. 269, 25668– 25676 [PubMed] [Google Scholar]
- 41.Kasza A., Petersen H. H., Heegaard C. W., Oka K., Christensen A., Dubin A., Chan L., Andreasen P. A. ( 1997) Eur. J. Biochem. 248, 270– 281 [DOI] [PubMed] [Google Scholar]
- 42.Leiper K., Croll A., Booth N. A., Moore N. R., Sinclair T., Bennett B. ( 1994) J. Clin. Pathol. 47, 214– 217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Conard J., Brosstad F., Lie Larsen M., Samama M., Abildgaard U. ( 1983) Haemostasis 13, 363– 368 [DOI] [PubMed] [Google Scholar]