Abstract
Lipolysis is the catabolic pathway by which triglycerides are hydrolyzed into fatty acids. Adipose triglyceride lipase (ATGL) and hormone-sensitive lipase (HSL) have the capacity to hydrolyze in vitro the first ester bond of triglycerides, but their respective contributions to whole cell lipolysis in human adipocytes is unclear. Here, we have investigated the roles of HSL, ATGL, and its coactivator CGI-58 in basal and forskolin-stimulated lipolysis in a human white adipocyte model, the hMADS cells. The hMADS adipocytes express the various components of fatty acid metabolism and show lipolytic capacity similar to primary cultured adipocytes. We show that lipolysis and fatty acid esterification are tightly coupled except in conditions of stimulated lipolysis. Immunocytochemistry experiments revealed that acute forskolin treatment promotes HSL translocation from the cytosol to small lipid droplets and redistribution of ATGL from the cytosol and large lipid droplets to small lipid droplets, resulting in enriched colocalization of the two lipases. HSL or ATGL overexpression resulted in increased triglyceride-specific hydrolase capacity, but only ATGL overexpression increased whole cell lipolysis. HSL silencing had no effect on basal lipolysis and only partially reduced forskolin-stimulated lipolysis. Conversely, silencing of ATGL or CGI-58 significantly reduced basal lipolysis and essentially abolished forskolin-stimulated lipolysis. Altogether, these results suggest that ATGL/CGI-58 acts independently of HSL and precedes its action in the sequential hydrolysis of triglycerides in human hMADS adipocytes.
Adipose tissue fat stores in humans are mainly dependent upon fatty acid (FA)2 supply, FA esterification to triglycerides (TG), and TG breakdown, or lipolysis. Adipose tissue lipolysis is governed by three lipases. Adipose triglyceride lipase (ATGL) and hormone-sensitive lipase (HSL) both have the capacity to initiate TG degradation by cleaving the first ester bond, but HSL is unique in its capacity to break down the second ester bond, converting diglycerides (DG) to monoglycerides (1–3). The non-rate-limiting monoglyceride lipase completes lipolysis by cleaving the last ester bond from a monoglyceride molecule, leading to glycerol release (4). Adipose tissue lipolysis has received much attention over the past 10 years because of its altered regulation in obesity (5).
HSL resides freely in the cytosol and can associate with lipid droplets (LD). It is regulated by hormones such as catecholamines, insulin, and natriuretic peptides. Catecholamines bind to β-adrenoceptors on adipocyte cell membranes and activate cyclic AMP-dependent protein kinase. Similarly, natriuretic peptides bind to type A receptors and activate cyclic GMP-dependent protein kinase (6). The protein kinase action in stimulated lipolysis is 2-fold: 1) phosphorylation of HSL, leading to its translocation from the cytosol to LD (7, 8), and 2) phosphorylation of perilipin A (6, 9, 10), the predominant perilipin isoform in adipocytes, enhancing interaction between HSL and LD. The importance of HSL activity in stimulating complete lipolysis is indisputable, particularly given its unique capacity to hydrolyze DG. However, lipolysis is not exclusively dependent upon HSL because HSL null mice revealed residual TG lipase activity in adipose tissue (2, 11). Another adipose tissue lipase was identified (3, 12, 13). ATGL, also known as desnutrin or patatin-like phospholipase domain-containing protein 2, shows affinity toward TG only (3, 14). ATGL is activated by CGI-58, an esterase/thioesterase/lipase subfamily protein devoid of TG hydrolase enzymatic activity (15, 16). The role of HSL and ATGL has been investigated in murine fat cell lipolysis, but the relative importance of these lipases in basal and protein kinase A-stimulated human fat cell lipolysis has remained elusive.
Increased fat mass is associated with defects in adipose tissue metabolism. In obesity, resistance to catecholamine-induced lipolysis is observed (17–19). This inhibition of lipolysis may be naturally occurring as an adaptive protective mechanism to minimize FA release and its deleterious consequences on metabolism. Indeed, decreased expression of HSL and ATGL has been observed in isolated adipocytes and differentiated preadipocytes of obese subjects and adipose tissue of insulin-resistant subjects, respectively (20–23). However, by virtue of its mass, adipose tissue basal lipolysis elevates circulating levels of FAs in obese subjects, thereby increasing the risk of insulin resistance. Therefore, the use of pharmacological lipid-lowering agents that act through inhibition of lipolysis has been a promising research avenue leading to the development of several series of HSL inhibitors (24).
Herein, we sought to examine the respective contributions of HSL and ATGL to lipolysis and re-esterification in fat cells derived from human adipose tissue derived-multipotent stem cells (termed hMADS cells). These cells, which exhibit at a clonal level normal karyotype, self-renewal ability, and no tumorigenicity, are able to differentiate into functional adipocytes (25, 26). We investigated the localization of HSL and ATGL in basal and stimulated lipolytic conditions and studied lipase activities and whole cell lipolysis in adipocytes with altered expression levels of HSL, ATGL, and its coactivator CGI-58. Our results provide novel insights into ATGL localization and its critical role with coactivator CGI-58 in DG provision to HSL during basal and stimulated lipolysis.
EXPERIMENTAL PROCEDURES
Cell Culture
hMADS cells were maintained in proliferation medium (Dulbecco's modified Eagle's medium low glucose, 10% fetal bovine serum, 2 mm l-glutamine, 10 mm Hepes buffer, 50 units/ml of penicillin, 50 μg/ml of streptomycin, supplemented with 2.5 ng/ml of human fibroblast growth factor 2). The cells were inoculated in 6-well plates at a density of 44,000 cells/ml and kept at 37 °C in 5% CO2. Six days post-seeding, fibroblast growth factor 2 was removed from proliferation medium. On the next day (day 0), the cells were incubated in differentiation medium (DM; serum-free proliferation medium/Ham's F-12 medium containing 10 μg/ml of transferrin, 5 μg/ml of insulin, 0.2 nm triiodothyronine, 100 μm 3-isobutyl-1-methylxanthine, 1 μm dexamethasone, and 100 nm rosiglitazone). At day 3, dexamethasone and 3-isobutyl-1-methylxanthine were omitted from DM, and at day 9 rosiglitazone was also omitted. The experiments were carried out between days 12 and 15. Pharmacological treatment of cells was performed during functional measurements (3 h) with the adenylate cyclase activator forskolin (FK) and/or specific HSL inhibitor Bay (22, 27). Primary culture of differentiated preadipocytes was performed as previously described (28).
Determination of mRNA Levels
Total RNA was extracted using the RNeasy total RNA mini kit (Qiagen). RNA concentration and purity were assessed spectrophotometrically with a NanoDrop (Digitalbio). After treatment with DNase I (Invitrogen) and reverse transcription of 1 μg of total RNA with SuperScript II (Invitrogen), real time quantitative PCR was performed with ABI PRISM 7500 (Applied Biosystems). For each primer pair, a standard curve was obtained using serial dilutions of hMADS cDNA prior to mRNA quantitation. 18 S rRNA was used as a control to normalize gene expression.
Adenoviral Transduction
Adenoviruses were produced at the Gene Therapy Laboratory of Nantes. They contained, in tandem, the green fluorescent protein (GFP) gene and the full-length human ATGL or HSL cDNA downstream of separate cytomegalovirus promoters (29). An adenovirus only containing the GFP gene was used as control. On day 12 of differentiation, the cells were infected for 6 h at a multiplicity of infection (m.o.i.) of 400 unless stated otherwise. Gene expression, flux measurements, and maximal activities were determined 72 h post-transduction.
RNA Interference
RNA interference was achieved by small interfering RNA. Briefly, on day 7 of differentiation, hMADS cells were detached from culture dishes with 0.25% trypsin/EDTA (Invitrogen) and counted. Control (siGFP; Ambion) or gene-specific small interfering RNAs for HSL (Ambion), ATGL (Ambion), and CGI-58 (Qiagen) were delivered into adipocytes at 100 pmol/2 million cells (1100 volts, 20 ms) with a microporator (Digitalbio). The adipocytes were then mixed with DM and reseeded onto Biocoat collagen multiwell plates (BD Biosciences). Gene expression, flux measurements, and enzymatic activities were assessed 6 days post-microporation. For each gene, two separate small interfering RNAs were tested. The targeted sequences, flanked with dTdT overhangs, for data presented are: GFP, 5′-GCA GCA CGA CUU CUU CAA G; HSL, 5′-AGG ACA ACA UAG CCU UCU U; ATGL, 5′-AGU UCA UUG AGG UAU CUA A; and CGI-58, 5′-GGC CUG AUU UCA AAC GAA A.
Lipolytic Flux Measurement
Whole cell lipolysis was investigated using two approaches: glycerol and nonesterified fatty acids (NEFAs) release in culture media, and radioactive lipolytic flux after inhibition of fatty acid re-esterification. For the former, lipolysis was assessed in DM lacking differentiation agents but supplemented with 2% BSA. The medium was changed, and following 3 h of treatment, glycerol and NEFAs released in the medium were measured using commercially available kits (free glycerol reagent from Sigma and NEFA-C from Wako Chemicals). To circumvent complications in cultured cells caused by important re-esterification flux of FAs, we performed whole cell lipolysis measurements in the presence of Triacsin C (Sigma), an inhibitor of acyl-CoA synthase. Because acyl-CoA synthase is a core component of NEFA determination kits, we combined the use of Triacsin C to a radioactive lipolytic flux approach. Briefly, lipolysis was carried out following an overnight loading period of hMADS cells with DM lacking differentiation agents but supplemented with 2% BSA and [3H-9,10]oleic acid (OA) (final concentration, 50 μm, 8 μCi/ml; Amersham Biosciences). On the next morning, loading medium was discarded, and the cells were washed twice with phosphate-buffered saline with 1% BSA (Sigma) to remove residual loading medium and nonincorporated [3H-9,10]OA. Fresh DM supplemented with 2% BSA and Triacsin C (10 μm) was added to initiate lipolysis. The medium (50 μl) was sampled at time 0 to determine baseline levels of [3H-9,10]OA. Following 3 h of drug treatment, the medium was sampled for counting of released [3H-9,10]OA. The cells were washed twice with phosphate-buffered saline and scraped with extraction buffer (10 mm Tris-HCl, pH 7.4, 0.25 m sucrose, 1 mm EDTA, 1 mm dithiothreitol) for protein normalization of lipolysis. Protein concentrations were determined with Bio-Rad protein assay using BSA as standard.
Lipase Activity
In vitro enzymatic activities were performed as described (30). Briefly, triolein or cholesterol oleate was emulsified with phospholipids by sonication. Fat-depleted BSA (Sigma) was used as a FA acceptor. Cytosolic fractions of hMADS adipocytes were incubated for 30 min at 37 °C with the different substrates. Hydrolysis was stopped, and radiolabeled oleic acid released was measured using a scintillation counter (Tri-Carb 2100TR; Packard). To determine HSL-independent TG lipase activity, the selective HSL inhibitor Bay was added during assays.
Immunoblotting
Western blots were performed on cytosolic and fat cake fractions of hMADS cells. The fractions of interest were separated with four series of differential centrifugation at 21,000 × g (4 °C). After the initial centrifugation step (40 min), which served to pellet organelles, the cytosol was aspirated with a syringe, leaving the fat cake behind. Each fraction was spun again and the cytosol was aspirated from the fat cake to yield clean respective fractions. Each fraction was transferred to a clean tube, and the fat cake was resuspended in a small volume of extraction buffer. Unless noted otherwise, equivalent amounts of protein present in samples were resolved in 8% SDS-PAGE. The separated proteins were transferred to a nitrocellulose membrane (Hybond ECL; GE Healthcare). The blots were incubated and immunoblotted with polyclonal anti-hHSL (1:12,000; gift from Dr. Cecilia Holm, Lund University, Lund, Sweden), anti-hATGL (1:1000; Cell Signaling), and anti-hCGI-58 (1:1000; Tebu-Bio). Normalization of cytosolic and fat cake fractions was performed with anti-β-actin (1:1000; Cell Signaling), and anti-vimentin (1:1000; Euromedex), respectively. Bound immunoglobulins were detected with a horseradish peroxidase-labeled goat anti-rabbit or anti-mouse IgG conjugate. Revelation was performed using enhanced chemiluminescence (ECL Plus; GE Healthcare) and densitometry analysis of bands with ImageQuant TL V2005 (GE Healthcare).
Immunocytochemistry
To examine localization of HSL and ATGL in the same cell, we developed a sequential double immunohistochemical staining procedure. Briefly, the cells were fixed and permeabilized as previously described (31). Then slides were incubated with the primary antibody against the first antigen (chicken anti-HSL, 1:500) overnight at 4 °C in incubation buffer (permeabilization buffer with 5% fetal calf serum). The slides were washed three times with permeabilization buffer over a 15-min period and then incubated for 1 h with Alexa Fluor 488 goat anti-chicken (A11039) in incubation buffer at room temperature. The slides were washed as before and then incubated overnight at 4 °C with the primary antibody against the second antigen (rabbit anti-ATGL at 1:300) in incubation buffer. The slides were again washed and incubated for 1 h with Alexa Fluor 568 donkey anti-rabbit (A10042) at 1:1500 and Hoechst 33258 at 1:2000 in incubation buffer. For each primary antibody, control experiments were performed, and antibodies were tested using Western blot methodology. All of the primary antibodies were detecting one single band corresponding to the predicted molecular mass of the protein. Specificity of secondary antibodies was tested in the control experiments. Alexa Fluor 568 donkey anti-rabbit antibody was not staining cells incubated with chicken anti-HSL and Alexa Fluor 488 Goat anti-Chicken antibodies. Similarly, Alexa Fluor 488 goat anti-chicken antibody gave no signal in cells incubated with rabbit anti-ATGL and 568 donkey anti-rabbit antibodies. The chicken anti-HSL antibody was a kind gift from Dr. Cecilia Holm. The rabbit anti-ATGL antibody was from Cell Signaling. Alexa fluorophore-conjugated secondary antibodies were all from Invitrogen Molecular Probes.
Microscopy and Image Analysis
Immunofluorescence images were made with an Axio Observer.Z1 inverted microscope (Carl Zeiss MicroImaging) using a 63×/1.40 oil Plan-Apochromat lens and an Axiocam MRm camera. The following filters were used for indicated fluorophores: Alexa 488, filter 489038; and Alexa 568, filter 489043. No cross-bleeding was observed between red and green channels. Optical sections were taken at the calculated optimal z axis interval (Nyquist theory) beginning at the surface of the cell. At least 50 sections were made of each cell. Image stacks were deconvolved with the constrained iterative maximum likelihood algorithm, giving results with confocal-like properties. Automatic correction for Z-stacks was included in the deconvolution algorithm. This function corrected for bleaching and lamp flicker. At least 10 cells were analyzed for each condition. The intensity distribution within the colocalization region (extent of colocalization) was evaluated by Pearson's correlation coefficient (r2) for each optical section (after deconvolution), and a mean of Pearson's coefficient was calculated. Statistical analysis was done using unpaired t test. A straight line was drawn across the cell image, and distribution of signal in each channel along the line was evaluated in different optical sections. Matching profiles of green and red channels were regarded as colocalization.
Statistical Analysis
Statistical significance was determined by one-way ANOVA (Dunnett's post hoc test), two-way ANOVA (Bonferroni correction), or Pearson's correlation coefficient. Group differences were considered significant for p < 0.05 (*), p < 0.01 (**), and p < 0.001 (***).
RESULTS
The hMADS Cell Line, an Appropriate Model to Study Human Fat Cell Metabolism
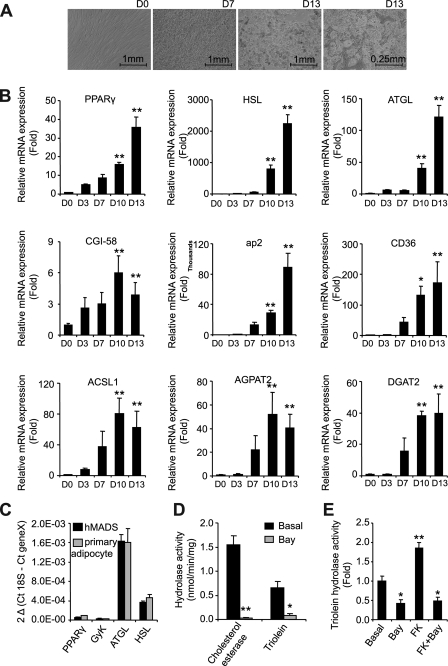
To study human adipocyte metabolism, we chose to use the hMADS cell line (25, 26). It offers the advantages of being of human white adipose tissue origin, while being less variable than primary culture of preadipocytes. During differentiation, visual examination of the cells clearly demonstrated a phenotypical progression from fibroblast-type cells at day 0 to round-shaped cells by day 7 (Fig. 1A). Significant lipid accumulation occurred between days 7 and 15. Day 13 was established as the optimal day for handling cells, after which they became overly fragile. Differentiation yield was estimated at 80–85%. We followed differentiation kinetics with real time reverse transcription PCR to validate the use of this model for investigation of fat cell metabolism (Fig. 1B). Expression levels of PPARγ, lipases, FA transporters, and esterification enzymes increased throughout differentiation, indicating a concerted up-regulation of FA metabolism genes in hMADS adipocytes. We then assessed gene expression in both hMADS adipocytes and primary culture of differentiated preadipocytes at day 13 of differentiation to further validate these cells as an acceptable model of human adipocytes (Fig. 1C). Expression levels of PPARγ, HSL, and ATGL were comparable between the two models. Glycerol kinase expression was also examined given the importance of lipid turnover in cell culture models. Low levels of expression were observed in both cases (32). Next, we measured lipid hydrolase activity of HSL and ATGL in hMADS adipocytes. In white adipose tissue, cholesterol esterase hydrolase activity is entirely attributed to HSL (11). In hMADS adipocytes, the Bay compound totally inhibited this activity, showing that it is a highly potent HSL inhibitor (Fig. 1D). Using triolein as a substrate in the presence and absence of Bay demonstrates that HSL accounts for 85% of total TG hydrolase activity, whereas the remaining 15% is attributed to ATGL, the predominant non-HSL lipase represented in the cytosolic fraction (Fig. 1D). The respective contributions of HSL and ATGL to TG hydrolase activity in hMADS cells are comparable with what was previously observed in other human models such as primary culture of preadipocytes and adipose tissue (22). In vitro activation of triolein hydrolase activity by protein kinase A is attributed to HSL (33). Treatment of cells with the adenylate cyclase activator FK, alone or in combination with the HSL inhibitor Bay, illustrates that this was also the case for hMADS adipocytes (Fig. 1E). These data support the use of hMADS cells for studies of human fat cell metabolism.
FIGURE 1.
hMADS cells as a model to study human adipocyte metabolism. A, representative micrographs of hMADS cells at days 0, 7, and 13 of differentiation. B, gene expression of fatty acid metabolism genes throughout hMADS differentiation, expressed as fold induction from day 0. C, gene expression of differentiation markers in hMADS and primary adipocytes at day 13 of differentiation, expressed as 2ΔCt. GyK, glycerol kinase. D, cholesterol esterase and triolein hydrolase activity of hMADS cells in the presence and absence of Bay (10 μm) during the assay. E, triolein hydrolysis activity of hMADS cells pretreated with Bay (10 μm) and/or FK (1 μm). The data are presented as the means ± S.D. (n = 4). For one-way ANOVA, *, p < 0.05, significantly different from day 0 or basal condition.
Whole Cell Lipolysis and Re- esterification
To further evaluate TG breakdown in adipocytes, we next measured basal and FK-stimulated lipolysis in hMADS adipocytes. The robust release of NEFA and glycerol by FK treatment was totally abrogated by the addition of Bay, suggesting a strict HSL dependence of the process (Fig. 2A). However, the NEFA to glycerol ratio under basal conditions was close to 1, indicating an important re-esterification of FA during lipolysis. In an attempt to circumvent re-esterification, we measured lipolysis in the presence of acyl-CoA synthase inhibitor, Triacsin C. It involved an overnight loading period of cells with [3H-9,10]OA bound to BSA, followed 16 h later by a washing period to eliminate nonincorporated [3H-9,10]OA. Lipolysis was initiated with fresh medium in the presence of Triacsin C and drugs as indicated. Because [3H-9,10]OA and glycerol release are not directly comparable, the results were expressed as fold induction over untreated cells (Fig. 2B). The similar stimulation observed for [3H-9,10]OA and glycerol release during basal and FK-stimulated lipolysis shows that the re-esterification process was fully abolished. Subsequent experiments were therefore performed by measuring [3H-9,10]OA release in the presence of Triacsin C to evaluate complete FA release without esterification as a confounding variable. This approach revealed an HSL-independent FK-stimulated lipolysis that was previously masked by FA re-esterification (Fig. 2, A versus B). This lipolytic event was likely the result of ATGL activation. Clearly, important re-esterification fluxes take place in hMADS adipocytes, a phenomenon also observed in primary cultures of adipocytes. The re-esterification to lipolysis ratio was 0.86 ± 0.02 and was tightly coupled over a wide range of lipolytic fluxes (Fig. 2C). FK treatment successfully uncoupled re-esterification from lipolytic flux, whereas Bay addition restored coupling (Fig. 2D). Altogether, these data suggest a more important re-esterification of DG than monoglycerides in hMADS adipocytes.
FIGURE 2.
Lipolysis and re-esterification fluxes in hMADS adipocytes. A, release of lipolytic products NEFA and glycerol with/without Bay (10 μm) and/or FK (1 μm). B, [3H-9,10]OA and glycerol release in the presence of Triacsin C (10 μm) with/without Bay (10 μm) and/or FK (1 μm). C, correlation of [3H-9,10]OA release and re-esterification fluxes. Re-esterification was calculated as follows: [([3H-9,10]OA release with Triacsin C - [3H-9,10]OA release without Triacsin C)/[3H-9,10]OA release with Triascin C]. D, [3H-9,10]OA release and re-esterification fluxes of hMADS with/without Bay (10 μm) and/or FK (1 μm). All of the lipolytic and re-esterification fluxes were measured over 3 h. The data are presented as the means ± S.E. (n = 4–5). For one-way ANOVA, * or §, p < 0.05, significantly different from own basal; for two-way ANOVA, #, p < 0.05, significantly different from basal condition.
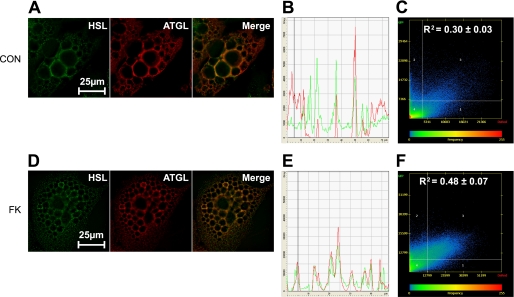
HSL and ATGL Colocalization following FK Stimulation
We investigated intracellular localization of HSL and ATGL in adipocytes in basal and FK-stimulated states. In the basal state, HSL was mainly localized to the cytosol, whereas ATGL was localized both in the cytoplasm and around big central LD (Fig. 3, A and B). Some colocalization of HSL and ATGL was observed in the basal state (Fig. 3C). After stimulation with FK, fragmentation of LD was observed. HSL and ATGL were translocated to small peripheral LD (Fig. 3, D and E). Although staining profiles for HSL and ATGL differed to some extent prior to FK stimulation, they matched each other closely after stimulation. Consequently, the colocalization coefficient increased by 60% (Fig. 3F). The staining pattern of ATGL following FK stimulation suggests a translocation to LD and concomitant colocalization with HSL and therefore a potential implication of ATGL in stimulated lipolysis of hMADS adipocytes.
FIGURE 3.
Intracellular localization of HSL and ATGL before and after FK stimulation. Control (CON) and FK-treated cells were stained for ATGL and HSL and microscopy images were analyzed as described under “Experimental Procedures.” A and D, immunofluorescence images of one representative optical section after deconvolution in control and FK-treated cells. B and E, distribution of the staining across the cell for one representative optical section in control and FK-treated cells. C and F, colocalization scatterplots evaluated by Pearson colocalization coefficients as means from at least 150 optical sections from five different cells in control (0.30 ± 0.03) and FK-treated (0.48 ± 0.07) cells. Colocalization coefficients for control and treated cells differed significantly (p < 0.0001).
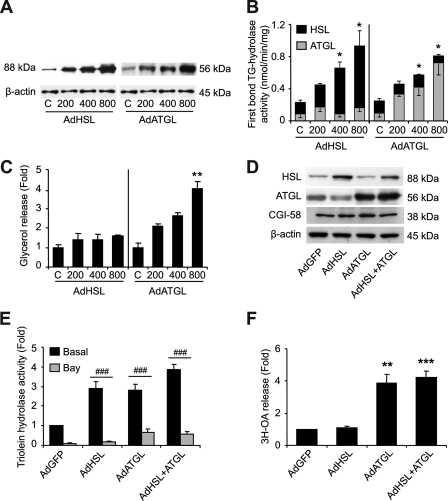
Increased ATGL Content Modulates Basal Rates of Lipolysis in Fat Cells
We altered HSL and ATGL balance in hMADS adipocytes to gain insight into their respective contributions to basal TG hydrolase activity and whole cell lipolysis. We first increased HSL and ATGL expression and content independently by adenoviral transductions using increasing m.o.i. (Fig. 4A). Triolein hydrolysis rates were determined in the absence or presence of Bay. From these data, we calculated the capacity of each lipase to hydrolyze a single ester bond. These computations revealed that the first ester bond TG hydrolase activity increased in proportion to the content of HSL or ATGL (Fig. 4B). Because a greater fraction of ATGL was found on LD than HSL (17 ± 2% versus 9 ± 1%, p < 0.05) under the control and adenoviral transduction conditions (supplemental Fig. S1), ATGL activity was corrected to account for this difference. However, changes in TG hydrolase activity did not translate into alteration of whole cell lipolysis, because basal lipolysis was accentuated with ATGL overexpression, but not HSL (Fig. 4C). These results suggest that ATGL and HSL action in whole cell lipolysis is sequential despite the high in vitro capacity of HSL to initiate TG degradation. We next compared single (HSL or ATGL) with dual lipase (HSL and ATGL) overexpression at a given m.o.i. HSL and ATGL protein content were equally increased in single and dual lipase overexpression (Fig. 4D). As expected, HSL overexpression increased cytosolic Bay-inhibited TG hydrolase activity (Fig. 4E). ATGL overexpression enhanced cytosolic ATGL hydrolase activity but also potentiated HSL hydrolase activity when compared with AdGFP, independently of HSL content. These results support the notion that ATGL content is limiting HSL action. Single ATGL and dual lipase overexpression showed that basal fat cell lipolysis is dependent upon ATGL but not HSL content (Fig. 4F).
FIGURE 4.
Assessment of HSL and ATGL overexpression in hMADS adipocytes. hMADS cells infected with adenoviruses containing the GFP gene alone or in tandem with human HSL and/or ATGL cDNA. m.o.i.s of HSL or ATGL ranged from 200 to 800 but were supplemented with GFP alone for a total m.o.i. of 800 in all conditions. A, Western blots of HSL and ATGL normalized to β-actin. B, theoretical HSL and ATGL first bond TG hydrolase activity of hMADS cells. Assuming that all three ester bonds of TG substrate (triolein) are cleaved during the in vitro assay (50), the activity of HSL was calculated as follows: [(TG hydrolase activity without Bay − TG hydrolase activity with Bay)/3]. The activity of ATGL is calculated from TG hydrolase activity in the presence of Bay. C, glycerol release. D, Western blots of HSL, ATGL, and CGI-58 in cytosolic fractions of hMADS adipocytes normalized to β-actin for single and dual adenoviral transductions of lipases at 400 m.o.i. E, cytosolic triolein hydrolase activity in the presence and absence of Bay (10 μm) during assays following single and double adenoviral transductions of lipases at 400 m.o.i. F, basal [3H-9,10]OA release with Triascin C (10 μm) following single and dual adenoviral transductions of lipases at 400 m.o.i. The data are presented as the means ± S.E. The experiments were performed in duplicate. For one-way ANOVA (n = 3), *, p < 0.05, significantly different from own control or AdGFP; for two-way ANOVA (n = 6), #, p < 0.05, significantly different from AdGFP condition.
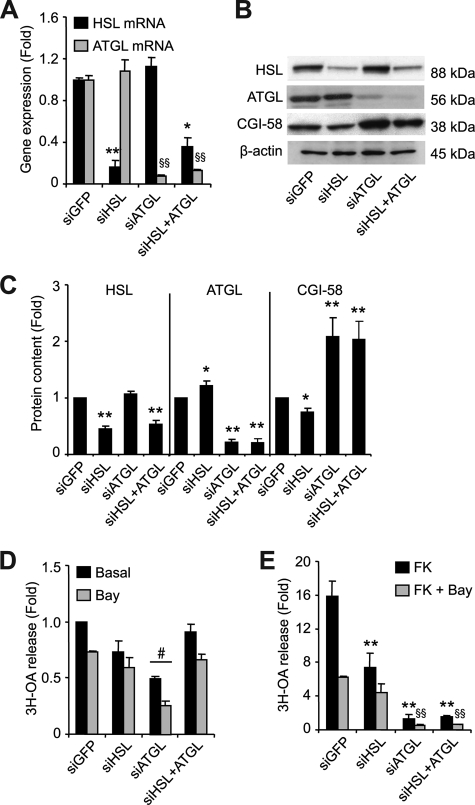
Gene Silencing of Lipases Reveals an Important Role for ATGL in Stimulated Lipolysis
We next reduced lipase expression using gene silencing. HSL and ATGL gene expression were reduced by 80–90% (Fig. 5A). Silencing of the lipases did not affect differentiation, as assessed by PPARγ gene expression (data not shown). Western blot analysis clearly shows reduced HSL and ATGL protein content following their respective targeted silencing. Gene silencing led to two interesting observations. First, ATGL protein content increased slightly but significantly (22%, p < 0.05) with HSL silencing (Fig. 5, B and C). Second, CGI-58 protein content increased by over 100% (p < 0.01) with ATGL silencing (Fig. 5, B and C). These adaptations were observed in both the cytosolic and fat cake fractions and did not involve cellular redistribution (supplemental Fig. S2). HSL TG hydrolase activity was reduced by 60%, whereas ATGL TG hydrolase activity was reduced by 50% (data not shown). The efficient silencing of the lipases prompted us to analyze lipolysis. Basal lipolysis was reduced in cells with decreased ATGL expression (Fig. 5D). Surprisingly, dual silencing of HSL and ATGL did not significantly alter basal lipolysis. FK-stimulated lipolysis was, however, severely reduced. Diminution of HSL expression resulted in a 53% decrease, whereas that of ATGL was more drastic and resulted in a 92% decrease in FK-stimulated lipolysis (Fig. 5E). These findings emphasize the rate-limiting nature of ATGL action not only in basal but also in FK-stimulated fat cell lipolysis.
FIGURE 5.
Assessment of HSL and ATGL gene silencing in hMADS adipocytes. Control (siGFP) or gene-specific small interfering RNAs for HSL and/or ATGL were delivered into suspended adipocytes with a microporator and reseeded in DM. A, gene expression of HSL and ATGL expressed as fold induction over siGFP. B, Western blots of HSL, ATGL, and CGI-58 in cytosolic fractions of hMADS cells. C, densitometry analysis of Western blots normalized to β-actin in cytosolic fractions (n = 10). D, basal [3H-9,10]OA release with Triascin C (10 μm) in the presence and absence of Bay (10 μm). E, FK-stimulated [3H-9,10]OA release with Triacsin C (10 μm) in the presence and absence of Bay (10 μm) expressed as fold induction over basal levels. The data are presented as the means ± S.E. The experiments were performed in duplicate. Unless stated otherwise, for one-way ANOVA (n = 3), * or §, p < 0.05, significantly different from corresponding basal; for two-way ANOVA (n = 6), #, p < 0.05, significantly different from siGFP condition.
Importance of CGI-58 in the Regulation of ATGL for Basal and FK-stimulated Lipolysis
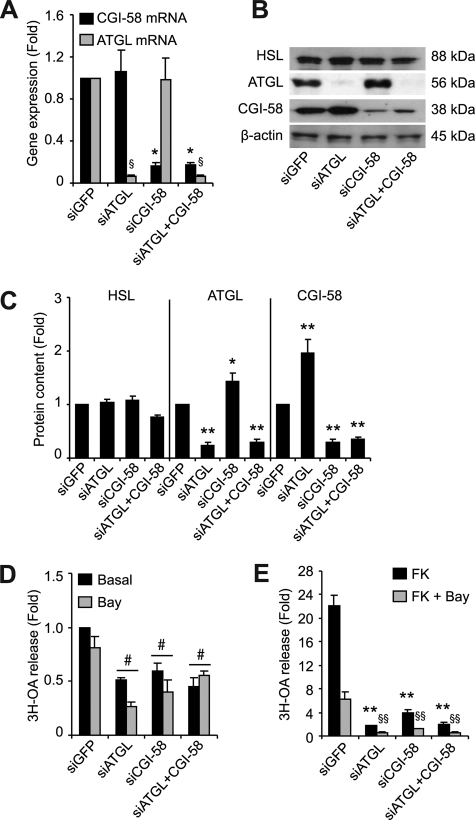
To further elucidate the role and regulation of ATGL in basal and FK-stimulated lipolysis, we reduced the expression of its coactivator CGI-58. Gene silencing of CGI-58 alone or with ATGL was equally efficient in reducing CGI-58 expression (Fig. 6A) and did not influence HSL or PPARγ expression (data not shown). Western blot analyses demonstrate a clear reduction of ATGL or CGI-58 protein levels following their respective gene silencing (Fig. 6B). As previously noted in Fig. 5B, ATGL silencing more than doubled CGI-58 protein content in the cytosolic (Fig. 6, B and C) and fat cake fraction (supplemental Fig. S2) of hMADS adipocytes. Similarly, ATGL protein content increased by almost 50% with silencing of CGI-58 (Fig. 6, B and C, and supplemental Fig. S2). TG hydrolase activity was unchanged by the absence of CGI-58 (data not shown). However, basal and FK-stimulated lipolysis were reduced by 40 and 82%, respectively (Fig. 6, D and E), revealing the importance of CGI-58 in the control of lipolysis. Dual silencing of ATGL and CGI-58 did not further reduce lipolysis.
FIGURE 6.
Assessment of CGI-58 gene silencing in hMADS adipocytes. Control (siGFP) or gene-specific small interfering RNAs for ATGL and/or CGI-58 were delivered into suspended adipocytes with a microporator and reseeded in DM. Gene expression and functional measurements were assessed on day 13, 6 days post-microporation. A, gene expression of ATGL and CGI-58 expressed as fold induction over siGFP. B, Western blot analyses in cytosolic fractions of hMADS cells. C, densitometry analysis of Western blots normalized to β-actin in cytosolic fractions (n = 10). D, basal [3H-9,10]OA release with Triascin C (10 μm) in the presence and absence of Bay (10 μm). E, FK-stimulated [3H-9,10]OA release with Triacsin C (10 μm) in the presence and absence of Bay (10 μm) expressed as fold induction over basal levels. The data are presented as the means ± S.E. The experiments were performed in duplicate. Unless stated otherwise, for one-way ANOVA (n = 3), * or §, p < 0.05, significantly different from corresponding siGFP; for two-way ANOVA (n = 6), #, p < 0.05, significantly different from siGFP condition.
DISCUSSION
Obesity is characterized by an expanded fat mass and significant FA release in the bloodstream. A defect in human adipose tissue lipolysis, which may be primary, has been shown in obesity (22, 23, 34). Moreover, the link between circulating FA and insulin resistance is well documented (35); therefore pharmacological tools targeting adipose tissue lipases in the treatment of insulin resistance are of interest (24). To gain insight into the molecular mechanisms governing adipose tissue lipolysis and the pathological and physiological implications of the lipases, we investigated the respective contributions of HSL, ATGL, and cofactor CGI-58 in basal and FK-stimulated human fat cell lipolysis. We used hMADS adipocytes, which originate from human white adipose tissue and have essential fat cell characteristics (25, 26). We further validated the use of this model by showing a concerted up-regulation of genes involved in FA metabolism throughout differentiation as well as similarities to differentiated preadipocytes (22). The hMADS adipocyte cell line is a valuable tool for studying human fat cell lipolysis, particularly in light of differences between human and rodent adipose tissue lipolysis (5). It complements recent developments in lipolysis that largely originate from rodent models or cell types other than fat cells (10, 16, 36–39).
Re-esterification of FA, estimated from the ratio of released NEFA and glycerol (40), is often overlooked in adipose tissue metabolism. This is a critical issue because the balance between lipolysis and re-esterification determines the net efflux of FA from the fat cells. In mature adipocytes, two of every three FAs are re-esterified under basal conditions (41, 42), but the implication of lipolytic flux or lipase content in this process is poorly examined. Re-esterification and lipolysis were tightly coupled in hMADS adipocytes and remained unperturbed by changes in lipases content. A transient reduction in DAG stores did, however, challenge re-esterification. In support of this hypothesis, it has been shown that newly synthesized DG are preferentially hydrolyzed during stimulated lipolysis (43). Conversely, it would be of interest to determine whether re-esterification is facilitated by a larger adipose tissue DG pool, which may temporarily be present in obesity as a result of impaired FK-stimulated lipolysis and HSL expression (22, 23).
The recent discovery of ATGL has led to a reassessment of the roles of lipases in fat cell lipolysis. Both HSL and ATGL have the in vitro capacity to cleave the first FA bond of a TG molecule, which raises questions about their respective roles in basal and FK-stimulated lipolysis in a whole adipocyte context. Our results demonstrate an HSL TG hydrolase capacity exceeding that of ATGL, as previously reported for human adipose tissue (14). However, whole cell basal lipolysis measurements clearly demonstrate the governing nature of ATGL. First, increased lipolysis in response to adenoviral transduction of ATGL indicates that, unlike HSL, ATGL regulates basal lipolysis independently of the perilipin barrier on LDs. A model detailing this interaction in murine adipocytes has previously been proposed (44, 45). Second, alteration of basal lipolysis with overexpression and silencing of ATGL, but not HSL, demonstrate the sequential action of these lipases. This is in line with decreased glycerol and FA release with ATGL knockdown in 3T3 L1 (46) and the lack of accentuated lipolysis in human HSL transgenic mice (47).
We previously advocated a prominent role for ATGL in basal lipolysis and for HSL in stimulated conditions (22). The combination of approaches and findings reported here indicate that ATGL is also governing FK-stimulated lipolysis. First, our immunocytochemistry results demonstrate the translocation of ATGL from the cytosol to smaller LD following FK stimulation. This increased its colocalization with HSL under stimulated conditions. These findings are in slight contrast with those from Granneman et al. (31), who showed ATGL translocation to LD in murine adipocytes but without alteration of HSL and ATGL colocalization. Thus, there may be some species differences in the intracellular trafficking of lipases during fat cell lipolysis. Second, although TG hydrolase activity in FK-stimulated conditions is clearly dependent upon HSL, measuring lipolysis in the absence of re-esterification revealed an important HSL-independent contribution to stimulated lipolysis. Lastly, abrogation of FK-stimulated lipolysis with ATGL or CGI-58 gene silencing confirmed ATGL as the rate-limiting enzyme in FK-stimulated lipolysis. Together, our results illustrate the importance of ATGL in DG provision to HSL in basal and FK-stimulated adipocyte lipolysis despite the capacity of HSL to cleave the first TG ester bond.
CGI-58 has recently been shown to exert lysophosphatidic acid acyltransferase activity (48). This characteristic may appear to conflict with its role as an activator of ATGL. However, both functions appear to be independent of each other because CGI-58 overexpression increases total phospholipids yet reduces TG levels. Nonetheless, gene silencing of CGI-58 evidenced the great dependence of ATGL on CGI-58 in basal and FK-stimulated human adipocyte lipolysis, despite the more modest activation of ATGL activity by CGI-58 in human compared with the mouse (Ref. 38 and data not shown). The importance of ATGL and CGI-58 in both basal and FK-stimulated lipolysis contrasts with two current models detailing the interaction between ATGL and its coactivator (44, 45). One model postulates that ATGL is not associated with CGI-58 under basal conditions but rather requires FK-dependent phosphorylation of perilipin for release and association of CGI-58 with ATGL. In another model, CGI-58 binds and activates ATGL on LD via interaction with perilipin A in the basal state but not following FK-dependent phosphorylation of perilipin. Our results in human adipocytes highlight species differences in whole cell dynamics of the lipolytic machinery. Other factors may be important for the control of lipolysis and lipase activity such as ADRP (adipose differentiation-related protein) and other members of the lipid-binding PAT (perilipin, adipophilin, Tip47 (tail-interacting protein 47)) protein family (39, 44, 49). The need for addressing their role in human fat cells is warranted.
Increased CGI-58 content with ATGL silencing is intriguing. Western blot analysis of CGI-58 in the fat cake fraction of adipocytes demonstrates that CGI-58 is not redistributed from LD to the cytosol following ATGL silencing. Stability of CGI-58 may be improved when interaction between CGI-58 and ATGL is reduced, as would be the case during ATGL gene silencing. Increased ATGL protein content as a result of CGI-58 silencing further supports this hypothesis.
In conclusion, despite the strong TG hydrolase activity of HSL toward TG in vitro, ATGL governs basal and FK-stimulated lipolysis in human fat cells. ATGL action is critical in providing DG to HSL, such that the latter can perform its unique action in DG degradation. ATGL acts independently of perilipin and requires CGI-58 for both basal and FK-stimulated lipolysis in human hMADS adipocytes. Our data therefore suggest that adipose tissue ATGL could be targeted, in addition to HSL, to reduce FA release as a strategy to combat the metabolic syndrome.
Supplementary Material
Acknowledgments
We are indebted to Elisabeth Vatan, Carine Valle, and Balbine Roussel (INSERM U858) for technical expertise. We are grateful to Jean José Maoret (Institut Fédératif de Recherche Biomédical de Toulouse, IFR150). We thank the Vector Core of the University Hospital of Nantes, supported by the Association Française contre les Myopathies for the production of adenovirus vectors.
This work was supported by INSERM, YSL BEAUTE/BRI, the Swedish Research Council, and the Commission of the European Communities Integrated Project HEPADIP Contract LSHM-CT-2005-018734 and Collaborative Project ADAPT Contract HEALTH-F2–2008-2011 00. This work was also supported by funds from the Natural Sciences and Engineering Research Council of Canada (to V. B.) and The Netherlands Organization for Scientific Research Grant NWO Rubicon 820.07.025 (to J. J.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1 and S2.
- FA
- fatty acid
- ATGL
- adipose triglyceride lipase
- Bay
- 4-isopropyl-3-methyl-2-[1-(3-(S)-methyl-piperidin-1-yl)-methanoyl]-2H-isoxazol-5-one
- DG
- diglyceride(s)
- FK
- forskolin
- GFP
- green fluorescent protein
- hMADS cells
- human adipose tissue-derived multipotent stem cells
- HSL
- hormone-sensitive lipase
- LD
- lipid droplet
- NEFA
- nonesterified fatty acid(s)
- TG
- triglyceride(s)
- DM
- differentiation medium
- BSA
- bovine serum albumin
- OA
- oleic acid
- ANOVA
- analysis of variance
- PPAR
- peroxisome proliferator-activated receptor
- m.o.i.
- multiplicity of infection.
REFERENCES
- 1.Belfrage P., Jergil B., Strålfors P., Tornqvist H. ( 1978) Adv. Exp. Med. Biol. 101, 113– 126 [DOI] [PubMed] [Google Scholar]
- 2.Haemmerle G., Zimmermann R., Hayn M., Theussl C., Waeg G., Wagner E., Sattler W., Magin T. M., Wagner E. F., Zechner R. ( 2002) J. Biol. Chem. 277, 4806– 4815 [DOI] [PubMed] [Google Scholar]
- 3.Zimmermann R., Strauss J. G., Haemmerle G., Schoiswohl G., Birner-Gruenberger R., Riederer M., Lass A., Neuberger G., Eisenhaber F., Hermetter A., Zechner R. ( 2004) Science 306, 1383– 1386 [DOI] [PubMed] [Google Scholar]
- 4.Fredrikson G., Tornqvist H., Belfrage P. ( 1986) Biochim. Biophys. Acta 876, 288– 293 [DOI] [PubMed] [Google Scholar]
- 5.Langin D., Arner P. ( 2006) Trends Endocrinol. Metab. 17, 314– 320 [DOI] [PubMed] [Google Scholar]
- 6.Sengenes C., Bouloumie A., Hauner H., Berlan M., Busse R., Lafontan M., Galitzky J. ( 2003) J. Biol. Chem. 278, 48617– 48626 [DOI] [PubMed] [Google Scholar]
- 7.Egan J. J., Greenberg A. S., Chang M. K., Wek S. A., Moos M. C., Jr., Londos C. ( 1992) Proc. Natl. Acad. Sci. U.S.A. 89, 8537– 8541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Strålfors P., Belfrage P. ( 1985) FEBS Lett. 180, 280– 284 [DOI] [PubMed] [Google Scholar]
- 9.Miyoshi H., Perfield J. W., 2nd, Souza S. C., Shen W. J., Zhang H. H., Stancheva Z. S., Kraemer F. B., Obin M. S., Greenberg A. S. ( 2007) J. Biol. Chem. 282, 996– 1002 [DOI] [PubMed] [Google Scholar]
- 10.Miyoshi H., Souza S. C., Zhang H. H., Strissel K. J., Christoffolete M. A., Kovsan J., Rudich A., Kraemer F. B., Bianco A. C., Obin M. S., Greenberg A. S. ( 2006) J. Biol. Chem. 281, 15837– 15844 [DOI] [PubMed] [Google Scholar]
- 11.Osuga J., Ishibashi S., Oka T., Yagyu H., Tozawa R., Fujimoto A., Shionoiri F., Yahagi N., Kraemer F. B., Tsutsumi O., Yamada N. ( 2000) Proc. Natl. Acad. Sci. U.S.A. 97, 787– 792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jenkins C. M., Mancuso D. J., Yan W., Sims H. F., Gibson B., Gross R. W. ( 2004) J. Biol. Chem. 279, 48968– 48975 [DOI] [PubMed] [Google Scholar]
- 13.Villena J. A., Roy S., Sarkadi-Nagy E., Kim K. H., Sul H. S. ( 2004) J. Biol. Chem. 279, 47066– 47075 [DOI] [PubMed] [Google Scholar]
- 14.Mairal A., Langin D., Arner P., Hoffstedt J. ( 2006) Diabetologia 49, 1629– 1636 [DOI] [PubMed] [Google Scholar]
- 15.Lass A., Zimmermann R., Haemmerle G., Riederer M., Schoiswohl G., Schweiger M., Kienesberger P., Strauss J. G., Gorkiewicz G., Zechner R. ( 2006) Cell Metab. 3, 309– 319 [DOI] [PubMed] [Google Scholar]
- 16.Schweiger M., Schreiber R., Haemmerle G., Lass A., Fledelius C., Jacobsen P., Tornqvist H., Zechner R., Zimmermann R. ( 2006) J. Biol. Chem. 281, 40236– 40241 [DOI] [PubMed] [Google Scholar]
- 17.Bougnères P., Stunff C. L., Pecqueur C., Pinglier E., Adnot P., Ricquier D. ( 1997) J. Clin. Invest. 99, 2568– 2573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jensen M. D., Haymond M. W., Rizza R. A., Cryer P. E., Miles J. M. ( 1989) J. Clin. Invest. 83, 1168– 1173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jocken J. W., Goossens G. H., van Hees A. M., Frayn K. N., van Baak M., Stegen J., Pakbiers M. T., Saris W. H., Blaak E. E. ( 2008) Diabetologia 51, 320– 327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Berndt J., Kralisch S., Klöting N., Ruschke K., Kern M., Fasshauer M., Schön M. R., Stumvoll M., Blüher M. ( 2008) Exp. Clin. Endocrinol. Diabetes 116, 203– 210 [DOI] [PubMed] [Google Scholar]
- 21.Jocken J. W., Langin D., Smit E., Saris W. H., Valle C., Hul G. B., Holm C., Arner P., Blaak E. E. ( 2007) J. Clin. Endocrinol. Metab. 92, 2292– 2299 [DOI] [PubMed] [Google Scholar]
- 22.Langin D., Dicker A., Tavernier G., Hoffstedt J., Mairal A., Rydén M., Arner E., Sicard A., Jenkins C. M., Viguerie N., van Harmelen V., Gross R. W., Holm C., Arner P. ( 2005) Diabetes 54, 3190– 3197 [DOI] [PubMed] [Google Scholar]
- 23.Large V., Reynisdottir S., Langin D., Fredby K., Klannemark M., Holm C., Arner P. ( 1999) J. Lipid Res. 40, 2059– 2066 [PubMed] [Google Scholar]
- 24.Wang M., Fotsch C. ( 2006) Chem. Biol. 13, 1019– 1027 [DOI] [PubMed] [Google Scholar]
- 25.Rodriguez A. M., Elabd C., Delteil F., Astier J., Vernochet C., Saint-Marc P., Guesnet J., Guezennec A., Amri E. Z., Dani C., Ailhaud G. ( 2004) Biochem. Biophys. Res. Commun. 315, 255– 263 [DOI] [PubMed] [Google Scholar]
- 26.Rodriguez A. M., Pisani D., Dechesne C. A., Turc-Carel C., Kurzenne J. Y., Wdziekonski B., Villageois A., Bagnis C., Breittmayer J. P., Groux H., Ailhaud G., Dani C. ( 2005) J. Exp. Med. 201, 1397– 1405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lowe D. B., Magnuson S., Qi N., Campbell A. M., Cook J., Hong Z., Wang M., Rodriguez M., Achebe F., Kluender H., Wong W. C., Bullock W. H., Salhanick A. I., Witman-Jones T., Bowling M. E., Keiper C., Clairmont K. B. ( 2004) Bioorg. Med. Chem. Lett. 14, 3155– 3159 [DOI] [PubMed] [Google Scholar]
- 28.Tiraby C., Tavernier G., Lefort C., Larrouy D., Bouillaud F., Ricquier D., Langin D. ( 2003) J. Biol. Chem. 278, 33370– 33376 [DOI] [PubMed] [Google Scholar]
- 29.He T. C., Zhou S., da Costa L. T., Yu J., Kinzler K. W., Vogelstein B. ( 1998) Proc. Natl. Acad. Sci. U.S.A. 95, 2509– 2514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Holm C., Olivecrona G., Ottosson M. ( 2001) Assays of Lipolytic Enzymes in Methods in Molecular Biology ( Ailhaud G. ed) pp. 97– 119, Humana, Totowa, NJ: [DOI] [PubMed] [Google Scholar]
- 31.Granneman J. G., Moore H. P., Granneman R. L., Greenberg A. S., Obin M. S., Zhu Z. ( 2007) J. Biol. Chem. 282, 5726– 5735 [DOI] [PubMed] [Google Scholar]
- 32.Mazzucotelli A., Viguerie N., Tiraby C., Annicotte J. S., Mairal A., Klimcakova E., Lepin E., Delmar P., Dejean S., Tavernier G., Lefort C., Hidalgo J., Pineau T., Fajas L., Clément K., Langin D. ( 2007) Diabetes 56, 2467– 2475 [DOI] [PubMed] [Google Scholar]
- 33.Khoo J. C., Aquino A. A., Steinberg D. ( 1974) J. Clin. Invest. 53, 1124– 1131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hellström L., Langin D., Reynisdottir S., Dauzats M., Arner P. ( 1996) Diabetologia 39, 921– 928 [DOI] [PubMed] [Google Scholar]
- 35.Boden G. ( 1997) Diabetes 46, 3– 10 [PubMed] [Google Scholar]
- 36.Brasaemle D. L., Levin D. M., Adler-Wailes D. C., Londos C. ( 2000) Biochim. Biophys. Acta 1483, 251– 262 [DOI] [PubMed] [Google Scholar]
- 37.Miyoshi H., Perfield J. W., 2nd, Obin M. S., Greenberg A. S. ( 2008) J. Cell. Biochem. 105, 1430– 1436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schweiger M., Schoiswohl G., Lass A., Radner F. P., Haemmerle G., Malli R., Graier W., Cornaciu I., Oberer M., Salvayre R., Fischer J., Zechner R., Zimmermann R. ( 2008) J. Biol. Chem. 283, 17211– 17220 [DOI] [PubMed] [Google Scholar]
- 39.Yamaguchi T., Omatsu N., Morimoto E., Nakashima H., Ueno K., Tanaka T., Satouchi K., Hirose F., Osumi T. ( 2007) J. Lipid Res. 48, 1078– 1089 [DOI] [PubMed] [Google Scholar]
- 40.Vaughan M. ( 1962) J. Biol. Chem. 237, 3354– 3358 [PubMed] [Google Scholar]
- 41.Reshef L., Olswang Y., Cassuto H., Blum B., Croniger C. M., Kalhan S. C., Tilghman S. M., Hanson R. W. ( 2003) J. Biol. Chem. 278, 30413– 30416 [DOI] [PubMed] [Google Scholar]
- 42.Wang T., Zang Y., Ling W., Corkey B. E., Guo W. ( 2003) Obes. Res. 11, 880– 887 [DOI] [PubMed] [Google Scholar]
- 43.Edens N. K., Leibel R. L., Hirsch J. ( 1990) J. Lipid Res. 31, 1351– 1359 [PubMed] [Google Scholar]
- 44.Brasaemle D. L. ( 2007) J. Lipid Res. 48, 2547– 2559 [DOI] [PubMed] [Google Scholar]
- 45.Granneman J. G., Moore H. P. ( 2008) Trends Endocrinol. Metab. 19, 3– 9 [DOI] [PubMed] [Google Scholar]
- 46.Kershaw E. E., Hamm J. K., Verhagen L. A., Peroni O., Katic M., Flier J. S. ( 2006) Diabetes 55, 148– 157 [PMC free article] [PubMed] [Google Scholar]
- 47.Lucas S., Tavernier G., Tiraby C., Mairal A., Langin D. ( 2003) J. Lipid Res. 44, 154– 163 [DOI] [PubMed] [Google Scholar]
- 48.Ghosh A. K., Ramakrishnan G., Chandramohan C., Rajasekharan R. ( 2008) J. Biol. Chem. 283, 24525– 24533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Listenberger L. L., Ostermeyer-Fay A. G., Goldberg E. B., Brown W. J., Brown D. A. ( 2007) J. Lipid Res. 48, 2751– 2761 [DOI] [PubMed] [Google Scholar]
- 50.Raclot T., Holm C., Langin D. ( 2001) Biochim. Biophys. Acta 1532, 88– 96 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.