Abstract
Skin maintenance and healing after wounding requires complex epithelial-mesenchymal interactions purportedly mediated by growth factors and cytokines. We show here that, for wound healing, transforming growth factor-β-activated kinase 1 (TAK1) in keratinocytes activates von Hippel-Lindau tumor suppressor expression, which in turn represses the expression of platelet-derived growth factor-B (PDGF-B), integrin β1, and integrin β5 via inhibition of the Sp1-mediated signaling pathway in the keratinocytes. The reduced production of PDGF-B leads to a paracrine-decreased expression of hepatocyte growth factor in the underlying fibroblasts. This TAK1 regulation of the double paracrine PDGF/hepatocyte growth factor signaling can regulate keratinocyte cell proliferation and is required for proper wound healing. Strikingly, TAK1 deficiency enhances cell migration. TAK1-deficient keratinocytes displayed lamellipodia formation with distinct microspike protrusion, associated with an elevated expression of integrins β1 and β5 and sustained activation of cdc42, Rac1, and RhoA. Our findings provide evidence for a novel homeostatic control of keratinocyte proliferation and migration mediated via TAK1 regulation of von Hippel-Lindau tumor suppressor. Dysfunctional regulation of TAK1 may contribute to the pathology of non-healing chronic inflammatory wounds and psoriasis.
Wound healing is a highly dynamic process that involves complex interactions of extracellular matrix molecules, soluble mediators, various resident cells, and infiltrating leukocyte subtypes. The immediate goal in repair is to achieve tissue integrity and homeostasis. The healing process involves three phases that overlap in time and space, namely inflammation, re-epithelialization, and tissue remodeling. Re-epithelialization is accomplished by increased keratinocyte proliferation and guided migration of the keratinocytes over the granulation tissue. Such processes require ordered changes in keratinocyte behavior and phenotype, which are dictated by the interplay of keratinocytes with dermal fibroblasts, i.e. epithelial-mesenchymal communication. This complex interplay demands the integration of diverse signals through a network of soluble factors exerting autocrine and paracrine activity from the wound microenvironment, culminating in appropriate cellular responses (1, 2). Aberrations to this signaling network may impair or enhance cell migration and proliferation, leading to insufficient or excessive wound repair and life-threatening consequences such as tumor growth and metastasis. Therefore, to understand the effect of any molecule in normal cellular function, studies into its role in this signaling network and how they culminate to an appropriate cell response become fundamental and necessary.
Transforming growth factor-β (TGF-β)4-activated kinase 1 (TAK1) belongs to the MAPK kinase kinase family. This serine/threonine kinase is a key intermediate in inflammatory cytokines tumor necrosis factor-α (TNF-α) and interleukin 1 (IL-1) (3, 4) as well as TGF-β (5)-mediated signaling pathways. Activated TAK1 has the capacity to stimulate its downstream MAPK and NFκB-inducing kinase-IκB kinase cascades (6). The former activates c-Jun N-terminal kinase (JNK) and p38 MAPK while the latter activates NF-κB (3, 7, 8). A deficiency in TAK1 results in impaired TNF-α- and IL-1-stimulated JNK activity, p38 phosphorylation, and IκBα degradation (7, 9). Studies of keratinocyte-specific TAK1 knock-out (TAK1-KO) mice confirmed the role of TAK1 in skin inflammation. These TAK1-KO mice died by postnatal day 7 and developed intra-epidermal micro-abscesses (10, 11). The TAK1-KO mice displayed abnormal epidermis with impaired differentiation and increased cellular proliferation; however, no significant difference in proliferation index was observed in culture of these mutant keratinocytes in vitro. Nevertheless, the latter suggests a crucial role of the underlying dermis in mitigating some effects of epidermal TAK1. Although the role of TAK1 in inflammatory response is well established, the role of TAK1 and its mechanism of action in keratinocyte proliferation and migration remain unknown.
Herein, we show that the deficiency in TAK1 resulted in increased cell proliferation and migration. We provide evidence of a double paracrine mechanism that make a pivotal contribution to the enhanced cell proliferation in TAK1-deficient epidermis. This study also reveals a novel homeostatic role of TAK1 in controlling cell migration. These aberrant phenotypes, as a consequence of TAK1 deficiency, are mediated via the dysregulated expression of von Hippel-Lindau tumor suppressor.
EXPERIMENTAL PROCEDURES
Reagents and Antibodies
The reagents and antibodies were as follows: Anti-TAK1 and anti-pVHL for immunohistochemistry (Upstate Biotechnology); anti-TAK1 for immunoblot (Cell Signaling); anti-cdc42-, anti-Rac1, and anti-RhoA (Cytoskeleton), anti-cytokeratin 10 (CK10), involucrin, and Ki67 (Novacastra); neutralizing anti-PDGF-BB antibody (Peprotech); 4′,6-diamidino-2-phenylindole and Vectashield mounting medium (Vector Laboratories); AlexaFluor® 488 goat anti-mouse IgG (Molecular Probes, Invitrogen); anti-Sp1 antibody and goat anti-rabbit IgG horseradish peroxidase (Santa Cruz Biotechnology); Collagen Type I Rat Tail (BD Biosciences); human primary fibroblasts, keratinocytes, and culture medium (Cascade Biologics); transfection reagent FuGENE HD (Roche Applied Science); and double-promoter lentivirus-based siRNA vector and pFIV Packaging kit were from System Biosciences. All restriction enzymes and DNA/RNA-modifying enzymes were from Fermentas. Kinase inhibitors were from Merck. Otherwise, chemicals were from Sigma-Aldrich.
Lentivirus siRNA Constructs and Transduction
The sequences of TAK1 and control siRNA were shown in supplemental Table S1. The siRNAs were into the pFIV-Hl/U6 siRNA vector according to the manufacturer's protocol. Positive clones were identified by PCR and by DNA sequencing. Production of pseudoviral particles and transduction of cells were as described by the manufacturer. Following transduction, the cells were enriched with 350 ng/ml puromycin for a week.
Transient Transfection and Transactivation Assay
TAK1 cDNA was subcloned into pCMV5 mammalian expression vector (Stratagene). An ∼732-bp proximal promoter of the human VHL was PCR-amplified from human genomic DNA using Pfu polymerase (12). The resulting fragment was subcloned into the pGL-3 Basic luciferase reporter vector (Promega, Madison, WI). Site-directed mutagenesis was performed using QuikChange site-directed mutagenesis kit (Stratagene). The sequence of primers is available in supplemental Table S1. Keratinocytes were cotransfected with a luciferase reporter driven by the VHL promoter construct, cDNAs encoding for various kinases and pEF1-β-galactosidase as a control of transfection efficiency (13). The constitutively active (ca) protein kinase A, MEK1, and MEKK1 were from Clontech. The caMKK7 was from Cell BioLabs. The various caSEK1, caJNK, caSGK, and caIKK expression vectors were from E. Nishida (Kyoto University, Japan), R. J. Davis (University of Massachusetts Medical School, Worcester), D. J. Templeton (University of Virginia Medical School, Charlottesville), and D. V. Goeddel (Tularik, Inc.). TAK1 was as previously described (5, 14). After transfection, cells were cultured for 48 h prior to lysis. Luciferase activity was measured using the Promega luciferase assay on a Microbeta Trilux (PerkinElmer Life Sciences). β-Galactosidase activity was measured in the cell lysate by a standard assay using 2-nitrophenyl-β-d-galactopyranoside as a substrate.
Total RNA Isolation and Real-time qPCR
Total RNA was isolated from cells using an Aurum total RNA kit (Bio-Rad) following the supplier's protocol. Total RNA (2.5 μg) was reverse-transcribed with oligo(dT) primers using RevertAidTM H Minus M-MuLV. Real-time PCR was performed with KAPA SYBR Fast qPCR (KAPABiosystem). Melt curve analysis was included to assure that only one PCR product was formed. Expression was normalized to the control gene ribosomal L27, which did not change under any of the experimental conditions studied. The sequence of primers is available in supplemental Table S1. Interferon response detection was done as recommended by the manufacturer (System Biosciences).
Organotypic Skin Culture
Organotypic skin cultures (OTCs) were performed as previously described (15).
Immunofluorescence and Immunohistochemistry
OTCs were fixed with 4% paraformaldehyde in PBS for 2 h at room temperature, washed twice with PBS, and embedded in Tissue-Tek OCT compound medium (Sakura, Leica). Eight-micron cryosections were processed for immunofluorescence as described previously (16). Apoptotic cells were detected using the TUNEL assay according to the manufacturer's protocol (Roche Applied Science). As positive control for TUNEL assay, the section was pre-treated with DNase I. The slides were counterstained with 4′,6-diamidino-2-phenylindole and mounted for microscopic observation. Immunohistochemistry on the indicated wound section was performed with anti-TAK1 or anti-pVHL antibodies (Upstate Biotechnology). The signal was amplified using the ABC-peroxidase method (Vector Laboratories) and revealed using 3,3′-diaminobenzidine for TAK1 (brown) or in the presence of nickel for pVHL (dark blue). Images were taken using MIRAX MIDI (Carl Zeiss) with a Plan-Apochromatic 20×/0.8 objective and MIRAX Scan software.
In Situ Proximity Ligation Assay
KCTRL and KTAK1-B cells were subcultured onto glass chamber slides (Lab-Tek). The following day, the cells were fixed with methanol:acetone (1:1) at −20 °C for 10 min. The slides were washed twice with PBS. The slides were blocked with 1% bovine serum albumin in PBS for 2 h at room temperature and were incubated with primary rabbit anti-pVHL and mouse anti-Sp1 antibodies overnight at 4 °C. The slides were washed as above. DuolinkTM in situ PLA was performed as recommended by the manufacturer (OLink Biosciences). As negative control, no primary antibody was used. Images were taken using LSM510 META confocal laser scanning microscope (Carl Zeiss).
Protein Array Analysis
Phosphoprotein and growth factor array membranes were processed according to the manufacturer's protocol (RayBiotech). These arrays were used to compare conditioned media from 2-week-old OTCs reconstructed using KCTRL and KTAK1-B keratinocytes. Protein spots were detected by chemiluminescence. Signal intensities were quantified using ImageJ analysis software and were normalized with the mean intensity of the positive controls on each membrane.
ELISA and Integrin-mediated Cell Adhesion Assays
The concentration of growth factors was measured using sandwich ELISA (R&D Systems) according to the manufacturer's instruction. The integrin-mediated cell adhesion assay was performed as recommended by the manufacturer (Chemicon).
Immunoblot Analysis
Total protein was extracted from cells with ice-cold lysis buffer (20 mm Na2H2PO4, 250 mm NaCl, 1% Triton X-100, 0.1% SDS). Equal amounts of protein extracts were resolved by SDS-PAGE and electrotransferred onto polyvinylidene difluoride membranes. Membranes were processed according to standard procedure, and proteins were detected by chemiluminescence. A Coomassie Blue-stained membrane was used to check for equal loading and transfer.
GTPases Activation Assay for cdc42, Rac1, and RhoA
GTPases activation assays were carried out as previously described (17). Purified recombinant GST-p21 binding domain of PAK was used to measure active cdc42 and Rac1, whereas GST-Rho binding domain of rhotekin was use to measure active RhoA.
Protein-Protein and ChIP
In vivo protein-protein cross-link and chromatin immunoprecipitation (ChIP) were carried out as previously described except anti-pVHL and anti-Sp1 antibodies were used (18). Primers used for ChIPs are available in supplemental Table S1.
RESULTS
Elevated TAK1 Expression in Wound Epithelia
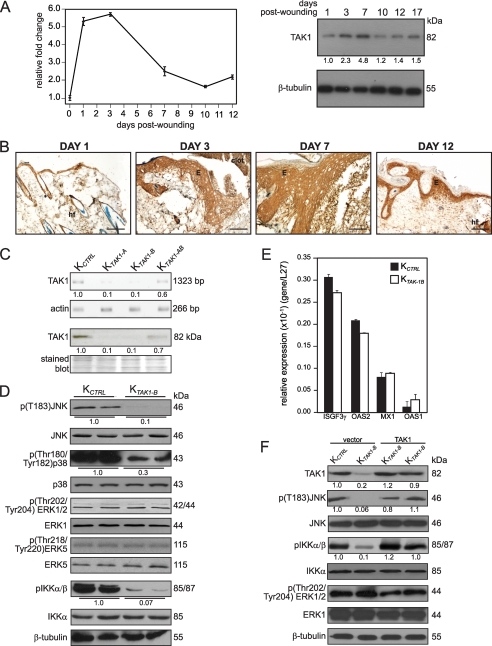
Immediately after injury, an acute inflammation response ensures immune competence and orchestrates later wound healing events including re-epithelialization. The major goal of re-epithelialization, of which cell migration and proliferation are dominant cellular events, is the reconstitution of the epithelium as a functional barrier. Although the role of TAK1 activation in inflammation is well established, little is known about its role in cell migration, which is crucial to the re-establishment of the wounded area. In the first instance, we examined the expression profile of TAK1 during the healing of mouse skin full-thickness excisional wound. We found that, during this healing, the expression profile of TAK1 mRNA and protein peaked at day 3–7 post-wounding, as shown by quantitative real-time PCR (qPCR) and immunoblot analysis (Fig. 1A). Immunohistochemistry staining with anti-TAK1 revealed that the expression of TAK1 was elevated in the wound epithelia (Fig. 1B), compared with only a basal level in unwounded skin. Immunostaining performed using pre-immune IgG did not display any staining (supplemental Fig. S1A).
FIGURE 1.
TAK1 expression is induced during skin wounding. A, TAK1 mRNA and protein expression profiles during wound healing determined using qPCR (left panel) and immunoblotting (right panel). Ribosomal protein L27 was used as a normalizing housekeeping gene. β-Tubulin showed equal loading and transfer. B, immunohistochemical analysis for TAK1 in skin wound biopsies. Mice skin wound biopsies at the indicated days of post-wounding were cryosectioned and stained with antibodies against TAK1. Representative pictures from wound epithelia were shown. E, epidermis; WB, wound bed; hf, hair follicle. Scale bar, 20 μm. C, lentivirus-mediated knockdown of TAK1 in keratinocytes. Human keratinocytes were transduced with a lentiviral vector harboring a control or with two different TAK1 (TAK1-A and TAK1-B) siRNAs. Expression of TAK1 in either control (KCTRL) or TAK1 siRNA (KTAK1-A or KTAK1-B) transduced keratinocytes was assessed by qPCR and immunoblotting. Values below each band represent the mean -fold differences in expression level with respect to control from five independent experiments (n = 5). Keratinocytes were transduced with TAK1-B siRNA (KTAK1-B) were used for subsequent studies. D, expression of phosphorylated and total JNK, p38, ERK1/2, ERK5, and IKKα/β in KCTRL and KTAK1-B. Reduced levels of p(Thr183)JNK, p(Thr180/Tyr182)p38, and pIKKα/β confirmed that endogenous TAK1 expression was suppressed. Values below each band represent the mean -fold differences in expression level with respect to KCTRL, which was assigned the value of one. E, expression level of 2′,5′-oligoadenylate synthetase isoforms 1 and 2 (OAS1 and OAS2), interferon-induced myxovirus resistance 1 (MX1), and interferon-stimulated transcription factor 3γ (ISGF3γ) mRNAs in KCTRL and KTAK1-B as determined by qPCR. Ribosomal protein L27 was used as a normalizing housekeeping gene. F, expression of phosphorylated and total JNK, IKKα/β, and ERK1/2 in KCTRL and KTAK1-B transfected with either expression vector or vector containing TAK1 cDNA. β-Tubulin showed equal loading and transfer. Values below each band represent the mean -fold differences in expression level with vector-transfected KCTRL, which was assigned the value of one.
To investigate whether the up-regulation of TAK1 was similarly observed in human, we performed a retrospective examination of human wounds. The TAK1 mRNA level was elevated in human ulceric skin biopsies compared with normal skin as determined by qPCR (supplemental Fig. S1B). These observations underscore an important role of TAK1 during re-epithelialization of rodents and human skin wounds.
TAK1 Deficiency Enhances Keratinocyte Migration
Cell migration over the provisional wound bed is essential for re-epithelialization of injured tissues. As expression of TAK1 was increased during re-epithelialization of the wound, we questioned whether TAK1 deficiency affects keratinocyte migration. Thus, we generated human keratinocytes whose endogenous TAK1 expression was suppressed by lentivirus-mediated RNA interference. The efficiency of siRNA knockdown of TAK1 expression in human keratinocytes was assessed by qPCR and immunoblot analysis. Keratinocytes transduced with either TAK1-A (KTAK1-A) or TAK1-B (KTAK1-B) sequences showed ∼90% reduction in the expression of TAK1 mRNA when compared with unrelated control siRNA (KCTRL) (Fig. 1C). Consistent with the mRNA expression, immunoblot analysis showed negligible level of TAK1 protein in both KTAK1-A and KTAK1-B (Fig. 1C). For subsequent analysis, KTAK1-B keratinocytes were used. The expression of phosphorylated JNK, p38, and IKKα/β were down-regulated in KTAK1-B cells, consistent with the role of TAK1 in the activation of these signaling pathways. There was no difference in phosphorylation of other MAPKs, such as ERK1/2 and ERK5 (Fig. 1D). The induction of interferon responses has been reported as a challenge to the specificity of some RNA interference approaches (19). To test whether gene silencing induced by the lentiviral-mediated RNA interference was associated with interferon-response induction, we measured the expression of some key interferon response genes by qPCR and the ability to rescue the KTAK1-B phenotype by the re-introduction of an expression vector harboring TAK1 cDNA that has a silent third-codon point mutation within the target region. No induction of 2′,5′-oligoadenylate synthetase isoforms 1 and 2, interferon-induced myxovirus resistance 1, and interferon-stimulated transcription factor 3γ was detected in the KTAK1-B when compared with either wild-type untransduced cells or KCTRL (Fig. 1E). Similarly, the re-introduction of TAK1 into KTAK1-B (TAK1-transfected KTAK1-B) restored the expression of phosphorylated JNK and IKKα/β but had no effect on ERK1/2 (Fig. 1F). Our results showed that gene silencing was not associated with nonspecific interferon-response induction, i.e. off-target effect, and that the deficiency of TAK1 attenuated the activation of JNK, p38, and IKKα/β.
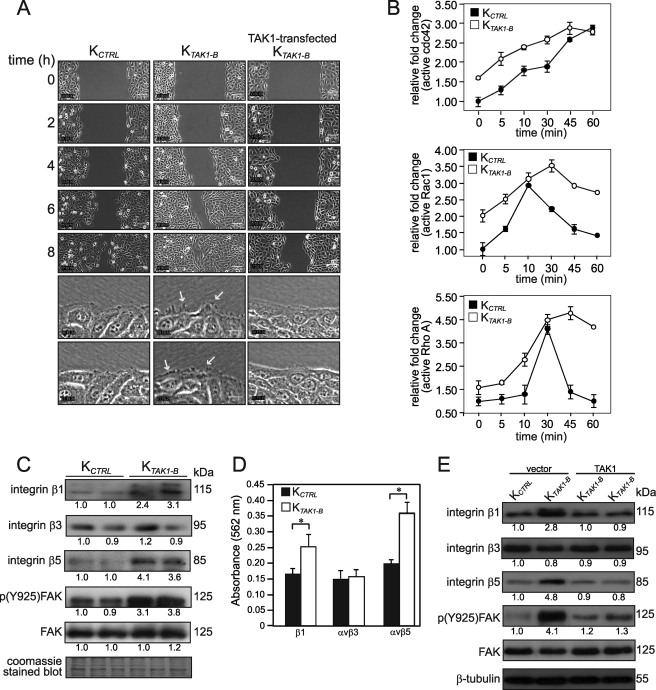
Next, we investigate the effect of TAK1 deficiency in keratinocyte migration using in vitro scratch wound assay. Interestingly, our live-imaging results showed that KTAK1-B closed the in vitro wound 3–4 h ahead of KCTRL (Fig. 2A and supplemental videos S1 and S2). Cell proliferation and migration contributed to the closure of the in vitro wound. To more precisely determine the contribution of cell migration toward wound closure, we performed similar experiments in the presence of mitomycin C. Consistent with the above observation, KTAK1-B closed the in vitro wound by 10 h in the presence of mitomycin C, a 2-h delay as compared with vehicle-treated KTAK1-B. In contrast, KCTRL failed to close the wound even by the end of the 12-h experimental period (supplemental Fig. S2 and videos S3 and S4). Close examination of the migratory front revealed that KTAK1-B displayed more extended lamellipodia with numerous microspike-like features when compared with KCTRL (Fig. 2A and supplemental videos S5 and S6). TAK1-transfected KTAK1-B cells have a delayed migration when compared with KTAK1-B and were similar to KCTRL (Fig. 2A and supplemental video S7).
FIGURE 2.
TAK1 deficiency enhances cell migration. A, time-lapsed images of wounded cultures of control (KCTRL), TAK1-knockdown (KTAK1-B), and TAK1-transfected KTAK1-B (top five panels). Scale bar, 100 μm. See supplemental videos S1, S2, and S7, respectively. Higher magnification images from video microscopy showing the migratory front of KCTRL, KTAK1-B, and TAK1-transfected KTAK1-B (bottom two panels). Arrows indicate the focal adhesion points and microspike-like extensions in lamellipodia during migration. Scale bar, 20 μm. Also see supplemental videos S5 and S6. B, graphs showed relative -fold change of active cdc42 (top), Rac1 (middle), and RhoA (bottom) in KCTRL and KTAK1-B exposed for the indicated time periods (minutes) with serum. Values represent the mean -fold change in active cdc42, Rac1, and RhoA relative to level at zero min of KCTRL (mean ± S.D.; n = 5). C, TAK1 modulates the expression of integrins and activation of focal adhesion kinase (FAK). Immunoblot analyses of integrins β1, β3, and β5, total FAK, and phosphorylated FAK p(Y925)FAK in KCTRL and KTAK1-B. Coomassie Blue-stained blot showed equal loading and transfer. D, integrin-mediated cell adhesion assay showed elevated expression of integrins β1 and avβ5 in TAK1-deficient keratinocytes. Statistical analysis was determined using two-tailed Mann-Whitney test; *, p < 0.01. E, re-introduction of TAK1 into KTAK1-B attenuated the expression of integrins and phosphorylation of FAK. Immunoblot analyses of integrins β1, β3, and β5, total FAK, and phosphorylated FAK p(Y925)FAK in vector- or TAK1-transfected KCTRL and KTAK1-B transfected with either expression vector or vector harboring TAK1 cDNA. β-Tubulin showed equal loading and transfer.
Our earlier results demonstrated that TAK1-deficient cells migrated faster than the KCTRL. The small Rho GTPases, cdc42, Rac1, and RhoA, are pivotal intracellular mediators crucial for the formation of lamellipodia, cytoskeleton network, and cell migration. Rac1 plays a role in the protrusion of lamellipodia and in forward movement, whereas cdc42 maintains cell polarity, including lamellipodia activity at the leading wound edge (20). To gain further insight into the mechanism, we examined the activation of small Rho-GTPases, specifically cdc42, Rac1, and RhoA in KCTRL and KTAK1-B cells stimulated with serum. Consistent with the enhanced migration of KTAK1-B, an elevated and sustained activation of cdc42, Rac1, and RhoA was observed (Fig. 2B and supplemental Fig. S3). KCTRL responded with a reduced activation of the three Rho GTPases. Notably, an elevated basal expression of active cdc42, Rac1, and RhoA was observed in KTAK1-B when compared with KCTRL before serum stimulation, thus we hypothesized that TAK1 likely exerted its action upstream of Rho GTPases activation. Integrin ligation and clustering triggers the subsequent activation of the GTPases: cdc42, Rac1, and RhoA. Immunoblot analysis revealed higher expression level of integrins β1 and β5, but not β3, in KTAK1-B when compared with KCTRL (Fig. 2C). Consistent with increased integrin ligation, we also detected enhanced activation, i.e. phosphorylation of focal adhesion kinase (FAK) in KTAK1-B (Fig. 2C). The observation was further confirmed using integrin-mediated adhesion assay, which showed significant increase in integrins β1 and β5 expression in KTAK1-B (Fig. 2D). As expected, the expression of integrins β1 and β5 and phosphorylated FAK in TAK1-transfected KTAK1-B returned to the similar level observed in KCTRL (Fig. 2E). Altogether, our results showed that the deficiency in TAK1 enhances keratinocyte migration associated with an elevated expression of integrins β1 and β5 and exacerbated activation of cdc42, Rac1, and RhoA.
TAK1-deficient Epidermis Showed Impaired Differentiation and Increased Proliferation
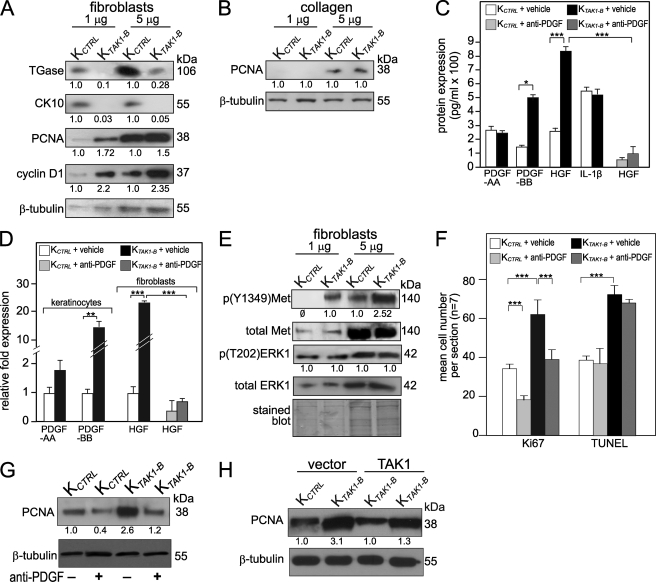
Cell proliferation also plays important role during re-epithelialization of the wound. The epidermis of TAK1-KO mice displayed increased cell proliferation, but no significant difference was observed in in vitro culture of these mutant keratinocytes, thus suggesting the involvement of epithelial-mesenchymal communications. To understand the role of TAK1 in cell proliferation, we constructed and examined OTCs using either KCTRL or KTAK1-B keratinocytes with underlying human primary dermal fibroblasts. Our results showed that the overall epidermal thickness between KCTRL and KTAK1-B OTCs was similar, however, the suprabasal layer of the KTAK1-B epidermis was significantly thinner and appeared as discontinuous patches when compared with KCTRL OTC (KTAK1-B versus KCTRL: 38.3 ± 0.7 μm versus 101.7 ± 0.4 μm) (supplemental Fig. S4). Immunofluorescence performed using differentiation markers, keratin 10 (CK10) and involucrin, showed that KTAK1-B OTCs had an impaired epidermal differentiation where the expression of differentiation markers were dysregulated when compared with KCTRL OTCs (supplemental Fig. S4). This observation was further confirmed by immunoblot analysis using CK10 and transglutaminase 1, a terminal differentiation marker (Fig. 3A). KTAK1-B OTCs showed more Ki67-positive proliferating cells (KTAK1-B versus KCTRL: 8.6 ± 2.8 versus 4.8 ± 1.2 labeled cells per microscopic field) as compared with the KCTRL OTCs (supplemental Fig. S4). These were further corroborated by immunoblotting with cyclin D1 and PCNA as proliferation markers (Fig. 3A). To gain further insight, we examined cell proliferation in OTCs constructed using either KTAK1-B or KCTRL keratinocytes with collagen, i.e. absence of underlying fibroblasts denoted as KTAK1-B/col and KCTRL/col, respectively (Fig. 3B). In contrast to KTAK1-B OTCs constructed with fibroblasts, no significant difference in cell proliferation was observed in KTAK1-B/col when compared with KCTRL/col, which was further confirmed by immunoblot analysis using anti-PCNA antibody (Fig. 3, compare A with B, and compare supplemental Fig. S4 with S5A). Our results point toward a homeostatic role of keratinocyte TAK1 that safeguard against epidermal hyperproliferation in a non autonomous manner.
FIGURE 3.
TAK1 regulates keratinocyte proliferation via a double paracrine mechanism. Immunoblot analysis of KCTRL and KTAK1-B epidermis from OTCs constructed either normal fibroblasts (A) or collagen only (B). 1 and 5 μg of protein lysates were used. Cytokeratin 10 (CK10) and transglutaminase I (TGase) were used a late and terminal differentiation makers, respectively. PCNA and cyclin D1 are proliferation markers; cleaved caspase 3 is an apoptotic marker. β-Tubulin served as a loading control. Values below indicate the mean relative -fold change with respect to KCTRL of three independent OTCs. Protein (C) and mRNA expression (D) levels of PDGF-AA and PDGF-BB, HGF, and IL-1β as determined by ELISA and qPCR, respectively. Conditioned serum-free OTC medium from KCTRL and KTAK1-B OTCs with fibroblasts, treated with either PBS (vehicle) or neutralizing anti-PDGF antibody, were used for ELISA measurement. qPCR was performed using RNA extracted from epidermis and dermal fibroblasts of indicated OTCs. Epidermis was physically separated from OTC after a 20-min treatment with Dispase. Fibroblasts embedded in collagen were isolated after collagenase treatment. PDGF and HGF mRNAs were not detected in fibroblasts and keratinocytes, respectively. Values are mean ± S.D. of three independent experiments (n = 3). Statistical analysis was determined using two-tailed Mann-Whitney test; *, p < 0.05; **, p < 0.01; and ***, p < 0.001. E, immunoblot analysis of KCTRL and KTAK1-B epidermis from OTCs constructed with underlying fibroblasts. The expression of activated, i.e. phosphorylated HGF receptor, Met p(Y1349)Met was increased in KTAK1-B epidermis. No difference was detected for total Met, total ERK1, and phosphorylated p(T202)ERK1. F, KTAK1-B OTCs with underlying fibroblasts were cultured in medium supplemented with anti-PDGF (200 nm) or vehicle (PBS). Mean numbers of proliferating and apoptotic cells were numerated after detection by anti-Ki67 antibody or TUNEL, respectively. Values were mean from five standardized microscopic fields per section, performed on seven sections from three independent OTCs. KCTRL OTCs with underlying fibroblasts served as control. Immunoblot analysis of epidermis from KCTRL and KTAK1 OTCs treated with either PBS (−) or anti-PDGF antibody (+) (G) or vector- and TAK1-transfected KCTRL and KTAK1-B OTCs (H). A cell proliferation marker proliferating cell nuclear antigen (PCNA) was used. β-Tubulin served as a loading control. Values below each band represent the mean -fold differences in expression level with KCTRL, which was assigned the value of one.
TAK1 Regulates Cell Proliferation via Double Paracrine Mechanism
Conceivably, this non-autonomous effect of TAK1 on cell proliferation is likely mediated by the changes in the production and secretion of mitogenic or anti-mitogenic factors by the fibroblasts upon stimulation by KTAK1-B-derived factors. To understand the underlying mechanism of TAK1 deficiency in cell proliferation, an unbiased protein antibody array was done. Inflammatory and growth factor arrays were used to compare conditioned media from OTCs composed of either KTAK1-B or KCTRL with normal primary fibroblasts. A total of 76 distinct proteins was screened, and the results showed changes in the protein expression level of platelet-derived growth factor (PDGF)-BB and hepatocyte growth factor (HGF), which were further confirmed by ELISA measurement of the OTC culture medium (Figs. 3C and S5B). No difference in PDGF-AA, IL-1β, and TGF-β1 were detected (Figs. 3C and S5B). Real-time PCR detected a 13.4-fold increase in the expression of PDGF-B mRNA in KTAK1-B when compared with KCTRL (Fig. 3D). PDGF mRNA was not detected in the fibroblasts. An elevated HGF mRNA was detected in the fibroblasts extracted from KTAK1-B OTCs and was undetectable in the keratinocytes (Fig. 3D). Notably, KTAK1-B OTCs treated with anti-PDGF-BB-neutralizing antibody showed reduced HGF production in the culture medium (Fig. 3C) and in the fibroblasts from KTAK1-B OTCs (Fig. 3D), indicating a PDGF/HGF epithelial-mesenchymal communication. To further verify the above observation, we examined the activation status of HGF receptor, Met, in the epidermal keratinocytes. Immunoblot showed an increase in phosphorylation of Met in KTAK1-B as compared with KCTRL (Fig. 3E). The increased proliferation seen in KTAK1-B OTCs was not an effect of enhanced ERK activity, which remained unchanged in comparison to KCTRL OTCs (Fig. 3E).
To underscore the importance of PDGF/HGF signaling in the increased epidermal proliferation observed in TAK1-deficient epidermis. We cultured KCTRL and KTAK1-B OTCs in medium supplemented with anti-PDGF antibody. We hypothesized that, if PDGF signaling played a major role in manifesting the phenotype of KTAK1-B OTCs, the effect can be neutralized with anti-PDGF antibody. Indeed, the addition of neutralizing anti-PDGF lowered the number of Ki67-positive proliferating cells in KCTRL and KTAK1-B OTCs when compared with vehicle-treated KCTRL OTC (Figs. 3F and S5C). This was again confirmed by immunoblot analysis using anti-PCNA antibody (Fig. 3G). Similarly, epidermis of TAK1-transfected KTAK1-B OTCs showed a reduced expression of PCNA when compared with vector-transfected KTAK1-B OTCs but was similar to KCTRL OTCs (Fig. 3H). Altogether, our results indicate that the double paracrine PDGF/HGF signaling plays a pivotal role in the action of TAK1 in cell proliferation.
TAK1 Suppresses Sp1-mediated Signaling via Increased Expression of von Hippel-Lindau Tumor Suppressor
Our results showed that the absence of TAK1 enhances cell migration and proliferation associated with concomitant increased expressions of integrins β1 and β5 and PDGF-B/HGF, respectively. Next, we sought to investigate the mechanism by which TAK1 negatively regulate their expressions.
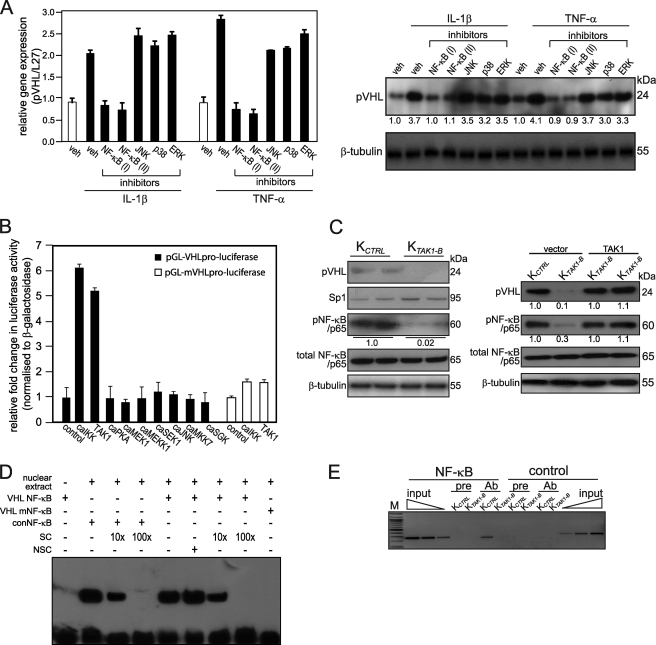
Earlier studies showed that von Hippel-Lindau tumor suppressor protein (pVHL)-Sp1 interaction suppresses PDGF-B expression (21). We hypothesize that TAK1 directly increases the expression of pVHL, which sequesters Sp1 and consequently repressed PDGF-B expression. We first examined the expression of pVHL mRNA and protein levels in KCTRL treated with either IL-1β or TNF-α in the presence of various kinase inhibitors. With the exception of two different NF-κB inhibitors, BAY 11–7082 and SN50, all other inhibitors did not attenuate IL-1β- or TNF-α-induced increase in pVHL mRNA and protein expression when compared with vehicle-treated KCTRL (Fig. 4A). Next, we studied the role of different kinases in regulating the promoter of the human VHL gene. The human VHL promoter was isolated by PCR, and a proximal 732-bp fragment containing the transcription initiation site was subcloned into a luciferase reporter gene (12). This VHL promoter reporter construct was co-transfected with expression vectors encoding constitutively active (ca) kinases into KCTRL. The activation of TAK1 stimulates the activity of downstream mediators, including IKKα/β, MKK7, and JNK (22, 23). Transient transfection studies showed that IKKα and TAK1 stimulated the VHL promoter activity (Fig. 4B). Additional experiments ruled out the possible involvement of MKK7, JNK, protein kinase A, MEK1, MEKK1, and SEK1 signals in regulating human VHL expression. The human VHL gene promoter contains a putative NF-κB binding site (12). Site-directed mutagenesis of the putative NF-κB binding site in the VHL promoter abolished the stimulating effect of IKK and TAK1 (Fig. 4B).
FIGURE 4.
TAK1 regulates pVHL gene expression via NF-κB. A, pVHL mRNA (left panel) and protein (right panel) expression in KCTRL treated for 24 h with either DMSO (vehicle) or specific inhibitors of the indicated kinases in the presence of with IL-1β or TNF-α. The various kinase inhibitors were NF-κB(I), BAY 11–7082; NF-κB(II), SN50; JNK, 1,9-pyrazoloanthrone; p38, [2-(4-chlorophenyl)-4-(4-fluorophenyl)-5-pyridin-4-yl-1,2-dihydropyrazol-3-one]; and ERK1/2, PD98059. Ribosomal protein L27 was used as a normalizing housekeeping gene. β-Tubulin showed equal loading and transfer. B, transactivation assay in keratinocytes co-transfected with a luciferase reporter gene driven by the human VHL promoter (pGL-VHLpro-luciferase), cDNA encoding for indicated constitutively active (ca) kinases and pEF1-β-galactosidase as control of transfection efficiency. Luciferase activity was measured, and normalized reporter activity is shown as -fold induction as compared with reporter construct transfected-KCTRL (control). VHL promoter reporter construct, whose NF-κB site was mutated, is denoted as pGL-mVHLpro-luciferase. Data are mean ± S.E., n = 3. C, immunoblot analysis of KCTRL and KTAK1-B epidermis (left panel) or vector- and TAK1-transfected KCTRL and KTAK1-B (right panel) using antibodies against pVHL, Sp1, total, and phosphorylated NF-κB(p65). β-Tubulin served as a loading control. Values below each band represent the mean -fold differences in expression level with respect to KCTRL from five independent experiments (n = 5). D, electrophoretic mobility shift assay of human VHL NF-κB binding site. Radiolabeled VHL NF-κB binding sequence was incubated with nuclear extract isolated from KCTRL. NSC denotes nonspecific competitor, a scrambled NF-κB binding sequence. SC denotes non-radiolabeled consensus NF-κB sequence (conNF-κB). As positive control, conNF-κB was used. Mutated VHL NF-κB site is denoted by VHL mNF-κB. E, pVHL is a direct NF-κB target gene. Chromatin immunoprecipitation were done in KCTRL and KTAK1-B using pre-immune IgG (pre) or antibody against p65 subunit of NF-κB (Ab). Promoter region with the NF-κB binding site was immunoprecipitated and specifically amplified in KCTRL using Ab. No amplified signal was obtained in KTAK1-B or using pre-immune IgG. A control region upstream of NF-κB binding site served as negative control. M, 100-bp DNA marker.
Next, we examined the protein expression level of pVHL, Sp1, and NF-κB/p65 in KTAK1-B and KCTRL by immunoblot analysis. In concordance with the role of TAK1 in NF-κB activation, the phosphorylation NF-κB(p65) was reduced in TAK1-deficient keratinocytes. Interestingly, pVHL was barely detectable in the KTAK1-B keratinocytes when compared with KCTRL (Fig. 4C). No difference in total NF-κB(p65) and Sp1 expression was observed (Fig. 4C). As expected, TAK1-transfected KTAK1-B showed increased expression of pVHL and phosphorylated NF-κB(p65) when compared with vector-transfected KTAK1-B (Fig. 4C).
To determine if human pVHL, is a target gene of TAK1/NF-κB, we performed electrophoretic mobility shift assay and ChIP. Specific protein-DNA complexes were detected in electrophoretic mobility shift assay for VHL NF-κB binding site, which was effectively competed by unlabeled consensus NF-κB but not by a nonspecific competitor oligonucleotide (Fig. 4D). Mutation to the VHL NF-κB binding sequence (VHL mNF-κB) eliminated its interaction with nuclear extract from KCTRL. As positive control, the labeled consensus NF-κB (conNF-κB) oligonucleotide was used as a probe, which was specifically competed by the consensus NF-κB (Fig. 4D). Similarly, ChIP done on KCTRL and KTAK1-B using anti-NF-κB(p65) antibody showed that NF-κB(p65) specifically bound to this site in KCTRL but not in KTAK1-B (Fig. 4E). No immunoprecipitation and amplification were seen with pre-immune IgG and with a control sequence upstream of the NF-κB site on the promoter gene of VHL (Fig. 4E). Taken together, we concluded that TAK1/IKK/NF-κB signaling is necessary for the induction of pVHL expression.
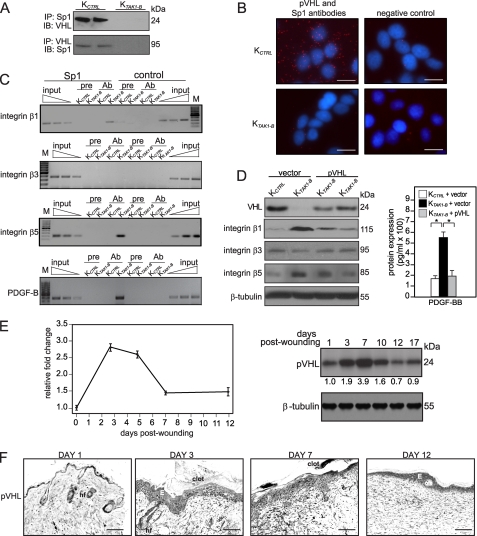
In silico analysis of the promoters of human PDGF-B and integrins β1, β3, and β5 genes revealed putative Sp1 binding elements (21, 24–26). In the first instance, we verified the interaction between pVHL and Sp1 in KTAK1-B and KCTRL by in vivo protein-protein cross-link as previously described (18). Our result showed that interaction between pVHL and Sp1 was only observed in KCTRL (Fig. 5A). To further strengthen this observation, in situ proximity ligation assay (PLA) was performed using anti-pVHL and anti-Sp1 antibodies. PLA allows for the detection and quantification of interacting proteins. Our results showed a significant number of interacting pVHL-Sp1 pairs in the cytoplasm of the KCTRL when compared with KTAK1-B (Fig. 5B).
FIGURE 5.
TAK1 suppresses Sp-1-mediated signaling via increased expression of pVHL. A, in vivo co-immunoprecipitation of pVHL and Sp1. Immunoprecipitation (IP) was performed using Sp1 antibody, followed by immunoblot (IB) analysis with pVHL antibody, and vice versa. B, detection of pVHL-Sp1 complex in KCTRL and KTAK1-B using DUOlinkTM in situ PLA with anti-pVHL and anti-Sp1 antibodies. PLA signals are shown in red and the nuclei in blue. The nuclei image has been acquired in one z-plane. Negative control without primary antibody. Scale bar, 50 μm. C, pVHL inhibits Sp1-mediated PDGF-B and integrin β1 and β5, but not β3, gene expression. Chromatin from KCTRL and KTAK1-B was immunoprecipitated with either a Sp1 antibody (Ab) or preimmume IgG (pre). Enrichment of a DNA fragment encompassing the Sp1 binding sites was evaluated by PCR. Aliquots of the extracts were also used before immunoprecipitation (input). No amplified signal was obtained in using pre-immune IgG. A control region upstream of NF-κB binding site served as negative control. D, ectopic expression of pVHL in KTAK1-B attenuates the expression of integrins β1 and β5 and PDGF-B. Immunoblot analyses (left panel) and ELISA for PDGF-BB in condition medium (right panel) of KCTRL and KTAK1-B cells transfected with either empty expression vector or vector harboring cDNA of pVHL. E, pVHL expression is induced during skin wounding. pVHL mRNA and protein expression profiles during wound healing were determined using qPCR (left panel) and immunoblotting (right panel). Ribosomal protein L27 was used as a normalizing housekeeping gene. β-Tubulin showed equal loading and transfer. F, immunohistochemical analysis for pVHL in skin wound biopsies. Mice skin wound biopsies at indicated days of post-wounding were cryosectioned and stained with antibodies against pVHL. 3,3′-Diaminobenzidine with nickel (dark blue) was used as substrate. Representative pictures from wound epithelia are shown. E, epidermis; WB, wound bed; hf, hair follicle. Scale bar, 20 μm.
Next, we performed ChIPs using anti-Sp1 antibody. Our results showed that Sp1 was bound to the Sp1 binding sites in integrins β1 and β5 and PDGF-B, but not integrin β3 (Fig. 5C). Conceivably, the reduced pVHL expression in KTAK1-B allowed free Sp1 to bind and transactivate these genes. To strengthen these results, we performed ChIPs using anti-Sp1 antibody of these promoters in KTAK1-B stably expressing pVHL (pVHL-transfected KTAK1-B). The expression of pVHL in transfected KTAK1-B was comparable to KCTRL as determined by immunoblot analysis (Fig. 5D). Consistent with the aforementioned results, these pVHL-transfected KTAK1-B cells expressed a reduced expression of integrins β1 and β5 and PDGF-BB (Fig. 5D). To further verify the results, we performed ChIP using anti-Sp1 antibody with these cells. The sequences spanning the Sp1 binding site were significantly reduced in the immunoprecipitates obtained from pVHL-transfected KTAK1-B when compared with vector-transfected KTAK1-B (supplemental Fig. S6). In addition, no signal was seen with pre-immune serum, and no binding was detected in the control sequence.
The cellular role of pVHL can be hypoxia-inducible factor (HIF)-dependent or -independent manner. In silico analysis of the promoters of human PDGF-B and integrin β1 and β5 genes failed to identify the putative HIFα binding site. Nonetheless, to eliminate this possibility, we first performed immunoblot analysis of HIFα using KCTRL and KTAK1-B epidermis. Our results showed no significant change in the expression of HIFα between KCTRL and KTAK1-B (supplemental Fig. S7A) Furthermore, cobalt chloride-simulated hypoxia condition, which increased HIFα expression, delayed KCTRL migration, suggesting that HIFα is not responsible for the observed phenotype of KTAK1-B (supplemental Fig. S7A, compare supplemental videos S1, S2, and S8).
Finally, we examined the expression profile of pVHL mRNA and protein during wound healing. We found that, during the healing of the mouse skin full-thickness excisional wound, the expression profile of pVHL mRNA was similar to that of TAK1, peaking at days 3–7 post-wounding, as shown by qPCR and immunoblot analysis (Fig. 5E). Immunohistochemical analysis revealed that pVHL was strongly expressed in the wound epithelia, similar to TAK1 (Fig. 5F). No primary anti-pVHL antibody served as negative control (supplemental Fig. S7B). Altogether, our results showed that TAK1/NF-κB directly up-regulates the expression of tumor suppressor pVHL, which represses PDGF-B and selected integrin gene expression via the Sp1 binding element in their cognate promoter during wound healing.
DISCUSSION
Following injury, the restoration of its functional integrity is of utmost importance to the survival of the organism. The regeneration and maintenance of epithelium to close the wound is dictated by epithelial-mesenchymal interactions and purportedly mediated by the action of central players, such as chemokines and growth factors. This communication is crucial for preventing either insufficient or excess wound repair. Previous work has shown that mice with a keratinocyte-specific deletion of TAK1 exhibit severe skin inflammation and display abnormal epidermis with impaired differentiation, increased cell proliferation, and apoptosis (10, 11). In this study, we reveal that keratinocyte-specific TAK1 regulates epidermal proliferation via a double paracrine mechanism and characterize a novel unsuspected role of TAK1 in cell migration. We show that pVHL, whose expression is up-regulated by TAK1/IKK/NF-κB signaling, interacts and sequesters transcription factor Sp1, which is necessary for PDGF-B and integrins β1 and β5 expression.
The formation of normal epidermal tissue requires a continuous exchange of signals with the underlying dermal fibroblasts (15, 28). Keratinocyte-specific TAK1-knock-out mice displayed enhanced epidermal proliferation, which was not observed in monolayer culture, suggesting that dermal fibroblasts have pivotal contribution. We reveal a double paracrine PDGF/HGF signaling that plays an important role in TAK1-mediated epidermal proliferation. For any cells to respond productively to PDGF, they must possess corresponding cell surface receptors, either the α-PDGF or β-PDGF receptors. Although keratinocytes contribute to PDGF activity by their ample capacity to secreted PDGF, they do not express any transmembrane receptor recognizing any classic PDGF isoforms; thus target cells for these ligands in skin are restricted to cells of mesenchymal origin, e.g. dermal fibroblasts (29, 30). We further show that NF-κB, a downstream mediator of TAK1, regulates the expression of pVHL. The pVHL protein interacts and sequesters transcription factor Sp1, which is required for PDGF-B expression (21). A similar mechanism was also exploited by pVHL to repress VEGF gene expression (31). PDGF-B was previously reported to increase HGF production in the fibroblasts to stimulate keratinocyte proliferation (32). Indeed, PDGF-BB secreted by keratinocytes acts as a paracrine factor to induce HGF production by fibroblasts, which in return, enhance epidermal proliferation (Fig. 6). This study underscores the fact that TAK1 in the epidermal keratinocytes fulfils a homeostatic role during epidermal formation, modulating cell proliferation via a double paracrine fashion, involving the underlying dermal fibroblasts. The paracrine mechanism described herein emphasizes the importance of epithelial-mesenchymal communication in the regulation of epidermal proliferation, which was not observed in TAK1-knock-out monolayer culture.
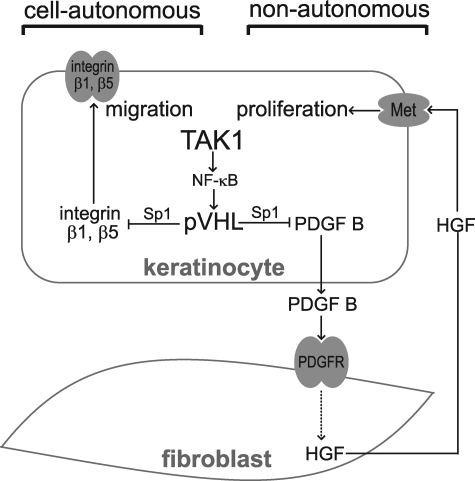
FIGURE 6.
TAK1 plays a homeostatic role in regulating cell migration and proliferation. Epidermal TAK1 regulates keratinocyte proliferation by a double paracrine mechanism through release of PGDF-B, which induces HGF in the fibroblasts. TAK1 activates NF-κB to stimulate the expression of pVHL. This facilitates pVHL-Sp1 interaction and sequesters Sp1 from promoting PDGF-B expression. In response to PDGF, the fibroblasts increase the release of HGF, which acts via its receptor, Met, to stimulate keratinocytes proliferation. Via a similar mechanism, TAK1 negatively regulates the expression of integrins β1 and β5 in a cell-autonomous manner.
Cell migration during re-epithelialization is equally crucial for efficient wound closure. In this study, we reveal a novel homeostatic role of TAK1 in the control of keratinocyte migration. TAK1-deficient keratinocytes migrate faster, associated with enhanced integrin β1 and β5 expression, activation of FAK, and small Rho GTPases when compared with its wild-type counterpart. The elevated active Rho GTPases expression profile supports a faster migration rate and persistent lamellipodia formation. The functions of pVHL can be mediated via HIFα-dependent or -independent mechanisms. Our findings suggested that hypoxia retarded keratinocyte migration. However, hypoxia-mediated migration is complex and context-dependent. It can be modulated by extracellular matrix, matricellular proteins, matrix metalloproteinases, and other signaling pathways (33). It can also differ among keratinocytes isolated from donors of different age (34). However, our findings suggest that the mechanism underlying the phenotype of TAK1-deficient keratinocytes is HIFα-independent. Interestingly, TAK1 employs similar regulatory mechanism, i.e. via pVHL to modulate integrin β1 and β5 expression. In support of a role in cell migration for pVHL, it was reported that highly aggressive breast cancer expressed either no pVHL or low pVHL level (35). High pVHL expression also resulted in a decrease of tubulin turnover indicating a role for pVHL in cellular processes such as migration, polarization, and cell-cell interactions (36).
The development, maintenance, and regeneration of the epidermal integrity are likely to involve the concerted effort of numerous signaling pathways, including epithelial-mesenchymal communications. One of such communications is the IL-1/keratinocyte growth factor/granulocyte macrophage-colony stimulating factor double paracrine mechanism (37). Given the pivotal role of TAK1 in propagating the effect of numerous inflammatory cytokines, such as TNF-α, the PDGF/HGF mechanism described herein complements the IL-1/keratinocyte growth factor/granulocyte macrophage-colony stimulating factor mechanism, particularly during wound repair. TAK1 may also modulate cell migration indirectly via HGF in vivo, when epithelia-mesenchymal interaction is evident. It was recently shown that HGF-mediated cell migration involves a PAK-LIMK pathway (38). It is conceivable that both mechanisms are utilized for epidermal regeneration during wound healing.
Psoriasis is a non-contagious chronic inflammatory skin disease characterized by hyperproliferative epidermal growth, a phenotype that was similarly observed in mice with a keratinocyte-specific deletion of TAK1 (10, 11). A recent report suggests that TAK1 deletion causes dysregulation of reactive oxygen species in keratinocytes, which is causally associated with skin inflammation (39). Reactive oxygen species has been associated with psoriasis (40, 41). Our study reveals the underlying mechanism by which TAK1 regulates epidermal proliferation via a double paracrine mechanism involving the underlying dermal fibroblasts. We showed that TAK1 in the keratinocytes directly up-regulated the expression of pVHL, reduced PDGF-B expression, and consequently diminished HGF/c-Met signaling. A deficiency in TAK1 resulted in reduced NF-κB activation and pVHL expression. Lending support, analysis of skin biopsies of patients with psoriasis showed that pVHL, a direct target of TAK1, is underexpressed in psoriatic skin and highly expressed in healthy skin (42). Furthermore, PDGF receptor expression was greatly elevated in psoriatic fibroblasts, rendering them highly responsive to PDGF and PDGF-induced HGF production (27). Altogether, our findings herein and those of others suggest that TAK1 deficiency may contribute to the etiology of psoriasis.
Supplementary Material
Acknowledgments
We thank Dr. Samuel Ko and Anna Teo (Carl Zeiss, Singapore Pte Ltd.) for their expertise in image acquisition using the MIRAX MIDI.
This work was supported in part by the Ministry of Education, Singapore (ARC 8/06) and Nanyang Technological University, Singapore (Grants RG 127/05 and 158/06).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S7, Table S1, and Videos S1–S8.
- TGF-β
- transforming growth factor-β
- ChIP
- chromatin immunoprecipitation
- CK10
- cytokeratin 10
- ELISA
- enzyme-linked immunosorbent assay
- HGF
- hepatocyte growth factor
- HIFα
- hypoxia-inducible factor α
- IKK
- IκB kinase
- IL-1
- interleukin 1
- JNK
- c-Jun N-terminal kinase
- siRNA
- small interference RNA
- KCTRL
- control siRNA keratinocytes
- KTAK1-B
- TAK1 siRNA keratinocytes
- KGF
- keratinocyte growth factor
- MAPK
- mitogen-activated protein kinase
- MEK1
- MAPK kinase 1
- MEKK1
- MAPK kinase kinase 1
- MKK7
- MAPK kinase 7
- OTC
- organotypic skin culture
- PDGF
- platelet-derived growth factor
- PLA
- proximity ligation assay
- pVHL
- von Hippel-Lindau tumor suppressor
- qPCR
- real-time quantitative PCR
- SEK1
- MAPK kinase 4
- SGK
- serum and glucocorticoid-inducible kinase
- TAK1
- TGF-β-activated kinase 1
- TNF-α
- tumor necrosis factor-α
- KO
- knock out
- ca
- constitutively active
- PBS
- phosphate-buffered saline
- TUNEL
- terminal deoxynucleotidyl transferase-mediated dUTP nick end labeling
- ERK
- extracellular signal-regulated kinase
- GST
- glutathione S-transferase.
REFERENCES
- 1.Werner S., Grose R. ( 2003) Physiol. Rev. 83, 835– 870 [DOI] [PubMed] [Google Scholar]
- 2.Fusenig N. E. ( 1994) in The Keratinocyte Handbook ( Leigh I., Lane B., Watt F. eds) pp. 71– 94, Cambridge University Press, London [Google Scholar]
- 3.Ninomiya-Tsuji J., Kishimoto K., Hiyama A., Inoue J., Cao Z., Matsumoto K. ( 1999) Nature 398, 252– 256 [DOI] [PubMed] [Google Scholar]
- 4.Sato S., Sanjo H., Takeda K., Ninomiya-Tsuji J., Yamamoto M., Kawai T., Matsumoto K., Takeuchi O., Akira S. ( 2005) Nat. Immunol. 6, 1087– 1095 [DOI] [PubMed] [Google Scholar]
- 5.Yamaguchi K., Shirakabe K., Shibuya H., Irie K., Oishi I., Ueno N., Taniguchi T., Nishida E., Matsumoto K. ( 1995) Science 270, 2008– 2011 [DOI] [PubMed] [Google Scholar]
- 6.Wang C., Deng L., Hong M., Akkaraju G. R., Inoue J., Chen Z. J. ( 2001) Nature 412, 346– 351 [DOI] [PubMed] [Google Scholar]
- 7.Takaesu G., Surabhi R. M., Park K. J., Ninomiya-Tsuji J., Matsumoto K., Gaynor R. B. ( 2003) J. Mol. Biol. 326, 105– 115 [DOI] [PubMed] [Google Scholar]
- 8.Yao J., Kim T. W., Qin J., Jiang Z., Qian Y., Xiao H., Lu Y., Qian W., Gulen M. F., Sizemore N., DiDonato J., Sato S., Akira S., Su B., Li X. ( 2007) J. Biol. Chem. 282, 6075– 6089 [DOI] [PubMed] [Google Scholar]
- 9.Shim J. H., Xiao C., Paschal A. E., Bailey S. T., Rao P., Hayden M. S., Lee K. Y., Bussey C., Steckel M., Tanaka N., Yamada G., Akira S., Matsumoto K., Ghosh S. ( 2005) Genes Dev. 19, 2668– 2681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Omori E., Matsumoto K., Sanjo H., Sato S., Akira S., Smart R. C., Ninomiya-Tsuji J. ( 2006) J. Biol. Chem. 281, 19610– 19617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sayama K., Hanakawa Y., Nagai H., Shirakata Y., Dai X., Hirakawa S., Tokumaru S., Tohyama M., Yang L., Sato S., Shizuo A., Hashimoto K. ( 2006) J. Biol. Chem. 281, 22013– 22020 [DOI] [PubMed] [Google Scholar]
- 12.Zatyka M., Morrissey C., Kuzmin I., Lerman M. I., Latif F., Richards F. M., Maher E. R. ( 2002) J. Med. Genet. 39, 463– 472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tan N. S., Michalik L., Noy N., Yasmin R., Pacot C., Heim M., Flühmann B., Desvergne B., Wahli W. ( 2001) Genes Dev. 15, 3263– 3277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu X. Y., Seh C. C., Cheung P. C. ( 2008) FEBS Lett. 582, 4023– 4031 [DOI] [PubMed] [Google Scholar]
- 15.Chong H. C., Tan M. J., Philippe V., Tan S. H., Tan C. K., Ku C. W., Goh Y. Y., Wahli W., Michalik L., Tan N. S. ( 2009) J. Cell Biol. 184, 817– 831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Michalik L., Desvergne B., Tan N. S., Basu-Modak S., Escher P., Rieusset J., Peters J. M., Kaya G., Gonzalez F. J., Zakany J., Metzger D., Chambon P., Duboule D., Wahli W. ( 2001) J. Cell Biol. 154, 799– 814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tan N. S., Icre G., Montagner A., Bordier-ten-Heggeler B., Wahli W., Michalik L. ( 2007) Mol. Cell. Biol. 27, 7161– 7175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.IJpenberg A., Tan N. S., Gelman L., Kersten S., Seydoux J., Xu J., Metzger D., Canaple L., Chambon P., Wahli W., Desvergne B. ( 2004) EMBO J. 23, 2083– 2091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bridge A. J., Pebernard S., Ducraux A., Nicoulaz A. L., Iggo R. ( 2003) Nat. Genet. 34, 263– 264 [DOI] [PubMed] [Google Scholar]
- 20.Nobes C. D., Hall A. ( 1999) J. Cell Biol. 144, 1235– 1244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rafty L. A., Khachigian L. M. ( 2002) J. Cell. Biochem. 85, 490– 495 [DOI] [PubMed] [Google Scholar]
- 22.Adhikari A., Xu M., Chen Z. J. ( 2007) Oncogene 26, 3214– 3226 [DOI] [PubMed] [Google Scholar]
- 23.Delaney J. R., Mlodzik M. ( 2006) Cell Cycle 5, 2852– 2855 [DOI] [PubMed] [Google Scholar]
- 24.Villa-Garcia M., Li L., Riely G., Bray P. F. ( 1994) Blood 83, 668– 676 [PubMed] [Google Scholar]
- 25.Cervella P., Silengo L., Pastore C., Altruda F. ( 1993) J. Biol. Chem. 268, 5148– 5155 [PubMed] [Google Scholar]
- 26.Lai C. F., Feng X., Nishimura R., Teitelbaum S. L., Avioli L. V., Ross F. P., Cheng S. L. ( 2000) J. Biol. Chem. 275, 36400– 36406 [DOI] [PubMed] [Google Scholar]
- 27.Krueger J. G., Krane J. F., Carter D. M., Gottlieb A. B. ( 1990) J. Invest. Dermatol. 94, 135S– 140S [DOI] [PubMed] [Google Scholar]
- 28.Cheng N., Bhowmick N. A., Chytil A., Gorksa A. E., Brown K. A., Muraoka R., Arteaga C. L., Neilson E. G., Hayward S. W., Moses H. L. ( 2005) Oncogene 24, 5053– 5068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Reuterdahl C., Sundberg C., Rubin K., Funa K., Gerdin B. ( 1993) J. Clin. Invest. 91, 2065– 2075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ansel J. C., Tiesman J. P., Olerud J. E., Krueger J. G., Krane J. F., Tara D. C., Shipley G. D., Gilbertson D., Usui M. L., Hart C. E. ( 1993) J. Clin. Invest. 92, 671– 678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mukhopadhyay D., Knebelmann B., Cohen H. T., Ananth S., Sukhatme V. P. ( 1997) Mol. Cell. Biol. 17, 5629– 5639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lederle W., Stark H. J., Skobe M., Fusenig N. E., Mueller M. M. ( 2006) Am. J. Pathol. 169, 1767– 1783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.O'Toole E. A., van, Koningsveld R., Chen M., Woodley D. T. ( 2008) J. Cell. Physiol. 214, 47– 55 [DOI] [PubMed] [Google Scholar]
- 34.Xia Y. P., Zhao Y., Tyrone J. W., Chen A., Mustoe T. A. ( 2001) J. Invest. Dermatol. 116, 50– 56 [DOI] [PubMed] [Google Scholar]
- 35.Zia M. K., Rmali K. A., Watkins G., Mansel R. E., Jiang W. G. ( 2007) Int. J. Mol. Med. 20, 605– 611 [PubMed] [Google Scholar]
- 36.Lolkema M. P., Mehra N., Jorna A. S., van Beest M., Giles R. H., Voest E. E. ( 2004) Exp. Cell Res. 301, 139– 146 [DOI] [PubMed] [Google Scholar]
- 37.Maas-Szabowski N., Shimotoyodome A., Fusenig N. E. ( 1999) J. Cell Sci. 112, 1843– 1853 [DOI] [PubMed] [Google Scholar]
- 38.Ahmed T., Shea K., Masters J. R., Jones G. E., Wells C. M. ( 2008) Cell Signal. 20, 1320– 1328 [DOI] [PubMed] [Google Scholar]
- 39.Omori E., Morioka S., Matsumoto K., Ninomiya-Tsuji J. ( 2008) J. Biol. Chem. 283, 26161– 26168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Trouba K. J., Hamadeh H. K., Amin R. P., Germolec D. R. ( 2002) Antioxid. Redox. Signal. 4, 665– 673 [DOI] [PubMed] [Google Scholar]
- 41.Young C. N., Koepke J. I., Terlecky L. J., Borkin M. S., Boyd S. L., Terlecky S. R. ( 2008) J. Invest. Dermatol. 128, 2606– 2614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tovar-Castillo L. E., Cancino-Díaz J. C., García-Vázquez F., Cancino-Gómez F. G., León-Dorantes G., Blancas-González F., Jiménez-Zamudio L., García-Latorre E., Cancino-Díaz M. E. ( 2007) Int. J. Dermatol. 46, 239– 246 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.