Abstract
Epidermal growth factor (EGF) exerts pleiotropic effects during oncogenesis, including the stimulation of cell migration and invasiveness. Although a number of traditional signaling proteins (e.g. Ras and Rho GTPases) have been implicated in EGF-stimulated cancer cell migration, less is known about the identity of those proteins functioning further downstream in this growth factor pathway. Here we have used HeLa carcinoma cells as a model system for investigating the role of tissue transglutaminase (TGase), a protein that has been linked to oncogenesis, in EGF-stimulated cancer cell migration and invasion. Treatment of HeLa cells with EGF resulted in TGase activation and its accumulation at their leading edges, whereas knocking down TGase expression, or treating cells with a TGase inhibitor, blocked EGF-stimulated cell migration and invasion. We show that EGF signaling through Ras and c-Jun N-terminal kinase is responsible for targeting TGase to the leading edges of cells and activating it. The requirement for EGF to properly localize and activate TGase can be circumvented by the expression of oncogenic Ras (G12V), whose ability to stimulate migration is also dependent on TGase. We further show that, in the highly aggressive breast cancer cell line MDAMB231, where EGF stimulation is unnecessary for migration and invasive activity, TGase is already at the leading edge and activated. These findings demonstrate that TGase plays a key role in cancer cell motility and invasiveness and represents a previously unappreciated participant in the EGF pathway that stimulates these processes in cancer cells.
The EGF2 receptor is a cell surface receptor tyrosine kinase that is expressed in a variety of normal cell lineages. Upon binding ligand, the EGF receptor becomes activated and initiates a highly regulated series of signaling events, which direct changes in gene transcription and various cellular responses that are vital for proper development and tissue homeostasis (1–3). In addition to its physiological functions, the EGF receptor has also been intimately associated with oncogenesis. Enhanced EGF receptor signaling, as a result of receptor overexpression and/or an autocrine stimulation of the receptor, is a hallmark of a variety of human tumors (2, 4, 5). Moreover, high levels of EGF receptor expression detected in brain, breast, pancreatic, and lung tumors are well correlated with poor prognosis, and therapeutic strategies targeting the EGF receptor itself or signaling events initiated by the receptor are currently being used as cancer treatments (4, 6, 7). These findings, coupled with the fact that ectopically expressing the EGF receptor in normal cell types is sufficient to induce ligand-dependent cellular transformation (8), strongly implicate EGF receptor signaling in cancer development. Thus, it is not surprising that a good deal of effort continues to be devoted toward understanding how EGF receptor activation gives rise to malignant transformation.
Exposing cancer cells to EGF can elicit or potentiate a host of phenotypes consistent with the oncogenic or transformed state. These include the excessive stimulation of cell cycle progression, protection against the apoptotic responses triggered by chemotherapy, and enhanced cell motility and invasive activity (8–10). Of particular interest has been to better understand the molecular mechanisms that underlie the ability of cancer cells to migrate and/or invade in response to EGF, because tumor cells that exhibit these traits are thought to have acquired an aggressive phenotype. By studying the actions of EGF in cancer cells, it is expected that new insights will be gained regarding the signaling events that are responsible for the development of highly aggressive tumors.
EGF-induced cell motility involves a multistep, cyclic process that integrates a highly coordinated activation and distribution of signaling molecules within cells with rearrangements of the actin cytoskeleton and plasma membrane (11). The cumulative effect of this process is the establishment of a polarized cellular morphology, where actin polymerization drives membrane protrusions at the front or leading edge of the cell followed by actin bundling and cell contraction at the rear. A few of the more extensively investigated signaling molecules that have been implicated in mediating the EGF-induced migration of cancer cells include members of the Ras superfamily of small GTPases, phosphatidylinositol 3-kinase (PI3K), and phospholipase Cγ (12–14). Extracellular-signal-regulated protein kinase (ERK) and JNK, two members of the mitogen-activated protein kinase (MAPK) family, which are best known for stimulating cell growth and differentiation (3), represent additional examples of signaling molecules that are also becoming appreciated for their roles in promoting cancer cell motility (15). For instance, interfering with the ability of EGF to stimulate ERK activity using the MEK inhibitors PD98059 or U0126 was shown to block the EGF-stimulated migration of LIM1215 colon cancer cells, as well as A549 lung carcinoma cells (16, 17). Likewise, incubating the breast carcinoma cell line MDAMB468 with the JNK inhibitor SP600125 rendered these cells insensitive to the migration-promoting actions of EGF (18). Although these findings collectively highlight the importance of Ras and Rho-family GTPases, and the MAPKs ERK and JNK, as playing significant roles in the EGF-signaling pathways stimulating cancer cell motility, there remains to be identified those proteins that are acting further downstream from this growth factor at the level of the leading edge and/or the cytoskeleton.
One candidate of potential interest is TGase, a unique multifunctional protein that exhibits an ability to bind and hydrolyze GTP, as well as catalyze an enzymatic transamidation reaction that results in the formation of covalent cross-links between two proteins or a protein and a polyamine (19). What makes this protein an interesting candidate to consider is that the overexpression and/or activation of TGase have been reported in highly aggressive and chemo-insensitive human brain, breast, ovarian, and pancreatic tumors (20–23). In addition, ectopically expressing TGase in the SKBR3 breast cancer cell line was sufficient to confer a survival advantage to these cells when challenged with the chemotherapeutic agent doxorubicin (24). Likewise, both the invasive and drug-resistant phenotypes of MDAMB231 breast carcinoma cells could be suppressed when the chronic TGase activity associated with these cells was inhibited by either knocking down TGase expression using RNAi or by incubating the cells with the TGase inhibitor monodansylcadaverine (MDC) (24–26).
Given these potential connections between TGase and different aspects of cancer progression, we investigated the possibility that it might be playing a role in the EGF-signaling pathways that contribute to cancer cell migration and invasiveness, using the human cervical carcinoma HeLa cell line as a model for studying these processes. We found that treating HeLa cells with EGF caused TGase to accumulate at the leading edges of migrating cells, as well as activated both its GTP-binding and transamidation activities. Introducing TGase-specific siRNAs into cells, or inhibiting the transamidation activity of TGase with MDC, blocked the ability of EGF to promote HeLa cell migration and invasion, indicating that TGase is in fact an essential participant in the EGF-signaling pathway responsible for these activities. We further determined that EGF can work through Ras and JNK to localize TGase to the leading edges of cells and to activate it. To our knowledge, these data indicate for the first time that EGF-promoted tumor cell aggressiveness is tightly coupled to its ability to activate TGase, and in particular demonstrate that TGase plays an essential role in the ability of EGF to stimulate cancer cell migration and invasive activity.
EXPERIMENTAL PROCEDURES
Materials
Cell culture reagents, EGF, Lipofectamine, Lipofectamine 2000, the control and TGase siRNAs, and the HA and Myc antibodies were from Invitrogen. RA, DAPI, doxorubicin, and MDC were from Sigma, whereas BPA was from Pierce. LY294002, PD98059, SP600125, SB202190, PP2, the Rho-kinase inhibitor, and nocodazole were from Calbiochem. The TGase and actin antibodies were from Neomarkers, whereas the antibodies that recognize the activated forms of AKT, JNK, ERK, and c-Jun were from Cell Signaling. The EGF receptor and the phospho-EGF receptor antibodies were from BD Transduction Laboratories.
Cell Culture
HeLa and MDAMB231 cell lines were grown in RPMI 1640 medium containing 10% fetal bovine serum (FBS), whereas HeLa cells stably expressing HA-tagged pcDNA3 vector or HA-tagged Ras (G12V) were selected and maintained in the same growth medium supplemented with 600 μg/ml G418. The pcDNA3 constructs encoding the various forms of Myc-tagged TGase were transfected into cells using Lipofectamine, whereas the control and TGase siRNAs were introduced into cells using Lipofectamine 2000. Where indicated, the cell cultures were incubated with 5 μm RA or 0.1 μg/ml EGF, in the presence and absence of 15 μm MDC or in the presence and absence of the small molecule inhibitors indicated under “Results.” The cells were then either fixed with 3.7% formaldehyde or lysed with cell lysis buffer (25 mm Tris, 100 mm NaCl, 1% Triton X-100, 1 mm EDTA, 1 mm dithiothreitol, 1 mm NaVO4, and 1 mm β-glycerol phosphate). Protein concentrations were determined using the Bio-Rad DC protein assay.
Western Blot Analysis
Equal concentrations of each cell lysate were resolved by SDS-PAGE, and then the proteins were transferred to polyvinylidene difluoride. The filters were incubated with the various primary antibodies diluted in TBST (20 mm Tris, 135 mm NaCl, and 0.02% Tween 20). The primary antibodies were then detected with horseradish peroxidase-conjugated secondary antibodies (Amersham Biosciences) followed by exposure to ECL reagent.
Immunofluorescence
Some of the cells fixed with 3.7% formaldehyde were permeabilized with phosphate-buffered saline containing 0.1% Triton X-100, and then blocked in phosphate-buffered saline containing 7% bovine serum albumin. Following blocking, the cells were incubated with the indicated primary antibodies for 2 h, rinsed with phosphate-buffered saline, and then incubated with either Oregon green- or rhodamine-conjugated secondary antibody (Molecular Probes) for an additional hour. Where indicated, rhodamine-conjugated phalloidin was used to stain actin, and DAPI was used to stain nuclei. The cells were then washed, mounted, and visualized using either the 40× or 63× objectives on a Zeiss Axioskop fluorescent microscope. Images were captured/processed using IPLAB.
Cell Migration (Scratch) Assay
Naive cells or cells transfected with control or TGase siRNAs were grown to confluence, at which time they were incubated with RPMI medium containing 0.2% FBS for 15 h. The medium was then replaced (±0.1 μg/ml EGF) for an additional 12 h, and then the cell cultures were scratched using a pipette tip. The cultures were rinsed to remove detached cells and then incubated with medium containing 0.2% FBS ± 0.1 μg/ml EGF, ±15 μm MDC, ±1 μm SP600125, ±6 μm PD98059, or ±10 μm LY294002. Fifteen hours later the cells were fixed and visualized by light microscopy. These assays were performed three times. Where indicated, immunofluorescence was performed on the in vitro migration assays. In these cases, the cells were fixed 3 h after scratching the wound and then stained with a TGase antibody, rhodamine-conjugated phalloidin, and DAPI.
Cell Invasion Assay
Parental HeLa cells or HeLa cells that had been transfected with control or TGase siRNAs and maintained in RPMI medium containing 0.2% FBS were seeded at 5000 cells/well ± 15 μm MDC in the upper chamber of a Millicell Culture Plate Insert (Millipore) pre-coated with Matrigel (BD Biosciences). RPMI medium containing 0.2% FBS ± 0.1 μg/ml EGF was added to the lower chamber, and the cultures were maintained for 1 day. The cells that accumulated on the lower side of the filter were fixed with methanol, stained with Giemsa stain, and then scored. The invasion assays were performed three times, and the results from each experiment were averaged.
Photoaffinity Labeling of TGase and Transamidation Assay
These assays were performed as previously described (24, 27).
Cell Viability Assay
One set of HeLa cells was incubated in medium containing 0.2% FBS ± 15 μm MDC or ± 0.15 μm doxorubicin. Another set of HeLa cells was transfected with control or TGase siRNAs and maintained in medium containing 0.2% FBS. Two days later, all of the cultures were trypsinized, incubated with trypan blue, and viewed using a light microscope. At least 300 cells were counted for each of the culturing conditions, and the ratio of viable cells (those cells that have not taken up trypan blue) to non-viable cells (those that have taken up trypan blue) was determined. These assays were performed three times, and the average percentage of cell death for each culturing condition was graphed.
RESULTS
EGF Activates TGase and Recruits It to the Leading Edge
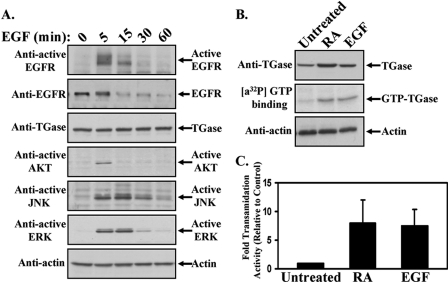
The overall goal of this study was to determine whether the suggested connections between TGase and cancer progression (20–25) reflect an important role for this protein in the EGF-signaling pathway that stimulates cancer cell migration and invasive activity. The human cervical carcinoma HeLa cell line was used to investigate this question, because these cells express both the EGF receptor and TGase (Fig. 1A, second and third panels from the top), as well as represent an established model system for studying cancer cell migration and invasion (10, 28). Fig. 1A, top panel, shows that treating serum-deprived HeLa cells with EGF for increasing lengths of time results in the transient activation of the EGF receptor, with the receptor being maximally activated within 5 min of EGF stimulation and reduced to near background levels by 60 min. To determine the effects of stimulating the activity of the EGF receptor on downstream signaling events, antibodies that recognize the phosphorylated forms of AKT, JNK, and ERK were used to readout the activation levels of these signaling proteins. The results in Fig. 1A demonstrate that EGF treatment stimulates the activities of AKT (fourth panel from the top), JNK (fifth panel from the top), and ERK (sixth panel from the top), although the duration of activation varied for these different signaling kinases. Maximal AKT activity was detectable only through 5 min of growth factor treatment, whereas the activation profiles of JNK and ERK were more extended and closely paralleled the activation levels of the EGF receptor.
FIGURE 1.
EGF induces TGase activation in HeLa cells. A, serum-starved HeLa cells were treated with EGF for the indicated times and then lysed. The cell extracts were immunoblotted with the indicated antibodies. B, HeLa cells were cultured in medium containing 0.2% FBS (low serum medium) without (Untreated) or with either RA or EGF. One day later the cells were lysed, and the extracts were immunoblotted with TGase and actin antibodies (top and bottom panels, respectively). The GTP-binding activities of TGase in the cell lysates were determined using a photoaffinity labeling assay with [α-32P]GTP (middle panel). C, the levels of TGase transamidation activity in the cell extracts were also determined by assaying the incorporation of BPA into lysate proteins. The assays were performed three times, and the results of each assay were quantified by densitometry, averaged together, and graphed as -fold increases in transamidation activity relative to the control. The error bars indicate ±S.D.
We next asked how EGF treatment would affect TGase activity. HeLa cells maintained in low serum medium in the absence or presence of either EGF or retinoic acid (RA) were collected, and then the GTP-binding and enzymatic transamidation activities of TGase in each sample were determined. Exposing the cells to EGF markedly enhanced the GTP-binding activity of TGase compared with control cells, as indicated by the photo-affinity incorporation of [α-32P]GTP (Fig. 1B, middle panel), without significantly altering its expression (Fig. 1B, top panel). Likewise, when the cell lysates were assayed for TGase-catalyzed transamidation activity, as readout by the incorporation of 5-(biotinamido)pentylamine (BPA) into lysate proteins, more than a 7-fold increase in enzymatic activity was detected in the EGF-treated cells compared with the non-treated control cells (Fig. 1C). It is also noteworthy that EGF appears to be comparable to RA, one of the best known and most potent activators of TGase (29–31), at inducing the GTP-binding and transamidation activities of TGase (Fig. 1B, top panel, and Fig. 1C, respectively).
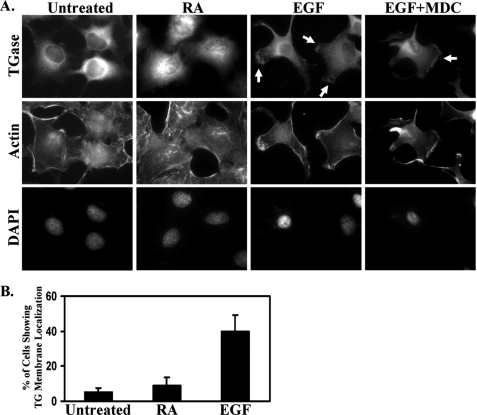
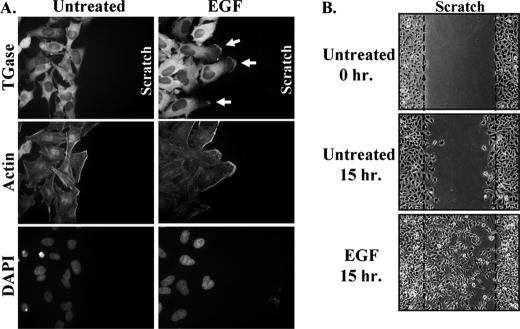
We then examined whether EGF also influenced the subcellular localization of TGase. Immunofluorescent analysis performed on HeLa cells cultured under low serum conditions revealed that TGase was expressed primarily throughout the cytoplasm, with its highest levels of expression being concentrated around the nucleus (Fig. 2A, top panel of the first column). RA treatment did not noticeably change the distribution of TGase within the cells (Fig. 2A, top panel of the second column). However, EGF caused a marked change in the cellular distribution of TGase, such that nearly 40% of the cells showed an accumulation of TGase at their plasma membranes (Fig. 2A, top panel of the third column and Fig. 2B). Interestingly, the localization of TGase detected along the surfaces of the cells following EGF stimulation was not uniform, but rather frequently appeared to be localized to discrete sites of actin-rich cell extensions (Fig. 2A, top and middle panel of the third column). Because these cell extensions exhibited a strong resemblance to the leading edge morphology observed in actively migrating cells (11), we explored the possibility that TGase might be enriched along the leading edges of HeLa cells exposed to EGF. As a means to induce directional cell movement and thereby define the leading edges of cells, we performed wound healing or scratch assays on confluent cultures of HeLa cells treated with or without EGF. Within 3 h of disrupting the cell monolayers by scratching them with a pipette tip, the EGF-stimulated HeLa cells, but not the control cells, began to form leading edges as indicated by the presence of cell extensions into the wound and the accumulation of F-actin along the surface of these cell extensions (Fig. 3A, compare the middle panels). Immunostaining the same cells with an anti-TGase antibody revealed that TGase was in fact consistently detected along the leading edges of EGF-stimulated HeLa cells at the sites of F-actin assembly (Fig. 3A, top right panel). This result, combined with the finding that exposing HeLa cells to EGF strongly induces cell motility as indicated by the near complete closure of the wound (Fig. 3B), suggested that TGase might be an important player in EGF-promoted cell migration.
FIGURE 2.
TGase accumulates at the plasma membranes of HeLa cells treated with EGF. A, HeLa cells were cultured in medium containing 0.2% FBS (low serum medium) without (Untreated) or supplemented with RA, EGF, or EGF plus MDC for a day and then were fixed. Immunofluorescence was performed on the samples using a TGase antibody, rhodamine-conjugated phalloidin (Actin), and DAPI (to stain nuclei). The resulting fluorescence images of the cells are shown. Membrane-localized TGase is indicated with arrows. B, the percentage of HeLa cells with membrane-localized TGase in untreated cells, versus RA- or EGF-treated cells, was determined. At least 250 cells were scored for each condition, and the results shown represent the findings from at least three independent experiments that were averaged together and graphed. The error bars indicate ±S.D.
FIGURE 3.
TGase is localized to the leading edges of actively migrating cells. Duplicate sets of scratch assays were performed on cultures of HeLa cells treated without (Untreated) or with EGF. A, one set of cultures was fixed 3 h after scratching the wound. Immunofluorescence was performed on these samples using a TGase antibody, rhodamine-conjugated phalloidin (Actin), and DAPI. The resulting fluorescence images shown are of cells along one edge of the wound. The location of the scratched wound is indicated (scratch) and membrane-localized TGase is denoted with arrows. B, a second set of cell cultures was either fixed immediately after scratching the wound (Untreated-0 h) or 15 h after scratching the wound (Untreated-15 h and EGF-15 h) and then visualized by light microscopy. Each panel represents a single image that shows both edges of a wound that was scratched in a monolayer of cells exposed to the indicated culturing conditions. The extent of wound closure for each condition is shown. The dashed lines represent the widths of the wounds at the start of the assay.
TGase Activity Is Essential for EGF-stimulated Cell Migration and Invasive Activity
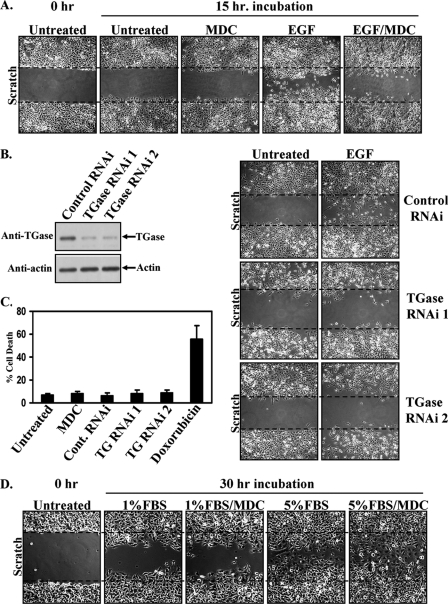
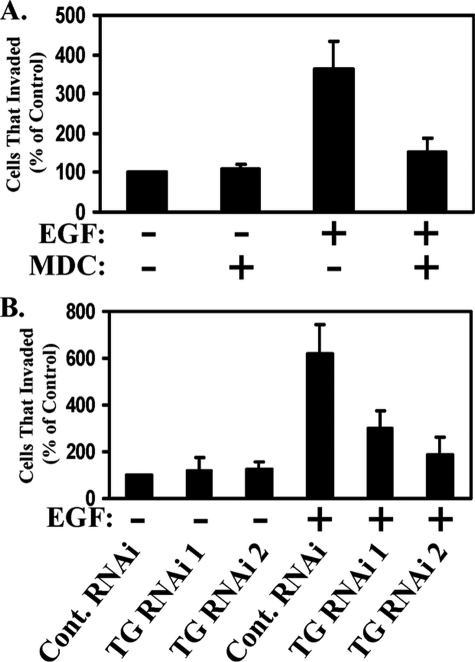
We then set out to directly assess the involvement of TGase in EGF-stimulated cell migration using the wound-healing/scratch assay. As shown in Fig. 4A, when HeLa cells were preincubated with the TGase transamidation inhibitor MDC, EGF failed to stimulate their ability to migrate. Consistent with these results, knocking down TGase expression using two different siRNAs (Fig. 4B, left side) effectively blocked EGF-stimulated HeLa cell migration (Fig. 4B, right side).
FIGURE 4.
TGase is essential for EGF-induced HeLa cell migration. A, scratch assays were performed on HeLa cells treated without (Untreated) or with EGF, ± MDC. Fifteen hours after scratching the wounds, the cells were fixed and visualized by light microscopy to determine the extent of wound closure. One untreated plate of cells was fixed immediately after scratching the wound (0 h-Untreated) to show the size of the wounds at the start of the assay. Each panel represents a single image that shows both edges of a wound that was scratched in a monolayer of cells exposed to the indicated culturing conditions. The widths of the initially scratched wounds are indicated with dashed lines. B, lysates of HeLa cells transfected with control-RNAi, TGase-RNAi 1, or TGase-RNAi 2 were immunoblotted with TGase and actin antibodies (left panels). Scratch assays were then performed on cells transfected with the same siRNAs and incubated without (Untreated) or with EGF. The cells were processed and presented as outlined in A (right panels). C, parental HeLa cells or HeLa cells transfected with control or TGase-siRNAs were incubated with medium containing 0.2% FBS (low serum medium) without (Untreated) or with MDC or doxorubicin. Two days later, cell viability assays were performed on the cell cultures, with the percentage of cell death being determined for each condition. The assays were performed at least three times, and the results were averaged together and graphed. The error bars indicate ±S.D. D, scratch assays were conducted on HeLa cells cultured in medium containing either 1% or 5% FBS, ± MDC. 30 h after scratching the wounds, the cells were fixed and visualized by light microscopy to determine the extent of wound closure. One plate of cells was fixed immediately after scratching the wound (0 h-Untreated) to show the size of the wounds at the start of the assay. Each panel represents a single image that shows both edges of a wound that was scratched in a monolayer of cells exposed to the indicated culturing conditions. The widths of the initially scratched wounds are indicated with dashed lines.
To further implicate TGase as an essential mediator of EGF-stimulated cell migration, two additional experiments were performed. Because in some cases interfering with TGase activity can make cells susceptible to apoptotic challenges (24, 25, 32), we first wanted to verify that the inability of EGF to stimulate cell migration, under conditions where TGase expression and/or its activity were inhibited in our experimental system, cannot be attributed to a nonspecific cell death response. Cell viability assays were carried out on HeLa cells that were cultured under precisely the same conditions used for the migration assays. As shown in Fig. 4C, neither treatment with MDC nor the introduction of TGase siRNAs into HeLa cells negatively affected cell viability, whereas exposing the cells to the chemotherapeutic agent doxorubicin caused nearly 60% of the cells to die.
We then asked whether TGase functions as a general mediator of HeLa cell migration or if its effects were more specific for EGF. To address this, the cells were subjected to a scratch assay that used medium containing either 1% or 5% fetal bovine serum (FBS) to stimulate cell motility. Fig. 4D shows that, while incubating the cell cultures with either amount of serum promoted cell migration, the cells treated with 5% FBS closed the wound much more efficiently than those cells treated with just 1% FBS. However, even the rate of cell migration induced with 5% FBS was considerably slower than that observed with EGF, requiring twice the time period to close the wound to a comparable extent as occurred with EGF treatment (i.e. 30 h for 5% FBS compared with 12–15 h for EGF). In addition, unlike what we found with EGF (see Fig. 4A), exposing the cells to MDC had no effect on the ability of either amount of serum (1% or 5% FBS) to stimulate cell migration (Fig. 4D). Collectively, these data firmly establish that TGase is not playing a general role in cell migration but rather is a specific component of the EGF-signaling pathway that stimulates the motility of these cancer cells.
Tumor cell invasiveness relies heavily on cell migration (11). Thus, we wanted to see whether TGase might also contribute to the ability of EGF to promote invasiveness. We examined this possibility by taking advantage of the in vitro Matrigel invasion assay. Fig. 5A shows that exposing HeLa cells to EGF caused more than a 3.5-fold increase in the number of cells that could transverse the Matrigel over the basal rate of invasion seen in the non-stimulated control cells. While incubating HeLa cells with MDC had little, if any, impact on the basal rate of cell invasion, treatment with the inhibitor severely compromised the invasive advantage afforded by EGF. Moreover, knocking down TGase expression by siRNA reduced the EGF-stimulated invasiveness of the cells by as much as 70% (Fig. 5B). These findings indicate that, in addition to being required for EGF-stimulated cell migration, TGase is also essential for EGF-stimulated invasiveness.
FIGURE 5.
TGase is important for the EGF-stimulated invasive activity in HeLa cells. A and B, graphs representing the invasive activities of HeLa cells exposed to various culturing conditions, as assessed using the Matrigel Transwell assay system. In A, the cells were incubated without or with EGF ± MDC for 1 day before being scored for invasiveness. In B, HeLa cells expressing control-RNAi, TGase-RNAi 1, or TGase-RNAi 2 were incubated without or with EGF for 1 day before being scored for invasiveness. These assays were performed at least three times, and the results were averaged together and graphed. The error bars indicate ±S.D.
TGase Activation Is Not Required for Its Recruitment to the Leading Edge
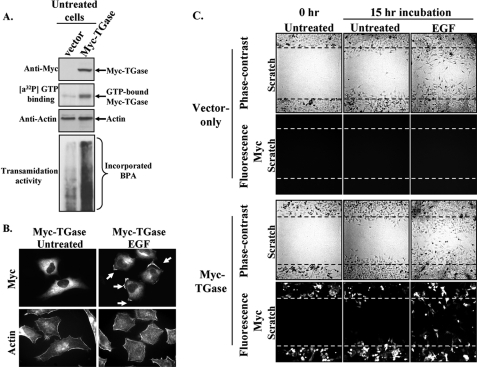
Given that the EGF-stimulated motility and invasive activities of HeLa cells are tightly coupled to the ability of this growth factor to promote TGase activation and its accumulation at the leading edges of cells, we wanted to better understand how EGF influences the activity and subcellular distribution of TGase. We initially focused our efforts on assessing whether TGase activation was required for its localization at the leading edges of cells. To address this question, we examined whether exogenously expressed Myc-tagged forms of wild-type TGase, or point mutants of TGase that were GTP-binding defective and/or transamidation-defective, in HeLa cells functioned similarly to endogenous TGase and accumulated along the plasma membranes of cells treated with EGF. As has been reported for several other cell lines (24, 32, 33), ectopically expressed wild-type TGase in serum-starved HeLa cells (Fig. 6A, top panel) exhibits both [α-32P]GTP-binding activity (Fig. 6A, second panel from the top) and the ability to catalyze the incorporation of BPA into cell extract proteins (Fig. 6A, bottom panel). Immunofluorescent analysis performed on these same transfectants revealed that, despite being capable of GTP-binding and transamidation activity, the ectopically expressed TGase was largely cytosolic in unstimulated cells. However, following their treatment with EGF, the Myc-tagged wild-type TGase could be readily detected in the plasma membranes of the transfectants (Fig. 6B, compare the two top panels).
FIGURE 6.
TGase accumulation at the leading edges of HeLa cells stimulated with EGF does not require its GTP-binding or enzymatic transamidation capabilities. Multiple sets of serum-starved (Untreated) or EGF-treated HeLa cells ectopically expressing the control vector (vector) or a Myc-tagged form of TGase (Myc-TGase) were lysed, fixed, or subjected to a scratch assay. A, the cell lysates were immunoblotted with Myc and actin antibodies (top panel and third panel from the top, respectively) and also subjected to GTP-binding and transamidation activity assays as readout using a photo-affinity labeling assay with [α-32P] GTP (second panel from the top), or by the incorporation of BPA into lysate proteins (bottom panel). B, immunofluorescence was performed on the fixed cells using a Myc antibody and rhodamine-conjugated phalloidin (Actin). Membrane-localized Myc-TGase is indicated with arrows. C, scratch assays performed on the transfectants were either fixed immediately after scratching the wound (Untreated-0 h) or 15 h after scratching the wound (Untreated-15 h and EGF-15 h). Immunofluorescence was performed on the transfectants using a Myc antibody, and then the cells were visualized by phase-contrast and fluorescent microscopy. Each panel represents a single image that shows both edges of a wound that was scratched in a monolayer of cells exposed to the indicated culturing conditions. The extent of wound closure for each condition is shown. The dashed lines represent the widths of the wounds at the start of the assay.
Determining that exogenously expressed TGase in HeLa cells exhibits functional activity even in the absence of EGF stimulation but is not targeted to cell membranes, afforded us an opportunity to examine whether localizing TGase to the leading edges of cells might be linked to the motility-promoting actions of this protein. Serum-deprived HeLa cell cultures transiently expressing either control vector or a Myc-tagged form of TGase were subjected to scratch assays in the presence or absence of EGF, and the resulting rates of cell migration were analyzed using phase-contrast and fluorescent microscopy. As anticipated, the vector-control cells only exhibited significant migratory capability when cultured in the presence of EGF (Fig. 6C, top two sets of panels). The bottom set of panels in Fig. 6C shows the phase-contrast and corresponding immunofluorescent images of cells that were transiently transfected with a Myc-tagged TGase construct and then subjected to a scratch assay. The cells in the scratch assay that ectopically expressed the Myc-tagged TGase protein were determined by immunostaining the transfectants with an anti-Myc antibody, followed by detection with fluorescence microscopy. The results of these experiments show that simply activating TGase by its overexpression in cells cannot mimic the migration-promoting actions of EGF and stimulate wound closure, thus lending further support to the idea that both the activation and appropriate subcellular distribution of TGase are necessary for its ability to mediate cell motility responses.
The analysis was expanded to determine whether previously established GTP binding-defective and/or transamidation-defective forms of TGase (33) could also be localized to the leading edges of cells in an EGF-dependent manner. This set of experiments was carried out essentially as described using the Myc-tagged wild-type TGase construct in Fig. 6B. Although the transient expression levels of the GTP binding-defective mutant (Myc-TGase R580L), the transamidation-defective mutant (Myc-TGase C277V), and the TGase double mutant that lacked both of these activities (Myc-TGase C277V/R580L), in HeLa cells were consistently less than that obtained with wild-type TGase, we found that each of the TGase mutants responded to EGF stimulation and accumulated at the leading edges of cells (data not shown). The finding that the transamidation activity of TGase is dispensable for its recruitment to the leading edge is also consistent with the results shown in the top panel of Fig. 2A. Here, the immunofluorescent images of cells that had been preincubated with the transamidation inhibitor MDC show that the inhibitor had no influence on the ability of EGF to localize TGase to the plasma membranes of cells. Taken together, these findings suggest that the GTP-binding capability and the enzymatic transamidation activity of TGase are neither necessary nor sufficient for its EGF-stimulated membrane localization.
EGF Signaling to JNK Is Required for the Recruitment of TGase to the Leading Edge
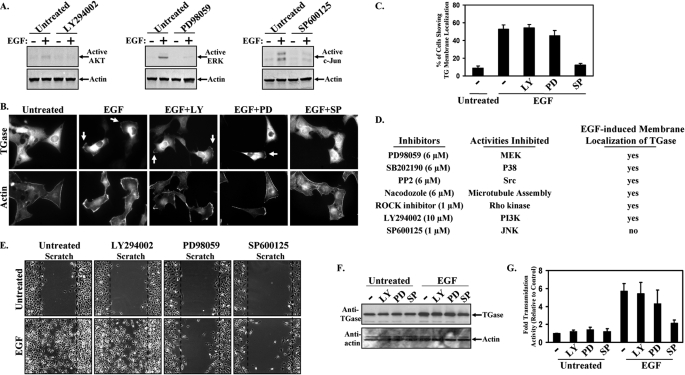
We then set out to delineate the EGF-stimulated signaling pathway responsible for the recruitment of TGase to the leading edges of cells. PI3K, as well as the MAPK family members ERK and JNK, represented some attractive candidates to consider, based on the increasing evidence that links these signaling proteins to EGF-mediated cell migration responses in a variety of cancer cell types (12, 15). The top panels in Fig. 7A show that, under conditions where EGF stimulates cell migration (i.e. 2–3 h of treatment), it activates ERK, JNK, and to a much weaker extent AKT. To examine the potential roles of these signaling proteins in localizing TGase to the plasma membranes of cells, we utilized inhibitors that specifically blocked their activation. Duplicate sets of HeLa cells were stimulated with EGF in the presence or absence of the PI3K inhibitor, LY294002, the JNK inhibitor, SP600125, or the MEK inhibitor (to block ERK activity), PD98059, and then one set of cells was lysed while the other set was fixed. To confirm that the inhibitors did indeed block the activities of PI3K, ERK, and JNK, the cell extracts collected were immunoblotted with antibodies that recognized the phosphorylated forms of AKT (to measure PI3K activity), ERK, and c-Jun (to measure JNK activity). The top panels in Fig. 7A show that the EGF-stimulated PI3K, ERK, and JNK activities were inhibited by LY294002, PD98059, and SP600125 treatment, respectively.
FIGURE 7.
JNK activity is necessary for EGF to fully activate TGase and to localize it to the leading edges of cells. Multiple sets of HeLa cell cultures were incubated in medium containing 0.2% FBS (low serum medium) without (Untreated) or with EGF ± various inhibitors including LY294002 (LY), PD98059 (PD), or SP600125 (SP). A, one set of cell cultures was lysed, and then the cell extracts were subjected to Western blot analysis using antibodies that recognize the activated forms of AKT, ERK, and JNK (top panels). The blots were reprobed with actin to confirm equal loading (bottom panels). B, another set of cell cultures was fixed and immunofluorescence performed on the samples using a TGase antibody and rhodamine-conjugated phalloidin (Actin). The resulting fluorescence images of the cells are shown. Membrane-localized TGase is indicated with arrows. C, the percentage of HeLa cells treated with ECF ± one of the indicated inhibitors showing membrane-localized TGase was determined. These assays were performed three times, and the results were averaged together and graphed. The error bars indicate ±S.D. D, a list of the inhibitors examined for their ability to block the EGF-stimulated accumulation of TGase at the leading edges of cells. The left column lists the name of each inhibitor and the concentration at which it was used. The middle column indicates the specific activity blocked by an inhibitor, whereas the right column indicates whether the EGF-stimulated recruitment of TGase to the leading edges of cells incubated with a given inhibitor still occurs (yes) or is blocked (no). E, a scratch assay was performed on another set of HeLa cell cultures exposed to the indicated inhibitors ± EGF. 15 h after scratching the wounds, the cells were fixed and visualized by light microscopy to determine the extent of wound closure. Each panel represents a single image that shows both edges of a wound that was scratched in a monolayer of cells exposed to the indicated culturing conditions. The widths of the wounds scratched at the start of the assay are indicated with dashed lines. F, a third set of cell cultures treated with the indicated inhibitors was lysed and immunoblotted with TGase and actin antibodies. G, the levels of transamidation activity present in the cell extracts were determined by assaying the incorporation of BPA into lysate proteins. The assays were performed at least three times, and the results of these assays were quantified by densitometry and graphed. The error bars indicate ±S.D.
From the fixed set of HeLa cells, the resulting cellular distribution of TGase was determined by immunostaining the cell cultures with an anti-TGase antibody. The top panel in Fig. 7B and the graph in Fig. 7C show that incubating the cells with LY294002 or PD98059 did not significantly alter the ability of EGF to target TGase to the leading edges of cells, indicating that neither PI3K nor ERK activity is required to mediate this process. On the other hand, incubating the cells with the JNK inhibitor, SP600125, resulted in a marked decrease in the amount of membrane-localized TGase following EGF stimulation. The effect of the JNK inhibitor at blocking the EGF-stimulated accumulation of TGase at cell membranes appears to be rather specific based on two additional pieces of evidence. First, as indicated in the Table in Fig. 7D, several additional cell signaling inhibitors, as well as the microtubule assembly inhibitor, nocodazole, were also examined and found to be ineffective at blocking the EGF-stimulated membrane localization of TGase. The second piece of evidence arguing for a direct connection between JNK activity and the subcellular localization of TGase was obtained from experiments in which we show that the ability of the JNK inhibitor to block the build-up of TGase along the leading edges of EGF-stimulated cells cannot be attributed to the fact that SP600125 acts as a general inhibitor of cell motility. In particular, we show that, although blocking ERK activation using PD98059 can abrogate the EGF-stimulated migration of HeLa cells just as effectively as the JNK inhibitor, SP600125 (Fig. 7E), treatment with PD98059 did not alter the recruitment of TGase to the cell surface in response to growth factor stimulation (see Fig. 7B).
EGF Signaling to JNK Is Required for the EGF-dependent Activation of TGase
Given the requirement for TGase activity in the EGF-stimulated migration of HeLa cells (see Fig. 4A), it was of interest to delineate the signaling pathway that is responsible for the growth factor-dependent activation of TGase. We again examined inhibitors for PI3K, JNK, and MEK to see whether one or more of these signaling kinases were necessary for the ability of EGF to activate TGase. Fig. 7F shows that none of these inhibitors affected EGF-stimulated TGase expression, indicating that any differences in the extent of EGF-induced TGase activity that accompany the inhibition of one or more of the above mentioned signaling kinases is not due to changes in the expression level of TGase. The whole cell extracts used for the Western blot analyses (Fig. 7F) were then assayed for TGase-catalyzed transamidation activity using the incorporation of BPA into lysate proteins as a measurement. Whereas the extent of EGF-stimulated transamidation activity was unchanged when treating the cells with LY294002, and only modestly diminished by treatment with PD98059, it was significantly affected by the JNK inhibitor SP600125 (Fig. 7G). Blocking JNK activity caused a reduction of ∼4-fold in EGF-stimulated transamidation activity, such that it approached the basal levels of TGase activity assayed in non-stimulated control cells. These findings highlight the importance of JNK activity for the EGF-stimulated localization of TGase to the leading edges of cells, as well as for the EGF-dependent activation of its transamidation activity.
TGase Is Constitutively Active and Membrane-localized in HeLa Cells Expressing Oncogenic Ras and in the Highly Aggressive Breast Cancer MDAMB231 Cell Line
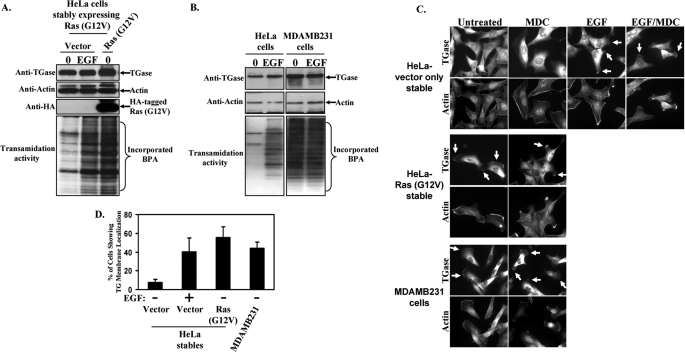
Our findings to this point suggested that the induction of TGase activity and its accumulation along the leading edges of cells are linked to the ability of EGF to stimulate HeLa cell motility and invasive activities. However, we wanted to see whether TGase activation and its plasma membrane localization were necessary for the migration responses elicited by other proteins that have been implicated in EGF-signaling activities, or that occur in other human cancer cell lines. Thus, we examined the involvement of TGase in mediating the migration-promoting actions of an oncogenic form of Ras introduced into HeLa cells, as well as explored the relationship between TGase and the highly aggressive/invasive phenotype exhibited by the human breast cancer cell line MDAMB231 (24, 25). Ras seemed to be a particularly relevant choice given that it functions as a major downstream target of the EGF receptor, and is the most frequently mutated oncogene in human cancers (34). HeLa cells stably expressing either the control vector or a dominant-active (oncogenic) form of Ras, Ras (G12V), were generated, and the expression of the Ras mutant protein was confirmed by Western blot analysis (Fig. 8A, third panel from the top). We then determined how Ras (G12V) expression affected TGase activation by comparing the levels of transamidation activity present in whole cell extracts from serum-starved, vector-only-expressing HeLa cells to serum-starved, Ras (G12V)-expressing cells. The bottom panel in Fig. 8A shows that the TGase-catalyzed transamidation activity detected in cells expressing the mutant form of Ras was indeed augmented relative to the vector-control cells (compare the first and third lanes). Moreover, the amount of TGase activity in the Ras (G12V)-expressing cells was comparable to that observed when the vector-control cells were stimulated with EGF, raising the possibility that one way the EGF receptor could promote the activation of TGase in HeLa cells is through its ability to signal to Ras.
FIGURE 8.
TGase is constitutively active and localized to the leading edges of HeLa cells expressing an activated form of Ras and in MDAMB231 breast cancer cells. Serum-starved HeLa cells stably expressing the control vector (vector) or an activated form of Ras (Ras (G12V)), as well as parental HeLa and MDAMB231 cells were treated without (0) or with EGF for a day and then were lysed. A, extracts of the HeLa stables were immunoblotted with TGase, actin, and HA antibodies (top, second from the top, and third from the top panels, respectively). The same cell extracts were then assayed for transamidation activity as determined by the incorporation of BPA into lysate proteins (bottom panel). B, the parental HeLa and MDAMB231 cell extracts were also immunoblotted with TGase and actin antibodies (top and middle panels, respectively) and assayed for transamidation activity (bottom panel). C, stable HeLa cell lines and parental MDAMB231 cells were incubated with serum-free medium (Untreated), or the indicated combinations of serum-free medium ± EGF and ± MDC for a day and then were fixed. Immunofluorescence was performed on the samples using a TGase antibody and rhodamine-conjugated phalloidin (Actin). The resulting fluorescent images of the cells are shown. Membrane-localized TGase is indicated with arrows. D, the percentage of serum-starved HeLa cells expressing the control vector or Ras (G12V), or the parental MDAMB231 cells showing membrane-localized TGase was determined. Vector-only-expressing HeLa cells stimulated with EGF were used as a positive control. The results of three independent experiments were averaged together and graphed. The error bars indicate ±S.D.
An analogous set of experiments was also performed to determine the levels of TGase expression and activation in the highly aggressive and invasive MDAMB231 breast cancer cells. Similar to previous findings by our laboratory and others (24, 25), a comparison of the TGase expression levels in HeLa and MDAMB231 cells shows that TGase expression is up-regulated in MDAMB231 cells (Fig. 8B, top panel). Analysis of the enzymatic transamidation activity associated with TGase expressed in MDAMB231 cells, as measured by the incorporation of BPA into lysate proteins, demonstrates that TGase is chronically activated in this cancer cell line (Fig. 8B, bottom panel). Interestingly, the treatment of MDAMB231 cells with EGF did not lead to a further enhancement in TGase activity, suggesting that TGase is already maximally activated in these cancer cells.
The resulting subcellular distribution of TGase within HeLa cells stably expressing either vector-alone or the Ras (G12V) construct, as well as in MDAMB231 cells exposed to various culture conditions, was assessed by immunostaining the cells with a TGase antibody. Representative fluorescent images taken of the different cell lines cultured under serum-deprived conditions are shown in the first column of Fig. 8C. Unlike the vector-control cells where TGase expression is primarily cytosolic (top panel), both HeLa cells expressing the Ras (G12V) mutant (middle panel) and the MDAMB231 cells (bottom panel) show a considerable accumulation of TGase at their plasma membranes. In fact, we found that nearly 60% of HeLa cells expressing Ras (G12V) and over 40% of the MDAMB231 cells showed some degree of membrane-localized TGase (Fig. 8D). Moreover, the cellular distribution of TGase within HeLa cells stably expressing the constitutively active form of Ras, as well as in MDAMB231 cells, was unchanged when the cells were exposed to the TGase inhibitor, MDC, and thus was indistinguishable from their control counterparts (those cells not exposed to MDC) (Fig. 8C, compare the first and second columns). This further demonstrates that the ability of TGase to be localized to the surface of cells does not require its transamidation capability.
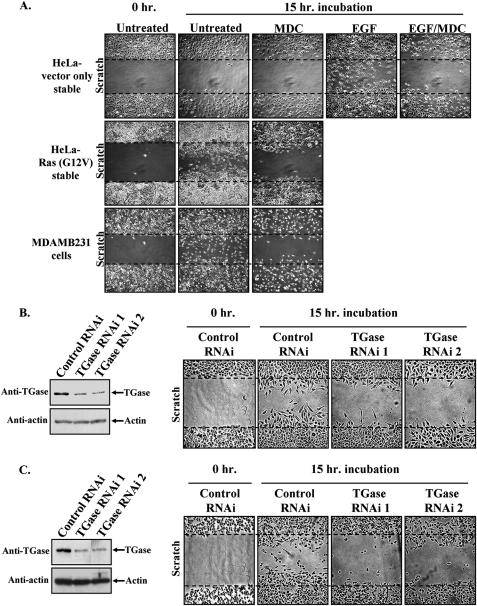
Because elevated levels of TGase activity, and its propensity to be localized to leading edges, are distinctive features exhibited by serum-deprived Ras (G12V)-expressing HeLa cells and MDAMB231 cells, we wondered whether these cell lines could migrate under serum-limiting conditions. This was examined by scratch assays as shown in Fig. 9A. Both the Ras (G12V)-expressing HeLa cells (middle panel) and the MDAMB231 cells (bottom panel) were able to efficiently promote wound closure when cultured in medium devoid of serum or EGF. Moreover, we found that the migration capabilities of each of these cell lines was sensitive to MDC treatment, suggesting that TGase activity is critical for the cell migration responses elicited by the expression of a constitutively activated form of Ras in HeLa cells, and as exhibited by the highly aggressive breast cancer cell line MDAMB231. This idea was further supported by the fact that knocking down TGase expression using siRNAs in the Ras(G12V)-expressing HeLa cells (Fig. 9B, left panel), or in MDAMB231 cells (Fig. 9C, left panel) also blocked the ability of these cells to migrate (Fig. 9, B and C, right panels). Based on these findings, we therefore conclude that enhanced TGase activity and the accumulation of TGase at the leading edges of cancer cells may serve as critical markers of tumor cell aggressiveness, including those tumors where their aggressive behavior is dependent on EGF receptor-signaling activities.
FIGURE 9.
Blocking the TGase activity associated with Ras (G12V)-expressing HeLa cells and MDAMB231 cells diminishes their ability to migrate. A, scratch assays were performed on the stable HeLa cell lines and MDAMB231 cells treated without (Untreated) or with EGF ± MDC. B and C, analogous scratch assays were also performed on Ras(G12V)-expressing HeLa cells and MDAMB231 in which TGase expression was knocked down by siRNA. The blot in B shows the amount of knockdown in the stable HeLa cell line, whereas the blot in C shows the amount of TGase knockdown achieved in the MDAMB231 cells. Fifteen hours after scratching the wounds, the cells were fixed and visualized by light microscopy to determine the extent of wound closure. One untreated plate of cells was fixed immediately after scratching the wound (0 h-Untreated) to show the size of the wounds at the start of the assay. Each panel represents a single image that shows both edges of a wound that was scratched in a monolayer of cells exposed to the indicated culturing conditions. The widths of the initially scratched wounds are indicated with dashed lines.
DISCUSSION
Determining the molecular mechanisms responsible for the aggressive phenotype of human cancer cells is particularly important, not only for furthering our understanding of cancer progression, but also for the development of novel intervention points for cancer therapies. One of the hallmark characteristics of malignant transformation is the ability of growth factors like EGF to stimulate cell migration and invasiveness (10, 27, 34). We have used the cervical carcinoma HeLa cell line as a model to identify participants in these EGF-stimulated activities. Here we show that TGase is one such protein that plays an essential role in the ability of EGF to stimulate HeLa cell migration and invasive activity. The initial indication that this was the case came from our findings that treating HeLa cells with EGF led to the activation of the GTP-binding and enzymatic transamidation activities of TGase, as well as the accumulation of TGase along the leading edges of actively migrating cells. We then showed that by either knocking down TGase expression in HeLa cells or by using MDC, a competitive transamidation inhibitor that binds at the active site of TGase, EGF was no longer capable of stimulating cell migration or invasion. Interestingly, although ectopically expressed TGase in HeLa cells exhibits both GTP-binding and transamidation activities, it is unable to mimic the actions of EGF and trigger a cell migration response. Thus, TGase activation is necessary but not sufficient for mediating the aggressive behavior exhibited by EGF-treated HeLa cells. Indeed, the EGF-stimulated recruitment of TGase to the leading edges of cells is likely to be an important step, along with TGase activation, in the ability of this growth factor to promote cell migration and invasive activity.
Certainly, a question of interest concerned how EGF signals the activation and recruitment of TGase to the leading edges of cells. Based on previous work where we demonstrated that the up-regulation of TGase expression and the activation of its GTP-binding and transamidation activities in various cell lineages stimulated with RA and/or EGF were dependent on PI3K activity (24, 35), we anticipated that PI3K would play a similar role in the EGF-mediated regulation of TGase function in HeLa cells. However, blocking PI3K activity with LY294002 did not affect the ability of EGF to stimulate TGase activation in HeLa cells. Rather, we found that EGF-dependent signaling to JNK, a member of the MAPK family, is essential for the activation of TGase as well as its recruitment to the leading edge, and consequently for EGF-stimulated cell migration. The role of JNK in EGF signaling to TGase appears to be highly specific, because inhibitors of several other signaling molecules, including other members of the MAPK family (i.e. ERK and p38), were ineffective at blocking EGF-induced TGase activity or its recruitment to the leading edges of cells. It is especially interesting that the same EGF-signaling pathway is responsible for both of these aspects of TGase regulation. This raises the possibility that the EGF-dependent accumulation of TGase at the leading edges of cells and the EGF-stimulated activation of TGase, are coupled events. It may be that TGase is maintained in an “inactive” conformational state, perhaps through some type of regulatory interaction with a binding partner while in the cytosol, but that EGF-dependent signals that recruit TGase to the cell surface also relieve this inhibitory constraint. It seems likely that EGF works through Ras to signal to JNK and regulate TGase function, as cells expressing oncogenic Ras can by-pass the need for EGF stimulation to recruit TGase to the leading edges of cells and activate its GTP-binding and enzymatic activities. Moreover, the requirement for EGF stimulation can be circumvented in MDAMB231 cells, where TGase exhibits constitutive activation and accumulation at their leading edges, and correspondingly, these cells are highly motile and invasive even in the absence of growth factor treatment.
We still do not fully understand how EGF signaling through Ras and JNK brings TGase to the leading edges of cells, although we assume it is the outcome of a JNK-catalyzed phosphorylation event, nor is it yet clear how TGase plays an essential role in EGF-stimulated cell migration and invasiveness. Such questions have also been challenging to answer for other proteins shown to accumulate at the leading edges of migrating cells upon their exposure to different extracellular stimuli and signaling cues (11, 14, 28, 36, 37). The EGF-stimulated recruitment of TGase to the leading edges of migrating HeLa cells may bring it into close proximity to a specific protein transamidation substrate or a set of protein substrates that need to be cross-linked in order for cell motility to occur. Although a number of signaling and cytoskeletal proteins have been identified in screens as putative TGase substrates (19), we have not yet been able to implicate any of these potential substrates in the TGase-dependent cell migration response in cancer cells. However, recently we have obtained a potentially interesting clue regarding how TGase might play a role in cell migration, stemming from the finding that it can form a stable complex with actin as measured by the co-immunoprecipitation of these proteins from HeLa cells as well as from certain human breast cancer cell lines.3 What is particularly intriguing about these findings is that while wild-type TGase is capable of forming a stable complex with actin, a transamidation-defective form of TGase in which the active-site cysteine residue has been changed to valine, is unable to be co-immunoprecipitated with actin. Although we have demonstrated that actin can act as a transamidation substrate of TGase in vitro, as indicated by the appearance of higher molecular weight (cross-linked) forms of actin when recombinant forms of actin and TGase are combined in the presence of Ca2+, we have found no indication that actin is similarly modified by TGase, either in migrating HeLa or MDAMB231 cells (data not shown). Moreover, the fact that the enzymatically competent form of TGase undergoes stable complex formation with actin, would seem to argue against actin serving as a transamidation substrate, otherwise once it was cross-linked to an acceptor protein, it would be expected to dissociate from TGase (i.e. the typical product release from an enzyme). Thus, these findings seem to suggest that the enzyme active site of TGase might be involved in directly binding to actin, perhaps enabling TGase to serve as a scaffold for the recruitment of other proteins that influence actin polymerization or other effects on actin dynamics in cells. Future efforts will be directed at further examining this intriguing possibility.
Increasing amounts of recent evidence have linked TGase to human cancer progression. For example, TGase overexpression and/or its aberrant activation have been noted in a significant percentage of the more aggressive brain, breast, prostate, ovarian, and pancreatic tumors and tumor-derived cell lines (20–22, 38). Moreover, inhibiting the chronic TGase activity found in some of these cancer cell lines (e.g. MDAMB231 cells) by either knocking down TGase expression using RNAi or incubating the cells with TGase inhibitors, has been shown here and in other studies to sensitize cancer cells to chemotherapeutic challenges, as well as limit their aggressive behavior (20, 24–26). Although additional work is needed to further establish the relationship between the EGF-dependent signaling activities and TGase in other types of cancer, our findings highlight TGase as a potentially important point for therapeutic intervention against those human malignancies whose aggressive phenotype is coupled to EGF receptor expression and activation.
Acknowledgment
We thank Cindy Westmiller for her expert secretarial assistance.
3 B. Li and M. Antonyak, unpublished data.
- EGF
- epidermal growth factor
- BPA
- 5-(biotinamido)pentylamine
- ERK
- extracellular-signal-regulated protein kinase
- MAPK
- mitogen-activated protein kinase
- MEK
- MAPK/ERK kinase
- MDC
- monodansylcadaverine
- PI3K
- phosphatidylinositol 3-kinase
- RA
- retinoic acid
- TGase
- tissue transglutaminase
- JNK
- c-Jun N-terminal kinase
- RNAi
- RNA interference
- siRNA
- small interference RNA
- HA
- hemagglutinin
- DAPI
- 4′,6-diamidino-2-phenylindole
- FBS
- fetal bovine serum.
REFERENCES
- 1.Citri A., Yarden Y. ( 2006) Nat. Rev. Mol. Cell Biol. 7, 505– 516 [DOI] [PubMed] [Google Scholar]
- 2.Henson E. S., Gibson S. B. ( 2006) Cell. Signal. 18, 2089– 2097 [DOI] [PubMed] [Google Scholar]
- 3.Katz M., Amit I., Yarden Y. ( 2007) Biochim. Biophys. Acta 1773, 1161– 1176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Laskin J. J., Sandler A. B. ( 2004) Cancer Treat. Rev. 30, 1– 17 [DOI] [PubMed] [Google Scholar]
- 5.Zandi R., Larsen A. B., Andersen P., Stockhausen M. T., Poulsen H. S. ( 2007) Cell. Signal. 19, 2013– 2023 [DOI] [PubMed] [Google Scholar]
- 6.Bublil E. M., Yarden Y. ( 2007) Curr. Opin. Cell Biol. 19, 124– 134 [DOI] [PubMed] [Google Scholar]
- 7.Li G., Wong A. J. ( 2008) Expert Rev. Vaccines 7, 977– 985 [DOI] [PubMed] [Google Scholar]
- 8.Moscatello D. K., Ramirez G., Wong A. J. ( 1997) Cancer Res. 57, 1419– 1424 [PubMed] [Google Scholar]
- 9.Antonyak M. A., Kenyon L. C., Godwin A. K., James D. C., Emlet D. R., Okamoto I., Tnani M., Holgado-Madruga M., Moscatello D. K., Wong A. J. ( 2002) Oncogene 21, 5038– 5046 [DOI] [PubMed] [Google Scholar]
- 10.Katz M., Amit I., Citri A., Shay T., Carvalho S., Lavi S., Milanezi F., Lyass L., Amariglio N., Jacob-Hirsch J., Ben-Chetrit N., Tarcic G., Lindzen M., Avraham R., Liao Y. C., Trusk P., Lyass A., Rechavi G., Spector N. L., Lo S. H., Schmitt F., Bacus S. S., Yarden Y. ( 2007) Nat. Cell Biol. 9, 961– 969 [DOI] [PubMed] [Google Scholar]
- 11.Sahai E. ( 2007) Nat. Rev. Cancer 7, 737– 749 [DOI] [PubMed] [Google Scholar]
- 12.Hill K., Welti S., Yu J., Murray J. T., Yip S. C., Condeelis J. S., Segall J. E., Backer J. M. ( 2000) J. Biol. Chem. 275, 3741– 3744 [DOI] [PubMed] [Google Scholar]
- 13.Piccolo E., Innominato P. F., Mariggio M. A., Maffucci T., Iacobelli S., Falasca M. ( 2002) Oncogene 21, 6520– 6529 [DOI] [PubMed] [Google Scholar]
- 14.Yip S. C., El-Sibai M., Coniglio S. J., Mouneimne G., Eddy R. J., Drees B. E., Neilsen P. O., Goswami S., Symons M., Condeelis J. S., Backer J. M. ( 2007) J. Cell Sci. 120, 3138– 3146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Huang C., Jacobson K., Schaller M. D. ( 2004) J. Cell Sci. 117, 4619– 4628 [DOI] [PubMed] [Google Scholar]
- 16.Hauck C. R., Sieg D. J., Hsia D. A., Loftus J. C., Gaarde W. A., Monia B. P., Schlaepfer D. D. ( 2001) Cancer Res. 61, 7079– 7090 [PubMed] [Google Scholar]
- 17.Pouliot N., Nice E. C., Burgess A. W. ( 2001) Exp. Cell Res. 266, 1– 10 [DOI] [PubMed] [Google Scholar]
- 18.Cui X., Kim H. J., Kuiatse I., Kim H., Brown P. H., Lee A. V. ( 2006) Cancer Res. 66, 5304– 5313 [DOI] [PubMed] [Google Scholar]
- 19.Lorand L., Graham R. M. ( 2003) Nat. Rev. Mol. Cell Biol. 4, 140– 156 [DOI] [PubMed] [Google Scholar]
- 20.Hwang J. Y., Mangala L. S., Fok J. Y., Lin Y. G., Merritt W. M., Spannuth W. A., Nick A. M., Fiterman D. J., Vivas-Mejia P. E., Deavers M. T., Coleman R. L., Lopez-Berestein G., Mehta K., Sood A. K. ( 2008) Cancer Res. 68, 5849– 5858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Singer C. F., Hudelist G., Walter I., Rueckliniger E., Czerwenka K., Kubista E., Huber A. V. ( 2006) Clin. Exp. Metastasis 23, 33– 39 [DOI] [PubMed] [Google Scholar]
- 22.Verma A., Guha S., Diagaradjane P., Kunnumakkara A. B., Sanguino A. M., Lopez-Berestein G., Sood A. K., Aggarwal B. B., Krishnan S., Gelovani J. G., Mehta K. ( 2008) Clin. Cancer Res. 14, 2476– 2483 [DOI] [PubMed] [Google Scholar]
- 23.Yuan L., Siegel M., Choi K., Khosla C., Miller C. R., Jackson E. N., Piwnica-Worms D., Rich K. M. ( 2007) Oncogene 26, 2563– 2573 [DOI] [PubMed] [Google Scholar]
- 24.Antonyak M. A., Miller A. M., Jansen J. M., Boehm J. E., Balkman C. E., Wakshlag J. J., Page R. L., Cerione R. A. ( 2004) J. Biol. Chem. 279, 41461– 41467 [DOI] [PubMed] [Google Scholar]
- 25.Mangala L. S., Fok J. Y., Zorrilla-Calancha I. R., Verma A., Mehta K. ( 2007) Oncogene 26, 2459– 2470 [DOI] [PubMed] [Google Scholar]
- 26.Kim D. S., Park S. S., Nam B. H., Kim I. H., Kim S. Y. ( 2006) Cancer Res. 66, 10936– 10943 [DOI] [PubMed] [Google Scholar]
- 27.Antonyak M. A., McNeill C. J., Wakshlag J. J., Boehm J. E., Cerione R. A. ( 2003) J. Biol. Chem. 278, 15859– 15866 [DOI] [PubMed] [Google Scholar]
- 28.Zuo X., Zhang J., Zhang Y., Hsu S. C., Zhou D., Guo W. ( 2006) Nat. Cell Biol. 8, 1383– 1388 [DOI] [PubMed] [Google Scholar]
- 29.Chiocca E. A., Davies P. J., Stein J. P. ( 1988) J. Biol. Chem. 263, 11584– 11589 [PubMed] [Google Scholar]
- 30.Piacentini M., Fesus L., Sartori C., Ceru M. P. ( 1988) Biochem. J. 253, 33– 38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sporn M. B., Roberts A. B. ( 1983) Cancer Res. 43, 3034– 3040 [PubMed] [Google Scholar]
- 32.Mann A. P., Verma A., Sethi G., Manavathi B., Wang H., Fok J. Y., Kunnumakkara A. B., Kumar R., Aggarwal B. B., Mehta K. ( 2006) Cancer Res. 66, 8788– 8795 [DOI] [PubMed] [Google Scholar]
- 33.Datta S., Antonyak M. A., Cerione R. A. ( 2007) Biochemistry 46, 14819– 14829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Malumbres M., Barbacid M. ( 2003) Nat. Rev. Cancer 3, 459– 465 [DOI] [PubMed] [Google Scholar]
- 35.Antonyak M. A., Boehm J. E., Cerione R. A. ( 2002) J. Biol. Chem. 277, 14712– 14716 [DOI] [PubMed] [Google Scholar]
- 36.Chen H. Y., Shen C. H., Tsai Y. T., Lin F. C., Huang Y. P., Chen R. H. ( 2004) Mol. Cell. Biol. 24, 10558– 10572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Huang C., Jacobson K., Schaller M. D. ( 2004) Cell Cycle 3, 4– 6 [PubMed] [Google Scholar]
- 38.Verma A., Wang H., Manavathi B., Fok J. Y., Mann A. P., Kumar R., Mehta K. ( 2006) Cancer Res. 66, 10525– 10533 [DOI] [PubMed] [Google Scholar]