Abstract
To investigate drug mechanisms of action and identify molecular targets for the development of rational drug combinations, we conducted synthetic small interfering RNA (siRNA)-based RNAi screens to identify genes whose silencing affects anti-cancer drug responses. Silencing of RRM1 and RRM2, which encode the large and small subunits of the human ribonucleotide reductase complex, respectively, markedly enhanced the cytotoxicity of the topoisomerase I inhibitor camptothecin (CPT). Silencing of RRM2 was also found to enhance DNA damage as measured by histone γ-H2AX. Further studies showed that CPT up-regulates both RRM1 and RRM2 mRNA and protein levels and induces the nuclear translocation of RRM2. The checkpoint kinase 1 (Chk1) was up-regulated and activated in response to CPT, and CHEK1 down-regulation by siRNA and small molecule inhibitors of Chk1 blocked RRM2 induction by CPT. CHEK1 siRNA also suppressed E2F1 up-regulation by CPT, and silencing of E2F1 suppressed the up-regulation of RRM2. Silencing of ATR or ATM and inhibition of ATM activity by KU-55933 blocked Chk1 activation and RRM2 up-regulation. This study links the known components of CPT-induced DNA damage response with proteins required for the synthesis of dNTPs and DNA repair. Specifically, we propose that upon DNA damage, Chk1 activation, mediated by ATM and ATR, up-regulates RRM2 expression through the E2F1 transcription factor. Up-regulation in RRM2 expression levels coupled with its nuclear recruitment suggests an active role for ribonucleotide reductase in the cellular response to CPT-mediated DNA damage that could potentially be exploited as a strategy for enhancing the efficacy of topoisomerase I inhibitors.
Two water-soluble DNA topoisomerase 1 (Top1)2 inhibitors, derived from camptothecin (CPT), are in clinical use; topotecan, for the treatment of ovarian and lung cancers, and irinotecan, for colorectal cancers. Further CPT derivatives and non-CPT Top1 inhibitors are in preclinical development as anticancer agents (1–3). Despite the fact that camptothecins are highly targeted agents with Top1 as their sole cellular target, the response of cancer cells to the inhibition of Top1 by camptothecins is highly variable and remains for the most part undefined (2, 4, 5). One of the critical mechanism for the antiproliferative activity of camptothecins is the generation of replication-associated DNA double-strand breaks by collisions between replication forks and drug-stabilized Top1 cleavage complexes (6, 7), which results in phosphorylation of histone H2AX (8) that can be detected as histone γH2AX foci (9, 10).
Synthetic siRNA-based RNAi screening is emerging as a powerful approach to revealing the determinants of cellular responses to drugs. Using a synthetic siRNA-based RNAi screen to identify genes whose silencing affects the activity of CPT, we found that RNAi against RRM1 and RRM2, both ribonucleotide reductase genes, markedly enhanced the cytotoxicity of CPT.
The ribonucleotide reductase (RNR) enzyme complex is essential for the de novo synthesis of deoxyribonucleotides (dNTPs) precursors for DNA synthesis. RNR catalyzes the reduction of ribonucleoside diphosphates to deoxyribonucleoside diphosphates and maintains a highly regulated and balanced pool of dNTPs for DNA replication and repair. A failure in the control of dNTP levels leads to cell death or genetic abnormalities (11, 12).
In mammals, RNR is an heterodimeric tetramer composed of two identical large subunits RRM1 and two identical small subunit RRM2 (11). Each RRM1 subunit contains an active site (controlling enzyme activity) and an allosteric site (controlling substrate specificity by binding nucleoside triphosphates). Each RRM2 subunits contains a non-heme (binuclear) iron center and a stable tyrosyl free radical. Both are essential for catalysis (11). Recently, an additional small subunit has been found, RRM2B (p53R2), which is induced by p53 and can substitute for RRM2 to form a highly active RNR complex involved in DNA repair (for review, see Ref. 11).
RNR activity is closely regulated during the cell cycle, peaking in S-phase. In yeast, expression of the large subunit RNR1 fluctuates more than 10-fold during the cell cycle, whereas the small subunit RNR2 mRNA levels show only a 2-fold change. In mammalian cells, RRM2 protein levels begin to rise in late G1 and reach their highest level during S-phase, whereas the levels of RRM1 remain relatively constant throughout the cell cycle. Fluctuations in RRM2 protein levels have been attributed to both transcriptional up-regulation during S-phase and proteasome-mediated degradation as cells enter mitosis (11).
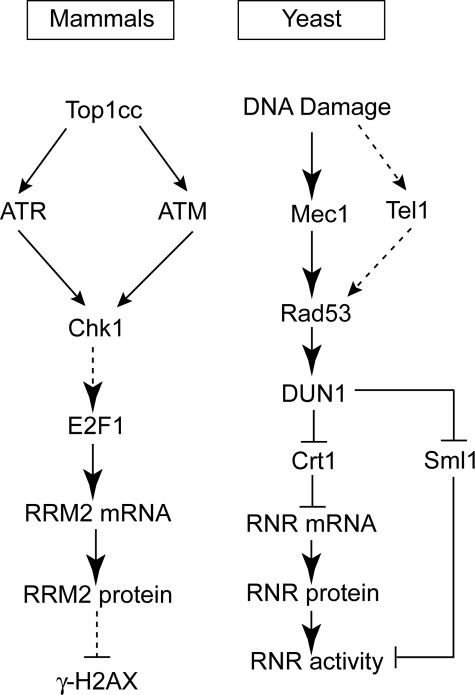
DNA damage also regulates RNR activity. In budding yeast RNR up-regulation (14, 15) depends on the protein kinases Mec1 and Rad53. Mec1 initiates Rad53 activation by phosphorylating Rad53, and Rad53 is further activated by autophosphorylation. Activated Rad53 up-regulates RNR by phosphorylating Dun1, another protein kinase (16). Activated (phosphorylated) Dun1 up-regulates RNR by at least two routes (see Fig. 7). The first is through phosphorylation of Sml1 (17, 18), which dissociates Sml1 from RNR and de-represses the activity of RNR (19, 20). The second route by which Dun1 up-regulates RNR is through phosphorylation of Crt1, which dissociates Crt1 from the RNR promoter and de-represses RNR gene transcription (21). Thus, in budding yeast, the DNA damage response kinases Mec1-Rad53-Dun1 act as positive regulators of RNR both at the transcriptional and post-transcriptional levels.
FIGURE 7.
Scheme of RRM2 induction after CPT treatment. Briefly, DNA damage induced by Top1-DNA covalent complex (Top1cc) induces the activation of Chk1, mediated by ATM and ATR, which up-regulates RRM2 expression through the transcription factor E2F1.
In mammalian cells evidence is limited regarding the regulation of RNR by the orthologs of the yeast Mec1 and Rad53, ATM/ATR and Chk1/Chk2, respectively (22, 23). Nevertheless, the mammalian Crt1 ortholog, Rfx1, has been found to bind to the RNR2 gene and block its transcription (14). Meanwhile, RNR genes contain E2F binding sites and could be activated at the transcription level by E2F1 overexpression in quiescent cells before the induction of S-phase (13, 24). E2F1 activity is also up-regulated in response to CPT (25). A recent study also provided evidence for RNR2 regulation by Chk1 for S-phase progression (23). Ataxia telangiectasia (AT) cells are also deficient in activating p53R2 compared with normal wild type cells (26, 27). Our present findings indicate that up-regulation of RRM2 transcription in response to DNA damage in human cells involves an ATR/ATM-Chk1-E2F1 pathway, which is reminiscent of the Mec1-Rad53-Dun1 pathway in budding yeast (see Fig. 7).
EXPERIMENTAL PROCEDURES
Cell Lines and Chemicals
MDA-MB-231 breast cancer cells and HCT-116 colorectal cancer cells were obtained from the Developmental Therapeutics Program (NCI, National Institutes of Health, nci.nih.gov) and were maintained in RPMI 1640 medium containing 10% fetal bovine serum. All siRNAs were obtained from Qiagen Inc. (Germantown, MD). CPT was obtained from Sigma. UCN-01 was obtained from the Developmental Therapeutics Program. CHIR124 was a kind gift from Chiron Corp. KU-55933 was a kind gift from KuDOS Pharmaceuticals Ltd. (Cambridge, UK). CPT and KU-55933 were prepared at 10 mm in DMSO. UCN-01 and CHIR124 were prepared at 1 mm in DMSO. Drug stock solutions were separated into aliquots at −20 °C. All drugs were diluted to desired concentrations in full medium immediately before each experiment. The final DMSO concentrations did not exceed 0.1%.
siRNA Screening
The CPT chemosensitization RNAi screen was performed using a library of synthetic siRNAs targeting ≈400 genes associated with cancer and using a previously described multiplexed siRNA screening strategy (see Martin et al. (28) for details). Multiplexes comprised six siRNAs corresponding to three unique gene targets (two siRNAs per gene). Multiplexes were evaluated at a final concentration of 60 nm (10 nm of each individual siRNA) in a 96-well-plate format. Only the interior 60 wells were used. Transfections were performed by precomplexing siRNA (6 pmol) with Oligofectamine lipid transfection reagent (Invitrogen) in 50 μl of serum-free RPMI in individual plate wells for 30 min at ambient temperature. MDA-MB-231 cells (4500) were added in 50 μl of RPMI supplemented with 10% fetal bovine serum to yield transfection mixtures consisting of 60 nm total siRNA in RPMI with 5% fetal bovine serum. This final mixture was incubated at ambient temperature for 45 min before being placed at 37 °C in a humidified atmosphere containing 5% CO2. The library was screened in duplicate (half intended to receive CPT treatment and half to establish siRNA multiplex basal activity). After 48 h the medium was removed, and 100 μl of fresh medium containing either CPT (≈EC50, 0.1% DMSO) or vehicle only (0.1% DMSO) was added, and the cells incubated for an additional 48 h at 37 °C. After this time, cell viability was assayed (Cell Titer Blue Reagent, Promega, Madison, WI). Plate median values were used for normalization. Multiplexes were assayed in duplicate. For deconvolution studies, the two siRNAs targeting a given gene were evaluated as a pair, each used at 10 nm, and compared with cells transfected with negative control siRNA (siNeg) (20 nm).
Gene-specific RNAi and Transfection
For RNA analysis, transfections were performed as described for screening, except that 2500 cells were seeded. Unless otherwise stated, after 48 h the medium was removed, 100 μl of fresh medium containing CPT (1 μm, 0.1% DMSO) was added, and the cells were incubated at 37 °C for 24–72 h depending on the end assay. For protein analysis, transfections were performed in 6-well plates with Oligofectamine reagent. Cells transfected with negative control siRNA were used for comparison. The target sequences for the experimental siRNAs are listed in supplemental Table 1.
Bromodeoxyuridine Incorporation
After 30 min of pulse-labeling with 50 μmol/liter bromodeoxyuridine (BrdUrd, Calbiochem), cells were collected and stained with fluorescein isothiocyanate-conjugated anti-BrdUrd antibody (BD Biosciences). Cells were resuspended in 500 μl of propidium iodide (PI) solution (50 μg/ml PI and 50 μg/ml RNase A) and analyzed with a FACScan flow cytometer (BD Biosciences).
RNA Analysis
Gene-specific transcript levels were measured using a branched DNA-based assay (QuantiGene Reagent System, Panomics, Fremont, CA) in single or multiplex formats. The single gene assay format has been described previously (28). In this study probes corresponding to RRM1, RRM2, or CTNNB1 (control) and human cyclophilin (PPIB) (for normalization) (Panomics) were used. The multiplex format (Quantigene® Plex assay, Panomics) was performed using a custom-designed panel consisting of 20 different XMAP® beads, each conjugated with a probe set corresponding to a different mRNA. The mRNAs assayed included genes associated with this study (RRM2, RRM1, E2F1, CHEK1, CHEK2, ATM, and ATR). Bead identity was used to identify each mRNA species, and the fluorescent signal from each bead (Bioplex, Bio-Rad) was used to quantify the amount of RNA.
Western Blot
Protein levels were measured by Western blot with corresponding specific primary antibodies, including those against RRM2, Chk1, and E2F1 (Santa Cruz, CA), phosphorylated-H2AX (γ-H2AX) (Upstate/Millipore), RRM1 (Chemicon/Millipore), glyceraldehyde-3-phosphate dehydrogenase, and phosphorylated-Chk1(Ser-317) (Cell Signaling). Shown are the representative data from separate experiments.
Immunofluorescence Assays
MDA-MB-231 cells plated in four-well chamber slides (Nalgene Nunc International, Rochester, NY) were used to study the subcellular localization of RRM2. After drug treatment, cells were fixed for 20 min with 4% paraformaldehyde in phosphate-buffered saline (PBS) (pH 7.4) and washed twice with PBS. After incubation for 20 min with 70% ethanol and washing with PBS, the cells were incubated in blocking buffer (8% bovine serum albumin in PBS) for 1 h before incubation for 2 h with primary antibodies against RRM2 (Santa Cruz, CA). Slides were incubated for an additional 1 h with the Alex488-conjugated secondary antibody (Alexa Fluor® 488 donkey anti-goat IgG, Molecular Probes/Invitrogen). After three washes in PBS, cells were stained with 0.5 μg/ml PI and 100 μg/ml RNaseA (Sigma) for 15 min in the dark. Finally, slides were washed with PBS for three times and mounted with Vectashield anti-fade mounting media (Vector Laboratories, Inc., Burlingame, CA). Images were taken using a Nikon Eclipse TE-300 confocal microscope.
Statistical Analyses
All the data are represented as mean values ± S.D. The significance of differences between means was assessed by the Student's t test, with p < 0.05 being considered statistically significant.
RESULTS
RRM1 and RRM2 Gene Silencing Enhances CPT Cytotoxicity
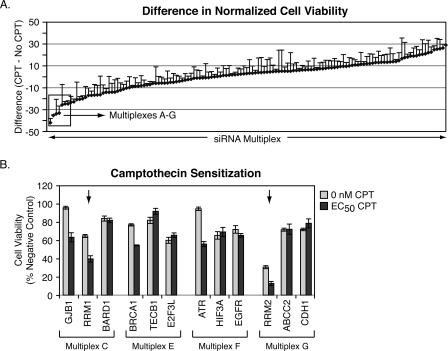
Early studies of CPT showed this compound was cytotoxic to breast cancer cell lines (29); however, adverse in vivo side effects and/or drug resistance due to overexpression of ABCG2 (30, 31) have limited the clinical development of the CPT derivatives as main line treatments for breast cancer. Recent studies have, however, reconsidered the potential of CPT in treatment of advanced breast cancer alone and in combination with other chemotherapeutic agents (32). A rational approach to determining additional pathways that could be used to improve the application of Top1 inhibitors in advanced breast cancer could be revealed by RNAi screening in an appropriate model system. The breast cancer MDA-MB-231 cell line is frequently used as a cellular model of triple-negative, basal-like breast cancer as it lacks expression of the estrogen receptor, progesterone receptor, and ERBB2 (HER2/Neu). A multiplex siRNA-based RNAi screen (28) was performed in the MDA-MB-231 to identify genes that modulated the activity of CPT (Fig. 1A). A duplicate screen showed good correlation (r = 0.8, data not shown). Deconvolution of the top sensitizing siRNA multiplexes identified RRM1 and RRM2 as genes whose silencing not only affected cell viability alone but also significantly enhanced the cytotoxicity of CPT (multiplexes C and G, Fig. 1B). As expected (33), BRCA1 and ATR knockdowns also produced CPT sensitization (multiplexes E and F, respectively). This screen also identified gap junction protein β1 (GJB1), a member of the connexin family of proteins, as an additional gene whose silencing enhances CPT toxicity. Notably, connexin proteins have been linked to mechanisms of cell death (34). However, this target was not pursued further in the context of this study.
FIGURE 1.
siRNA multiplex screen and follow-up deconvolution analysis. A, results from the CPT chemosensitization RNAi screen conducted in MDA-MB-231 breast cancer cells. The y axis represents the normalized activity of each siRNA multiplex in the presence of CPT (EC50) minus the normalized activity of each siRNA multiplex in the absence of CPT. Plate median values were used for normalization. B, deconvolution of the top seven sensitizing multiplexes (designate A–G), with values greater than 2 S.D. from the median, identified RRM1 and RRM2 as gene targets whose silencing significantly enhanced the cytotoxicity of CPT. Targets known to be associated with cellular response to CPT-induced DNA damage were also identified (BRCA1 and ATR) (33).
DNA Replication under RRM2 Knockdown
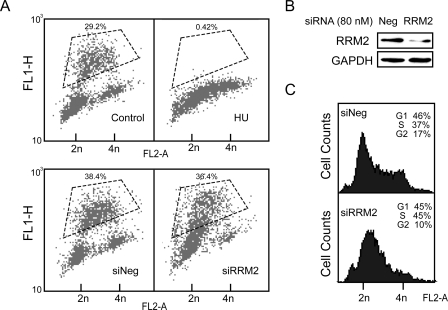
HU inactivates RNR by targeting specifically the RRM2 subunit (35). In contrast to RRM2 knockdown (Fig. 1B), HU is known to protect from the cytotoxicity of CPT (7, 36). To clarify the difference between the effects of HU and RRM2 silencing on CPT cytotoxicity, a BrdUrd incorporation assay was used to detect the DNA replication in cells treated with HU or with RRM2 siRNA. Five mm HU blocked DNA replication totally after 24 h of treatment (Fig. 2A). On the other hand, although the RRM2 protein level was knocked down with siRNA (Fig. 2B), only limited effects on cell cycle distribution were produced. siRNA against RRM2 produced an increase of S-phase cells but without fully arresting DNA synthesis as most cells in S-phase continued to incorporate BrdUrd (Fig. 2C). Thus, RRM2 knockdown, unlike HU, only partially reduced DNA synthesis.
FIGURE 2.
The effect of RRM2 knockdown on DNA replication and cell cycle. MDA-MB-231 cells were transfected with 80 nm RRM2 siRNA or treated with 5 mm HU for 24 h. A, representative FACScan analyses after pulse-labeling with BrdUrd. BrdUrd was added in the last 30 min of treatment. BrdUrd and PI incorporation are plotted on the y and x axes, respectively. Gated regions defined from the control cells without BrdUrd (data not shown) were used to calculate BrdUrd incorporation relative to total positive signal. B, knockdown of RRM2 protein levels with RRM2 siRNA; siNeg, negative control siRNA. GAPDH, glyceraldehyde-3-phosphate dehydrogenase. C, representative FACScan analyses with PI staining.
RRM2 Induction by CPT Treatment
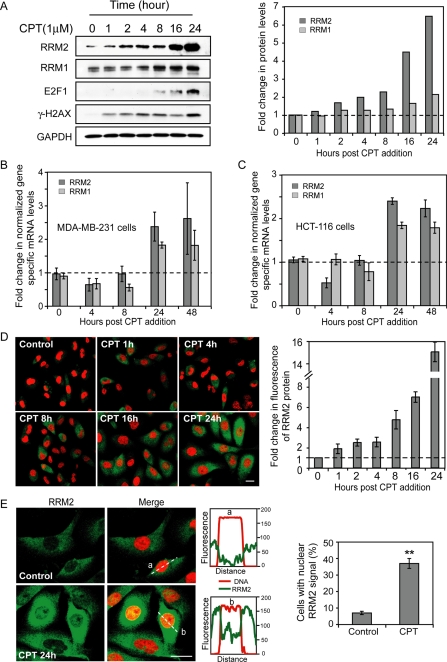
Because in yeast cells, the two RNR genes encoding the small (RNR2) and large (RNR3) subunits are both up-regulated at the transcriptional level in response to DNA damage (15), we hypothesized that CPT could also up-regulate the mammalian RNR genes, which may form part of the basis for the sensitization to CPT seen after RNAi against the RNR genes. To assess this possibility we examined the protein and mRNA levels of RRM1 and RRM2 after treatment of MDA-MB-231 cells with CPT. At the protein level both RRM2 and RRM1 were induced, and RRM2 showed the greatest increase after CPT addition (Fig. 3A). At the mRNA level, RRM2 expression was induced ∼2.5-fold, and RRM1 was induced about 1.8-fold, 24–48 h post-CPT addition (Fig. 3B). RRM1 and RRM2 induction was also observed in the human colon carcinoma cell line HCT-116 (Fig. 3C).
FIGURE 3.
Induction of RRM2 expression both at the protein and mRNA levels and nuclear translocation of RRM2 in response to CPT. A, time course of RRM2 protein expression after CPT treatment in MDA-MB-231 cells. Left, representative result of Western blots; right, corresponding quantitation of RRM2 protein levels. GAPDH, glyceraldehyde-3-phosphate dehydrogenase. B, RRM2 mRNA induction by CPT (1 μm for the indicated times) in MDA-MB-231cells. C, RRM2 induction in human colon cancer HCT-116 cells treated with 1 μm CPT. D, immunofluorescence detection of RRM2 in MDA-MB-231 cells treated with CPT (1 μm for the indicated times). Left, representative confocal microscope images (×40), bar = 8 μm; right, quantitation of average RRM2 fluorescence signal per cell; the level in untreated cells was defined as 1. E, CPT-induced translocation of RRM2 from cytoplasm to nucleus. Left, representative immunofluorescence confocal microscopy images (×100), bar = 8 μm; middle, quantitative distribution of the RRM2 and PI signals in two representative cells using the Nikon Eclipse TE-300 confocal microscope software as shown in the graphs; right, quantitation of the ratio of cells with nuclear RRM2 signals with or without CPT treatment.
Because the RRM2 induction after CPT treatment was more pronounced than the RRM1 induction, and because of the regulatory role of the RNR2 subunit in the DNA damage response of yeast, we choose to focus our subsequent studies on RRM2. Immunofluorescence assays were performed to further examine the RRM2 induction by CPT at different time points up to 24 h. The overall RRM2 fluorescence signal per cell rose with drug exposure time, and RRM2 induction was already detectable after 1 h of CPT treatment (Fig. 3D). Moreover, subcellular localization analyses showed translocation of RRM2 from cytoplasm to nucleus in CPT-treated cells. Compared with the untreated control, the population of cells with nuclear RRM2 signal after CPT treatment increased from 7 to 37% (Fig. 3E). Those data confirmed the induction of RRM2 expression in response to CPT and revealed nuclear translocation of RRM2, consistent with the potential role of RRM2 in the DNA repair of Top1-induced DNA damage.
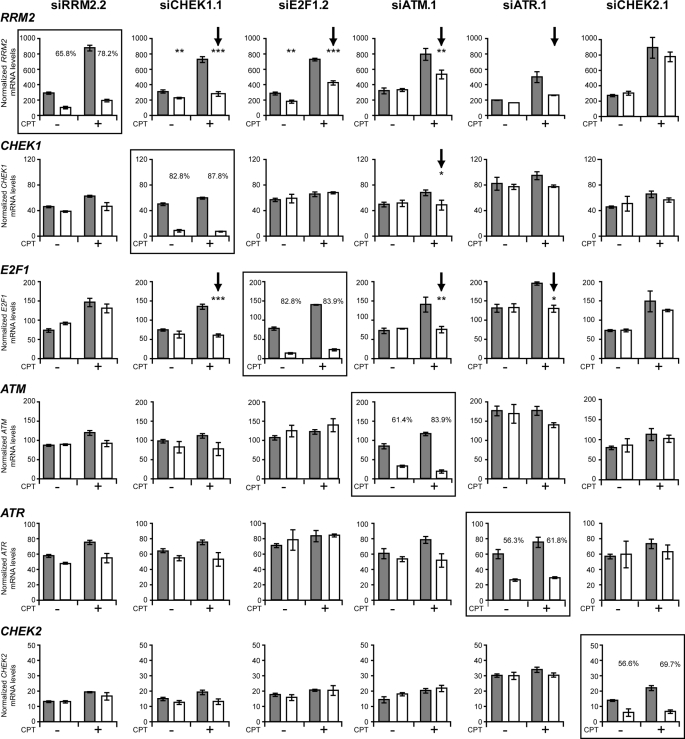
CHEK1, E2F1, ATM, or ATR Gene Knockdown Suppresses the Transcriptional Up-regulation of RRM2 after CPT Treatment
To probe the DNA damage response mechanisms underlying the transcriptional up-regulation of RRM2 induced by CPT, we investigated the effects of RNAi targeting RRM2, E2F1, CHEK1, CHEK2, ATM, and ATR on the expression of those genes in the absence and presence of CPT. Each siRNA showed high efficiency silencing of the corresponding target gene expression in the absence (and presence) of CPT. As seen in Fig. 4, besides RRM2, silencing of CHEK1, E2F1, ATM, or ATR all partially suppressed the up-regulation of RRM2 by CPT. However, CHEK2 silencing had no effect on the RRM2 induction. The silencing of CHEK1 also suppressed the up-regulation of E2F1 expression, and silencing of ATM and ATR blocked both CHEK1 and E2F1 up-regulation at transcription level. Together these results suggested that ATM and ATR regulate RMM2 via CHEK1 and E2F1.
FIGURE 4.
Gene expression after synthetic siRNA transfection of MDA-MB-231 breast cancer cells in the absence (−) and presence (+) of CPT. mRNA levels for each indicated gene were simultaneously measured in MDA-MB-231 cells transfected with either a control siRNA-siNeg (gray bars) or the stated experimental siRNA, indicated at the top of each column (open bars), using a custom designed multiplex branched DNA assay (Panomics Inc., Freemont, CA). The expression of each mRNA (RRM2, CHEK1, E2F1, ATM, ATR, and CHEK2) was normalized to cyclophilin B (PPIB) mRNA levels. Cells were treated with CPT (EC50) or vehicle only (DMSO) 48 h post-siRNA transfection, and RNA levels were assessed 24 h later. Data are shown as the mean ± S.D. of three independent transfections, except for siATR.1, where transfections were conducted in duplicate. t tests (two tailed, unpaired, equal variance) were used to compare siNeg versus siExperimental treatments; *, p < 0.05; **, p < 0.01; ***, p < 0.001. The framed histograms correspond to the homologous siRNA and mRNA; the percentage silencing siExperimental versus siNeg is shown for each gene target. Arrows indicate changes discussed under “Results.”
RRM2 Up-regulation Is Dependent on Chk1
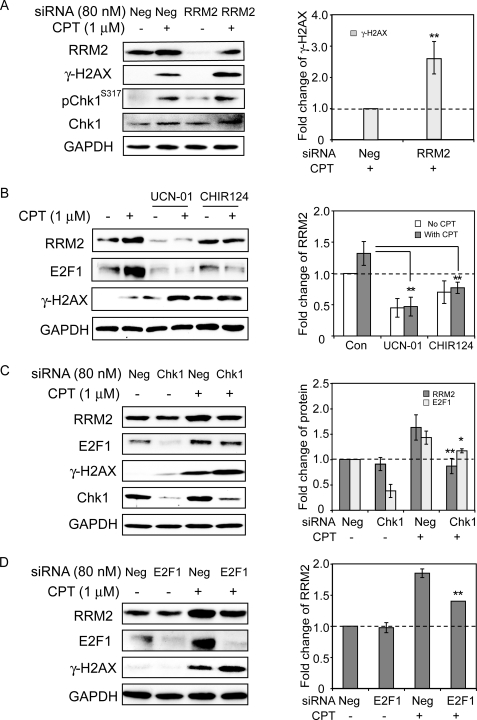
In yeast cells, Rad53 regulates the expression of the ribonucleotide reductase small subunit RNR2 in response to DNA damage (37–39), but in mammalian cells the regulation of RRM2 is still unclear. Of the mammalian homologues of Rad53, Chk1, and Chk2, only the silencing of CHEK1 suppressed the transcription up-regulation of RRM2 in response to CPT, whereas CHEK2 silencing had no effect (Fig. 4). Therefore, we investigated the relationship between Chk1 and RRM2 in MDA-MB-231 cells treated with CPT. Fig. 5 shows that Chk1 was activated by CPT. Indeed, phosphorylated-Chk1 (Chk1-Ser-317) increased upon CPT treatment (Fig. 5A). Both the Chk1 inhibitors UCN-01 and CHIR124 decreased the up-regulation of RRM2 in response to CPT, which suggested that Chk1 was involved in the up-regulation of RRM2 in response to CPT treatment (Fig. 5B). This was further confirmed by knockdown experiments with siRNA, which showed an attenuation of the up-regulation of RRM2 by CHEK1 knockdown (Fig. 5C). CHEK1 knockdown also enhanced the γ-H2AX response to CPT. Taken together, those experiments demonstrate that the up-regulation of RRM2 induced by CPT is Chk1-dependent and may have a protective effect as inactivation of Chk1 enhances CPT-induced DNA damage (9, 40).
FIGURE 5.
Chk1- and E2F1-dependent induction of RRM2 and suppression of γH2AX. A, inhibition of RRM2 induction and enhancement of γ-H2AX after RRM2 knockdown. One day after transfection with 80 nm RRM2 siRNA or negative control siRNA (siNeg), MDA-MB-231 cells were treated with 1 μm CPT for 24 h. Proteins were detected by Western blotting (left). Right, quantitation of γ-H2AX protein levels based on the data of two independent Western blotting experiments; the level in CPT-treated siNeg-transfected cells was defined as 1. GAPDH, glyceraldehyde-3-phosphate dehydrogenase. B, inhibition of RRM2 and E2F1 induction and activation of γ-H2AX by the Chk1 inhibitors UCN-01 or CHIR124. MDA-MB-231 cells were pretreated with UCN-01 (300 nm) or CHIR124 (100 nm) for 1 h before the addition of CPT for 24 h. Left, Western blotting. Right, quantitation of RRM2 protein levels based on three independent Western blotting experiments. C, Chk1 knockdown inhibits RRM2 up-regulation by CPT and activates γ-H2AX. One day after transfection with 80 nm Chk1 siRNA, MDA-MB-231 cells were treated with 1 μm CPT for 24 h. Left, Western blotting. Right, quantitation of RRM2 and E2F1 protein levels based on three independent Western blotting experiments. Statistical significant difference was shown between siChk1+CPT and siNeg+CPT. D, E2F1 knockdown reduces RRM2 up-regulation and activates γ-H2AX. One day after transfection with 80 nm E2F1 siRNA, MDA-MB-231 cells were treated with 1 μm CPT for 24 h. Left, Western blotting. Right, quantitation of RRM2 protein levels based on two independent Western blotting experiments. Statistical significant difference was shown between siE2F1+CPT and siNeg+CPT. Standard t tests were used for statistical analyses; *, p < 0.05; **, p < 0.01.
The Chk1-dependent RRM2 Up-regulation Is Mediated by E2F1
Because the RNR gene is one of the transcriptional targets of the replication- and stress-associated transcription factor E2F1 (13, 24) and because E2F1 has been involved in the DNA damage response through its phosphorylation by ATM and Chk2 (41, 42), we investigated in more details the role of E2F1 in the up-regulation of RRM2 in response to CPT. After our finding that E2F1 increased at the mRNA level under exposure to CPT (Fig. 4), we examined E2F1 response at the protein level. Fig. 5 (B–D) shows enhanced E2F1 protein signal in cells treated with CPT. To determine the effect of E2F1 on the RRM2 up-regulation, knockdown of E2F1 with siRNA was then introduced. We had already found that E2F1 silencing suppressed RRM2 transcriptional up-regulation in response to CPT (Fig. 4). E2F1 knockdown also blocked the up-regulation of RRM2 at the protein level (Fig. 5D). These data demonstrate that RRM2 up-regulation in response to CPT is regulated by E2F1.
The next set of experiments was done to examine the relationship between Chk1 and E2F1 in the RRM2 up-regulation. After siChk1 silencing, E2F1 base-line expression was reduced at the protein level (Fig. 5C), and the up-regulation of E2F1 by CPT was reduced both at the mRNA (Fig. 4) and protein levels (Fig. 5C). Together, our data indicate that the regulation of RRM2 expression is mediated by Chk1-dependent up-regulation of E2F1.
ATM and ATR Are Required for RRM2 Up-regulation during CPT Treatment
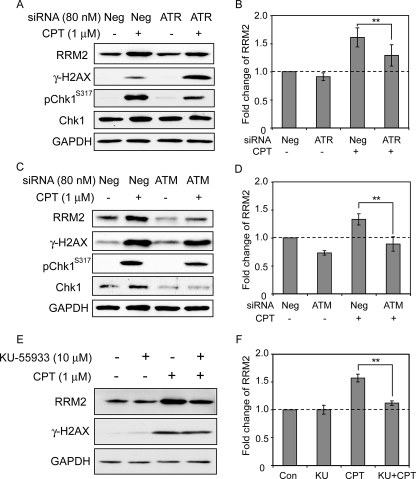
As the data above showed that Chk1 acted as a key operator for RRM2 up-regulation after CPT treatment, we next tested the role of ATR and ATM, the upstream regulatory kinases of Chk1. In yeast cells, Mec1 and Rad53 activate the downstream checkpoint kinase Dun1, which leads to the transcriptional induction of the RNR genes (21). The homologue of Mec1 in mammalian cells, ATR, is well known as the immediate upstream regulator of Chk1 in the DNA damage response (43, 44). Consistent with the possibility that ATR acted as an upstream regulator of Chk1, we found that ATR down-regulation by siRNA reduced RRM2 up-regulation (Fig. 6, A and B). This result is also consistent with ATR as one of the genes whose down-regulation by siRNA sensitized cells to CPT (Fig. 1).
FIGURE 6.
ATR- and ATM-dependent induction of RRM2 and impact on γ-H2AX induction. A, ATR knockdown reduces RRM2 up-regulation. Neg, negative control siRNA. B, quantitation of RRM2 protein levels based on data obtained in two independent experiments. C, ATM knockdown inhibits RRM2 up-regulation by CPT. D, quantitation of RRM2 protein levels based on data obtained in two independent experiments. E, inhibition of RRM2 induction by the ATM inhibitor, KU-55933. Cells were pretreated with KU-55933 (10 μm) for 1 h before the addition of CPT (1 μm for 24 h). Proteins were detected by Western blotting. F, quantitation of RRM2 protein levels based on data obtained in two independent experiments. KU refers to KU-55933. Standard t tests were used for statistical analyses; *, p < 0.05; **, p < 0.01.
Cross-talks are also likely between the upstream phosphatidylinositol 3-kinases (ATM, ATR, and DNA protein kinase) and their downstream kinases (Chk1 and Chk2) (44), and it is well established that ATM is rapidly and strongly activated upon cellular exposure to CPT (9, 45). Evidence that ATM could act upstream from Chk1 and affect RRM2 up-regulation was first obtained at an mRNA level. Indeed, as previously shown in Fig. 4, silencing of ATM suppressed the up-regulation of RRM2 at the transcriptional level as well as repressed the induction of CHK1 and E2F1. At the protein level, ATM down-regulation with siRNA was also found to decrease the RRM2 and Chk1 activations (Fig. 6, C and D). Furthermore, using the ATM inhibitor KU-55933 (46), we found that RRM2 induction was reduced by inhibition of ATM activity. Together, those data demonstrate that ATM inhibition attenuates the CPT-elicited RRM2 up-regulation, which indicates that both ATM and ATR function as upstream regulators of Chk1 and are involved in the RRM2 up-regulation in response to Top1-induced DNA damage.
RRM2 Up-regulation Limits DNA Damage Induction by CPT Measured by γ-H2AX
Phosphorylation of histone H2AX on serine 139 (referred to as γ-H2AX) is a sensitive marker for DNA damage (8), and we had previously shown that CPT as well as the indenoisoquinolines (non-CPT Top1 inhibitors) are potent inducers of replication-associated DNA double-strand breaks that can be identified with γ-H2AX (9, 47). Under conditions where RRM2 protein expression was attenuated by down-regulation of RRM2 by siRNA, γ-H2AX was significantly increased (Fig. 5A). Similarly, inhibition of Chk1 with UCN-01 or CHIR124 (Fig. 5B) or siRNA (Fig. 5C), inhibition of E2F1 with siRNA (Fig. 5D), and down-regulation of ATR by siRNA (Fig. 6A) all enhanced the γ-H2AX response to CPT. This γ-H2AX enhancement by down-regulation of RRM2 is consistent with the enhanced sensitivity of cells subjected to RNAi screening (Fig. 1). Together, these data suggest that RRM2 induction limits CPT-induced DNA damage and allows DNA repair.
DISCUSSION
A multiplex siRNA screen was performed in the breast cancer cell line MDA-MB-231 to identify genes that modulate the activity of CPT. This strategy has previously been used to streamline screen size and identify gene targets that significantly affect the growth of MDA-MB-231 cells (28). In this study the screening of 135 multiplexes corresponding to ≈400 genes identified several that significantly sensitized cells to CPT. A deconvolution of active multiplexes identified known and novel modulators of CPT activity. Included were BRCA1, ATR, RRM1, and RRM2 (Fig. 1). Because BRCA1 (48) and ATR (49) gene alterations have previously been established to sensitize mammalian cells to CPT (33), we focused our studies on the ribonucleotide reductase subunits, especially the small subunit RRM2. This screen also identified GJB1, a member of the connexin family, as an additional gene whose silencing enhances CPT toxicity. Notably, connexin proteins have been linked to mechanisms of cell death (34). However, this target was not pursued further in the context of this study.
The sensitizing effects of the RRM2 (and RRM1) knockdown (Fig. 1) was rather unexpected based on the prior studies that established that the RRM2 inhibitor HU protects from the cytotoxicity of CPT (7, 36). Our interpretation for this differential response to the chemical inhibitor (HU) and RRM2 knockdown is that DNA replication is only partially attenuated after the knockdown of RRM2, whereas it is completely blocked by HU (Fig. 2). As the DNA damages induced by CPT is mostly dependent on DNA replication in cancer cells, the total block of DNA replication by HU or aphidicolin prevents the conversion of CPT-induced Top1 cleavage complexes into DNA damage (50, 51). Thus, the maintained DNA synthesis under RRM2 knockdown allows the formation of replication-mediated DNA damage by CPT but incapacitates DNA repair. Thus, the greater damage in cells undergoing RRM2 knockdown, as demonstrated by enhanced γ-H2AX response to CPT (Fig. 4A), indicates the importance of RRM2 for DNA repair.
Our study shows transcriptional activation of RRM2 in response to CPT (Fig. 3), which to our knowledge has not been reported before. Because it is well established that CPT activates Chk1 after its phosphorylation by ATR (52, 53) (Fig. 6), we tested whether Chk1 activation was linked to RRM2 up-regulation. This turned out to be the case, as inhibition of Chk1 by siRNA or chemical inhibitors (UCN-01 and CHIR124) (10) blocked CPT-induced RRM2 induction measured at the mRNA (Fig. 4) and protein levels (Fig. 5, B and C). UCN-01 has independently been shown to inhibit the accumulation of RRM2 mRNA and to block RRM2 induction in response to hydroxyurea (14). Thus, Chk1 appears to be a key regulator for the induction of RRM2 in response to DNA damage in human cells.
The effect of Chk1 on RRM2 appears mediated by E2F1, a known transcription activator of RRM2, required for S-phase progression (13) (Fig. 5, B and C). Silencing of E2F1 reduced RRM2 induction both at the mRNA (Fig. 4) and protein levels (Fig. 5D). Thus, our results suggest that activation of Chk1 by Top1-mediated DNA damage induces E2F1-mediated transcriptional activation of RRM2 (Fig. 7). Whether the upstream function of Chk1 on E2F1 in the regulation of RRM2 is direct or indirect still remains to be investigated. Further studies are warranted to determine whether Chk1 might activate E2F1 by phosphorylation, by stabilizing E2F1 polypeptide levels, by transcriptional activation, or via pRB protein as in the ATM/Chk2 pathway (41, 42). Our data also show that both ATM and ATR are upstream kinases of Chk1 (Fig. 6). Thus, it is plausible that both ATR (52, 53) and ATM (this study) act as upstream activators of Chk1, which then up-regulates RRM2 through E2F1 activation (Fig. 7).
Our findings suggest a parallel framework between the well established regulatory pathways for RNR activation in response to DNA damage in budding yeast (17–21) and in mammalian cells (Fig. 7). It also reveals differential regulation of the small RNR subunits between mammalian and yeast cells. Indeed, in normal yeast cells the small RNR subunits RNR2 and RNR4 are concentrated in the nuclear compartment, whereas the large subunit, RNR1, is cytoplasmic. In response to DNA damage, yeast RNR2 and RNR4 translocate to the cytoplasm and bind RNR1 to form an active complex (54). In contrast, in mammalian cells both the large and small RNR subunits RRM1 and RRM2, respectively, are cytoplasmic in the absence of DNA damage, where they form active RNR tetramers that synthesize dNTPs, which are transported to the nucleus for DNA replication (55, 56). Our study shows RRM2 translocation from the cytoplasm to the nucleus in response to CPT (Fig. 3E). In agreement with our finding, RRM2 nuclear translocation has also been reported after UV treatment in human tumor cells (57). Thus, further investigations are warranted to elucidate the differential subcellular distribution of RNR between human and yeast cells.
We also demonstrate the importance of RNR regulation for the DNA repair of Top1-mediated DNA damage induced by CPT. Indeed, γH2AX formation is markedly enhanced in RRM2-downregulated cells (Fig. 5). We also found that inhibition of Chk1 either with siRNA or drugs enhanced γH2AX (Fig. 5), which is consistent with data obtained in other cells lines exposed simultaneously to CPT and Chk1 inhibitors (9, 45). Thus, the well known enhancement of γH2AX by Chk1 inhibition (9, 10, 40) could in part be related to defective induction of RRM2 by Chk1 inactivation (Figs. 5 and 6). This finding provides a novel mechanistic rationale for the observed synergism between inhibitors of Top1 and Chk1 (40, 58, 59).
Increasing evidence demonstrates the importance of the small ribonucleotide reductase subunit RRM2 for the cell survival in response to DNA damage. RRM2 protein levels increase after exposure to ionizing irradiation in human tumor cells and RRM2 overexpression confers resistance to infrared, which appears mediated by enhanced DNA repair during G2 (60). Our study shows a key role for RRM2 in response to Top1-mediated DNA damage induced by CPT, as inhibition of RRM2 expression enhanced the killing of MDA-MB-231 cells exposed to CPT (Fig. 1). The role of RRM2 in cell survival can be extended to other forms of DNA damage as short hairpin RNA-mediated reduction of RRM2 also resulted in an increased sensitivity to cisplatin in HCT116 cells (61). Whether the sensitization of cells to DNA damage by RRM2 reduction is solely due to reduction of dNTP synthesis is questionable. Indeed, although some studies have shown that RNR activity is closely related to RNR subunit expression (11), other data suggested a lack of tight correlation between RRM2 expression and RNR activity (62). Consistent with the latter, the potent RNR inhibitor hydroxyurea protects cells from the cytotoxicity induced by CPT or TPT (7, 36) (data not shown), which is the opposite to the sensitization obtained with RRM2 siRNAs (Fig. 1). The protective effect of hydroxyurea has been attributed to a lack of conversion of Top1 cleavage complexes into DNA double-strand breaks by prevention of replication fork collisions (6, 7).
RNR has been considered as an anti-cancer molecular target for some time; in particular, antisense approaches against RRM2 have been pursued, and more recently RRM2 has been tested as a target for siRNA-based therapy (63–66). This study suggests that combining treatment with Top1 inhibitors may be enhanced if rationally combined with approaches that inhibit the activity of the ribonucleotide reductase proteins.
Supplementary Material
Acknowledgments
Qiagen Inc. supplied some of the RNAi reagents used in this study to the NCI as part of a Collaborative Research Agreement.
This research was supported, in whole or in part, by the National Institutes of Health Intramural Research Program, Center for Cancer Research, NCI.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Table 1.
- Top1
- topoisomerase I
- ATM
- ataxia-telangiectasia (AT) mutated
- ATR
- ataxia-telangiectasia and Rad3-related
- CPT
- camptothecin
- Chk1
- checkpoint kinase 1
- E2F1
- E2F transcription factor 1
- RNR
- ribonucleotide reductase
- RRM2
- ribonucleotide reductase R2
- siRNA
- small interfering RNA
- BrdUrd
- bromodeoxyuridine
- PBS
- phosphate-buffered saline
- PI
- propidium iodide
- HU
- hydroxyurea.
REFERENCES
- 1.Bailly C. ( 2003) Crit. Rev. Oncol. Hematol. 45, 91– 108 [DOI] [PubMed] [Google Scholar]
- 2.Pommier Y. ( 2006) Nat. Rev. Cancer 6, 789– 802 [DOI] [PubMed] [Google Scholar]
- 3.Teicher B. A. ( 2008) Biochem. Pharmacol. 75, 1262– 1271 [DOI] [PubMed] [Google Scholar]
- 4.Goldwasser F., Shimizu T., Jackman J., Hoki Y., O'Connor P. M., Kohn K. W., Pommier Y. ( 1996) Cancer Res. 56, 4430– 4437 [PubMed] [Google Scholar]
- 5.Li T. K., Liu L. F. ( 2001) Annu. Rev. Pharmacol. Toxicol. 41, 53– 77 [DOI] [PubMed] [Google Scholar]
- 6.Hsiang Y. H., Lihou M. G., Liu L. F. ( 1989) Cancer Res. 49, 5077– 5082 [PubMed] [Google Scholar]
- 7.Holm C., Covey J. M., Kerrigan D., Pommier Y. ( 1989) Cancer Res. 49, 6365– 6368 [PubMed] [Google Scholar]
- 8.Bonner W. M., Redon C. E., Dickey J. S., Nakamura A. J., Sedelnikova O. A., Solier S., Pommier Y. ( 2008) Nat. Rev. Cancer 8, 957– 967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Furuta T., Takemura H., Liao Z. Y., Aune G. J., Redon C., Sedelnikova O. A., Pilch D. R., Rogakou E. P., Celeste A., Chen H. T., Nussenzweig A., Aladjem M. I., Bonner W. M., Pommier Y. ( 2003) J. Biol. Chem. 278, 20303– 20312 [DOI] [PubMed] [Google Scholar]
- 10.Seiler J. A., Conti C., Syed A., Aladjem M. I., Pommier Y. ( 2007) Mol. Cell. Biol. 27, 5806– 5818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nordlund P., Reichard P. ( 2006) Annu. Rev. Biochem. 75, 681– 706 [DOI] [PubMed] [Google Scholar]
- 12.Herrick J., Sclavi B. ( 2007) Mol. Microbiol. 63, 22– 34 [DOI] [PubMed] [Google Scholar]
- 13.DeGregori J., Kowalik T., Nevins J. R. ( 1995) Mol. Cell. Biol. 15, 4215– 4224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lubelsky Y., Reuven N., Shaul Y. ( 2005) Mol. Cell. Biol. 25, 10665– 10673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhou Z., Elledge S. J. ( 1993) Cell 75, 1119– 1127 [DOI] [PubMed] [Google Scholar]
- 16.Chen S. H., Smolka M. B., Zhou H. ( 2007) J. Biol. Chem. 282, 986– 995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhao X., Muller E. G., Rothstein R. ( 1998) Mol. Cell 2, 329– 340 [DOI] [PubMed] [Google Scholar]
- 18.Zhao X., Chabes A., Domkin V., Thelander L., Rothstein R. ( 2001) EMBO J. 20, 3544– 3553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Uchiki T., Dice L. T., Hettich R. L., Dealwis C. ( 2004) J. Biol. Chem. 279, 11293– 11303 [DOI] [PubMed] [Google Scholar]
- 20.Zhao X., Rothstein R. ( 2002) Proc. Natl. Acad. Sci. U.S.A. 99, 3746– 3751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huang M., Zhou Z., Elledge S. J. ( 1998) Cell 94, 595– 605 [DOI] [PubMed] [Google Scholar]
- 22.Elledge S. J., Zhou Z., Allen J. B., Navas T. A. ( 1993) BioEssays 15, 333– 339 [DOI] [PubMed] [Google Scholar]
- 23.Naruyama H., Shimada M., Niida H., Zineldeen D. H., Hashimoto Y., Kohri K., Nakanishi M. ( 2008) Biochem. Biophys. Res. Commun. 374, 79– 83 [DOI] [PubMed] [Google Scholar]
- 24.Chabes A. L., Björklund S., Thelander L. ( 2004) J. Biol. Chem. 279, 10796– 10807 [DOI] [PubMed] [Google Scholar]
- 25.Fusaro G., Wang S., Chellappan S. ( 2002) Oncogene 21, 4539– 4548 [DOI] [PubMed] [Google Scholar]
- 26.Eaton J. S., Lin Z. P., Sartorelli A. C., Bonawitz N. D., Shadel G. S. ( 2007) J. Clin. Invest. 117, 2723– 2734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yamaguchi T., Matsuda K., Sagiya Y., Iwadate M., Fujino M. A., Nakamura Y., Arakawa H. ( 2001) Cancer Res. 61, 8256– 8262 [PubMed] [Google Scholar]
- 28.Martin S. E., Jones T. L., Thomas C. L., Lorenzi P. L., Nguyen D. A., Runfola T., Gunsior M., Weinstein J. N., Goldsmith P. K., Lader E., Huppi K., Caplen N. J. ( 2007) Nucleic Acids Res. 35, e57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nieves-Neira W., Pommier Y. ( 1999) Int. J. Cancer 82, 396– 404 [DOI] [PubMed] [Google Scholar]
- 30.Brangi M., Litman T., Ciotti M., Nishiyama K., Kohlhagen G., Takimoto C., Robey R., Pommier Y., Fojo T., Bates S. E. ( 1999) Cancer Res. 59, 5938– 5946 [PubMed] [Google Scholar]
- 31.Doyle L. A., Ross D. D. ( 2003) Oncogene 22, 7340– 7358 [DOI] [PubMed] [Google Scholar]
- 32.O'Connor T., Rustum Y., Levine E., Creaven P. ( 2008) Cancer Chemother. Pharmacol. 61, 125– 131 [DOI] [PubMed] [Google Scholar]
- 33.Pommier Y., Barcelo J. M., Rao V. A., Sordet O., Jobson A. G., Thibaut L., Miao Z. H., Seiler J. A., Zhang H., Marchand C., Agama K., Nitiss J. L., Redon C. ( 2006) Prog. Nucleic Acid Res. Mol. Biol. 81, 179– 229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Decrock E., Vinken M., De Vuyst E., Krysko D. V., D'Herde K., Vanhaecke T., Vandenabeele P., Rogiers V., Leybaert L. ( 2009) Cell Death Differ. 16, 524– 536 [DOI] [PubMed] [Google Scholar]
- 35.Lassmann G., Thelander L., Gräslund A. ( 1992) Biochem. Biophys. Res. Commun. 188, 879– 887 [DOI] [PubMed] [Google Scholar]
- 36.Cheng M. F., Chatterjee S., Berger N. A. ( 1994) Oncol. Res. 6, 269– 279 [PubMed] [Google Scholar]
- 37.Allen J. B., Zhou Z., Siede W., Friedberg E. C., Elledge S. J. ( 1994) Genes Dev. 8, 2401– 2415 [DOI] [PubMed] [Google Scholar]
- 38.Koc A., Merrill G. F. ( 2007) Biochem. Biophys. Res. Commun. 353, 527– 530 [DOI] [PubMed] [Google Scholar]
- 39.Woolstencroft R. N., Beilharz T. H., Cook M. A., Preiss T., Durocher D., Tyers M. ( 2006) J. Cell Sci. 119, 5178– 5192 [DOI] [PubMed] [Google Scholar]
- 40.Furuta T., Hayward R. L., Meng L. H., Takemura H., Aune G. J., Bonner W. M., Aladjem M. I., Kohn K. W., Pommier Y. ( 2006) Oncogene 25, 2839– 2849 [DOI] [PubMed] [Google Scholar]
- 41.Lin W. C., Lin F. T., Nevins J. R. ( 2001) Genes Dev. 15, 1833– 1844 [PMC free article] [PubMed] [Google Scholar]
- 42.Stevens C., Smith L., La Thangue N. B. ( 2003) Nat. Cell Biol. 5, 401– 409 [DOI] [PubMed] [Google Scholar]
- 43.Liu Q., Guntuku S., Cui X. S., Matsuoka S., Cortez D., Tamai K., Luo G., Carattini-Rivera S., DeMayo F., Bradley A., Donehower L. A., Elledge S. J. ( 2000) Genes Dev. 14, 1448– 1459 [PMC free article] [PubMed] [Google Scholar]
- 44.Bartek J., Lukas J. ( 2003) Cancer Cell 3, 421– 429 [DOI] [PubMed] [Google Scholar]
- 45.Takemura H., Rao V. A., Sordet O., Furuta T., Miao Z. H., Meng L., Zhang H., Pommier Y. ( 2006) J. Biol. Chem. 281, 30814– 30823 [DOI] [PubMed] [Google Scholar]
- 46.Hickson I., Zhao Y., Richardson C. J., Green S. J., Martin N. M., Orr A. I., Reaper P. M., Jackson S. P., Curtin N. J., Smith G. C. ( 2004) Cancer Res. 64, 9152– 9159 [DOI] [PubMed] [Google Scholar]
- 47.Antony S., Agama K. K., Miao Z. H., Takagi K., Wright M. H., Robles A. I., Varticovski L., Nagarajan M., Morrell A., Cushman M., Pommier Y. ( 2007) Cancer Res. 67, 10397– 10405 [DOI] [PubMed] [Google Scholar]
- 48.Fedier A., Steiner R. A., Schwarz V. A., Lenherr L., Haller U., Fink D. ( 2003) Int. J. Oncol. 22, 1169– 1173 [PubMed] [Google Scholar]
- 49.Cliby W. A., Lewis K. A., Lilly K. K., Kaufmann S. H. ( 2002) J. Biol. Chem. 277, 1599– 1606 [DOI] [PubMed] [Google Scholar]
- 50.Pommier Y., Gupta M., Valenti M., Nieves-Neira W. ( 1996) Ann. N.Y. Acad. Sci. 803, 60– 73 [DOI] [PubMed] [Google Scholar]
- 51.Tsao Y. P., Russo A., Nyamuswa G., Silber R., Liu L. F. ( 1993) Cancer Res. 53, 5908– 5914 [PubMed] [Google Scholar]
- 52.Wang J. L., Wang X., Wang H., Iliakis G., Wang Y. ( 2002) Cell Cycle 1, 267– 272 [PubMed] [Google Scholar]
- 53.Flatten K., Dai N. T., Vroman B. T., Loegering D., Erlichman C., Karnitz L. M., Kaufmann S. H. ( 2005) J. Biol. Chem. 280, 14349– 14355 [DOI] [PubMed] [Google Scholar]
- 54.Lee Y. D., Elledge S. J. ( 2006) Genes Dev. 20, 334– 344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Engström Y., Rozell B., Hansson H. A., Stemme S., Thelander L. ( 1984) EMBO J. 3, 863– 867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Engström Y., Rozell B. ( 1988) EMBO J. 7, 1615– 1620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhou B., Liu X., Mo X., Xue L., Darwish D., Qiu W., Shih J., Hwu E. B., Luh F., Yen Y. ( 2003) Cancer Res. 63, 6583– 6594 [PubMed] [Google Scholar]
- 58.Shao R. G., Cao C. X., Shimizu T., O'Connor P. M., Kohn K. W., Pommier Y. ( 1997) Cancer Res. 57, 4029– 4035 [PubMed] [Google Scholar]
- 59.Tse A. N., Schwartz G. K. ( 2004) Cancer Res. 64, 6635– 6644 [DOI] [PubMed] [Google Scholar]
- 60.Kuo M. L., Hwang H. S., Sosnay P. R., Kunugi K. A., Kinsella T. J. ( 2003) Cancer J. 9, 277– 285 [DOI] [PubMed] [Google Scholar]
- 61.Lin Z. P., Belcourt M. F., Cory J. G., Sartorelli A. C. ( 2004) J. Biol. Chem. 279, 27030– 27038 [DOI] [PubMed] [Google Scholar]
- 62.Lin Z. P., Belcourt M. F., Carbone R., Eaton J. S., Penketh P. G., Shadel G. S., Cory J. G., Sartorelli A. C. ( 2007) Biochem. Pharmacol. 73, 760– 772 [DOI] [PubMed] [Google Scholar]
- 63.Desai A. A., Schilsky R. L., Young A., Janisch L., Stadler W. M., Vogelzang N. J., Cadden S., Wright J. A., Ratain M. J. ( 2005) Ann. Oncol. 16, 958– 965 [DOI] [PubMed] [Google Scholar]
- 64.Heidel J. D., Liu J. Y., Yen Y., Zhou B., Heale B. S., Rossi J. J., Bartlett D. W., Davis M. E. ( 2007) Clin. Cancer Res. 13, 2207– 2215 [DOI] [PubMed] [Google Scholar]
- 65.Klisovic R. B., Blum W., Wei X., Liu S., Liu Z., Xie Z., Vukosavljevic T., Kefauver C., Huynh L., Pang J., Zwiebel J. A., Devine S., Byrd J. C., Grever M. R., Chan K., Marcucci G. ( 2008) Clin. Cancer Res. 14, 3889– 3895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Stadler W. M., Desai A. A., Quinn D. I., Bukowski R., Poiesz B., Kardinal C. G., Lewis N., Makalinao A., Murray P., Torti F. M. ( 2008) Cancer Chemother. Pharmacol. 61, 689– 694 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.