Abstract
The peptide toxins of poisonous Amanita mushrooms are bicyclic octapeptides (amatoxins) or heptapeptides (phallotoxins). In Amanita bisporigera, α-amanitin and phallacidin are synthesized as 35- and 34-amino acid proproteins, respectively, in which the amino acid sequences found in the mature toxins are flanked by conserved amino acid sequences. The presence of invariant Pro residues immediately upstream of the toxin regions and as the last predicted amino acid in the toxin regions themselves suggests that a Pro-specific peptidase is responsible for the initial post-translational processing of the Amanita toxin proproteins. We purified an enzyme from the phalloidin-producing mushroom Conocybe albipes that cleaves a synthetic 22-mer phalloidin peptide to release the mature toxin peptide (AWLATCP). Mass spectrometric analysis of the purified protein combined with isolation and sequencing of the encoding gene indicates that the responsible processing enzyme is a member of the prolyl oligopeptidase (POP) subfamily of proteases (EC 3.4.21.26). The processing enzyme was able to use the chromogenic POP substrate benzyloxycarbonyl-Gly-Pro-p-nitroanilide and was inhibited by the specific POP inhibitor benzyloxycarbonyl-Pro-prolinal. Both Pro bonds in the proprotein are cleaved by the same enzyme, with the C-terminal Pro bond cleaved first or much faster than the N-terminal Pro bond. Transient accumulation of the N-terminal intermediate indicates that cleavage is not strongly processive. A synthetic peptide representing the phallacidin proprotein was also cleaved by the POP of C. albipes, but a precursor of amanitin (which is not made by C. albipes) was cleaved inefficiently.
Deadly poisonous mushrooms in the genera Amanita, Galerina, Lepiota, and Conocybe synthesize the cyclic peptide amatoxins and phallotoxins (1–4). Structurally, amatoxins (e.g. α-amanitin) are bicyclic octapeptides, and phallotoxins (e.g. phalloidin and phallacidin) are chemically related bicyclic heptapeptides. Both are hydroxylated and contain a Trp-Cys cross-bridge (tryptathionine), which has not been found in other natural products (5). Phallotoxins also contain one d amino acid (d-hydroxy-Asp or d-Thr). Despite their chemical similarity, amatoxins and phallotoxins have different modes of action; amatoxins are specific inhibitors of RNA polymerase II, and phallotoxins bind and stabilize F-actin (6, 7).
Unlike other known cyclic peptides from fungi, the amatoxins and phallotoxins are biosynthesized on ribosomes instead of nonribosomal peptide synthetases (8). In A. bisporigera, the genes for α-amanitin and phallacidin are translated as 35 and 34-amino acid proproteins, respectively (8). Although α-amanitin and phallacidin have only three amino acids in common, the upstream and downstream sequences of the proproteins are highly similar. Furthermore, the genome of A. bisporigera contains at least another 20 related sequences which collectively are characterized by conserved sequences flanking a hypervariable “toxin” region of 7–10 amino acids (8) (Fig. 1B). We refer to this family of genes as the MSDIN family for the first five highly conserved amino acids of the proproteins. Apparently, these poisonous mushrooms have evolved a mechanism of combinatorial peptide biosynthesis that uses the same biochemical template to synthesize a wide range of small, cyclic peptides.
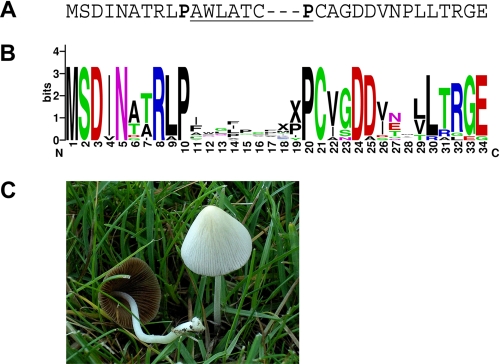
FIGURE 1.
A, The sequence of the phalloidin proprotein of A. phalloides (8). The amino acids of the mature toxin are underlined. The two cleaved Pro residues are shown in bold text. B, WebLogo (41) representation of 14 predicted MSDIN sequences from Amanita species (A. bisporigera, A. phalloides, or A. ocreata). C, C. albipes growing in cultivated lawn on the Michigan State University campus, June, 2008. In A, a 3-amino acid gap has been introduced before the C-terminal Pro because the toxin regions of some of the MSDIN sequences in the WebLogo alignment have 10 amino acids, whereas phalloidin has only 7. Also, the sequence of the phalloidin proprotein in A has been truncated from its actual 34 amino acids to 31 amino acids to facilitate its alignment with the sequences shown in B. This is because only genomic sequences are available for most of the MSDIN sequences shown in B, and based on cDNAs for AMA1 and PHA1, the other MSDIN sequences probably have an intron interrupting the antepenultimate codon. The sequences in B are GenBankTM accession numbers EU196139–EU196155. The length of the toxin region between the N-terminal invariant Pro and including the C-terminal invariant Pro varies from 7 to 10 amino acids in different members of the MSDIN family. One, two, or three X's were placed within the toxin region before the C-terminal conserved Pro residue in B for toxin peptides of nine, eight, or seven amino acids, respectively.
Among the conserved amino acids in the MSDIN gene family are two invariant Pro residues flanking the hypervariable region (Fig. 1B). All known amatoxins and phallotoxins contain one Pro, and A. phalloides is also known to make other cyclic peptides (of 6–10 amino acids), all of which contain at least one Pro residue (4). Post-translational processing of the MSDIN proproteins is predicted to involve cleavage at these Pro residues, resulting in one Pro remaining in the toxin and the other being removed (Fig. 1B).
The goal of the present work was to identify the enzyme that cleaves the MSDIN proproteins to release the peptides of the mature toxins. To identify the responsible peptidase, it was necessary to find a practical starting material. Most toxin-producing fungi in the genus Amanita are obligately ectomycorrhizal with their host plants, and they grow very slowly and do not produce toxins in culture. Galerina marginata produces toxins in culture but grows slowly (9). Some species of the mushroom genus Conocybe produce phallotoxins or amatoxins (1, 3). Conocybe species are abundant locally at certain seasons. In particular, Conocybe albipes (Fig. 1C), which produces the phallotoxin phalloidin, grows on lawns in the Midwest United States, and in the spring it is possible to collect large (>1 kg) quantities (10). For these reasons we chose C. albipes as a starting material for identification of the enzyme that processes the toxin proproteins. Here we report the purification of the enzyme that releases the mature heptapeptide from a phallotoxin precursor and show that it is a member of the prolyl oligopeptidase (POP)2 subfamily of serine proteases.
EXPERIMENTAL PROCEDURES
Purification of POP
Fruiting bodies (basidiocarps) of C. albipes were collected from lawns on the campus of Michigan State University during the spring and summer of 2008 and stored at −80 °C until use. Fifty grams of frozen mushroom were ground in liquid nitrogen in a mortar and pestle. The ground powder was resuspended in 200 ml of 50 mm Tris·HCl, pH 8.0, containing 10 ml of protease inhibitor mixture solution. The inhibitor mixture solution (10 ml) contained five tablets of Complete Mini EDTA-free protease inhibitor (Roche Applied Science) and 5 ml of a protease inhibitor mixture (Sigma P2714) that had been dissolved in 10 ml of deionized water. The sample was centrifuged in a Sorvall GSA rotor at 10,200 × g for 10 min at 4 °C. Ammonium sulfate was added to the supernatant to 30% saturation (16.6 gm/100 ml). After stirring at 4 °C for 30 min, the sample was centrifuged (10,200 × g, 10 min, 4 °C). Ammonium sulfate was then added to 60% saturation (the addition of 18.4 gm/100 ml) and centrifuged again. Ammonium sulfate was added to the supernatant to 80% saturation (the addition of 13.1 gm/100 ml), and the pellet from this final spin was redissolved in 5 ml of 20 mm NaH2PO4, pH 7.0. Ammonium sulfate was added to a final concentration of 1.7 m, and the sample was filtered through a 0.22-μm Millex filter (Millipore). Five ml was applied to a TosoHaas phenyl 5PW hydrophobic interaction column (7.5 mm × 7.5 cm) and eluted with a 30 min gradient from 20 mm NaH2PO4 + 1.7 m ammonium sulfate, pH 7.0, to 20 mm NaH2PO4, pH 7.0. The flow rate was 1 ml/min, and 1-ml fractions were collected. Protease inhibitor mixture solution (50 μl) was added to each fraction.
Fractions with activity (see below) were buffer-exchanged into 10 mm KH2PO4, pH 6.8, 0.02% sodium azide on an Econo-Pac 10DG column (Bio-Rad). The sample (5 ml) was then applied to a Bio-Rad CHT5-I hydroxyapatite column (10 mm × 6.4 cm) and eluted with a gradient from 10 mm KH2PO4, pH 6.8, 0.02% sodium azide to 0.35 m KH2PO4, pH 6.8, 0.02% sodium azide over 30 min at a flow rate of 1 ml/min. Protease inhibitor mixture solution (50 μl) was added to each fraction. Active fractions were pooled and buffer-exchanged into 25 mm Tris·HCl, pH 8.0. The sample (5 ml) was applied onto a TosoHaas DEAE-5PW column (7.5 mm × 7.5 cm) and eluted with a 40-min linear gradient from 25 mm Tris·HCl, pH 8.0, to 40% 25 mm Tris·HCl, pH 8.0, 0.6 m NaCl. Protease inhibitor mixture solution (50 μl) was added to each fraction.
POP Assays
The chromogenic POP substrate Z-Gly-Pro-pNA was purchased from Sigma-Aldrich and used at a final concentration of 1 mm. The POP inhibitor Z-Pro-prolinal was purchased from Biomol (Plymouth Meeting, PA) and used at 1 μm. All assays were done at 37 °C. Synthetic peptides (Table 1) representing proproteins of phalloidin, phallacidin, and α-amanitin were obtained from Bachem, Celtek, or the Michigan State University Research Technology Support Facility. The peptides were dissolved in deionized water at 1 mg/ml. Purified POP (50 μl of a 1-ml fraction (typically in 25 mm Tris·HCl, pH 8.0) was mixed with 5 μl of synthetic peptide (7 μl for the phalloidin precursor). At the end of the incubation 1 μl of 100 mm DTT was added, and the samples held for another 10 min at 21 °C and then stored at −80 °C. POP was inactivated by heating at 100 °C for 2 min (boiled control).
TABLE 1.
Synthetic peptides used in this study
| Name | Sequence |
|---|---|
| Phalloidin precursor (22-mer) | MSDINATRLPAWLATCPCAGDD |
| Phalloidin mature peptide | AWLATCP |
| Phallacidin precursor (17-mer) | ATRLPAWLVDCPCVGDD |
| Phallacidin mature peptide | AWLVDCP |
| α-Amanitin precursor (16-mer) | TRLPIWGIGCNPCVGD |
| α-Amanitin mature peptide | IWGIGCNP |
Analysis of Products
Peptides were separated on an Agilent Model 1100 HPLC equipped with a C18 column (Vydac 218TP54). Elution solution A was water + 0.1% trifluoroacetic acid, and solution B was acetonitrile + 0.075% trifluoroacetic acid. Flow rate was 1 ml/min with a gradient from 20% solution A to 60% solution B in 15 min. Detection was at 280 nm. Peaks were collected manually, evaporated by vacuum centrifugation, and transferred to autosampler vials.
LC-MS/MS was performed on a Micromass Q-TOF Ultima API instrument equipped with a Waters CapLC capillary HPLC. Spectra were analyzed with MassLynx software (Waters). MS/MS fragmentation of the most abundant molecule in each fraction was performed. The spectra were converted with the MaxEnt 3 module (MassLynx) into singly charged spectra. The spectra were then submitted to the PepSeq module of MassLynx for peptide sequence prediction. The molecular mass was set to be the mass shown on the spectrum. Minimum mass S.D. was set to 0.025. Sequence display threshold (% probability) was set to 1. Peptide identity was further confirmed with the Proteins module of MassLynx. The putative peptide sequences that matched the obtained mass spectra were used to calculate potential fragments. The MS/MS spectra of the peptides were then loaded and used to search against the calculated fragments. When hits were seen throughout the calculated fragmentation, the predicted sequences were taken to reflect the true peptide sequences.
Isolation of CaPOP1 from C. albipes
Total RNA was prepared using the RNeasy plant RNA extraction kit (Qiagen, Valencia, CA). First-strand cDNAs for 5′-RACE and 3′-RACE were synthesized using 1 μg of total RNA with Moloney murine leukemia virus reverse transcriptase (Clontech). Degenerate primers (5′-GGRACBACDCKRTCRTC-3′ and 5′-GGBGSDTCBAAYGGHGG-3′) were based on five predicted Basidiomycetes POP gene sequences; R stands for A or G; B stands for C, G, or T; K stands for G or T; Y stands for C or T. A 275-bp PCR fragment was cloned, and analysis of its sequence by BLASTX indicated strong similarity to known POP genes. This fragment was used as a template for RACE primer design. Using the SMART RACE kit (Clontech), 5′-RACE and 3′-RACE were performed using primers 5′-CTCCGCAATCGCAGCTCCAAAGGTC-3′ and 5′-GGACGAGTGACTACGGCAACCCAGATG-3′, respectively. Sequences generated from the RACE reactions were used to assemble a full-length cDNA of CaPOP1.
Proteomics
Aliquots (0.5 ml) of the fractions with POP activity were dialyzed against 2 liters of deionized water and lyophilized. SDS-PAGE was performed on 7.5% Ready Gels (Bio-Rad). Protein bands were visualized with ProtoBlue Safe (National Diagnostics, Atlanta, GA). Gel bands were subjected to in-gel tryptic digestion (11), and the extracted peptides were injected with a Waters nanoAcquity Sample Manager onto a Waters Symmetry C18 peptide trap (5 μm, 180 μm × 20 mm) at 4 μl/min in 5% acetonitrile, 0.1% formic acid. The bound peptides were then eluted onto a Waters BEH C18 nanoAcquity column (1.7 μm, 100 μm × 100 mm) over 35 min with a gradient of 5% B to 35% B in 21 min, ramping to 90% B from 21–24 min and back to constant 5% B at 24.1 min using a Waters nanoAcquity Ultra Performance LC (buffer A = 99.9% water, 0.1% formic acid; buffer B = 99.9% acetonitrile, 0.1% formic acid). The flow rate was 700 nl/min, ramping to 800 nl/min at 21 min and back to 700 nl/min at 35 min.
Eluted peptides were sprayed into a ThermoFisher LTQ Linear Ion trap mass spectrometer outfitted with a MICHROM Bioresources ADVANCE nano-spray source. The top five ions in each survey scan were then subjected to data-dependent zoom scans followed by low energy collision-induced dissociation, and the resulting MS/MS spectra were converted to peak lists using BioWorks Browser Version 3.3.1 (ThermoFisher) using the default LTQ instrument parameters. Peak lists were searched using the Mascot searching algorithm, Version 2.2, against a data base of the complete proteome of Laccaria bicolor downloaded from the Joint Genome Institute (www.jgi.doe.gov) to which the sequence of CaPOP1 had been added (12). The Mascot output was then analyzed using Scaffold to probabilistically validate protein identifications using the ProteinProphet computer algorithm (13). Assignments validated above the Scaffold 95% confidence filter were considered true. Mascot parameters for all databases were: up to 2 missed tryptic sites; fixed modification of carbamidomethyl cysteine; variable modification of oxidation of methionine; peptide tolerance of ±200 ppm; MS/MS tolerance of 0.8 Da; peptide charge state limited to +2/+3.
RESULTS
Purification of the Processing Enzyme
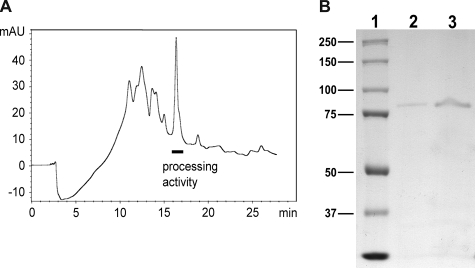
Using a synthetic 22-mer peptide corresponding to the proprotein of phalloidin from A. bisporigera as substrate (Table 1), we purified in three steps from C. albipes an enzyme capable of releasing the heptapeptide corresponding to phalloidin. The cleavage enzyme was very sensitive to proteolysis, necessitating the inclusion of a mixture of protease inhibitors in all of the purification buffers. After the final step (Fig. 2A), the cleavage activity appeared as a single band by SDS-PAGE (Fig. 2B). The apparent molecular mass of the purified protein was ∼80 kDa (Fig. 2B).
FIGURE 2.
A, final purification step (anion exchange chromatography) of processing enzyme activity from C. albipes. Elution position of activity with phalloidin peptide precursor and activity with Z-Gly-Pro-pNA is indicated. B, SDS-PAGE of purified processing enzyme activity. Lane 1, size markers (kDa). Lane 2, 0.1 μg of purified cleavage enzyme. Lane 3, 0.5 μg of enzyme. mAU, milliabsorbance units.
At each step of the purification, activity against the chromogenic POP substrate Z-Gly-Pro-pNA and cleavage activity were co-eluted (data not shown), suggesting that the cleavage enzyme is a POP. We isolated a gene encoding a POP from C. albipes using degenerate PCR primers based on putative POP genes from other fungi. Sequencing and RACE (5′ and 3′) were used to obtain the sequence of the full-length cDNA and, hence, protein. The C. albipes POP cDNA (CaPOP1) transcript has a 5′-untranslated region of 84 nucleotides and a 3′-untranslated region (stop codon to poly(A) tail) of 209 nucleotides. The deduced protein has 733 amino acids and a molecular mass of 83.0 kDa, a pI of 6.3, and no predicted signal peptide (14). CaPOP1 has the conserved catalytic triad (Ser, Asp, His) as well as all of the amino acids of the “pocket.” However, not all residues of the “flexible side chains” are conserved (15, 16) (Fig. 3).
FIGURE 3.
Primary amino acid sequence of prolyl oligopeptidase from C. albipes (CaPOP1) deduced from an encoding cDNA. Underlined peptides were identified by MS/MS proteomics of the purified protein. Amino acids of the catalytic triad (Ser-581, Asp-665, and His-701) are double-underlined and in bold. The amino acids of the pocket, as defined by Fülöp et al. (15), are Trp-623, Phe-503, Val-668, Val-608, Tyr-627, and Asn-582. CaPOP1 has been submitted to GenBankTM with accession number FJ906819.
The sequence of CaPOP1 shows highest overall amino acid identity to hypothetical proteins from the mushrooms L. bicolor and Coprinopsis cinerea, with expect scores of 0.0 and overall identity of 65 and 59%, respectively. The sequence of CaPOP1 is also strongly similar to many other putative or known POPs from animals, plants, and bacteria. It has 40% overall identity with porcine POP (expect score 3e-154) (15). Orthologs of CaPOP1 were found in all fully sequenced Basidiomycetes (available at the Department of Energy Joint Genome Institute, the Broad Institute, and GenBankTM) including A. bisporigera (8).
Identification of the Cleavage Enzyme
The purified cleavage enzyme (Fig. 2B) was digested with trypsin and subjected to LC MS/MS. The results were compared against a full in silico tryptic proteome of L. bicolor to which was added the sequence of CaPOP1. Twenty-two unique peptides giving 39% coverage that matched CaPOP1 were obtained (Fig. 3). The log (e) score was −230.5 (17). The next best match in the L. bicolor proteome was one peptide and a log(e) value of −2.6. This result unambiguously identifies the cleavage enzyme as the product of CaPOP1.
Kinetics and Mechanism of Cleavage
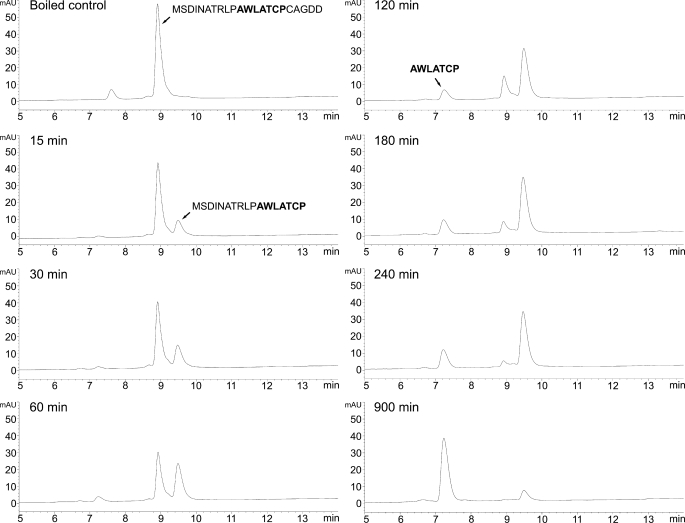
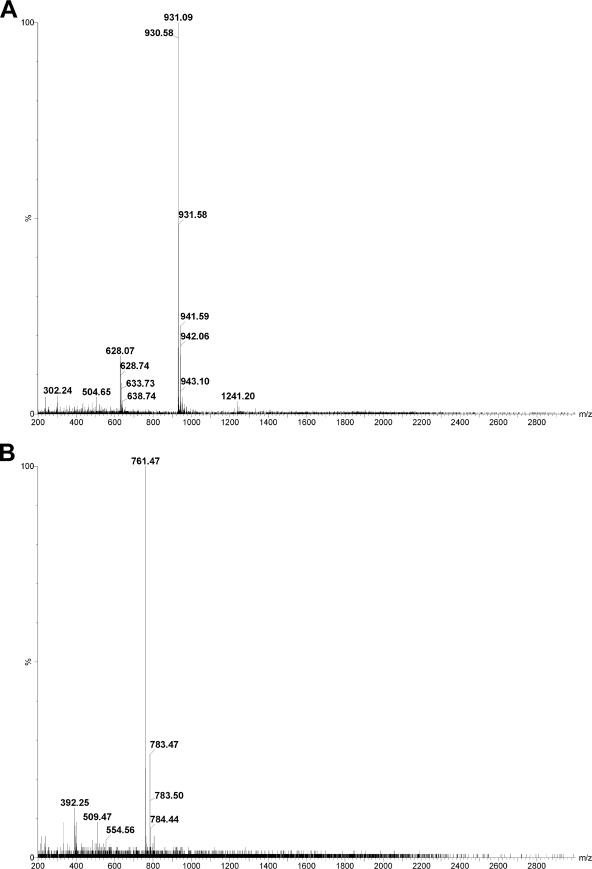
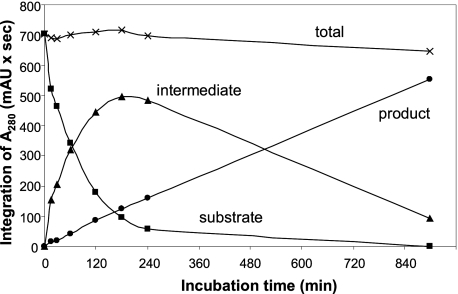
A time course of cleavage of the synthetic phalloidin peptide is shown in Fig. 4. Peaks were identified by continuous mass monitoring of the LC effluent and by manual collection of the individual UV absorbing peaks. Mass spectra of the intermediate eluting at 9.5 min and the product eluting at 7.2 min are shown in Fig. 5, A and B. All peptide sequences were confirmed by MS/MS using the PepSeq module of MassLynx. Peptide identities were further confirmed with the Proteins module of MassLynx.
FIGURE 4.
Time course of digestion of synthetic phalloidin peptide with purified CaPOP1. Peptides were analyzed by reverse phase HPLC (see “Experimental Procedures”) with monitoring of absorbance at 280 nm. mAU, milliabsorbance units.
FIGURE 5.
A, MS of the compound eluting at 9.5 min in Fig. 4 (phalloidin intermediate). B, MS of the compound eluting at 7.2 min in Fig. 4 (phalloidin mature peptide).
The sequence of the final product indicates that the C. albipes enzyme cuts on the carboxyl end of Pro, like other POPs. During the cleavage of the synthetic proprotein to the final 7-mer product, only one intermediate was observed which consisted of the N-terminal peptide produced by cleavage at the C-terminal Pro. Continuous selective ion monitoring in this and other experiments found no evidence for the C-terminal peptide (i.e. cleavage after the N-terminal Pro only). There was also no evidence for the cyclized form of the intermediate or the final product.
Quantitation of the products formed during the time course indicates that the same POP enzyme cleaves at both Pro residues of the phalloidin precursor and that the enzyme hydrolyzes the C-terminal Pro bond before the N-terminal Pro bond (Fig. 6). Alternatively, the enzyme initially cleaves randomly, but cleaves the C-terminal Pro bond much faster than the N-terminal. The transient appearance of an intermediate indicates that CaPOP1 does not act processively, i.e. the intermediate is released from the enzyme between cleavage reactions (Figs. 5 and 8).
FIGURE 6.
Quantitation of the reaction of processing enzyme (data taken from the experiment shown in Fig. 4). Purified CaPOP1 was incubated with a synthetic 22-mer phalloidin peptide (Table 1) for the indicated times. Products were quantitated by their absorbance at 280 nm.
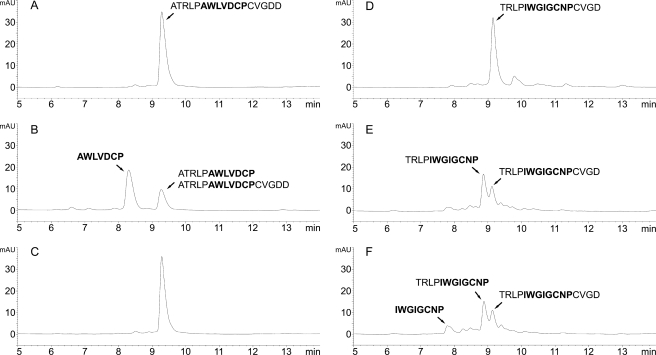
The amino acid sequence of phalloidin (AWLATCP) differs from that of phallacidin (AWLVDCP) by two amino acids. A synthetic 17-mer peptide based on phallacidin was also cleaved efficiently by CaPOP1 (Fig. 7, A and B). The specific POP inhibitor Z-Pro-prolinal blocks cleavage of the phallacidin precursor peptide completely (Fig. 7C); the inhibitor also blocks cleavage of the phalloidin precursor (data not shown).
FIGURE 7.
Cleavage of phallacidin and amanitin synthetic precursor peptides. A, inactivated CaPOP1 incubated for 2 h with the 17-mer phallacidin precursor (Table 1). B, active CaPOP1 incubated for 2 h with the phallacidin precursor. The intermediate of the reaction with the phallacidin precursor could not be resolved from the uncleaved peptide by HPLC but could be unambiguously identified by MS. C, as in panel B but with the addition of the POP inhibitor Z-Pro-prolinal at 1 μm. D–F, processing of the 16-mer α-amanitin precursor (Table 1) by CaPOP1. D, boiled control. E, 1-h incubation. F, 2-h incubation. The same batch of purified enzyme was used for all of the experiments shown in this figure, which was different from the one used in the experiment shown in Fig. 4. mAU, milliabsorbance units.
Using the same batch of CaPOP1 and identical assay conditions, a 16-mer peptide based on the proprotein of α-amanitin was cleaved less efficiently than the phallacidin precursor (Fig. 7, D–F); even after 2 h, only a small amount of the mature amanitin 8-mer had accumulated (Fig. 7F). The mass spectrum of the mature amanitin cleavage product, eluted at 7.8 min, is shown in supplemental Fig. S1.
DISCUSSION
The sequences of the proproteins of amatoxins, phallotoxins, and other members of the MSDIN gene family suggest that a Pro-specific peptidase catalyzes the initial post-translational processing of this group of natural products (8). Combined biochemical and molecular genetic approaches were used to demonstrate that, in the phalloidin-producing mushroom C. albipes, the initial processing step is catalyzed by a member of the POP subfamily of serine proteases (EC 3.4.21.26; Merops family S9A). The same enzyme cleaves at the carboxyl side of both flanking Pro residues to release the peptide of the mature toxin. The enzyme cleaves initially, or much faster, at the C-terminal Pro and is non-processive, evidenced by transient accumulation of the N-terminal intermediate.
POPs or DNA sequences encoding POP-like proteins have previously been identified in many animals, plants, bacteria, and other Basidiomycetes, including Basidiomycetes in the Agaricales (i.e. gilled mushrooms). All available Basidiomycetes genomes have at least one gene strongly similar to mammalian POPs and to CaPOP1, but based on a survey of more than 20 completed Ascomycetes fungal genomes, this taxonomic group seems to lack POP genes. POPs have been especially well studied in mammals, where they have been shown to hydrolyze several classes of Pro-containing peptides including many peptide hormones (18–23). Changes in human blood serum levels of POP have been associated with depression, mania, schizophrenia, and response to lithium (24). A POP inhibitor reverses scopolamine-induced amnesia in rats (18). POPs are also being investigated as a treatment for celiac-sprue disease, an autoimmune disorder triggered by Pro-rich peptides in certain cereal glutens (16, 23). The results presented here indicate a novel function of POP, which is to process the precursor proproteins of the toxic cyclic peptides of amatoxin and phallotoxin-producing mushrooms.
Small, modified, and biologically active peptides synthesized on ribosomes are known from bacteria, spiders, snakes, cone snails, plants, and amphibian skin (25–29). Like the toxins of Amanita species and other mushrooms, these peptides are synthesized as proproteins and, thus, must undergo at least one post-translational proteolytic cleavage. The cone snail toxins (conotoxins) are cleaved at dibasic amino acids (Arg and Lys), and only one cleavage is necessary to release the mature conotoxin (30). The precursors of the patellamides and other cyanobactins, which are head-to-tail cyclic peptides made by symbiotic cyanobacteria, must be cleaved twice to release the mature peptides, and this reaction is predicted to be catalyzed by an endopeptidase of the subtilisin family (Merops S8) that is encoded within the patellamide operon (28). The mechanism of cyclization of the patellamides is not known, although theoretical calculations suggest that it could be spontaneous (31). The cyclotides are true head-to-tail cyclic peptides that are larger (28–37 amino acids) than the mushroom toxins. Cyclotides are cleaved from their proproteins by an Asn endopeptidase, which simultaneously catalyzes cyclization (25, 32). In our experiments we saw no evidence of cyclized phalloidin heptapeptide or amanitin octapeptide, and therefore, cyclization of the MSDIN family of compounds is probably not catalyzed by POP. In the biosynthesis of microcin J25, which is a “head-to-backbone” cyclic peptide, the precursor proprotein must be proteolytically cleaved once to release the mature peptide. Cleavage and cyclization are catalyzed by two clustered genes, mcjB and mcjC, and the processing requires ATP (33). Another family of small, bacterial cyclic peptides, the autoinducing peptides of staphylococci, which are thiolactones, are processed by a membrane-bound cysteine endopeptidase (34, 35). In being catalyzed by a Pro-specific peptidase, processing of the MSDIN peptides of poisonous mushrooms occurs by a different mechanism than any previously described.
It is known from studies on other POPs that the specificity of POP enzymes is controlled by more than simply the presence of an internal Pro residue. Mammalian POP has been reported to cleave sometimes at other amino acids, such as Val, Ala, or Cys (20, 36). Conversely, POPs often do not cleave at all possible Pro residues (20, 37). For example, POP of lamb kidney can cleave peptides after a double Pro but not between adjacent Pro residues (38). Some members of the extended MSDIN family have additional predicted Pro residues within the hypervariable region (Ref. 8; GenBankTM accession numbers EU196139–EU196155). Furthermore, some additional cyclic peptides purified from A. phalloides have internal, nonadjacent Pro residues, such as CyAB (cyclo[Ser-Phe-Phe-Phe-Pro-Ile-Pro]) and CyAD (cyclo[Met-Leu-Gly-Phe-Leu-Pro-Leu-Pro]) (4). Insofar as these compounds are processed from their proproteins like the amatoxins and phallotoxins, then the responsible POP must not cut at all possible Pro residues. Another interesting situation is found with the cyclic decapeptide antamanide, also made by A. phalloides, which contains four Pro residues in two pairs (cyclo[Phe-Phe-Val-Pro-Pro-Ala-Phe-Phe-Pro-Pro]) (4).
Our experiments indicate that CaPOP1 cleaves two phallotoxin precursors more efficiently than an α-amanitin precursor (Fig. 7). This might be related to the fact that C. albipes only produces phallotoxins. However, it cannot be excluded that this is due to different lengths of the synthetic peptides used (Table 1). The extra amino acids of the phalloidin precursor (22-mer) compared with the phallacidin and α-amanitin precursors (Table 1) might provide essential amino acids to promote interaction with CaPOP1. Further studies with synthetic derivatives of the MSDIN family and with the full-length proproteins will be necessary to establish the detailed substrate specificities of CaPOP1 and other MSDIN-processing POPs from other toxin-producing fungi.
Yoshimoto et al. (39) found POP activity in 19 of 22 species of mushroom (Agaricales) tested, and of the species that could be cultured, POP activity was always intracellular. However, the same authors later purified an extracellular POP from the common mushroom Agaricus bisporus (40). In our experiments CaPOP1 was very sensitive to proteolytic degradation during purification, which is atypical of extracellular fungal enzymes. It also lacks any detectable signal peptide. These observations suggest that CaPOP1 is intracellular, which is consistent with CaPOP1 having a role in posttranslational processing of the phalloidin proprotein. Our studies do not, however, unequivocally establish that CaPOP1 is the enzyme that processes the phalloidin precursor in vivo. The role of POP in toxin non-producing mushrooms is unknown, although an extracellular location in the case of A. bisporus would suggest a role in saprophytic scavenging. Although many naturally occurring peptides, such as mammalian hormones, can be cleaved by POPs in vitro, the in vivo substrates of POPs in animals, plants, and bacteria are unknown in most cases (18).
In conclusion, in this paper we have shown that the POP of the phalloidin-producing mushroom C. albipes can efficiently and precisely cleave the precursor of phalloidin, releasing the heptapeptide of the mature toxin. Subsequent steps in the biosynthesis of phalloidin must include cyclization, hydroxylation, epimerization of l-Thr to d-Thr, and formation of the tryptathionine cross-bridge.
Supplementary Material
Acknowledgments
We thank Joe Lekyam and the Michigan State University Research Technology Support Facility for synthesis of peptides, DNA sequencing, LC-MS/MS, and proteomics analysis. We also thank Lalita Patel for experimental assistance.
This work was supported by Chemical Sciences, Geosciences, and Biosciences Division, Office of Basic Energy Sciences, Office of Science, United States Department of Energy Grant DE-FG02-91ER20021 (to the Plant Research Laboratory).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. S1.
The nucleotide sequence(s) reported in this paper has been submitted to the Gen-BankTM/EBI Data Bank with accession number(s) FJ906819.
- POP
- prolyl oligopeptidase
- Z-
- benzyloxycarbonyl
- pNA
- p-nitroanilide
- LC
- liquid chromatography
- MS/MS
- tandem mass spectrometry
- RACE
- rapid amplification of cDNA ends
- HPLC
- high performance liquid chromatography.
REFERENCES
- 1.Brady L. R., Benedict R. G., Tyler V. E., Stuntz D. E., Malone M. H. ( 1975) Lloydia 38, 172– 173 [PubMed] [Google Scholar]
- 2.Enjalbert F., Cassanas G., Rapior S., Renault C., Chaumont J.-P. ( 2004) Mycologia 96, 720– 729 [DOI] [PubMed] [Google Scholar]
- 3.Hallen H. E., Watling R., Adams G. C. ( 2003) Mycol. Res. 107, 969– 979 [DOI] [PubMed] [Google Scholar]
- 4.Wieland T. ( 1986) Peptides of Poisonous Amanita Mushrooms, Springer-Verlag New York Inc., New York [Google Scholar]
- 5.May J. P., Perrin D. M. ( 2007) Pept. Sci. 88, 714– 724 [Google Scholar]
- 6.Bushnell D. A., Cramer P., Kornberg R. D. ( 2002) Proc. Natl. Acad. Sci. U. S. A. 99, 1218– 1222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wulf E., Deboben A., Bautz F. A., Faulstich H., Wieland T. ( 1979) Proc. Natl. Acad. Sci. U. S. A. 76, 4498– 4502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hallen H. E., Luo H., Scott-Craig J. S., Walton J. D. ( 2007) Proc. Natl. Acad. Sci. U. S. A. 104, 19097– 19101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Muraoka S., Shinozawa T. ( 2000) J. Biosci. Bioeng. 89, 73– 76 [DOI] [PubMed] [Google Scholar]
- 10.Hutchinson L. J., Madzia S. E., Barron G. L. ( 1996) Can. J. Bot. 74, 431– 434 [Google Scholar]
- 11.Shevchenko A., Wilm M., Vorm O., Mann M. ( 1996) Anal. Chem. 68, 850– 858 [DOI] [PubMed] [Google Scholar]
- 12.Perkins D. N., Pappin D. J., Creasy D. M., Cottrell J. S. ( 1999) Electrophoresis 20, 3551– 3567 [DOI] [PubMed] [Google Scholar]
- 13.Nesvizhskii A. I., Keller A., Kolker E., Aebersold R. ( 2003) Anal. Chem. 75, 4646– 4658 [DOI] [PubMed] [Google Scholar]
- 14.Emanuelsson O., Brunak S., von Heijne G., Nielsen H. ( 2007) Nat. Protoc. 2, 953– 971 [DOI] [PubMed] [Google Scholar]
- 15.Fülöp V., Böcskei Z., Polgár L. ( 1998) Cell 94, 161– 170 [DOI] [PubMed] [Google Scholar]
- 16.Shan L., Mathews I. I., Khosla C. ( 2005) Proc. Natl. Acad. Sci. U. S. A. 102, 3599– 3604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Craig R., Beavis R. C. ( 2004) Bioinformatics 20, 1466– 1467 [DOI] [PubMed] [Google Scholar]
- 18.Brandt I., Scharpé S., Lambeir A. M. ( 2007) Clin. Chim. Acta 377, 50– 61 [DOI] [PubMed] [Google Scholar]
- 19.Cunningham D. F., O'Connor B. ( 1997) Biochim. Biophys. Acta 1343, 160– 186 [DOI] [PubMed] [Google Scholar]
- 20.García-Horsman J. A., Männistö P. T., Venäläinen J. I. ( 2007) Neuropeptides 41, 1– 24 [DOI] [PubMed] [Google Scholar]
- 21.Gass J., Khosla C. ( 2007) Cell Mol. Life Sci. 64, 345– 355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Polgár L. ( 2002) Cell. Mol. Life Sci. 59, 349– 362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Szeltner Z., Polgár L. ( 2008) Curr. Protein Pept. Sci. 9, 96– 107 [DOI] [PubMed] [Google Scholar]
- 24.Williams R. S. ( 2005) Prog. Neuropsychopharmacol. Biol. Psychiatry 29, 1029– 1037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Craik D. J., Cemazar M., Daly N. L. ( 2007) Curr. Opin. Drug Discov. Devel. 10, 176– 184 [PubMed] [Google Scholar]
- 26.Escoubas P. ( 2006) Mol. Divers. 10, 545– 554 [DOI] [PubMed] [Google Scholar]
- 27.Olivera B. M. ( 2006) J. Biol. Chem. 281, 31173– 31177 [DOI] [PubMed] [Google Scholar]
- 28.Schmidt E. W., Nelson J. T., Rasko D. A., Sudek S., Eisen J. A., Haygood M. G., Ravel J. ( 2005) Proc. Natl. Acad. Sci. U. S. A. 102, 7315– 7320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Simmaco M., Mignogna G., Barra D. ( 1998) Biopolymers 47, 435– 450 [DOI] [PubMed] [Google Scholar]
- 30.Milne T. J., Abbenante G., Tyndall J. D., Halliday J., Lewis R. J. ( 2003) J. Biol. Chem. 278, 31105– 31110 [DOI] [PubMed] [Google Scholar]
- 31.Milne B. F., Long P. F., Starcevic A., Hranueli D., Jaspars M. ( 2006) Org. Biomol. Chem. 4, 631– 638 [DOI] [PubMed] [Google Scholar]
- 32.Saska I., Gillon A. D., Hatsugai N., Dietzgen R. G., Hara-Nishimura I., Anderson M. A., Craik D. J. ( 2007) J. Biol. Chem. 282, 29721– 29728 [DOI] [PubMed] [Google Scholar]
- 33.Duquesne S., Destoumieux-Garzón D., Zirah S., Goulard C., Peduzzi J., Rebuffat S. ( 2007) Chem. Biol. 14, 793– 803 [DOI] [PubMed] [Google Scholar]
- 34.Novick R. P., Geisinger E. ( 2008) Annu. Rev. Genet. 42, 541– 564 [DOI] [PubMed] [Google Scholar]
- 35.Qiu R., Pei W., Zhang L., Lin J., Ji G. ( 2005) J. Biol. Chem. 280, 16695– 16704 [DOI] [PubMed] [Google Scholar]
- 36.Nomura K. ( 1986) FEBS Lett. 209, 235– 237 [DOI] [PubMed] [Google Scholar]
- 37.Moriyama A., Nakanishi M., Sasaki M. ( 1988) J. Biochem. 104, 112– 117 [DOI] [PubMed] [Google Scholar]
- 38.Koida M., Walter R. ( 1976) J. Biol. Chem. 251, 7593– 7599 [PubMed] [Google Scholar]
- 39.Yoshimoto T., Sattar A. K., Hirose W., Tsuru D. ( 1988) J. Biochem. 104, 622– 627 [DOI] [PubMed] [Google Scholar]
- 40.Sattar A. K., Yamamoto N., Yoshimoto T., Tsuru D. ( 1990) J. Biochem. 107, 256– 261 [DOI] [PubMed] [Google Scholar]
- 41.Crooks G. E., Hon G., Chandonia J. M., Brenner S. E. ( 2004) Genome Res. 14, 1188– 1190 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.