Abstract
tRNA guanine transglycosylase (TGT) enzymes are responsible for the formation of queuosine in the anticodon loop (position 34) of tRNAAsp, tRNAAsn, tRNAHis, and tRNATyr; an almost universal event in eubacterial and eukaryotic species. Despite extensive characterization of the eubacterial TGT the eukaryotic activity has remained undefined. Our search of mouse EST and cDNA data bases identified a homologue of the Escherichia coli TGT and three spliced variants of the queuine tRNA guanine transglycosylase domain containing 1 (QTRTD1) gene. QTRTD1 variant_1 (Qv1) was found to be the predominant adult form. Functional cooperativity of TGT and Qv1 was suggested by their coordinate mRNA expression in Northern blots and from their association in vivo by immunoprecipitation. Neither TGT nor Qv1 alone could complement a tgt mutation in E. coli. However, transglycosylase activity could be obtained when the proteins were combined in vitro. Confocal and immunoblot analysis suggest that TGT weakly interacts with the outer mitochondrial membrane possibly through association with Qv1, which was found to be stably associated with the organelle.
Queuosine (Q3; (7-{[(4,5-cis-dihydroxy-2-cyclo-penten-1-yl)-amino]methyl}-7-deazaguanosine) is a modified 7-deazaguanosine molecule found at the wobble position of transfer RNA that contains a GUN anticodon sequence: tRNATyr, tRNAAsn, tRNAHis, and tRNAAsp (1). The Q-modification is widely distributed in nature in the tRNA of eubacteria, plants, and animals; a notable exception being yeast and plant leaf cells (2, 3). Interestingly, Q-modification has also been detected in aspartyl tRNA from mitochondria of rat (4) and opossum (5). In most eukaryotes, the Q molecule can be further modified by the addition of a mannosyl group to Q-tRNAAsp and a galactosyl group to Q-tRNATyr (1).
Eubacteria are unique in their ability to synthesize Q. As part of this biosynthetic process, the eubacterial tRNA guanine transglycosylase (TGT) enzyme inserts the Q precursor molecule, 7-aminomethyl-7-deazaguanine (preQ1) into tRNA, which is then converted to Q by two further enzymatic steps at the tRNA level (6). Eukaryotes by contrast salvage queuosine from food and enteric bacteria either as the free base (referred to as queuine) or as queuosine 5′-phosphate subsequent to normal tRNA turnover (7). A Q-related molecule, archaeosine, is found at position 15 of the D loop of most archaeal tRNA, where it functions to stabilize the tRNA structure (8). The enzyme involved in archaeosine biosynthesis is structurally and mechanistically related to the eubacterial TGT but with adaptations necessitated by the differences imposed by its unique substrate and tRNA specificity (9, 10).
The crystal structure of the Zymononas mobilis (Z. mobilis) TGT has been determined and revealed the enzyme to be an irregular (β/α)8 TIM barrel with a C-terminal zinc-binding subdomain (11). Insight into the residues involved in catalysis came from mutational and kinetic analysis of the recombinant Escherichia coli enzyme (12) and from the Z. mobilis TGT structure as an RNA-bound intermediate complexed to the final preQ1-modified RNA product (13). This work showed the essential role of Asp-280 (Z. mobilis numbering) as the active site nucleophile. Asp-102, which was originally ascribed the role of active site nucleophile, functions as a general acid/base during catalysis (12, 10). Although, the E. coli and Z. mobilis TGT enzymes are monomeric in solution (14), at high protein concentrations the E. coli enzyme can oligomerize (15), and structural data from the Z. mobilis TGT has shown the formation of a 2:1 complex with tRNA; a possible functional requirement for catalysis (10).
In contrast to the eubacterial enzyme, which is a single protein species, purification of the eukaryotic TGT suggested that the catalytically active enzyme is a heterodimeric molecule: subunits of 60 and 43 kDa in rabbit erythrocytes (16), 66 and 32 kDa in bovine liver (17), 60 and 34.5 kDa in rat liver (18), and a homodimer of two 68-kDa proteins in wheat germ (16, 19). A partial amino acid sequence was recovered from two of these active enzyme preparations. The identity of the proteins from bovine liver (17) could not be assigned at the time of publication. However, our searches show that the peptides from the larger 65-kDa subunit are identical to asparaginyl tRNA synthetase, and those of the smaller 32-kDa subunit correspond to 2,4-dienoyl CoA reductase. A highly pure preparation from rabbit reticulocytes (20) gave peptides with homology to the immunophilin p59, human elongation factor 2 (EF2), and a deubiquitinating enzyme, USP14. It is noteworthy that none of the peptide sequences obtained showed similarity to the eubacterial TGT. The results do suggest, however, that in eukaryotes the TGT activity could be embedded in a multisubunit complex.
Most recently, Deshpande and Katze (21) identified a cDNA clone encoding a putative TGT catalytic subunit. Cloning the cDNA into a mammalian expression plasmid reconstituted TGT activity in GC3/c1 cells, which are known to be naturally deficient in Q-containing tRNA (22). In this study, we identify for the first time the composition of the eukaryotic tRNA guanine transglycosylase, reconstitute the catalytic activity in vitro, and examine the intracellular distribution of the active subunits.
EXPERIMENTAL PROCEDURES
cDNA Cloning
The E. coli TGT was PCR-amplified from genomic DNA using the primer pair ETF (5′-gcgcatatgaaatttgaactggacaccacc-3′) and ETR (5′-cacctcgagttaatcaacgttcaaaggtggtattc-3′) and cloned into the pET15b plasmid (Novagen) using NdeI and XhoI to generate the ETGT:pET15b plasmid (His tag). The cDNA clones for Qv0 (NM029128; IMAGE: 30105859) and Qv2 (BC017628; IMAGE: 4505816) were purchased from the IMAGE consortium. Primers were designed for the AUG translation start and TGA stop site of Qv0 to search for additional related proteins by reverse transcription-PCR (RT-PCR) leading to the discovery of Qv1, below.
Full-length mouse TGT (fTGT) and Qv1 were reverse-transcribed from total kidney RNA of 4-week-old male mice using a fTGT-specific reverse primer FTR (5′-cacctcgagtcatgtgagcatgattcccacagag-3′) and a QTRTD1 reverse primer QR (5′-cacctcgagtgcaaacatctgtctgcaaatgagttc-3′; note the stop codon was converted to an Ala; underlined) according to the Superscript III protocol (Invitrogen) for gene-specific primers. First-strand products were separated from reaction components using a nucleotide removal kit (Qiagen). TGT and Qv1 were amplified by PCR using the aforementioned reverse primers and the forward primers FTF (5′-gacgaattcatggcggcggtaggcagcccaggttc-3′) and QF (5′-cgacatatgatgaagctgagtctcatcaaagtcg-3′), respectively. The fTGT cDNA was cloned into the EcoRI and XhoI restriction sites of pGEX6P1 (Invitrogen) to give the plasmid fTGT:pGEX6P1 (GST tag), whereas Qv1 was cloned into the NdeI and XhoI restriction sites of the pET21a plasmid (Novagen) to produce the plasmid Qv1:pET21a (His tag). A truncated version of the mouse TGT (tTGT) was also produced, missing the coding sequence for the first 16 amino acids. This sequence was amplified from the fTGT:pGEX6P1 plasmid using the primer pair tTF (5′-gaccatatgcggctggtcgctgagtgcagtc-3′) and tTR (5′-cacgtcgactcatgtgagcatgattcccacaag-3′). The PCR product was cloned into the NdeI and SalI restriction sites of the pET15b plasmid to yield tTGT:pET15b (His tag). DNA sequencing of all constructs was performed in the forward and reverse direction. The Qv1 sequence had not been previously described and was therefore deposited in the EMBL nucleotide data base under GenBankTM accession number FM985972.
For the generation of mammalian expression constructs, the TGT and Qv1 cDNA were cloned into the EcoRI and XhoI sites of pcDNA3.1/Myc-His (Invitrogen), pcDNA3.1/HA (modified plasmid), pCMV.Myc, and pCMV.HA (Clontech).
Recombinant Protein Expression
BL21(DE3) tgt::Kmr cells (whose tgt gene was disrupted by the insertion of a group II intron, supplemental Fig. S1) were transformed with ETGT: pET15b, fTGT:pGEX6P1, tTGT:pET15b, and Qv1:pET21a plasmids and cultured in LB medium. Induction of protein expression was performed with 0.1 mm isopropyl-1-thio-β-d-galactopyranoside in 2xYT broth, at 18 °C overnight. His-tagged E. coli TGT, tTGT, and Qv1 proteins were purified by nickel-charged HiTrap chromatography, whereas the GST-tagged fTGT was isolated on a GSTrap FF prepacked column by cleavage of the tag using PreScission Protease (GE Healthcare) according to the manufacturer's instructions.
Antisera Production and Clean-up
Antisera to TGT and Qv1 were raised in New Zealand White rabbits (Harlan, UK) and purified by NAb Protein A spin columns (Pierce) according to the manufacturer's protocol. Antisera were further purified by counterselection against purified TGT and Qv1 protein bound to Ultralink Biosupport resin (Pierce). Coupling of TGT and Qv1 to resin was carried out overnight in 0.1 m MOPS, 0.6 m sodium citrate, pH 7.5, at room temperature. The resin was quenched with 3 m ethanolamine, washed with 1 m NaCl, and equilibrated with 10 mm Tris, pH 7.4. Antisera were eluted with a 10 mm Tris, pH 7.4 buffer containing 1 mm NaCl.
tRNA Guanine Transglycosylase Assay
When required, bulk tRNA was isolated from E. coli according to published protocols (23). To assess transglycosylase activity, enzyme (2 μg) was added to 150 μl of assay buffer (50 mm Tris-HCl, pH 7.5, 20 mm NaCl, 5 mm MgCl2, 1 mm dithiothreitol, and 2 units of SUPERasin (Ambion) RNase inhibitor mixture) containing 0.1 absorbance unit of Baker's yeast tRNA (Roche Applied Science) or E. coli tRNA, and 1.8 μm [8-14C]guanine. The reaction was incubated for 2 h at 37 °C before quenching with 450 μl of acid phenol:chloroform, pH 4.5 (Invitrogen) and spun for 3 min at 16,000 × g to separate aqueous and organic layers. The upper (aqueous) phase was loaded onto a 0.5-ml DEAE cellulose (Whatman) spin column and spun at 0.1 × g for 30 s. The flow-through was reloaded on the column another three times. The DEAE was then washed with 2 ml of wash buffer (20 mm Tris-HCl, pH 7.5, 10 mm MgCl2, 200 mm NaCl). Bound tRNA was eluted with 1 ml of elution buffer (20 mm Tris-HCl, pH 7.5, 10 mm MgCl2, 1 m NaCl). Eluate was mixed with 10 volumes of Ecoscint A (National Diagnostics) and scintillation counted.
[14C]Guanine Displacement Assay
tRNA labeled with [8-14C]guanine in the wobble position of anticodon loop (tRNA*) was produced as described previously (24). A 150-μl reaction containing 5 absorbance units of tRNA* and 4 μg of enzyme in assay buffer was incubated at 37 °C for 2 h. Reactions were directly loaded onto a DEAE spin column and spun at 0.1 × g for 30 s at 4 °C. The column was washed with 2 ml of wash buffer. Eluate and wash were combined, added to 10 volumes Ecoscint A, and counted by scintillation.
RT-PCR Analysis of QTRTD1 Splice Variants
Total RNA was extracted from adult male mice and reverse-transcribed using a QTRTD1 reverse primer QSR (5′-gacgtcatctcttggttgcaaccagttg-3′) capable of amplifying all three splice variants or an 18S ribosomal subunit reverse primer 18SR (5′-tgccttccttggatgtgg-3′) according to the Superscript III protocol for gene-specific primers (Invitrogen). First-strand products were separated from reaction components using a nucleotide removal kit (Qiagen). The QTRTD1 splice variants and the 18S subunit were amplified by PCR using the aforementioned reverse primers and the forward primers QSF (5′-ccagctcacactctcatccctagca-3′) and 18SF (5′-cgtctgccctatcaactttc-3′), respectively.
RNA Blot Analyses
Total RNA was size-fractionated, transferred to nylon membrane, and probed using radioactively labeled cDNA for TGT and Qv1 retrieved from the pGEX6P1 and pET21a vectors, respectively. As a loading control, the membrane was stained with methylene blue to visualize the 18S and 28S ribosomal bands.
Cell Culture, Transfection, and Subcellular Fractionation
COS-7 monkey kidney cells were from the European Collection of Cell Cultures. Cells were maintained in Dulbecco's modified Eagle's medium supplemented with 10% (v/v) fetal bovine serum, 2 mm l-glutamine, 1 mm sodium pyruvate, and 50 units of penicillin/streptomycin in an atmosphere of 5% CO2 at 37 °C. Cells were transfected in OptiMEM (Invitrogen) using Lipofectamine 2000 (Invitrogen) in a ratio of 2 μl to 1 μg of DNA. Total, cytoplasmic, nuclear, membrane, and mitochondrial fractions were isolated using kits (Pierce) as instructed by the manufacturer.
Immunocytochemistry
Cells were seeded at 1 × 105 in 4-well chamber slides (BD-Falcon). Mitochondrial staining was carried out with 100 nm Mitotracker Red CMXRos (Invitrogen) for 15 min before fixation with 4% (w/v) paraformaldehyde in phosphate-buffered saline. Cells transfected with Myc- and HA-tagged plasmids were incubated with 1:500 mouse monoclonal anti-Myc antisera (Clontech) and rabbit polyclonal anti-HA antisera (Clontech), respectively. Anti-mouse Alexa Fluor 488 and anti-rabbit Alexa Fluor 635 (Invitrogen) secondary antibody were used at 1:1000 dilution. For confocal analysis, antisera raised against TGT and Qv1 were used at 1:500 dilution before being probed with 1:1000 anti-rabbit Alexa Fluor 488 (Invitrogen). Nuclei were stained with DAPI (4′,6′-diamidino-2-phenylindole). The coverslips were mounted using Prolong Gold (Invitrogen). Images were taken using the 405-, 488-, and 633-nm laser lines on an Olympus FluoView TM FV1000 confocal laser-scanning microscope (Olympus).
RESULTS
Eukaryotes Contain Two Genes Whose Products Are Homologous to the Eubacterial tgt
The E. coli TGT protein was chosen as bait sequence in protein BLAST (pblast) and translated nucleotide BLAST (tblastn) searches of the mouse nonredundant and EST data bases.
Initial searches identified a mouse TGT (NM021888), transcribed from the queuine tRNA ribosyltransferase (QTRT1) gene on chromosome 9, which is equivalent to the previously identified human TGT catalytic subunit (21). Further searches, using both the E. coli and mouse TGT proteins as bait, revealed two additional related mouse members of the family (NM029128 and BC017628), and a third was subsequently identified in male mouse kidney (FM985972) by RT-PCR (see “Experimental Procedures”). The three latter proteins arise from alternative spliced transcripts of the queuine tRNA ribosyltransferase domain containing 1 (QTRTD1) gene, which is composed of nine exons and is transcribed from the minus strand of chromosome 16 (supplemental Fig. S2).
In accordance with standard convention (25), the three spliced products have been assigned the names QTRTD1 variant_0 (Qv0), which is missing exon 5, QTRTD1 variant_1 (Qv1), which contains all nine exons, and QTRTD1 variant_2 (Qv2), which in addition to containing all nine exons has retained one-third of intron 6, exploiting a later downstream cryptic donor site to engage with exon 7.
The three QTRTD1 splice variants were also identifiable within the Alternative Splicing Data Base (ASD, 26), although Qv1 was present only as a 5′ and 3′ partial clone. Interestingly, four clones in the ASD reveal the existence of exonic isoforms for exon 2 because of alternative splice donor events. In all cases, the extension of exon 2 generates a truncated peptide of only 68 amino acids. These are unlikely to be functional and are not considered further.
QTRTD1 Variants Are Distantly Related to the Eukaryotic and Eubacterial TGTs
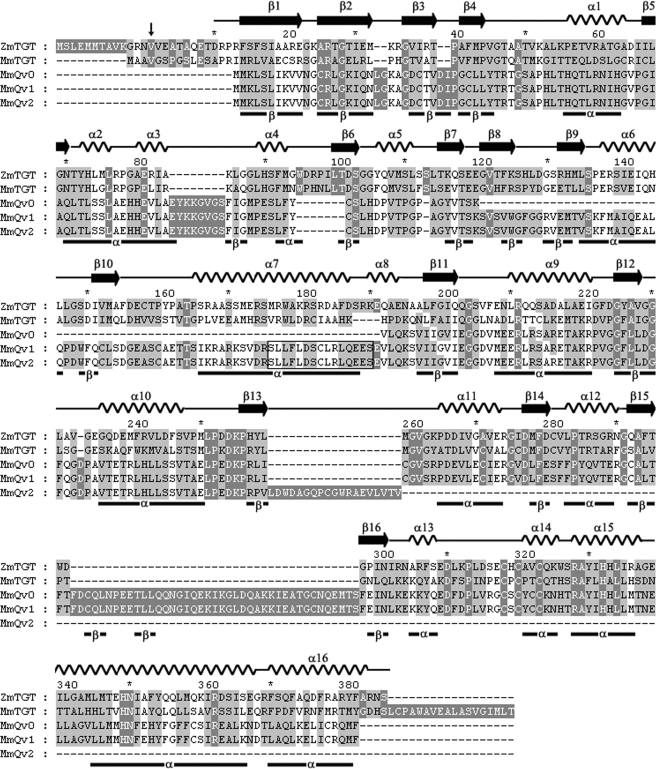
Alignment of the mouse and Z. mobilis TGT protein sequences (Fig. 1) revealed a high degree of amino acid conservation (42% identity), and it is therefore expected that the eukaryotic TGT adopts a similar (β/α)8 fold (13). It is notable from the alignment that the residues involved in catalysis (residue numbering adopted from the Z. mobilis TGT) are highly conserved; the two essential active site aspartates (Asp-102, Asp-280), residues involved in substrate recognition (Ser-103, Asp-156, Gln-203, Gly-230, Leu-231), residues involved in tRNA recognition (Arg-286, Arg-289), and residues responsible for coordinating the zinc ion (Cys-318, Cys-320, Cys-323, and His-349).
FIGURE 1.
Protein sequence alignment of TGT and QTRTD1 splice variants. Alignment of the Z. mobilis TGT (ZmTGT) with the mouse TGT (MmTGT) and mouse QTRTD1 splice variants (MmQv0, MmQv1, and MmQv2) using the multiple alignment program GeneDoc (44). Conserved amino acids are shown in white on a dark gray background. The numbering refers to the Z. mobilis TGT sequence beginning at the fourth possible start codon GTG (Val), denoted by an arrow, as described previously (14). The secondary structure of the Z. mobilis TGT is shown above the sequences (11), whereas the predicted secondary structure elements of the QTRTD1 proteins, generated using the Porter program (27), are presented below the sequences.
The three QTRTD1 splice variants Qv0, Qv1, and Qv2 generate protein products of 345, 416, and 271 amino acids, respectively. Aligning the QTRTD1 splice variants with the TGT proteins shows a low overall degree of conservation (Fig. 1). For example, mouse TGT and Qv1 share 23% sequence identity and 41% homology. The potential secondary structure of the QTRTD1 variants was examined using the computer application Porter (27), the results of which are shown below the alignment blocks. Manual adjustments were made to conserve the positioning of the secondary structure elements with respect to the Z. mobilis TGT. It is highly probable given the conserved alternating pattern of secondary elements that the mammalian QTRTD1 variants share a similar (β/α)8 structure to the TGT proteins.
Assuming the fidelity of our alignment, the residues known to be important for catalysis in the TGT proteins are not conserved in the N-terminal two-thirds of the QTRTD1 proteins. However, there is a distinct conservation of residues in the C-terminal subdomain involved in zinc binding (Cys-318, Cys-320, Cys-323, and His-349) and a conserved substitution of the active site nucleophile Asp-280 to a glutamic acid residue. A further interesting feature of the alignment is the almost complete conservation of residues in the C terminus of Qv1 and Qv2 with an established role in dimer formation in the Z. mobilis TGT protein (Trp/His-326, Ala-329, Tyr-330, His-333, Leu-334, Glu-339, Leu-341, Leu-345) and a reciprocal presence of dimer-forming residues in the N-terminal β1-α1 region (Ala-49, Thr-50, Lys-52, Leu-74, Pro-78, Phe-92) of mouse TGT (10).
The splicing pattern of the QTRTD1 gene gives rise to significant insertions and deletions. The Qv0 sequence is missing a 71-amino acid segment (Lys-120 to Val-191) relative to Qv1 and Qv2. The purpose of this region is at present unclear. However, it is intriguing that it contains a consensus leucine-rich nuclear export signal (Fig. 1, boxed region, SLLFLDSCLRLQEES), which not only adheres to the accepted form Φ-X2–3-Φ-X2–3-Φ-X-Φ (Φ = L, I, V, F, M; X is any amino acid), but the region is rich in glutamate, aspartate, and serine and occurs at the end of a putative α-helix at the border of secondary structure elements; features typical of a leucine-rich export signal (28, 29).
An insertion of 41 amino acids in Qv0 and Qv1 is seen relative to the TGT proteins (at Asp-297). This occurs within the zinc-binding subdomain and immediately C-terminal to the last helix (α12 in Z. mobilis TGT) of the (β/α)8 motif. Qv2 is notable in that although it has the longest coding sequence its transcript produces the shortest protein because of the presence of an early stop site, consequently leading to a complete loss of the terminal zinc-binding domain.
Qv1 Is the Predominant Splice Variant of QTRTD1 in Adult Mouse Tissue, and Its mRNA Expression Level Mirrors That of TGT
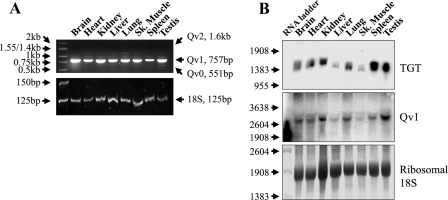
The expression pattern of splice variants is often specific to a particular developmental stage, tissue, or disease state. To determine the expression pattern of the QTRTD1 splice variants in adult mouse, primers were designed to exon 2 and exon 7, which could amplify all three splice variants, yielding amplicons of 551 bp, 757 bp, and 1.6 kb for Qv0, Qv1, and Qv2, respectively (Fig. 2A). It was observed that the Qv1 transcript is present in adult mouse brain, heart, kidney, liver, lung, skeletal muscle, spleen, and testis (upper panel). By contrast, no product could be detected for Qv0 or Qv2. Amplification from the 18S ribosomal subunit (125 bp) acted as a loading control (lower panel).
FIGURE 2.
mRNA expression pattern of TGT and QTRTD1 in adult mouse tissues. Total RNA was extracted from adult male mouse tissue. A, RT-PCR analysis was performed to evaluate the expression of the three splice variants of QTRTD1; the expected sizes are shown on the right hand side of the figure (upper panel). The 18S ribosomal subunit served as an internal standard (lower panel). B, RNA blot analysis was performed using the cDNA for fTGT (upper panel) and Qv1 (middle panel). Note that the first lane of the Qv1 blot contains a nonspecific hybridization signal. Staining of the transfer membrane with methylene blue, to visualize the 18S ribosomal subunit, was used as a loading control (lower panel). RNA ladder denotes RNA molecular weight markers.
We next examined the mRNA expression of TGT and Qv1 in mouse by Northern blot analysis (Fig. 2B). The message for both proteins was found to exist in all tissues examined, giving a hybridization band at 1405 bp for TGT (upper panel) and 3125 bp for Qv1 (middle panel), which reasonably correlates with the predicted sizes of 1334 bp and 3031 bp for the TGT and Qv1 cDNA sequences, respectively. In the case of Qv1 a second fainter hybridizing band, migrating above the main species, was observable in all tissues with the exception of testis.
Unexpectedly, the blot analysis revealed a correlation between the relative tissue expression levels of the TGT and Qv1 transcripts. These data indicate that the transcription of both proteins may be coordinately regulated, and by extension that the proteins may function in a cooperative manner. A search of eukaryotic species for the expression of TGT and Qv1 demonstrated that both proteins are invariably found together (results not shown), further hinting their cooperative function. The only eukaryotic species found not to contain a Qv1 homologue was the Baker's yeast, Saccharomyces cerevisiae. Curiously, Baker's yeast is also the only eukaryote from which the TGT gene was absent.
Qv1 and TGT Co-associate in Vivo and Partition to the Cytoplasm
The observation that the transcript levels of TGT and Qv1 correlate in mouse tissue and that the proteins are found together in eukaryotes led us to hypothesize that they may account for the heterodimeric tRNA guanine transglycosylase activity observed from protein purification studies.
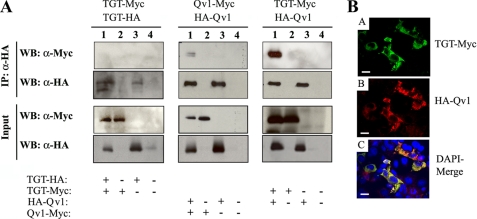
Constructs that add an N- or C-terminal Myc or HA epitope were developed for TGT and Qv1. Initial transient transfection studies showed that the N-terminal tags interfered with either the expression or the stability of the TGT protein and, therefore, could not be used. This was not the case with the Qv1 constructs. To examine the ability of the proteins to self-associate or co-associate, combinations of N- and C-terminally tagged forms of both proteins were transiently transfected into COS-7 cells, and lysates were immunoprecipitated using HA-specific monoclonal antibody (Fig. 3A). Our results showed that TGT protein, which had been tagged by a Myc epitope (TGT-Myc) or HA epitope (TGT-HA) on the C terminus, is unable to self-associate (left hand upper panel, lane 1). A weak self-association was, however, observed for Qv1 (middle upper panel, lane 1) that had been tagged on the C terminus with a Myc epitope (Qv1-Myc) and on the N terminus with an HA epitope (HA-Qv1). By contrast, an intense signal, indicative of strong interaction, was observed by immunoprecipitation of the TGT-Myc and HA-Qv1 proteins (right hand upper panel, lane 1).
FIGURE 3.
TGT and Qv1 co-immunoprecipitate and co-localize to the cytoplasm. COS-7 cells were transiently transfected with constructs that express TGT with a C-terminal Myc (TGT-Myc) or HA epitope (TGT-HA) and with constructs that express Qv1 with an N-terminal HA epitope (HA-Qv1) or a C-terminal Myc epitope (Qv1-Myc), as indicated above each panel. A, lysates were immunoprecipitated (IP) and immunoblotted (WB) using antibodies to Myc (α-Myc) or HA tag (α-HA). Total cell lysates Western-blotted directly served as transfection controls (Input). B, COS-7 cells co-transfected with TGT-Myc and HA-Qv1 plasmids were double-labeled with anti-HA and anti-Myc antibodies and analyzed by confocal microscopy. TGT (green in upper panel) and Qv1 (red in middle panel) localize to the cytoplasm. The merge (lower panel) of TGT and Qv1 in the presence of the DNA-selective DAPI stain (blue channel) indicates that the proteins co-segregate to the cytoplasm (yellow) and are excluded from the nucleus. Scale bar equals 20 μm.
Confocal microscopy of TGT-Myc and HA-Qv1 showed similar intracellular localization (Fig. 3B). A conspicuous feature of the confocal image is that the TGT-Myc (upper panel) and HA-Qv1 (middle panel) proteins are absent from the nucleus, as visualized by DAPI stain (lower panel). These data support the possibility that these proteins interact, as suggested earlier by the co-immunoprecipitation experiments.
Interaction of TGT and Qv1 Is Required for tRNA Guanine Transglycosylase Activity
There are no reports describing the successful overexpression of eukaryotic TGT in E. coli. Indeed, it has been stated that the cDNA for the human TGT catalytic subunit is toxic to E. coli (21), and it has been shown that expression of E. coli TGT in a naturally Q-deficient E. coli stain, B105, results in loss of viability (30).
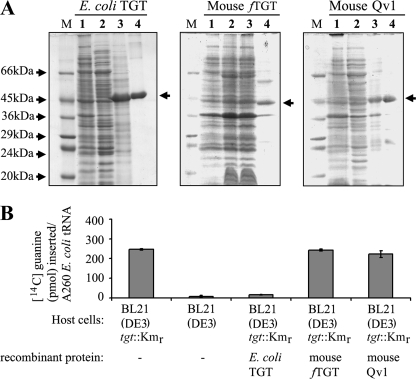
Despite these reservations, we developed a strain of E. coli (BL21 (DE3) tgt::Kmr) that are deficient in Q to overproduce the mouse TGT and Qv1 proteins; thereby negating the possible complication of chromosomally derived TGT contaminating the purified protein preparation (supplemental Fig. S1). BL21 (DE3) tgt::Kmr cells were transformed with plasmids used for the expression of E. coli TGT, mouse full-length TGT (fTGT), and mouse Qv1 and protein recovered as described under “Experimental Procedures.” SDS-PAGE analysis of the proteins (Fig. 4A) showed that E. coli TGT migrates at 46 kDa (left hand panel, lane 4), the fTGT migrates at 44 kDa (middle panel, lane 4), and Qv1 at 45 kDa (right hand panel, lane 4). Note that the hexahistidine tag (∼1.5 kDa) was not removed from E. coli TGT and Qv1 proteins, whereas the GST tag was removed from the fTGT protein.
FIGURE 4.
Expression of mouse TGT or Qv1 in tgt mutant E. coli fails to reconstitute Q biosynthesis. A, purification of recombinant E. coli TGT (left hand panel), full-length mouse TGT (fTGT, middle panel), and mouse Qv1 (right hand panel) from BL21(DE3) tgt::Kmr cells. Note that the hexahistidine tag was not removed from E. coli TGT or Qv1. Lane 1, uninduced E. coli; lane 2, isopropyl-1-thio-β-d-galactopyranoside-induced E. coli grown overnight; lane 3, insoluble pellet fraction; lane 4, purified protein from nickel-agarose (polyhistidine-tagged proteins) and glutathione-agarose (GST-tagged protein). M denotes protein molecular weight markers. B, evaluation of the queuosine content of tRNA isolated from BL21(DE3) or BL21(DE3) tgt::Kmr cells harboring no expression construct or constructs producing the proteins indicated. The presence of queuosine in tRNA inhibits the insertion of [14C]guanine by the E. coli TGT enzyme.
If the mammalian enzymes were catalytically active, they should have the capacity to insert preQ1 into bacterial tRNA (a known substrate for the eukaryotic enzyme (31)) and thereby restore Q-modification to tRNA. Once Q is incorporated into tRNA it can no longer function as a substrate for guanine insertion by the E. coli TGT enzyme (32). We therefore isolated bulk tRNA from BL21 (DE3) cells and BL21 (DE3) tgt::Kmr cells that were either nontransformed or expressing the E. coli TGT, fTGT, or Qv1 recombinant proteins (Fig. 4B). As expected, the E. coli TGT readily inserted radiolabeled guanine into tRNA from the BL21 (DE3) tgt::Kmr cells, but not tRNA from BL21 (DE3) cells. Overexpression of recombinant E. coli TGT in the BL21 (DE3) tgt::Kmr cells resulted in the tRNA no longer being accepted as a substrate for guanine insertion, verifying the presence of the Q-modification in tRNA. By contrast, the expression of fTGT or Qv1 did not restore Q-modification to tRNA as it remained a substrate for the insertion of radiolabeled guanine.
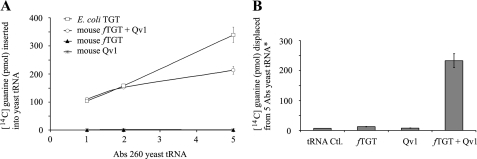
The isolated fTGT and Qv1 proteins were then assayed in vitro, individually and together, for tRNA guanine transglycosylase activity, by examining their ability to insert radiolabeled guanine into yeast tRNA (Fig. 5A). Purified E. coli TGT served as a control. It was observed that neither fTGT nor Qv1 alone could functionally insert guanine into tRNA. However, when added together in a 1:1 molar ratio, tRNA guanine transglycosylase activity was established, which was comparable to that of the E. coli TGT protein. To verify that the inability of fTGT and Qv1 to individually insert guanine was not attributed to its being a nonphysiological substrate, the ability of the enzymes to insert their natural substrate queuine into tRNA was analyzed. To do this, the E. coli TGT enzyme was first used to precharge radiolabeled guanine into tRNA (tRNA*), and the ability of fTGT and Qv1 to displace the guanine by queuine was assayed (Fig. 5B). Again, neither fTGT nor Qv1 could insert queuine into tRNA*. However, combining the two proteins resulted in clear queuine insertion and displacement of the radiolabeled guanine base.
FIGURE 5.
Co-incubation of TGT and Qv1 in vitro reconstitutes tRNA guanine transglycosylase activity. A, ability of radiolabeled guanine to be inserted into yeast tRNA (naturally queuosine-deficient) was examined for fTGT and Qv1 individually or together. The E. coli TGT served as a positive control for the assay. B, ability of fTGT and Qv1, either individually or together, to insert queuine into tRNA was assessed by the displacement of radiolabeled guanine that had been preloaded onto yeast tRNA.
Qv1 and TGT Co-localize to the Mitochondria
The distinct cytoplasmic co-localization of TGT and Qv1 observed earlier by confocal microscopy of transfected tagged constructs prompted us to examine the intracellular distribution of the endogenous proteins. Antisera specific to TGT and Qv1 were raised in rabbits. Counter-selecting the antisera against the opposing recombinant protein bound to support resin ensured their specificity (supplemental Fig. S3).
Confocal analysis of COS-7 cells showed that the TGT protein has a distinct perinuclear wisp-like appearance (Fig. 6A, panels A–C) that is curiously similar to the pattern observed for Qv1 (panels D–F). The pattern was suspected to arise from mitochondrial localization. Therefore, cells were co-stained using MitoTracker Red, a dye that recognizes actively respiring mitochondria. Our analysis clearly showed mitochondrial proximity for TGT (Fig. 6B, panels A–C) and Qv1 (panels D–F).
FIGURE 6.
TGT and Qv1 co-localize to the mitochondria in COS-7 cells. A, antisera specific to TGT (left hand side, red channel) and Qv1 (right hand side, red channel) were used for confocal analysis of COS-7 cells. DNA-selective DAPI stain (blue channel) was used to distinguish the nucleus. B, cells were probed with antisera specific to TGT (left hand side, green channel) and Qv1 (right hand side, green channel) and co-stained with MitoTracker Red to identify the mitochondria and DAPI stain. C, immunoblot analysis of cell fractions using TGT-specific and Qv1-specific antisera. UB denotes an upper band observed in the nuclear fractions of the TGT immunoblots. D, cells transfected with TGT-Myc were labeled with anti-Myc antibodies (upper panel, green channel) and DAPI stain (left hand side) or co-stained with MitoTracker Red (right hand side). Scale bar equals 20 μm.
Immunoblot analysis was performed on total cell lysate and the cytoplasmic, nuclear, membrane, and mitochondrial fractions of COS-7 cells. Interestingly, the endogenous TGT and Qv1 protein both gave a molecular mass of ∼62 kDa, which contrasts to the earlier size of ∼45 kDa observed for the recombinant enzymes. With respect to their intracellular distribution, the TGT protein was found to be cytoplasmic and nuclear in location. Quite surprisingly, no detectable signal was obtained in the mitochondrial fraction, even after prolonged exposure. The Qv1 protein, by contrast, showed a clear signal in the mitochondrial fraction, a weak cytoplasmic signal, and no detectable signal in the nucleus.
Proteins that localize to the mitochondria typically contain an N-terminal amphipathic signal sequence, not discernable in our Qv1 protein. It is noteworthy that translation from the start of exon 1 generates a 42-amino acid peptide continuous with the Qv1 reading frame. Analysis of this extension using the MitoProtII program gives a 0.9585 probability of export to the mitochondria (33). However, in silico genome walking of EST clones failed to identify an upstream translational start codon.
The above data indicated to us that although Qv1 stably associates with the mitochondria, the TGT protein is only loosely bound. We suspect that TGT dissociates upon purification of the mitochondria. To verify this possibility, the TGT-Myc construct was transfected into COS-7 cells and the intracellular distribution examined. A perinuclear wisp-like pattern, identical to that of the endogenous protein, was observed (Fig. 6D, panels A–C) which co-localized with Mitotracker Red stain (panels D–F); thus, verifying the innate capacity of the TGT protein to associate with the mitochondria.
The interaction of TGT and Qv1 was also examined by size-exclusion chromatography. Because of the difficulty in obtaining sufficient amounts of fTGT (∼60 μg per liter) we examined the possibility of removing the first 16 amino acids, which do not form part of the (β/α)8 core, to help enhance protein expression (supplemental Fig. S4A). The truncated version of TGT (tTGT) was found to express comparatively well (∼1 mg per liter). Importantly, the transglycosylase activity of tTGT was found to be similar to that of fTGT (supplemental Fig. S4B). With gel filtration, TGT gave a single peak corresponding to 26 kDa (supplemental Fig. S5A) whereas Qv1 eluted at a molecular mass of ∼37.5 kDa (supplemental Fig. S5B), suggesting that both TGT and Qv1 are monomers in solution. The fact that both proteins migrate at a significantly smaller size by gel filtration relative to SDS-PAGE (∼45 kDa) could indicate that the proteins have a tendency to interact with the column matrix. It is interesting, however, that previous gel filtration studies on Superdex 200 of bovine liver tRNA transglycosylase gave sizes corresponding to 65 and 32 kDa (17). Likewise, the enzyme activity from rat liver gave a 60- and 34.5-kDa band on denaturing gel electrophoresis and a major peak of enzymatic activity at ∼30–38 kDa following FPLC chromatography on Superose-6 (18).
To examine the capacity of the TGT and Qv1 proteins to interact, equimolar amounts of both proteins were added together and gel-filtration chromatography performed. Only two peaks were evident at 26 and 37.5 kDa (supplemental Fig. S5C) indicating that TGT and Qv1 do not spontaneously associate in a stable form in solution. We conclude that the TGT subunit associates with the external membrane of the mitochondria and through direct or indirect interaction with the Qv1 subunit creates an active tRNA guanine transglycosylase complex.
DISCUSSION
This study is the first to identify the eukaryotic complex responsible for Q formation in tRNA as a heteromer of TGT and Qv1. Both proteins are found in diverse animal tissues, and their homologues are widely distributed in eukaryotic species, in agreement with the near ubiquitous distribution of their reaction product, Q-modified tRNA (1).
The TGT subunit of the eukaryotic enzyme shows a marked conservation of catalytic residues with respect to its eubacterial counterparts. It is surprising, therefore, that it displays no inherent catalytic activity in isolation when presented with either guanine or queuine as substrate. However, it may be anticipated that it is the TGT subunit that performs the core transglycosylation step during catalysis, given the clear sequence divergence of Qv1 from the active eubacterial enzymes.
A number of potential functions for the Qv1 partner subunit are conceivable. Crystal structure data have shown that Z. mobilis TGT dimerizes in the presence of a tRNA stem loop (13, 10). The dimer was found to accommodate only one tRNA molecule, and, as a result of restrictions imposed by close packing of the proteins, only one subunit was capable of forming a strong interaction with the U33G34U35 anticodon. The second subunit was exempt from anticodon recognition, instead forming interactions with the anticodon stem loop, where it served as a platform to fix the bound tRNA (10). The Qv1 subunit of the eukaryotic complex could be similarly engaged during the catalytic process. A need for a partner subunit may also arise from the fundamental eukaryotic requirement to salvage the queuosine 5′-phosphate nucleoside after normal tRNA turnover, by cleavage of the 7-deazapurine-N-glycosidic bond (7). Indeed, these two processes are not mutually exclusive and theoretically could occur in tandem.
Proceeding from the above argument, the stable association of Qv1 with mitochondria raises some confounding issues. First, although a conceivable mechanism for Q-modification of cytosolic tRNA may be reached, an explanation of how the mitochondrial encoded aspartyl tRNA (a known substrate for Q-modification) is processed, escapes explanation at present. Second, the means by which Qv1 locates to the mitochondria is uncertain and requires further investigation. Our preliminary data suggest that neither N- nor C-terminally tagged Qv1 associates with the mitochondria4 signifying that the endogenous protein exploits additional processes. In likelihood, a mitochondria-targeting sequence directs Qv1 localization, as suggested from our theoretical translation of the upstream noncoding region. Third, the interaction of TGT and Qv1 must necessarily occur through exposed domains on both proteins, with obvious limitations inherent to Qv1. Drawing on observations from the Z. mobilis dimer, Qv1 shows an almost complete conservation of residues known to form the eubacterial dimer interphase within its C-terminal zinc-binding domain, whereas the eukaryotic TGT shows a reciprocal conservation of dimer-forming residues in the N-terminal β1-α1 region. We suggest that Qv1 and TGT may utilize these respective domains for their association.
Interestingly, the gel-filtration studies showed no stable interaction between recombinant TGT and Qv1. It is possible that substrate (tRNA) could be involved in bridging the two proteins during the catalytic reaction as was observed for the Z. mobilis TGT in the presence of RNA (10). Alternatively, additional proteins may be required, such as those found during protein purification studies of the mammalian tRNA guanine transglycosylase activity (17, 20). A further possibility is the involvement of a post-translational phosphorylation event. Previous studies on the purified rat tRNA guanine transglycosylase activity have shown it to be a substrate for and activated by protein kinase C (PKC, 18). The report argues that the transglycosylase enzyme exists as a heterodimer of ∼104 kDa, composed of a regulatory 60-kDa subunit, thought to be the PKC substrate, and a smaller 34.5-kDa catalytic subunit (18).
There is a notable difference in size between the recombinant TGT and Qv1 (44 and 45 kDa, respectively) relative to that of the endogenous proteins (both 62 kDa) on SDS-PAGE. Although no concrete explanation for this difference can be given at present, it is possible that additional upstream sequence exists for both proteins. The small sizes observed for the recombinant proteins by gel filtration, 26 kDa for an N-terminally truncated TGT (missing the first 16 amino acids) and 37.5 kDa for Qv1 may signify that the proteins are interacting nonspecifically with the gel-filtration resin. It is noteworthy that proteins corresponding to molecular masses of 32–35 kDa have been observed previously in gel-filtration studies of rat (18) and bovine liver (17) tRNA guanine transglycosylase activity.
Because TGT and Qv1 are found so extensively in eukaryotic species, it is interesting to speculate on their evolutionary origin. The divergence of Qv1 from the eubacterial enzymes could indicate that it is the older of the two, arising at the time when eubacterial and archaeo-eukaryotic lineages divided. The similarity of the eubacterial and eukaryotic TGT proteins, on the other hand, suggests the eukaryotic enzyme was acquired later in evolution, most probably as a result of horizontal transfer from the pro-mitochondria (34).
The need for the eukaryotic TGT to engage with a mitochondria-localized partner for catalysis inevitably infers that the Q-modification of tRNA occurs in the cytoplasm and not in the nucleus. In agreement with this conclusion, Q-modification is a terminal step in tRNA maturation, occurring only after removal of the precursor tRNA termini and of intervening intronic sequences (35). Furthermore, previous studies using Xenopus laevis oocytes have shown that nuclear microinjection of a cloned yeast tRNATyr gene does not lead to appreciable amounts of Q-modification (35) in comparison to the efficient Q-modification of yeast tRNAAsp when microinjected into the cytoplasm (36, 37). Incidentally, our studies invariably demonstrate the exclusion of Qv1 from the nucleus, which may be dependent on a canonical leucine-rich export consensus present in the protein.
No attempt was made to functionally characterize the Qv0 and Qv2 proteins, because they are not major forms in adult mice. A role of Qv0 in development may be inferred from the fact it was found in an embryonic germ cell library. On the other hand, the Qv2 EST was derived from a retinal library, and given that it has lost the zinc-binding domain, it may be speculated that it is catalytically inactive.
The results of this study open the way for an in-depth analysis of the eukaryotic tRNA guanine transglycosylase and of the physiological role that Q plays. Studies to date have shown clear changes in Q-tRNA levels during development (38) and differentiation (39, 40), and a loss of Q-tRNA during malignancy (41), which may relate to the stage of the disease (42). In addition, the absence of Q has been shown to be lethal in mice when tyrosine is withdrawn from the diet (43). Undoubtedly, additional roles for this ubiquitous dietary-derived component are awaiting discovery.
Supplementary Material
Acknowledgments
We thank Dr. Tim Mantle and Dr. Ken Mok for helpful advice and discussions and Dr. Gavin McManus for assistance with gel-filtration chromatography.
This work was supported by Programme Grant 05-IN3-I761 and Research Frontiers Grant BIMF318 from the Science Foundation Ireland.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S5.
The nucleotide sequence(s) reported in this paper has been submitted to the Gen-BankTM/EBI Data Bank with accession number(s) FM985972.
4 V. P. Kelly and P. Hayes, unpublished observations.
- Q
- queuosine
- TGT
- tRNA guanine transglycosylase
- Qv1
- queuine tRNA guanine transglycosylase domain containing 1 variant 1
- preQ1
- 7-aminomethyl-7-deazaguanine
- fTGT
- full-length mouse TGT
- tTGT
- truncated mouse TGT
- HA
- hemagglutinin
- MOPS
- 4-morpholinepropanesulfonic acid
- DAPI
- 4′,6-diamidino-2-phenylindole
- EST
- expressed sequence tag
- GST
- glutathione S-transferase
- RT-PCR
- reverse transcription-PCR.
REFERENCES
- 1.Nishimura S. ( 1983) Prog. Nucleic Acid Res. Mol. Biol. 28, 49– 73 [DOI] [PubMed] [Google Scholar]
- 2.Kasai H., Oashi Z., Harada F., Nishimura S., Oppenheimer N. J., Crain P. F., Liehr J. G., von Minden D. L., McCloskey J. A. ( 1975) Biochemistry. 14, 4198– 4208 [DOI] [PubMed] [Google Scholar]
- 3.Katze J. R., Basile B., McCloskey J. A. ( 1982) Science. 216, 55– 56 [DOI] [PubMed] [Google Scholar]
- 4.Randerath E., Agrawal H. P., Randerath K. ( 1984) Cancer Res. 44, 1167– 1171 [PubMed] [Google Scholar]
- 5.Mörl M., Dörner M., Pääbo S. ( 1995) Nucleic Acids Res. 23, 3380– 3384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Iwata-Reuyl D. ( 2003) Bioorg. Chem. 31, 24– 43 [DOI] [PubMed] [Google Scholar]
- 7.Gündüz U., Katze J. R. ( 1984) J. Biol. Chem. 259, 1110– 1113 [PubMed] [Google Scholar]
- 8.Gregson J. M., Crain P. F., Edmonds C. G., Gupta R., Hashizume T., Phillipson D. W., McCloskey J. A. ( 1993) J. Biol. Chem. 268, 10076– 10086 [PubMed] [Google Scholar]
- 9.Ishitani R., Nureki O., Fukai S., Kijimoto T., Nameki N., Watanabe M., Kondo H., Sekine M., Okada N., Nishimura S., Yokoyama S. ( 2002) J. Mol. Biol. 318, 665– 677 [DOI] [PubMed] [Google Scholar]
- 10.Stengl B., Reuter K., Klebe G. ( 2005) Chembiochem. 6, 1926– 1939 [DOI] [PubMed] [Google Scholar]
- 11.Romier C., Reuter K., Suck D., Ficner R. ( 1996) EMBO J. 15, 2850– 2857 [PMC free article] [PubMed] [Google Scholar]
- 12.Garcia G. A., Kittendorf J. D. ( 2005) Bioorg. Chem. 33, 229– 251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xie W., Liu X., Huang R. H. ( 2003) Nat. Struct. Biol. 10, 781– 788 [DOI] [PubMed] [Google Scholar]
- 14.Reuter K., Ficner R. ( 1995) J. Bacteriol. 177, 5284– 5288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Garcia G. A., Koch K. A., Chong S. ( 1993) J. Mol. Biol. 231, 489– 497 [DOI] [PubMed] [Google Scholar]
- 16.Howes N. K., Farkas W. R. ( 1978) J. Biol. Chem. 253, 9082– 9087 [PubMed] [Google Scholar]
- 17.Slany R. K., Müller S. O. ( 1995) Eur. J. Biochem. 230, 221– 228 [DOI] [PubMed] [Google Scholar]
- 18.Morris R. C., Brooks B. J., Eriotou P., Kelly D. F., Sagar S., Hart K. L., Elliott M. S. ( 1995) Nucleic Acids Res. 23, 2492– 2498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Walden T. L., Jr., Howes N., Farkas W. R. ( 1982) J. Biol. Chem. 257, 13218– 13222 [PubMed] [Google Scholar]
- 20.Deshpande K. L., Seubert P. H., Tillman D. M., Farkas W. R., Katze J. R. ( 1996) Arch. Biochem. Biophys. 326, 1– 7 [DOI] [PubMed] [Google Scholar]
- 21.Deshpande K. L., Katze J. R. ( 2001) Gene 265, 205– 212 [DOI] [PubMed] [Google Scholar]
- 22.Gündüz U., Elliott M. S., Seubert P. H., Houghton J. A., Houghton P. J., Trewyn R. W., Katze J. R. ( 1992) Biochim. Biophys. Acta 1139, 229– 238 [DOI] [PubMed] [Google Scholar]
- 23.Yang W. K., Novelli G. D. ( 1971) Methods Enzymol. 20, 44– 55 [Google Scholar]
- 24.Hoops G. C., Townsend L. B., Garcia G. A. ( 1995) Biochemistry 34, 15381– 15387 [DOI] [PubMed] [Google Scholar]
- 25.Eppig J. T., Blake J. A., Bult C. J., Kadin J. A., Richardson J. E. ( 2007) Nucleic Acids Res. 35, D630– D637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Thanaraj T. A., Stamm S., Clark F., Riethoven J. J., Le Texier V., Muilu J. ( 2004) Nucleic Acids Res. 32, D64– D69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pollastri G., McLysaght A. ( 2005) Bioinformatics 21, 1719– 1720 [DOI] [PubMed] [Google Scholar]
- 28.la Cour T., Kiemer L., Mølgaard A., Gupta R., Skriver K., Brunak S. ( 2004) Protein Eng. Des. Sel. 17, 527– 536 [DOI] [PubMed] [Google Scholar]
- 29.Kutay U., Güttinger S. ( 2005) Trends Cell Biol. 15, 121– 124 [DOI] [PubMed] [Google Scholar]
- 30.Dineshkumar T. K., Thanedar S., Subbulakshmi C., Varshney U. ( 2002) Microbiology 148, 3779– 3787 [DOI] [PubMed] [Google Scholar]
- 31.Shindo-Okada N., Okada N., Ohgi T., Goto T., Nishimura S. ( 1980) Biochemistry 19, 395– 400 [DOI] [PubMed] [Google Scholar]
- 32.Okada N., Nishimura S. ( 1979) J. Biol. Chem. 254, 3061– 3066 [PubMed] [Google Scholar]
- 33.Claros M. G., Vincens P. ( 1996) Eur. J. Biochem. 241, 779– 786 [DOI] [PubMed] [Google Scholar]
- 34.Anantharaman V., Koonin E. V., Aravind L. ( 2002) Nucleic Acids Res. 30, 1427– 1464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nishikura K., De Robertis E. M. ( 1981) J. Mol. Biol. 145, 405– 420 [DOI] [PubMed] [Google Scholar]
- 36.Haumont E., Droogmans L., Grosjean H. ( 1987) Eur. J. Biochem. 168, 219– 225 [DOI] [PubMed] [Google Scholar]
- 37.Carbon P., Haumont E., De Henau S., Keith G., Grosjean H. ( 1982) Nucleic Acids Res. 10, 3715– 3732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Singhal R. P., Kopper R. A., Nishimura S., Shindo-Okada N. ( 1981) Biochem. Biophys. Res. Commun. 99, 120– 126 [DOI] [PubMed] [Google Scholar]
- 39.Shindo-Okada N., Terada M., Nishimura S. ( 1981) Eur. J. Biochem. 115, 423– 428 [DOI] [PubMed] [Google Scholar]
- 40.Chen Y. L., Wu R. T. ( 1994) Cancer Res. 54, 2192– 2198 [PubMed] [Google Scholar]
- 41.Baranowski W., Dirheimer G., Jakowicki J. A., Keith G. ( 1994) Cancer Res. 54, 4468– 4471 [PubMed] [Google Scholar]
- 42.Dirheimer G., Baranowski W., Keith G. ( 1995) Biochimie 77, 99– 103 [DOI] [PubMed] [Google Scholar]
- 43.Marks T., Farkas W. R. ( 1997) Biochem. Biophys. Res. Commun. 230, 233– 237 [DOI] [PubMed] [Google Scholar]
- 44.Nicholas K. B., Nicholas H. B., Jr., Deerfield D. W., II ( 1997) EMBNEW.NEWS 4, 14 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.