Abstract
Maintenance of genomic stability ensures faithful transmission of genetic information and helps suppress neoplastic transformation and tumorigenesis. Although recent progress has advanced our understanding of DNA damage checkpoint regulations, little is known as to how DNA repair, especially the RAD51-dependent homologous recombination repair pathway, is executed in vivo. Here, we reveal novel properties of the BRCA2-associated protein PALB2 in the assembly of the recombinational DNA repair machinery at DNA damage sites. Although the chromatin association of PALB2 is a prerequisite for subsequent BRCA2 and RAD51 loading, the focal accumulation of the PALB2·BRCA2·RAD51 complex at DSBs occurs independently of known DNA damage checkpoint and repair proteins. We provide evidence to support that PALB2 exists as homo-oligomers and that PALB2 oligomerization is essential for its focal accumulation at DNA breaks in vivo. We propose that both PALB2 chromatin association and its oligomerization serve to secure the BRCA2·RAD51 repair machinery at the sites of DNA damage. These attributes of PALB2 are likely instrumental for proficient homologous recombination DNA repair in the cell.
Fanconi anemia is a rare disease in which patients are prone to the development of childhood aplastic anemia and cancer as well as other congenital defects. Cellular phenotypes of FA4 patients are also characterized by their hypersensitivity toward DNA-cross-linking agents, such as mitomycin C (MMC) or cisplatin. Accordingly, MMC treatment greatly induces aberrant chromosomal structures in cells derived from FA patients, including chromosome breakage and chromatin interchanges. Thus, genomic instability is considered as one of the fundamental causes responsible for the clinical and cellular phenotypes observed among FA patients.
In human cells two major repair pathways are employed to repair DSBs, namely the homologous recombination (HR) and the non-homologous end-joining pathways. The use of the sister chromatid as information donor during repair renders HR a largely faithful mechanism (1), whereas non-homologous end-joining often leads to genetic mutations because of the gain or loss of genetic information (2).
Mounting evidence suggests a functional connection between the 13 FA-complementation group genes (FA-A, B/FAAP95, C, D1/BRCA2, D2, E, F, G/XRCC9, I, J/BACH1, L/PHF9/FAAP43, M/Hef/FAAP250, and N/PALB2) and the DNA repair pathway (3). Recent studies revealed that eight of the FA proteins form a complex to facilitate the ubiquitylation of FANCD2 and FANCI; however, mechanistically how they affect DNA repair remains elusive. Importantly, the identification of the FANCJ/BACH1, FANCD1/BRCA2, and FANCN/PALB2 proteins as components of the HR machinery further support the notion that FA mutations result in DNA repair defects (3–7).
Genetics and biochemical studies have shown that the FANCD1 product, BRCA2, facilitates the assembly of RAD51 onto ssDNA substrates, forming a nucleoprotein filament (8–10) that catalyzes DNA strand invasion and D-loop formation. Accordingly, abrogation of FANCD1/BRCA2 function abolishes focal accumulation of RAD51 at DNA breaks. The recent identification of FANCN/PALB2 as the Partner and Localizer of BRCA2 (11) indicated that, much like the damage-signaling pathway, a hierarchical relationship exist for the HR pathway. PALB2 is essential for the focal accumulation of BRCA2 and RAD51 at DSBs. Moreover, PALB2 depletion compromised HR repair and cell survival in response to genotoxic stress (11). Similarly, HR defects and hypersensitivity to cross-linking agents are restored in FANCN/PALB2 patient cells by reconstitution or spontaneous reversion of PALB2, indicating that PALB2 dysfunction is responsible for this FA subtype (12). Moreover, inactivation of PALB2 has also been implicated in breast cancer predisposition, as truncation mutations of PALB2 are found in familial breast cancer cases with intact BRCA1 and BRCA2 (13–15). PALB2 mutations are also associated with an elevated frequency of prostate and colorectal cancers, although the role of PALB2 in the suppression of these cancer types requires further exploration (14, 16). Nevertheless, these human genetic studies provide strong evidence to support that PALB2 plays a critical role in HR repair and is important for the maintenance of genomic integrity and tumor suppression.
Given the intimate relationship between PALB2 and HR repair, we decided to examine mechanistically how PALB2 regulates the BRCA2-RAD51-dependent DNA repair events. Interestingly, we found an oligomerization domain on PALB2 and provide evidence to support that PALB2 focal accumulation at the site of DNA damage requires its oligomerization property. Together with its chromatin associating ability, PALB2 initiates recombinational repair at DSBs via the coordination of BRCA2 and RAD51 association with chromatin and the concentration of the repair complex at sites of DNA breaks.
EXPERIMENTAL PROCEDURES
Antibodies
Monoclonal antibodies against the FLAG epitope (M2) were purchased from Sigma. Rabbit polyclonal anti-RAD51 (D51), anti-BRCA2 (C25), and anti-phopsho-H2AX antibodies were described previously (17). Rabbit polyclonal anti-PALB2 antibodies were generated by immunizing rabbits with GST-PALB2 F6 (residues 611–764) recombinant protein expressed and purified from Escherichia coli. Goat anti-ATR antibody (C-17) and mouse anti-CHK1 were purchased from Santa Cruz, and rabbit anti-p-CHK1 S317 antibody was purchased from Upstate. Both β-actin and β-tubulin antibodies were purchased from Sigma.
Cell Cultures
Cell lines of human origin were maintained in RPMI supplemented with 10% fetal bovine serum and 1% penicillin and streptomycin. Mouse embryonic fibroblasts deficient of ATM, 53BP1, H2AX, MDC1, and RNF8 were maintained in Dulbecco's modified Eagle's medium supplemented with 15% fetal bovine serum and 1% penicillin and streptomycin (18–21). Cell lines were maintained in 37 °C incubator with 5% CO2.
Constructs
pOZC-PALB2 (a gift from Dr. David Livingston (Dana-Farber Cancer Institute)) was subcloned into the entry vector pDONR201 (Invitrogen). Mutations or deletions of PALB2 were generated using site-directed mutagenesis (QuikChange, Stratagene). All plasmid transfection into mammalian cells were performed using Lipofectamine 2000.
Retrovirus Production and Infection
pDONR201-PALB2 constructs were transferred into a gateway-compatible pEF1A-HA-FLAG retroviral vector. Virus supernatant was collected 48 h after the co-transfection of pEF1A vectors and pcl-ampho into BOSC23 cells. EUFA1341 cells were infected with viral supernatant in the presence of Polybrene. Cells were selected in growth media containing 2 μg/ml puromycin. Protein expression in transduced cells was confirmed by Western blot and immunofluoresence staining using anti-FLAG antibodies.
RNA Interference
SmartPool siRNA targeting human ATR and CHK1 were purchased from Dharmacon. A non-targeting siRNA was used as the control. U2OS cells were seeded at 30% confluency for 24 h before double siRNA transfection using Oligofectamine (Invitrogen). Forty-eight hours after the second siRNA transfection, cells were subjected to ionizing radiation (10 Gy) and collected for further analysis.
Chromatin Fractionation, Immunoprecipitation, and Pulldown Experiments
Cells were lysed in NETN (20 mm Tris-HCl, pH 8, 100 mm NaCl, 1 mm EDTA, 0.5% Nonidet P-40) buffer. After centrifugation, the pellets were washed extensively with NETN and boiled in 2× Laemmli buffer to extract chromatin-associated proteins.
For immunoprecipitation or pulldown experiments, cell extracts prepared using NETN buffer were incubated with either S-agarose (EMD Biosciences) or GST fusion proteins immobilized on glutathione beads for 2 h at 4 °C. Beads were washed with NETN buffer, and proteins were eluted by boiling in 2× Laemmli buffer. Samples were resolved by SDS-PAGE and transferred to polyvinylidene difluoride membrane. Immunoblotting was subsequently performed with the antibodies as indicated.
Immunofluoresence Staining
Cells grown on coverslips were mock-treated or treated with 10 Gy of γ-irradiation. Cells were then pre-extracted with buffer containing 0.5% Triton X and fixed using 3% paraformaldehyde solution. Immunostaining experiments were performed using anti-FLAG (M2), anti-γH2AX, BRCA2, RAD51, and PALB2 antibodies. Cells were mounted onto glass slides in 4′,6-diamidino-2-phenylindole-containing antifade. Immunofluorescent analyses and image capturing were performed on a Nikon Eclipse 800 microscope.
MMC Sensitivity Assay
1 × 103 cells were seeded onto a 60-mm dish in triplicate. Different concentrations of MMC were added 24 h after cell seeding. Cells were incubated for 14 days, and the resulting colonies were fixed and stained with Coomassie Blue. Number of colonies was counted using a GelDoc with Quantity One software (Bio-Rad). Results were the averages of data obtained from three independent experiments.
Gene Conversion Assay
1 × 106 cells were electroporated with 15 μg of pDR-GFP plasmid together with 5 μg of pCBASce plasmid at 270 V, 975 microfarads using a Bio-Rad Gene Pulsar II. Cells were plated onto 60-mm dishes and incubated in culture media for 48 h before fluorescence-activated cell sorter analyses. Cells were analyzed in a BD Biosciences FACScan on a green (FL1) versus orange (FL2) fluorescence plot. Results were the averages of data obtained from three independent experiments.
RESULTS
Role of PALB2 in HR Repair Complex Assembly
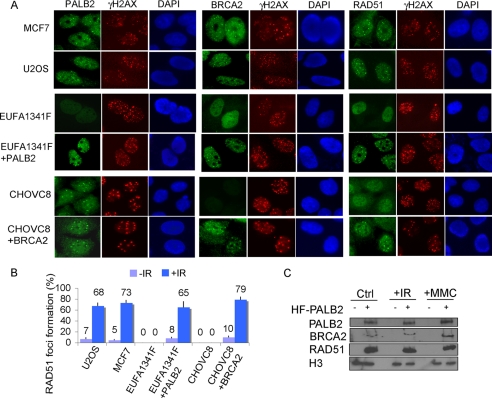
PALB2 was previously ascribed a role as a scaffold protein and is essential for BRCA2 relocalization in response to DNA damage (11). Given that BRCA2 is required for RAD51 accumulation at DNA breaks, we asked whether the three proteins might function in a linear and hierarchical pathway. Discernible foci that colocalized with those of γH2AX were observed for PALB2, BRCA2, and RAD51 in U2OS and MCF7 cells after ionizing radiation treatment (IR) (Fig. 1, A and B). In contrast, both BRCA2 and RAD51 foci were absent in the PALB2-deficient fibroblast EUFA1341, indicating that PALB2 function is required for the focal accumulation of both repair proteins at DSBs. To further test whether PALB2 is indeed responsible for the DSB recruitment of BRCA2 and RAD51, we reconstituted EUFA1341 cells with PALB2 (EUFA1341+PALB2) and examined BRCA2 and RAD51 IR-induced foci formation (IRIF). Results indicated that BRCA2 and RAD51 IRIF was restored and can be readily observed in EUFA1341+PALB2 cells. On the other hand, the retention of PALB2 at DNA breaks was unaffected by the BRCA2 status, as PALB2 foci persisted in both BRCA2-deficient VC8 Chinese hamster ovary (CHO) cells and a derivative that has been reconstituted with wild-type BRCA2 (Fig. 1A). This is in contrast to RAD51, whose foci were observed only in CHO VC8 cells reconstituted with BRCA2 (Fig. 1B). These data suggest that PALB2 facilitates the accumulation of RAD51 at DNA lesions by targeting BRCA2 to chromatin.
FIGURE 1.
PALB2 controls focal accumulation of BRCA2 and RAD51 at DNA damage site. A, cells were irradiated with 10 Gy of ionizing radiation and recovered for 5 h. Immunofluorescence staining was performed using antibodies as indicated. C, PALB2-deficient EUFA1341 cells and its derivative (EUFA1341+PALB2) were treated with 10 Gy of ionizing radiation and recovered for 5 h or 1 mm MMC for 24 h. Cell lysates were separated by fractionation to retrieve both nuclear and chromatin-associated proteins. The presence of PALB2, BRCA2, and RAD51 was examined by Western blot analyses as indicated. B, RAD51 foci formation was examined in indicated cell lines using anti-RAD51 antibody. DAPI, 4′,6-diamidino-2-phenylindole.
The above cytological observations suggest a linear relationship between PALB2, BRCA2, and RAD51 in response to DNA damage. To biochemically test if the PALB2 is responsible for the recruitment of BRCA2·RAD51 complex to chromatin, EUFA1341 or EUFA1341+PALB2 were treated with either IR or DNA cross-linking agent MMC. Chromatin fractionation indicated that only in the presence of PALB2, a significant portion of BRCA2 and RAD51 associated with chromatin after DNA damage (Fig. 1C). Together, these results indicate that PALB2 is crucial for the recruitment of BRCA2 and RAD51 to chromatin and is important for their focal accumulation at sites of DNA breaks.
Recruitment of the PALB2·BRCA2·RAD51 Repair Complex to DSBs Can Occur Independently of Several Known DNA Damage Repair or Checkpoint Pathways
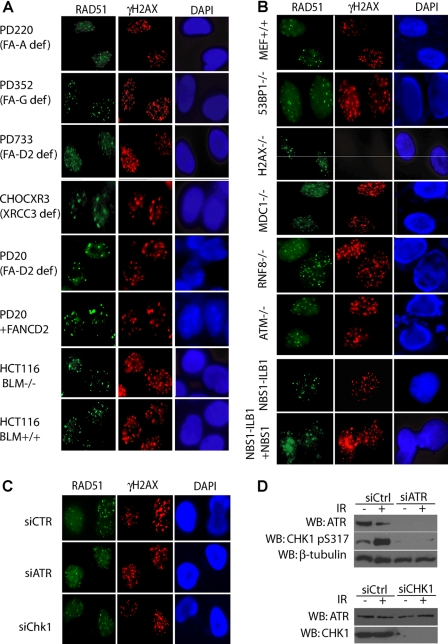
In an attempt to identify possible upstream signaling molecules that facilitate the accumulation of the PALB2·BRCA2·RAD51 complex at DNA damage sites, we examined its DNA damage-induced focus formation in cells deficient for various components in DNA damage repair or checkpoint pathways (Fig. 2). Because of the limited sensitivity of our PALB2 and BRCA2 antibodies, RAD51 focus formation was exploited as a marker to assess the focal accumulation of the PALB2·BRCA2·RAD51 complex in response to DNA damage.
FIGURE 2.
RAD51 focus formation is not dependent on major DNA repair and DNA damage checkpoint pathway components. Cells deficient in various components in DNA repair (A) or DNA damage signaling and checkpoint pathways (B and C) were examined for the formation of ionizing radiation induced RAD51 foci. Cells were all collected 5 h after radiation (10 Gy), and immunostaining experiments were performed with anti-RAD51 and anti-γH2AX antibodies. D, expression levels of ATR and CHK1 after siRNA knockdown were monitored by Western blot (WB) analyses. DAPI, 4′,6-diamidino-2-phenylindole. siCTR, control siRNA.
We first investigated cell lines deficient for various DNA repair components. These included the FANCA-deficient (PD220), FANCG-deficient (PD352), and FANCD2-deficient (PD733) cells. As shown in Fig. 2A, RAD51 IRIF was not drastically changed by deficiencies in these FA genes. Examination of FANCD2 deficient (PD20) and its derivative cell line that has been reconstituted with FANCD2 (PD20+FANCD2) further confirmed that RAD51 IRIF could form in FANCD2-deficient cells (Fig. 2A). Likewise, cells deficient in the BLM helicase (HCT116 BLM−/−) also did not reveal any exceedingly large change in RAD51 focal accumulation as compared with its normal counterpart (Fig. 2A).
Next we tested whether the localization of PALB2·BRCA2·RAD51 complex might be regulated by any of the known DNA damage checkpoint pathway components (Fig. 2B). In a series of mouse embryonic fibroblasts generated from different knock-out mice, we observed that RAD51 was able to localize to γH2AX-containing foci in ATM−/−, 53BP1−/−, H2AX−/−, MDC1−/−, and −/− mouse embryonic fibroblasts and their respective wild-type counterparts after ionizing radiation. RAD51 foci formation was also detected in cells deficient for NBS1 (Fig. 2B). Similarly, RAD51 IRIF was observed after depletion of ATR or CHK1 in U2OS cells (Fig. 2, C and D).
We noted that there were changes in the percentages of cells displaying RAD51 foci in some of the deficient cells examined (data not shown); however, none of these deficiencies led to complete loss of RAD51 foci as observed in PALB2- and BRCA2-deficient cells. Although it is likely that RAD51 focus formation may be altered in some of these cell lines and, therefore, suggest that some of these DNA damage repair or checkpoint proteins may regulate RAD51 focal formation, our results indicated that none of them is essential for the formation of RAD51 foci after DNA damage.
PALB2 Focal Accumulation to DSBs Requires Its Oligomerization
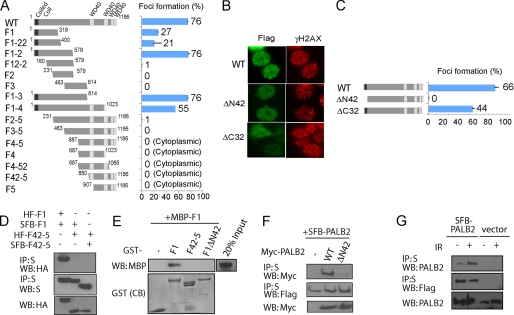
To elucidate how PALB2 is recruited to damaged chromatin, HEK293T was transiently transfected with plasmids encoding wild type and a series of overlapping S-tag, FLAG epitope, and streptavidin-binding peptide tripe-tagged (dubbed as SFB-tag)-PALB2 deletion mutants spanning the entire coding sequence of PALB2 (Fig. 3A). Cells were exposed to IR, and indirect immunofluorescence staining was used to visualize PALB2 localization. Like endogenous PALB2, SFB-tagged PALB2 became concentrated at foci that colocalized with γH2AX (Fig. 3, A and B). Interestingly, such co-localization with γH2AX was observed only in PALB2 deletion mutants that retained the N terminus (F12, F13, and F14), indicating that the PALB2 N terminus mediates PALB2 accumulation at DNA lesions. Further deletions of flanking residues from either side of the F12 fragment affected PALB2 foci formation. Note that F12-2 demonstrated a reduction in the percentage of cells with damage-induced foci, indicating a possible foci formation domain within residues 1–160 of PALB2 (Fig. 3A). A domain search identified a coiled-coil domain runs from residues 9 to 42 of PALB2. Notably, deletion of the coiled-coil domain (ΔN42) resulted in severely diminished IRIF resembling the phenotype of mutant F12-2, indicating that the PALB2 N terminus plays an important role in mediating its retention at DNA damage sites (Fig. 3, B and C).
FIGURE 3.
PALB2 focus formation at DSBs requires its N terminus. A, HEK293T cells transfected with plasmids encoding SFB-tagged wild type (WT) or deletion mutants of PALB2 were exposed to 10 Gy of ionizing radiation. Cells were fixed, and immunostaining was performed with anti-FLAG and anti-pH2AX antibodies. The percentage of cells showing foci overlapping with γH2AX was plotted. B, foci accumulation of wild type and PALB2 internal deletion mutants. The percentage of foci positive cells was plotted in C. D, co-immunoprecipitation (IP) experiments using hemagglutinin (HA)-FLAG-tagged and SFB-tagged PALB2 F1 and F42–5 fragments were performed (5% input was showed). WB, Western blot. E, recombinant GST and MBP-tagged PALB2 fragments were subjected to pulldown assays. CB, Coomassie Blue staining. F, Myc-tagged wild-type and ΔN42 PALB2 mutant were subjected to co-immunoprecipitation experiments along with wild-type SFB-tagged PALB2 (5% input was showed). G, 293T cells stably expressing SFB-PALB2 or vector alone were mock-treated or irradiated. Cell lysates were subjected to S beads pulldown (5% input was showed). The amounts of SFB-tagged and endogenous PALB2 presented in the precipitates were determined by Western blotting analyses using anti-FLAG or anti-PALB2 antibodies.
Because coiled-coil domain is often involved in protein oligomerization, we tested whether PALB2 has intrinsic oligomerization property. The SFB-PALB2 N terminus specifically immunoprecipitated HA-PALB2 F1 fragment but not the C-terminal F42–5 fragment (Fig. 3D). In addition, the C-terminal F42–5 fragments did not form oligomers (Fig. 3D). These data suggest that PALB2 oligomerizes via its N terminus. Next we tested if the deletion of the coiled-coil domain could abolish the PALB2 oligomerization. Bacterially expressed and purified GST- or MBP-tagged PALB2 fragments were used in pulldown experiments. Although the GST-PALB2 F1 fragment successfully pulled down MBP-PALB2 F1 in vitro, deletion of the coiled-coil domain (F1ΔN42) abolished this interaction (Fig. 3E). Accordingly, the Myc-ΔN42 deletion mutant also failed to interact with wild-type SFB-PALB2, whereas oligomerization occurred between SFB- and Myc-tagged wild-type PALB2 in vivo (Fig. 3F). Interestingly, we observed an enhanced oligomerization between SFB-PALB2 and endogenous PALB2 after DNA damage (Fig. 3G), implying that the oligomerization status of PALB2 may be modulated and, thus, account for its focal localization after DNA damage. Taken together, our data suggest that PALB2 N terminus, which harbors a coiled-coil domain required for its oligomerization, is important for its focal concentration at DNA damage sites.
PALB2 C Terminus WD40 Repeats Mediate Its Interaction with BRCA2
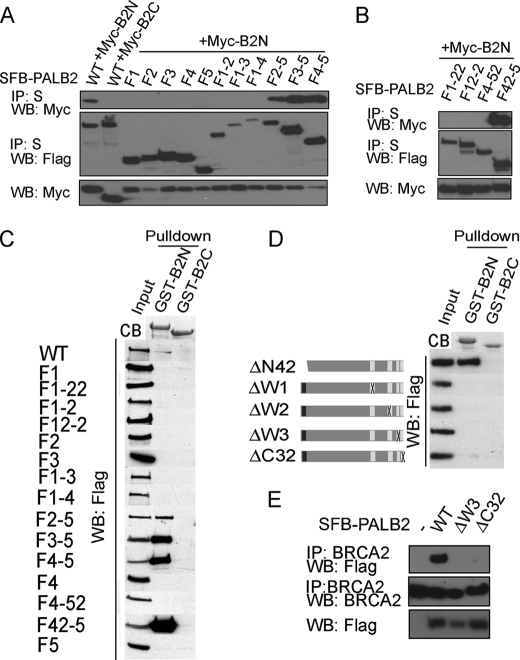
Results from a previous report (Xia et al. (11)) indicated that the interaction between PALB2 and BRCA2 is critical for HR. The PALB2 binding domain has been mapped to the very N terminus of BRCA2 (residues 10–40) (11). To further understand how PALB2 facilitates the BRCA2-dependent HR function in vivo, we sought to identify the region on PALB2 responsible for its interaction with BRCA2. A series of SFB-tagged PALB2 deletion mutants were tested for their ability to interact with Myc-BRCA2 N terminus that harbors the PALB2 binding domain (Ref. 11; B2N, residues 1–305 of BRCA2). Results indicated that the PALB2-BRCA2 interaction required a region of PALB2 which encompasses four WD40 domains (i.e. residues 850–1186, Fig. 4, A and B). The same results were obtained in pulldown experiments (Fig. 4C) when bacterially expressed GST-B2N and control GST fusion protein encoding the BRCA2 C terminus (B2C, residue 3120–3418) were used. These data indicate that PALB2 C-terminal WD40 domains are required for BRCA2 interaction.
FIGURE 4.
PALB2 interacts with BRCA2 via its C-terminal WD40 repeats. A and B, SFB-tagged wild type and deletion mutants of PALB2 were expressed in HEK293T and subjected to co-immunoprecipitation (IP) with Myc-B2N or Myc-B2C. C, pulldown assays using GST-B2N or control GST-B2C purified from E. coli. Western blotting (WB) using anti-FLAG antibody was performed to verify the interaction between wild type or mutants of PALB2 with BRCA2. D, internal deletion mutants lacking each of the four WD40 domains at the C terminus of PALB2 and the N-terminal mutant of PALB2 with deletion of the coiled-coil domain were subjected to pulldown assays similar to that described in C. E, extracts prepared from 293T cells expressing SFB-tagged wild-type, ΔW3, or ΔC32 mutants of PALB2 were subjected to immunoprecipitation using anti-BRCA2 antibody. Western blotting experiments were conducted with anti-BRCA2 or anti-FLAG antibodies. B2N, BRCA2 N terminus; B2C, BRCA2 C terminus; CB, Coomassie Blue stain.
To ask whether all four of the WD40 domains are needed for the observed PALB2/BRCA2 interaction, deletion mutants lacking each of the WD40 domains (ΔW1, ΔW2, ΔW3, and ΔC32-deleted C-terminal 32 residues containing the 4th WD40 domain) or ΔN42 were examined. Results showed that the interaction between PALB2 and BRCA2 was abolished by deletion in any of the four PALB2 WD40 domains, whereas the deletion of the N-terminal coil-coiled domain had no effect on the protein complex formation (Fig. 4, D and E). Our results suggest that the physical interaction between PALB2 and BRCA2 requires the PALB2 C terminus and that all four WD40 domains therein are indispensable in this regard.
The HR Function of PALB2 Requires Its Oligomerization Domain and Its BRCA2-interacting Motif
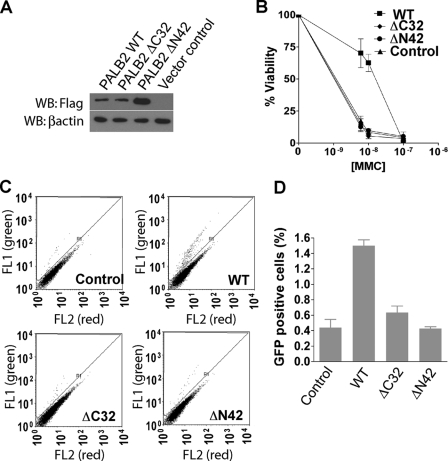
Cells deficient in PALB2 exhibit hypersensitivity to DNA cross-linking agents and display HR defects. To explore the physiological relevance of the PALB2 domains that mediate DNA damage-induced focus formation and BRCA2 complex formation, we first set out to determine whether mutants that lack the above domains, namely ΔC32 and ΔN42, can restore the DNA damage hypersensitivity phenotype of PALB2-deficient cells. EUFA1341 cells transduced with wild-type PALB2, the ΔC32 or the ΔN42 mutant (Fig. 5A), or vector alone were treated with different doses of the DNA cross-linking agent MMC (Fig. 5B). A clonogenic assay indicated that only cells reconstituted with wild-type PALB2, but not those with the ΔC32 or ΔN42 mutant, restored MMC resistance (Fig. 5B), suggesting that PALB2 function requires both oligomerization and BRCA2 interacting domains.
FIGURE 5.
PALB2 elevates cell survival following DNA damage by restoring homologous recombination repair. A, EUFA1341 cells were transduced with vector control or HA-FLAG-tagged wild-type PALB2 (WT), the C-terminal deletion mutant (ΔC32), and the N-terminal deletion mutant (ΔN42). PALB2 expression was confirmed by Western blot (WB) as indicated. B, clonogenic assay after MMC treatment indicated that both the N-terminal and C-terminal regions of PALB2 are required for cell survival after MMC treatment. Results are the averages of three independent experiments. C and D, gene conversion in EUFA1341 reconstituted with wild-type or mutant PALB2. GFP-positive cells indicative of gene conversion (R1) were quantified by flow cytometry analysis. Results are the average of three independent experiments and were presented as mean ± S.E.
We also performed a gene conversion assay to examine HR efficiency in these cell lines. In accordance with results from MMC sensitivity (Fig. 5B), re-introduction of wild-type PALB2 restored HR, whereas the ΔC32 or ΔN42 variants are defective in this regard (Fig. 5, C and D). From these results, we concluded that PALB2 function in HR is contingent upon its oligomerization property and its association with BRCA2.
DISCUSSION
PALB2 was previously implicated in the recruitment of BRCA2 to DNA breaks (11). Dysregulation of PALB2 function resulted in the failure of DNA damage-induced BRCA2 relocalization to DSBs and impaired HR repair (11). However, mechanistically how PALB2 is recruited to DNA breaks and how it mediates recombinational repair remain largely unexplored. In this study we have examined how PALB2 orchestrates DNA repair in response to DNA damage. PALB2 localizes to chromatin and assembled as oligomers at the site of DNA damage, which then serves as an anchor for the loading of BRCA2 and RAD51 to allow the activation of the error-free HR repair. Accordingly, disruption of either the PALB2 oligomerization domain or the BRCA2 binding motif resulted in defective HR and increased cellular sensitivity to DNA-damaging agents.
Previous studies have shown that PALB2 affects the stability and association of BRCA2 with certain nuclear structure in the cell and is essential for BRCA2 focal accumulation upon DNA damage (11). Here we provide evidence to support that PALB2 not only stabilizes the association of BRCA2 at the chromatin but is also a prerequisite for BRCA2 binding to chromatin structure (Fig. 1C). Indeed, we found that PALB2 associate with chromatin constitutively irrespective of treatments with DNA damaging reagent, as does BRCA2 and RAD51 (Fig. 1C).
Our observation underscores a possible intrinsic DNA binding activity of PALB2 in HR functioning. Notably, identification of DNA binding activity among the PALB2·BRCA2·RAD51 complex is not unprecedented, as RAD51 was previously shown to display single and double strand DNA binding activities (22). Likewise, a ssDNA binding domain (DBD) on BRCA2 has also been identified (23). It has been proposed that the BRCA2 DBD helps mediate the assembly of the RAD51-ssDNA nucleoprotein filament via the displacement of the single-strand DNA-binding protein RPA from the HR substrate (24). However, a recent report has provided evidence that the BRCA2 DBD might be dispensable for HR repair. Specifically, BRCA2 mutants lacking the DBD were shown to restore HR repair capacity and resistance to a PARP1 inhibitor to BRCA2-deficient cells (25). This observation is congruent with the hypothesis that PALB2 might possess DNA binding activity that could substitute BRCA2 DBD activity. In this study we observed a linear hierarchical relationship in the recruitment of the three HR repair proteins to DNA damage-induced foci in the sequence of 1) PALB2, 2) BRCA2, and 3) RAD51 (Fig. 1, A and B). Being the upstream co-factor, PALB2 might serve as the initial recognition module that in turn facilitates the loading and accumulation of the BRCA2·RAD51 complex at DSBs. Once the PALB2·BRCA2·RAD51 repair complex is loaded onto the damaged DNA, additional DNA binding activities of BRCA2, RAD51, and others then play important roles in mediating subsequent steps during HR repair processes.
We note that the deletion of the PALB2 N-terminal coiled-coil domain alone disrupted its focus forming ability in vivo (Fig. 3). Coiled-coil domains are modules that promote protein-protein interaction or protein oligomerization (26). We found that PALB2 exists as a dimeric or multimeric structure via its coiled-coil domain (Fig. 3, D–G). We proposed that upon DNA damage, the constitutively chromatin-associated PALB2 oligomerize so as to facilitate its accumulation at sites of DSBs to initiate HR repair by recruiting the BRCA2·RAD51 repair complex. Indeed, the PALB2 deletion mutant lacking the oligomerization domain not only is defective in foci formation at DSBs, but it also demonstrated deficits in homologous recombination and MMC sensitivity (Fig. 5). Interestingly, we and others have also found that PALB2 interacts with BRCA1 via its coiled-coil domain, forming a three-way complex with BRCA2 to allow optimal HR repair (27, 44). Given that PALB2 foci formation does not require BRCA1, our observation that the PALB2 coiled-coil motif mediates its oligomerization suggests that this particular property might regulate sustained localization of PALB2 at DSBs in vivo. We are currently testing this hypothesis.
It has been illustrated that protein oligomerization stabilizes protein-DNA association in DNA damage signaling, where oligomerization of Rad9 is required for sustain checkpoint signaling (28). Indeed, many of the DNA repair protein have been shown to exist as dimer or oligomers. Oligomerization of RAD51 to form the nucleofilament has been demonstrated by electronic microscopy (29). Structural studies also implicated a dimerization property for BRCA2 (23). Together with the PALB2 oligomerization property revealed in the current study and also its association with BRCA1, we proposed that the DNA repair complex exists as arrays of protein homo- and hetero-oligomers at site of DSBs upon DNA damage, where the quaternary structural configurations of the oligomers mediate the stabilization of the complex at the DSBs to facilitate subsequent repair processes.
Although our study shows that oligomerization of PALB2 is required for its focal accumulation at DSBs and, thus, the subsequent recruitment of the BRCA2·RAD51 complex to DNA lesions, mechanistically how PALB2 locates DNA breaks has yet to be clarified. It was previously shown that ATR-ATRIP senses DNA damage via recognition of RPA-coated ssDNA (30). The PALB2·BRCA2·RAD51 repair complex may utilize a similar system to recognize DSBs, as the replacement of RPA from ssDNA by RAD51 nucleofilament is the first step for DNA repair (8–10). Another possibility is that structural changes of the chromatin surrounding DNA breaks may signal for the recruitment of PALB2. A number of chromatin remodeling enzymes have been found to play an early role in response to DNA damage to facilitate the recruitment of signaling molecules as well as DNA repair proteins (31). The remodeled chromatin structure might enhance PALB2 access to DNA lesions. This may occur in parallel to the known role of chromatin remodeling in the DNA damage signaling cascade (32, 33).
Alignment of PALB2 homologues (data not shown) shows that both the coiled-coil domain and the WD40 repeats are highly conserved among species. This suggests that both domains are significant for PALB2 function. Indeed, apart from the oligomerization property mediated by the coiled-coil motif, we have also refined the BRCA2 binding motif on PALB2 to its WD40 repeats (Fig. 4). WD40 repeats are known to mediate multiprotein complex assemblies as well as protein-DNA association. WD repeats are thought to organize as circularized β-propeller structure and serves as a rigid scaffold for protein interactions. The fact that deletion of any of the four WD40 domains on the PALB2 C terminus disrupted its interaction with BRCA2 indicated that the tertiary structure of the β-propeller is required for their interaction. We observed that disruption in the PALB2/BRCA2 interaction led to HR deficiency as observed in PALB2 FA patient cells (Fig. 5). It suggests that the essential role of PALB2 in HR is primarily via its ability in recruiting the BRCA2·RAD51 complex to DSBs. Because both coiled-coil and WD40 motifs are structural scaffolds and are both required for PALB2 function in HR, the PALB2 structure should provide insightful information to explain how PALB2 organizes the repair complex at DSBs.
DNA damage checkpoints play key roles in ensuring genomic stability by delaying cell cycle progression to allow sufficient time for DNA repair (34, 35). One would expect that cross-talk between the DNA repair machinery and cell cycle checkpoint pathways might enable an elaborate and precise coordination of cellular events essential for cell survival and proliferation. It remains largely elusive as to what the connections between DNA repair processes and DNA damage checkpoints are.
Recent studies demonstrated that human CtBP-interacting protein and its orthologues are involved in the generation of ssDNAs through resection of DSBs (36–41). The formation of RPA-ssDNA complexes could then lead to the recruitment and activation of ATR-ATRIP, which in turns control the intra-S checkpoint (30). Similarly RPA-ssDNA complexes may also be required for HR repair. Thus, RPA-coated ssDNA might represent a common signal for DNA repair and cell cycle checkpoint control. ATR and CHK1 depletion was reported to dramatically reduce RAD51 IRIF and HR repair (42, 43). However, we still detected RAD51 IRIF after depletion of ATR or CHK1 by siRNA in our study. The discrepancies in these observations might largely be because of the very different cell types used in these studies. Nevertheless, these differences point out that the relationship between cell cycle checkpoints and DNA repair is multifactorial and may involve many feedback loops and converging/diverging points. In agreement with this idea, PALB2 depletion also resulted in compromised intra-S checkpoint defect (11), suggesting an intimate relationship between checkpoint control and HR repair.
We believe that cross-talk between the DNA repair machinery and cell cycle checkpoint control not only couples the initiation of DNA repair and checkpoint activation but also serves as means to coordinate between the resumption of cell cycle progression upon completion of DNA repair. Future studies will reveal mechanistically how this coordination is achieved in the cell.
Acknowledgments
We thank Prof. David Livingston for the pOZC-PALB2, Prof. Maria Jasin for the pDR-GFP and pCBASce plasmids, and Prof. Johan P de Winter for EUFA1341 cell. S. M. Sy thanks all colleagues in the Chen laboratory for insightful discussion and technical assistance, especially Dr. Jun Huang.
This work was supported, in whole or in part, by National Institutes of Health Grant CA089239 (to J. C.).
- FA
- Fanconi anemia
- ssDNA
- single strand DNA
- DSBs
- double strand breaks
- HR
- homologous recombination
- IRIF
- ionizing radiation-induced focus formation
- MMC
- Mitomycin C
- SFB
- S-tag, FLAG epitope, and streptavidin-binding peptide-tag
- siRNA
- small interfering RNA
- Gy
- gray
- GST
- glutathione S-transferase
- CHO
- Chinese hamster ovary
- DBD
- DNA binding domain
- RPA
- replication protein A
- ATR
- atoxia telangiectaia mutated and Rad3-related
- ATRIP
- ATR-interacting protein.
REFERENCES
- 1.Bartek J., Lukas C., Lukas J. ( 2004) Nat. Rev. Mol. Cell Biol. 5, 792– 804 [DOI] [PubMed] [Google Scholar]
- 2.Kennedy R. D., D'Andrea A. D. ( 2006) J. Clin. Oncol. 24, 3799– 3808 [DOI] [PubMed] [Google Scholar]
- 3.Wang W. ( 2007) Nat. Rev. Genet. 8, 735– 748 [DOI] [PubMed] [Google Scholar]
- 4.Zhang H., Tombline G., Weber B. L. ( 1998) Cell 92, 433– 436 [DOI] [PubMed] [Google Scholar]
- 5.Gudmundsdottir K., Ashworth A. ( 2006) Oncogene 25, 5864– 5874 [DOI] [PubMed] [Google Scholar]
- 6.Cantor S. B., Bell D. W., Ganesan S., Kass E. M., Drapkin R., Grossman S., Wahrer D. C., Sgroi D. C., Lane W. S., Haber D. A., Livingston D. M. ( 2001) Cell 105, 149– 160 [DOI] [PubMed] [Google Scholar]
- 7.Litman R., Peng M., Jin Z., Zhang F., Zhang J., Powell S., Andreassen P. R., Cantor S. B. ( 2005) Cancer Cell 8, 255– 265 [DOI] [PubMed] [Google Scholar]
- 8.Pellegrini L., Venkitaraman A. ( 2004) Trends Biochem. Sci. 29, 310– 316 [DOI] [PubMed] [Google Scholar]
- 9.Lord C. J., Ashworth A. ( 2007) Nat. Struct. Mol. Biol. 14, 461– 462 [DOI] [PubMed] [Google Scholar]
- 10.Thorslund T., West S. C. ( 2007) Oncogene 26, 7720– 7730 [DOI] [PubMed] [Google Scholar]
- 11.Xia B., Sheng Q., Nakanishi K., Ohashi A., Wu J., Christ N., Liu X., Jasin M., Couch F. J., Livingston D. M. ( 2006) Mol. Cell 22, 719– 729 [DOI] [PubMed] [Google Scholar]
- 12.Xia B., Dorsman J. C., Ameziane N., de Vries Y., Rooimans M. A., Sheng Q., Pals G., Errami A., Gluckman E., Llera J., Wang W., Livingston D. M., Joenje H., de Winter J. P. ( 2007) Nat. Genet. 39, 159– 161 [DOI] [PubMed] [Google Scholar]
- 13.Rahman N., Seal S., Thompson D., Kelly P., Renwick A., Elliott A., Reid S., Spanova K., Barfoot R., Chagtai T., Jayatilake H., McGuffog L., Hanks S., Evans D. G., Eccles D., Easton D. F., Stratton M. R. ( 2007) Nat. Genet. 39, 165– 167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Erkko H., Xia B., Nikkilä J., Schleutker J., Syrjäkoski K., Mannermaa A., Kallioniemi A., Pylkäs K., Karppinen S. M., Rapakko K., Miron A., Sheng Q., Li G., Mattila H., Bell D. W., Haber D. A., Grip M., Reiman M., Jukkola-Vuorinen A., Mustonen A., Kere J., Aaltonen L. A., Kosma V. M., Kataja V., Soini Y., Drapkin R. I., Livingston D. M., Winqvist R. ( 2007) Nature 446, 316– 319 [DOI] [PubMed] [Google Scholar]
- 15.Tischkowitz M., Xia B., Sabbaghian N., Reis-Filho J. S., Hamel N., Li G., van Beers E. H., Li L., Khalil T., Quenneville L. A., Omeroglu A., Poll A., Lepage P., Wong N., Nederlof P. M., Ashworth A., Tonin P. N., Narod S. A., Livingston D. M., Foulkes W. D. ( 2007) Proc. Natl. Acad. Sci. U.S.A. 104, 6788– 6793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Foulkes W. D., Ghadirian P., Akbari M. R., Hamel N., Giroux S., Sabbaghian N., Darnel A., Royer R., Poll A., Fafard E., Robidoux A., Martin G., Bismar T. A., Tischkowitz M., Rousseau F., Narod S. A. ( 2007) Breast Cancer Res. 9, R83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen J., Silver D. P., Walpita D., Cantor S. B., Gazdar A. F., Tomlinson G., Couch F. J., Weber B. L., Ashley T., Livingston D. M., Scully R. ( 1998) Mol. Cell 2, 317– 328 [DOI] [PubMed] [Google Scholar]
- 18.Ward I. M., Minn K., van Deursen J., Chen J. ( 2003) Mol. Cell. Biol. 23, 2556– 2563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Celeste A., Difilippantonio S., Difilippantonio M. J., Fernandez-Capetillo O., Pilch D. R., Sedelnikova O. A., Eckhaus M., Ried T., Bonner W. M., Nussenzweig A. ( 2003) Cell 114, 371– 383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lou Z., Minter-Dykhouse K., Franco S., Gostissa M., Rivera M. A., Celeste A., Manis J. P., van Deursen J., Nussenzweig A., Paull T. T., Alt F. W., Chen J. ( 2006) Mol. Cell 21, 187– 200 [DOI] [PubMed] [Google Scholar]
- 21.Huen M. S., Huang J., Yuan J., Yamamoto M., Akira S., Ashley C., Xiao W., Chen J. ( 2008) Mol. Cell. Biol. 28, 6104– 6112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shinohara A., Ogawa H., Ogawa T. ( 1992) Cell 69, 457– 470 [DOI] [PubMed] [Google Scholar]
- 23.Yang H., Jeffrey P. D., Miller J., Kinnucan E., Sun Y., Thoma N. H., Zheng N., Chen P. L., Lee W. H., Pavletich N. P. ( 2002) Science 297, 1837– 1848 [DOI] [PubMed] [Google Scholar]
- 24.Shin D. S., Chahwan C., Huffman J. L., Tainer J. A. ( 2004) DNA Repair 3, 863– 873 [DOI] [PubMed] [Google Scholar]
- 25.Edwards S. L., Brough R., Lord C. J., Natrajan R., Vatcheva R., Levine D. A., Boyd J., Reis-Filho J. S., Ashworth A. ( 2008) Nature 451, 1111– 1115 [DOI] [PubMed] [Google Scholar]
- 26.Burkhard P., Stetefeld J., Strelkov S. V. ( 2001) Trends Cell Biol. 11, 82– 88 [DOI] [PubMed] [Google Scholar]
- 27.Zhang F., Ma J., Wu J., Ye L., Cai H., Xia B., Yu X. ( 2009) Curr. Biol. 19, 524– 529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Usui T., Foster S. S., Petrini J. H. ( 2009) Mol. Cell 33, 147– 159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sung P., Robberson D. L. ( 1995) Cell 82, 453– 461 [DOI] [PubMed] [Google Scholar]
- 30.Zou L., Elledge S. J. ( 2003) Science 300, 1542– 1548 [DOI] [PubMed] [Google Scholar]
- 31.Murr R., Loizou J. I., Yang Y. G., Cuenin C., Li H., Wang Z. Q., Herceg Z. ( 2006) Nat. Cell Biol. 8, 91– 99 [DOI] [PubMed] [Google Scholar]
- 32.Harper J. W., Elledge S. J. ( 2007) Mol. Cell 28, 739– 745 [DOI] [PubMed] [Google Scholar]
- 33.Huen M. S., Chen J. ( 2008) Cell Res. 18, 8– 16 [DOI] [PubMed] [Google Scholar]
- 34.Motoyama N., Naka K. ( 2004) Curr. Opin. Genet. Dev. 14, 11– 16 [DOI] [PubMed] [Google Scholar]
- 35.Myung K., Datta A., Kolodner R. D. ( 2001) Cell 104, 397– 408 [DOI] [PubMed] [Google Scholar]
- 36.Uanschou C., Siwiec T., Pedrosa-Harand A., Kerzendorfer C., Sanchez-Moran E., Novatchkova M., Akimcheva S., Woglar A., Klein F., Schlögelhofer P. ( 2007) EMBO J. 26, 5061– 5070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Takeda S., Nakamura K., Taniguchi Y., Paull T. T. ( 2007) Mol. Cell 28, 351– 352 [DOI] [PubMed] [Google Scholar]
- 38.Sartori A. A., Lukas C., Coates J., Mistrik M., Fu S., Bartek J., Baer R., Lukas J., Jackson S. P. ( 2007) Nature 450, 509– 514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Penkner A., Portik-Dobos Z., Tang L., Schnabel R., Novatchkova M., Jantsch V., Loidl J. ( 2007) EMBO J. 26, 5071– 5082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Limbo O., Chahwan C., Yamada Y., de Bruin R. A., Wittenberg C., Russell P. ( 2007) Mol. Cell 28, 134– 146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chen L., Nievera C. J., Lee A. Y., Wu X. ( 2008) J. Biol. Chem. 283, 7713– 7720 [DOI] [PubMed] [Google Scholar]
- 42.Sørensen C. S., Hansen L. T., Dziegielewski J., Syljuåsen R. G., Lundin C., Bartek J., Helleday T. ( 2005) Nat. Cell Biol. 7, 195– 201 [DOI] [PubMed] [Google Scholar]
- 43.Collis S. J., Barber L. J., Clark A. J., Martin J. S., Ward J. D., Boulton S. J. ( 2007) Nat. Cell Biol. 9, 391– 401 [DOI] [PubMed] [Google Scholar]
- 44.Sy S. M., Huen M. S., Chen J. ( 2009) Proc. Natl. Acad. Sci. U. S. A. 106, 7155– 7160 [DOI] [PMC free article] [PubMed] [Google Scholar]