Abstract
The autosomal dominant dementia familial encephalopathy with neuroserpin inclusion bodies is characterized by the accumulation of ordered polymers of mutant neuroserpin within the endoplasmic reticulum of neurones. We show here that intracellular neuroserpin polymers activate NF-κB by a pathway that is independent of the IRE1, ATF6, and PERK limbs of the canonical unfolded protein response but is dependent on intracellular calcium. This pathway provides a mechanism for cells to sense and react to the accumulation of folded structures of mutant serpins within the endoplasmic reticulum. Our results provide strong support for the endoplasmic reticulum overload response being independent of the unfolded protein response.
An increasing number of disorders are recognized to result from the aggregation and tissue deposition of misfolded proteins. Indeed, their shared mechanism provides the basis for a new class of disorder, the conformational diseases (1). These include Alzheimer, Huntington, and Parkinson diseases as well as the amyloidoses and serpinopathies. The serpinopathies are characterized by the aggregation and tissue deposition of members of the serine protease inhibitor or serpin superfamily of proteins (2). Point mutations of serpins such as α1-antitrypsin, antithrombin, and α1-antichymotrypsin result in a sequential linkage between the exposed mobile reactive center loop of one molecule and β-sheet A of another (3, 4). The resulting ordered polymers then accumulate as inclusion bodies within the lumen of the endoplasmic reticulum (ER)5 (1, 2). This is associated with disease due to loss of function (i.e. the reduction in active protein) and/or a toxic gain of function (i.e. the cytotoxicity of protein aggregates).
One of the most striking serpinopathies is the autosomal dominant dementia familial encephalopathy with neuroserpin inclusion bodies or FENIB (5). This results from one of four naturally occurring point mutations in the neuroserpin gene: S49P, S52R, H338R, or G392E (6). The mutant neuroserpin proteins form ordered polymers that accumulate in the ER of neurones within the cerebral cortex, hippocampus, and substantia nigra (5–7). The resulting inclusions lead to progressive dementia, with the age of onset of disease being inversely proportional to the rate at which the mutants form polymers in vitro and the number of intra-cerebral inclusions (6).
The accumulation of misfolded proteins within the lumen of the ER activates the PERK (PKR-like ER kinase), IRE1 (inositol-requiring kinase 1), and ATF6 (activating transcription factor 6) limbs of the unfolded protein response (UPR). This pathway serves to attenuate protein translation and increase the production of molecular chaperones to promote polypeptide folding and remove terminally misfolded proteins by ER-associated degradation (ERAD) (8). The UPR is increasingly being implicated in the pathogenesis of human disease (9). It is striking that accumulation of mutant Z α1-antitrypsin polymers within the ER of hepatocytes does not elicit the UPR (10) but does activate NF-κB (11, 12). This has been explained by the activation of the ER overload response (EOR) (13), a stress signaling pathway that links the accumulation of folded proteins within the ER with the activation of NF-κB. The term EOR was originally coined to describe the NF-κB response to ER accumulation of viral proteins (14), but was subsequently extended to include other proteins retained within the ER despite achieving native or near native conformations (15, 16). However, NF-κB activation in response to ER dysfunction has been shown to require PERK signaling, and so the very existence of an EOR pathway distinct from the UPR remains controversial (17).
Here we have used wild-type and mutants of neuroserpin to investigate the consequences of ordered protein accumulation within the ER. We demonstrate that neuroserpin polymers activate NF-κB by a calcium-dependent pathway that is independent of the IRE, ATF6, and PERK limbs of the canonical UPR. These data provide strong support for a signaling pathway that directly links the activation of NF-κB with the accumulation of ordered proteins within the ER.
EXPERIMENTAL PROCEDURES
Antibodies and Reagents
The rabbit polyclonal anti-neuroserpin antibody was produced by Abcam (Cambridge, UK) using purified recombinant wild-type neuroserpin as the antigen (18). Rabbit polyclonal anti-GAPDH and donkey polyclonal anti-rabbit IgG (Texas Red) antibodies were also from Abcam. Goat polyclonal anti-rabbit IgG (horseradish peroxidase) and rabbit polyclonal anti-mouse IgG (horseradish peroxidase) antibodies were from Sigma-Aldrich. Rabbit polyclonal anti-Phospho-eIF2α (S51) from Cell Signaling was a kind gift from Dr. Shiu-Wan Chan (Faculty of Life Sciences, University of Manchester, UK). All pharmacological cell-permeable inhibitors were purchased from Calbiochem (Merck Chemicals Ltd., Nottingham, UK). Unless stated otherwise, reagents for cell culture were purchased from Sigma-Aldrich.
Culture of Stable PC12 Tet-On Cells Lines Expressing Neuroserpin
The cells were cultured in Dulbecco's modified Eagle's medium supplemented with 10% v/v heat-inactivated horse serum, 5% v/v Tet Approved Fetal Bovine Serum (BD Biosciences), 10 mm HEPES, 1× non-essential amino acids, 0.2 unit/ml bovine insulin, 200 μg/ml Geneticin, and 100 μg/ml Hygromycin B (selective antibiotics from Invitrogen), and incubated at 37 °C and 5% v/v CO2 in a humidified incubator. Neuroserpin expression was typically induced with 10 μg/ml doxycycline (BD Biosciences).
Mouse Embryonic Fibroblast (MEF) eIF2α-S51A Cell Line
Immortalized wild-type and eIF2α S51A/A MEFs were transduced with pBABE retrovirus encoding WT, S52R, G392E, and ΔNS neuroserpin using previously described protocols (19, 20). Briefly, cells were selected for stable transgene expression by selection with 2.5 μg/ml puromycin, with surviving cells considered as stable expressing pools. Several stable pools expressing neuroserpin were established, and these were then used in the functional studies. All results described are the average of at least three independent repeats.
SDS and Non-denaturing PAGE and Western Blot Analysis
Cell lysates and culture media were analyzed in 10% w/v SDS-PAGE or 7.5% w/v acrylamide non-denaturing gels as described previously (21).
Metabolic Labeling and Immunoprecipitation
Metabolic labeling experiments with 35S-Met/Cys were performed as described previously (21). PC12 cells were treated with 200 μm leupeptin, 25 μm lactacystin, or 3 μm brefeldin A from Calbiochem (CN Biosciences, Nottingham, UK) for 1 h prior to starvation and these agents were present in all subsequent steps until harvesting.
Reverse Transcription-PCR and XBP1 Splicing Assay
Total RNA was isolated from mammalian cells using the recommended protocol in the Qiagen RNeasy Isolation kit. (Qiagen, UK). The concentration of RNA was determined, and 1 μg of total RNA was used as a template for first strand cDNA synthesis using the manufacturer's recommended protocols (Promega, Madison, WI). This was then used to semi-quantify the relative levels of neuroserpin and β-actin mRNA expression and the unspliced to spliced ratio of XBP1. The primers were RT_NS_S (5′-TCT CCA TTG AGT ATT GCT CTT GC), RT_NS_AS (5′-TCT CCT TGC TGA TAC ATC ATT GG), Actin_S (5′-CTT CGC GGG CGA CGA TGC), Actin_AS (5′-TGG TGG TGA AGC TGT AGC C), RtXBP1S (5′-AAA CAG AGT AGC AGC ACA GAC TGC), and RtXBP1AS (5′-TCC TTG TGG GTA GAC CTC TGG GAG). Optimal PCR conditions were used as described previously (22).
Luciferase Assay
Transfections were performed in 6-well plates that had been pre-coated with 0.1 mg/ml poly-l-lysine. Typically, 2 μg of either p(5×)ATF6-luc (firefly) or pELAM1-luc (firefly) and 50 ng of pRL-TK (Renilla) transfection efficiency control reporter plasmids (all kind gifts from Dr. Timothy Weaver, Cincinnati Children's Hospital Medical Center, Cincinnati, OH) were mixed with 6.25 μl of Lipofectamine 2000 (Invitrogen) in serum-free Opti-MEM culture medium (Amersham Biosciences) following the manufacturer's protocol. The cells were lysed post-transfection using the Dual-Luciferase Reporter Assay (Promega) recommended protocol in 1× passive lysis buffer. Both firefly and Renilla luciferase activity were measured using a Glomax Luminometer (Promega), and firefly luciferase activity was calculated relative to Renilla transfection efficiency. All measurements were performed in triplicate.
Ca2+ Flux Measurements
PC12 cells were resuspended at a concentration of 1 × 106 cells/ml and loaded with Fluo-4 NW dye assay buffer (Invitrogen) according to the manufacturer's recommend protocol for 30 min at 37 °C and then 15 min at room temperature. The cells were then loaded onto a FACSCalibur cell cytometer (BD Biosciences) and read at 488 nm over time. Calcium was mobilized by the addition of 1 μm thapsigargin (TG) and the measurements continued to a relative increase in fluorescence that was stable over time. Control cells were pre-treated with 2 μg/ml tunicamycin for 16 h, or the calcium store was depleted by the addition of 1 μm TG for 30 min and recovery for 1 h in fresh media before analysis. All experiments were performed in triplicate and analyzed with FlowJo software (BD Biosciences).
Statistical Analysis
When comparing within a series, for example when analyzing the effects of a drug on each cell line, a paired t test was used. When comparing data between series, then two-way ANOVA with a Bonferroni post test was used.
RESULTS
Mutants of Neuroserpin Accumulate within the ER of PC12 Cells
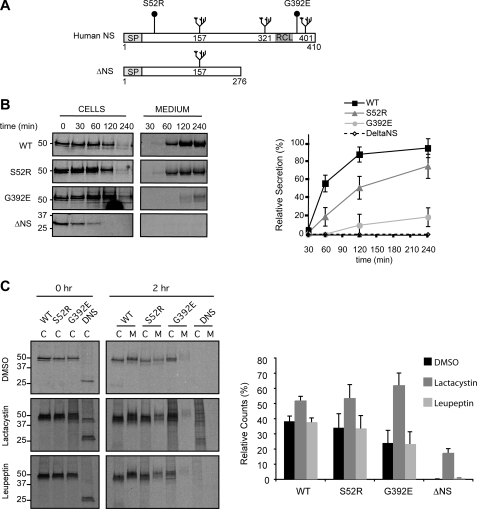
To study the cellular responses to the expression of mutant neuroserpin, we used rat pheochromocytoma (PC12) Tet-On cell lines that conditionally expressed wild-type human neuroserpin (WT) or the S52R and G392E mutants that are associated with moderate and severe forms of the dementia FENIB, respectively (23) (Fig. 1A). In addition, we generated a stable cell line that expressed a novel misfolding mutant of neuroserpin, with deletion of the C-terminal 134 amino acids, which include the reactive loop (ΔNS) (Fig. 1A). This mutant is equivalent to the null Hong Kong variant of the serpin α1-antitrypsin, which cannot form ordered polymers and is an efficient activator of the UPR and a potent substrate for ERAD (24). We first assessed the presence of neuroserpin in these cells lines by SDS-PAGE and Western blot analysis (supplemental Fig. S1). All four PC12 cell lines expressed neuroserpin when the cells were treated with increasing concentrations of doxycycline. WT, S52R, and G392E neuroserpin migrated with a molecular mass of 50 kDa, whereas the truncated version of neuroserpin, ΔNS, migrated at 28 kDa. Our previous work showed that in these cell lines WT neuroserpin was mainly stored in secretory granules at the tips of neurites, characteristic of a protein secreted through the regulated secretory pathway (23). In contrast, S52R and G392E neuroserpin formed inclusions within the ER that were found to be endoglycosidase H-sensitive polymers (21, 23). ΔNS neuroserpin displayed diffuse ER staining, but no signal was detected within the Golgi complex (results not shown), suggesting that none of the misfolded mutant protein was trafficked out of the ER. We next quantified neuroserpin secretion from each cell line by metabolic labeling with [35S]methionine/cysteine followed by immunoprecipitation of neuroserpin, SDS-PAGE, and autoradiography. WT neuroserpin was almost completely secreted into the culture medium by 4 h, compared with 78 and 18% of S52R and G392E neuroserpin, respectively (Fig. 1B). ΔNS neuroserpin was not detected in the culture medium despite being cleared from the cell lysates within 1 h of chase (Fig. 1B).
FIGURE 1.
Characterization of PC12 Tet-On cell lines expressing neuroserpin variants. A, schematic representation of neuroserpin variants used in this study. Full-length human neuroserpin is 410 amino acids long, with N-linked glycans at positions 157, 321, and 401, a signal peptide (SP) and reactive center loop (RCL) as indicated. The polymerogenic mutants have amino acid substitutions at positions 52 (Ser to Arg) and 392 (Gly to Glu), respectively, and ΔNS has a premature stop codon at position 276. We generated PC12 Tet-On cell lines that stably expressed these constructs under the control of the tetracycline response element in the pTRE-Tight expression vector. B, secretion of neuroserpin variants. PC12 cells expressing WT, S52R, G392E, and ΔNS neuroserpin were metabolically labeled with 35S-Met/Cys for 15 min and chased for the times indicated. Neuroserpin was immunoprecipitated from cell lysates and culture medium, resolved by 10% w/v SDS-PAGE, and quantified by autoradiography using a PhosphorImager. The graph shows average ± S.D. for three independent repeats. C, intracellular degradation of neuroserpin variants. PC12 cells treated with DMSO (vehicle), 25 μm lactacystin (irreversible inhibitor of the proteasome), or 200 μm leupeptin (inhibitor of lysosomal proteases) were metabolically labeled and analyzed as described in B. The histograms show the amount of radiolabeled neuroserpin remaining after the chase normalized to the initial amount of neuroserpin for each condition, and values are averages ± S.E. of three independent repeats. Control experiments in PC12 cells with leupeptin and lactacystin showed the efficacy of the inhibitor treatments in blocking processing of pro-cathepsin D to mature cathepsin D and the degradation of cyclin B1 respectively (data not shown).
We predicted that ΔNS neuroserpin would be a substrate for ERAD. To test this hypothesis we investigated the degradation of each neuroserpin variant by pulse-chase in the presence or absence of lactacystin (which irreversibly inhibits the proteasome (25)) or leupeptin (which inhibits lysosomal proteases (26, 27)). Treatment with lactacystin increased intracellular WT, S52R, G392E, and ΔNS neuroserpin by 1.3-, 1.6-, 2.7-, and 9.5-fold, respectively (Fig. 1C, middle panels and graph). In contrast, treatment with leupeptin had no effect on the intracellular levels of any neuroserpin species, suggesting lysosomal protease digestion plays a limited role in the degradation of mutant neuroserpin (Fig. 1C, bottom panels and graph).
Taken together, these data demonstrate that WT neuroserpin traffics normally through the secretory pathway and that S52R and G392E neuroserpin are secreted less efficiently and partially retained within the ER of PC12 cell lines. ΔNS neuroserpin is synthesized in the ER but rapidly cleared by ERAD without a detectable fraction being secreted into the media.
Accumulation of Neuroserpin Polymers within the ER Does Not Activate the UPR
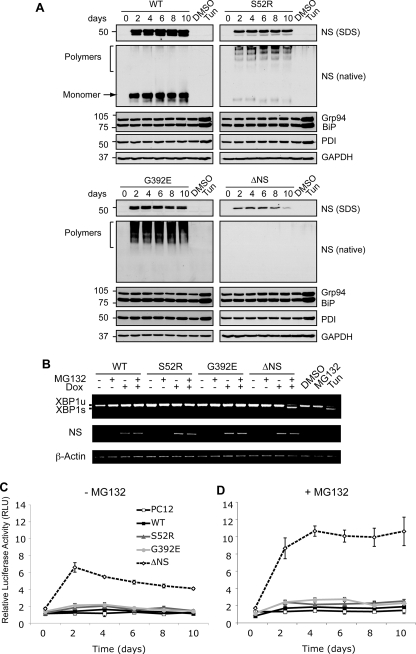
We next characterized the nature of the proteins retained within the ER up to 10 days after induction of expression by non-denaturing PAGE and Western blot analysis of PC12 cell lysates (Fig. 2A). WT neuroserpin migrated as a single monomeric band, whereas S52R and G392E neuroserpin formed the high molecular mass ladders that are characteristic of serpin polymers (Fig. 2A, native gel panels, arrow, and bracket). A proportion of S52R neuroserpin migrated as a monomeric band (Fig. 2A, S52R, native, arrow). Although we could detect the presence of ΔNS neuroserpin by Western blot after SDS-PAGE, we did not see any signal after non-denaturing PAGE, probably due to lower steady-state levels of this mutant protein and/or the inability of this misfolded protein to be resolved by non-denaturing PAGE.
FIGURE 2.
Accumulation of neuroserpin polymers within the ER does not activate the UPR. A, polymers of neuroserpin do not up-regulate ER luminal chaperones. Neuroserpin expression was induced in PC12 cells with 10 μg/ml doxycycline for 10 days, and cell lysates were resolved by 10% w/v SDS- and 7.5% w/v/non-denaturing PAGE. Western blot analysis for neuroserpin of SDS-PAGE (NS, SDS) revealed a 50-kDa band in WT, S52R, and G392E neuroserpin-expressing cells and 28 kDa in ΔNS neuroserpin cells. On non-denaturing PAGE (NS, native), WT neuroserpin migrated as a single monomer band (arrow), whereas S52R and G392E neuroserpin formed high molecular mass ladders that are characteristic of polymers (bracket). There was no detectable signal for ΔNS on non-denaturing PAGE. The expression levels of ER luminal chaperones regulated by the UPR (glucose regulated protein 94 (Grp94), immunoglobulin heavy chain-binding protein (BiP), and protein disulfide isomerase (PDI)) were determined in the same membranes by Western blotting. Treatment with 2 μg/ml tunicamycin (Tun) for 16 h compared with vehicle alone (DMSO) was used as a positive control for UPR induction, and protein loading was assessed by Western blot analysis for GAPDH. B, polymers of neuroserpin do not activate IRE1. The expression of WT, S52R, G392E, and ΔNS neuroserpin was induced for 4 days with 10 μg/ml doxycycline (Dox), and cells were either harvested or treated 16 h prior to RNA isolation with 100 nm MG132, a reversible inhibitor of the proteasome (46). XBP1 mRNA was amplified by PCR and resolved by 2% w/v TBE agarose gel electrophoresis. Unspliced XBP1 (XBP1u) and spliced XBP1 (XBP1s) products migrated at 486 and 457 bp, respectively. Neuroserpin (NS) and β-actin cDNAs were amplified to demonstrate transgene induction and as loading controls, respectively. PC12 cells treated with 2 μg/ml tunicamycin (Tun) were used as a positive control for UPR-induced XBP1 mRNA splicing. C and D, polymers of neuroserpin do not activate ATF6. Neuroserpin expression was induced in PC12 cell lines with 10 μg/ml doxycycline for 10 days. Twenty-four hours prior to lysis the cells were co-transfected with a plasmid encoding firefly luciferase under the control of a UPR response element (p5×ATF6-Luc) and with the transfection efficiency reporter pRL-TK Renilla luciferase. The graphs show firefly luciferase normalized to Renilla luciferase as averages ± S.D. of three repeats, and values are expressed in relative light units (RLU). Cells were mock-treated (C) or treated with 100 nm MG132 (D) for 16 h prior to harvesting.
Because the mutants of neuroserpin accumulated within the ER, we next assessed their effect on the UPR. None of the proteins activated the UPR as assessed by expression levels of glucose-regulated protein 94, immunoglobulin heavy chain-binding protein (BiP), and protein disulfide isomerase (Fig. 2a). In contrast, tunicamycin, a canonical UPR stimulus that inhibits N-linked glycosylation, caused an increase in the steady-state levels of all these ER luminal chaperones (Fig. 2A, DMSO versus Tun). Nuclear lysates were prepared from the same samples and analyzed for the up-regulation of CAAT/enhancer-binding protein homologous protein, a transcription factor specifically activated by the UPR (22, 28). There was no detectable CAAT/enhancer-binding protein homologous protein expression in any of the PC12 cell lines expressing neuroserpin (data not shown).
The apparent absence of a UPR even in response to ΔNS, the ERAD substrate, might represent an issue of sensitivity. We therefore assessed the activation of the UPR by the splicing of X-box-binding protein 1 (XBP1) mRNA, which reports activation of IRE1, a proximal UPR sensor in the ER membrane. When the PC12 cell lines were induced to express wild-type or mutant neuroserpin with 10 μg/ml doxycycline for 4 days, no XBP1 mRNA splicing was detected (Fig. 2B, XBP1u). In contrast, cells treated with 2 μg/ml tunicamycin generated a faster migrating species corresponding to spliced XBP1 mRNA (Fig. 2B, Tun, XBP1u, and XBP1s). We did detect XBP1 mRNA splicing in cells expressing ΔNS neuroserpin when the cells were treated with the proteasome inhibitor MG132 for 16 h prior to harvesting (Fig. 2B, ΔNS+/+).
This raised the possibility that the methods we had used to detect UPR activation were insufficiently sensitive to detect subtle perturbations caused by the expression of mutant neuroserpin. To address this concern, we next assessed the activation of ATF6 using a highly sensitive reporter construct (pAT6(5×)-Luc, also known as the pUPRE-Luc (29)). In these experiments, co-transfection with pRL-TK 24 h prior to cell collection reported transfection efficiency (Fig. 2, C and D). Only PC12 cells expressing ΔNS activated ATF6 (p < 0.001, ΔNS versus WT, two-way ANOVA with Bonferroni post test (Fig. 2C)). This up-regulation was increased when the cells were treated with 100 nm MG132 for 16 h prior to harvesting, whereas the other neuroserpin variants failed to activate ATF6 after proteasome inhibition (p < 0.001, ΔNS-MG132 versus ΔNS+MG132, two-way ANOVA with Bonferroni post test (Fig. 2D)).
These data show that the ER accumulation of neuroserpin polymers did not activate the UPR and that this failure was not due to defective UPR signaling, because an appropriate activation of ATF6 and XBP1 splicing response occurred during accumulation of ΔNS neuroserpin.
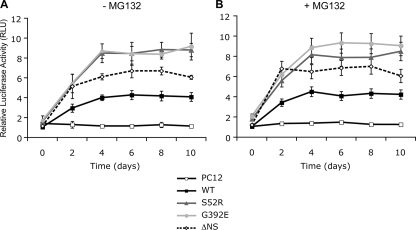
ER Accumulation of Neuroserpin Polymers Activates NF-κB
It has been reported that the accumulation of Z α1-antitrypsin in the ER of hepatocytes can activate NF-κB (11, 12). We therefore asked whether the accumulation of polymeric neuroserpin could also activate this transcription factor. The stable PC12 Tet-On cell lines expressing neuroserpin variants were induced with 10 μg/ml doxycycline and subsequently co-transfected with the NF-κB reporter pELAM1-Luc and the transfection efficiency reporter pRL-TK (Fig. 3). We found low levels of NF-κB activation in cells expressing WT and ΔNS neuroserpin (p < 0.001, WT and ΔNS versus PC12, two-way ANOVA with Bonferroni post test (Fig. 3A)). The expression of ER-retained S52R and G392E neuroserpin, however, resulted in much higher levels of NF-κB activation than WT (p < 0.001, S52R and G392E versus WT, two-way ANOVA with Bonferroni post test (Fig. 3A)). There was no further increase in NF-κB activation following the addition of MG132 (Fig. 3B), suggesting that NF-κB activation does not report the accumulation of ERAD substrates. The addition of conditioned media from PC12 cells expressing S52R and G392E neuroserpin to parental PC12 cells for 10 days did not activate NF-κB.
FIGURE 3.
The ER accumulation of neuroserpin polymers activates NF-κB. Neuroserpin expression was induced in PC12 cell lines with 10 μg/ml doxycycline, and cells were harvested at 2-day intervals for up to 10 days. Twenty-four hours prior to collection cells were co-transfected with a plasmid encoding a NF-κB-driven luciferase reporter (pELAM1-Luc) and with the transfection efficiency reporter pRL-TK Renilla luciferase. The graphs show firefly luciferase normalized to Renilla luciferase as averages ± S.D. of three repeats, and values are expressed in relative light units (RLU). NF-κB was measured in cells mock-treated (A) or treated with 100 nm MG132 (B) for 16 h prior to harvesting.
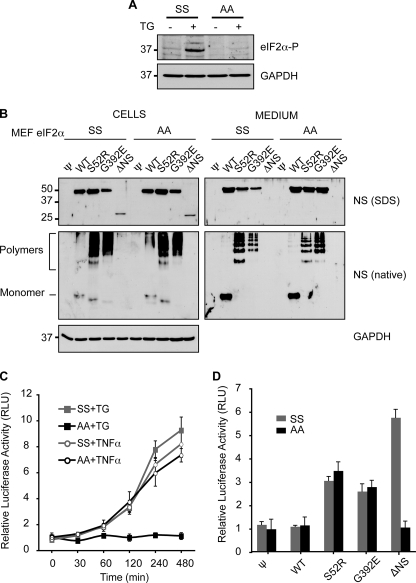
NF-κB Activation by Neuroserpin Polymers Is Independent of eIF2α Phosphorylation
In addition to IRE1 and ATF6, UPR signaling occurs via the ER-localized kinase PERK (30, 31), which has previously been shown to activate NF-κB by the phosphorylation of eIF2α (17, 32, 33). We investigated whether polymers of neuroserpin could activate NF-κB independently of the PERK branch of the UPR by using eIF2αA/A knock-in cells, in which mutation of serine 51 to alanine renders eIF2α unable to be phosphorylated by PERK (32). Both eIF2αS/S and eIF2αA/A MEFs were treated with 1 μm thapsigargin for 30 min to confirm that only the eIF2αS/S cells could phosphorylate serine 51 when the UPR is activated (Fig. 4A). The eIF2αS/S and eIF2αA/A MEF cells were next transduced with parental pBABE VSV-G pseudotyped retrovirus alone (ψ) or encoding WT, S52R, G392E, or ΔNS neuroserpin. Stable pools of neuroserpin transgene-expressing cells were selected with puromycin, and neuroserpin expression levels were assessed in all the cell pools by SDS-PAGE and Western blot analysis. The steady-state levels of all neuroserpin variants were similar in cell lysates and culture media for both eIF2αS/S and eIF2αA/A cells (Fig. 4B, Cells and Medium). As a control, TNF-α, which activates NFκB via phosphorylation of IκB, activated the NF-κB reporter in both eIF2αS/S and eIF2αA/A MEF cells (p < 0.005, both MEF lines untreated versus 8-h TNF-α, t test (Fig. 4C)). In contrast, only the eIF2αS/S MEF cell line activated NF-κB in response to the UPR activating agent thapsigargin, confirming the role of eIF2α phosphorylation in UPR-induced NFκB activation but not in TNFα signaling (p < 0.005, eIF2αS/S untreated versus 8 h thapsigargin, t test, p > 0.05 eIF2αA/A untreated versus 8-h thapsigargin (Fig. 4C)). The stable pools of neuroserpin transgene-expressing cells were then co-transfected with the NF-κB and control luciferase reporters, and the relative levels of NF-κB activity were determined. The empty virus control cells and cells expressing WT neuroserpin did not activate the NF-κB reporter (p > 0.05, t test, Fig. 4d, ψ and WT). ΔNS neuroserpin-expressing cells were able to activate NF-κB in the eIF2αS/S MEF cells, but this signal was abrogated in eIF2αA/A cell lines (p < 0.001, ψ versus ΔNS in eIF2αS/S, p > 0.05 in eIF2αA/A cells, t test; Fig. 4D, ΔNS), suggesting that in this case NF-κB activation is mediated through the PERK limb of the UPR. In contrast, the S52R and G392E neuroserpin polymer-forming cell lines were able to activate NF-κB in both eIF2αS/S and eIF2αA/A MEF cell lines (Fig. 4D, S52R and G392E). These data provide clear evidence that the accumulation of ordered protein polymers within the ER can activate NF-κB independently of the canonical UPR signaling pathway.
FIGURE 4.
Neuroserpin polymers expressed in eIF2α S51A/A MEF cells can still activate NF-κB. Wild-type eIF2α S51S/S and PERK signaling-deficient eIF2α S51A/A MEF cells were transduced with pBABE retrovirus alone (ψ) or encoding WT, S52R, G392E, or ΔNS neuroserpin. A, only wild-type eIF2α S51S/S cells can phosphorylate eIF2α. eIF2α S51S/S and eIF2α S51A/A MEF cells were treated as indicated with 1 μm thapsigargin (TG) for 30 min and allowed to recover for 3 h in fresh medium. Cell lysates were resolved by 10% w/v SDS-PAGE and phosphorylated-eIF2α detected by Western blot analysis. GAPDH was used as a protein loading control. B, neuroserpin is similarly expressed and handled in eIF2α S51S/S and eIF2α S51A/A MEF cells. Cell lysates and culture media from puromycin-selected stable pools of MEF cells expressing neuroserpin variants were resolved by 10% w/v SDS or 7.5% w/v non-denaturing PAGE (native), followed by Western blot analysis for neuroserpin or GAPDH as a protein loading control. C, eIF2α S51A/A MEF cells fail to activate NF-κB upon UPR induction with thapsigargin. eIF2α S51S/S and eIF2α S51A/A MEF cells were co-transfected with the NF-κB (pELAM1-Luc) and pRL-TK reporter plasmids as described in Fig. 3. Cells were either treated with 1 μm thapsigargin (TG) for 30 min with recovery in fresh medium or 200 ng/ml TNF-α for the indicated times prior to lysis and luciferase assay. The graph shows firefly luciferase normalized to Renilla luciferase as averages ± S.D. of three repeats, and values are expressed in relative light units (RLU). D, eIF2α S51A/A MEF cells expressing polymer-forming mutants of neuroserpin can activate NF-κB. eIF2α S51S/S and eIF2α S51A/A cells expressing WT or mutant neuroserpin were co-transfected with the NF-κB (pELAM1-Luc) and pRL-TK reporter plasmids as described in Fig. 3, and the normalized activity of NF-κB was expressed in RLUs. The data are averages ± S.D. of six independent experiments.
Polymer-associated NF-κB Activation Is Attenuated by Sequestering Intracellular Calcium
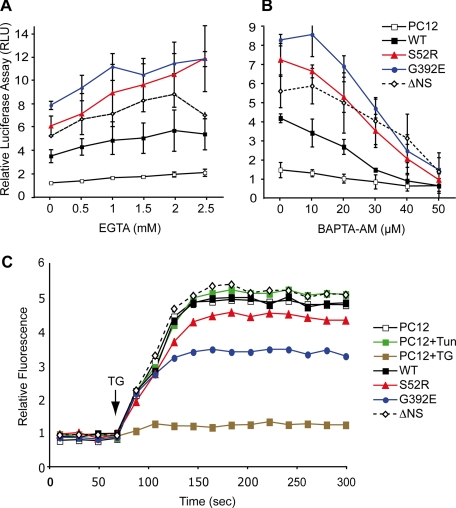
Activation of NF-κB has previously been shown to be dependent on calcium (13, 34, 35). We therefore treated the PC12 cell lines induced to express neuroserpin with chelators of either extracellular (EGTA, pH 7.4) or intracellular (BAPTA-AM) calcium. Increasing concentrations of EGTA had no effect on the activity of NF-κB in any of the cell lines (Fig. 5A). In contrast, chelating intracellular calcium resulted in a concentration-dependent reduction in NF-κB signaling in PC12 cells expressing WT, S52R, G392E, and ΔNS neuroserpin (Fig. 5B). These agents induced only minimal cellular toxicity and had no effect on the ability of the mutants to form polymers (data not shown). To control for possible off-pathway toxic effects of the acetoxymethyl (AM) moiety liberated during the loading of cells with BAPTA-AM, we treated cells with another AM-ester, BCECF-AM, according to the manufacturer's instructions. We found that BCECF-AM, which does not chelate intracellular calcium, failed to affect activation of the NFκB reporter (data not shown). Cells expressing the polymer-forming mutants of neuroserpin also showed a reduced ability to mobilize calcium when treated with 1 μm thapsigargin (p < 0.01, PC12 versus G392E, two-way ANOVA with Bonferroni post test (Fig. 5C)), suggesting that polymer accumulation affects ER calcium homeostasis. A similar difference of intracellular calcium increase in response to thapsigargin was seen between WT and G392E neuroserpin-expressing cells when assayed in calcium-free Opti-MEM medium (data not shown). This suggests that the effect of G392E neuroserpin is primarily on the release of ER calcium stores, rather than modifying store-operated calcium influx from the extracellular space. Taken together these data show that increased NF-κB activity in response to the accumulation of neuroserpin polymers within the ER is dependent on increased Ca2+ flux to the cytosol.
FIGURE 5.
The activation of NF-κB by neuroserpin polymers depends on intracellular Ca2+. Neuroserpin expression in PC12 cells was induced for 5 days with 10 μg/ml doxycycline, cells were co-transfected with NF-κB (pELAM1-Luc) and pRL-TK reporter plasmids as described in Fig. 3, and the normalized activity of NF-κB was expressed in RLU. The culture medium was supplemented 16 h prior to harvesting with increasing concentrations of EGTA, pH 7.4 (A), a membrane-impermeable extracellular Ca2+ chelator or BAPTA-AM (B), a membrane-permeable intracellular Ca2+ chelator. The values are averages ± S.D. of three independent repeats. C, expression of neuroserpin in PC12 cell lines was induced for 5 days with 10 μg/ml doxycycline, or PC12 cells were treated with either 2 μg/ml tunicamycin (Tun) for 16 h or 1 μm thapsigargin (TG) for 30 min. All cells were subsequently loaded with Fluo-4 (Molecular Probes, Invitrogen). Intracellular Ca2+ mobilization was measured with an emission optical density of 488 nm over time with the addition of 1 μm thapsigargin (arrow), which induces Ca2+ efflux into the cytoplasm and excites the Fluo-4 chromophore. The data were collected using a FACSCalibur (BD Biosciences) flow cytometer and analyzed with FlowJo; a representative dataset of three independent repeats is shown.
DISCUSSION
The serpinopathies are unusual among the conformational diseases in that they result from the retention of ordered polymers, rather than unfolded proteins, within the ER of the cell of synthesis (1, 2). This is most strikingly displayed in the dementia FENIB that is characterized by the intra-neuronal retention of polymers of neuroserpin as inclusions or Collin's bodies (5, 6). We have assessed the effect of these intracellular inclusions in cell lines that conditionally express WT neuroserpin, mutants that cause moderate (S52R) and severe (G392E) forms of FENIB, and a truncated misfolding version of neuroserpin that is a substrate for ERAD (ΔNS).
In our PC12 cell lines, WT neuroserpin was normally trafficked through the secretory pathway, as we have previously shown (23), whereas the truncated ΔNS neuroserpin was rapidly degraded by ERAD with a half-life of ∼30 min. This is similar to the cellular handling of the homologous Null Hong Kong variant of α1-antitrypsin (24). The S52R and G392E mutants of neuroserpin that underlie FENIB (6) were retained as polymers within the ER as shown before (21). Intracellular levels of all neuroserpin mutants increased when cells were treated with the proteasome inhibitors lactacystin and MG132 (Fig. 1C and results not shown). This indicates that there is a soluble misfolding fraction of S52R and G392E mutant neuroserpin that can be degraded by ERAD (Fig. 6). In contrast, the inhibition of lysosomal degradation had little effect on the clearance of S52R and G392E neuroserpin. This suggests that autophagy may not be as important in the degradation of neuroserpin polymers in contrast to polymers of Z α1-antitrypsin (36, 37).
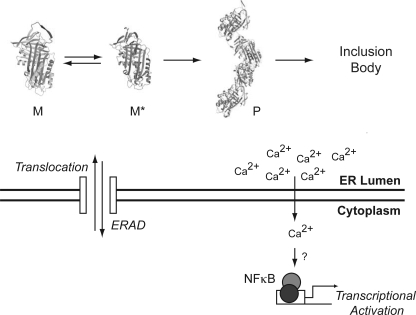
FIGURE 6.
Molecular mechanism of NF-κB activation by serpin polymers. Newly synthesized serpin molecules enter the ER by co-translational translocation. Correctly folded molecules (native protein) are packaged into ER-to-Golgi transport vesicles for secretion (not shown), whereas folding intermediates and misfolded species are retained in the ER, where they can be targeted into the ERAD pathway. Some mutant serpins monomers (M) form homogenous ordered polymers (P) via partial RCL insertion intermediates (M*), which escape the ER quality control system and form inclusion bodies. These inclusion bodies are in effect “insoluble” to the cell and may no longer be seen by the canonical quality control systems. Accumulated polymers of mutant serpins can elicit the specific activation of the transcription factor NF-κB independently of the UPR by increasing cytosolic Ca2+.
The retention of polymers of S52R and G392E neuroserpin within the ER had no effect on the expression levels of the UPR-dependent ER chaperones glucose-regulated protein 94, BiP, or protein disulfide isomerase (Fig. 2A). These chaperones were unaffected when intracellular concentrations of mutant proteins were increased by proteasome inhibition (results not shown). The expression of truncated ΔNS neuroserpin similarly had no effect, suggesting that the expression levels of these ER chaperones were insensitive to low level accumulation/misfolding of proteins within the ER. Truncated ΔNS neuroserpin was able, however, to elicit the splicing of XBP1 mRNA when its intracellular levels were raised by proteasome inhibition, whereas polymers of mutant neuroserpin had no effect on XBP1 mRNA splicing even when their levels were increased with the same treatment (Fig. 2B). This lack of UPR activation upon neuroserpin polymer accumulation was also apparent when characterized by a more sensitive assay, based on the pAT6(5x)-Luc or UPRE-driven luciferase reporter plasmid (Fig. 2C).
Although the accumulation of polymeric neuroserpin within the ER did not cause splicing of XBP1 mRNA or activation of ATF6, it did activate NF-κB (Fig. 3). This activation of NF-κB was similar to that previously reported for cell and mouse models of α1-antitrypsin deficiency (11, 12). Previous work has shown that the activation of the UPR can induce NF-κB signaling through PERK (17, 33). We therefore used MEF cells deficient in the PERK signaling branch of the UPR to assess polymer activation of NF-κB in more detail. Polymers of neuroserpin, but not the ERAD substrate ΔNS neuroserpin, were able to activate NF-κB in MEF eIF2αA/A cells (Fig. 4). These data confirm that neuroserpin polymers can activate NF-κB by a pathway that is independent of the canonical UPR (Fig. 6). This is reminiscent of an ER to nucleus signaling pathway that has been termed the EOR (13, 14, 38). The EOR was initially described in the context of adenovirus E3/19K protein and ΔF508 mutant of cystic fibrosis transmembrane conductance regulator accumulation and has been proposed as the signaling pathway that operates upon accumulation of α1-antitrypsin polymers within hepatocytes (11, 12). Our results support the existence of a UPR-independent ER stress-signaling pathway that activates NF-κB. The term “ER overload response” (13), however, may not be the most accurate to describe serpin polymer-mediated activation of NF-κB. This is because the overload of any protein that causes physical disruption of the ER would be associated with a reduction in ER efficiency and/or perturbance of Ca2+ homeostasis and the induction of ER stress (39, 40). Furthermore, there were no morphological changes in the ER caused by polymer accumulation in our PC12 cell lines that would justify the term “overload” (data not shown). Moreover, the term EOR does not distinguish between misfolded proteins, such as cystic fibrosis transmembrane conductance regulator (15) or correctly folded proteins, such as viral inclusions (14) or polymers of neuroserpin and α1-antitypsin. We therefore suggest that “ordered protein response” (OPR) would be a more appropriate name for this pathway to contrast with the unfolded protein response (UPR).
The EOR is associated with mobilization of intracellular calcium (35, 41–44). We therefore assessed the effect of extrinsic and intrinsic calcium chelators on the UPR (ΔNS neuroserpin) and OPR (polymer-mediated) activation of NF-κB. Extracellular chelation of calcium had little effect on either UPR- or OPR-mediated activation of NF-κB. In contrast, signaling of NF-κB was attenuated by the chelation of intracellular Ca2+. This was most striking for the cell lines expressing S52R and G392E neuroserpin. The intracellular capacity for calcium flux in the two polymer-forming cell lines was significantly reduced when Ca2+ was mobilized from the ER by the addition of 1 μm thapsigargin. These results suggest that ER Ca2+ regulation is involved in the polymer-mediated activation of NF-κB in the OPR. Importantly neither secreted neuroserpin, nor extrinsic factors secreted by the neuroserpin cell lines into the culture medium, had any effect on the activity of NF-κB in parental PC12 cells. Thus, this cell-autonomous signal must derive from the site of intracellular polymer accumulation. Therefore, the activation of NF-κB by mutant polymer accumulation in PC12 cells is mediated by a calcium-dependent process involved in transducing the signal from the ER to the nucleus.
In summary, we have dissected the signaling mediators that activate the transcription factor NF-κB following the accumulation of polymers of neuroserpin within the ER. This work supports the existence of a UPR-independent ER-to-nucleus signal transduction pathway that corresponds to the EOR, but for accuracy, we propose to name it the ordered protein response or OPR. This pathway is also likely to be activated by the accumulation of ordered polymers of mutant α1-antitypsin and in other serpinopathies. Transient activation of NF-κB in response to ER-ordered polymers might regulate key pathways induced during cellular stress, such as proliferation, metabolic activity, and differentiation (45). However, chronic activation of NF-κB may contribute to the cell death that underlies the accumulation of polymers of neuroserpin and α1-antitypsin to cause dementia and cirrhosis, respectively.
Supplementary Material
Acknowledgments
We are grateful to Dr. Timothy Weaver (Cincinnati Children's Hospital Medical Center, Cincinnati, OH) for the pATF6(5×)-Luc, pELAM1-Luc, and pRL-TK luciferase reporters. M. J. D. wishes to thank all members of the Lomas laboratory and Dr. Karin Römisch for help and guidance.
This work was supported, in whole or in part, by National Institutes of Health Grants RO1-DK042394, RO1-HL052173, and PO1-HL057346 (to R. J. K.). This work was also supported by the Medical Research Council (MRC, UK) and Papworth National Health Service Trust.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. 1.
- ER
- endoplasmic reticulum
- FENIB
- familial encephalopathy with neuroserpin inclusion bodies
- PERK
- PKR-like ER kinase
- ATF6
- activating transcription factor 6
- IRE1
- inositol-requiring kinase 1
- UPR
- unfolded protein response
- ERAD
- ER-associated degradation
- EOR
- ER overload response
- GAPDH
- glyceraldehyde-3-phosphate dehydrogenase
- WT
- wild type
- TG
- thapsigargin
- ANOVA
- analysis of variance
- BAPTA-AM
- 1,2-bis(2-aminophenoxy)ethane-N,N,N′,N′-tetraacetic acid tetrakis(acetoxymethyl ester)
- OPR
- ordered protein response
- RLU
- relative light unit(s)
- MEF
- mouse embryonic fibroblast.
REFERENCES
- 1.Carrell R. W., Lomas D. A. ( 1997) Lancet 350, 134– 138 [DOI] [PubMed] [Google Scholar]
- 2.Lomas D. A., Carrell R. W. ( 2002) Nat. Rev. Genet. 3, 759– 768 [DOI] [PubMed] [Google Scholar]
- 3.Gooptu B., Hazes B., Chang W. S., Dafforn T. R., Carrell R. W., Read R. J., Lomas D. A. ( 2000) Proc. Natl. Acad. Sci. U.S.A. 97, 67– 72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sivasothy P., Dafforn T. R., Gettins P. G., Lomas D. A. ( 2000) J. Biol. Chem. 275, 33663– 33668 [DOI] [PubMed] [Google Scholar]
- 5.Davis R. L., Shrimpton A. E., Holohan P. D., Bradshaw C., Feiglin D., Collins G. H., Sonderegger P., Kinter J., Becker L. M., Lacbawan F., Krasnewich D., Muenke M., Lawrence D. A., Yerby M. S., Shaw C. M., Gooptu B., Elliott P. R., Finch J. T., Carrell R. W., Lomas D. A. ( 1999) Nature 401, 376– 379 [DOI] [PubMed] [Google Scholar]
- 6.Davis R. L., Shrimpton A. E., Carrell R. W., Lomas D. A., Gerhard L., Baumann B., Lawrence D. A., Yepes M., Kim T. S., Ghetti B., Piccardo P., Takao M., Lacbawan F., Muenke M., Sifers R. N., Bradshaw C. B., Kent P. F., Collins G. H., Larocca D., Holohan P. D. ( 2002) Lancet 359, 2242– 2247 [DOI] [PubMed] [Google Scholar]
- 7.Davis R. L., Holohan P. D., Shrimpton A. E., Tatum A. H., Daucher J., Collins G. H., Todd R., Bradshaw C., Kent P., Feiglin D., Rosenbaum A., Yerby M. S., Shaw C. M., Lacbawan F., Lawrence D. A. ( 1999) Am J. Pathol. 155, 1901– 1913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ron D., Walter P. ( 2007) Nat. Rev. Mol. Cell Biol. 8, 519– 529 [DOI] [PubMed] [Google Scholar]
- 9.Marciniak S. J., Ron D. ( 2006) Physiol. Rev. 86, 1133– 1149 [DOI] [PubMed] [Google Scholar]
- 10.Graham K. S., Le A., Sifers R. N. ( 1990) J. Biol. Chem. 265, 20463– 20468 [PubMed] [Google Scholar]
- 11.Hidvegi T., Schmidt B. Z., Hale P., Perlmutter D. H. ( 2005) J. Biol. Chem. 280, 39002– 39015 [DOI] [PubMed] [Google Scholar]
- 12.Lawless M. W., Greene C. M., Mulgrew A., Taggart C. C., O'Neill S. J., McElvaney N. G. ( 2004) J. Immunol. 172, 5722– 5726 [DOI] [PubMed] [Google Scholar]
- 13.Pahl H. L., Baeuerle P. A. ( 1995) EMBO J. 14, 2580– 2588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pahl H. L., Sester M., Burgert H. G., Baeuerle P. A. ( 1996) J. Cell Biol. 132, 511– 522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Knorre A., Wagner M., Schaefer H. E., Colledge W. H., Pahl H. L. ( 2002) Biol. Chem. 383, 271– 282 [DOI] [PubMed] [Google Scholar]
- 16.Pahl H. L. ( 1999) Physiol. Rev. 79, 683– 701 [DOI] [PubMed] [Google Scholar]
- 17.Deng J., Lu P. D., Zhang Y., Scheuner D., Kaufman R. J., Sonenberg N., Harding H. P., Ron D. ( 2004) Mol. Cell. Biol. 24, 10161– 10168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Belorgey D., Crowther D. C., Mahadeva R., Lomas D. A. ( 2002) J. Biol. Chem. 277, 17367– 17373 [DOI] [PubMed] [Google Scholar]
- 19.Bartz S. R., Vodicka M. A. ( 1997) Methods 12, 337– 342 [DOI] [PubMed] [Google Scholar]
- 20.Lu P. D., Jousse C., Marciniak S. J., Zhang Y., Novoa I., Scheuner D., Kaufman R. J., Ron D., Harding H. P. ( 2004) EMBO J. 23, 169– 179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Miranda E., Römisch K., Lomas D. A. ( 2004) J. Biol. Chem. 279, 28283– 28291 [DOI] [PubMed] [Google Scholar]
- 22.Marciniak S. J., Yun C. Y., Oyadomari S., Novoa I., Zhang Y., Jungreis R., Nagata K., Harding H. P., Ron D. ( 2004) Genes Dev. 18, 3066– 3077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Miranda E., MacLeod I., Davies M. J., Pérez J., Römisch K., Crowther D. C., Lomas D. A. ( 2008) Hum. Mol. Genet. 17, 1527– 1539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sifers R. N., Brashears-Macatee S., Kidd V. J., Muensch H., Woo S. L. ( 1988) J. Biol. Chem. 263, 7330– 7335 [PubMed] [Google Scholar]
- 25.Fenteany G., Standaert R. F., Lane W. S., Choi S., Corey E. J., Schreiber S. L. ( 1995) Science 268, 726– 731 [DOI] [PubMed] [Google Scholar]
- 26.Libby P., Goldberg A. L. ( 1978) Science 199, 534– 536 [DOI] [PubMed] [Google Scholar]
- 27.Seglen P. O. ( 1983) Methods Enzymol. 96, 737– 764 [DOI] [PubMed] [Google Scholar]
- 28.Ma Y., Brewer J. W., Diehl J. A., Hendershot L. M. ( 2002) J. Mol. Biol. 318, 1351– 1365 [DOI] [PubMed] [Google Scholar]
- 29.Shen J., Prywes R. ( 2005) Methods 35, 382– 389 [DOI] [PubMed] [Google Scholar]
- 30.Harding H. P., Zhang Y., Ron D. ( 1999) Nature 397, 271– 274 [DOI] [PubMed] [Google Scholar]
- 31.Harding H. P., Zhang Y., Bertolotti A., Zeng H., Ron D. ( 2000) Mol. Cell 5, 897– 904 [DOI] [PubMed] [Google Scholar]
- 32.Scheuner D., Song B., McEwen E., Liu C., Laybutt R., Gillespie P., Saunders T., Bonner-Weir S., Kaufman R. J. ( 2001) Mol. Cell 7, 1165– 1176 [DOI] [PubMed] [Google Scholar]
- 33.Jiang H. Y., Wek S. A., McGrath B. C., Scheuner D., Kaufman R. J., Cavener D. R., Wek R. C. ( 2003) Mol. Cell. Biol. 23, 5651– 5663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Biswas G., Anandatheerthavarada H. K., Zaidi M., Avadhani N. G. ( 2003) J. Cell Biol. 161, 507– 519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lilienbaum A., Israël A. ( 2003) Mol. Cell. Biol. 23, 2680– 2698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Palmer E. A., Kruse K. B., Fewell S. W., Buchanan S. M., Brodsky J. L., McCracken A. A. ( 2003) J. Cell Sci. 116, 2361– 2373 [DOI] [PubMed] [Google Scholar]
- 37.Teckman J. H., Perlmutter D. H. ( 2000) Am. J. Physiol. Gastrointest Liver Physiol. 279, G961– G974 [DOI] [PubMed] [Google Scholar]
- 38.Pahl H. L., Baeuerle P. A. ( 1997) Trends Biochem. Sci. 22, 63– 67 [DOI] [PubMed] [Google Scholar]
- 39.Michalak M., Robert Parker J. M., Opas M. ( 2002) Cell Calcium 32, 269– 278 [DOI] [PubMed] [Google Scholar]
- 40.Molinari M., Helenius A. ( 2000) Science 288, 331– 333 [DOI] [PubMed] [Google Scholar]
- 41.Pahl H. L., Baeuerle P. A. ( 1996) FEBS Lett. 392, 129– 136 [DOI] [PubMed] [Google Scholar]
- 42.Kuang E., Wan Q., Li X., Xu H., Liu Q., Qi Y. ( 2005) J. Cell. Physiol. 204, 549– 559 [DOI] [PubMed] [Google Scholar]
- 43.Steffan N. M., Bren G. D., Frantz B., Tocci M. J., O'Neill E. A., Paya C. V. ( 1995) J. Immunol. 155, 4685– 4691 [PubMed] [Google Scholar]
- 44.Trushin S. A., Pennington K. N., Algeciras-Schimnich A., Paya C. V. ( 1999) J. Biol. Chem. 274, 22923– 22931 [DOI] [PubMed] [Google Scholar]
- 45.Perkins N. D. ( 2007) Nat. Rev. Mol. Cell Biol. 8, 49– 62 [DOI] [PubMed] [Google Scholar]
- 46.Rock K. L., Gramm C., Rothstein L., Clark K., Stein R., Dick L., Hwang D., Goldberg A. L. ( 1994) Cell 78, 761– 771 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.