Abstract
Synthesis of a protective cyst wall is required for survival outside of the host and for infection of Giardia lamblia. Little is known of gene regulation of the cyst wall proteins (CWPs) during differentiation into dormant cysts. WRKY homologues constitute a large family of DNA-binding proteins in plants that are involved in several key cellular functions, including disease resistance, stress response, dormancy, and development. A putative wrky gene has been identified in the G. lamblia genome. We found that wrky expression levels increased significantly during encystation. The epitope-tagged WRKY was translocated into the nuclei during encystation. Recombinant WRKY specifically bound to its own promoter and the encystation-induced cwp1 and cwp2 promoters. WRKY contains several key residues for DNA binding, and mutation analysis revealed that its binding sequences are similar to those of the known plant WRKY proteins and that two of them are positive cis-acting elements of the wrky and cwp2 promoters. Overexpression of WRKY increased the cwp1-2 and myb2 mRNA levels, and these gene promoters were bound by WRKY in vivo. Interestingly, the wrky and cwp1-2 genes were up-regulated by ERK1 (extracellular signal-related kinase 1) overexpression, suggesting that WRKY may be a downstream component of the ERK1 pathway. In addition, a WRKY mutant that cannot enter nuclei and an ERK1 mutant lacking the predicted kinase domain showed decreased cwp1-2 gene expression. Our results suggest that the WRKY family has been conserved during evolution and that WRKY is an important transactivator of the cwp1-2 genes during G. lamblia differentiation into dormant cysts.
Giardia lamblia is an intestinal protozoan parasite responsible for outbreaks of waterborne diarrhea (1, 2). Children with chronic diarrhea from its infection face the risk of malnutrition and delayed mental development (3). G. lamblia has two life cycle stages in response to different inhospitable environments: a pathogenic trophozoite form and a dormant infectious cyst form (4, 5). The cysts are protectively walled and resistant to hypotonic lysis by fresh water and gastric acid and are responsible for transmission of giardiasis. Despite the importance of the cyst wall in giardial pathophysiology, regulation of its synthesis during encystation is poorly understood. During encystation, genes encoding cyst wall structural proteins (Cwp1, Cwp2, and Cwp3) and enzymes in the cyst wall polysaccharide biosynthetic pathway are coordinately induced (6–11).
G. lamblia also raises great biological interest for understanding the progress of eukaryotic evolution, since it has been proposed as an early branching eukaryote (12, 13). The lack of clear giardial homologs to many cellular components for DNA synthesis, transcription and RNA processing suggests their divergence or their functional redundancy with other proteins in some pathways (14). Many aspects of giardial gene transcription are unusual. G. lamblia has a highly divergent TATA-binding protein and lacks 8 of the 12 general transcription initiation factors (15, 16). Unusually short 5′-flanking regions (<65 bp) with no consensus TATA boxes or other cis-acting elements identified in late branching eukaryotic promoters are sufficient for the expression of some genes (6, 7, 9, 17–20). Instead, AT-rich sequences have been found around the transcription start sites of many genes, functionally similar to the initiator element in late branching eukaryotes (6, 7, 9, 17–20). They are essential for promoter activity and play a predominant role in determining the positions of the transcription start sites (17–19). Specific regulatory regions have been identified in the encystation-induced cwp2 promoter, including a positive cis-acting element (−23 to −10 relative to the translation start site) required for encystation-specific promoter activity and a negative cis-acting element (−64 to −23 region) required for promoter activity in vegetative cells (21). The latter region also contains a region (−61 to −52) that controls sterol-mediated decrease in the cwp2 transcription (22).
There is little understanding of the molecular mechanisms governing transcriptional regulation of the cyst wall biosynthetic pathway. Few transcription factors have been characterized to date in G. lamblia (23–25). A Myb family transcription factor (Myb2) is encystation-induced and is involved in coordinating up-regulation of the cwp1–3 genes (23, 26). Two GARP (named from the maize GOLDEN2, Arabidopsis response regulator proteins and the Chlamydomonas Psr1 protein) family transcription factors may be involved in transcriptional regulation of many different genes, including the encystation-induced cwp1 gene and constitutive ran gene (24). An ARID (AT-rich interaction domain) family transcription factor can bind to specific AT-rich Inr sequences and function as an important transactivator in the regulation of the cwp1 gene (25).
WRKY proteins comprise a large family of transcription factors in plants involved in many physiological processes during growth and development; during dormancy; and during response to biotic and abiotic stress, disease resistance, and senescence (27–31). More than 70 members of the WRKY family found in Arabidopsis thaliana have diverse roles in response to various pathogen and environmental conditions (27). The mRNA levels of these wrky genes also increased in response to these changing environments (30, 32). The wrky gene family has been found in many plants but has not been identified to date in yeast or animal genomes (33). The WRKY domain consists of ∼60 amino acid residues and has a conserved WRKYGQK sequence followed by a zinc finger-like motif (27). All known WRKY proteins contain either one or two WRKY domains. They can be classified on the basis of both the number of WRKY domains and the features of their zinc finger-like motifs (27). WRKY proteins with two WRKY domains belong to group I. WRKY proteins with one WRKY domain belong to group II or III (27). The group I or II WRKY domains have C2H2 zinc finger-like motifs, but the group III WRKY domains have C2HC zinc finger-like motifs (27). All of these WRKY domains have similar sequence specificities for DNA binding (27). The similarity between members of the WRKY family is limited to their WRKY domains (27). Putative transactivation domains and nuclear localization signals have been identified outside of the WRKY domains (27).
WRKY transcription factors have a high binding affinity to a DNA sequence designated the W box, (c/t)TGAC(c/t). W box-dependent binding activity requires both the invariable WRKY amino acid signature and the cysteine and histidine residues of the WRKY domain, which tetrahedrally coordinate a zinc atom (34). Several defense-related genes in plants have multiple copies of W boxes in their promoters that are recognized by WRKY proteins and necessary for the inducible expression of these genes (35–37). Many wrky genes in plants have W boxes in their own promoters, suggesting a positive or negative autoregulation of the wrky genes (36, 38). WRKY factors have been identified as downstream components of a defense- and pathogen-induced mitogen-activated protein kinase (MAPK)2 signaling pathway in plants (39, 40). A putative wrky gene has been identified in G. lamblia genome (33). The WRKY domains of the giardial WRKY protein are highly similar to those of the group I WRKY protein (33).
Because WRKY proteins play a regulatory role in plant development and dormancy (27, 28, 41), we asked whether giardial WRKY can influence gene expression during Giardia differentiation into dormant cysts. We found that the expression levels of the giardial wrky gene increased during encystation and that the WRKY protein localized to both nuclei and cytosol during vegetative growth but mainly to the nuclei during encystation. Interestingly, WRKY can bind specifically to the (g/a)GTCA(g/a) sequence in the wrky and cwp2 promoter, which is similar to the antisense sequence of the W box in plants. WRKY can also bind specifically to the cwp1 promoter. We also found that the WRKY binding sites were positive cis-acting elements of the wrky and cwp2 promoters. We used chromatin immunoprecipitation (ChIP) assays to confirm the binding of WRKY to the wrky, cwp1, cwp2, and myb2 gene promoters. A member of MAPK family, ERK1 (extracellular signal-related kinase 1), has been identified to exhibit a significantly increased kinase activity during early encystation (42). We also found that the wrky, cwp1, cwp2, and myb2 mRNA levels were increased by the ERK1 overexpression, suggesting that WRKY may be a downstream component of the MAPK/ERK1 pathway. In addition, a WRKY mutant that cannot enter nuclei and an ERK1 mutant lacking the predicted kinase domain showed decreased cwp1, cwp2, and myb2 gene expression relative to the respective wild type cell lines. Our results suggest that WRKY can function as a transactivator of the cwp1 and cwp2 genes to regulate G. lamblia differentiation into dormant cysts.
EXPERIMENTAL PROCEDURES
G. lamblia Culture
Trophozoites of G. lamblia WB (ATCC 30957), clone C6, were cultured in modified TYI-S33 medium (43) and encysted, as previously described (8). Cyst count was performed on the late log/early stationary phase cultures (1.5 × 106 cells/ml) during vegetative growth, as previously described (44).
Isolation and Analysis of the wrky Gene
The G. lamblia genome data base (available on the World Wide Web) (14, 45) was searched with the amino acid sequences of the WRKY domain of Arabidopsis WRKY2 (GenBankTM accession number NM_125010) using the BLAST program (46). This search detected one putative homologue for WRKY that has been reported previously (GenBankTM accession number XM_001708755; open reading frame 9237 in the G. lamblia genome data base) (33). The WRKY coding region with 324 nucleotides of 5′-flanking regions was cloned, and the nucleotide sequence was determined. The wrky gene sequence in the data base was essentially correct. To isolate the cDNA of the wrky gene, we performed RT-PCR with wrky-specific primers using total RNA from G. lamblia. For RT-PCR, 5 μg of DNase-treated total RNA from vegetative and 24-h encysting cells was mixed with oligo(dT)12–18 and Superscript II RNase H− reverse transcriptase (Invitrogen). Synthesized cDNA was used as a template in subsequent PCR with primers WRKYF (ATGAAAGAAGGATCCCTGCA) and WRKYR (AGAGTAAACGTTAACCATCGG). Genomic and RT-PCR products were cloned into pGEM-T easy vector (Promega) and sequenced (Applied Biosystems). RT-PCR analysis of cwp1 (U09330) and ran (U02589) gene expression was performed using primers cwp1F (ATGATGCTCGCTCTCCTT) and cwp1R (TCAAGGCGGGGTGAGGCA) and primers ranF (ATGTCTGACCCAATCAGC) and ranR (TCAATCATCGTCGGGAAG), respectively.
RNA Extraction, RT-PCR, and Quantitative Real Time PCR Analysis
Total RNA was extracted from the G. lamblia cell line at the differentiation stages indicated in the legends to Figs. 2, 7, and 8, using TRIzol reagent (Invitrogen). For RT-PCR, 5 μg of DNase-treated total RNA was mixed with oligo(dT)12–18 and Superscript II RNase H− reverse transcriptase (Invitrogen). Synthesized cDNA was used as a template in subsequent PCR. Semiquantitative RT-PCR analysis of wrky (XM_001708755; open reading frame 9237), wrky-ha, cwp1 (U09330; open reading frame 5638), cwp2 (U28965; open reading frame 5432), myb2 (AY082882; open reading frame 8722), ran (U02589; open reading frame 15869), and 18 S ribosomal RNA (M54878; open reading frame r0019) gene expression was performed using primers wrkyF (ATGAAAGAAGGATCCCTGCAT) and wrkyR (TGGTGGTTTTTCTGATGCTAC), wrkyHAF (TGAGAACATTAACTTGACGAATGCA) and HAR (AGCGTAATCTGGAACATCGTATGGGTA), cwp1F and cwp1R, cwp2F (ATGATCGCAGCCCTTGTTCTA) and cwp2R (CCTTCTGCGGACAATAGGCTT), myb2F (ATGTTACCGGTACCTTCTCAGC) and myb2R (GGGTAGCTTCTCACGGGGAAG), ranF and ranR, and 18SrealF (AAGACCGCCTCTGTCAATCAA) and 18SrealR (GTTTACGGCCGGGAATACG), respectively. For quantitative real time PCR, SYBR Green PCR master mixture was used (Kapa Biosystems). PCR was performed using an Applied Biosystems PRISMTM 7900 Sequence Detection System (Applied Biosystems). Specific primers were designed for detection of the wrky, wrky-ha, cwp1, cwp2, myb2, ran, and 18 S ribosomal RNA genes: wrkyrealF (ATTGACGCAGGGCATTCG) and wrkyrealR (CGAAGGGATTTTGCTGGTAGAC); wrkyHAF and HAR; cwp1realF (AACGCTCTCACAGGCTCCAT) and cwp1realR (AGGTGGAGCTCCTTGAGAAATTG); cwp2realF (TAGGCTGCTTCCCACTTTTGAG) and cwp2realR (CGGGCCCGCAAGGT); myb2realF (TCCCTAATGACGCCAAACG) and myb2realR (AGCACGCAGAGGCCAAGT); ranrealF (TCGTCCTCGTCGGAAACAA) and ranrealR (AACTGTCTGGGTGCGGATCT); 18SrealF and 18SrealR. Two independently generated stably transfected lines were made from each construct, and each of these cell lines was assayed three separate times. The results are expressed as relative expression level over control. Student's t tests were used to determine statistical significance of differences between samples.
FIGURE 2.
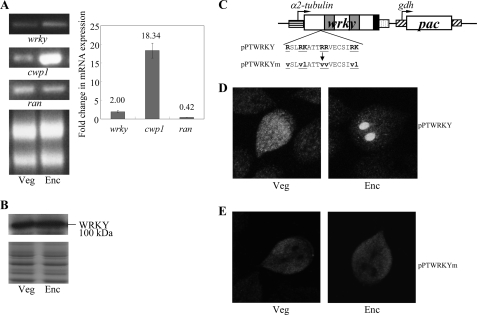
Analysis of wrky gene expression. A, RT-PCR and quantitative real time PCR analysis of wrky gene expression. RNA samples were prepared from G. lamblia wild-type non-transfected WB cells cultured in growth (Veg) or encystation medium and harvested at 24 h (Enc). RT-PCR was performed using primers specific for wrky, cwp1, and ran genes. Ribosomal RNA loading controls are shown in the bottom panel. Representative results are shown on the left. Real time PCR was preformed using primers specific for wrky, cwp1, ran, and 18 S rRNA genes. Transcript levels were normalized to 18 S rRNA levels. -Fold changes in mRNA expression are shown as the ratio of transcript levels in encysting cells relative to vegetative cells. Results are expressed as the means ± S.E. of at least three separate experiments (right). B, WRKY protein levels in different stages. The wild-type non-transfected WB cells were cultured in growth (Veg) or encystation medium for 24 h (Enc) and then subjected to SDS-PAGE and Western blot. The blot was probed by anti-WRKY antibody. Representative results are shown. Equal amounts of proteins loaded were confirmed by SDS-PAGE and Coomassie Blue staining. C, diagrams of the pPTWRKY plasmid. The pac gene (open box) is under the control of the 5′- and 3′-flanking regions of the gdh gene (striated box). The wrky gene is under the control of the 5′-flanking region of the α2-tubulin gene (open box) and the 3′-flanking region of the ran gene (dotted box). The filled black box indicates the coding sequence of the HA epitope tag. In pPTWRKYm, a stretch of basic amino acids between residues 315 and 331 is mutated as shown in underlined letters. D and E, localization of WRKY and its mutant. The pPTWRKY or pPTWRKYm stable transfectants were cultured in growth (Veg; left panels) or encystation medium for 24 h (Enc; right panels) and then subjected to immunofluorescence analysis using anti-HA antibody for detection. The product of pPTWRKY localizes to the nuclei and cytosol in a vegetative trophozoite and to the nuclei in an encysting trophozoite, respectively (D). The product of pPTWRKYm localizes to the cytosol in both vegetative and encysting trophozoites (E).
FIGURE 7.
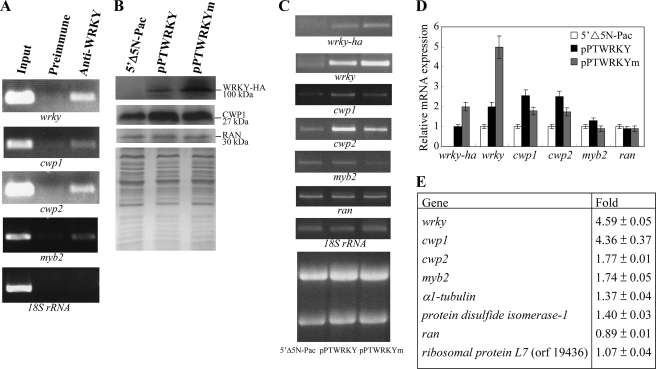
Induction of cwp1 and cwp2 gene expression in the WRKY overexpressing cell line. A, recruitment of WRKY to the cwp1 and cwp2 promoters. The non-transfected WB cells were cultured in encystation medium for 24 h and then subjected to ChIP assays. Anti-WRKY antibody was used to assess binding of WRKY to endogenous gene promoters. Preimmune serum was used as a negative control. Immunoprecipitated chromatin was analyzed by PCR using primers that amplify the 5′-flanking region of specific genes. At least three independent experiments were performed. Representative results are shown. Immunoprecipitated products of WRKY yielded more PCR products of wrky, cwp1, cwp2, and myb2 promoters, indicating that WRKY was bound to these promoters. The 18 S ribosomal RNA gene promoter was used as a negative control for our ChIP analysis. B, overexpression of WRKY increased the levels of CWP1. The 5′Δ5N-Pac, pPTWRKY, and pPTWRKYm stable transfectants were cultured in encystation medium for 24 h and then subjected to SDS-PAGE and Western blot. The blot was probed by anti-HA and anti-CWP1 antibody. Equal amounts of proteins loaded were confirmed by detection of giardial RAN protein. Representative results are shown. C, RT-PCR analysis of gene expression in the WRKY- and WRKYm-overexpressing cell line. The 5′Δ5N-Pac, pPTWRKY, and pPTWRKYm stable transfectants were cultured in encystation medium for 24 h and then subjected to RT-PCR analysis. PCR was preformed using primers specific for wrky-ha, wrky, cwp1, cwp2, myb2, ran, and 18 S ribosomal RNA genes. Ribosomal RNA loading controls are shown in the bottom panel. D, quantitative real time PCR analysis of gene expression in the WRKY- and WRKYm-overexpressing cell line. Real time PCR was preformed using primers specific for wrky, cwp1, cwp2, myb2, ran, and 18 S ribosomal RNA. Transcript levels were normalized to 18 S ribosomal RNA levels. -Fold changes in mRNA expression are shown as the ratio of transcript levels in the pPTWRKY or pPTWRKYm cell line relative to the 5′Δ5N-Pac cell line. Results are expressed as the means ± S.E. of at least three separate experiments. E, up-regulation of cwp1, cwp2, and myb2 in the WRKY-overexpressing cell line. Microarray data were obtained from the 5′Δ5N-Pac and pPTWRKY cell lines. -Fold changes are shown as the ratio of transcript levels in the pPTWRKY cell line relative to the 5′Δ5N-Pac cell line. Results are expressed as the means ± S.E. of at least three separate experiments.
FIGURE 8.
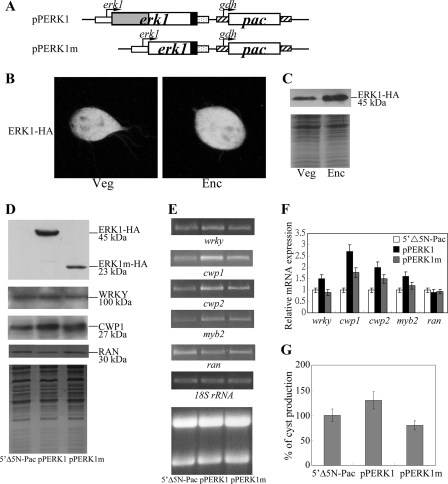
Induction of wrky and cwp1-2 gene expression in the ERK1-overexpressing cell line. A, diagrams of the pPERK1 and pPERK1m plasmids. The pac gene (open box) expression cassette is the same as in Fig. 2C. The erk1 gene is under the control of its own 5′-flanking region (open boxes) and the 3′-flanking region of the ran gene (dotted box). ERK1m does not contain the predicted kinase domain (residues 26–202) (gray box). The filled box indicates the coding sequence of the HA epitope tag. B, cytosolic localization of ERK1. The pPERK1 stable transfectants were cultured in growth medium (Veg) or encystation medium, harvested at 24 h (Enc), and then subjected to immunofluorescence analysis using anti-HA antibody for detection. The left and right panels show that the product of pPERK1 localizes to the cytosol and flagella in vegetative and encysting trophozoites, respectively. C, ERK1 protein levels in pPERK1 stable transfectants. The pPERK1 stable transfectants were cultured in growth medium (Veg) or encystation medium and harvested at 24 h (Enc). HA-tagged ERK1 protein was detected using an anti-HA antibody by Western blot analysis. Coomassie-stained total protein loading control is shown below. D, overexpression of ERK1 increased the levels of WRKY and CWP1. The 5′Δ5N-Pac, pPERK1, and pPERK1m stable transfectants were cultured in growth medium and then subjected to SDS-PAGE and Western blot. The blot was probed by anti-HA, anti-WRKY, and anti-CWP1 antibody. Equal amounts of proteins loaded were confirmed by detection of giardial RAN protein. Representative results are shown. E, RT-PCR analysis of gene expression in the ERK1- and ERK1m-overexpressing cell line. The 5′Δ5N-Pac, pPERK1, and pPERK1m stable transfectants were cultured in growth medium and then subjected to RT-PCR analysis. PCR was preformed using primers specific for wrky, cwp1, cwp2, myb2, ran, and 18 S ribosomal RNA genes. Ribosomal RNA loading controls are shown in the bottom panel. F, quantitative real time PCR analysis of gene expression in the ERK1- and ERK1m-overexpressing cell line. Real time PCR was preformed using primers specific for wrky, cwp1, cwp2, myb2, ran, and 18 S ribosomal RNA. Transcript levels were normalized to 18 S ribosomal RNA levels. -Fold changes in mRNA expression are shown as the ratio of transcript levels in pPERK1 or pPERK1m cell line relative to the 5′Δ5N-Pac cell line. Results are expressed as the means ± S.E. of at least three separate experiments. G, overexpression of ERK1 increased cyst formation. The 5′Δ5N-Pac, pPERK1, and pPERK1m stable transfectants were cultured in growth medium. Cyst count was performed on the late log/early stationary phase cultures (1.5 × 106 cells/ml). The sum of total cysts is expressed as relative expression level over control. Values are shown as means ± S.E.
Plasmid Construction
All constructs were verified by DNA sequencing with a BigDye Terminator 3.1 DNA sequencing kit and an Applied Biosystems 3100 DNA analyzer (Applied Biosystems). Plasmid 5′Δ5N-Pac was a gift from Dr. Steven Singer and Dr. Theodore Nash (47). The wrky gene was amplified with oligonucleotides WRKYBF (GGGGCGTGATCAATGAAAGAAGGATCCCTGCAT) and WRKYMR (GGCGACGCGTAGAGTAAACGTTAACCATCGG), digested with BclI/MluI, and ligated in place of the BamHI/MluI-excised e2f1 gene in pPE2F1.3 The resulting plasmid, pPTWRKY, contained the wrky gene controlled by the α2-tubulin promoter with an HA tag fused at its C terminus. For constructing pPTWRKYm, a PCR with oligonucleotide WRKYBF and WRKYmR (CTCTGTGGTGGTtaatacGATGCTACACTCGACtactacCGTTGTAGCtaatacCAGGGAtacTGGCTCCGAAAC; mutated nucleotides are in lowercase) generated a 0.9-kb product. Another PCR with primers WRKYmF (GTTTCGGAGCCAgtaTCCCTGgtattaGCTACAACGgtagtaGTCGAGTGTAGCATCgtattaACCACCACAGAG; mutated region is shown in lowercase) and WRKYMR generated a 1.7-kb PCR product. A second run of PCR with the above two products and primers WRKYBF and WRKYMR generated a 2.7-kb PCR product that was digested with BclI and MluI and ligated in place of the BamHI/MluI-excised e2f1 gene in pPE2F1.3 The resulting plasmid, pPTWRKYm, contains a wrky gene with a mutation of the coding region of a stretch of basic amino acids between residues 315 and 331, which is located N-terminal to the WRKY domain. An NheI/ClaI fragment containing the luciferase gene, the 32-bp ran promoter, and two copies of a 19-bp tet operator sequence from pPop2N (48) was replaced by the NheI/ClaI-excised luciferase gene and cwp1 promoter from pPW1 (24), resulting in pPW1N. The 324-bp 5′-flanking region of the genomic wrky gene was amplified with oligonucleotides WRKY5NF (GGCGGCTAGCCTCGTAGCCCATCATGAGCGT) and WRKY5NR (GGCGCCATGGACTTTTTAACTAATAGATTGG), digested with NheI/NcoI, and ligated in place of the NheI/NcoI-excised cwp1 promoter sequence in pPW1N. The resulting plasmid, pPWRKY5, contained the luciferase gene under the control of the wrky promoter. The 324-bp 5′-flanking region of the genomic wrky gene was amplified with oligonucleotides WRKY5NF and WRKY5mNR (GGCGCCATGGACTTTTTAACTAATAGATTGGAGCGAAATTCGGGTACAGTCCTCTGAGTGCGTCTCTTGGAAAATCAGcgcagtgGGGTA; mutated nucleotides are in lowercase), digested with NheI/NcoI, and ligated in place of the NheI/NcoI-excised cwp1 promoter sequence in pPW1N. The resulting plasmid, pPWRKY5m, contained the luciferase gene under the control of the wrky promoter with a mutation on the WRKY binding site. The 393-bp 5′-flanking region of the genomic cwp2 gene was amplified with oligonucleotides CWP25NF (GGCGGCTAGCTTTGGCCGCCAATTCACACAG) and CWP25NR (GGCGCCATGGTTTATTTTCCCAGCCACTGTT), digested with NheI/NcoI, and ligated in place of the NheI/NcoI-excised cwp1 promoter sequence in pPW1N. The resulting plasmid, pPCWP25, contained the luciferase gene under the control of the cwp2 promoter. The 393-bp 5′-flanking region of the genomic cwp2 gene was amplified with oligonucleotides CWP25NF and CWP25mNR (GGCGCCATGGTTTATTTTCCCAGCCACTGTTGAGCTGCTGTTATCTcgcagtgTCTACAGCATCAGTCTA; mutated nucleotides are in lowercase), digested with NheI/NcoI, and ligated in place of the NheI/NcoI-excised cwp1 promoter sequence in pPW1N. The resulting plasmid, pPCWP25m, contained the luciferase gene under the control of the cwp2 promoter with a mutation on the WRKY binding site. The erk1 gene and its 300-bp 5′-flanking region was amplified from genomic DNA with oligonucleotides ERK1NF (GGCGGCTAGCTCTCGGGTGCCGTTAGAATAC) and ERK1MR (GGCGACGCGTCATCCACACAGACTCCGGGAT), digested with NheI/MluI, and ligated in place of the NheI/MluI-excised luciferase gene in pPop2NHA (48). The resulting plasmid, pPERK1, contained the erk1 gene controlled by its own promoter with an HA tag fused at its C terminus. For constructing pPERK1m, a PCR with oligonucleotide ERK1NF and ERK1mR (CCACCCCAGTATGATCTCCAAGGCCTTTGTCACCTT) generated a 0.3-kb product. Another PCR with primers ERK1mF (AAGGTGACAAAGGCCTTGGAGATCATACTGGGGTGG) and ERK1MR generated a 0.5-kb PCR product. A second run of PCR with the above two products and primers ERK1NF and ERK1MR generated a 0.8-kb PCR product that was digested with NheI and MluI and ligated in place of the NheI/MluI-excised luciferase gene in pPop2NHA. The resulting plasmid, pPERK1m, contains an erk1 gene lacking the coding sequence for the predicted kinase domain (nucleotides 76–606, residues 26–202).
Transfection, Luciferase Assay, and Western Blot Analysis
Cells transfected with the pP series plasmid containing the pac gene were selected and maintained with 54 μg/ml puromycin. The luciferase activity was determined as described (9). After stable transfection with specific constructs, luciferase activity was determined in vegetative cells at late log/stationary phase (1.5 × 106 cells/ml) or in 24-h encysting cells, as described (9), and was measured with an Optocomp I luminometer (MGM Instruments). Two independently generated stably transfected lines were made from each construct, and each of these lines was assayed three separate times. Western blots were probed with anti-V5-horseradish peroxidase (Invitrogen) or anti-HA monoclonal antibody (Covance, Princeton, NJ; 1:5000 in blocking buffer) or anti-WRKY (see below; 1:5000 in blocking buffer) and detected with peroxidase-conjugated goat anti-mouse IgG (Pierce; 1:5000) or peroxidase-conjugated goat anti-rabbit IgG (Pierce; 1:5000) and enhanced chemiluminescence (GE Healthcare).
Expression and Purification of Recombinant WRKY Protein
The genomic wrky gene was amplified using oligonucleotides WRKYF and WRKYR. The product was cloned into the expression vector pCRT7/CT-TOPO (Invitrogen) in frame with the C-terminal His and V5 tag to generate plasmid pWRKY. For constructing the pWRKYΔC, The genomic wrky gene was amplified using oligonucleotides WRKYF and WRKYdR (GGATTCTATTTGATCGTAGCT). The product was cloned into the expression vector pCRT7/CT-TOPO (Invitrogen) in frame with the C-terminal His and V5 tag. The resulting plasmid, pWRKYΔC, encodes a wrky gene with a C-terminal (residues 586–915) deletion. The pWRKY or pWRKYΔC plasmid was freshly transformed into Escherichia coli BL21 (DE3) pLysE (QIAexpressionist; Qiagen). An overnight preculture was used to start a 250-ml culture. E. coli cells were growth to an A600 of 0.5 and then induced with 1 mm isopropyl-d-thiogalactopyranoside (Promega) for 4 h. Bacteria were harvested by centrifugation and sonicated in 10 ml of buffer A (50 mm sodium phosphate, pH 8.0, 300 mm NaCl) containing 10 mm imidazole and protease inhibitor mixture (Sigma). The samples were centrifuged, and the supernatant was mixed with 1 ml of a 50% slurry of nickel-nitrilotriacetic acid Superflow (Qiagen). The resin was washed with buffer A containing 20 mm imidazole and eluted with buffer A containing 250 mm imidazole. Fractions containing WRKY or WRKYΔC were pooled, dialyzed in 25 mm HEPES, pH 7.9, 20 mm KCl, and 15% glycerol, and stored at −70 °C. Protein purity and concentration were estimated by Coomassie Blue and silver staining compared with serum albumin. WRKY was purified to apparent homogeneity (>95%).
Generation of Anti-WRKY Antibody
Purified WRKY protein was used to generate rabbit polyclonal antibodies through a commercial vendor (Angene, Taipei, Taiwan).
Immunofluorescence Assay
The pPTWRKY or pPTWRKYm stable transfectants were cultured in growth medium under puromycin selection. Cells cultured in growth medium or encystation medium for 24 h were harvested, washed in phosphate-buffered saline, and attached to glass coverslips (2 × 106 cells/coverslip) and then fixed and stained (9). Cells were reacted with anti-HA monoclonal antibody (1:300 in blocking buffer; Molecular Probes) and anti-mouse ALEXA 488 (1:500 in blocking buffer; Molecular Probes) as the detector. The ProLong antifade kit with 4′,6-diamidino-2-phenylindole (Invitrogen) was used for mounting. WRKY was visualized using a Leica TCS SP2 spectral confocal system.
Electrophoretic Mobility Shift Assay
Double-stranded oligonucleotides specified throughout were 5′-end-labeled as described (18). Binding reaction mixtures contained the components described (25). Labeled probe (0.02 pmol) was incubated for 15 min at room temperature with 5 ng of purified WRKY protein in a 20-μl volume supplemented with 0.5 μg of poly(dI-dC) (Sigma). Competition reactions contained a 200-fold molar excess of cold oligonucleotides. In an antibody supershift assay, 0.8 μg of an anti-V5-horseradish peroxidase antibody (Bethyl Laboratories) was added to the binding reaction mixture. The mixture was separated on a 6% acrylamide gel by electrophoresis.
ChIP Assays
The WB clone C6 cells were inoculated into encystation medium (5 × 107 cells in 45 ml of medium) and harvested after 24 h in encystation medium under drug selection and washed in phosphate-buffered saline. ChIP was performed as described previously (26) with some modifications. Formaldehyde was then added to the cells in phosphate-buffered saline at a final concentration of 1%. Cells were incubated at room temperature for 15 min, and reactions were stopped by incubation in 125 mm glycine for 5 min. After phosphate-buffered saline washes, cells were lysed in luciferase lysis buffer (Promega) and protease inhibitor (Sigma) and then vortexed with glass beads. The cell lysate was sonicated on ice and then centrifuged. Chromatin extract was incubated with protein G plus/protein A-agarose (Merck) for 1 h. After removal of protein G plus/protein A-agarose, the precleared lysates were incubated with 2 μg of anti-WRKY antibody or preimmune serum for 2 h and then incubated with protein G plus/protein A-agarose (Merck) for 1 h. The beads were washed with low salt buffer (0.1% SDS, 1% Triton X-100, 2 mm EDTA, 20 mm Tris-HCl, pH 8.0, 150 mm NaCl) twice, high salt buffer (0.1% SDS, 1% Triton X-100, 2 mm EDTA, 20 mm Tris-HCl pH 8.0, 500 mm NaCl) once, LiCl buffer (0.25 m LiCl, 1% Nonidet P-40, 1% sodium deoxycholate, 1 mm EDTA, 10 mm Tris-HCl, pH 8.0) once, and TE buffer (20 mm Tris-HCl, 1 mm EDTA, pH 8.0) twice. The beads were resuspended in elution buffer containing 50 mm Tris-HCl, pH 8.0, 1% SDS, and 10 mm EDTA at 65 °C for 4 h. To prepare DNA representing input DNA, 2.5% of precleared chromatin extract without incubation with anti-WRKY was combined with elution buffer. Eluted DNA was purified by the QIAquick PCR purification kit (Qiagen). Purified DNA was subjected to PCR followed by agarose gel electrophoresis. Primers 18SF (CCAAAAAAGTGTGGTGCAGG) and 18SR (GCCGGGCGCGGGCGCCGCGG) were used to amplify the 18 S ribosomal RNA gene promoter as a control for our ChIP analysis. Primers wrky5F (GACGTTTTTGAACCCAAGTATGTATTTA) and wrky5R (TGCGTCTCTTGGAAAATCAGTCT), cwp15F (CAACGGCTTACTAAATCATTCTCTTG) and cwp15R (TTCTGTGTTTCTTGATCTGAGAGTTGT), cwp25F (GAGCATGGGTTCTGACAAATAGTG) and cwp25R (CATCAGTCTACTGTTTCTTTTTAGTTCATATCT), and myb25F (TGCACTGTAGCGTTTCCATTTG) and myb25R (ACTTACCCGTAATGGCGTTGAC) were used to amplify wrky, cwp1, cwp2, and myb2 gene promoters within the −148 to −49 region and the −133 to −51 region, respectively.
Microarray Analysis
RNA purified was quantified by A260 nm by an ND-1000 spectrophotometer (Nanodrop Technology) and analyzed by a Bioanalyzer 2100 (Agilent Technologies) with an RNA 6000 Nano LabChip kit. RNA from the pPTWRKY cell line was labeled by Cy5, and RNA from the 5′Δ5N-Pac cell line was labeled by Cy3. 0.5 μg of total RNA was amplified by a Low RNA Input Fluor Linear Amp kit (Agilent Technologies) and labeled with Cy3 or Cy5 (CyDye; PerkinElmer Life Sciences) during the in vitro transcription process. 0.825 μg of Cy-labeled cRNA was fragmented to an average size of about 50–100 nucleotides by incubation with fragmentation buffer at 60 °C for 30 min. Correspondingly fragmented labeled cRNA was then pooled and hybridized to a G. lamblia oligonucleotide microarray (Agilent Technologies) at 60 °C for 17 h. After washing and drying by nitrogen gun blowing, microarrays were scanned with an Agilent microarray scanner (Agilent Technologies) at 535 nm for Cy3 and 625 nm for Cy5. Scanned images were analyzed by Feature Extraction version 9.1 software (Agilent Technologies), and image analysis and normalization software was used to quantify signal and background intensity for each feature; data were substantially normalized by the rank consistency filtering LOWESS method.
RESULTS
Identification and Expression of the wrky Gene
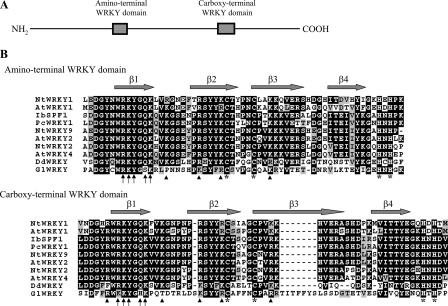
To identify genes encoding novel WRKY proteins from G. lamblia, we performed BLAST searches (46) against the G. lamblia genome data base (available on the World Wide Web) (14, 45) using the amino acid sequences of the WRKY domain of Arabidopsis WRKY2 (GenBankTM accession number NM_125010) as a query sequence. This search detected one putative homologue for WRKY that has been reported recently (GenBankTM accession number XM_001708755; open reading frame 9237 in the G. lamblia genome data base) (33). Comparison of genomic and cDNA sequences showed that the wrky gene contained no introns. The deduced giardial WRKY protein contains 915 amino acids with a predicted molecular mass of ∼98.88 kDa and a pI of ∼7.53. It has two WRKY-like domains, as predicted by Pfam (available on the World Wide Web) (49). One is near the N terminus (residues 343–400), and the other one is near the C terminus (residues 585–653) (Fig. 1A).
FIGURE 1.
Domain architecture of WRKY protein and alignment of the WRKY domains. A, schematic representation of the giardial WRKY protein. The gray boxes indicate the WRKY domains. B, alignment of the WRKY domains. The WRKY domains from members of the plant WRKY family, including WRKYs from A. thaliana (At), D. discoideum (Dd), G. lamblia (Gl), Ipomoea batatas (Ib), Nicotiana tabacum (Nt), and Petroselinum crispum (Pc), are analyzed by ClustalW 1.83 (59). GenBankTM accession numbers for NtWRKY1, AtWRKY1, IbSPF1, PcWRKY1, NtWRKY9, AtWRKY2, NtWRKY2, AtWRKY4, DdWRKY, and GlWRKY are AF096298, AF442390, D30038, U48831, AB063576, NM_125010, AF096299, NM_101262, XM_638694, and XM_001708755, respectively. Black boxes, gray boxes, and hyphens indicate identical amino acids, conserved amino acids, and gaps in the respective proteins, respectively. The gray arrows indicate the β1–β4 strands in the C-terminal WRKY domain of the Arabidopsis WRKY4 protein (50). The arrows indicate the key residues contacting the bases in the WRKYGQK region. The arrowheads indicate basic residues that make contact with the backbone. The cysteines and histidines of zinc finger-like motifs in G. lamblia WRKY are all positionally conserved with those in plant WRKYs (asterisks).
The structure of the WRKY domain includes four β strands (β1–β4) (Fig. 1B) (50). Structural studies of the C-terminal WRKY domain of Arabidopsis WRKY4 show that the four β strands form an antiparallel β sheet and that the β1 strand containing the conserved WRKYGQK sequence contacts the major groove (50). A C-terminal zinc-binding pocket is formed by the conserved cysteine and histidine residues at one end of the β sheet (50). The Arg, Lys, Tyr, Gln, and the second Lys of WRKYGQK are the key residues contacting the bases (Fig. 1B, arrows) (50). The Trp of WRKYGQK is important in forming the structural core of the WRKY domain by making hydrophobic contact with one of the histidines of the zinc-binding motif and stabilizing the zinc coordination (50). The presence of Gly in WRKYGQK allows the β1 strand to be kinked and able to enter deeply into the major groove (50). Five other basic residues in the β1–β3 strands contact the backbone, but only four are present in the N-terminal WRKY domain (Fig. 1B, arrowheads) (50). As aligned in Fig. 1B, the sequences of the two WRKY-like domains in the giardial WRKY are highly similar to those found in the original group I WRKY family in plants. The similarity between the giardial WRKY and group I WRKY family is limited to their WRKY domains (data not shown). The conserved WRKYGQK sequence in plant WRKY domains is changed to WRKYGSK and WKKYGHK in the two giardial WRKY domains (Fig. 1B). Only four of the five basic residues that make contact with backbone are present in the C-terminal WRKY domain (Fig. 1B, arrowheads). A 7-residue insertion is present in the middle of the zinc-binding motif of the C-terminal WRKY domain (Fig. 1B). Giardial genes frequently have unique amino acid inserts (23, 51, 52). In addition, all of the cysteines and histidines of the zinc-binding motifs in giardial WRKY domains are positionally conserved with those in plant WRKY domains (Fig. 1B).
Encystation-induced Expression of the wrky Gene
RT-PCR and quantitative real time PCR analysis of total RNA showed that the wrky transcript was present in vegetative cells and increased significantly in 24-h encysting cells (Fig. 2A). As controls, we found that the mRNA levels of the cwp1 and ran genes increased and decreased significantly during encystation, respectively (Fig. 2A). The products of the cwp1 and ran genes are the component of the cyst wall and the ras-related nuclear protein (6, 18). To determine the expression of WRKY protein, we generated an antibody specific to the full-length WRKY. Western blot analysis confirmed that this antibody recognized WRKY at a size of ∼100 kDa (Fig. 2B). WRKY was expressed in vegetative cells, and its levels increased significantly during encystation (Fig. 2B).
Localization of the WRKY Protein
To determine the expression of WRKY protein, we prepared construct pPTWRKY, in which the wrky gene is controlled by the α2-tubulin promoter with an HA tag fused at its C terminus (Fig. 2C) and stably transfected it into G. lamblia. The HA-tagged WRKY was detected in both nuclei and cytosol during vegetative growth and exclusively to the two nuclei during encystation (Fig. 2D and Fig. S1A for nuclei stained with 4′,6-diamidino-2-phenylindole), indicating that WRKY may play an important role during encystation. We further identified the portion of WRKY that is sufficient to direct the protein to the nuclei. No typical nuclear localization signal was predicted in WRKY using the PSORT software (available on the World Wide Web) (53). Mutation of a stretch of basic amino acids between residues 315 and 331, which is located N-terminal to the WRKY domain (pPTWRKYm) (Fig. 2C), resulted in loss of nuclear localization in both vegetative and encysting cells (Fig. 2E), suggesting that these basic residues may play some role in the exclusively nuclear localization.
Identification of the WRKY Binding Sites
The nuclear localization of WRKY suggested that it might also function as a transcription factor in G. lamblia. To test its DNA binding activity, we expressed WRKY with a C-terminal V5 tag in E. coli and purified it to >95% homogeneity, as assessed in a silver-stained gel (Fig. S1B). An anti-V5-horseradish peroxidase antibody specifically recognized the recombinant V5-tagged WRKY in Western blots (Fig. 3A).
FIGURE 3.
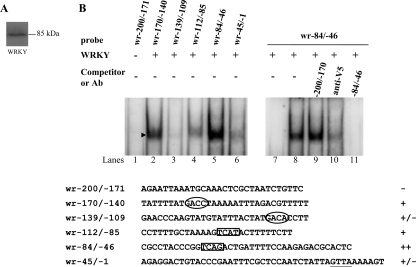
DNA-binding ability of WRKY revealed by electrophoretic mobility shift assays. A, Western blot analysis of recombinant WRKY protein with a V5 tag at its C terminus purified by affinity chromatography. The WRKY protein is detected by anti-V5 antibody. B, detection of WRKY binding sites. Electrophoretic mobility shift assays were performed using purified WRKY and various 32P-end-lableled oligonucleotide probes from the wrky 5′-flanking region as described. Components in the binding reaction mixtures are indicated above the lanes. The arrowhead indicates the shifted complex. The putative W boxes with a GTCA or TGAC core sequence are framed or circled, respectively. The WRKY binding specificity for wr−84/−46 probe was confirmed by competition and supershift assays. Some reaction mixtures contained a 200-fold molar excess of cold oligonucleotides wr−84/−46 or wr−200/−170 or 0.8 μg of anti-V5-horseradish peroxidase antibody, as indicated above the lanes. ++, +, +/−, and − represent strong binding, moderate binding, weak binding, and no binding, respectively.
In plants, WRKY factors have a high binding affinity to a DNA sequence designated the W box, (c/t)TGAC(c/t), which mediates rapid response to elicitor and is present in their own promoters (27). To determine whether purified WRKY binds DNA, we performed electrophoretic mobility shift assays with double-stranded DNA sequences from its own promoter. We searched the giardial wrky promoter region and found four putative WRKY binding sites with a TGAC core sequence or with an antisense sequence of the core sequence, GTCA. They are aTGACc, aTGACa, aGTCAt, and gGTCAg, in the probes wr−170/−140 (−170 to −140 relative to the translation start site of the wrky gene), −139/−109, −112/−85, and −84/−46, respectively (Fig. 3B). Incubation of a labeled double-stranded DNA probe wr−84/−46 with WRKY resulted in the formation of a shifted band (Fig. 3B, lane 5). WRKY did not bind to either single strand of the wr−84/−46 probe (data not shown). WRKY did not bind to the wr−200/−171 probe, which contains no putative WRKY binding site (Fig. 3B, lane 1). We also found that WRKY bound moderately to the wr−170/−140 and wr−112/−85 probe (Fig. 3B, lanes 2 and 4), but it bound very weakly to the wr−139/−109 probe (Fig. 3B, lane 3). Therefore, the sequences aTGACc, aGTCAt, and gGTCAg, but not aTGACa (antisense of tGTCAt), are needed for binding of WRKY.
The binding specificity was confirmed by competition and supershift assays (Fig. 3B, lanes 7–11). The formation of the shifted wr−84/−46 band was almost totally competed by a 200-fold molar excess of unlabeled wr−84/−46, but not by the same excess of a nonspecific competitor, wr−200/−171 (Fig. 3B, lanes 9 and 11). The intensity of the shifted wr−84/−46 band was decreased significantly by the addition of anti-V5-horseradish peroxidase antibody (Fig. 3B, lane 10).
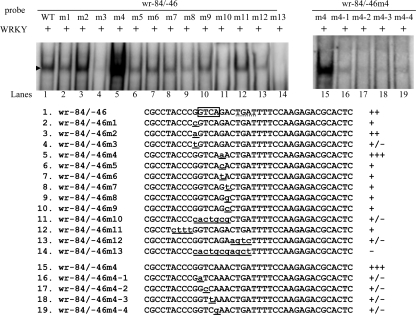
We further determined whether WRKY specifically recognized the gGTCAg sequence of the wr−84/−46 probe. We found that mutation of either side of the sequence flanking the GTCA core sequence from G to A significantly increased the binding of WRKY (wr−84/−46m2 and m4) (Fig. 4, lanes 3 and 5). Mutation of the first G to C or mutation of the sixth G to C or T slightly decreased binding (wr−84/−46m1, m5, and m6) (Fig. 4, lanes 2, 6, and 7). However, mutation of the first G to T (the wr−84/−46m3 probe) significantly decreased binding (Fig. 4, lane 4). Therefore, the sequence (a/g)GTCA(a/t/g/c) or cGTCA(a/g) is needed for binding of WRKY. In addition, both sides of the sequences flanking the core sequence are G/A, better than T/C for binding, suggesting that (a/g)GTCA(a/g), the antisense sequence of plant WRKY binding sites, (c/t)TGAC(c/t), is the best binding sequence for WRKY (27). Mutation of the seventh A to T, G, or C (the wr−84/−46m7–9 probes) decreased binding slightly (Fig. 4, lanes 8–10), suggesting that this nucleotide is not important for binding. Scanning mutagenesis of the 4-bp GTCA core sequence in the wr−84/−46m4 probe showed that any single substitution within the GTCA sequence decreased the binding significantly but did not completely eliminate the binding (Fig. 4, lanes 15–19). We also found that WRKY did not bind to the poly(A) probe (AAAAAAAAAAAAAAAAAAAAAAA), poly(A) + 1T probe (AAAAAAAAAAATAAAAAAAAAAA), or poly(A) + TC probe (AAAAAAAAAAATCAAAAAAAAAA) (data not shown), suggesting that the WRKY binding site is different from the ARID1 binding site (25).
FIGURE 4.
Mutation analysis of the wr−84/−46 probe sequence containing the putative WRKY binding site. The sequences flanking the GTCA core sequence in oligonucleotide wr−84/−46 were mutated as described (left). The sequences of the GTCA core sequence in oligonucleotide wr−84/−46m4 were mutated as described (right). Base changes in the mutants are shown in underlined lowercase letters. Components in the binding reaction mixtures are indicated above the lanes. The arrowhead indicates the shifted complex. +, +/−, and − represent moderate binding, weak binding, and no binding, respectively. ++ and +++ represent strong binding.
We searched the 200-bp 5′-flanking regions of 38 giardial genes and found that 23 genes contain (a/g)GTCA(a/t/g/c) or cGTCA(a/g) or their respective antisense sequence, including encystation-induced cwp2 and myb2 genes (Table 1). The products of the cwp2 and myb2 genes are the component of the cyst wall and the transcriptional activator of cwp1, cwp2, and cwp3 genes, respectively (6, 26). Interestingly, the cwp2, phosphoenolpyruvate carboxykinase, and wrky genes have three copies of WRKY binding sites. This suggests that WRKY may participate in transcriptional regulation of the encystation-induced cwp2 gene and many other giardial genes.
TABLE 1.
Presence of potential WRKY binding sequences in 23 giardial gene promoters
| Genesa | Coding strands | ||
|---|---|---|---|
| cwp2 (U28965) | GGTCAA−38b | GTGACT−107 | AGTCAC−178 |
| Phosphoenolpyruvate carboxykinase (AF308935) | GGTCAA−45 | GGTCAA−167 | CTGACT−188 |
| wrky (NM_125010) | GGTCAG−70 | AGTCAT−95 | ATGACC−159 |
| α-Giardin (X52485) | GGTCAA−49 | TTGACC−141 | |
| Cysteine protease (U83277) | GGTCAA−47 | ATGACC−179 | |
| Cysteine sulfurtransferase (AF311744) | AGTCAC−35 | AGTCAG−138 | |
| Encystation-specific cysteine protease (AF293408) | AGTCAT−157 | CGTCAG−161 | |
| glp3 (EAA38047) | TTGACG−121 | GTGACT−181 | |
| Glutamate dehydrogenase (M84604) | CTGACC−41 | GGTCAA−135 | |
| myb2 (AY082882) | CGTCAA−73 | AGTCAA−137 | |
| Triose-phosphate isomerase (L02120) | ATGACC−174 | CTGACT−180 | |
| Adenylate kinase (U19901) | ATGACC−109 | ||
| α2-Giardin (M34550) | TTGACG−120 | ||
| α1-Tubulin (J04648) | CGTCAA−95 | ||
| Fibrillarin (L28115) | ATGACC−126 | ||
| g6pi-a (AF071897) | CGTCAA−58 | ||
| g6pi-b (AF071896) | AGTCAG−184 | ||
| glp1 (EAA38934) | TTGACT−28 | ||
| glp4 (EAA39659) | GTGACC−136 | ||
| Guanine phosphoribosyltransferase (U43180) | AGTCAG−91 | ||
| myb1 (AF257483) | CTGACG−61 | ||
| Protein-disulfide isomerase-1 (U64730) | CGTCAA−23 | ||
| Pyruvate:flavodoxin oxidoreductase (L27221) | GGTCAA−29 |
a The other 15 genes searched for those sequences but not listed were as follows: 18 S ribosomal RNA (M54878), actin (L29032), α2-tubulin (AF331826), ATPase (U18938), axoneme-associated protein GASP-180 (AF400249), calmodulin (AF359239), caltractin (U42428), chaperonin 60 (AF029695), cwp1 (U09330), cwp3 (AY061927), cytidine triphosphate synthetase (U30328), glp2 (EAA42633), glyceraldehyde 3-phosphate dehydrogenase (M88062), ran (U02589), and signal receptor α subunit (AF072125).
b Numbers indicate positions of the last nucleotide relative to the translation start site of the gene.
We also found that WRKY can bind to the wr−45/−1 probe, which does not contain the TGAC/GTCA core sequence but contains the aGTTAa (which is similar to aGTCAa) sequence (Fig. 3B, lane 6). Mutation of the aGTTAa sequence to gACCGg resulted in no binding of WRKY (data not shown), suggesting that WRKY can bind to aGTTAa, although weakly. Mutation of the putative binding sequence from gGTCAag to cactgca (wr−84/−46m10 probe) did not completely eliminate the binding (Fig. 4, lane 11), suggesting the presence of another binding region in the wr−84/−46 probe. Further mutation of the 11 nucleotides, gGTCAagctga, to cactgcgagct can completely eliminate the binding, suggesting that WRKY can bind, although weakly, to another region in the wr−84/−46 probe, cTGATt (antisense of aATCAg, aATCAg, is similar to aGTCAg).
Studies suggest that WRKY proteins can bind to the DNA major groove using the first β strand containing the WRKYGQK residues to enter the DNA major groove and to contact the DNA bases (50). To investigate how WRKY binds to DNA, we used distamycin A, a minor groove binder, as a competitive inhibitor of WRKY binding (54). The binding of WRKY to DNA did not change with increasing concentrations of distamycin A (data not shown), suggesting that WRKY binds to the major groove but not the minor groove of DNA sequences.
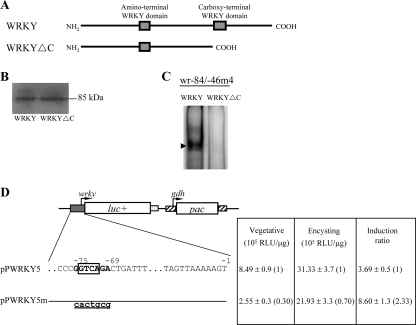
The C-terminal WRKY domains of the plant group I WRKY proteins are known to be important for DNA binding (34). To understand whether the C-terminal WRKY domain of giardial WRKY (residues 585–653) is also important for DNA binding (Fig. 5A), the C-terminal region (residues 585–915) was deleted, and the resulting WRKYΔC was expressed in E. coli and purified. We found that the purified WRKYΔC did not bind to wr−84/−46m4 (Fig. 5C), indicating that the C-terminal WRKY domain is important for DNA binding. Similar levels of wild-type WRKY and WRKYΔC were added to the binding reaction mixture (Fig. 5B).
FIGURE 5.
Analysis of the DNA-binding domain of WRKY and its binding site. A, diagrams of the WRKY and WRKYΔC proteins. Gray boxes indicate the WRKY domains. WRKYΔC does not contain the C-terminal WRKY-like domain and C-terminal region (residues 585–915). B, Western blot analysis of recombinant WRKY and WRKYΔC proteins. The WRKY or WRKYΔC protein with a V5 tag at its C terminus was purified by affinity chromatography and then detected by anti-V5-horseradish peroxidase antibody in Western blots. Note that the predicted molecular mass of WRKY and WRKYΔC is ∼99 and 65 kDa, respectively. However, both WRKY and WRKYΔC run at ∼100 kDa. C, loss of DNA binding ability of WRKYΔC. Electrophoretic mobility shift assays were performed using purified WRKY and WRKYΔC and wr−84/−46 probe. The arrowhead indicates the shifted complex. D, mutation analysis of the WRKY binding site in the wrky promoter region. In the pPWRKY5 construct, a firefly luciferase gene (luc+; open box) is flanked by the 5′-flanking region of the wrky gene and 3′-flanking region of the ran gene (dotted boxes). The pac gene (open box) expression cassette is the same as in Fig. 2C. Numbers of the 5′-flanking region of the wrky gene are relative to the translation start site (+1). The WRKY binding sequences are in boldface type. The GTCA core sequence is framed. The mutated sequence in the construct pPWRKY5m is shown in underlined lowercase letters. After stable transfection with these constructs, luciferase activity was measured in vegetative cells and 24-h encysting cells as described under “Experimental Procedures.” Values are shown as means ± S.E. in the right panel. The induction ratio was obtained by dividing the activity in the encysting cells by the activity in the vegetative cells of each construct.
The WRKY Binding Sequences Are Sufficient for Transcriptional Activation
We further investigated the ability of the WRKY binding site to regulate its own promoter functions by mutation analysis. The 5′-flanking region −324/−1 of the wrky gene was sufficient for up-regulation of the luciferase reporter gene during encystation (construct pPWRKY5, induction ratio ∼3.69) (Fig. 5D). Mutation from −75 to −69 of the wrky promoter, which spans a WRKY binding site (in the region of the wr−84/−46 probe) resulted in significant decreases of luciferase activity to ∼30 and ∼70% of the wild-type value in vegetative and encysting cells, respectively, and a ∼2.3-fold increase in the induction ratio to ∼8.6 (Fig. 5D). These results indicate that one of multiple WRKY binding sites (−75 to −69 region) in the wrky promoter functions as a positive cis-acting element in both vegetative and encysting stages.
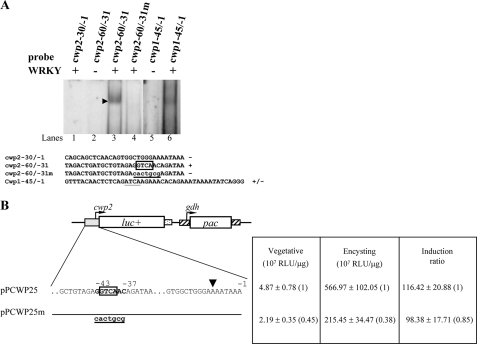
Because the protein level and nuclear localization of WRKY increased during encystation, we tested the hypothesis that it may regulate the transcription of genes required for encystation. Interestingly, we found that the cwp2 gene promoter has a gGTCAa sequence in the −43 to −38 region (Table 1). Incubation of a labeled double-stranded DNA probe cwp2−60/−31 with WRKY resulted in the formation of a shifted band (Fig. 6A, lane 3). Mutation of the sequence aGTCAac to cactgcg can completely eliminate the binding of WRKY (Fig. 6A, lane 4). WRKY did not bind to the cwp2−30/−1 probe, which contains no putative WRKY binding site (Fig. 6A, lane 1). We also found that WRKY can bind weakly to the cwp1−45/−1 probe, which does not contain the TGAC/GTCA core sequence but contains the gATCAa (which is similar to gGTCAa) sequence (Fig. 6A, lane 6), suggesting that WRKY can bind to the cwp1 promoter, although weakly.
FIGURE 6.
Binding of WRKY to the cwp1 and cwp2 promoters. A, WRKY binding site was required for the binding. Electrophoretic mobility shift assays were performed using purified WRKY and a 32P-end-lableled oligonucleotide probe from the 5′-flanking region of the cwp1 and cwp2 genes as described. The arrowhead indicates the shifted complex. The putative GTCA or ATCA core sequence in W box is framed and underlined, respectively. The mutated sequence is shown in underlined lowercase letters. + and − represent DNA binding and no binding, respectively. B, mutation analysis of the WRKY binding site in the cwp2 promoter region. Data are presented as in Fig. 5D, except that the luciferase gene is flanked by the 5′-flanking region of the cwp2 gene. The transcription start site is indicated by an arrowhead (6).
We further investigated the ability of the WRKY binding sites to regulate the cwp2 promoter function by mutation analysis. The 5′-flanking region −393/−1 of the cwp2 gene was sufficient for up-regulation of the luciferase reporter gene during encystation (construct pPCWP25, induction ratio ∼116) (Fig. 6B). Mutation from −43 to −37 of the cwp2 promoter, which spans a WRKY binding site (in the region of the cwp2−60/−31 probe), resulted in significant decreases of luciferase activity to ∼45 and ∼38% of the wild-type value in vegetative and encysting cells, respectively, and a slight decrease in the induction ratio to ∼98 (Fig. 6B). These results indicate that one of multiple WRKY binding sites (−43 to −37 region) in the cwp2 promoter functions as a positive cis-acting element in both vegetative and encysting stages.
Recruitment of WRKY to the wrky, cwp1, cwp2, and myb2 Promoters
We further used ChIP assays to study the association of WRKY with specific promoters in the WRKY-overexpressing cell line. We found that WRKY was associated with its own promoter and the cwp1, cwp2, and myb2 promoters during encystation (Fig. 7A) or during vegetative growth (data not shown). However, WRKY was not associated with the 18 S ribosomal RNA gene promoter, which has no WRKY binding site (Fig. 7A).
Overexpression of WRKY Induces the Expression of cwp1, cwp2, and myb2 Genes
To study the role of WRKY in G. lamblia, we expressed wrky constitutively by the α2-tubulin gene promoter (pPTWRKY; Fig. 2C) and observed its gene expression. A ∼100-kDa protein was detected (Fig. 7B), which is matched to the predicted molecular mass of WRKY (∼99 kDa) with the HA tag (∼1 kDa). Similar to the expression pattern of the endogenous WRKY protein, the levels of the WRKY-HA protein increased significantly during encystation (data not shown; also see Fig. 2B). We found that WRKY overexpression resulted in a significant increase of the CWP1 protein levels during encystation (Fig. 7B). RT-PCR and quantitative real time PCR analysis showed that the mRNA levels of the endogenous wrky plus vector-expressed wrky in the WRKY-overexpressing cell line increased by ∼2-fold (p < 0.05) (Fig. 7, C and D) relative to the control cell line, which expressed only the puromycin selection marker (5′Δ5N-Pac) (Fig. 2C, right part of pPTWRKY) (47). The mRNA levels of the endogenous cwp1, cwp2, and myb2 genes in the WRKY-overexpressing cell line increased by ∼1.3–2.6-fold (p < 0.05) relative to the 5′Δ5N-Pac control cell line (Fig. 7D). The results suggest that the overexpressed WRKY can transactivate the cwp1, cwp2, and myb2 genes.
To further understand the function of giardial WRKY, we observed the effect of overexpression of a WRKY mutant (WRKYm) that cannot enter nuclei (Fig. 2, C and E). We found that the levels of WRKYm protein increased significantly compared with that of wild type WRKY during encystation (Fig. 7B). We further analyzed whether the transcript levels of the WRKYm were changed. As shown by RT-PCR and quantitative real time PCR analysis, the levels of HA-tagged wrkym mRNA increased by ∼2.5-fold (p < 0.05) compared with that of wild type HA-tagged wrky during encystation (Fig. 7, C and D). This suggests a negative autoregulation of the wrky gene. We did not detect any HA-tagged wrky transcripts in the 5′Δ5N-Pac control cell line (Fig. 7C). As a control, similar levels of intensity of the giardial RAN protein (∼30 kDa) were detected by anti-human RAN antibody (Fig. 7B), and similar mRNA levels of the ran and 18 S ribosomal RNA genes were detected (Fig. 7, C and D). In addition, the levels of the CWP1 protein and of the cwp1, cwp2, and myb2 mRNA decreased (for mRNA, by ∼30%) in the WRKYm-overexpressing cell line relative to the wild-type WRKY-overexpressing cell line (Fig. 7, B–D). Similar results were obtained during vegetative growth (Fig. S1, C and D). The results suggest a loss of transactivation activity of WRKYm. Although WRKYm was expressed at higher levels, its inactivity may be due to its inability to enter the nucleus.
Oligonucleotide microarray assays confirmed the up-regulation of the cwp1, cwp2, and myb2 gene expression in the WRKY-overexpressing cell line to ∼1.74- to ∼4.59-fold the levels in the control cell line (Fig. 7E). Two genes listed in Table 1, α1-tubulin and protein sulfide isomerase 1, were also up-regulated by WRKY overexpression (Fig. 7E). As a control, similar mRNA levels of the ran and ribosomal protein L7 (open reading frame 19436) genes were detected (Fig. 7E).
Overexpression of ERK1 Increases the Expression of wrky, cwp1, cwp2, and myb2 Genes
Plant WRKYs have been identified as downstream components of defense- and pathogen-induced MAPK signaling cascades (39, 40). In previous studies, a member of MAPK family, ERK1, has been identified to exhibit a significantly increased kinase activity during early encystation (42). To determine whether giardial WRKY is regulated by the MAPK pathway, we prepared construct pPERK1 in which the erk1 gene is controlled by its own promoter with an HA tag fused at its C terminus (Fig. 8A) and stably transfected it into G. lamblia. The HA-tagged ERK1 was detected in cytosol and flagella during both vegetative growth and encystation (Fig. 8B). A ∼45-kDa protein was detected (Fig. 8C), which is similar to the predicted molecular mass of ERK1 (43.96 kDa) with the HA tag (∼1 kDa). The levels of the ERK1 protein increased significantly during encystation (Fig. 8C). We found that ERK1 overexpression resulted in a significant increase of the WRKY and CWP1 protein levels (Fig. 8D). RT-PCR and quantitative real time PCR analysis showed that the mRNA levels of the endogenous wrky, cwp1, cwp2, and myb2 genes in the ERK1-overexpressing cell line increased by ∼1.5–2.7-fold (p < 0.05) (Fig. 8, E and F) relative to the control cell line, which expressed only the puromycin selection marker (5′Δ5N-Pac) (Fig. 8A, right part of pPERK1) (47).
To further understand whether giardial WRKY is regulated by the MAPK pathway, we constructed a pPERK1m plasmid that encodes a mutant ERK1 (ERK1m) lacking the predicted kinase domain (residues 26–202) (Fig. 8A). We found that deletion of the predicted kinase domain resulted in decreased levels of the WRKY and CWP1 proteins (Fig. 8D). We also found a significant decrease of the wrky, cwp1, cwp2, or myb2 mRNA levels in the ERK1m-overexpressing cell line relative to the wild-type ERK1 overexpressing cell line (Fig. 8, E and F). As a control, similar levels of intensity of the giardial RAN protein (∼30 kDa) were detected by anti-human RAN antibody (Fig. 8D), and similar mRNA levels of the ran and 18 S ribosomal RNA genes were detected (Fig. 8, E and F). The results suggest that WRKY may be a downstream component of a MAPK/ERK1 signaling pathway.
We further asked whether the levels of cyst formation increased with the increase of the CWP1 in the ERK1-overexpressing cell line. We found that the cyst number in the ERK1-overexpressing cell line (pPERK1) increased by ∼1.3-fold (p < 0.05) (Fig. 8G) relative to the control cell line, which expresses only the puromycin selection marker (5′Δ5N-Pac) (Fig. 8A, right part of pPERK1) (47), indicating that the overexpressed ERK1 can increase the cyst formation. We also found that deletion of the predicted kinase domain decreased cyst number by ∼40% (p < 0.05) relative to the wild-type ERK1-overexpressing cell line (Fig. 8G).
DISCUSSION
The WRKY protein family is a group of transcriptional regulators that perform a variety of cellular functions in plants (27). It has not been identified to date in yeast or animal genomes (33). However, it has been recently found in non-plant eukaryotic organism Dictyostelium discoideum and G. lamblia (33). Similarly, putative GARP transcription factor genes have been recently identified only in plants and G. lamblia but not in yeast or animals (24). Although divergent in sequence, giardial WRKY is clearly recognizable as a member of the WRKY family. This suggests that the WRKY family may have evolved before divergence of G. lamblia from the main eukaryotic line of descent but may have been lost from lineages leading to the yeast and animals. To date, giardial WRKY is the first WRKY transcription factor identified in early diverging protozoan parasites. Further genomic analyses will reveal whether WRKY family proteins are shared with other eukaryotic lineages, such as Trichomonas vaginalis, Entamoeba histolytica, Plasmodium falciparum, and Trypanosoma brucei.
G. lamblia colonizes the intestinal tract, and it must encyst to survive outside and infect new hosts (1). The giardial cyst wall proteins are key components of the cyst wall that are synthesized during Giardia differentiation into dormant cysts (1). However, regulation of cwp gene expression during encystation is poorly understood. The giardial promoters defined to date, including the encystation-induced cwp1–3 gene promoters, contain A/T-rich Inr-like elements and are relatively short (17–19). Previously, we have identified three transcription factors whose expression increased significantly during encystation: Myb2, ARID1, and a GARP-like protein (23–26). The Myb2 protein can bind to specific sequences in the cwp1–3 promoters and may contribute to the transcriptional activation of these genes (23, 26). The ARID1 and GARP-like proteins can bind to specific sequences in the cwp1 gene (24, 25). These target sequences are all positive cis-acting elements, and ARID1 can transactivate the cwp1 gene (23–25).
A number of WRKY proteins in plants can function as transcriptional activators and be involved in development, dormancy, biotic and abiotic stress, disease resistance, and senescence (27, 28, 30). Therefore, in this study, we tried to investigate the role of the giardial WRKY in regulating encystation-specific cwp genes. We found that the giardial WRKY can bind to specific plant W box-like sequences. The cwp2 and wrky core promoter region contained multiple WRKY binding sites. Mutation of one of the WRKY binding sites in the cwp2 promoter resulted in decreases of activity in both vegetative and encysting cells, indicating that WRKY may also be a transcriptional activator in the regulation of the giardial cwp2 gene. We also found that the constitutively overexpressed WRKY increased the levels of the cwp2 mRNA (and also the cwp1 and myb2 mRNA; see below for discussion) in G. lamblia. ChIP assays confirmed the association of WRKY with its own promoter and the cwp2 promoter (and also the cwp1 and myb2 promoters; see below for discussion). The expression of the plant wrky genes was induced in response to changing environments, including development, dormancy, biotic and abiotic stress, disease resistance, and senescence (30, 32). We also found that the giardial WRKY localized exclusively to the two nuclei, and its expression increased significantly during encystation, suggesting that it may also play a role in this differentiation. No likely nuclear localization signals except some basic amino acids are evident in WRKY. We found an important role of the basic residues located N-terminal to the WRKY domains in the exclusively nuclear localization. Similarly, nuclear localization signals have been identified outside of the plant WRKY domains (27). We also found a loss of transactivation activity of WRKYm on the expression of the cwp1 and cwp2 genes. Although WRKYm was expressed at higher levels, its inactivity may be due to its inability to enter nucleus.
Many important transcription factors involved in developmental regulation and in stress response have an autoregulation mechanism, including mammalian c-Myb and plant WRKY (38, 55). Positive autoregulation has been found in Myb2 involving Myb2 binding sites in its own promoter region to maintain high levels of Myb2 protein during encystation (23, 26). We found that mutation of one of the WRKY binding sites in the wrky promoter resulted in decreases of activity in both vegetative and encysting cells, indicating that this binding site is a positive cis-acting element. Because we only mutated one of multiple WRKY binding sites in the wrky, its own promoter, it is possible that other WRKY binding sites may be important for its positive or negative autoregulation. We found that mutation of a putative nuclear localization signal of WRKY (WRKYm) resulted in loss of nuclear localization. The levels of WRKYm mRNA and protein increased significantly compared with that of wild type WRKY, suggesting a negative autoregulation of the wrky gene. It has been shown that plant WRKY6 may be negatively autoregulated by inhibiting the activity of its own promoter and other closely related WRKY factor gene promoters (38).
The plant WRKY proteins have been found to act downstream of a MAPK pathway that is defense- and pathogen-induced (39, 40). One giardial MAPK, ERK1, exhibits a significantly increased kinase activity during early encystation (42). We found that the expression of the giardial ERK1 increased significantly during encystation, and overexpression of ERK1 resulted in an increase of the wrky and cwp2 mRNA levels, suggesting that the MAPK signaling cascades may modulate the amount and capability of WRKY for transactivation and/or DNA binding during encystation. The localization of overexpressed ERK1 in cytosol and flagella is different from that of a previous report (42), in which localization of ERK1 in the median body and outer edges of adhesive disc can be observed using the homemade anti-ERK1 antibody. It is possible that cytosolic ERK1 may function to activate unidentified protein kinases in the ERK1 pathway, and then cytosolic WRKY may be modified and translocated to nuclei upon activation. The potential mechanisms involved in this regulation remain to be investigated. We also found that deletion of the predicted kinase domain of ERK1 resulted in decreased levels of the WRKY and CWP1 mRNA and proteins, suggesting that WRKY may be a downstream component of a MAPK/ERK1 signaling pathway.
The giardial WRKY contains two WRKY-like domains, similar to the plant group I WRKY proteins, suggesting that group I may be the ancestral form of the WRKY domain (27, 30, 33). Although the two giardial WRKY-like domains are similar to those of the plant WRKY family, they still have divergent characteristics. From the results with Arabidopsis WRKY4, β1 strand containing the conserved WRKYGQK sequence contacts the major groove, and the most important residues of WRKY contacting the target sequence bases are predicted to be Arg, Lys, Tyr, Gln, and the second Lys of WRKYGQK (50). Five other basic residues in the β1–β3 strands contact the backbone (Fig. 1B). The presence of Trp and Gly of WRKYGQK allows stabilization of the zinc coordination and deep entrance of the β1 strand into the major groove, respectively (50). The C-terminal conserved cysteines and histidines are the zinc-coordinating residues (50). Most of these residues are conserved in the giardial WRKY (Fig. 1B). However, in the C-terminal WRKY domain, the Arg and Gln of WRKYGQK are changed to Lys and His, respectively, only four of the five basic residues that make contact with backbone are present, and a 7-residue insertion is present in the middle of the zinc-binding motif (Fig. 1B). It is interesting that a change of the Trp, Lys, Tyr, or the second Lys of WRKYGQK resulted in a complete loss of binding of plant WRKY (34). However, a change of Arg, Gly, or Gln only partially reduced DNA binding of plant WRKY (34). This result is correlated with the presence of DNA binding activity of the giardial WRKY with the change of the Arg and Gln residues in the C-terminal WRKY domain.
Although divergent from plant WRKY proteins, giardial WRKY still maintains the ability to bind DNA in a sequence-specific manner. The best binding sequence for giardial WRKY is (a/g)GTCA(a/g). Its antisense sequence is the plant W box (c/t)TGAC(c/t) (27). This indicates that the binding sites of the members of the WRKY family have been conserved during evolution. In addition, giardial WRKY was found to possess a moderate binding affinity for another sequence, (a/g)GTCA(t/c) or cGTCA(a/g) (Fig. 4, lanes 2, 6, and 7), and a low binding affinity for the sequence aGTTAa or aATCAg (whose core sequence is similar to GTCA) (Figs. 3B, lane 6, and 4, lanes 11–14). The non-conservative substitutions of residues that contact the major groove and phosphodiester backbone might explain why giardial WRKY tolerates more target sequence. Since the specificity determined by a 6-bp sequence does not seem high enough for choosing target genes on the giardial genome, it is possible that additional transcription factors interacting with WRKY help to increase the specificity for transcriptional regulation.
In plants, WRKY factors have a high binding affinity to the W box, which may appear as a monomeric form or two tandem repeats in forward or reverse orientation (36, 56). Giardial WRKY can bind to the gGTCAgacTGATt sequence (TGAT is the antisense sequence of ATCA) in the wrky 5′-flanking region (−75 to −63 relative to the translation start site), which contains two binding sites in reverse orientation (Fig. 4, lanes 11–14). Two tandem repeats of WRKY binding sites (the −180 to −174 region) are also present in the same orientation in the triosephosphate isomerase gene promoter (Table 1). In plants, multiple copies of W boxes (an average of 4.3 copies/promoter) have been found in all members of the defense response gene promoters (57). We searched the 200-bp 5′-flanking regions of 38 giardial genes and found that 23 genes contain at least one of the WRKY binding sites (Table 1). Interestingly, the cwp2, phosphoenolpyruvate carboxykinase, and wrky promoters, contain three copies of WRKY binding sites, and seven other gene promoters, including the myb2 promoter, contain two copies of WRKY binding sites. The presence of multiple WRKY binding sites in the encystation-induced cwp2 and myb2 gene promoters and its own promoter suggest that WRKY may play an important role in regulation of encystation and in its autoregulation. The presence of the WRKY binding sites in other gene promoters suggests that WRKY may be involved in other physiological response by transcriptional regulation of many different genes.
Our results showed that constitutively expressed WRKY increased the expression of cwp2 gene by ∼2.5-fold (Fig. 7D). However, the cwp2 promoter could be increased ∼116-fold during encystation (Fig. 6B). Mutation analysis of the cwp2 promoter has provided evidence for involvement of one WRKY binding site during vegetative growth and encystation (Fig. 6B). It should be noted that, during encystation, the levels of cwp2 promoter activity decreased more when this WRKY binding site was mutated, suggesting that this site could be more important during encystation. WRKY may need to cooperate with some other transcription factors to transactivate the cwp2 gene during encystation. In plants, three pathogen-induced WRKY proteins can form both homocomplexes and heterocomplexes and enhance or reduce their DNA binding activity (58). Since only one putative homologue for WRKY has been identified in G. lamblia to date, it is possible that the giardial WRKY protein regulates specific target genes by interacting with itself or with other classes of DNA-binding proteins that occupy directly adjacent binding sites within the target promoter region. Interestingly, in the cwp2 promoter, ARID1 binding site/Inr (−8 to −1 region) and Myb2 binding sites (−22 to −17, −40 to −35, and −45 to −15 regions) are very close to one WRKY binding site (−43 to −38 region). Further studies will be required to elucidate whether giardial WRKY functions as an activator via association with some encystation-specific cofactors, such as ARID1 or Myb2, in the promoter context of encystation-induced genes.
Our results also showed that constitutively expressed WRKY increased the expression of cwp1 and myb2 genes. We found that WRKY can bind weakly to the cwp1−45/−1 probe in vitro, which does not contain the TGAC/GTCA core sequence but contains the gATCAa (which is similar to gGTCAa) sequence (Fig. 6A, lane 6), suggesting that WRKY can bind to the cwp1 promoter, although weakly. We also detected the association of WRKY with the cwp1 promoter in vivo, suggesting that WRKY may activate the cwp1 promoter directly. We also found the presence of the WRKY binding sites in the myb2 promoter and an association of WRKY with the myb2 promoter in vivo, suggesting that WRKY may activate the myb2 promoter directly. However, WRKY may also activate the cwp1 promoter through an indirect mechanism. This activation is most probably through an interaction of WRKY and Myb2 and then through a direct binding of Myb2 to the cwp1 promoter.
Our study provides evidence for the involvement of WRKY in DNA binding and transactivation of the cwp1-2 genes in the early diverging protozoan G. lamblia. WRKY gene expression has been found to increase when plants go dormant in response to dry environment (28). We have also found the important role of the giardial WRKY in induction of formation of the cyst, which is the dormant stage. Our studies provide new insights into the evolution of eukaryotic DNA binding domain and transcriptional mechanisms during differentiation.
Supplementary Material
Acknowledgments
We thank Dr. Chien-Kuo Lee, Dr. Tsai-Kun Li, Dr. Shin-Hong Shiao, and Dr. Nei-Li Chan for helpful comments; Yi-Li Liu and I-Ching Huang for technical support in DNA sequencing; and Li-Hsin Su, Pei-Wei Chiu, Yi-Ting Wang, and Sheng-Fung Chuang for technical support in PCR and cell culture. We also thank Dr. Kwang-Jen Hsiao and Dr. Chen-Jee Hong from Taipei Veterans General Hospital and Mei-Ying Liu from the National Yang-Ming University for providing facilities to carry out the work. We are also very grateful to the researchers and administrators of the G. lamblia genome data base for providing genome information.
This work was supported by National Science Council Grants NSC 94-2320-B-002-093 and NSC 96-2320-B-002-040-MY3 and National Health Research Institutes Grants NHRI-EX96-9510NC, NHRI-EX97-9510NC, and NHRI-EX98-9510NC in Taiwan and was also supported in part by the Department of Medical Research of National Taiwan University Hospital.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. S1.
3 Y. C. Huang and C. H. Sun, unpublished data.
- MAPK
- mitogen-activated protein kinase
- ChIP
- chromatin immunoprecipitation
- RT
- reverse transcription
- HA
- hemagglutinin
- CWP
- cell wall protein.
REFERENCES
- 1.Adam R. D. ( 2001) Clin. Microbiol. Rev. 14, 447– 475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wolfe M. S. ( 1992) Clin. Microbiol. Rev. 5, 93– 100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Celiksöz A., Aciöz M., Deðerli S., Cinar Z., Elaldi N., Erandaç M. ( 2005) Pediatr. Int. 47, 567– 571 [DOI] [PubMed] [Google Scholar]
- 4.Eichinger D. ( 2001) Curr. Opin. Microbiol. 4, 421– 426 [DOI] [PubMed] [Google Scholar]
- 5.Gillin F. D., Reiner D. S., McCaffery J. M. ( 1996) Annu. Rev. Microbiol. 50, 679– 705 [DOI] [PubMed] [Google Scholar]
- 6.Luján H. D., Mowatt M. R., Conrad J. T., Bowers B., Nash T. E. ( 1995) J. Biol. Chem. 270, 29307– 29313 [DOI] [PubMed] [Google Scholar]
- 7.Mowatt M. R., Luján H. D., Cotten D. B., Bowers B., Yee J., Nash T. E., Stibbs H. H. ( 1995) Mol. Microbiol. 15, 955– 963 [DOI] [PubMed] [Google Scholar]
- 8.Sun C. H., McCaffery J. M., Reiner D. S., Gillin F. D. ( 2003) J. Biol. Chem. 278, 21701– 21708 [DOI] [PubMed] [Google Scholar]
- 9.Knodler L. A., Svärd S. G., Silberman J. D., Davids B. J., Gillin F. D. ( 1999) Mol. Microbiol. 34, 327– 340 [DOI] [PubMed] [Google Scholar]
- 10.Van Keulen H., Steimle P. A., Bulik D. A., Borowiak R. K., Jarroll E. L. ( 1998) J. Eukaryot. Microbiol. 45, 637– 642 [DOI] [PubMed] [Google Scholar]
- 11.Luján H. D., Mowatt M. R., Nash T. E. ( 1997) Microbiol. Mol. Biol. Rev. 61, 294– 304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sogin M. L., Gunderson J. H., Elwood H. J., Alonso R. A., Peattie D. A. ( 1989) Science 243, 75– 77 [DOI] [PubMed] [Google Scholar]
- 13.Baldauf S. L. ( 2003) Science 300, 1703– 1706 [DOI] [PubMed] [Google Scholar]
- 14.Morrison H. G., McArthur A. G., Gillin F. D., Aley S. B., Adam R. D., Olsen G. J., Best A. A., Cande W. Z., Chen F., Cipriano M. J., Davids B. J., Dawson S. C., Elmendorf H. G., Hehl A. B., Holder M. E., Huse S. M., Kim U. U., Lasek-Nesselquist E., Manning G., Nigam A., Nixon J. E., Palm D., Passamaneck N. E., Prabhu A., Reich C. I., Reiner D. S., Samuelson J., Svard S. G., Sogin M. L. ( 2007) Science 317, 1921– 1926 [DOI] [PubMed] [Google Scholar]
- 15.Best A. A, Morrison H. G., McArthur A. G., Sogin M. L., Olsen G. J. ( 2004) Genome Res. 14, 1537– 1547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Seshadri V., McArthur A. G., Sogin M. L., Adam R. D. ( 2003) J. Biol. Chem. 278, 27804– 27810 [DOI] [PubMed] [Google Scholar]
- 17.Elmendorf H. G., Singer S. M., Pierce J., Cowan J., Nash T. E. ( 2001) Mol. Biochem. Parasitol. 113, 157– 169 [DOI] [PubMed] [Google Scholar]
- 18.Sun C. H., Tai J. H. ( 1999) J. Biol. Chem. 274, 19699– 19706 [DOI] [PubMed] [Google Scholar]
- 19.Yee J., Mowatt M. R., Dennis P. P., Nash T. E. ( 2000) J. Biol. Chem. 275, 11432– 11439 [DOI] [PubMed] [Google Scholar]
- 20.Holberton D. V., Marshall J. ( 1995) Nucleic Acids Res. 23, 2945– 2953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Davis-Hayman S. R., Hayman J. R., Nash T. E. ( 2003) Int. J. Parasitol. 33, 1005– 1012 [DOI] [PubMed] [Google Scholar]
- 22.Worgall T. S., Davis-Hayman S. R., Magana M. M., Oelkers P. M., Zapata F., Juliano R. A., Osborne T. F., Nash T. E., Deckelbaum R. J. ( 2004) J. Lipid Res. 45, 981– 988 [DOI] [PubMed] [Google Scholar]
- 23.Sun C. H., Palm D., McArthur A. G., Svärd S. G., Gillin F. D. ( 2002) Mol. Microbiol. 46, 971– 984 [DOI] [PubMed] [Google Scholar]
- 24.Sun C. H., Su L. H., Gillin F. D. ( 2006) Mol. Biochem. Parasitol. 146, 45– 57 [DOI] [PubMed] [Google Scholar]
- 25.Wang C. H., Su L. H., Sun C. H. ( 2007) J. Biol. Chem. 282, 8905– 8914 [DOI] [PubMed] [Google Scholar]
- 26.Huang Y. C., Su L. H., Lee G. A., Chiu P. W., Cho C. C., Wu J. Y., Sun C. H. ( 2008) J. Biol. Chem. 283, 31021– 31029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Eulgem T., Rushton P. J., Robatzek S., Somssich I. E. ( 2000) Trends Plant Sci. 5, 199– 206 [DOI] [PubMed] [Google Scholar]
- 28.Pnueli L., Hallak-Herr E., Rozenberg M., Cohen M., Goloubinoff P., Kaplan A., Mittler R. ( 2002) Plant J. 3, 319– 330 [DOI] [PubMed] [Google Scholar]
- 29.Sun C., Palmqvist S., Olsson H., Borén M., Ahlandsberg S., Jansson C. ( 2003) Plant Cell 15, 2076– 2092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ulker B., Somssich I. E. ( 2004) Curr. Opin. Plant Biol. 7, 491– 498 [DOI] [PubMed] [Google Scholar]
- 31.Yoda H., Ogawa M., Yamaguchi Y., Koizumi N., Kusano T., Sano H. ( 2002) Mol. Genet. Genomics 267, 154– 161 [DOI] [PubMed] [Google Scholar]
- 32.Dong J., Chen C., Chen Z. ( 2003) Plant Mol. Biol. 51, 21– 37 [DOI] [PubMed] [Google Scholar]
- 33.Zhang Y., Wang L. ( 2005) BMC Evol. Biol. 5, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Maeo K., Hayashi S., Kojima-Suzuki H., Morikami A., Nakamura K. ( 2001) Biosci. Biotechnol. Biochem. 65, 2428– 2436 [DOI] [PubMed] [Google Scholar]
- 35.Rushton P. J., Torres J. T., Parniske M., Wernert P., Hahlbrock K., Somssich I. E. ( 1996) EMBO J. 15, 5690– 5700 [PMC free article] [PubMed] [Google Scholar]
- 36.Eulgem T., Rushton P. J., Schmelzer E., Hahlbrock K., Somssich I. E. ( 1999) EMBO J. 18, 4689– 4699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Du L. Q., Chen Z. X. ( 2000) Plant J. 24, 837– 847 [DOI] [PubMed] [Google Scholar]
- 38.Robatzek S., Somssich I. E. ( 2002) Genes Dev. 16, 1139– 1149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wan J., Zhang S., Stacey G. ( 2004) Mol. Plant Pathol. 5, 125– 135 [DOI] [PubMed] [Google Scholar]
- 40.Asai T., Tena G., Plotnikova J., Willmann M. R., Chiu W. L., Gomez-Gomez L., Boller T., Ausubel F. M., Sheen J. ( 2002) Nature 415, 977– 983 [DOI] [PubMed] [Google Scholar]
- 41.Johnson C. S., Kolevski B., Smyth D. R. ( 2002) Plant Cell 14, 1359– 1375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ellis J. G., 4th., Davila M., Chakrabarti R. ( 2003) J. Biol. Chem. 278, 1936– 1945 [DOI] [PubMed] [Google Scholar]
- 43.Keister D. B. ( 1983) Trans. R. Soc. Trop. Med. Hyg. 77, 487– 488 [DOI] [PubMed] [Google Scholar]
- 44.Su L. H., Lee G. A., Huang Y. C., Chen Y. H., Sun C. H. ( 2007) Mol. Biochem. Parasitol. 156, 124– 135 [DOI] [PubMed] [Google Scholar]
- 45.McArthur A. G., Morrison H. G., Nixon J. E., Passamaneck N. Q., Kim U., Hinkle G., Crocker M. K., Holder M. E., Farr R., Reich C. I., Olsen G. E., Aley S. B., Adam R. D., Gillin F. D., Sogin M. L. ( 2000) FEMS Microbiol. Lett. 189, 271– 273 [DOI] [PubMed] [Google Scholar]
- 46.Altschul S. F., Madden T. L., Schäffer A. A., Zhang J., Zhang Z., Miller W., Lipman D. J. ( 1997) Nucleic Acids Res. 25, 3389– 3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Singer S. M., Yee J., Nash T. E. ( 1998) Mol. Biochem. Parasitol. 92, 59– 69 [DOI] [PubMed] [Google Scholar]
- 48.Chen Y. H., Su L. H., Sun C. H. ( 2008) Int. J. Parasitol. 38, 1305– 1317 [DOI] [PubMed] [Google Scholar]
- 49.Finn R. D, Tate J., Mistry J., Coggill P. C., Sammut S. J., Hotz H. R., Ceric G., Forslund K., Eddy S. R., Sonnhammer E. L., Bateman A. ( 2008) Nucleic Acids Res. 36, D281– D288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yamasaki K., Kigawa T., Inoue M., Tateno M., Yamasaki T., Yabuki T., Aoki M., Seki E., Matsuda T., Tomo Y., Hayami N., Terada T., Shirouzu M., Tanaka A., Seki M., Shinozaki K., Yokoyama S. ( 2005) Plant Cell 17, 944– 956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Abel E. S., Davids B. J., Robles L. D., Loflin C. E., Gillin F. D., Chakrabarti R. ( 2001) J. Biol. Chem. 276, 10320– 10329 [DOI] [PubMed] [Google Scholar]
- 52.Svärd S. G., Rafferty C., McCaffery J. M., Smith M. W., Reiner D. S., Gillin F. D. ( 1999) Mol. Biochem. Parasitol. 98, 253– 264 [DOI] [PubMed] [Google Scholar]
- 53.Nakai K., Kanehisa M. ( 1992) Genomics 14, 897– 911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Coll M., Frederick C. A., Wang A. H., Rich A. ( 1987) Proc. Natl. Acad. Sci. U.S.A. 84, 8385– 8389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Nicolaides N. C., Gualdi R., Casadevall C., Manzella L., Calabretta B. ( 1991) Mol. Cell. Biol. 11, 6166– 6176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yang P., Chen C., Wang Z., Fan B., Chen Z. ( 1999) Plant J. 18, 141– 149 [Google Scholar]
- 57.Maleck K., Levine A., Eulgem T., Morgan A., Schmid J., Lawton K. A., Dangl J. L., Dietrich R. A. ( 2000) Nat. Genet. 26, 403– 410 [DOI] [PubMed] [Google Scholar]
- 58.Xu X., Chen C., Fan B., Chen Z. ( 2006) Plant Cell 18, 1310– 1326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Chenna R., Sugawara H., Koike T., Lopez R., Gibson T. J., Higgins D. G., Thompson J. D. ( 2003) Nucleic Acids Res. 31, 3497– 3500 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.