Abstract
Prolectin, a previously undescribed glycan-binding receptor, has been identified by re-screening of the human genome for genes encoding proteins containing potential C-type carbohydrate-recognition domains. Glycan array analysis revealed that the carbohydrate-recognition domain in the extracellular domain of the receptor binds glycans with terminal α-linked mannose or fucose residues. Prolectin expressed in fibroblasts is found at the cell surface, but unlike many glycan-binding receptors it does not mediate endocytosis of a neoglycoprotein ligand. However, compared with other known glycan-binding receptors, the receptor contains an unusually large intracellular domain that consists of multiple sequence motifs, including phosphorylated tyrosine residues, that allow it to interact with signaling molecules such as Grb2. Immunohistochemistry has been used to demonstrate that prolectin is expressed on a specialized population of proliferating B cells in germinal centers. Thus, this novel receptor has the potential to function in carbohydrate-mediated communication between cells in the germinal center.
Membrane-bound mammalian glycan-binding receptors, often referred to as lectins, are believed to play multiple distinct roles in the immune system, decoding information in complex oligosaccharide structures on cell surfaces and soluble glycoproteins (1, 2). A host of glycan-binding receptors on dendritic cells and macrophages function in pathogen recognition, often resulting in uptake of microbes through endocytic mechanisms. Examples include the mannose receptor, DC-SIGN,3 langerin, and the macrophage galactose receptor. Glycan-binding receptors can also recognize glycans found on the surfaces of mammalian cells. Some of these receptors, such as the selectins, mediate adhesion between leukocytes and endothelia (3, 4). A small number of receptors, notably members of the siglec family, bind mammalian-type glycans and have been shown to have potential signaling functions (5). While multiple glycan-binding receptors have been described on cells of the myeloid lineage, the complement of such receptors on lymphocytes is much more restricted. The best characterized examples are the T-cell adhesion molecule L-selectin (4) and the B-cell receptor CD22, also designated siglec-2 (5).
Genomic screening for potential glycan-binding receptors has usually been undertaken by initially searching for the presence of one of the several types of structural domains that are known to support sugar-binding activity (6). Knowledge of the structures of multiple families of modular carbohydrate-recognition domains (CRDs) has facilitated identification of proteins with potential sugar-binding activity and can lead to predictions of what types of ligands might be bound. Although the human genome has been extensively screened with profile-recognition algorithms that identify common sequence motifs associated with CRDs, refinements to the genome sequence and improvements in gene-recognition algorithms occasionally result in detection of novel proteins that contain putative CRDs.
We describe a previously undetected glycan-binding receptor identified by re-screening of the human genome and provide characterization of its molecular and cellular properties. Based on its expression in a specialized population of proliferating B cells in germinal centers, we propose that it be designated prolectin. Our results suggest that prolectin functions in carbohydrate-mediated communication between cells in the germinal center.
EXPERIMENTAL PROCEDURES
Prolectin Cloning, Expression, and Purification
The full-length cDNA was amplified from a spleen cDNA library (Clontech) using 40 cycles of PCR with Advantage 2 polymerase mix from Takara and forward primer CCCTGGCTGCCACTTGTCAGGTTC and reverse primer GGGCTTCAACAGGAACATTTCCGC (Invitrogen). The amplified cDNA was isolated by gel electrophoresis and cloned into vector pCRII-TOPO (Invitrogen).
The portion of the cDNA encoding the extracellular domain of prolectin was inserted into the expression vector T5T and expressed in Escherichia coli strain BL21(DE3) following the procedure used for DC-SIGN (7). Inclusion bodies isolated by sonication were dissolved in guanidine hydrochloride in the presence of a small amount of 2-mercaptoethanol and renatured by dilution into loading buffer (0.5 m NaCl, 25 mm Tris-Cl, pH 7.8, 25 mm CaCl2) followed by extensive dialysis against the same buffer. Protein from 6 liters of bacterial culture, in a final volume of 500 ml of loading buffer, was isolated on a 10-ml column of mannose-Sepharose (8), which was washed with loading buffer and eluted with 2-ml fractions of eluting buffer (0.5 m NaCl, 25 mm Tris-Cl, pH 7.8, 2.5 mm EDTA). Aliquots (25 μl) of fractions were examined by SDS-PAGE (9).
Sugar Binding and Glycan Array Analysis
For glycan array analysis, modified primers were used to append a biotinylation tag Gly-Leu-Asn-Asp-Ile-Phe-Glu-Ala-Gln-Lys-Ile-Glu-Trp-His-Glu after the C-terminal cysteine residue of the CRD. The modified cDNA was inserted into vector T5T, co-expressed with plasmid birA, which encodes biotin ligase (Avidity), and induced in the presence of biotin (10). The monomeric, biotinylated protein, purified on a 10-ml column of mannose-Sepharose as described above for the extracellular domain, was complexed with Alexa-488-labeled streptavidin (Invitrogen) by incubation overnight at a ratio of ∼2 mol of CRD to 1 mol of streptavidin subunit. The complex was isolated on a 1-ml column of mannose-Sepharose, which was washed with loading buffer and eluted with 0.5-ml aliquots of elution buffer. The protein was tested against version 3.1 of the glycan array of the Consortium for Functional Glycomics using the standard protocol. The extracellular domain of prolectin was used to coat polystyrene wells, which were used for solid-phase binding competition assays with 125I-Man-BSA (E-Y Laboratories) as the reporter ligand (7). Oligosaccharide ligands were obtained from Carbosynth Ltd.
Analysis in Transfected Fibroblasts
The full-length cDNA was cloned into vector pVcos and used to transfect Ψ-cre packaging cells (11). The resulting pseudoviruses were harvested and used to infect Rat-6 fibroblasts, which were subjected to selection in 400 μg/ml G418. Analysis of endocytic activity with 125I-Man-BSA was conducted as previously described (11). Receptor was isolated from cells by solubilization in lysis buffer (150 mm NaCl, 25 mm Tris-Cl, pH 7.8, 2 mm CaCl2) containing 1% Triton X-100 and protease and phosphatase inhibitors (Set 1 and Set 2 from Calbiochem). After brief sonication and incubation for 30 min on ice, debris was removed by centrifugation for 15 min at 100,000 × g, and the supernatant was applied to a 1-ml column of mannose-Sepharose, which was washed with lysis buffer containing 0.1% Triton X-100 and eluted in 0.5-ml fractions with elution buffer containing 0.1% Triton X-100. Fractions were precipitated by addition of trichloroacetic acid to 10% for 10 min on ice, and precipitates were centrifuged for 5 min at 18,000 × g, washed twice with 0.5 ml of ethanol:ether (1:1), and dissolved in sample buffer for gel electrophoresis.
Rabbit polyclonal antibodies to prolectin were generated using 1 mg of the bacterially expressed extracellular domain as antigen following the speedy 28-day rabbit polyclonal antibody program from Eurogentec. For affinity purification, an affinity column was constructed with bacterially expressed CRD immobilized on Affi-Gel 10 (Bio-Rad Laboratories). Monoclonal antibodies to phosphotyrosine were obtained from Amersham Biosciences (4G10) and BIOMOL International (PY20), and polyclonal antibody to Grb2 was purchased from Cell Signaling Technology. Alkaline phosphatase-conjugated protein A (Calbiochem) and goat anti-mouse IgG (Jackson ImmunoResearch), visualized with 5-bromo-4-chloro-3-indolyl nitroblue tetrazolium phosphatase substrate (Calbiochem), were used to counterstain blots.
Immunohistochemistry and Immunofluorescence
Formalin-fixed, paraffin-embedded tissue array slides of human normal organs were provided by SuperBioChips. The antibodies used were: mouse anti-Ki67, clone MIB-1 (1:80 dilution), mouse anti-CD68, clone KP1 (1:40), mouse anti-CD21 (1:33), and negative control mouse IgG1 (all Dako); affinity-purified rabbit anti-prolectin (10 μg/ml); and negative control normal rabbit IgG (gift from David Guiliano, Imperial College London). Slides were deparaffinized in xylene and rehydrated through a graded ethanol series. Heat-mediated antigen retrieval was performed using antigen unmasking solution (Vector Laboratories), except in the case of anti-CD21, where retrieval was performed using trypsin enzymatic antigen retrieval solution (Abcam). Sections were blocked with 10% normal goat serum (Vector Laboratories), and incubated for 90–120 min with primary antibody in 2% serum.
For immunofluorescence, sections were incubated with Alexa Fluor 488 goat anti-rabbit IgG and Alexa Fluor 594 goat anti-mouse IgG (Invitrogen) and mounted using Vectashield mounting medium with 4′,6-diamidino-2-phenylindole (Vector Laboratories). For immunohistochemistry, sections were incubated with biotinylated goat anti-rabbit IgG or anti-mouse IgG (Vector Laboratories), followed by quenching of endogenous peroxidase by 15-min incubation in 3% H2O2 in phosphate-buffered saline. Antigen was visualized by the peroxidase method using the Vectastain elite ABC and DAB substrate kits, and counterstained with Vector Hematoxylin QS (Vector Laboratories). Sections were dehydrated, cleared, and mounted with VectaMount permanent mounting medium (Vector Laboratories). Microscopy was performed on a Nikon Eclipse E400 microscope equipped with a DXM1200 digital camera. Images were captured with Lucia GF software, version 4.60, and were merged in Corel PhotoPaint 12.
RESULTS
The Prolectin Gene Encodes a Novel Receptor Containing a C-type CRD
A previously undescribed C-type CRD was identified in an entry denoted FLJ45910 protein (GenBankTM accession EAW84437) in the human genome annotation submitted by Celera Genomics. The domain contained key residues that typically form a Ca2+-dependent sugar-binding site in C-type glycan-binding receptors (12). On further examination, the genomic region encoding this predicted gene was found to correspond in part to a gene recently given symbol CLEC17A (GeneID 388512) by the HUGO Gene Nomenclature Committee. The predicted protein from this interpretation of the genome data (UniProtKB/Swiss-Prot accession number Q6ZS10) lacks the C-terminal portion of the CRD.
Based on the predicted cDNA sequence from the Celera annotation, primers were designed for PCR amplification of a cDNA encoding the putative receptor, and receptor mRNA was detected in libraries from multiple tissues. After this cDNA was cloned, the sequence of an IMAGE clone (13) that has an extended 5′ region was deposited in GenBankTM (GenBankTM accession number BC140848 for the cDNA, and UniProtKB/Swiss-Prot accession number B2RTX0 for the deduced amino acid sequence). Cloning with primers in this region confirmed the presence of a longer transcript which matched exactly the sequence of the IMAGE clone in the data base. A full-length cDNA, including this region, was used as a basis for further experiments.
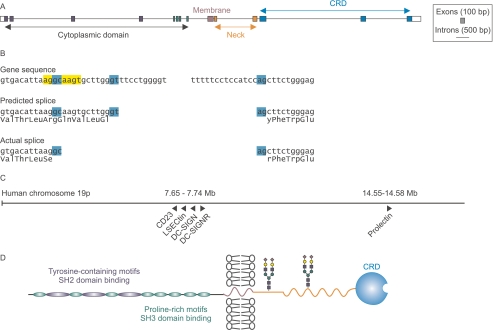
Using the full-length cDNA as a guide, the complete prolectin gene structure, shown in Fig. 1A, was deduced. Comparison of the gene and cDNA sequences revealed that there must be an unusual GC splice donor site that is utilized instead of the nearby canonical GT sequence used in the Celera annotation. This splice results in a deletion of four amino acids in the actual sequence compared with the predicted sequence (Fig. 1B). The sequence flanking the GC splice donor site, AGGCAAGT, exactly matches the ideal sequence for binding to the U2-type spliceosome, which is believed to compensate for the absence of the canonical GT donor site (14). The length of the introns near the 3′ and 5′ ends of the gene, combined with the unusual splice site, are probably responsible for the fact that the prolectin cDNA was not correctly identified by any of the gene prediction algorithms.
FIGURE 1.
Prolectin gene structure and protein organization. A, scale diagram of the prolectin gene. For clarity, exons are shown expanded 3-fold relative to introns. B, details of unusual GC splice site at the 5′ end of the final intron. The predicted GT splice site in the Celera annotation of the genome, GenBankTM accession EAW84437, leads to an insertion of 12 bases corresponding to insertion of 4 amino acids in the predicted protein sequence, but it leaves the reading frame intact so that the conserved residues of the CRD are present. C, scale diagram showing the relative positions of glycan-binding receptors containing C-type CRDs encoded on human chromosome 19. D, distribution of functional domains in prolectin. Predicted N-glycosylation sites are indicated with hypothetical glycans in stick figures.
The prolectin gene is located on the long arm of human chromosome 19, which contains a cluster of genes for several other receptors that utilize C-type CRDs, including the dendritic cell receptor DC-SIGN (Fig. 1C). However, the new gene is located roughly 7 mega bases away from this gene cluster.
The complete prolectin cDNA sequence encodes a type II transmembrane protein with an extended intracellular N-terminal domain and a C-terminal extracellular C-type CRD (Fig. 1D). The cytoplasmic domain contains repeated motifs that match patterns for binding to intracellular signaling molecules. There are at least three potential SH2-binding sites and eight potential SH3-binding sequences (15). Such a large cytoplasmic domain with so many potential interactions is unprecedented for a member of the C-type lectin family. These results suggest that the receptor has novel properties expected for a sugar-binding receptor with signaling functions.
The Extracellular Domain of Prolectin Binds Two Classes of Cell Surface Glycans
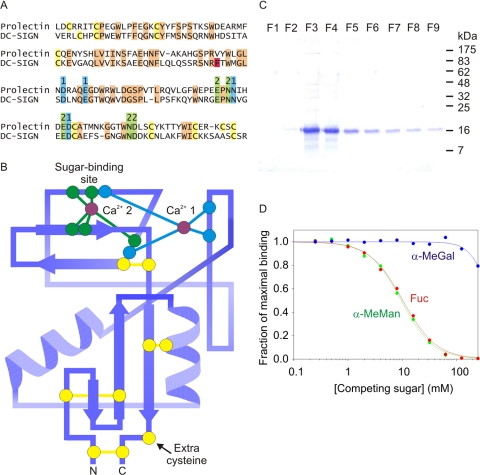
Based on the presence of a characteristic pattern of amino acids forming the conserved Ca2+-binding site in C-type CRDs such as DC-SIGN, it was predicted that the protein encoded by this novel gene would bind mannose and related sugars (Fig. 2, A and B) (12, 16). The extracellular domain was therefore produced in a bacterial expression system and tested for binding to immobilized mannose. The protein was found to bind tightly to the resin and was eluted with EDTA (Fig. 2C). Binding was confirmed in a solid-phase assay, in which receptor extracellular domain immobilized in polystyrene wells was probed with iodinated, mannose-conjugated serum albumin (125I-Man-BSA). Competition with monosaccharides revealed similar binding affinities for mannose, fucose, and N-acetylglucosamine, with much weaker binding to galactose (Fig. 2D and Table 1).
FIGURE 2.
Characterization of the CRD in prolectin. A, sequence comparison between the CRDs of DC-SIGN and prolectin, with cysteine residues highlighted in yellow, ligands for Ca2+ 1 and 2 highlighted in blue and green, and other conserved residues shaded orange. Phe325 of DC-SIGN, which forms a critical part of the secondary binding site, is highlighted in red. B, diagram of the proposed topology of the CRD from prolectin, based on the structure of the CRD from DC-SIGN (Protein Data Base entry 1K9I). Conserved residues are highlighted in the same colors as in A. C, gel showing purification of the extracellular domain of prolectin expressed in E. coli. Aliquots of fractions eluted from a mannose-Sepharose affinity column with EDTA were run on the gel, which was stained with Coomassie Blue. D, examples of solid-phase binding competition assays using monosaccharides to inhibit binding of 125I-Man-BSA (radiolabeled, mannose-conjugated bovine serum albumin) to the extracellular domain of prolectin immobilized in polystyrene wells.
TABLE 1.
Summary of prolectin competition binding data
KI values are shown relative to the value for αMe-Man run in parallel in each assay. The average KI for αMe-Man was 9.9 ± 1.2 mm.
| Ligand | Relative KI |
|---|---|
| αMe-Man | 1.0 |
| αMe-Glc | 3.2 ± 0.1 |
| αMe-Gal | >30 |
| Man | 1.2 ± 0.1 |
| GlcNAc | 1.2 ± 0.1 |
| Fuc | 1.1 ± 0.1 |
| Lea | 0.22 ± 0.01 |
| Lex | 0.72 ± 0.08 |
| Man9 | 0.064 ± 0.04 |
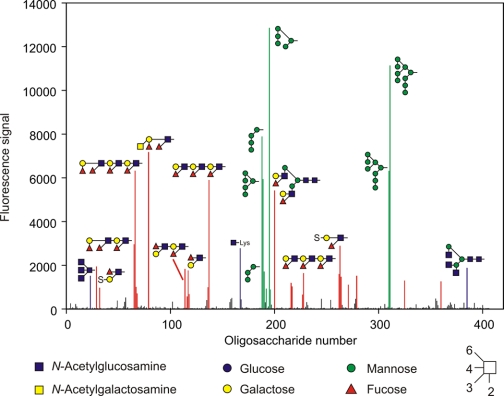
A more detailed analysis of sugar-binding specificity was undertaken by probing a glycan array containing more than 400 oligosaccharide ligands covalently immobilized on glass. CRD expressed with a C-terminal biotinylation tag was bound to fluorescently labeled streptavidin, providing a tetravalent complex that was screened for binding to the array. The ligands detected on the array fall largely into two groups, displaying either terminal mannose or terminal fucose residues (Fig. 3). Closer examination of the mannose-containing ligands indicates that the strongest signals are obtained with structures containing terminal Manα1–2Manα1–2Man sequences and that the fucose-containing ligands include terminal Lewisa, Lewisx, and Lewisy structures. Probing of the array at lower protein concentration resulted in reduction of all the signals, indicating that no one of these categories of ligand binds with dramatically higher affinity than others. The observed binding to both high mannose and Lewis-type structures was also confirmed in competition assays (Table 1). These assays indicate that the Lewisa trisaccharide is a better ligand for prolectin than is Lewisx, suggesting that the apparent preference for Lewisx-containing structures on the array is a result of the disposition of these structures, often in multiple copies, on larger oligosaccharides.
FIGURE 3.
Probing a glycan array with a tetrameric complex of Alexa 488-labeled streptavidin with biotin-tagged CRD from prolectin. The protein concentration was 0.45 mg/ml. Glycans that are bound by the probe are colored based on the terminal residues present: red for fucose, green for mannose, and blue for N-acetylglucosamine. Structures of the ligands that give the strongest signals are indicated using symbol nomenclature. A complete list of all the glycans on the array is provided in supplemental Table S1.
The profile of results is similar to that obtained for the dendritic cell receptor DC-SIGN, but there are significant differences between the preferred ligands for the two proteins (16). Binding of oligosaccharides to C-type CRDs usually involves docking of a monosaccharide into the primary binding site at a conserved Ca2+, with enhanced affinity for the oligosaccharide resulting from secondary contacts between other sugars in the oligosaccharide and residues nearby on the protein surface (12, 16). A phenylalanine residue in DC-SIGN that makes key contacts with portions of high mannose oligosaccharides and a valine residue that contributes to the specificity of DC-SIGN for Lewisx and Lewisa structures are not conserved in prolectin, suggesting that the secondary binding sites in the two receptors must be different. This conclusion is consistent with the observed differences in selectivity for different ligands within broadly similar classes.
Prolectin Is Expressed at the Cell Surface
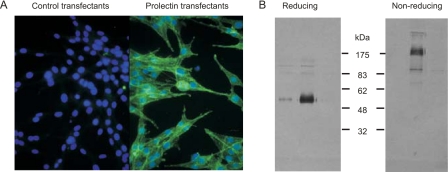
The properties of prolectin were examined by stable expression of a full-length cDNA in Rat-6 fibroblasts, cells that display no detectable surface glycan-binding receptors but which have been shown to support activity of multiple exogenous receptors containing C-type CRDs (11, 16, 17). Immunofluorescence staining revealed that prolectin appears on the cell surface of the transfectants (Fig. 4A), and receptor isolated from the fibroblasts was found to form disulfide-linked oligomers with molecular mass in excess of 175 kDa (Fig. 4B). Based on the previously characterized pattern of disulfide bonds within C-type CRDs (18, 19), a single free cysteine residue would be present in the CRD (Fig. 2B). Interchain disulfide bonding of this cysteine could lead to dimer formation, but the larger size of the oligomers observed would require formation of additional interchain disulfide bonds involving one of the cysteine residues in the transmembrane domain of the receptor. Dimerization of dimers brought about in this way would lead to tetramer formation, which would be consistent with the observed size of the disulfide-linked oligomer observed on gels.
FIGURE 4.
Expression of prolectin in fibroblasts. A, immunofluorescence of transfectants and control cells with anti-prolectin antibodies followed by Alexa 488-labeled secondary antibodies. Nuclei were stained with 4′,6-diamidino-2-phenylindole. B, gel electrophoresis of intact prolectin isolated from transfected fibroblasts. Aliquots of three peak fractions from mannose-Sepharose affinity column were run on gels in the presence and absence of reducing agent. Protein was detected by blotting the gel and probing with antibodies to prolectin followed by alkaline phosphatase-labeled protein A.
Oligomer formation is a common feature of glycan-binding receptors and can be an important determinant of the way that they interact with glycans on cell surfaces (12, 20, 21). Many receptors form dimers, and DC-SIGN forms tetramers, but they are noncovalent and the neck sequence that supports oligomer formation in DC-SIGN is unrelated to the sequence of the neck in prolectin (22).
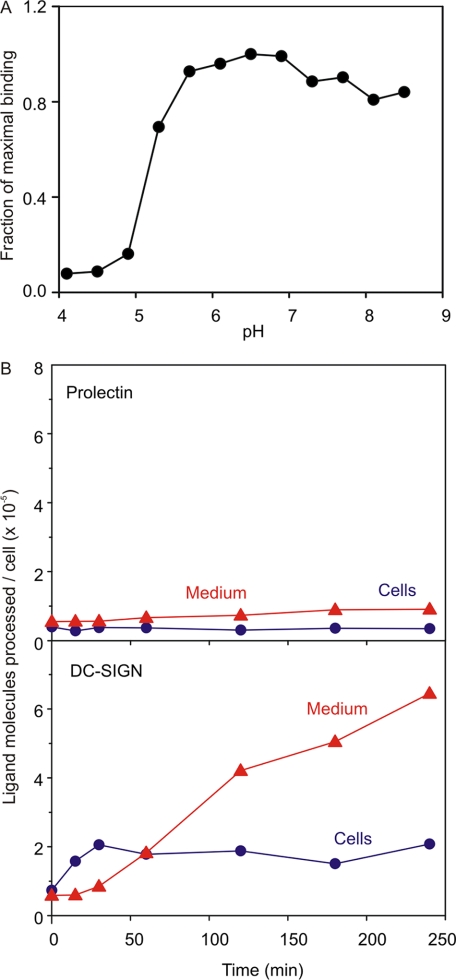
Because many oligomeric type II receptors that contain C-type CRDs mediate endocytosis (11, 16, 17), it was of interest to see if prolectin supports endocytic activity. The pH profile of ligand binding in the solid-phase assay showed that ligand release occurred at low pH, but that half-maximal release occurred at pH 5.2 (Fig. 5A). This value is substantially lower than the typical value for recycling endocytic receptors such as DC-SIGN and the hepatic asialoglycoprotein receptor (16, 23). When 125I-Man-BSA was incubated with the transfected fibroblasts, no uptake and processing of ligand was observed, providing a sharp contrast with DC-SIGN transfectants assayed in parallel (Fig. 5B).
FIGURE 5.
Trafficking of prolectin expressed in fibroblasts. A, pH dependence of ligand binding. Binding of 125I-Man-BSA to immobilized extracellular domain of prolectin was assayed at various pHs in the presence of 2 mm Ca2+. B, uptake and degradation of 125I-Man-BSA by fibroblasts expressing prolectin or DC-SIGN. Degradation was measured as acid-soluble fragments appearing in the medium.
The Cytoplasmic Domain of Prolectin Communicates with Signaling Molecules
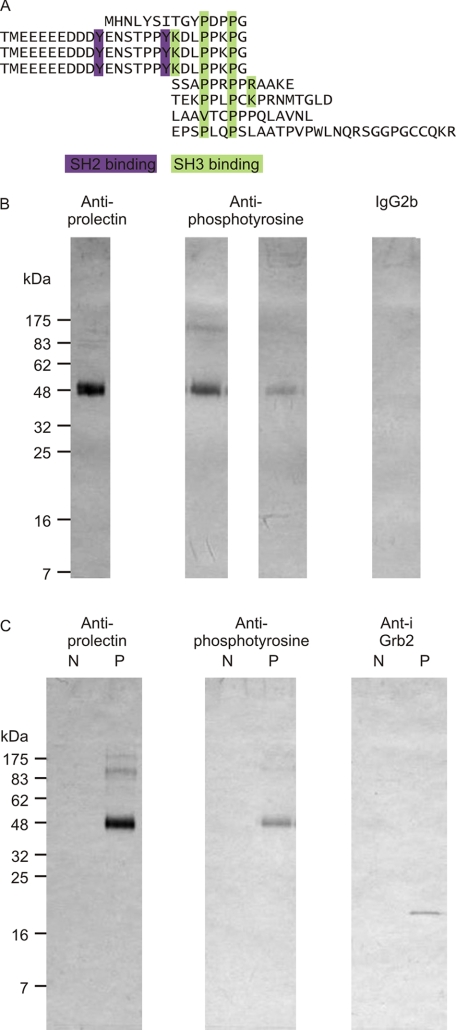
Although prolectin does not support endocytosis, examination of the cytoplasmic domain suggested the alternative possibility that it has signaling capacity, because of the presence of numerous sequences predicted to bind SH2 and SH3 domains (Fig. 6A). In the oligomeric protein, these sequences would form large clusters of potential binding sites for adapter molecules that could link the receptor to intracellular signaling pathways. The SH2-binding sequences contain tyrosine residues that would be expected to be targets for tyrosine kinases and antibody probing of blots of prolectin purified from the fibroblasts confirmed the presence of phosphotyrosine (Fig. 6B).
FIGURE 6.
Potential signaling interactions of prolectin. A, potential binding motifs for SH2 and SH3 domains in the cytoplasmic domain of prolectin. B, prolectin phosphorylation. Aliquots of prolectin-containing fractions from mannose-Sepharose columns of extracts from transfected fibroblasts were separated on 17.5% SDS-polyacrylamide gels, which were blotted and probed with antibody to the CRD of prolectin, two different anti-phosphotyrosine monoclonal antibodies (left, 4G10; right, PY20) and a control mouse IgG2b antibody. C, cytoplasmic tail interactions. Aliquots of elution fractions from mannose-Sepharose columns of extracts from fibroblasts transfected with empty vector (N) and with vector containing the prolectin cDNA (P) were separated on 17.5% SDS-polyacrylamide gels, which were blotted and probed with antibody to the CRD of prolectin, anti-phosphotyrosine (monoclonal antibody 4G10), and affinity-purified polyclonal antibody to Grb2.
The sequence context of the phosphorylated tyrosine residues suggests potential interactions with specific classes of cytoplasmic signaling molecules (24). In particular, the ubiquitous adapter protein Grb2 contains an SH2 domain that binds the target sequence YXNX (25). Interestingly, Grb2 also contains class I SH3 domains that interact with the target SH3-binding motifs identified in prolectin. Consistent with these predictions, antibody blotting of the receptor-containing fractions from the mannose-Sepharose affinity column demonstrated that Grb2 co-purifies with prolectin (Fig. 6C), confirming that prolectin interacts with intracellular signaling machinery either directly or indirectly.
Prolectin Is Expressed on Dividing B Cells of Germinal Centers
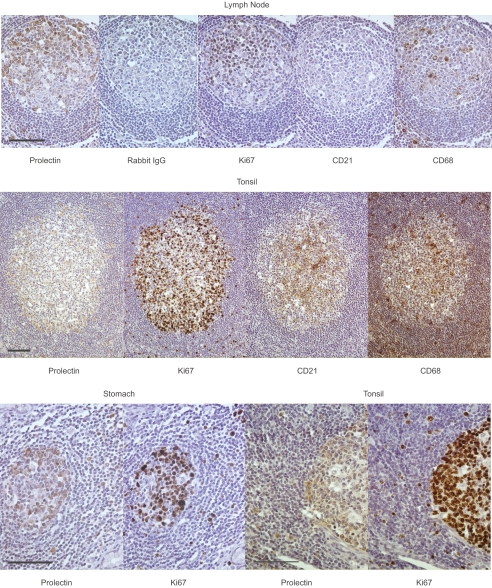
Immunohistochemistry using the affinity-purified polyclonal rabbit antibodies was used to screen an array of human tissues to define sites of expression. The results revealed a very restricted pattern of staining, with the major site of expression being germinal centers in various tissues, including lymph nodes, tonsils, stomach, intestine, appendix, and spleen (Fig. 7). Antibodies selective for follicular dendritic cells or tingible body macrophages found in the light zone of the germinal centers do not co-localize with prolectin. Thus, unlike other C-type lectins such as DC-SIGN and the mannose receptor, prolectin is not found on dendritic cells or macrophages. In contrast, Ki67 antigen found in nuclei of dividing B cells shows extensive overlap with the prolectin staining, suggesting a novel localization of this receptor in B cells.
FIGURE 7.
Immunohistochemical localization of prolectin. Germinal centers from multiple tissues were probed with polyclonal antibodies to prolectin, control rabbit IgG, or monoclonal antibodies specific for dividing cell nuclear antigen Ki67, dendritic cell marker CD21, and macrophage marker CD68. Antigens were visualized with peroxidase immunochemistry. Scale bars, 100 μm.
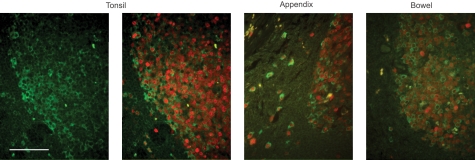
Co-localization of prolectin with the dividing B cell marker Ki67 was confirmed using immunofluorescence (Fig. 8). In germinal centers from multiple tissues, essentially all prolectin-expressing cells are also Ki67-positive. Based on their location and reaction with the anti-Ki67 antibody, the clustered, doubly stained cells in the dark zone appear to be centroblasts. Prolectin expression thus defines a sub-population of dividing B cells of the dark zone. In the surrounding mantle zone, the occasional Ki67-positive cells have a similar morphology to the prolectin-positive cells in the dark zone and all of these cells co-express prolectin.
FIGURE 8.
Comparison of prolectin and Ki67 antigen distribution. Germinal centers were stained with antibody to prolectin and counterstained with Alexa 488 secondary antibody (green). In the three images on the right, sections were also stained with Ki67 antibody and counterstained with Alexa 594 secondary antibody (red). Scale bar, 100 μm.
DISCUSSION
Although prolectin has some similarity to other glycan-binding receptors containing C-type CRDs, it has several properties that distinguish it from other known members of this group. Importantly, the fact that prolectin does not display endocytic activity is consistent with its expression in B cells rather than macrophages or dendritic cells. This pattern of expression and lack of endocytic activity makes it unlikely that prolectin functions as a primary recognition receptor in the innate immune response, a role that is commonly ascribed to DC-SIGN, langerin, the mannose receptor, and other receptors that contain C-type CRDs (1, 26–28). Similarly, in contrast to the selectin cell adhesion molecules, prolectin bears multiple signaling motifs in the cytoplasmic domain, so that it can interact directly with signaling pathways within lymphocytes, although it may also have a role in cell adhesion. Thus, prolectin does not readily fall into any of the previously described categories of C-type lectins, and it does not have any obvious paralogs in mammalian genomes.
Despite these differences, the glycan array results suggest that that there is likely to be overlap in potential ligands for prolectin and DC-SIGN, because both receptors display a dual specificity for mannose- and fucose-terminated glycans. Common types of ligands would include surface glycans on other cells bearing Lewisa, Lewisx, and Lewisy epitopes (16, 29). It has been proposed that these structures are presented to DC-SIGN on intercellular adhesion molecule 3 (ICAM-3) on T cells, leading to adhesion between the T cells and dendritic cells (30). By analogy, one function of prolectin may be to participate in interaction of the B cells on which it is expressed with T cells or other cells in the germinal center. However, the potential ability of prolectin to initiate signaling in response to a glycan ligand indicates that it probably has additional roles in communication between B cells and other cells in germinal centers.
The pattern of expression of prolectin within the germinal centers may provide some additional clues about its potential functions. Following somatic mutation of antibody genes in centroblasts of the dark zone, centrocytes are known to migrate to the light zone where selection of cells with enhanced affinity for antigen occurs and cells with dysfunctional immunoglobulin genes undergo apoptosis (31). The disposition of clusters containing many of the prolectin-positive cells near the edge of the dark zone, and the presence of individual cells in the adjacent regions of the mantle zone suggests that cells expressing prolectin might be in transit into or out of the germinal center and that the receptor may play a role in migration of cells between these compartments during antibody affinity maturation.
Supplementary Material
Acknowledgments
We thank David Guiliano, Niki Gounaris, and Murray Selkirk of the Division of Molecular and Cellular Biology, Imperial College, for providing access to the fluorescence microscope and David Smith, Department of Biochemistry, Emory University School of Medicine, for performing the glycan array analysis.
This work was supported by Grant 075565 from the Wellcome Trust (to M. E. T. and K. D.). This work was also supported, in part, by National Institutes of Health Grant GM062116 to the Consortium for Functional Glycomics.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Table S1.
- DC-SIGN
- dendritic cell-specific intercellular adhesion molecule 3-grabbing non-integrin
- CRD
- carbohydrate-recognition domain
- 125I-Man-BSA
- iodinated, mannose-conjugated bovine serum albumin.
REFERENCES
- 1.Weis W. I., Taylor M. E., Drickamer K. ( 1998) Immunol. Rev. 163, 19– 34 [DOI] [PubMed] [Google Scholar]
- 2.van Kooyk Y., Rabinovich G. A. ( 2008) Nat. Immunol. 9, 593– 601 [DOI] [PubMed] [Google Scholar]
- 3.Vestweber D., Blanks J. E. ( 1999) Physiol. Rev. 79, 181– 213 [DOI] [PubMed] [Google Scholar]
- 4.Rosen S. D. ( 2004) Annu. Rev. Immunol. 22, 129– 156 [DOI] [PubMed] [Google Scholar]
- 5.Crocker P. R., Paulson J. C., Varki A. ( 2007) Nat. Rev. Immunol. 7, 255– 266 [DOI] [PubMed] [Google Scholar]
- 6.Drickamer K., Taylor M. E. ( 2003) Methods Enzymol. 362, 560– 567 [DOI] [PubMed] [Google Scholar]
- 7.Mitchell D. A., Fadden A. J., Drickamer K. ( 2001) J. Biol. Chem. 276, 28939– 28945 [DOI] [PubMed] [Google Scholar]
- 8.Fornstedt N., Porath J. ( 1975) FEBS Lett. 57, 187– 191 [DOI] [PubMed] [Google Scholar]
- 9.Laemmli U. K. ( 1970) Nature 227, 680– 685 [DOI] [PubMed] [Google Scholar]
- 10.Powlesland A. S., Ward E. M., Sadhu S. K., Guo Y., Taylor M. E., Drickamer K. ( 2006) J. Biol. Chem. 281, 20440– 20449 [DOI] [PubMed] [Google Scholar]
- 11.Stambach N. S., Taylor M. E. ( 2003) Glycobiology 13, 401– 410 [DOI] [PubMed] [Google Scholar]
- 12.Weis W. I., Drickamer K. ( 1996) Annu. Rev. Biochem. 65, 441– 473 [DOI] [PubMed] [Google Scholar]
- 13.Lennon G., Auffray C., Polymeropoulos M., Soares M. B. ( 1996) Genomics 33, 151– 152 [DOI] [PubMed] [Google Scholar]
- 14.Thanaraj T. A., Clark F. ( 2001) Nucleic Acids Res. 29, 2581– 2593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Puntervoll P., Linding R., Gemünd C., Chabanis-Davidson S., Mattingsdal M., Cameron S., Martin D. M., Ausiello G., Brannetti B., Costantini A., Ferrè F., Maselli V., Via A., Cesareni G., Diella F., Superti-Furga G., Wyrwicz L., Ramu C., McGuigan C., Gudavalli R., Letunic I., Bork P., Rychlewski L., Küster B., Helmer-Citterich M., Hunter W. N., Aasland R., Gibson T. J. ( 2003) Nucleic Acids Res. 31, 3625– 3630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Guo Y., Feinberg H., Conroy E., Mitchell D. A., Alvarez R., Blixt O., Taylor M. E., Weis W. I., Drickamer K. ( 2004) Nat. Struct. Mol. Biol. 11, 591– 598 [DOI] [PubMed] [Google Scholar]
- 17.Coombs P. J., Graham S. A., Drickamer K., Taylor M. E. ( 2005) J. Biol. Chem. 280, 22993– 22999 [DOI] [PubMed] [Google Scholar]
- 18.Feinberg H., Mitchell D. A., Drickamer K., Weis W. I. ( 2001) Science 294, 2163– 2166 [DOI] [PubMed] [Google Scholar]
- 19.Wurzburg B. A., Tarchevskaya S. S., Jardetzky T. S. ( 2006) Structure 14, 1049– 1058 [DOI] [PubMed] [Google Scholar]
- 20.Drickamer K. ( 1999) Curr. Opin. Struct. Biol. 9, 585– 590 [DOI] [PubMed] [Google Scholar]
- 21.Dam T. K., Brewer C. F. ( 2008) Biochemistry 47, 8470– 8476 [DOI] [PubMed] [Google Scholar]
- 22.Feinberg H., Guo Y., Mitchell D. A., Drickamer K., Weis W. I. ( 2005) J. Biol. Chem. 280, 1327– 1335 [DOI] [PubMed] [Google Scholar]
- 23.Wragg S., Drickamer K. ( 1999) J. Biol. Chem. 274, 35400– 35406 [DOI] [PubMed] [Google Scholar]
- 24.Pawson T., Scott J. D. ( 1997) Science 278, 2075– 2080 [DOI] [PubMed] [Google Scholar]
- 25.Chardin P., Cussac D., Maignan S., Ducruix A. ( 1995) FEBS Lett. 369, 47– 51 [DOI] [PubMed] [Google Scholar]
- 26.van Kooyk Y., Geijtenbeek T. B. H. ( 2003) Nat. Rev. Immunol. 3, 697– 709 [DOI] [PubMed] [Google Scholar]
- 27.de Witte L., Nabatov A., Pion M., Fluitsma D., de Jong M. A., de Gruijl T., Piguet V., van Kooyk Y., Geijtenbeek T. B. ( 2007) Nat. Med. 13, 367– 371 [DOI] [PubMed] [Google Scholar]
- 28.Taylor P. R., Gordon S., Martinez-Pomares L. ( 2005) Trends Immunol. 26, 104– 110 [DOI] [PubMed] [Google Scholar]
- 29.Appelmelk B. J., van Die I., van Vliet S. J., Vandenbroucke-Grauls C. M., Geijtenbeek T. B., van Kooyk Y. ( 2003) J. Immunol. 170, 1635– 1639 [DOI] [PubMed] [Google Scholar]
- 30.Geijtenbeek T. B., Torensma R., van Vliet S. J., van Duijnhoven G. C., Adema G. J., van Kooyk Y., Figdor C. G. ( 2000) Cell 100, 575– 585 [DOI] [PubMed] [Google Scholar]
- 31.Allen C. D., Okada T., Cyster J. G. ( 2007) Immunity 27, 190– 202 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.