Abstract
The cell surface receptor tyrosine kinase HER2/neu enhances tumor metastasis. Recent studies suggest that deregulated microRNA (miRNA) expression promotes invasion and metastasis of cancer cells; we therefore explored the possibility that HER2/neu signaling induces the expression of specific miRNAs involved in this process. We identified a putative oncogenic miRNA, miR-21, whose expression is correlated with HER2/neu up-regulation and is functionally involved in HER2/neu-induced cell invasion. We show that miR-21 is up-regulated via the MAPK (ERK1/2) pathway upon stimulation of HER2/neu signaling in breast cancer cells, and overexpression of other ERK1/2 activators such as RASV12 or ID-1 is sufficient to induce miR-21 up-regulation in HER2/neu-negative breast cancer cells. Furthermore, the metastasis suppressor protein PDCD4 (programmed cell death 4) is down-regulated by miR-21 in breast cancer cells expressing HER2/neu. Our data reveal a mechanism for HER2/neu-induced cancer cell invasion via miRNA deregulation. In addition, our results identify miR-21 as a potential therapeutic target for the prevention of breast cancer invasion and metastasis.
The HER2/neu (c-erbB-2) proto-oncogene encodes a transmembrane protein-tyrosine kinase growth factor receptor, p185HER2, which is a member of the human epidermal growth factor receptor family. HER2/neu overexpression is found in about 30% of human breast cancers and several other cancer types. HER2/neu overexpression is associated with a poor clinical outcome, including a positive correlation with metastasis (1, 2). The involvement of HER2/neu in metastasis is supported by studies demonstrating that HER2/neu increases the metastatic potential of human and murine cancer cell lines (3) and induces lung metastasis in transgenic animal models (4). Additionally, HER2/neu signaling up-regulates genes that play important roles in cell invasion and metastasis, such as cyclooxygenase-2, CXCR4, and matrix metalloproteinases (5–7). Given the complex signaling network initiated by HER2/neu overexpression in cancer cells, it is likely that HER2/neu regulates additional unidentified players involved in these processes.
miRNAs4 constitute a class of 21 or 22 nucleotides noncoding RNAs that play an important role in development and cellular processes. Aberrant expression of miRNAs is associated with cancer (8), suggesting that some miRNAs can function as tumor suppressor genes or oncogenes. miRNAs may also cooperate with the loss of tumor suppressors or overexpression oncogenes in cancer cells to contribute to a fully malignant phenotype. Up-regulation of several miRNAs in breast cancer cells, such as miR-21 and miR-10b, can increase cell invasion and metastasis (9, 10). HER2/neu signaling activates a variety of transcription factors, such as AP-1, Myc, and NF-κB that alter miR-21 and other miRNA transcription (8, 11, 12). We therefore hypothesize that HER2/neu signaling may induce the expression of specific miRNAs, which contribute to the increased metastatic potential of HER2/neu-overexpressing cancer cells.
Here we describe a putative oncogenic miRNA, miR-21, whose expression is correlated with HER2/neu up-regulation. We found that HER2/neu signaling up-regulates miR-21 via the MAPK (ERK1/2) pathway and that its increased expression promotes cell invasion. Furthermore, miR-21 suppressed expression of the metastasis suppressor PDCD4 in HER2/neu-expressing breast cancer cells. Our data show a new mechanism by which HER2/neu induces cell invasion via miR-21 deregulation in breast cancer cells. This raises the possibility that anti-miR-21 therapies might exhibit a synergistic effect with other anti-HER2/neu therapeutics, for example Herceptin, in treating HER2/neu-associated breast cancers.
EXPERIMENTAL PROCEDURES
Cell Lines and DNA Constructs
Human breast cancer cell line SKBR3 (ATCC) was cultured in McCoy's 5A medium with 10% fetal bovine serum. BT-474 (ATCC) was cultured in RPMI 1640 with 10% fetal bovine serum. The HeLa (ATCC) cell line and its HER2/neu transfectant were maintained in Dulbecco's modified Eagle's medium with 10% fetal bovine serum. The human breast cancer cell line MDA-MB-435 transfected with a neo vector or a HER2-expressing vector were gifts from Dr. Mien-Chie Hung (M. D. Anderson Cancer Center, Houston, TX) and have been characterized and described previously (6). These cells were maintained in Dulbecco's modified Eagle's medium/F-12 supplemented with 10% fetal bovine serum and 400 μg/ml geneticin (Invitrogen). pEBS7 and pEBS7-HER2/neu vectors were gifts from Dr. Mikala Egeblad (13). Ets-1 small hairpin RNA-expressing vector was obtained from SuperArray Bioscience Corp. (Frederick, MD). Replication-defective mouse stem cell (MSCV-IRES-GFP) retrovirus encoding control vector, activated H-Ras (G12V), activated myristoylated AKT, Myc, or ID-1 were prepared as previously described (14). Nontransformed human MCF10A breast epithelial cells were infected with each of the recombinant retrovirus to generate stable cell populations. Greater than 80% of MCF10A stable cell populations expressed cis-linked GFP fluorescence marker as judged by fluorescence microscopy. The wild type ERK2 and dominant negative ERK2 were gift from Dr. Mien-Chie Hung (M. D. Anderson Cancer Center) and have been characterized and described previously (15). To construct the mCherry miR-21 and mCherry miR-155 sensor vectors, the GFP gene in the pSicoR vector (16) was replaced with the mCherry gene using the Age1 and BsrG1 sites. The oligonucleotides with either two complementary miR-21-binding sites (5′-GTACATCAACATCAGTCTGATAAGCTACCCGGGTCAACATCAGTCTGATAAGCTAG-3′) or miR-155-binding sites (5′-GTACACCCCTATCACGATTAGCATTAACCCGGGCCCCTATCACGATTAGCATTAAG-3′) were cloned downstream the mCherry gene using the BsrGI and EcoRI sites to produce the mCherry miR-21 sensor vector and the mCherry miR-155 sensor vector, respectively. To construct the miR-21 expression vector, 550 bp including the miR-21 pri-cursor was amplified by PCR from human genomic DNA with the forward primer 5′-AGTTTTCTTGCCGTTCTGTAAGT-3′ and the reverse primer 5′-GTCATGAAGACTATCCCCATTTC-3′. The final PCR product was ligated into a PCR2.1 TOPO TA cloning vector (Invitrogen) according to the manufacturer's instructions. The resulting vector was sequence verified and subsequently digested with BamHI and XhoI to release the DNA fragment, which was inserted into a modified pSicoR vector downstream from the GFP gene.
Microarray Analysis
1,544 modified oligonucleotides with C6 5′ amino linkers (IDT) were printed in triplicate using a microarrayer on house-made poly-l-lysine slides (17). The probes were reverse complements of annotated (18) and predicted miRNAs (19), with some probes shortened to standardize melting temperature. Probe sequences are listed in supplemental Table S1. Before hybridization, the arrays were prehybridized, and probes were UV cross-linked. miRNA microarrays were performed essentially as described (20) with the following modifications. RNA was extracted with TRIzol (Invitrogen) according to the manufacturer's protocol. 5 μg of total RNA was added to a ligation reaction containing either 250 ng of 5′-phospate-citidyl-uridyl-Cy3-3′ or 1 μg of 5′- phosphate-citidyl-citidyl-uridyl-Dy647-3′ (Dharmacon), and labeling was allowed to proceed overnight on ice. Two samples labeled with different dyes were then combined and precipitated with ethanol. Precipitated RNA was resuspended in 120 μl of hybridization buffer and hybridized for 2 h on arrays using a 1.7 × 2.8-cm Gene Frame (ABgene, Rochester, NY). The arrays were washed and then immediately scanned using a GenePix 4000B scanner (Axon Instruments, Sunnyvale, CA). The data were extracted, and local background was subtracted using Genepix Pro-4 software (Axon Instruments). The data were processed in Excel to calculate the median intensity for each set of triplicate probes in each channel. The displayed data (see Fig. 1B) summarizes the results from three arrays in which labeling of each sample was done with a different fluorophore on each array, known as dye swapping, to eliminate dye bias. The mean fluorescence for each probe from the two arrays was normalized to the maximum mean fluorescence for each sample. Only the 330 probes on the array corresponding to annotated human miRNAs are shown.
FIGURE 1.
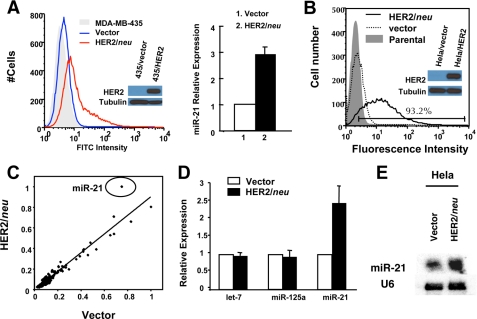
Ectopic expression of HER2/neu induces miR-21 up-regulation. A, MDA-MB-435 cells transfected with neo vector (blue line) or MDA-MB-435 transfected with HER2/neu-expressing vector (red line) were stained with fluorescein isothiocyanate (FITC)-conjugated anti-human HER2/neu antibody (left panel), and the miR-21 expression level in the MDA-MB-435 cells was assessed by using qRT-PCR (right panel). The cell lysates were separated by SDS-PAGE and analyzed by Western blot using anti-HER2/neu antibody or anti-tubulin antibody. B, HER2/neu expression in HeLa cells transfected with different vectors. HeLa cell line (gray histogram), HeLa cell line transfected with pEBS7 empty vector (dashed line), or HeLa cell line transfected with pEBS7-HER2/neu (black line) were stained with fluorescein isothiocyanate-conjugated anti-human HER2/neu antibody. The cell lysates were separated by SDS-PAGE and analyzed by Western blot using anti-HER2/neu antibody or anti-tubulin antibody. C, miRNA microarray analysis of the effect of HER2/neu overexpression. RNA from HeLa cells transfected with pEBS7 and pEBS7-HER2/neu were labeled with different fluorophores and competitively hybridized on microarrays. Each spot in the scatter plot represents the normalized mean fluorescence for one microRNA probe from two dye swapped arrays. D and E, RNA used in B was analyzed by qRT-PCR to assay expression of miR-21 (D) and in a separate experiment by Northern blot analysis under the same conditions (E). The mean and standard error from triplicate experiments are indicated.
Antibodies and Reagents
Fluorescein isothiocyanate-conjugated anti-human HER2/neu antibody was obtained from Abcam Inc. (Cambridge, MA). AKT, phospho-AKT (Ser473), and hemagglutinin tag (clone 6E2) are from Cell Signaling (Danvers, MA). Ras (Ab-1) is from Thermo Scientific (Waltham, MA). Myc (clone Y69) is from Epitomics (Burlingame, CA). β-Actin (clone AC-15) is from Sigma. HER2/neu antibody for Western blot is from Calbiochem (Gibbstown, NJ). The HER2/neu agonist antibody anti-HER2/neu ScFv-TNF-α (S147Y) was a gift from Dr. Sherie Morrison (UCLA, Los Angeles) and has been characterized and described previously (21). The Akt inhibitor LY294002 and ERK1/2 inhibitor U0126 were obtained from Cell Signaling.
Real Time PCR
For real time PCR of primary miR-21 transcripts (pri-miR-21) and Ets-1, total RNA was prepared using the mirVana miRNA isolation kit (Ambion, Austin, TX) and quantified by Nanodrop (Wilmington, DE). Reverse transcription was performed with a Superscript III first strand synthesis system for real time PCR (Invitrogen). Real time PCR was performed using 7900HT Fast real time PCR system (Applied Biosystems, Foster City, CA). The pri-miR-21 was amplified using the forward primer 5′-CATTGTGGGTTTTGAAAAGGTTA-3′ and the reverse primer 5′-CCACGACTAGAGGCTGACTTAGA-3′, and the specificity of the pri-miR-21 amplification was validated previously (22). The human Ets-1 gene was amplified using the forward primer 5′-AGGAGATGGGGAAAGAGGAA-3′ and the reverse primer 5′-AGCGGTACACGTAGCGTTTC-3′. The HPRT1 gene was amplified using the forward primer 5′-TGACACTGGCAAAACAATGCA-3′ and the reverse primer 5′-GGTCCTTTTCACCAGCAAGCT-3′, which was validated previously (23) and served as an internal control during the pri-miR-21 and Ets-1 mRNA amplification. The PCR was established with the Power SYBR Green PCR Master Mix (Applied Biosystems), and only one PCR product was generated in these reactions. PCR-based detection of mature miR-21 and other microRNAs was performed by the TaqMan miRNA assays (Applied Biosystems) as described previously (24). The PCR results were normalized with U6 RNA as an internal control and then expressed as relative expression compared with untreated samples.
Fluorescence-activated Cell Sorter Analysis
For detection of HER2/neu expression, the cells were treated with fluorescein isothiocyanate-conjugated anti-ErbB2 antibody (Abcam) at 4 °C for 30 min and analyzed with a LSRII flow cytometer (BD, Franklin Lakes, NJ). For detection of mCherry expression, the cells were directly analyzed with the LSRII flow cytometer.
Northern Blot
For detection of miR-21 by Northern blotting, RNA was extracted by using the TRIzol reagent according to the manufacturer's protocol (Invitrogen). 10 μg of total RNA was electrophoretically separated on a 10% polyacrylamide denaturing gel. Subsequently RNA was transferred to a Hybond-N+ membrane (Amersham Biosciences) by using a semi-dry Transblot electrophoresis apparatus (Bio-Rad). The RNA was cross-linked to the membrane by using UV radiation. Hybridization was carried out by using ULTRAHybOligo solution according to the manufacturer's instructions (Ambion). The probe sequence was complementary to the mature form of miR-21 and was labeled with γ-32P. After washing, the membranes were imaged using phosphorimaging.
Western Blot
Western blot analysis of PDCD4 expression was performed as previously described (25). In brief, the protein concentration was determined by protein assay kit (Bio-Rad), and the samples were separated in 9% SDS-polyacrylamide gels. After probing with a primary antibody, the membrane was incubated with secondary antibodies labeled with IRDye 800CW. Finally; signal intensity was determined with the Odyssey infrared imaging system and associated software (LI-COR Biosciences, Lincoln, NE). Relative expression of PDCD4 was normalized against the internal control β-actin.
Transfection
Transfection of MDA-MB-435 and BT-474 cells with anti-miR-21 or a control inhibitor (Dharmacon, Chicago, IL) was performed using the DharmaFECT small interfering RNA transfection reagents (Dharmacon) according to the manufacturer's protocol. Transfection of HeLa cells and BT-474 cells with DNA plasmids was performed using Lipofectamine 2000 according to the manufacturer's protocol (Invitrogen).
Virus Preparation
Lentiviruses were generated essentially as previously described (16). Briefly, 4 μg of lentiviral vector and 4 μg of a mixture of packaging vectors were cotransfected in 293T cells using FuGENE 6 (Roche Applied Science). The supernatants were collected 48–96 h after transfection, filtered through a 0.45-μm filter, and used directly to infect cancer cells.
Invasion Assay
The invasive potential of cells was measured in 24-well Matrigel-coated invasion chambers following the manufacturer's instructions (Millipore, Billerica, MA). After the cells were incubated for 24–48 h at 37 °C in a humidified incubator with 5% CO2, the noninvading cells that remained on the upper surface of the membrane were removed by scraping. Invading cells were stained using 4′,6-diamidino-2-phenylindole and counted in three randomly selected fields in the center of each membrane under fluorescence microscopy at medium power fields (100× or 200×).
Immunofluorescence Staining Assay
To detect the effect of HER2/neu on the direct miR-21 target PDCD4 expression at the cellular level, both MDA-MB-435/HER2 MDA-MB-435/Neo cells were directly immunostained with anti-PDCD4 antibody. Alternatively, MDA-MB-435/HER2 cells were transfected with anti-miR-21, or MDA-MB-435/Neo cells were transfected with miR-21 expression vector, followed by immunostaining with anti-PDCD4 antibody. The transfected cells were seeded on cover slides 24 h after transfection and fixed with 3% paraformaldehyde as described previously (26). After incubation with primary PDCD4 antibody (Rockland, Gilbertsville, PA), second antibody conjugated with Alexa Fluor 560 nm (Invitrogen) was used to reveal PDCD4 signals. The images were taken under a fluorescent microscope (Olympus, Center Valley, PA).
RESULTS
Ectopic Expression of HER2/neu Induces miR-21 Up- regulation
Overexpression of HER2/neu enhances local tumor invasion and lung metastasis of breast cancer cells (4). Previously, miR-21 and other MicroRNAs have been shown to increase the metastatic potential of breast cancers (9, 10), and HER2/neu signaling activates several transcription factors that have been shown to regulate miR-21 expression in various cancer cell lines (11, 22). We therefore explored the possibility that HER2/neu signaling induces the expression of miR-21 and/or other microRNAs, which could in turn contribute to the increased metastatic potential of these cancer cells.
We first examined whether miR-21 level is up-regulated in the HER2/neu-transfected MDA-MB-435 human breast cancer cell line, because overexpression of HER2/neu significantly enhances invasion and metastasis in this cell line without altering its cell growth rates, anchorage-independent growth, and other indices of tumorigenic potential (3). We observed that HER2/neu expression was sufficient to up-regulate mature miR-21 levels 2.9 ± 0.3-fold in MDA-MB-435 cells (Fig. 1A).
To verify that HER2/neu is sufficient to induce miR-21 up-regulation in other cancer cells and identify other HER2/neu-dependent microRNAs, we assessed the changes in miRNA expression in the HeLa/HER2 cells compared with vector control cells by using miRNA microarrays. We chose HeLa cells because of the fact that some cervical carcinoma tumors overexpress HER2/neu (27), and the transfection of HER2/neu expression vector is more feasible and efficient in this cell line.
A HeLa cell line was generated that stably expresses HER2/neu (Fig. 1B), and surprisingly, only a single signature from the array was significantly elevated in the HeLa/HER2 cells: the highly conserved miR-21 miRNA (Fig. 1C). This observation was validated using qRT-PCR, which showed that mature miR-21 expression in HeLa/HER2 cells was increased 2.5 ± 0.5-fold compared with control cells (Fig. 1D). Northern blot analysis further confirmed these analyses, showing a moderate but significant up-regulation of miR-21 expression (Fig. 1E). Our results demonstrate that HER2/neu overexpression up-regulated miR-21 significantly in both MDA-MB-435 and HeLa cells.
HER2/neu Signaling Induced miR-21 Up-regulation with Distinct Kinetics in Different Cancer Cell Lines
HER2/neu overexpression may stabilize the miR-21 level in breast cancer cells, thus resulting in higher miR-21 level compared with control cells. To examine whether HER2/neu signaling is able to induce de novo miR-21 expression, we followed the kinetics of miR-21 induction by assessing the relative abundance of its primary transcript and mature sequence upon HER2/neu signaling in different cancer cells. A HER2/neu agonist antibody (anti-HER2/neu ScFv-TNF-α (S147Y)) was used to induce HER2/neu signaling in various HER2/neu-expressing cells lines (21). Anti-HER2/neu ScFv-TNF-α (S147Y) (HER2/neu agonist) is a ScFv antibody specific for human HER2/neu fused to a mutant TNF-α with abrogated biological activity. This HER2/neu agonist has previously been shown to form a homotrimeric structure via the noncovalent binding of the mutant TNF-α and is therefore able to cross-link multiple HER2/neu receptors, thereby potently stimulating the tyrosine phosphorylation of HER2/neu receptor and the downstream AKT and MAPK signaling (21).
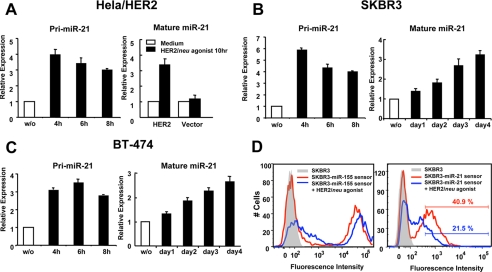
HER2/neu agonist treatment induced a rapid and substantial pri-miR-21 up-regulation in HeLa/HER2 (Fig. 2A), SKBR3 (Fig. 2B), and BT-474 (Fig. 2C) cells after 4 h, and the level of pri-miR-21 slightly decreased after 8 h of stimulation. In contrast, the kinetics of mature miR-21 processing in these cells is highly distinct. HER2/neu agonist treatment of HeLa/HER2 cells for 10 h caused a 3.4 ± 0.4-fold up-regulation of mature miR-21 expression as compared with untreated HeLa/HER2 cells. In contrast, no significant miR-21 up-regulation was observed in vector control cells treated with the HER2/neu agonist as compared with untreated cells (Fig. 2A). In contrast to HeLa/HER2 cells, up-regulation of mature miR-21 was delayed in the HER2/neu-overexpressing breast cancer cell lines SKBR3 (Fig. 2B) and BT-474 (Fig. 2C). Mature miR-21 was increased 3.2 ± 0.2- and 2.6 ± 0.2-fold in SKBR3 and BT-474 cells, respectively, stimulated with HER2/neu agonist for 4 days. These results suggest that HER2/neu signaling induces rapid transcription of pri-miR-21; however, processing to mature miR-21 is less efficient in SKBR3 and BT-474 breast cancer cells. Recently it has been shown that the expression of DICER1 is low in several HER2/neu-positive tumors (28), which may explain the delay in achieving increased mature miR-21 expression in SKBR3 and BT-474 cells.
FIGURE 2.
HER2/neu signaling induced miR-21 up-regulation with distinct kinetics in different cancer cell lines. Real time PCR analysis of miR-21 pri-cursor (pri-miR-21) and mature miR-21 expression in HeLa-HER2/neu or HeLa-vector cells (A), SKBR3 cells (B), and BT-474 cells (C) stimulated with 100 nm HER2/neu agonist at varying time points. D, SKBR3 cells infected with lentivirus carrying a mCherry gene with two complementary miR-21 or miR-155 binding sites in the 3′-untranslated region (mCherry miR-21 sensor or mCherry miR-155 sensor, respectively) were stimulated with 100 nm HER2/neu agonist for 4 days, and the fluorescent signal was detected by flow cytometry. The mean and standard error from triplicate experiments are indicated.
Given the significant but moderate level of miR-21 up-regulation, we wanted to determine whether a ∼3-fold change in miR-21 expression would mitigate a change in target mRNA expression. To study the potential functional consequence of miR-21 up-regulation in response to HER2/neu signaling, we took advantage of a lentivirus construct encoding a fluorescent reporter gene (mCherry) containing two exact miR-21-binding sites in its 3′-untranslated region (mCherry miR-21 sensor). A similar construct with miR-155-binding sites (mCherry miR-155 sensor) was used as a control vector because miR-155 levels do not change significantly in response to HER2/neu expression (data not shown). SKBR3 cells infected with the mCherry miR-155 sensor exhibited strong reporter expression, indicating that the miR-155 level is very low or undetectable in HER2/neu-overexpressing breast cancer cells (Fig. 2D, left panel). Upon treatment with the HER2/neu agonist, the mCherry miR-155 sensor exhibited a slight increase of the reporter signal (Fig. 2D, left panel), probably because of the widespread activation of transcription factors induced by HER2/neu signaling. In contrast, SKBR3 cells infected with the mCherry miR-21 sensor exhibited a moderate reporter signal, indicating that the mCherry mRNA sequence is silenced by endogenous miR-21 expression in these cells (Fig. 2D, right panel). After treatment with HER2/neu agonist, the miR-21 sensor exhibited an even greater repression (50%) of the reporter signal (Fig. 2D, right panel), indicating that the ∼3-fold miR-21 expression level change induced by the HER2/neu signaling is sufficient to repress target gene expression and may therefore have a functional consequence in cancer cells by repressing endogenous target mRNAs.
HER2/neu Regulates miR-21 Expression via the MAPK (ERK1/2) Pathway
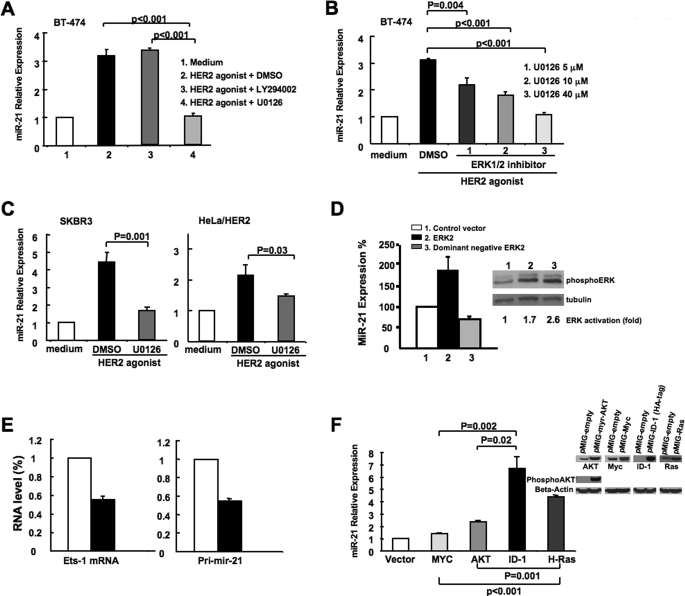
The RAS-MEK1/2-ERK1/2 and phosphatidylinositol 3-kinase-AKT pathways are two major signaling cascades downstream of activated HER2/neu. To test which pathway is responsible for the miR-21 up-regulation, we treated HER2/neu-overexpressing cells BT-474 with an inhibitor of ERK1/2 (U0126) or AKT (LY294002) and subsequently stimulated these cells for 4 days with HER2/neu agonist. Vehicle-treated cells up-regulated miR-21 levels by 3.2 ± 0.2-fold after stimulation, whereas the ERK1/2 inhibitor completely blocked miR-21 induction by the HER2/neu agonist (Fig. 3A) in a dose-dependent manner (Fig. 3B). In contrast, the AKT inhibitor (LY294002) did not reduce HER2/neu-dependent miR-21 up-regulation (Fig. 3A). We also observed that treatment with the ERK1/2 inhibitor repressed the miR-21 induction by HER2/neu agonist in SKBR3 cells and HER2/neu transfected HeLa cells (Fig. 3C). Finally, miR-21 level increased 100% in 293T cells transfected with a wild type ERK2 but decreased 30% in 293T cells transfected with a dominant negative ERK2 (Fig. 3D). These findings indicate that ERK1/2 is a key mediator for the HER2/neu-dependent miR-21 up-regulation and that this regulatory mechanism is conserved among several different cell types.
FIGURE 3.
HER2/neu regulates miR-21 expression via the MAPK (ERK1/2) pathway. A, BT-474 cells were treated with 100 nm HER2/neu agonist in the presence of 40 μm Me2SO, LY294002 or U0126. After 4 days, RNA was collected, and miR-21 expression was analyzed by qRT-PCR. B, BT-474 cells were treated with 100 nm HER2/neu agonist in the presence of 5, 10 and 40 μm U0126, and miR-21 expression was assessed as described for A. C, SKBR3 and HeLa-HER2/neu cells were treated with 100 nm HER2/neu agonist in the presence of Me2SO or 40 μm U0126, and miR-21 expression was determined as described for A. D, 293T cells were transfected with either control vector, wild type ERK2 or dominant negative ERK2. After 2 days, RNA was collected from each sample, and the mature miR-21 level was analyzed by real time PCR. The cell lysates were separated by SDS-PAGE and analyzed by Western blot using anti-phospho-ERK antibody or anti-tubulin antibody. The intensity of phospho-ERK was normalized with the intensity of tubulin to obtain the fold activation for ERK. E, BT-474 cells were transfected with an expression plasmid encoding both GFP and a Ets-1-targeting small hairpin RNA or both GFP and a scrambled small hairpin RNA sequence. After 7 days, green fluorescent-positive cells were sorted, and Ets-1 and pri-miR-21 transcript levels were determined by real time PCR. The values for the control samples were taken as reference. F, RNA was collected from MCF10A cells infected with recombinant retrovirus expressing a control vector (pMIG-empty), activated H-Ras (G12V), activated myristoylated AKT, Myc, and ID-1-hemagglutinin tag. miR-21 expression was analyzed by qRT-PCR. The mean and standard error from triplicate experiments are indicated. The cell lysates were separated by SDS-PAGE and analyzed by Western blot using the respective antibodies (anti-phospho-AKT, anti-Myc, anti-hemagglutinin tag, and anti-RAS) to demonstrate overexpression of the corresponding proteins.
We next examined which transcription factors activate miR-21 transcription. The miR-21 promoter contains a highly conserved region spanning 300 bp in which two STAT3-binding sites are present, and interleukin-6 treatment can induce miR-21 expression via STAT3 in myeloma cells (22). Three putative binding sites for AP-1 have also been identified in the miR-21 promoter, which can mediate miR-21 transcription in HeLa cells stimulated with phorbol 12-myristate 13-acetate (11). In addition to STAT3 and AP-1, we also found two consensus ETS-1-binding sequences within this highly conserved promoter region (supplemental Fig. S1). Because ETS-1 is known to be an ERK1/2-dependent transcription factor (29), we investigated whether the protein is involved in HER2/neu-associated miR-21 transcription in breast cancer cells. Indeed, knocking down Ets-1 by RNA interference inhibited pri-mIR-21 transcription by more than 40% in HER2/neu-overexpressing BT-474 cells (Fig. 3E), suggesting that Ets-1 is involved in the miR-21 transcription. Consistent with these results, mutation of a single Ets-1 binding site substantially decreased miR-21 promoter activity by more than 50% in HeLa cells stimulated with phorbol 12-myristate 13-acetate (11). Taken together, our data indicate that multiple transcription factors can regulate miR-21 transcription, working either independently or in synergy to respond to different stimuli.
To verify that the MEK1/2-ERK1/2 is indeed the key regulatory pathway for miR-21 up-regulation, we overexpressed the MEK1/2-ERK1/2 pathway activators H-RAS (G12V) or ID-1 (30, 31) in HER2/neu-negative MCF10A breast cancer cells. Overexpression of H-Ras (G12V) or ID-1 significantly induced miR-21 activation by 4.4 ± 0.1- and 6.7 ± 1-fold respectively (Fig. 3F). In contrast, overexpression of other oncogenes such as activated AKT or Myc that do not directly activate the MEK1/2-ERK1/2 pathway only induced low miR-21 up-regulation (2.6 ± 0.2- and 1.4 ± 0.1-fold, respectively) (Fig. 3F). A degree of cross-talk between RAS-MEK1/2-ERK1/2 and phosphatidylinositol 3-kinase-AKT signaling pathways exists in human breast cancers (32) and may explain the modest increase of miR-21 expression induced in cells that overexpress activated AKT. Taken together, our results suggest that the MEK1/2-ERK1/2 pathway predominantly regulates miR-21 expression in breast cancer cells and that miR-21 may be functionally involved in the H-Ras (G12V) and ID-1-associated oncogenic phenotypes.
miR-21 Is Crucial for HER2/neu-induced Cancer Cell Invasion
Overexpression of HER2/neu has been shown to significantly enhance invasion and metastasis in MDA-MB-435 cells without altering other tumorigenic features (3). Because some invasive-like phenotypes in other cancer cell lines result from increased cell growth and transformation potential, we reasoned that a HER2/neu-transfected MDA-MB-435 cell line would be an excellent model to investigate the mechanism of HER2/neu-mediated cell invasion specifically. Given that miR-21 is up-regulated by HER2/neu in this cell line (Fig. 1A) and that miR-21 has been shown to promote cell invasion and metastasis in breast cancer (3, 9), we were motivated to examine whether miR-21 function contributes to HER2/neu-driven invasive activities.
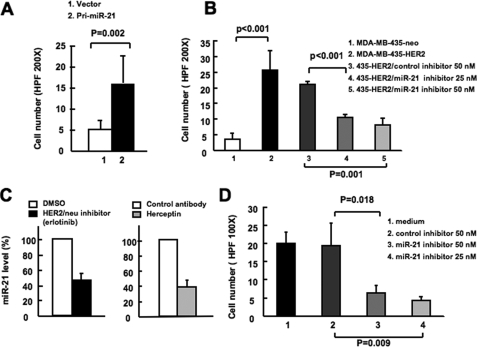
To test the activity of miR-21 in cell invasion in MDA-MB-435 cells, we ectopically expressed the 550-bp genomic sequence containing the miR-21 pri-cursor (pri-miR-21, 250 bp both upstream and downstream of the ∼50-bp miR-21 hairpin sequence) in MDA-MB-435 cells using a lentiviral vector, and we observed a 2-fold increase in mature miR-21 level in MDA-MB-435/pri-miR-21 cells (data not shown). The MDA-MB-435/pri-miR-21 cells exhibited a 3-fold increased ability to invade through Matrigel as compared with control cells in a Matrigel-coated Boyden chamber assay (Fig. 4A). This suggests that miR-21 up-regulation contributes to the invasiveness of HER2/neu-transfected MDA-MB-435 cells in vitro. Furthermore, functional inhibition of miR-21 activity by a miR-21 inhibitor at either 50 or 25 nm profoundly abrogated the invasive activity in MDA-MB-435/HER2 cells, whereas the invasive activity remained unchanged in cells transfected with a control inhibitor (Fig. 4B). These results indicate that miR-21 is a crucial player in HER2/neu-mediated cancer cell invasion and that modest expression level changes may result in a biological outcome.
FIGURE 4.
miR-21 is crucial for HER2/neu induced cancer cell invasion. A, MDA-MB-435 cells were infected with lentivirus carrying a miR-21 pri-cursor (pri-miR-21) or control vector, and cell invasion was assessed by using a Matrigel transwell assay. B, MDA-MB-435/HER2/neu cells were transfected with miR-21 inhibitor or control inhibitor. After 24 h, cell invasion was assessed by using Matrigel transwell assay. C, in the right panel, BT-474 cells were treated with either isotype control antibody or Herceptin antibody for 7 days. In the left panel, BT-474 cells were treated with either Me2SO or erlotinib for 2 day. The mature miR- 21 expression was detected by qRT-PCR. D, BT-474 cells were transfected with miR-21 inhibitor or control inhibitor. After 48 h, cell invasion was assessed using Matrigel transwell assay. The mean and standard error from triplicate experiments are indicated.
To evaluate the role of miR-21 in HER2/neu-mediated cell invasion in a more natural context, we performed similar experiments in BT-474 cells, which endogenously express high levels of both HER2/neu and miR-21 (data not shown). We first examined whether HER2/neu expression correlates with the miR-21 level in BT-474 cells. Upon treatment of these cells with the HER2/neu antagonist Herceptin or the HER2/neu kinase inhibitor erlotinib, the mature miR-21 levels decreased 62 and 54%, respectively (Fig. 4C). These results indicate that HER2/neu signaling is necessary for maintaining the high miR-21 level in BT-474 cells. More importantly, the invasive activity was significantly repressed in BT-474 cells transfected with miR-21 inhibitor (Fig. 4D), suggesting that HER2/neu-induced miR-21 up-regulation is crucial for the invasive activity of BT-474 cells. Together with the observations in MDA-MB-435/HER2 cells, these results suggest that miR-21 plays a crucial role in HER2/neu-mediated invasive phenotype in cancer cells.
HER2/neu Down-regulates PDCD4 Expression via Up-regulation of miR-21
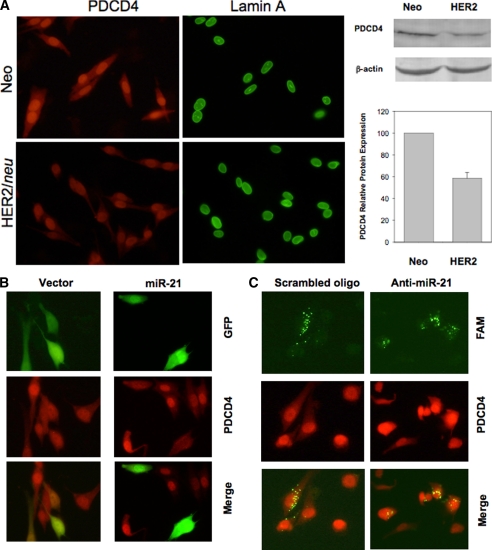
We and other groups have previously shown that PDCD4, a potent inhibitor for breast cancer cell invasion (33), is a direct target for miR-21 (9, 34, 35). Consistently, we found that ectopic expression of PDCD4 repressed the invasive activity of MDA-MB-435/HER2 cells (supplemental Fig. S2), suggesting that the PDCD4 level can influence the invasiveness of this cell line. Because HER2/neu induced miR-21 up-regulation in MDA-MB-435 cells, we next examined whether HER2/neu can modulate PDCD4 level via miR-21 in this cell line. Because PDCD4 is a predominantly nuclear protein (36), we therefore detected the nuclear PDCD4 expression using an immunofluorescence staining assay. As expected, immunofluorescence staining revealed that the PDCD4 signal was much weaker in MDA-MB-435/HER2 cells than in vector control (Neo) cells (Fig. 5A, left panel). We also performed a Western blot analysis and quantify the PDCD4 expression in both cell lines, and the PDCD4 expression was 45% decreased in the in MDA-MB-435/HER2 cells compared with vector control (Neo) cells (Fig. 5A, right panel).
FIGURE 5.
HER2/neu down-regulates PDCD4 expression via miR-21. A, detection of PDCD4 expression in MDA-MB-435/HER2/neu and MDA-MB-435/Neo cells by immunofluorescence microscopy. Nuclear structure protein lamin A represented a positive control in this experiment. B, miR-21 down-regulates PDCD4 in MDA-MB-435/Neo cells. C, anti-miR-21 increased PDCD4 level in MDA-MB-435/HER2/neu. In B and C, cells were first transfected with either miR-21 expression vector or carboxyfluorescein (FAM)-labeled anti-miR-21 and then immunostained with PDCD4 antibody, as described under “Experimental Procedures.”
To further determine the effect of miR-21 on PDCD4, we ectopically expressed miR-21 in MDA-MB-435/Neo cells. Because miR-21 expression vector was tagged with GFP, the GFP-positive represented overexpression of miR-21. Of interest, we detected a lower level of red signal (PDCD4) in miR-21 overexpression cells compared with nontransfected cells (Fig. 5B). In contrast, the vector control did not have such an effect on PDCD4 expression (Fig. 5B). Finally, we transfected MDA-MB-435/HER2 cells with locked nucleic acid antisense oligonucleotide specific to miR-21 (anti-miR-21) labeled with carboxyfluorescein (FAM). As shown in Fig. 5C, both scrambled and anti-miR-21 were localized in the cytoplasma. However, whereas the scrambled oligonucleotide had no effect on PDCD4 (red signal), anti-miR-21 clearly up-regulated PDCD4 expression. Therefore, these results suggest that HER2/neu signaling can indirectly regulate PDCD4 expression via induction of miR-21 expression. In addition, down-regulation of PDCD4 by miR-21 may contribute to the HER2/neu-mediated cell invasion and metastasis in breast cancer cells (Fig. 6).
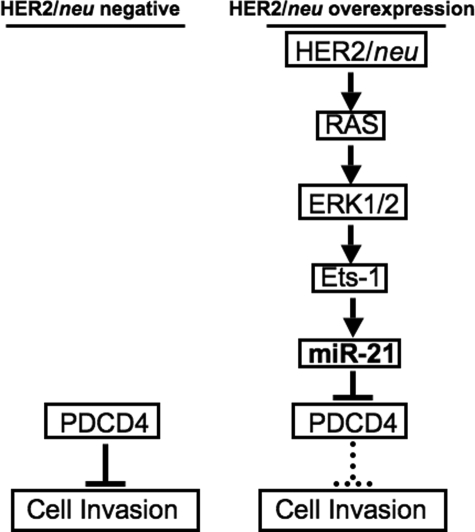
FIGURE 6.
Regulation of cell invasion via miR-21 in HER2/neu overexpression cancer cells. In the normal cells without overexpression of HER2/neu or other oncogenes associated with Ras pathway, the level of endogenous PDCD4 restricts the cell invasiveness. However, in the cancer cells with HER2/neu overexpressing, HER2/neu activates Ets-1 via the Ras/ERK1/2 pathway and consequently initiates the miR-21 transcription. miR-21 inhibits the translation of metastasis suppressor protein PDCD4 and/or other unidentified targets to promote the invasiveness of the HER2/neu-overexpressing cancer cells.
DISCUSSION
Deregulated miRNA expression can promote tumorigenesis (8) and contribute to cancer phenotypes, including invasion and metastasis (9, 10); however, how specific oncogenic pathways regulate miRNA expression in cancer cells is not well understood. In the present study, we have shown that an oncogenic receptor tyrosine kinase (RTK), HER2/neu, can initiate signaling that results in miR-21 up-regulation in various cancer cells. Although complete ablation of an oncogenic microRNA has striking phenotypes in vivo (37), moderate (∼2-fold) changes in an oncogenic microRNA can result in lymphoproliferative disease and autoimmunity (38). Our data show that even a 2–3-fold level of miR-21 up-regulation mediated by HER2/neu is sufficient to increase the invasiveness of cancer cells. To our knowledge, this is the first report to demonstrate that HER2/neu signaling can induce the expression of specific miRNAs, which in turn play crucial roles in the invasive phenotype of cancer cells. Because the signaling pathways induced by other RTKs, for example PDGFR and IGF1-R, partially overlap with those induced by HER2/neu, and overexpression of these RTKs is also associated with tumor progression (39, 40). It will be worthwhile to investigate how other RTKs deregulate miRNA expression and whether miR-21 overexpression is a common feature of RTK signaling.
Intriguingly, miR-21 has been shown to exhibit distinct oncogenic activities in different cell lines. miR-21 functions as an anti-apoptotic factor in human glioblastoma cell lines (41) and enhances the proliferative capacity of MCF-7 breast cancer cells (42) and hepatocellular cancer cell lines (43). Additionally, miR-21 promotes cell invasion and metastasis of Colo206f colorectal and MDA-MB-231 breast cancer cell lines (9, 34). Because miRNAs function via repression of target gene expression, and miR-21 has a variety of mRNA targets including PTEN, TPM-1, PDCD4, and Maspin (9, 25, 34, 43) that have been validated in distinct cell lines, it is likely that miR-21 exhibits distinct biological roles based on the “expression profile” of the miR-21 targets in a given cell line. Our observation supported this hypothesis because we found that HER/neu induced miR-21 down-regulated PDCD4 (Fig. 5) but not other published miR-21 targets in MDA-MB-435 breast cancer cells (data not shown), and ectopic expression of miR-21 did not affect MDA-MB-435 cell proliferation (data not shown) but significantly enhanced their invasiveness (Fig. 4A). The diverse interplay between HER2/neu, miR-21, and the cell-type-specific expression of targets could dictate the HER2/neu-induced oncogenic phenotypes in different cancer cells. Thus, a miR-21 target expression profile may be useful as a marker for predicting the phenotypes of the various HER2/neu-overexpressing cancers in a clinical setting.
In the present study, we demonstrated that miR-21 is induced via the MEK-ERK pathway downstream of HER2/neu. We tested another MEK-ERK pathway component, activated H-RAS (G12V), and found that it also significantly increased miR-21 levels in HER2/neu-negative breast cancer cells (Fig. 3F). Taken together our results suggest that miR-21 is an important downstream player involved in the HER2/neu-RAS-MEK-ERK signaling pathways associated with tumorigenesis. In addition to H-Ras (G12V), ID-1, an inhibitor of basic helix-loop-helix transcription factors, promotes the development of invasive breast cancer (44), and inhibition of ID-1 expression compromises the invasiveness and metastasis of breast cancer cells (45). Although the mechanism by which the MEK-ERK pathway activates ID-1 remains unclear, in our studies we found that overexpression of ID-1 induced the highest miR-21 up-regulation of the oncogenes we examined (Fig. 3F). Because up-regulation of miR-21 is associated with cell invasion, it is possible that miR-21 is required for the ID-1-induced metastasis in breast cancer cells. Taken together, miR-21 overexpression may be a common feature of oncogene signaling pathways associated with MEK-ERK pathway activation.
Here we show that HER2/neu can induce miR-21 up-regulation in a variety of cancer cell lines. However, miR-21 has been previously shown to be significantly up-regulated in many human breast cancer specimens and breast cancer cell lines independent of their HER2/neu expression status (45, 46). Because miR-21 is induced via the MEK-ERK pathway, it is likely that miR-21 is broadly up-regulated in breast cancer cells overexpressing oncogenes or receptors that activate this pathway. Indeed, expression of multiple members of the epidermal growth factor receptor family (HER1, HER2, or HER3) is present in most breast cancer cells, and these receptors have all been shown to activate the MEK-ERK pathway (47, 48). This may explain why miR-21 is frequently up-regulated in many breast cancer cells. This also raises the possibility that targeting miR-21 expression as a therapy may be of benefit not just in HER2/neu-overexpressing tumors but also in a broader subsets of patients. In summary, the identification of HER2/neu-induced miR-21 activation provides new mechanistic insight into how HER2/neu overexpression promotes invasion and metastasis in cancers.
Supplementary Material
Acknowledgments
We thank Dr. Mien-Chie Hung (M. D. Anderson Cancer Center) for the helpful suggestions and reagents provided for this manuscript. We also thank Danielle Lynch and Joan Brugge for MCF10A cells expressing the ecotropic receptor, Dr. Leonard Kusdra for the technical help of MCF10A cells expressing various oncogenes, and Dr. Gerard Evan (University of California, San Francisco) for helpful suggestions for this manuscript.
This work was supported, in whole or in part, by National Institutes of Health Grants P50-CA58207 and K08-CA104032. This work was also supported by funds from the Susan G. Komen Foundation and the Sandler Program in Biological Sciences (to A. G.). We thank the Sandler Family Foundation for the Sandler postdoctoral research fellowship award (to T. H. H.) for supporting this research.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Table S1 and Figs. S1 and S2.
- miRNA
- microRNA
- MAPK
- mitogen-activated protein kinase
- ERK
- extracellular signal-regulated kinase
- MEK
- MAPK/ERK kinase
- GFP
- green fluorescent protein
- qRT
- quantitative real time
- ScFV
- single chain Fv
- TNF
- tumor necrosis factor
- RTK
- receptor tyrosine kinase.
REFERENCES
- 1.Slamon D. J., Godolphin W., Jones L. A., Holt J. A., Wong S. G., Keith D. E., Levin W. J., Stuart S. G., Udove J., Ullrich A., et al. ( 1989) Science 244, 707– 712 [DOI] [PubMed] [Google Scholar]
- 2.Yu D., Hung M. C. ( 2000) Oncogene 19, 6115– 6121 [DOI] [PubMed] [Google Scholar]
- 3.Tan M., Yao J., Yu D. ( 1997) Cancer Res. 57, 1199– 1205 [PubMed] [Google Scholar]
- 4.Guy C. T., Webster M. A., Schaller M., Parsons T. J., Cardiff R. D., Muller W. J. ( 1992) Proc. Natl. Acad. Sci. U.S.A. 89, 10578– 10582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Subbaramaiah K., Norton L., Gerald W., Dannenberg A. J. ( 2002) J. Biol. Chem. 277, 18649– 18657 [DOI] [PubMed] [Google Scholar]
- 6.Li Y. M., Pan Y., Wei Y., Cheng X., Zhou B. P., Tan M., Zhou X., Xia W., Hortobagyi G. N., Yu D., Hung M. C. ( 2004) Cancer Cell 6, 459– 469 [DOI] [PubMed] [Google Scholar]
- 7.Bosc D. G., Goueli B. S., Janknecht R. ( 2001) Oncogene 20, 6215– 6224 [DOI] [PubMed] [Google Scholar]
- 8.He L., Thomson J. M., Hemann M. T., Hernando-Monge E., Mu D., Goodson S., Powers S., Cordon-Cardo C., Lowe S. W., Hannon G. J., Hammond S. M. ( 2005) Nature 435, 828– 833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhu S., Wu H., Wu F., Nie D., Sheng S., Mo Y. Y. ( 2008) Cell Res. 18, 350– 359 [DOI] [PubMed] [Google Scholar]
- 10.Ma L., Teruya-Feldstein J., Weinberg R. A. ( 2007) Nature 449, 682– 688 [DOI] [PubMed] [Google Scholar]
- 11.Fujita S., Ito T., Mizutani T., Minoguchi S., Yamamichi N., Sakurai K., Iba H. ( 2008) J. Mol. Biol. 378, 492– 504 [DOI] [PubMed] [Google Scholar]
- 12.Taganov K. D., Boldin M. P., Chang K. J., Baltimore D. ( 2006) Proc. Natl. Acad. Sci. U.S.A. 103, 12481– 12486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Egeblad M., Jäättelä M. ( 2000) Int. J. Cancer 86, 617– 625 [DOI] [PubMed] [Google Scholar]
- 14.Goga A., Yang D., Tward A. D., Morgan D. O., Bishop J. M. ( 2007) Nat. Med. 13, 820– 827 [DOI] [PubMed] [Google Scholar]
- 15.Yang J. Y., Zong C. S., Xia W., Yamaguchi H., Ding Q., Xie X., Lang J. Y., Lai C. C., Chang C. J., Huang W. C., Huang H., Kuo H. P., Lee D. F., Li L. Y., Lien H. C., Cheng X., Chang K. J., Hsiao C. D., Tsai F. J., Tsai C. H., Sahin A. A., Muller W. J., Mills G. B., Yu D., Hortobagyi G. N., Hung M. C. ( 2008) Nat. Cell Biol. 10, 138– 148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ventura A., Meissner A., Dillon C. P., McManus M., Sharp P. A., Van Parijs L., Jaenisch R., Jacks T. ( 2004) Proc. Natl. Acad. Sci. U.S.A. 101, 10380– 10385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.DeRisi J. L., Iyer V. R., Brown P. O. ( 1997) Science 278, 680– 686 [DOI] [PubMed] [Google Scholar]
- 18.Griffiths-Jones S., Grocock R. J., van Dongen S., Bateman A., Enright A. J. ( 2006) Nucleic Acids Res. 34, D140– 144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Berezikov E., Guryev V., van de Belt J., Wienholds E., Plasterk R. H., Cuppen E. ( 2005) Cell 120, 21– 24 [DOI] [PubMed] [Google Scholar]
- 20.Thomson J. M., Parker J., Perou C. M., Hammond S. M. ( 2004) Nat. Methods 1, 47– 53 [DOI] [PubMed] [Google Scholar]
- 21.Huang T. H., Morrison S. L. ( 2006) J. Pharmacol. Exp. Ther. 316, 983– 991 [DOI] [PubMed] [Google Scholar]
- 22.Löffler D., Brocke-Heidrich K., Pfeifer G., Stocsits C., Hackermller J., Kretzschmar A. K., Burger R., Gramatzki M., Blumert C., Bauer K., Cvijic H., Ullmann A. K., Stadler P. F., Horn F. ( 2007) Blood 110, 1330– 1333 [DOI] [PubMed] [Google Scholar]
- 23.Vandesompele J., De Preter K., Pattyn F., Poppe B., Van Roy N., De Paepe A., Speleman F. ( 2002) Genome Biol. 3, RESEARCH0034/1– 12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen C., Ridzon D. A., Broomer A. J., Zhou Z., Lee D. H., Nguyen J. T., Barbisin M., Xu N. L., Mahuvakar V. R., Andersen M. R., Lao K. Q., Livak K. J., Guegler K. J. ( 2005) Nucleic Acids Res. 33, e179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhu S., Si M. L., Wu H., Mo Y. Y. ( 2007) J. Biol. Chem. 282, 14328– 14336 [DOI] [PubMed] [Google Scholar]
- 26.Wu F., Chiocca S., Beck W. T., Mo Y. Y. ( 2007) Mol. Cancer Ther. 6, 1823– 1830 [DOI] [PubMed] [Google Scholar]
- 27.Chavez-Blanco A., Perez-Sanchez V., Gonzalez-Fierro A., Vela-Chavez T., Candelaria M., Cetina L., Vidal S., Dueñas-Gonzalez A. ( 2004) BMC Cancer 4, 59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Blenkiron C., Goldstein L. D., Thorne N. P., Spiteri I., Chin S. F., Dunning M. J., Barbosa-Morais N. L., Teschendorff A. E., Green A. R., Ellis I. O., Tavaré S., Caldas C., Miska E. A. ( 2007) Genome Biol. 8, R214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Paumelle R., Tulasne D., Kherrouche Z., Plaza S., Leroy C., Reveneau S., Vandenbunder B., Fafeur V. ( 2002) Oncogene 21, 2309– 2319 [DOI] [PubMed] [Google Scholar]
- 30.Smalley K. S. ( 2003) Int. J. Cancer 104, 527– 532 [DOI] [PubMed] [Google Scholar]
- 31.Ling M. T., Wang X., Ouyang X. S., Lee T. K., Fan T. Y., Xu K., Tsao S. W., Wong Y. C. ( 2002) Oncogene 21, 8498– 8505 [DOI] [PubMed] [Google Scholar]
- 32.Moelling K., Schad K., Bosse M., Zimmermann S., Schweneker M. ( 2002) J. Biol. Chem. 277, 31099– 31106 [DOI] [PubMed] [Google Scholar]
- 33.Nieves-Alicea R., Colburn N. H., Simeone A. M., Tari A. M. ( 2009) Breast Cancer Res. Treat. 114, 203– 209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Asangani I. A., Rasheed S. A., Nikolova D. A., Leupold J. H., Colburn N. H., Post S., Allgayer H. ( 2008) Oncogene 27, 2128– 2136 [DOI] [PubMed] [Google Scholar]
- 35.Frankel L. B., Christoffersen N. R., Jacobsen A., Lindow M., Krogh A., Lund A. H. ( 2008) J. Biol. Chem. 283, 1026– 1033 [DOI] [PubMed] [Google Scholar]
- 36.Böhm M., Sawicka K., Siebrasse J. P., Brehmer-Fastnacht A., Peters R., Klempnauer K. H. ( 2003) Oncogene 22, 4905– 4910 [DOI] [PubMed] [Google Scholar]
- 37.Ventura A., Young A. G., Winslow M. M., Lintault L., Meissner A., Erkeland S. J., Newman J., Bronson R. T., Crowley D., Stone J. R., Jaenisch R., Sharp P. A., Jacks T. ( 2008) Cell 132, 875– 886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Xiao C., Srinivasan L., Calado D. P., Patterson H. C., Zhang B., Wang J., Henderson J. M., Kutok J. L., Rajewsky K. ( 2008) Nat. Immunol. 9, 405– 414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hellawell G. O., Turner G. D., Davies D. R., Poulsom R., Brewster S. F., Macaulay V. M. ( 2002) Cancer Res. 62, 2942– 2950 [PubMed] [Google Scholar]
- 40.Carvalho I., Milanezi F., Martins A., Reis R. M., Schmitt F. ( 2005) Breast Cancer Res. 7, R788– 795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chan J. A., Krichevsky A. M., Kosik K. S. ( 2005) Cancer Res. 65, 6029– 6033 [DOI] [PubMed] [Google Scholar]
- 42.Si M. L., Zhu S., Wu H., Lu Z., Wu F., Mo Y. Y. ( 2007) Oncogene 26, 2799– 2803 [DOI] [PubMed] [Google Scholar]
- 43.Meng F., Henson R., Wehbe-Janek H., Ghoshal K., Jacob S. T., Patel T. ( 2007) Gastroenterology 133, 647– 658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Desprez P. Y., Lin C. Q., Thomasset N., Sympson C. J., Bissell M. J., Campisi J. ( 1998) Mol. Cell. Biol. 18, 4577– 4588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fong S., Itahana Y., Sumida T., Singh J., Coppe J. P., Liu Y., Richards P. C., Bennington J. L., Lee N. M., Debs R. J., Desprez P. Y. ( 2003) Proc. Natl. Acad. Sci. U.S.A. 100, 13543– 13548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sempere L. F., Christensen M., Silahtaroglu A., Bak M., Heath C. V., Schwartz G., Wells W., Kauppinen S., Cole C. N. ( 2007) Cancer Res. 67, 11612– 11620 [DOI] [PubMed] [Google Scholar]
- 47.Filardo E. J., Quinn J. A., Bland K. I., Frackelton A. R., Jr. ( 2000) Mol. Endocrinol. 14, 1649– 1660 [DOI] [PubMed] [Google Scholar]
- 48.Migliaccio A., Piccolo D., Castoria G., Di Domenico M., Bilancio A., Lombardi M., Gong W., Beato M., Auricchio F. ( 1998) EMBO J. 17, 2008– 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.