Abstract
Bacteria in the distal human gut have evolved diverse abilities to metabolize complex glycans, including the capacity to degrade these compounds as nutrients and to assemble their component sugars into new polymers such as extracellular capsules. The human gut bacterium Bacteroides thetaiotaomicron is well endowed with the ability to metabolize both host- and diet-derived glycans. Its genome contains 88 different polysaccharide utilization loci (PULs) for complex glycan catabolism and eight different gene clusters for capsular polysaccharide biosynthesis. Here, we investigate one of the prominent mechanisms by which this gut symbiont regulates many PULs involved in host mucin O-glycan degradation; namely, transcriptional regulation via the concerted interactions of cell-envelope-localized TonB-dependent transporters, extra-cytoplasmic function σ factors and anti-σ factors, which participate together in a regulatory pathway termed trans-envelope signaling. Unexpectedly, we found that several different trans-envelope signaling switches involved in PUL-mediated O-glycan degradation also modulate capsular polysaccharide synthesis. A novel regulatory pathway, which is dependent on expression of O-glycan-targeting outer membrane proteins, governs this coordinated regulation of glycan catabolism and capsule synthesis. This latter finding provides a new link in the dynamic interplay between complex glycan metabolism, microbial physiology, and host responses that occurs during colonization of the gut.
The adult human distal gut is home to a microbial consortium (microbiota) that plays critical roles in nutrient digestion (1). This community of ∼10–100 trillion microbes is generally dominated by two bacterial divisions (phyla), the Bacteroidetes and the Firmicutes, and harbors a collective genome, the microbiome, that is several orders of magnitude larger than the human genome (2–4). An important function encoded within the microbiome is production of degradative enzymes that target the chemically diverse pool of complex glycans that are constantly inundating the gut. Because many of the complex dietary polysaccharides that we consume transit our proximal gastrointestinal tract intact, these glycan degrading (glycolytic) activities augment human digestive physiology by providing catabolic functions we have not evolved. Thus, the microbiota allows us to assimilate otherwise indigestible dietary nutrients, and it is probable that competition for these nutrients is a major factor that shapes the composition and metabolism of this community (2).
In addition to being glycan consumers, many members of the microbiota possess the ability to synthesize new polysaccharides de novo. Many of these newly synthesized molecules take the form of secreted capsular polysaccharides, composed of myriad sugars combined via an array of glycosidic linkages. Some gut bacteria have the ability to reversibly alter their capsule composition, and this phenomenon is important for evasion of host adaptive immunity (5, 6). Expression of the zwitterionic capsular polysaccharide A from the human gut symbiont Bacteroides fragilis reduces the host immune response toward pathogenic bacteria by suppressing inflammatory pathways (7). Bacterial production of capsular polysaccharides is associated with increased resistance to phage, complement, and antimicrobial peptides (8–10). Extracellular polysaccharides are mediators of bacterial interactions with both other microbes and biotic/abiotic surfaces during the process of biofilm formation (11). Thus, variable expression of capsular polysaccharides is a trait that likely influences the dynamic relationship between microbiota members as well as how some of these species interact with their hosts.
Among the species prominently represented in the distal gut microbiota, members of the Bacteroidetes are particularly adept at degrading a variety of glycans present in our diet (starches, fructans, pectins, and hemicelluloses) and in the gut mucosa (O- and N-linked glycans and glycosaminoglycans) (12). Comparative and functional genomic studies in several Bacteroidetes species have revealed that these Gram-negative bacteria deploy similar sets of extracellular proteins, termed Sus-like systems, to bind and degrade diverse glycan structures (13, 14). Sus-like systems are encoded by polysaccharide utilization loci (PULs)3; these clusters ubiquitously encode homologs of a TonB-dependent oligosaccharide transporter (SusC) and an outer membrane lipoprotein (SusD) involved in glycan binding and also typically encode glycolytic enzymes (i.e. glycoside hydrolases, polysaccharide lyases, and carbohydrate esterases) (15). Molecular genetic analyses of several PULs in the human gut symbiont, Bacteroides thetaiotaomicron, have confirmed that these systems are subject to activation via a variety of different transcription factors, which respond to the monosaccharide content and linkage information contained in glycans that they target as growth substrates (14, 16). Thus, we hypothesize that each Bacteroidetes Sus-like system is a regulated, substrate-specific system for targeting a distinct glycan. The presence of dozens of systems in each species equips it to handle many different complex carbohydrate substrates (13).
B. thetaiotaomicron dedicates ∼18% of its 6.26-Mb genome to 88 individual PULs. In a recent study involving gnotobiotic mice, we determined that a subset of these loci are deployed during foraging on host mucin O-glycans (14). This attribute enables B. thetaiotaomicron to persist in the distal intestine when dietary glycan intake is scarce and also to be transmitted from a mother to her newborn pups when they consume a milk diet that is naturally devoid of complex, difficult to digest dietary polysaccharides. B. thetaiotaomicron expresses at least 16 different PULs in response to mucin O-glycans, and at least some of these systems are transcriptionally activated by chemically distinct glycosidic cues that may exist within the same glycan molecule (e.g. the disaccharides Gal-β1,3-GalNAc or Gal-β1,4-GlcNAc). The need to recognize the many different linkages and sugar combinations present in O-glycans may explain much of the seeming redundancy in PUL expression toward these substrates, which are composed of five principal monosaccharides that can be linked together into dozens of different heterogeneous structures (17), each of which could be recognized by a different PUL.
A second notable feature of the response of B. thetaiotaomicron to mucin O-glycans is transcriptional activation of PULs by extra-cytoplasmic function σ (ECF-σ) factor/anti-σ factor pairs. To date, this class of regulators has not been implicated in controlling PULs that target any other class of glycans (14). These transcription factors are predicted to form a signal transduction pathway by coupling with specialized SusC-like proteins (“signal transducing SusC-like transporters”) that import mucin-derived oligosaccharides into the periplasm (Fig. 1A). Upon recognition of substrate by a signal transducing SusC-like transporter, these three proteins are thought to work together to activate PUL transcription via a mechanism termed “trans-envelope signaling”; protein contacts made in both the periplasm (transporter/anti-σ interaction) and at the cytoplasmic face of the inner membrane (anti-σ/ECF-σ interaction) transduce a signal across the entire bacterial cell envelope that results in activation of the ECF-σ transcription factor (18).
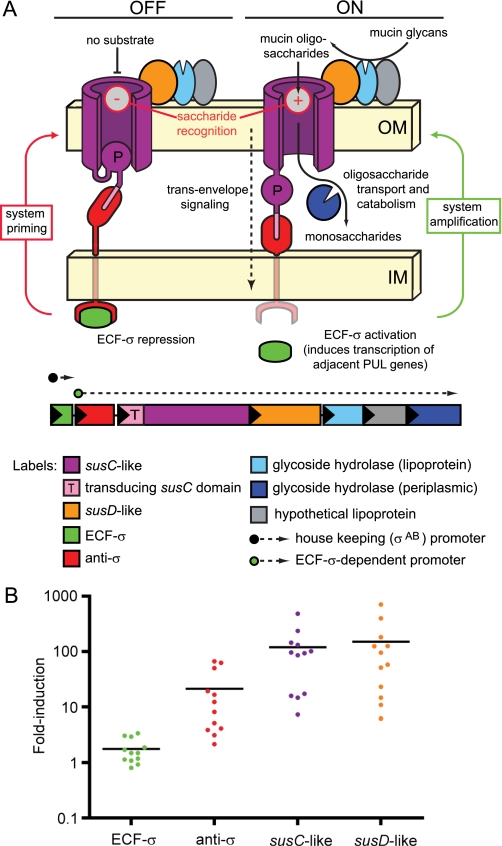
FIGURE 1.
Positive feedback model of PUL regulation. A, schematic of a hypothetical Sus-like system regulated by trans-envelope signaling and the PUL at which it is encoded (note that gene number and enzyme content is variable between different PULs). The left half of the Fig. illustrates the sequence of events leading to system priming in the “off” signaling state: (i) initial expression of ECF-σ (green) from a housekeeping σAB promoter (dashed arrow with a black circular base) results in a small amount of PUL gene expression via the ECF-σ-dependent transcript; (ii) simultaneous production of anti-σ (red), which physically interacts with both the SusC-like transporter (purple) and the cytoplasmic ECF-σ, causes the system to be held in the default off state unless glycan substrates are present. The right half illustrates events leading to amplified expression (“on” state) of the Sus-like system: (i) a cognate substrate (e.g. mucin O-glycan) is bound and then transported through the transducing SusC-like transporter; (ii) substrate transport initiates an allosteric signal that is transmitted from the SusC-like protein to the anti-σ factor via periplasmic contacts and then to the ECF-σ factor via cytoplasmic contacts; (iii) propagation of the trans-envelope signal results in ECF-σ activation and increased expression of the ECF-σ-dependent promoter(s) (dashed arrow with a green circular base) located upstream of nearby genes encoding structural and enzymatic components of the Sus-like system. B, comparison of induction levels for 12 trans-envelope signaling systems that respond to host glycans. The -fold induction (plotted on the y axis) relative to MM-glucose is shown for genes encoding ECF-σ factors (green data points), anti-σ factors (red data points), transducing SusC-like proteins (purple data points), and SusD-like proteins (orange data points) from 12 different PULs that are regulated by trans-envelope signaling. Data points represent individual genes, and horizontal black bars represent average -fold induction for each gene class in GeneChip expression experiments. The y axis is shown on log10 scale to allow visualization of differences in ECF-σ expression.
These putative trans-envelope signaling switches would operate in a manner analogous to the ferric dicitrate (Fec) signaling system of Escherichia coli (18). However, the number of these putative trans-envelope signaling switches in B. thetaiotaomicron is extraordinarily large; i.e. the ferric dicitrate system is uniquely represented in E. coli, whereas B. thetaiotaomicron has 26 such systems. We previously found that B. thetaiotaomicron activates at least 12 of its 26 putative switches in the presence of mucin O-glycan components, raising two key questions about how this signal transduction mechanism integrates the information contained in extracellular O-glycans. First, do simultaneously activated systems act exclusively of one another to sense various O-glycans, or can cross-talk occur between systems? Second, do trans-envelope signaling switches exert global effects on the B. thetaiotaomicron transcriptome that extend beyond regulation of the local PUL genes to which they are linked?
In this report we explore each of these questions about how B. thetaiotaomicron mobilizes its various “utensils” for dining on glycans available in its host habitat through a series of molecular genetic and biochemical experiments. Our results validate the proposed trans-envelope signaling mechanism and reveal that only minor amounts of cross-talk occur between simultaneously active switches. We also show for several different sensory systems that B. thetaiotaomicron coordinates its glycan catabolism and capsular polysaccharide (CPS) biosynthesis. Thus, a component of this species adaptation to life in the gut is co-regulation of nutrient intake and surface antigenicity, a finding that sheds light in the quest to understand the elaborate web of interactions between our diet, our gut microbiota, and our human physiology.
EXPERIMENTAL PROCEDURES
Bacterial Strains and Transcriptional Analyses
Bacterial strains and plasmids are summarized in supplemental Table S1. B. thetaiotaomicron was routinely grown in Tryptone-yeast extract-glucose medium (19) or on brain-heart infusion (BD Biosciences) agar supplemented with 10% horse blood (Colorado Serum Co.). Antibiotics were added as appropriate: erythromycin (25 μg ml−1), gentamicin (200 μg ml−1), tetracycline (2 μg ml−1), and 5-fluoro-2′-deoxyuridine (200 μg ml−1). Minimal medium (MM)-glucose cultures were grown as previously described in either test tubes or in a batch fermentor (5-ml and 800-ml culture volumes, respectively) (14). B. thetaiotaomicron mutant strains were constructed by either suicide plasmid insertion (denoted as Ω mutants) or by unmarked gene deletion (denoted as Δ mutants). Each method was used exactly as described in previous publications (14, 20).
Transcriptional profiling was performed using custom Affymetrix GeneChips containing probesets representing >98% of 4779 predicted protein-coding genes in the B. thetaiotaomicron genome (21). GeneChip targets were prepared from whole bacterial RNA (15) and hybridized to the microarrays according to standard Affymetrix protocols. Data were normalized using Microarray Suite 5 software (Affymetrix) and processed using GeneSpringGX 7.3.1 software (Agilent) according to a previously described workflow (14), except that -fold change cutoffs of 2.5 and 5 were used as indicated in the analyses reported in this work. All GeneChip data used in this study are available from the Gene Expression Omnibus (GEO) data base (www.ncbi.nlm.nih.gov). Further details concerning bacterial growth conditions, experimental parameters, and the GEO accession numbers for each experiment are provided in supplemental Table S2. In addition to the software and analysis described above, we also used the program DNA-Chip Analyzer (dChip) software (22) to generate the heatmap image shown in supplemental Fig. S4.
Additional measurement of transcript levels was performed by quantitative reverse transcriptase-PCR using a Stratagene Mx3000P machine, previously described reaction conditions (15), and the primers listed in supplemental Table S3. In all cases, transcript abundance was assayed at 78 °C.
Yeast Two-hybrid Analysis
Protein interactions between trans-envelope signaling components were probed by yeast two-hybrid assay using the Matchmaker Library Construction kit (BD Biosciences), the manufacturer's instructions, and the oligonucleotides described in supplemental Table S3. Sequences encoding either the N-terminal domains of signal-transducing SusC paralogs or the entire ECF-σ controlled by trans-envelope signaling, were ligated in-frame with the GAL4 DNA binding domain in the vector pGBKT7. Sequences encoding either the N- or C-terminal domains of anti-σ factors were ligated in-frame with the GAL4 activation domain in the vector pGADT7-rec. Fusion vectors were transformed into Saccharomyces cerevisiae test strain AH109 and selected on medium lacking l-leucine and l-tryptophan (Leu-, Trp-), which corresponds to the nutritional selection genes present in pGBKT7 and pGADT7-rec, respectively. Strains containing potentially interacting fusion proteins were screened on synthetic defined medium lacking l-leucine, l-tryptophan, adenine, and l-histidine (Leu-, Trp-, Ade-, His-), which imposes the additional requirement of interaction between GAL4 activation and binding domain fusion proteins to recover adenine and l-histidine prototrophy. Identical cell suspensions were replica-plated onto both types of media, and growth was scored after incubation for 2 days at 30 °C.
Capsule Extraction and Monosaccharide Analysis
Capsular polysaccharides were extracted from wild-type and mutant strains grown in a 1.2-liter Bioflo 110 batch fermentor (New Brunswick Scientific; 800 ml culture volumes in MM-glucose (5)). Briefly, cultures were grown to mid-log phase (A600 for three biological replicates were 0.60, 0.59, and 0.62 for wild type; 0.60, 0.63, 0.61 for ΩBT3992, and 0.73, 0.76, and 0.73 for a ΩBT3992/ΔCPS2 double mutant). Bacterial cells were collected by centrifugation at 5000 × g for 20 min and immediately frozen at −80 °C. Each cell pellet was thawed by the addition of 50 ml of warm water. Phenol (90% w/w; 50 ml) was then added, and the mixture was held at 65 °C with gentle stirring for 1 h followed by incubation at 4 °C overnight. The extraction mixture was subsequently centrifuged at 1600 × g for 20 min to separate the aqueous and phenol phases, and the aqueous phase was collected and dialyzed (1-kDa cutoff) exhaustively against deionized distilled water. Dialyzed samples were lyophilized to dryness and dissolved in 20 ml of boiling double distilled H2O followed by continuous agitation overnight at 4 °C. Samples were subjected to two consecutive rounds of centrifugation to remove insoluble particulates, the first at 7,000 × g for 20 min and the second at 150,000 × g in an ultracentrifuge for 4 h (both steps at 4 °C). Finally, the cleared sample was lyophilized and dissolved in hot (95 °C) double distilled H2O at a concentration of 10 mg ml−1.
Neutral and acidic sugars were assayed in extracellular polysaccharide samples by high pH anion exchange chromatography with pulsed amperometric detection (HPAEC-PAD) after acid hydrolysis of the samples in 2 m trifluoroacetic acid (100 °C for 4 h). Thirteen individual peaks corresponding to different sugars were distinguished under the conditions used. Nine of these eluted identically to known standards, whereas four did not behave like any of the standards examined (fucose, N-acetylgalactosamine, N-acetylglucosamine, galactose, glucose, mannose, galacturonic acid, glucuronic acid, and iduronic acid). Because both of the unknown monosaccharides that were abundant in CPS2 eluted similarly to acidic sugar standards, we also ran a sample of acid-hydrolyzed alginic acid, which is composed of mannuronic and guluronic acids. The presence of N-acetylgalactosamine and N-acetylglucosamine, which were deacetylated during acid hydrolysis, was inferred from the presence of galactosamine and glucosamine, respectively.
Microscopic Imaging of Capsules
For India ink staining, B. thetaiotaomicron strains were grown to late exponential phase in MM-glucose (A600 values were 1.10, 1.16, and 1.04 for wild type, 1.10, 1.22, and 0.87 for ΩBT3992, and 1.10, 1.19, and 0.75 for ΩBT3992/ΔCPS2). A 20-μl aliquot of the fresh culture was mixed gently on a slide with an equivalent amount of India ink (Higgins Ink), a cover glass was added, and a paper towel was firmly pressed over the slide to blot away extra liquid. Cells were imaged at 630× magnification by phase contrast microscopy (Zeiss Axioskop 2 MOT plus). Capsule measurements were determined from micrographs using ImageJ 1.40g software (rsb.info.nih.gov). A total of 150 individual cells were measured for each bacterial strain analyzed, 50 from each of the three individual experiments. Four measurements were made per cell and averaged to generate a single average capsule width.
Samples for quick-freeze, deep-etch scanning electron microscopy were taken from fresh MM-glucose cultures grown to similar points in late exponential phase as for the India Ink staining experiments (A600 values of 1.2, 1.0, and 1.2 for wild type, ΩBT3992 and ΩBT3992/ΔCPS2). Samples were processed and imaged according to previously described methods (23–25). Briefly, a 5-ml suspension of the culture was gently centrifuged, and the resulting bacterial pellet was layered onto a thin slice of aldehyde-fixed rat lung (used for support during freezing). The pellet was put onto a liquid helium-cooled copper block in a homemade “Cryopress” quick-freeze machine. The frozen pellet was then freeze-fractured in a standard Balzer's freeze etching unit and immediately “deep-etched” for 2 min at −104 °C. The specimen was then rotary-replicated with ∼2 nm of platinum applied from an angle of 20° above the horizontal and “backed” with a ∼10-nm film of pure carbon. Replicas were separated from the bacteria by brief floatation on a solution of 3–6% sodium hypochlorite (as household bleach) and finally washed several times in double distilled H2O before being picked up on 75-mesh Formvar-coated microscope grids. Replicas were viewed in a JEOL transmission electron microscope operated at 100 kV and photographed at 50,000× initial magnification.
Promoter Mapping Using 5′-Rapid Amplification of cDNA Ends
The start of the BT1053 transcript was mapped using the 5′-RACE system for rapid amplification of transcript ends kit (Invitrogen) and a mixture of total cellular RNA purified from B. thetaiotaomicron grown in MM-glucose and MM-neutral O-glycans. The kit employs terminal deoxynucleotidyltransferase to add a polydeoxycytidine sequence to the 5′ ends of transcripts before PCR amplification using a gene-specific primer and a universal primer complementary to polydeoxycytidine. Therefore, we were unable to identify the precise transcriptional start site for BT1053 due to the possibility that this transcript may have initiated at a cytidine residue. However, we mapped the probable transcription start site to within two possible initiating nucleotides (see supplemental Fig. S1 for details).
Sequence Analysis and Alignment
Deduced protein sequences for the ORFs described in supplemental Fig. S2 were aligned using ClustalW in the MegaAlign component of the Lasergene software package (DNAStar). Signal sequences and transmembrane helices were predicted using the programs SignalP and Toppred, respectively (26, 27).
RESULTS AND DISCUSSION
A Positive Feedback Model of PUL Regulation
As noted in the Introduction, our previous custom GeneChip-based, whole genome transcriptional profiling of B. thetaiotaomicron grown in a variety of MM formulations containing purified host glycans revealed that this species expresses at least 12 PULs that are coupled to ECF-σ/anti-σ trans-envelope signaling switches to catabolize mucin O-glycans (herein referred to as “O-glycans”) (14). When we analyzed individual gene expression patterns in these PULs, we noted that the ECF-σ gene associated with each locus did not exhibit similarly increased levels of transcription as the remaining genes in the cluster; during bacterial growth under conditions that resulted in maximum expression of each PUL, the ECF-σ genes in these 12 loci only exhibited an average induction of 1.8-fold (Fig. 1B). In contrast, genes encoding the anti-σ, SusC-, and SusD-like proteins associated with these same 12 systems exhibited average induction values of 21-, 119-, and 150-fold, respectively. Thus, the host glycan components that stimulate each of these systems do not equivalently influence ECF-σ expression, suggesting that these transcription factors are not auto-regulated and may be transcribed by RNA polymerase containing a different σ factor. Coupled with the mechanistic model of transcriptional control postulated for trans-envelope signaling (see the Introduction and Fig. 1A), this observation suggests a positive feedback loop in which expression of each PUL-encoded Sus-like system is “primed” by housekeeping σ factor-directed transcription of the coupled ECF-σ gene. In other words, a consistent amount of each ECF-σ is expressed from each locus and subsequently activates expression of both the associated anti-σ factor and the structural and enzymatic functions involved in glycan processing. In the absence of an inducing glycan, a small amount of each system is produced in an “off” state. When a glycan is recognized by a particular SusC-like transporter, a trans-envelope signal is sent that results in PUL activation and an amplified catabolic response toward the target glycan from which the transported saccharide was derived. Thus, by constitutively producing small amounts of each Sus-like system, B. thetaiotaomicron constantly surveys its environment for the various glycans that these systems recognize, equipping the cell to generate a rapid response upon substrate detection.
To determine whether PUL-associated ECF-σ genes are indeed expressed from “housekeeping” promoters, we mapped the transcription start site of one such gene, BT1053, that is associated with a single, O-glycan-inducible PUL (BT1032–53). Transcription of this gene is initiated downstream of a promoter containing just 1-bp difference relative to the −7/−33 consensus established for the primary σ-factor, σAB, previously identified in Bacteroides species (28). Similar putative promoter sequences were identified upstream of 24 of 26 ECF-σ genes examined (supplemental Fig. S1A). In conjunction with the expression data described above, this finding supports the model that expression of each ECF-σ-regulated Sus-like system is indeed primed by housekeeping transcription. Moreover, we were able to locate examples of similar σAB promoter sequences upstream of several other classes of PUL-associated transcriptional activators (supplemental Fig. S1B), suggesting that expression of other Sus-like systems may be primed in a similar way as those controlled by trans-envelope signaling.
Yeast Two-hybrid Analysis of Protein-Protein Interactions Involved in Trans-envelope Signaling
Trans-envelope signaling has not been investigated in an organism such as B. thetaiotaomicron that harbors multiple different systems that may be simultaneously activated within the same cell (18). Therefore, we first sought to define the signaling specificities of multiple trans-envelope signaling systems as well as to validate the signaling mechanism of at least one individual system.
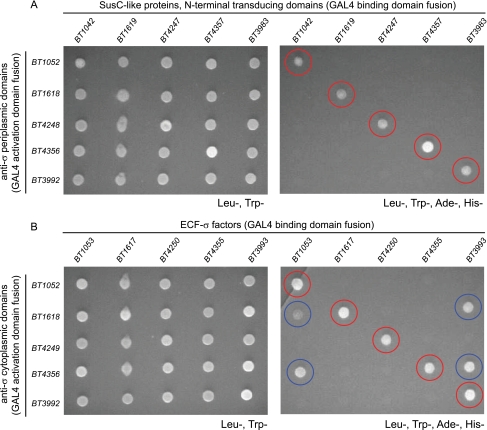
The predicted physical interactions between trans-envelope signaling protein components were examined for five ECF-σ-linked PULs that are simultaneously expressed at high levels in the distal guts of gnotobiotic mice monoassociated with B. thetaiotaomicron (14). A matrix of 50 different protein interactions were assayed for the 5 selected systems; 25 possible combinations for the SusC-like transporter/anti-σ interaction, which occurs in the periplasm, and 25 possible combinations for anti-σ/ECF-σ interaction, which occurs in the cytoplasm (Fig. 2). All 10 cognate interactions (i.e. those between protein components from the same PUL) were detected by yeast two-hybrid analysis. However, none of the remaining 20 non-cognate interactions was detected between periplasmic signaling partners, whereas just 4 non-cognate interactions were detected between cytoplasmic signaling partners. Thus, we were able to demonstrate protein interactions between both periplasmic and cytoplasmic signaling partners, and these interactions appear to be largely system-specific. Despite the evidence for signaling cross-talk provided by four non-cognate cytoplasmic interactions, additional experiments described below in which individual ECF-σ regulons were artificially activated via anti-σ mutation suggest that these non-cognate interactions between cytoplasmic signaling components do not result in biologically relevant cross-talk between PUL regulators.
FIGURE 2.
Protein interactions between trans-envelope signaling components. A, yeast two-hybrid assays of interactions between periplasmic trans-envelope signaling components (SusC-like protein N-terminal transducing domains and anti-σ C-terminal domains). B, yeast two-hybrid assays of interactions between cytoplasmic trans-envelope signaling components (ECF-σ factors and anti-σ N-terminal domains). Each grid represents a matrix of 25 different interactions tested for each signaling node. Panels on the left show growth of all possible interacting pairs under non-selective conditions that do not require protein interactions (Leu-, Trp-). Panels on the right show replicate growths of the same interacting partners shown on the left but under selective conditions (Leu-, Trp-, Ade-, His-); colony growth under this condition indicates protein interaction in the assay. Cognate interactions are highlighted with red circles, and non-cognate interactions are highlighted with blue circles. Gene names at the top and left of columns/rows indicate the source genes tested. The precise coding sequences used for yeast two-hybrid partner fusions are illustrated in supplemental Fig. S2. Colony growth was photographed after 2 days of growth at 30 °C.
Genetic Analysis of the Trans-envelope Signaling Mechanism
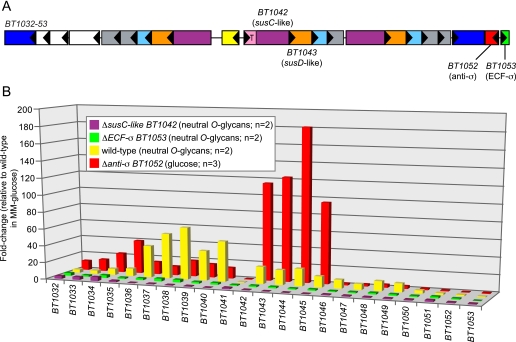
To further test the model of glycan-induced trans-envelope signaling proposed in Fig. 1A, we constructed a series of mutants in the genes encoding signaling components of one O-glycan-induced PUL (BT1032–53; see Fig. 3A for a map of this locus) and evaluated the effects of these perturbations on the ability of this PUL to be activated in response to O-glycans (Fig. 3B). This PUL was chosen for mechanistic analysis because it is highly induced in vitro in response to neutral O-glycans (14), which can be purified in abundance from porcine gastric mucosa, and because the system's anti-σ gene (BT1052) was not positioned upstream of the signal transducing susC-like transporter. This latter point is critical because it allowed construction of a plasmid insertion mutant in the anti-σ gene (BT1052) without the possibility of introducing polar effects on downstream genes that might also be involved in signaling.
FIGURE 3.
Molecular genetic analysis of trans-envelope signaling. A, schematic of the BT1032–53 PUL, which is induced in response to mucin O-glycans. Key genes that were subjected to mutational analysis are indicated with locus tag numbers, and all genes are labeled according to the key provided in Fig. 1; white boxes in this schematic represent genes with annotated functions other than those indicated in Fig. 1. B, a histogram showing the responses of 22 genes contained within the BT1032–53 PUL to different growth and/or bacterial genetic backgrounds. For wild-type B. thetaiotaomicron grown in a preparation of neutral O-glycans (14), most genes in this PUL show increased expression relative to a MM-glucose reference (yellow bars). Individual disruptions of genes encoding either the signal transducing SusC-like protein (BT1042, purple bars) or the ECF-σ associated with this system (BT1053, green bars) result in nearly complete loss of PUL activation during growth in the neutral O-glycan mixture. Conversely, de-repression of the trans-envelope signaling switch via anti-σ mutation (red bars) results in PUL expression during growth on glucose as a sole carbon source, which does not ordinarily cause induction of these genes. Values are based on normalized GeneChip expression data. Bars indicate -fold changes in expression relative to duplicate reference datasets derived from 5-ml cultures grown in MM-glucose. All bars represent the average response of each gene in two replicate datasets.
Growth of wild-type B. thetaiotaomicron in MM containing neutral O-glycans stimulated expression of this locus; all genes between BT1032 and BT1053, with the exception of an annotated integrase/tyrosine recombinase (BT1041), showed induction, suggesting that they are controlled by ECF-σBT1053. In contrast to wild type (yellow bars in Fig. 3B), two different mutants, a deletion of the ECF-σ gene BT1053 and a plasmid insertion in the signal transducing susC-like gene BT1042, eliminated the ability of this system to be induced during growth on neutral O-glycans (green and purple bars in Fig. 3B). Neither of these mutants exhibited noticeably altered growth rates in vitro when using neutral O-glycans as the sole carbon source, suggesting that redundancy in the B. thetaiotaomicron ability to use this host-derived substrate may compensate for loss of this one Sus-like system (data not shown).
To test whether the signaling defect observed with the BT1042 insertion mutant was due to specific loss of BT1042 or the result of a polar effect that silences the downstream genes (BT1043–45), we made a plasmid insertion in the adjacent downstream gene in the operon (BT1043) and evaluated the ability of this PUL to undergo normal activation. In response to neutral O-glycans, the ΩBT1043 mutant exhibited normal activation of “probe” genes in this PUL (BT1040 and BT1046) to levels that were similar to, or slightly greater than wild type (supplemental Fig. S3). Moreover, complementation of ΩBT1042 in trans with a wild-type copy of BT1042, but not the downstream BT1043–45 genes, restored induction of PUL genes to levels that were similar to wild type (data not shown). Thus, the only product of this operon required for signal transduction is BT1042. Together, these results demonstrate that the specialized TonB-dependent transporter associated with this system (BT1042) is required to propagate a signal that results in gene activation. The effect of this signal is ultimately mediated through a cognate transcription factor, ECF-σBT1053, which also needs to be present to activate PUL gene expression.
Multiple Sus-like systems involved in O-glycan catabolism, including the BT1032–53 system, are simultaneously expressed both in vitro during growth on O-glycans and in vivo in the distal mouse gut (14). The genetic experiments described above demonstrate that the trans-envelope signaling switch encoded within the BT1032–53 PUL is essential for its regulation. We also wished to determine whether loss of the positive-acting components of this particular trans-envelope signaling switch (BT1042 and BT1053) influenced other cellular responses to O-glycans, such as expression of other PULs. Therefore, we individually profiled our BT1042 and BT1053 mutants in MM-neutral O-glycans to see if loss of one trans-envelope signaling switch influenced expression of other PULs. As described in the supplemental material, expression of only one of nine PULs that were highly expressed in MM-neutral O-glycans was diminished in the BT1042 and BT1053 signaling mutants, and this was the BT1032–53 PUL to which these genes belonged (supplemental Fig. S4).
Our model predicts that active signaling from BT1042 through ECF-σBT1053 proceeds through the inner membrane-spanning anti-σBT1052. In the absence of O-glycans, this anti-σ should act as a repressor of ECF-σBT1053 and, upon O-glycan recognition, transmit the signal it receives from BT1042 to ECF-σBT1053, resulting in release of its transcriptional activity. Thus, inactivation of the anti-σBT1052 repressor should result in constitutive expression of genes controlled by ECF-σBT1053. Experiments with the E. coli ferric dicitrate (Fec) system have demonstrated that the cytoplasmic N-terminal domain of the anti-σFecR is initially required for ECF-σFecI maturation (29). With these thoughts in mind, we constructed a plasmid insertion mutant in BT1052 that resulted in a C-terminal truncation of the anti-σ gene (see supplemental Fig. S2B for the precise point of truncation). This mutant is expected to still express the N-terminal cytoplasmic domain of anti-σBT1052 required for ECF-σBT1053 maturation but is lacking the periplasmic domain required for coupling with SusCBT1042 in the periplasm. As anticipated, GeneChip profiling of this mutant during growth on glucose as a sole carbon source revealed expression of genes within the BT1032–53 PUL, even though no inducing O-glycans were present (red bars in Fig. 3B). This ability to artificially de-repress individual trans-envelope signaling switches in the absence of an external inducing glycan allowed us to probe the breadth of the signaling regulon associated with each system.
Global Regulation of Polysaccharide Metabolism by Trans-envelope Signaling
To define select trans-envelope signaling system transcriptional regulons, we constructed a series of five individual anti-σ factor disruptions aimed at causing de-repression of genes activated by each ECF-σ. The five disrupted anti-σ genes are components of the same five systems analyzed above by yeast two-hybrid analysis: BT1052, BT1618, BT3992, BT4356, and BT4249/BT4248 (the latter gene was originally annotated as two distinct divergent ORFs but was later confirmed to undergo site-specific recombination to generate a single full-length anti-σ gene (14); because the upstream BT4249 ORF was specifically targeted, this mutant is referred to as ΩBT4249).
Each anti-σ gene was disrupted using a plasmid insertion strategy that resulted in a truncated anti-σ gene and native expression of the 5′ end of the ORF, which encodes the cytoplasmic domain (truncation points are indicated in supplemental Fig. S2B). Each of the targeted anti-σ genes had previously exhibited increased expression during activation of its O-glycan-responsive PUL (Fig. 1B). Thus, we initially verified the effectiveness of our anti-σ disruptions by confirming induction of the remaining 5′ region of each gene using quantitative PCR. In each of the five mutant strains, anti-σ gene expression was increased 11–387-fold during growth in MM-glucose when compared with wild-type B. thetaiotaomicron, suggesting successful derepression (data not shown). Three mutants (ΩBT1618, ΩBT4249, and ΩBT4356), in which the disrupted anti-σ gene resides immediately upstream and in the same orientation as the adjacent sus-like genes, resulted in apparent polar effects on their downstream genes. This phenomenon, which was indicated by reduced or absent expression of downstream genes relative to the MM-glucose reference, was not anticipated due to the relatively large intergenic regions between the disrupted anti-σ and downstream sus-like genes (51, 114, and 161 bp). Nevertheless, it prohibited us from directly observing induction of the downstream genes.
To identify genes in the individual regulons associated with each of the trans-envelope signaling switches, we used our custom B. thetaiotaomicron GeneChips to generate whole-genome transcriptional profiles for each mutant strain as well as for the reference isogenic wild-type control. Each strain was grown to mid-exponential phase in MM-glucose, a condition that does not stimulate expression of any of these systems in wild-type B. thetaiotaomicron (three replicates were performed per strain; absorbance values at which cultures were harvested are provided in supplemental Table S2). Compared with wild type, a total of 295 genes exhibited expression changes ≥2.5-fold in 1 or more of the 5 different anti-σ mutants; 221 of these genes had higher expression than wild type, whereas 74 had lower expression (summarized in Table 1; for complete gene lists, plus -fold change values and their statistical significance, see supplemental Table S4). In the case of the two anti-σ disruptions where polar effects on downstream genes were not observed (ΩBT1052 and ΩBT3992), we were able to confirm increased expression of genes within the immediately adjacent PUL. Based on (i) autoregulation of the 5′ regions of the three anti-σ genes for which polarity was observed and (ii) previous evidence linking the ECF-σ genes associated with these three systems to PUL activation (14), we infer that similar regulation of adjacent genes also occurs in these systems.
TABLE 1.
Summary of genes with >2.5-fold expression changes in anti-σ mutants
| Gene categorya | ΩBT1052 (up/down) | ΩBT1052 ∷BT1052 (up/down) | ΩBT1618b (up/down) | ΩBT3992 (up/down) | ΩBT4249b (up/down) | ΩBT4356b (up/down) |
|---|---|---|---|---|---|---|
| CPS synthesis | 50/0 | 1/0 | 17/0 | 28/21 | 2/0 | 12/9 |
| PUL genes, total | 20/0 | 5/1 | 4/3 | 26/9 | 1/1 | 0/1 |
| PUL genes, unlinked to de-repressed PUL | 7/0 | 5/1 | 4/3 | 20/8 | 1/1 | 0/1 |
| CAZy, total | 12/0 | 1/2 | 0/1 | 12/3 | 1/1 | 1/1 |
| CAZy, unlinked to de-repressed PUL | 10/0 | 1/2 | 0/1 | 10/2 | 1/1 | 1/1 |
| Other functions | 8/3 | 24/3 | 12/8 | 12/10 | 6/2 | 7/7 |
| Total | 83/3 | 30/6 | 33/11 | 74/40 | 10/3 | 21/17 |
a Note that these categories are not mutually exclusive.
b These anti-σ disruptions were polar on downstream Sus-like outer membrane components. Therefore, they do not show de-repression of these linked PUL components. For a gene list, see supplemental Table S4.
Beyond regulation of adjacent PUL genes, other transcriptional changes were detected, especially in the non-polar BT1052 and BT3992 mutants (Table 1). These responses included induction of glycolytic functions that were not immediately linked to the PULs analyzed; a total of 12 genes encoding carbohydrate active enzymes (as defined in the CAZy data base (30)) were up-regulated, and 9 of these belonged to CAZy families with known relevance to O-glycan catabolism (Table 2). Thus, activation of individual ECF-σ regulons results in expression of glycan degrading functions that may augment the effectiveness of each PUL in substrate processing. Although we could not determine whether the activated ECF-σ factors directly or indirectly influence expression of these genes, the fact that their regulation occurs in MM-glucose medium reveals that their expression is linked to activation of the trans-envelope signaling switch per se and not to the sensing of glycan products liberated during O-glycan processing.
TABLE 2.
Induction of CAZy functions that are unlinked to de-repressed PULs
Activities that are relevant to mucin O-glycan degradation are shown in bold.
| Gene | CAZy family | Activities in group |
|---|---|---|
| BT0065 | Glycoside hydrolase, family 27 | α-N-Acetylgalactosaminidase, α-galactosidase |
| BT0456 | Glycoside hydrolase, family 20 | β-Hexosaminidase |
| BT0459 | Glycoside hydrolase, family 20 | β-Hexosaminidase |
| BT0460 | Glycoside hydrolase, family 20 | β-Hexosaminidase |
| BT0461 | Glycoside hydrolase, family 2 | β-Galactosidase, β-mannosidase, β-glucuronidase, β-glucosaminidase |
| BT0506 | Glycoside hydrolase, family 20 | β-Hexosaminidase |
| BT0683 | Glycoside hydrolase, family 97 | α-Glucosidase, α-galactosidase |
| BT1625 | Glycoside hydrolase, family 29 | α-l-Fucosidase |
| BT3169 | Glycoside hydrolase, family 31 | α-Glucosidase, α-xylosidase |
| BT3868 | Glycoside hydrolase, family 20 | β-Hexosaminidase |
| BT4395 | Glycoside hydrolase, family 84 | N-Acetyl-β-glucosaminidase, hyaluronidase |
| BT4690 | Glycoside hydrolase, family 13 | α-Glucosidase |
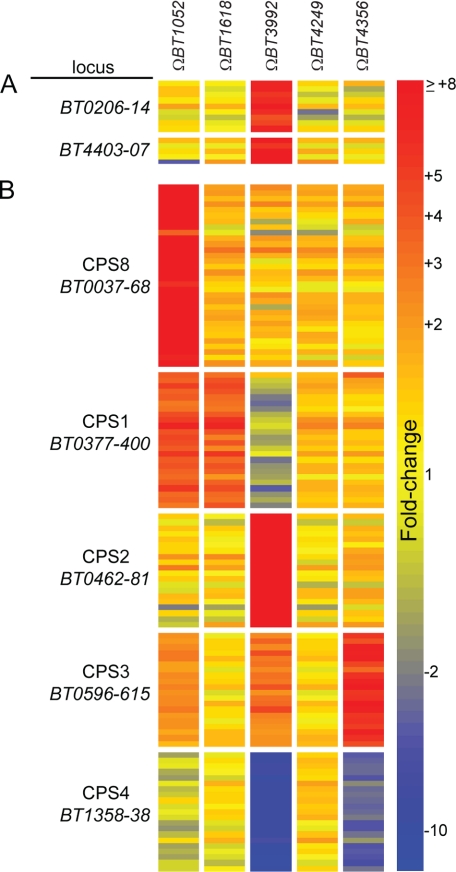
In addition to glycan degrading functions, some regulation of unlinked PUL genes was also observed (Table 1 and Fig. 4A). This phenomenon was most striking in the ΩBT3992 mutant, which caused up-regulation of genes in two different PULs. One PUL (BT0206–14) is not conspicuously paired with its own regulator. However, a second PUL (BT4403–07) is associated with its own ECF-σ/anti-σ gene pair, BT4402 and BT4403, respectively, and is likely regulated by its own dedicated trans-envelope signaling switch (the ECF-σ gene BT4402 is not listed with this PUL in Fig. 4 because it does not exhibit induction). Experiments described below suggest that the likely mechanism of increased BT4403–07 expression is via ECF-σBT3993 interaction with the BT4403–07 promoter(s) rather than anti-σBT3992 maturation of ECF-σBT4402 (i.e. cross-activation occurs at the level of ECF-σ/promoter and not via anti-σ/ECF-σ interactions). However, it is important to note that the average induction of the BT4403–07 PUL genes was only 16-fold in ΩBT3992 (-fold change values determined from normalized GeneChip intensities), whereas genes in the cognate BT3983–88 locus were up-regulated 336-fold in the same strain and dataset (supplemental Table S4).
FIGURE 4.
Other loci regulated by ECF-σ factors. A heatmap illustrating the transcriptional responses of genes in two PULs, BT0206–14 and BT4403–07 (A), and five B. thetaiotaomicron CPS synthesis loci (B) to mutation of five individual anti-σ factors and consequent de-repression of their corresponding ECF-σ-dependent regulons. Responses of individual genes within the locus tag range noted for each cluster are shown and are based on -fold change values determined from GeneChip datasets. Three biological replicates were performed for each anti-σ mutant and -fold changes quantified by referencing each gene to the average expression values observed in three datasets obtained from wild-type B. thetaiotaomicron grown in MM-glucose. -Fold change values are calibrated to the color bar shown at the right. Note that each of the PULs shown in panel A exhibits a specific response to disruption of anti-σBT3992, which activates ECF-σBT3993 but not other regulators. Also, four different CPS loci (CPS8, CPS1, CPS2, and CPS3) are activated differently in the various anti-σ mutants. One locus, CPS4, exhibits transcriptional repression under two conditions. Three remaining B. thetaiotaomicron VPI-5482 capsule loci (CPS5–7), which did not respond to any of the conditions tested, are not shown.
Regulation of Extracellular Capsule Expression by ECF-σ Factors
An unanticipated phenomenon was observed for all but one of the de-repressed ECF-σ regulons analyzed; artificial activation of these systems by anti-σ disruption in MM-glucose medium resulted in altered expression of some of the eight CPS gene clusters contained in B. thetaiotaomicron. These responses compose the largest class of regulated genes that are unlinked to the five trans-envelope signaling switches that we analyzed (Table 1). The B. thetaiotaomicron VPI-5482 type strain encodes 8 distinct CPS loci, each containing 15–32 genes that catalyze the transformation of activated precursor sugars (e.g. a nucleoside diphosphate-glucose), into various sugar derivatives, which are subsequently secreted and linked together to form a capsule (31, 32). Independent disruptions of four different anti-σ genes (BT1052, BT1618, BT3992, and BT4356) resulted in increased expression of four different CPS gene clusters (CPS8, CPS1, CPS2,and CPS3; Fig. 4B). Moreover, disruption of BT3992 and BT4356 resulted in reduced expression of CPS4, which ordinarily is the most highly expressed locus during in vitro growth and growth in vivo when dietary plant glycans are present (15, 21). Complementation of the ΩBT1052 mutant with a wild-type copy of BT1052, expressed under its native promoter, reduced expression of the two CPS loci (CPS8 and CPS1) that were activated in this mutant back to wild-type levels (Table 1 and supplemental Table S4), demonstrating that anti-σ disruption is causal to the observed CPS expression phenomenon.
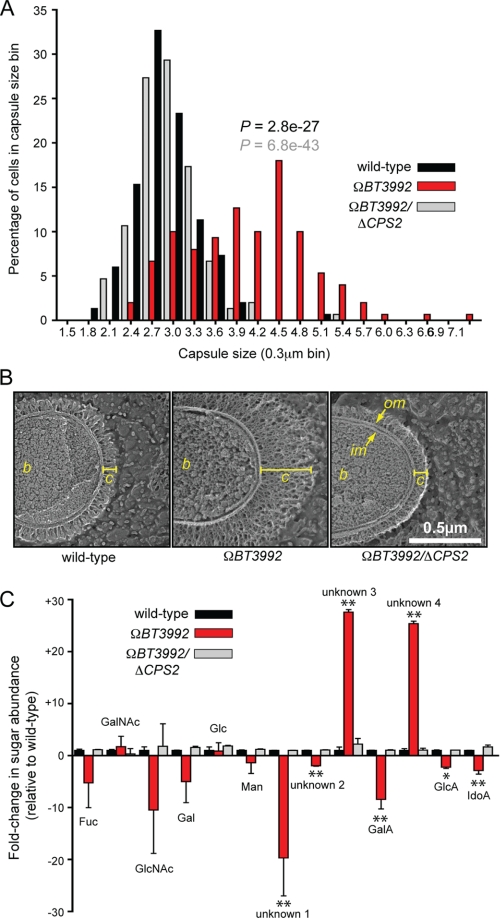
To further investigate the effects on capsule production, we selected the ΩBT3992 mutant for subsequent studies because it had the largest impact on capsule gene expression, an average 84-fold induction of the 21 genes comprising CPS2 (supplemental Table S4; -fold-change values were determined from normalized GeneChip intensities). We first analyzed extracellular capsule size in this mutant using two different techniques, standard light microscopy with India ink staining and quick- freeze, deep-etch scanning electron microscopy. Capsule width was measured in India ink-stained samples of wild-type B. thetaiotaomicron and compared with both the ΩBT3992 mutant and a ΩBT3992/ΔCPS2 double mutant (Fig. 5A); in this latter strain, ECF-σBT3993 was de-repressed, but the 21 genes comprising CPS2 were also deleted. The ΩBT3992 mutant, the only strain that retained elevated CPS2 expression, exhibited a significant increase in capsule size (average capsule width was 1.4 times greater than wild-type and 1.5 times greater than the ΩBT3992/ΔCPS2 double mutant; p values were 2.8e−27 and 6.8e−43, respectively, according to Student's t test; n = 150 individual cells scored/strain).
FIGURE 5.
Capsule production by the ΩBT3992 mutant. A, histogram plot of capsule sizes determined by India ink staining of the isogenic wild-type (black bars), ΩBT3992 (red bars), and the ΩBT3992/ΔCPS2 double mutant (gray bars) strains. Capsule sizes from a total of 150 individual cells were measured for each strain, 50 from each of three different experiments. p values are provided above the ΩBT3992 distribution and are color-coded based on the dataset (wild-type or double mutant) to which the ΩBT3992 distribution was compared. Representative images of India ink-stained encapsulated cells are provided in supplemental Fig. S5. B, quick-freeze, deep-etch scanning electron micrographs of wild-type and ΩBT3992 and ΩBT3992/ΔCPS2 double mutant cells, illustrating their capsule morphology. The capsule appears thicker in the ΩBT3992 mutant but of similar density as the other two strains. The width of each capsule (c) is indicated by a yellow line. The bacterial cell (b), inner membrane (im), and outer membrane (om) are labeled for reference. Magnification is 50,000×. C, analysis of monosaccharides present in wild-type and ΩBT3992 and ΩBT3992/ΔCPS2 double mutant capsules. A total of 13 different sugars were differentiated by HPAEC-PAD; nine of these eluted at the same time as known monosaccharide standards (see labels above histogram bars), whereas four compounds (labeled as unknown 1–4) did not correspond to any known standards used. Note the abundance of two uniquely represented and unknown sugars (unknowns 3 and 4) in the ΩBT3992 capsule. Total extracellular polysaccharides were extracted from cells grown in MM-glucose, and three biological replicates were performed for each strain. Values shown represent the average -fold increase of each sugar relative to the average present in wild-type cells, and error bars represent the S.D. Instances where sugar abundance in the ΩBT3992 mutant differed significantly from wild-type are indicated with asterisks (*, p ≤ 0.05; **, p ≤ 0.01 by Student's t test). Histogram bars are arranged along the x axis based on their elution points under the HPAEC-PAD conditions used (negative charge increases toward the right side of the graph). Representative HPAEC-PAD traces are provided in supplemental Fig. S6. Fuc, fucose; GalA, galacturonic acid; GlcA, glucuronic acid; IdoA, iduronic acid.
To gain a higher resolution view of the capsules expressed by these strains, we also examined their structures by quick-freeze, deep-etch scanning electron microscopy (Figs. 5B). Scanning electron microscopy micrographs confirmed the increased capsule size in the ΩBT3992 mutant and that this phenotype was diminished to near wild-type levels in the ΩBT3992/ΔCPS2 double mutant. Most notably, these high magnification views of the B. thetaiotaomicron capsule reveal the density and complexity of this bacterium's glycan-rich surface, an environment through which all assimilated nutrients, including the many other complex glycans that this species is capable of degrading, must pass to reach the cell surface.
Finally, we analyzed the compositions of the capsules produced by the wild-type and ΩBT3992 and ΩBT3992/ΔCPS2 strains. Extracellular polysaccharides extracted from bacterial cells grown to mid-exponential phase in MM-glucose were acid-hydrolyzed and assayed to determine their sugar content (Fig. 5C). Thirteen different sugars were detected; nine eluted identically to known monosaccharide standards, but four did not correspond to any standards used but were clearly distinguishable from the monosaccharide standards (see supplemental Fig. S6 for representative HPAEC-PAD traces). Compared with wild-type B. thetaiotaomicron, the sugars present in the ΩBT3992 capsule differed substantially. Two unidentified sugars that behaved similarly to acidic monosaccharides became more prominent with increased expression of CPS2, and these two sugars were diminished in the ΩBT3992/ΔCPS2 double mutant. Thus, in cells grown in vitro using glucose as a sole carbon source, the increased transcription of CPS2 clearly translates into a cellular phenotype; production of a larger surface capsule that differs biochemically from that usually expressed by wild-type B. thetaiotaomicron.
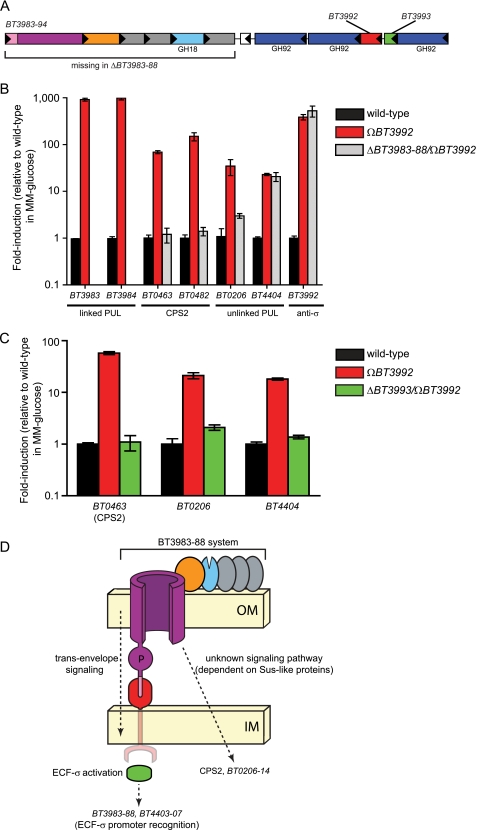
We attempted to culture B. thetaiotaomicron and another species with broad saccharolytic potential, Bacteroides ovatus, on a capsule extracted from the ΩBT3992 mutant. Neither species was able to use this extracted material as a sole carbon source. However, this lack of growth was not due to toxic inhibitors introduced during capsule extraction as the addition of glucose to minimal medium containing extracted CPS2 restored growth (data not shown). Based on these results, we concluded that the new cellular transcriptome expressed upon mutation of anti-σBT3992, which primarily involves increased expression of the O-glycan Sus-like system BT3983–88 (Fig. 6A), is associated with a regulated alteration of cell surface glycan chemistry. Possible physiologic reasons for the coordinated regulation of glycan metabolism with capsule structure are discussed below under “Prospectus.” To explore the basis of this coordination in more detail, we sought to characterize the regulatory pathway leading to increased CPS2 expression.
FIGURE 6.
Different regulatory pathways mediate CPS locus and PUL expression. A, a schematic of the BT3983–94 PUL. ORFs are color-coded according to the legend in Fig. 1, and predicted glycoside hydrolase (GH) activities are provided based on CAZy family designations. The white ORF in the middle of the locus is BT3989, a hypothetical gene that does not exhibit induction with the adjacent genes and may not be involved in PUL function. B, quantitative reverse transcriptase-PCR analysis of gene expression from three loci in wild-type (black bars) and ΩBT3992 (red bars) and ΩBT3992/ΔBT3983–88 (gray bars) strain backgrounds. Two genes, BT3983 and BT3984, encoding outer membrane protein components of the BT3983–94 Sus-like system, exhibit strong induction when anti-σBT3992 is disrupted. Note that these two transcripts are absent and, therefore, are not detectable in the ΩBT3992/ΔBT3983–88 double mutant. Similarly, two genes (BT0463 and BT0482) positioned near the beginning and end of CPS2, show induction in the ΩBT3992 strain. However, this induction is reduced to wild-type levels in the ΩBT3992/ΔBT3983–88 double mutant, indicating that expression of the BT3983–88 Sus-like components is required for CPS activation. A probe gene (BT0206) from one PUL (BT0206–14) that is unlinked to BT3992 is also dependent on expression of the BT3983–88 for its activation. Conversely, induction of a transcript (BT4404) from another unlinked PUL (BT4403–07) in both mutant strains indicates that the induction of unlinked genes is not always dependent on the presence of outer membrane components. Similarly, regulation of BT3992 is unaffected by loss of the Sus-like proteins. C, expression of genes in the ECF-σBT3993 regulon in isogenic wild-type (black bars) and ΩBT3992 (red bars) and ΩBT3992/ΔBT3993 (green bars) strains. Loss of ECF-σBT3993 eliminates activation of all genes in the regulon. Most notably, expression of BT4404, a component of the ECF-σ-linked BT4403–07 PUL, is reduced to wild-type levels. Thus, the cross-talk between these loci must occur at the level of ECF-σBT3993 activation of the BT4404 promoter and not at other, non-cognate signaling nodes, such as anti-σBT3992 activation of ECF-σBT4403, which would still be intact in the ΩBT3992/ΔBT3993 strain. D, a model summarizing the results of data presented in panels B and C. Note that the unknown signaling pathway that controls CPS2 and BT0206–14 expression is also dependent on the trans-envelope signaling pathway, which is required for expression of the BT3983–88 Sus-like system. IM, inner membrane; OM, outer membrane.
Regulation of CPS2 Synthesis Is Mediated by a Novel Pathway Requiring Sus-like Proteins
Two of five de-repressed ECF-σ regulons (disruptions of anti-σ genes BT1052 and BT3992) resulted in the strongest inductions of CPS loci (13- and 81-fold average induction of CPS8 and CPS2, respectively; Fig. 4 and supplemental Table S4). In contrast to the remaining two anti-σ disruptions (BT1618 and BT4356), which resulted in 4.1- and 6.5-fold changes, respectively, neither of these two mutants that caused the larger changes in CPS expression was associated with polar effects on their downstream sus-like genes. This observation led us to hypothesize that the transcriptional activation pathway(s) leading to CPS expression is dependent on the presence of components of the Sus-like outer membrane systems. Therefore, we reconstructed the anti-σ BT3992 gene insertion (ΩBT3992) that caused de-repression of CPS2 in a strain that also had a deletion of the entire BT3983–88 operon, which encodes the primary mucin O-glycan-responsive outer membrane system regulated by this trans-envelope signaling switch (see Fig. 6A for a schematic of this PUL). This double mutant (ΩBT3992/ΔBT3983–88) was grown to mid-log phase in MM-glucose medium, alongside the isogenic wild-type strain and the original ΩBT3992 single mutant, to determine whether loss of BT3983–88 influences CPS2 expression (Fig. 6B). Remarkably, quantitative reverse transcriptase-PCR analysis of transcript abundance revealed that the ΩBT3992/ΔBT3983–88 double mutant was unable to activate expression of probe genes (BT0463, BT0482, and BT0206) belonging to the CPS2 locus and the unlinked BT0206–14 PUL. In contrast, control assays indicated successful depression of the ECF-σBT3993 regulon in this double mutant; de-repression was confirmed by probing autoregulation of BT3992 as well as a gene (BT4404) from the unlinked BT4403–07 PUL, neither of which exhibited dependence on BT3983–88 for their expression.
Thus, a different signaling pathway, which requires expression of a Sus-like outer membrane system, controls CPS2 and BT0206–14 expression (Fig. 6D). The components of this pathway that ultimately influence these loci are unknown. However, we can eliminate the possibility that the mechanism responsible for activating these loci is simply the result of ECF-σBT3993 recognition of the CPS2 and BT0206–14 promoters; more likely, it involves another pathway that begins with expression of one or more proteins in the BT3983–88 system and ends with activation of the CPS2 promoter.
Finally, we probed expression of the CPS2 locus and the two different unlinked PULs that exhibited activation in the ΩBT3992 strain in an additional ΩBT3992/ΔBT3993 double mutant that also lacks ECF-σBT3993. Although this mutant expresses the same de-repressed truncated version of anti-σBT3992 as the single mutant described above, the absence of ECF-σBT3993 renders it incapable of activating gene expression through its cognate ECF-σ factor but could allow it to signal via non-cognate anti-σ/ECF-σ interactions. Like CPS2 and the BT0206–14 PUL, we found that the BT4403–07 PUL also requires ECF-σBT3993 for activation (Fig. 6C).
Therefore, the two unlinked PULs that show activation by ECF-σBT3993 each appear to be controlled by different routes; one system (BT0206–14) is regulated by a similar mechanism as CPS2 and is dependent on expression of Sus-like proteins, whereas the other system is likely induced by ECF-σBT3993 activation of a related promoter residing upstream of BT4403–07 (see Fig. 6D for a model). This latter case provides a likely example of ECF-σ/promoter cross-talk between two similar PULs and may reflect their recent duplication and diversification from a single ancestral locus (13). Future experiments will be required to explore each of these regulatory pathways in more detail, especially the unanticipated connection between expression of the BT3983–88 Sus-like system and the CPS2 and BT0206–14 loci.
Prospectus
Metabolism of complex carbohydrates is a fundamental force shaping the evolution of the human distal gut microbiota. In this competitive ecosystem, the needs of gut symbionts to efficiently harvest complex glycans as nutrients must be balanced with their simultaneous need to shield themselves from harmful environmental factors, including the innate and adaptive branches of the immune system, phage attacks, and antimicrobial peptides produced by the host and/or members of a microbial community. A common mechanism employed by many bacteria to protect against some of these environmental assaults is to produce polysaccharide capsules, which are often varied within subpopulations of the same species (e.g. by phase variation) to pre-adapt some lineages to future challenges (6). Although phase variation of microbial surface structures is frequently accomplished through random alterations in gene expression, such as promoter inversion, we show here that a broadly saccharolytic human gut symbiont, B. thetaiotaomicron, also modulates its surface by directly coordinating CPS production with its expression of Sus-like outer membrane proteins deployed during glycan acquisition. In turn, expression of Sus-like systems is primarily controlled by a variety of positive-acting transcription factors (e.g. ECF-σ factors and hybrid two-component systems, plus others), whose influence may be modulated by additional forms of regulation, such as catabolite repression and recombinational gene shuffling.4 Thus, B. thetaiotaomicron uses a complex signaling network to link the nutrient milieu of its gut environment with its extracellular capsule.
The immediate physiologic benefit obtained from coupling CPS synthesis to glycan catabolism remains obscure. However, the observation that B. thetaiotaomicron endures the metabolic cost of building a novel capsule structure encoded by CPS2 de novo, which differs chemically from the O-glycans targeted by the BT3983–88 Sus-like system and cannot be subsequently re-assimilated as a nutrient, supports the notion that this regulated cellular event plays some role in facilitating glycan acquisition. From this perspective, the high magnification scanning electron microscopy views of the B. thetaiotaomicron capsule are insightful; all cellular glycan catabolism by outer membrane-localized Sus-like systems must occur after substrates have transited the “forest” of the surface capsule, which can be several hundred nanometers thick. It is possible that one role of coordinated CPS and Sus-like system expression is to optimize the miscibility of target glycans and other nutrients within this capsule layer. A second possibility, which is not incompatible with the previous explanation, is that some capsule structures interfere with the glycan binding sites of certain Sus-like systems. Coordinated expression of a non-interfering capsule (e.g. CPS2 when BT3983–88 is expressed), which differs in its chemistry from the environmental glycans being targeted as nutrients, could alleviate this interference. Finally, work in the related gut symbiont B. fragilis has shown that at least one metabolic salvage pathway (l-fucose assimilation) serves as a non-de novo source of monosaccharides for CPS synthesis (33). B. fragilis adds l-fucose to several of its surface capsules but only does so when this sugar is salvageable from the growth environment. Although we show that expression of the CPS2 gene cluster by B. thetaiotaomicron directs synthesis of a capsule containing at least two prominent sugars when only glucose is present in the medium, it is possible that expression of this system in the context of actual O-glycan degradation, and the consequent availability of sugars other than glucose in the growth milieu, could result in additional linkages being added to the CPS2 polymer.
Taken together, the findings reported here reveal a link between some of the complex glycans encountered in the mammalian gut (mucin O-glycans) and the surface glycan structures of B. thetaiotaomicron, a representative of one of the two prominent bacterial phyla commonly inhabiting this human body habitat (2). In light of previous experiments, which observed shifts in CPS expression upon alteration of the types of dietary glycans being consumed by gnotobiotic mice colonized with B. thetaiotaomicron (15, 21), similar mechanisms may mediate coordination of glycan degradation and CPS synthesis in response to the presence of dietary and/or microbial carbohydrates. Additional studies in our laboratory have demonstrated that expression of at least one B. thetaiotaomicron CPS locus, CPS4, confers a fitness advantage in vivo in the distal gut of gnotobiotic mice (5). However, in gnotobiotic mice with an engineered and sustained CPS4-specific secretory IgA response, continued CPS4 expression becomes a disadvantage to the bacterium. Thus, a series of dynamic regulatory links likely couple the complex glycan milieu of the gut and the responses of the host immune system to the expressed surface carbohydrate repertoire of this and other gut Bacteroides in order to support sustained colonization. Future experiments will be required to further explore the molecular mechanisms underlying these regulatory pathways. The results should provide important insights into the complex and dynamic relationship between our diets, the structures and functions of our gut microbial communities, and our host physiology.
Supplementary Material
Acknowledgments
The University of California at San Diego Glycotechnology Core Resource skillfully performed all glycan analyses. We thank Andrew Goodman, Jeremiah Faith, Ansel Hsiao, Federico Rey, Henning Seedorf, Michelle Smith, and Justin Sonnenburg for many helpful suggestions during the course of these studies and Laura Kyro for assistance with graphics.
This work was supported, in whole or in part, by National Institutes of Health Grant DK30292. This work is also supported by the Missouri Life Sciences Trust Fund.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S6 and Tables S1–S5.
4 E. C. Martens and J. I. Gordon, manuscript in preparation.
- PUL
- polysaccharide utilization loci
- ECF-σfactor
- extra-cytoplasmic function σ factor
- CPS
- capsular polysaccharide
- MM
- Minimal medium
- HPAEC-PAD
- high pH anion exchange chromatography with pulsed amperometric detection
- CAZy
- carbohydrate active enzyme
- ORF
- open reading frame.
REFERENCES
- 1.Flint H. J., Bayer E. A., Rincon M. T., Lamed R., White B. A. ( 2008) Nat. Rev. Microbiol. 6, 121– 131 [DOI] [PubMed] [Google Scholar]
- 2.Ley R. E., Hamady M., Lozupone C., Turnbaugh P. J., Ramey R. R., Bircher J. S., Schlegel M. L., Tucker T. A., Schrenzel M. D., Knight R., Gordon J. I. ( 2008) Science 320, 1647– 1651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kurokawa K., Itoh T., Kuwahara T., Oshima K., Toh H., Toyoda A., Takami H., Morita H., Sharma V. K., Srivastava T. P., Taylor T. D., Noguchi H., Mori H., Ogura Y., Ehrlich D. S., Itoh K., Takagi T., Sakaki Y., Hayashi T., Hattori M. ( 2007) DNA Res. 14, 169– 181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Turnbaugh P. J., Hamady M., Yatsunenko T., Cantarel B. L., Duncan A., Ley R. E., Sogin M. L., Jones W. J., Roe B. A., Affourtit J. P., Egholm M., Henrissat B., Heath A. C., Knight R., Gordon J. I. ( 2009) Nature 457, 480– 484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Peterson D. A., McNulty N. P., Guruge J. L., Gordon J. I. ( 2007) Cell Host Microbe 2, 328– 339 [DOI] [PubMed] [Google Scholar]
- 6.Coyne M. J., Comstock L. E. ( 2008) J. Bacteriol. 190, 736– 742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mazmanian S. K., Round J. L., Kasper D. L. ( 2008) Nature 453, 620– 625 [DOI] [PubMed] [Google Scholar]
- 8.Booth S. J., Van Tassell R. L., Johnson J. L., Wilkins T. D. ( 1979) Rev. Infect. Dis. 1, 325– 336 [DOI] [PubMed] [Google Scholar]
- 9.Clements A., Gaboriaud F., Duval J. F., Farn J. L., Jenney A. W., Lithgow T., Wijburg O. L., Hartland E. L., Strugnell R. A. ( 2008) PLoS ONE 3, e3817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Llobet E., Tomás J. M., Bengoechea J. A. ( 2008) Microbiology 154, 3877– 3886 [DOI] [PubMed] [Google Scholar]
- 11.Branda S. S., Vik S., Friedman L., Kolter R. ( 2005) Trends Microbiol. 13, 20– 26 [DOI] [PubMed] [Google Scholar]
- 12.Salyers A. A., Vercellotti J. R., West S. E., Wilkins T. D. ( 1977) Appl. Environ. Microbiol. 33, 319– 322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xu J., Mahowald M. A., Ley R. E., Lozupone C. A., Hamady M., Martens E. C., Henrissat B., Coutinho P. M., Minx P., Latreille P., Cordum H., Van Brunt A., Kim K., Fulton R. S., Fulton L. A., Clifton S. W., Wilson R. K., Knight R. D., Gordon J. I. ( 2007) PLoS Biol. 5, e156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Martens E. C., Chiang H. C., Gordon J. I. ( 2008) Cell Host Microbe 4, 447– 457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bjursell M. K., Martens E. C., Gordon J. I. ( 2006) J. Biol. Chem. 281, 36269– 36279 [DOI] [PubMed] [Google Scholar]
- 16.D'Elia J. N., Salyers A. A. ( 1996) J. Bacteriol. 178, 7180– 7186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Karlsson N. G., Nordman H., Karlsson H., Carlstedt I., Hansson G. C. ( 1997) Biochem. J. 326, 911– 917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Koebnik R. ( 2005) Trends Microbiol. 13, 343– 347 [DOI] [PubMed] [Google Scholar]
- 19.Holdeman L. V., Cato E. D., Moore W. E. C. ( 1977) Anaerobe Laboratory Manual, Virginia Polytechnic Institute and State University Anaerobe Laboratory, Blacksburg, VA [Google Scholar]
- 20.Koropatkin N. M., Martens E. C., Gordon J. I., Smith T. J. ( 2008) Structure 16, 1105– 1115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sonnenburg J. L., Xu J., Leip D. D., Chen C. H., Westover B. P., Weatherford J., Buhler J. D., Gordon J. I. ( 2005) Science 307, 1955– 1959 [DOI] [PubMed] [Google Scholar]
- 22.Li C., Wong W. H. ( 2001) Proc. Natl. Acad. Sci. U.S.A. 98, 31– 36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Heuser J. E., Salpeter S. R. ( 1979) J. Cell Biol. 82, 150– 173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Heuser J. E. ( 1981) Trends Biochem. Sci. 6, 64– 68 [Google Scholar]
- 25.Heuser J. E. ( 1989) Prog. Clin. Biol. Res. 295, 71– 83 [PubMed] [Google Scholar]
- 26.Claros M. G., von Heijne G. ( 1994) Comput. Appl. Biosci. 10, 685– 686 [DOI] [PubMed] [Google Scholar]
- 27.Gardy J. L., Spencer C., Wang K., Ester M., Tusnády G. E., Simon I., Hua S., deFays K., Lambert C., Nakai K., Brinkman F. S. ( 2003) Nucleic Acids Res. 31, 3613– 3617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vingadassalom D., Kolb A., Mayer C., Rybkine T., Collatz E., Podglajen I. ( 2005) Mol. Microbiol. 56, 888– 902 [DOI] [PubMed] [Google Scholar]
- 29.Ochs M., Veitinger S., Kim I., Welz D., Angerer A., Braun V. ( 1995) Mol. Microbiol. 15, 119– 132 [DOI] [PubMed] [Google Scholar]
- 30.Cantarel B. L., Coutinho P. M., Rancurel C., Bernard T., Lombard V., Henrissat B. ( 2009) Nucleic Acids Res. 37, 233– 238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xu J., Bjursell M. K., Himrod J., Deng S., Carmichael L. K., Chiang H. C., Hooper L. V., Gordon J. I. ( 2003) Science 299, 2074– 2076 [DOI] [PubMed] [Google Scholar]
- 32.Coyne M. J., Chatzidaki-Livanis M., Paoletti L. C., Comstock L. E. ( 2008) Proc. Natl. Acad. Sci. U.S.A. 105, 13099– 13104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Coyne M. J., Reinap B., Lee M. M., Comstock L. E. ( 2005) Science 307, 1778– 1781 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.