Abstract
Cry toxins produced by the bacterium Bacillus thuringiensis are effective biological insecticides. Cadherin-like proteins have been reported as functional Cry1A toxin receptors in Lepidoptera. Here we present data that demonstrate that a coleopteran cadherin is a functional Cry3Aa toxin receptor. The Cry3Aa receptor cadherin was cloned from Tenebrio molitor larval midgut mRNA, and the predicted protein, TmCad1, has domain structure and a putative toxin binding region similar to those in lepidopteran cadherin B. thuringiensis receptors. A peptide containing the putative toxin binding region from TmCad1 bound specifically to Cry3Aa and promoted the formation of Cry3Aa toxin oligomers, proposed to be mediators of toxicity in lepidopterans. Injection of TmCad1-specific double-stranded RNA into T. molitor larvae resulted in knockdown of the TmCad1 transcript and conferred resistance to Cry3Aa toxicity. These data demonstrate the functional role of TmCad1 as a Cry3Aa receptor in T. molitor and reveal similarities between the mode of action of Cry toxins in Lepidoptera and Coleoptera.
The mode of action of Bacillus thuringiensis insecticidal Cry toxins has been extensively studied in lepidopteran larvae (1). Our current understanding is that the major factors that contribute to Cry toxicity in insects include solubilization and activation of the crystalline toxin as well as interactions between toxin and midgut receptors. In lepidopterans, several insect midgut proteins have been proposed as Cry toxin receptors (2). Cry1A receptor functionality has been demonstrated for cadherin proteins from Bombyx mori (3, 4), Manduca sexta (5, 6), Ostrinia nubilalis (7), and Heliothis virescens (8). The specific toxin-binding region in lepidopteran cadherins has been localized proximal to the cell membrane insertion site (9–11). Mutations in toxin binding motifs of lepidopteran cadherin genes are genetically linked to Cry1Ac resistance in H. virescens (12, 13), Helicoverpa armigera (14, 15), and Pectinophora gossypiella (9, 16).
Interactions between Cry toxins and cadherin receptors and the implications for toxicity have been studied in Lepidoptera more extensively than in any other insect order (2). According to the model proposed by Bravo et al. (17), Cry toxin binding to cadherin is followed by toxin oligomerization. Toxin oligomers reportedly are intermediates required for effective pore formation and ultimately toxicity (18). A fragment of the BtR1 cadherin from M. sexta, corresponding to repeat 12 and containing a critical toxin-binding region, enhanced the activity of Cry1A toxins in Lepidoptera (19) by promoting toxin oligomerization (20). However, other studies in Lepidoptera have found that cadherin fragments can reduce Cry1A toxicity (11, 21). An alternative model suggests that Cry toxin binding to cadherin receptors activates intracellular pathways leading to cell death (22). Notably, in both models, cadherin is a critical contact point for Cry toxins that is pivotal for intoxication.
In contrast to the lepidopteran model, relatively little is known about the steps involved in the intoxication process by coleopteran-specific Cry toxins. The solubility of one such toxin, Cry3Aa, was considered a determining factor for toxicity in the coleopteran midgut (23–25). Cry3Aa toxin binding assays and ligand blots with brush border membrane vesicles (BBMV)2 from Tenebrio molitor larval midgut demonstrated binding to a single high affinity (Kd = 17.5 nm) site on an unidentified 144-kDa BBMV protein (26). An ADAM metalloprotease in Leptinotarsa decemlineata (27) and a cadherin-like protein in Diabrotica virgifera virgifera (28) were proposed as putative Cry toxin receptor proteins. However, neither study demonstrated functional interactions between putative receptor and toxin. More recently, a partial D. virgifera virgifera cadherin fragment corresponding to cadherin repeat (CR) domains 8–10 was reported to bind activated Cry3Aa and Cry3Bb toxins and enhance toxin activity in several beetles, suggesting that cadherin plays a functional role in B. thuringiensis intoxication in beetles (29). Another study reported the oligomerization of Cry3Aa toxin after incubation with L. decemlineata BBMV, although the proteins involved in oligomerization were not identified (30).
The high degree of structural similarity among Cry1, -2, -3, and -4 toxins implies that they share a similar mode of action in insects (2, 31). To test this hypothesis, we evaluated whether a cadherin in the midgut of a B. thuringiensis-sensitive coleopteran is a functional receptor for coleopteran-specific Cry3Aa toxin. Our data indicate that Cry3Aa binds specifically to a cadherin-like protein from T. molitor. A peptide containing the predicted toxin binding region from T. molitor cadherin bound Cry3Aa toxin specifically and promoted Cry3Aa oligomerization in solution. More importantly, we demonstrate that reduced levels of the TmCad1 (T. molitor cadherin) transcript in actively feeding larvae correlate with a reduction in Cry3Aa toxicity. These results support the hypothesis that T. molitor cadherin is a functional Cry3Aa receptor analogous to lepidopteran cadherin receptors and highlight a common interface for Cry toxins in two major insect orders.
EXPERIMENTAL PROCEDURES
Insect Larvae, Bacterial Strains, and Toxin Purification
T. molitor larvae were from a laboratory colony reared on a diet of 50% rolled oats, 47.5% wheat flour, and 2.5% brewers' yeast at 60% relative humidity, 28 °C, no photoperiod.
Cry3Aa protoxin was purified from sporulated cultures of B. thuringiensis var. tenebrionis following methods described elsewhere (32). Purified Cry3Aa protoxin was quantified using the Coomassie Plus® protein assay (Pierce) with bovine serum albumin (BSA) as a standard.
BBMV Preparation
The isolation of T. molitor BBMV was performed as previously described (33). Purified BBMV were quantified as described previously for toxin quantification and were stored at −80 °C. N-Aminopeptidase activity was determined by the substrate leucine-p-nitroanilide and was used as a marker for brush border enzyme enrichment in the BBMV preparations. N-Aminopeptidase activities were enriched 3–5-fold in the BBMV preparations compared with initial midgut homogenates.
Cloning of TmCad1 cDNA
A partial cDNA encoding a T. molitor cadherin was identified among randomly selected clones from a T. molitor larval midgut cDNA library (34). Gene-specific primers were designed based on this sequence, and the complete coding sequence (TmCad1) was obtained from larval midgut mRNA by 5′- and 3′-rapid amplification of cDNA ends using the GeneRacer™ kit from Invitrogen and SuperTaq™ Plus DNA polymerase (Ambion, Austin, TX). Tm1 and Tm2 nucleotide primers (Table 1) were designed from the TmCad1 sequence in the sense orientation and were used with the GeneRacer 3′-primer to amplify the 3′-end. Similarly, PCR primers were designed in the antisense orientation (Tm3, Tm4, Tm5, Tm6, Tm7, Tm8, Tm9, and Tm10; Table 1) and were used with the GeneRacer 5′-primer and GeneRacer 5′-nested primer to amplify the missing 5′ cDNA fragments. PCR products were gel-purified and inserted into pCR2.1-TOPO or pCR4-TOPO cloning vectors (Invitrogen). DNA sequencing was performed using the GenomeLab™ DTCS Quick Start Kit on a CEQ8000 DNA sequencer (Beckman). Nucleotide primers (Tm11, Tm12, Tm13, Tm14, and Tm15) were designed from known TmCad1 and used to sequence missing internal regions of the subcloned cDNA.
TABLE 1.
Nucleotide primers used to obtain full-length TmCad1 cDNA, obtain missing internal cDNA fragments, generate partial rTmCad1p toxin-binding region fragment for protein expression, synthesize dsRNA, and amplify TmCad1 and RPS6 quantitative real-time PCR products
| Primer | Orientation | Position | Primer DNA Sequence |
|---|---|---|---|
| Tm1 | Sense | 4538–4563 | 5′-TGAAAGCGTGGTTGATCGGTGTTTCG-3′ |
| Tm2 | Sense | 4648–4676 | 5′-TCCAGTACCAAATTCGGGTCGCAAGAG-3′ |
| Tm3 | Antisense | 4152–4179 | 5′-GGCATCAGCTTTGTGATTTTCCGGCTCT-3′ |
| Tm4 | Antisense | 4018–4042 | 5′-TGTCCAGGTCGAGGTTAGATGGAGT-3′ |
| Tm5 | Antisense | 4055–4079 | 5′-TCTCCGGATTGCGTATTCATGGTAA-3′ |
| Tm6 | Antisense | 3864–3893 | 5′-TCAAACACTGGAGATTCGTCGTTCTGGTCT-3′ |
| Tm7 | Antisense | 3788–3811 | 5′-GCTTGTCAGCGTTAGATGACTGAA-3′ |
| Tm8 | Antisense | 3734–3753 | 5′-GAGCGGTTGTTTAAGGGTGA-3′ |
| Tm9 | Antisense | 2906–2929 | 5′-TGTCACCTTCATCGTCATCTTTCC-3′ |
| Tm10 | Antisense | 1388–1412 | 5′-TCATCGTTGCATATCATTTAGGTTGA-3′ |
| Tm11 | Sense | 1830–1853 | 5′-CGACGCAGATTTGGAGTTCTCGAT-3′ |
| Tm12 | Antisense | 2267–2290 | 5′-CAACCCAGTCGGGAGTGTTCTCAT-3′ |
| Tm13 | Sense | 377–404 | 5′-TCAAGAACTTGGACGACGAACATCCGAC-3′ |
| Tm14 | Antisense | 883–909 | 5′-GGCATCCACCGTAGCGAAGTTGTTCTC-3′ |
| Tm15 | Antisense | 1023–1044 | 5′-AATGTCTTCAAGGATCAGCAGT-3′ |
| Tm16 | Sense | Adapter | 5′-CACCGAGCACGAGGACACTGACAT-3′ |
| Tm17 | Antisense | 4526–4548 | 5′-CTACCACGCTTTCAAAATTGCTTCCA-3′ |
| Tm18 | Sense | 1982–2001 | 5′-AGGAAACACAACCTGGCAAC-3′ |
| Tm19 | Antisense | 2454–2473 | 5′-CCATGACAGGAACATTGTCG-3′ |
| Tm20 | Sense | 1982–2001 | 5′-taatacgactcactatagggAGGAAACACAACCTGGCAAC-3′ |
| Tm21 | Antisense | 2215–2234 | 5′-taatacgactcactatagggTCCATGTCAGCGTCAGTAGC-3′ |
| RPS6f | Sense | 401–420 | 5′-GGCCCAAGCGAGCATCTAAC-3′ |
| RPS6r | Antisense | 567–584 | 5′-GAGCGCCAACCTGTGACG-3′ |
Sequence Analysis
Multiple sequence alignments were constructed using ClustalW version 1.83 from EMBL-EBI (available on the World Wide Web). The TmCad1 sequence and predicted amino acid sequence were compared with those in public data bases, and all sequences with significant relatedness were identified.
Percent identity scores were calculated from single pairwise alignments using AlignX from Vector NTI Advance™ version 9.1.0 (Invitrogen) and from the ClustalW sequence alignment. Phylogenetic data were obtained from alignments (DIALIGN version 2.2.1) and used to generate protein distances from the algorithm protdist (Phylip version 3.6a3) and bootstrap analysis of 100 replicates using the Jones, Taylor, and Thorton model of amino acid substitution (35). Phylogenetic trees were made using a PHYLIP drawgram with branch lengths made from the Kitsch distance matrix.
Cloning, Expression, and Purification of the Predicted TmCad1 Toxin Binding Region
Primers Tm16 and Tm17 were used for PCR with KOD high fidelity DNA polymerase (EMD Biosciences, San Diego, CA) to amplify a 582-bp region from the TmCad1 cDNA (nucleotides 3,963–4,548). The generated amplicon was gel-purified and inserted into the Escherichia coli expression vector pET151-D-TOPO® (Invitrogen) to generate the pET151-rTmCad1p construct. Insertion of the correct sequence into the expression vector was confirmed by sequencing in both directions.
For expression, BL21 Star™ (DE3) E. coli were transformed with pET151-rTmCad1p, and cultures were grown as previously described (9). Expressed rTmCad1p peptide containing a histidine tag at the amino terminus was purified using Ni2+ affinity chromatography. Protein was extracted from E. coli inclusion bodies, and purification was performed under hybrid denaturing/native conditions, as described elsewhere (9). Eluted fractions containing rTmCad1p were pooled and dialyzed against 0.01 m Tris-HCl, pH 8.0, 0.01% Triton X-100®. For most applications (unless noted otherwise), rAcTEV protease (Invitrogen) was used to remove 27 of the 33 additional amino acid residues at the amino terminus of rTmCad1p, including the His6 tag and a V5 epitope. Purification of rAcTEV protease-treated rTmCad1p was performed according to the manufacturer's recommendations. Purified rTmCad1p was analyzed by SDS-PAGE in 10–20% Tricine gels (Invitrogen), and protein concentration was determined as for toxin quantification. Concentration and buffer exchange of rTmCad1p was with Centricon® centrifugal filters (Millipore Corp., Bedford, MA).
Binding of Cry3Aa to Purified rTmCad1p
Dot blot overlay binding assays were performed using previously described procedures adapted for Cry3Aa (9, 11). For toxin immunodetection, rabbit Cry3Aa antiserum was provided by AgDia (Elkhart, IN).
For in-gel binding assays, rTmCad1p was labeled with a fluorescent dye (IR800TmCad1p), using the IRDye® 800CW protein labeling kit (LI-COR Biosciences, Lincoln, NE). The infrared dye forms a stable ester conjugate with the peptide and has an emission maximum of 789 nm. Binding assays were performed according to the manufacturer's recommendations (LI-COR Biosciences). Briefly, ∼1–3 μg of rTmCad1p with an intact His6 tag, Cry3Aa protoxin, or BSA was combined with SDS sample buffer without reducing reagent or heat, and proteins were separated by 10% BisTris SDS-PAGE using MOPS buffer (Invitrogen). After electrophoresis, gels were fixed in 50% isopropyl alcohol for 15 min and washed three times in deionized water for 5 min each. Duplicate gels were either stained with Coomassie Blue (Imperial Protein Stain; Pierce) or incubated with 60 ng of IRTmCad1p in 10 ml of Odyssey blocking buffer (LI-COR Biosciences) containing 50 mm CaCl2, with or without a 500-fold molar excess of unlabeled rTmCad1p, in complete darkness for 1 h at room temperature with gentle shaking. Gels were washed three times with buffer (0.002 m imidazole-buffered saline with 0.02% Tween 20; KPL, Gaithersburg, MD) and were scanned at 700 and 800 nm on an Odyssey Imager using version 1.2.15 Odyssey software (LI-COR Biosciences).
Cry3Aa Oligomerization Assays
Purified Cry3Aa protoxin was labeled with NHS-LC-biotin (Pierce), as described elsewhere (37). Labeled protoxin (1.2 μg) was incubated for 1 h at room temperature with T. molitor BBMV (7.8 μg) in 100 μl (final volume) of phosphate-buffered saline alone or with either purified rTmCad1p (1.4 μg) or unlabeled Cry3Aa protoxin (24 μg). Unbound Cry3Aa protoxin was removed from BBMV by centrifugation (16,100 × g for 10 min) and washing pellets twice in 100 μl of ice-cold binding buffer. BBMV pellets were solubilized in sample buffer (38) and heat-denatured at 95 °C for 5 min prior to separation on 10% SDS-polyacrylamide gels. After electrophoresis, proteins were transferred to polyvinylidene difluoride overnight at 4 °C. Filters were blocked for 1 h at room temperature in phosphate-buffered saline, pH 7.5, 0.1% Tween 20 (PBS-T), 3% BSA. After blocking, biotinylated Cry3Aa protoxin was detected by streptavidin conjugated to horseradish peroxidase and enhanced chemiluminescence (Western Pico; Pierce).
Knockdown of TmCad1 Using RNA Interference
Initial template DNA was obtained by amplifying genomic DNA with primers Tm18 and Tm19 (Table 1). After sequence confirmation, a template for in vitro transcription was generated from the PCR product using gene-specific primers tailed with the T7 polymerase promoter sequence (Tm20 and Tm21). dsRNA was prepared with the Ambion MEGAscript high yield transcription kit (Applied Biosystems/Ambion, Austin, TX) according to the manufacturer's protocols. Purified dsRNA was stored at −80 °C until injected into T. molitor larvae.
Cohorts of T. molitor larvae (∼1 month old) were injected with ∼0.5 μl of filter-sterilized injection buffer (0.1 mm sodium phosphate, pH 7.2, 5 mm potassium chloride, 0.2% (v/v) green food color, n = 50) or injection buffer mixed 1:1 with dsRNA (∼470 ng/larva; n = 38). Fifty additional larvae were used as noninjected control group. Larvae surviving the injection process were reared on diet without toxin under standard rearing conditions until initiation of bioassays.
Quantitative Real Time PCR
Four days postinjection, larvae were evaluated for knockdown of TmCad1. Two larval guts from each of the control or injection groups were dissected into RNAlater®, and total RNA was isolated (RNeasy minikit; Invitrogen). Quantitative real-time PCR was performed on each template using forward (Tm18) and reverse (Tm19) primers (Brilliant II SYBR Green QRT-PCR master mix kit, Mx3000P thermocycler; Stratagene/Agilent, Santa Clara, CA). Relative -fold calculations were made with duplicates for each treatment group within the MX3000P software using RPS6 (ribosomal protein S6) to normalize gene expression (RPS6f forward primer and RPS6r reverse primer). Data were analyzed by one-way analysis of variance with Holm-Sidak pairwise multiple comparisons for statistically significant differences (p < 0.05).
Bioassay of Control and Injected T. molitor Larvae
Nine days post-injection, larvae from the control (either uninjected or buffer-injected) and dsRNA-injected treatments were randomly divided into two groups for each treatment. One half of each treatment group was placed on untreated control diet (rearing diet without rolled oats), and the other half was placed on diet containing 14% (w/v) Cry3Aa protoxin (mixed with a ceramic mortar/pestle). Individual larvae were placed into separate wells of a 96-well microtiter plate with 25 mg of either control or Cry3Aa-treated diet. Plates were sealed with air-permeable membranes (Breathe-Easier, Boston, MA) and placed in a controlled chamber at 60% relative humidity, 26 °C, total darkness. Larval mortality was evaluated after 10 days.
RESULTS
Identification and Cloning of TmCad1
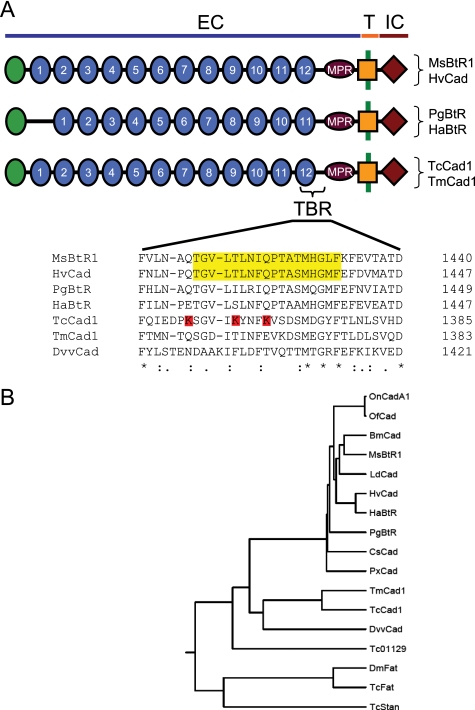
Because Cry1A toxins mediate toxicity in lepidopterans through interaction with a midgut cadherin, we designed a strategy to isolate a midgut cadherin from a B. thuringiensis-sensitive coleopteran. BLAST analysis (39) of a T. molitor larval midgut EST data base identified a clone with sequence identity to the M. sexta cadherin B. thuringiensis receptor (21). Gene-specific primers were designed based on the EST sequence, and 5′- and 3′-rapid amplification of cDNA end products were generated. A cDNA containing the entire TmCad1 coding sequence was obtained by reverse transcription-PCR and confirmed that our results from rapid amplification of cDNA ends were consistent with a single, continuous cadherin cDNA. TmCad1 cDNA consisted of 5,095 bp, with an open reading frame of 4,881 bp encoding a 1,626-amino acid protein (Fig. 1). Both the partial EST and the full-length TmCad1 cDNA were obtained from T. molitor midgut RNA, the target tissue for Cry intoxication. The predicted protein sequence, TmCad1, has a pI of 4.13 and molecular mass of 179,341 Da. Similar to lepidopteran cadherin B. thuringiensis receptor proteins, extracellular, transmembrane, and intracellular domains were found in TmCad1 (TMHMM Server version 2.0) and 12 extracellular repeat domains (Motif Scan of the PROSITE data base, available on the World Wide Web) (Fig. 2A).
FIGURE 1.
cDNA sequence and deduced open reading frame of TmCad1 with the position of rTmCad1p highlighted in gray (residues 1,322–1,516). The various domains of TmCad1 are underlined, including the membrane signal peptide with a double underline, the 12 cadherin repeat regions with solid arrows, the transmembrane domain with dashed arrows, and the cytoplasmic domain with dotted arrows.
FIGURE 2.
Domain structure and phylogenetic analysis of insect cadherins. A, schematic showing conserved domain structure of insect cadherin Cry receptors and alignment of the critical toxin binding region. Cadherins include those from M. sexta (MsBtR1, AAG37912) (21) H. virescens (HvCad, AAK85198) (13), P. gossypiella (PgBtR, AAP30715) (16), H. armigera (HaBtR, ABF69362) (43), T. molitor (TmCad1), and the putative T. castaneum ortholog (TcCad1; XP_971388). The extracellular (EC), transmembrane (T), and intracellular (IC) domains are illustrated. A Cry toxin-binding region (TBR) is found in the cadherin repeat region (numbered) closest to the membrane-proximal region (MPR). The critical toxin-binding sequence of lepidopteran cadherins is aligned with the corresponding region from TmCad1, TcCad1, and the D. virgifera virgifera cadherin (DvvCad, AAV88529). The Cry1A critical toxin binding regions in M. sexta (47) and H. virescens (11) cadherins are highlighted in yellow; lysine residues in the corresponding region of TcCad1 that are potentially disruptive to toxin binding are highlighted in red. B, phylogenetic tree analysis of insect cadherins using PHYLIP version 3.6a3. Protein sequences used in analysis are from M. sexta (MsBtR1), B. mori (BmCad, BAA99404), H. virescens (HvCad), H. armigera (HaBtR), P. gossypiella (PgBtR), Lymantria dispar (LdCad, AAL26896), Ostrinia furnacalis (OfCad, ABL10442), O. nubilalis (OnCadA1, AAT37678), Plutella xylostella (PxCad, ABU41413), Chilo suppressalis (CsCad, AAM78590), T. castaneum (TcCad1), T. molitor (TmCad1), D. virgifera virgifera (DvvCad), the additional T. castaneum cadherins (Tc01129, XP_971786), Starry night (TcStan, XP_968232), and Fat protein (TcFat, XP_971084), and Drosophila melanogaster Fat protein (DmFat, NP_477497).
Comparison of the putative toxin-binding region of TmCad1 with the Tribolium castaneum genome annotations revealed one ortholog, Tc00222, renamed TcCad1 (XP_ 971388). Sequence alignments of putative Cry receptor regions in cadherins from lepidopterans M. sexta (MsBtR1), H. virescens (HvCad), P. gossypiella (PgBtR), and H. armigera (HaBtR) with the coleopteran cadherins, TmCad1 and TcCad1, indicated a high degree of similarity in the predicted critical toxin binding region (Fig. 2A). Notably, three lysines (residues 1,359, 1,364, and 1,368) were exclusive to the T. castaneum sequence and were not found in MsBtR1, HvCad, PgBtR, HaBtR, or TmCad1.
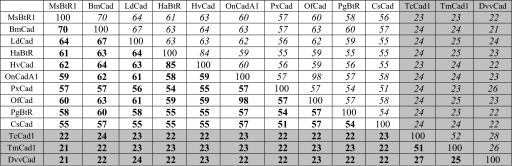
In a comparison of the full-length lepidopteran and coleopteran cadherin sequences, identity was higher within orders (Table 2). These groups also were retained in phylogenetic analyses (Fig. 2B), suggesting that TmCad1 is an ortholog of TcCad1. Moreover, of all of the cadherin-like proteins encoded in the T. castaneum genome, TcCad1 shares the greatest identity with the lepidopteran B. thuringiensis cadherin receptors. TmCad1 shares >50% identity with TcCad1 and 23–25% identity with lepidopteran cadherin-like proteins. Interestingly, the putative D. virgifera virgifera cadherin (DvvCad), purported to be a B. thuringiensis toxin receptor (28, 29), is only 25 and 27% identical to TmCad1 and TcCad1, respectively, similar to values with lepidopteran cadherins (21–24%). Key residues in this putative toxin binding site located in the CR domain immediately adjacent to the membrane-proximal region were not conserved in DvvCad (Fig. 2A). Our alignment agrees with Park et al. (29) in that a Cry3 toxin binding domain is not conserved in this CR domain of DvvCad, as it is in T. molitor and other lepidopteran cadherins. The lepidopteran cadherins were highly conserved and shared 54–98% identity among members within this order.
TABLE 2.
Percent identity scores from full-length sequence alignments of insect cadherins
Cadherin sequences include M. sexta (MsBtR1, AAG37912), B. mori (BmCad, BAA99404), L. dispar (LdCad, AAL26896), H. armigera (HaBtR, ABF69362), H. virescens (HvCad, AAK85198), O. nubilalis (OnCadA1, AAT37678), P. xylostella (PxCad, ABU41413), O. funarcalis (OfCad, ABL10442), P. gossypiella (PgBtR, AAP30715), C. suppressalis (CsCad, AAM78590), T. castaneum (TcCad1, XP_971388), T. molitor (TmCad1, DQ988044), and D. virgifera virgifera (DvvCad, AAV88529). Scores in boldface type were calculated from single pairwise alignments using AlignX from Vector NTI Advance 9.1.0. Italicized scores were calculated from the multiple alignment of protein sequences using ClustalW version 1.83 from EMBL-EBI (available on the World Wide Web). Gray-highlighted cells indicate coleopteran sequences.
Specific Binding of Cry3Aa to the Predicted Toxin-binding Region in TmCad1 Induces Toxin Oligomerization
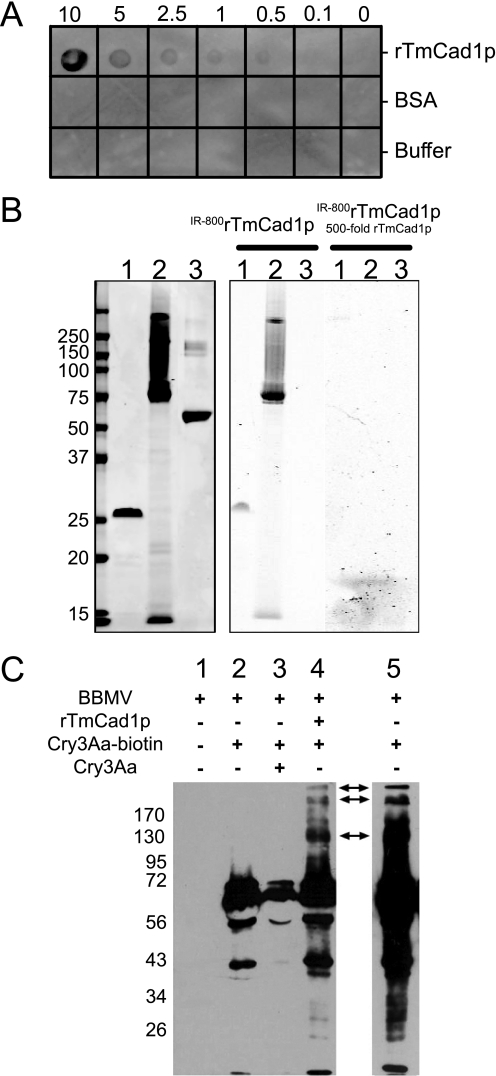
To test the binding of Cry3Aa to TmCad1, we cloned and expressed in E. coli the TmCad1 region homologous to the reported Cry1A receptor region in lepidopteran cadherins (10, 11). Binding of the purified peptide (rTmCad1p) to Cry3Aa was tested under native (dot blot) and denaturing (in-gel assay) methods. In dot blot assays, rTmCad1p bound Cry3Aa but not BSA, indicating specificity for binding to this peptide (Fig. 3A). Toxin binding was detected with a minimum amount of 0.5 μg of peptide. In an in-gel binding assay, IR-labeled rTmCad1p bound to unlabeled peptide and Cry3Aa protoxin but not to BSA (Fig. 3B, right), and binding was completely inhibited by the addition of a 500-fold molar excess of rTmCad1p peptide to the labeled toxin, indicating specificity in the peptide-toxin interaction. These results are evidence that the peptide interacts specifically with itself and with Cry3Aa.
FIGURE 3.
Purified rTmCad1p binds Cry3Aa and promotes oligomerization. A, dot blot overlay binding assay of 0.1–10 μg of rTmCad1p incubated with Cry3Aa toxin and detected with Cry3Aa antiserum. B, Coomassie-stained protein gel scanned at 700 nm (left) and in-gel toxin binding with IR800TmCad1p and competition assay with unlabeled rTmCad1p scanned at 800 nm (right). Lane 1, rTmCad1p; lane 2, Cry3Aa; lane 3, BSA. C, detection of Cry3Aa oligomerization using Western blotting. BBMV proteins (7.8 μg; lane 1) were incubated with 1.2 μg of biotinylated Cry3Aa protoxin alone (lane 2) in the presence of a 24-fold excess of unlabeled Cry3Aa protoxin (lane 3) or with 1.4 μg of rTmCad1p (lane 4). After incubation, unbound Cry3Aa toxin was washed by centrifugation, and final pellets were used for electrophoresis and transferring to polyvinylidene difluoride filters. Bound Cry3Aa toxin was detected with streptavidin conjugated to horseradish peroxidase and enhanced chemiluminescence (Western Pico; Pierce). Lane 5 represents the sample in lane 2 after a 20-fold longer exposure to film than in the other lanes. The arrows indicate the position of putative Cry3Aa dimers, trimers, and tetramers.
In the Lepidoptera model, interactions between Cry1A toxin and cadherin proteins result in toxin oligomerization (1). There is mounting evidence that toxin oligomers are crucial for toxicity (18, 40). Similarly, because the D. virgifera virgifera CR domains 8–10 cadherin peptide enhanced the activity of Cry3 toxins in several beetles, it was suggested that cadherins may promote the formation of prepore toxin oligomers, which thereby enhances toxicity (29). To evaluate whether TmCad1 can promote Cry3Aa oligomerization, we incubated biotinylated Cry3Aa protoxin with rTmCad1p and T. molitor BBMV proteins and detected the formation of toxin oligomers (Fig. 3C). Biotinylated Cry3Aa protoxin bound to T. molitor BBMV (Fig. 3C, lane 2), and the binding was specific, as determined by competition with unlabeled protoxin (Fig. 3C, lane 3). Incubation of biotinylated Cry3Aa protoxin with T. molitor BBMV proteins and rTmCad1p resulted in the formation of increased molecular mass Cry3Aa oligomers (Fig. 3C, lane 4). The molecular mass of the oligomers corresponds to that of dimers (∼130 kDa), trimers (∼195 kDa), and tetramers (∼260 kDa) of the 65-kDa protoxin. Oligomers were also detected when Cry3Aa protoxin was incubated with BBMV proteins, although a much longer exposure of blots to film was necessary for detection (Fig. 3C, lane 5), denoting lower levels of oligomerization than in the presence of rTmCad1 peptide. These results support the role of TmCad1 in promoting Cry3Aa toxin oligomerization.
RNA Interference Knockdown of TmCad1 Results in Decreased Susceptibility to Cry3Aa Toxin
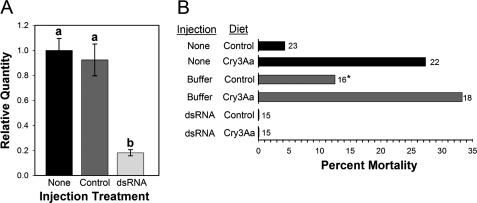
To test the role of TmCad1 as functional Cry3Aa receptor in vivo, T. molitor larvae were injected with either buffer or TmCad1-specific dsRNA. The amount of TmCad1 transcript in each treatment group was analyzed and compared with that of the noninjected control larvae using quantitative real time PCR (Fig. 4A). Four days post-injection, the amount of TmCad1 transcript was significantly reduced in larvae injected with TmCad1 dsRNA (∼80% less transcript than the basal level found in noninjected larvae). This knockdown was stable in larvae 2 weeks post-injection (data not shown).
FIGURE 4.
Knockdown of TmCad1 enhances survival of T. molitor on Cry3Aa. A, reduction in TmCad1 expression in larvae. RNA was obtained from T. molitor larvae, including controls either noninjected or buffer-injected, or larvae injected with dsRNA, and the relative amount of TmCad1 transcript in each treatment group was compared, normalized to the expression of RPS6. Data with the same letter are not statistically different (p < 0.05). B, bioassay of larvae from treatment groups (either controls or dsRNA-injected) with control or Cry3Aa-treated diets, as indicated on the left. The number of larvae in each bioassay group is indicated to the right of each bar. *, two larvae from the buffer-injected control group on control diet escaped at day 8 and were removed from the final calculation.
Injected larvae from both treatment groups were evaluated 9 days postinjection for their ability to survive Cry3Aa toxin-treated diets (Fig. 4B). After 10 days, there was no mortality in the dsRNA-injected groups fed either a control or Cry3Aa-treated diet. This sharply contrasted with the mortality observed in noninjected/control larvae on the B. thuringiensis-treated diet. About 27 and 33% of noninjected and buffer-injected larvae, respectively, died on the B. thuringiensis-treated diet. In comparison, only 4 and 12% mortalities were observed in the same treatments when larvae were fed control diets. Therefore, lower levels of TmCad1 expression in larvae directly correlated with survival on the Cry3Aa-treated diet, demonstrating the functional role of this cadherin as a putative Cry3Aa receptor.
DISCUSSION
Considering that coleopterans are some of the most damaging pests to field crops, forests, and stored products, research to develop effective biopesticides against beetle pest species is highly relevant and urgent. Most coleopteran-specific Cry toxins have limited host range and efficacy that prevent their widespread use in agriculture. Recently, transgenic corn expressing the coleopteran Cry toxin Cry3Bb has been developed to control coleopteran rootworms (Diabrotica spp.). Although treated refugia are part of the mandated resistance management for Cry3Bb corn, the threat of resistance to this transgenic technology is significant (41). Recent reports (42, 43) of field-evolved resistance to B. thuringiensis crops in lepidopteran pests highlight the need to counter this threat, particularly where toxins are not completely effective at controlling target insects. Understanding the mode of action of coleopteran-specific B. thuringiensis toxins will aid in the development of novel B. thuringiensis biopesticides with increased efficacy as well as in the development of resistance detection and management strategies.
Cadherins are Cry toxin receptors in lepidopteran larvae (3, 5–8) and also have been reported as putative receptors for mosquitocidal Cry toxins (44). In this report, we present evidence to support the hypothesis that a cadherin protein is a functional Cry toxin receptor in coleopteran larvae, the first demonstration of Cry3Aa receptor functionality for a coleopteran cadherin. In a previous report of a cadherin-like gene from D. virgifera virgifera (DvvCad), biochemical evidence was not provided to support the hypothesis that DvvCad acts as a functional receptor for Cry toxins (28). Another study demonstrated that a partial DvvCad fragment bound Cry3Aa and Cry3Bb and slightly enhanced Cry3 toxicity when together they were fed to insect larvae (29). Interestingly, this region differs from the Cry3Aa toxin binding region reported here for TmCad1. The Cry3Aa toxin binding region in TmCad1 corresponds to the reported membrane-proximal region in DvvCad (28, 29). The DvvCad-potentiating fragment (CR domains 8–10) corresponds to CR domains 9–11 in TmCad1, suggesting that beetle cadherins may possess different or multiple Cry3 toxin binding domains. Alternatively, there may be another cadherin homolog in D. virgifera virgifera. Our data indicate that there is little sequence conservation in DvvCad, corresponding to toxin binding region 3 for M. sexta (10) and TmCad1. The membrane-proximal region was not tested for DvvCad, and CR domains 9–11 were not tested for TmCad1. It is possible that these unique sites may be important for differences in susceptibility to different Cry3 toxins.
Another report indicated that a membrane ADAM metalloprotease from L. decemlineata is a receptor for Cry3Aa toxin (27). That study reported the presence of an ADAM recognition motif in domain II of Cry3Aa, and a peptide containing this sequence motif prevented Cry3Aa toxin pore formation in BBMV from L. decemlineata. In another study (45), inhibition of L. decemlineata ADAM metalloproteinase activity resulted in increased pore formation. However, the significance of Cry3Aa-ADAM interaction in toxin oligomerization or receptor interaction has not been delineated.
Lepidopteran cadherins function as receptors for Cry1A toxins by promoting toxin oligomerization (46) and/or by activation of intracellular death pathways (22). The TmCad1 peptide containing the toxin binding region, amino acid residues 1,322–1,516, was demonstrated to bind both rTmCad1p and Cry3Aa protoxin. In addition, TmCad1p promoted the formation of Cry3Aa toxin oligomers, similar to those found when the toxin was incubated with BBMV. These toxin oligomers may represent important prepore oligomeric structures, similar to those demonstrated to insert efficiently into membranes in Lepidoptera and promote pore formation (46) and toxicity (18). Cry3Aa oligomeric structures also have been reported after incubation of Cry3Aa protoxin with L. decemlineata BBMV (30).
As expected from the predicted functional Cry3Aa receptor role of TmCad1, reduction of TmCad1 expression in T. molitor by RNA interference resulted in increased survival on a Cry3Aa toxin-treated diet. Interestingly, reduced expression of TmCad1 did not result in observable fitness costs that adversely affected larval health. Furthermore, larvae injected with dsRNA suffered no mortality on control diets. Fitness costs also were not observed in some laboratory-selected Cry-resistant lepidopteran strains with cadherin mutations (36). Although the specific function of Cry toxin-binding cadherins in the insect gut is unknown, it is possible that its primary function may occur in a developmental stage other than that used for knockdown. Alternatively, analogous proteins may rescue the specific function carried out by Cry-binding cadherins in these insects, as has been suggested in cadherin-mutant insects that do not express the toxin binding cadherin and are toxin-insensitive.
Our study reports binding of Cry3Aa to a specific cadherin receptor protein from T. molitor and is the first demonstration that this protein functions as a receptor for the toxin within Coleoptera. The finding that lepidopteran and coleopteran insects share significant similarity in Cry toxin receptor regions further supports the hypothesis that cadherins are a common target in insects for Cry toxins (Fig. 5). Further research is necessary to establish the existence of additional conserved steps in the mode of action of Cry toxins in Coleoptera and Lepidoptera. A thorough comparative analysis of potential toxin binding regions within beetle cadherins also is needed. To date, low efficacy and the lack of knowledge regarding the mode of action of Cry toxins in coleopterans have limited the commercialization of coleopteran-specific Cry toxins. Characterization of the molecules directly involved in the mode of action of Cry toxins in Coleoptera will provide the tools necessary to increase the efficacy of Cry-based biopesticides against economically important beetles.
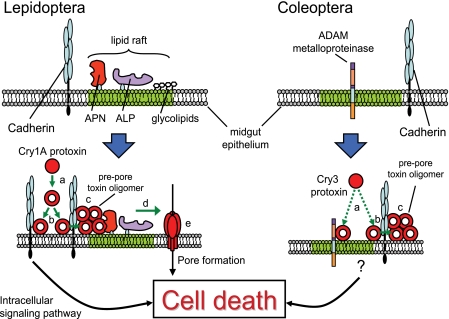
FIGURE 5.
Similarities between the Cry toxin mode of action in Lepidoptera and Coleoptera. Cadherins are key receptors for Cry toxins in both orders. In Lepidoptera, Cry1A toxins are protease-activated (a) and bind to cadherin receptors on the apical surface of the midgut membrane (b). Cell death results from either the activation of an intracellular signaling pathway or through the formation of toxin oligomers (c) that bind to N-aminopeptidase (APN) and/or alkaline phosphatase (ALP) (d) to insert in the membrane and form pores that result in cytotoxicity due to osmotic imbalance (e). In Coleoptera, Cry3 protoxin interacts with a gut-specific ADAM metalloproteinase. Toxin or protoxin binding to cadherin results in toxin oligomerization and eventually cell death. Further research is necessary to characterize post-binding steps in the coleopteran model.
Acknowledgments
We thank Sheila Prabhakar for a partial sequence of cadherin from a midgut EST library and Francisco Avila (AgDia) for the Cry3Aa toxin antiserum. We are grateful to Juan Ferre, Algimantas Valaitis, and Bruce Tabashnik for helpful comments on a previous version of the manuscript.
This is Contribution 08-145-J from the Kansas Agricultural Experimental Station.
The amino acid sequence of this protein can be accessed through NCBI Protein Database under NCBI accession number DQ988044.
- BBMV
- brush border membrane vesicle(s)
- ADAM
- a disintegrin and metalloprotease
- CR
- cadherin repeat
- EST
- expressed sequence tag
- rTmCad1p
- recombinant T. molitor cadherin-1 peptide
- BSA
- bovine serum albumin
- Tricine
- N-[2-hydroxy-1,1-bis(hydroxymethyl)ethyl]glycine
- BisTris
- 2-[bis(2-hydroxyethyl)amino]-2-(hydroxymethyl)propane-1,3-diol
- MOPS
- 4-morpholinepropanesulfonic acid
- dsRNA
- double-stranded RNA.
REFERENCES
- 1.Bravo A., Gill S. S., Soberón M. ( 2007) Toxicon 49, 423– 435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pigott C. R., Ellar D. J. ( 2007) Microbiol. Mol. Biol. Rev. 71, 255– 281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tsuda Y., Nakatani F., Hashimoto K., Ikawa S., Matsuura C., Fukada T., Sugimoto K., Himeno M. ( 2003) Biochem. J. 369, 697– 703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hara H., Atsumi S., Yaoi K., Nakanishi K., Higurashi S., Miura N., Tabunoki H., Sato R. ( 2003) FEBS Lett. 538, 29– 34 [DOI] [PubMed] [Google Scholar]
- 5.Hua G., Jurat-Fuentes J. L., Adang M. J. ( 2004) Insect Biochem. Mol. Biol. 34, 193– 202 [DOI] [PubMed] [Google Scholar]
- 6.Zhang X., Candas M., Griko N. B., Rose-Young L., Bulla L. A., Jr. ( 2005) Cell Death Differ. 12, 1407– 1416 [DOI] [PubMed] [Google Scholar]
- 7.Flannagan R. D., Yu C. G., Mathis J. P., Meyer T. E., Shi X., Siqueira H. A., Siegfried B. D. ( 2005) Insect Biochem. Mol. Biol. 35, 33– 40 [DOI] [PubMed] [Google Scholar]
- 8.Jurat-Fuentes J. L., Adang M. J. ( 2006) Biochemistry 45, 9688– 9695 [DOI] [PubMed] [Google Scholar]
- 9.Fabrick J. A., Tabashnik B. E. ( 2007) Insect Biochem. Mol. Biol. 37, 97– 106 [DOI] [PubMed] [Google Scholar]
- 10.Hua G., Jurat-Fuentes J. L., Adang M. J. ( 2004) J. Biol. Chem. 279, 28051– 28056 [DOI] [PubMed] [Google Scholar]
- 11.Xie R., Zhuang M., Ross L. S., Gomez I., Oltean D. I., Bravo A., Soberon M., Gill S. S. ( 2005) J. Biol. Chem. 280, 8416– 8425 [DOI] [PubMed] [Google Scholar]
- 12.Jurat-Fuentes J. L., Gahan L. J., Gould F. L., Heckel D. G., Adang M. J. ( 2004) Biochemistry 43, 14299– 14305 [DOI] [PubMed] [Google Scholar]
- 13.Gahan L. J., Gould F., Heckel D. G. ( 2001) Science 293, 857– 860 [DOI] [PubMed] [Google Scholar]
- 14.Xu X., Wu Y. ( 2008) J. Invertebr. Pathol. 97, 27– 32 [DOI] [PubMed] [Google Scholar]
- 15.Xu X., Yu L., Wu Y. ( 2005) Appl. Environ. Microbiol. 71, 948– 954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Morin S., Biggs R. W., Sisterson M. S., Shriver L., Ellers-Kirk C., Higginson D., Holley D., Gahan L. J., Heckel D. G., Carrière Y., Dennehy T. J., Brown J. K., Tabashnik B. E. ( 2003) Proc. Natl. Acad. Sci. U.S.A. 100, 5004– 5009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bravo A., Gómez I., Conde J., Muñoz-Garay C., Sánchez J., Miranda R., Zhuang M., Gill S. S., Soberón M. ( 2004) Biochim. Biophys. Acta 1667, 38– 46 [DOI] [PubMed] [Google Scholar]
- 18.Jiménez-Juárez N., Muñoz-Garay C., Gómez I., Saab-Rincon G., Damian-Almazo J. Y., Gill S. S., Soberón M., Bravo A. ( 2007) J. Biol. Chem. 282, 21222– 21229 [DOI] [PubMed] [Google Scholar]
- 19.Chen J., Hua G., Jurat-Fuentes J. L., Abdullah M. A., Adang M. J. ( 2007) Proc. Natl. Acad. Sci. U.S.A. 104, 13901– 13906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pacheco S., Gómez I., Gill S. S., Bravo A., Soberón M. ( 2009) Peptides 30, 583– 588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dorsch J. A., Candas M., Griko N. B., Maaty W. S., Midboe E. G., Vadlamudi R. K., Bulla L. A., Jr. ( 2002) Insect Biochem. Mol. Biol. 32, 1025– 1036 [DOI] [PubMed] [Google Scholar]
- 22.Zhang X., Candas M., Griko N. B., Taussig R., Bulla L. A., Jr. ( 2006) Proc. Natl. Acad. Sci. U.S.A. 103, 9897– 9902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Carroll J., Convents D., Van Damme J., Boets A., Van Rie J., Ellar D. J. ( 1997) J. Invertebr. Pathol. 70, 41– 49 [DOI] [PubMed] [Google Scholar]
- 24.Carroll J., Li J., Ellar D. J. ( 1989) Biochem. J. 261, 99– 105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Koller C. N., Bauer L. S., Hollingworth R. M. ( 1992) Biochem. Biophys. Res. Commun. 184, 692– 699 [DOI] [PubMed] [Google Scholar]
- 26.Belfiore C. J., Vadlamudi R. K., Osman Y. A., Bulla L. A., Jr. ( 1994) Biochem. Biophys Res. Commun. 200, 359– 364 [DOI] [PubMed] [Google Scholar]
- 27.Ochoa-Campuzano C., Real M. D., Martínez-Ramírez A. C., Bravo A., Rausell C. ( 2007) Biochem. Biophys. Res. Commun. 362, 437– 442 [DOI] [PubMed] [Google Scholar]
- 28.Sayed A., Nekl E. R., Siqueira H. A., Wang H. C., Ffrench-Constant R. H., Bagley M., Siegfried B. D. ( 2007) Insect Mol. Biol. 16, 591– 600 [DOI] [PubMed] [Google Scholar]
- 29.Park Y., Abdullah M. A., Taylor M. D., Rahman K., Adang M. J. ( 2009) Appl. Environ. Microbiol. 75, 3086– 3092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rausell C., García-Robles I., Sánchez J., Muñoz-Garay C., Martínez-Ramírez A. C., Real M. D., Bravo A. ( 2004) Biochim. Biophys. Acta 1660, 99– 105 [DOI] [PubMed] [Google Scholar]
- 31.Gómez I., Pardo-López L., Muñoz-Garay C., Fernandez L. E., Pérez C., Sánchez J., Soberón M., Bravo A. ( 2007) Peptides 28, 169– 173 [DOI] [PubMed] [Google Scholar]
- 32.Luo K., Banks D., Adang M. J. ( 1999) Appl. Environ. Microbiol. 65, 457– 464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wolfersberger M. G., Luthy P., Maurer A., Parenti P., Sacchi V. F., Giordana B., Hanozet G. M. ( 1987) Comp. Biochem. Physiol. A 86, 301– 308 [Google Scholar]
- 34.Prabhakar S., Chen M. S., Elpidina E. N., Vinokurov K. S., Smith C. M., Marshall J., Oppert B. ( 2007) Insect Mol. Biol. 16, 455– 468 [DOI] [PubMed] [Google Scholar]
- 35.Felsenstein J. ( 1989) Science 246, 941– 942 [DOI] [PubMed] [Google Scholar]
- 36.Gassmann A. J., Carrière Y., Tabashnik B. E. ( 2009) Annu. Rev. Entomol. 54, 147– 163 [DOI] [PubMed] [Google Scholar]
- 37.Jurat-Fuentes J. L., Adang M. J. ( 2001) Appl. Environ. Microbiol. 67, 323– 329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Laemmli U. K. ( 1970) Nature 227, 680– 685 [DOI] [PubMed] [Google Scholar]
- 39.Altschul S. F., Madden T. L., Schäffer A. A., Zhang J., Zhang Z., Miller W., Lipman D. J. ( 1997) Nucleic Acids Res. 25, 3389– 3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Soberón M., Pardo-López L., López I., Gómez I., Tabashnik B. E., Bravo A. ( 2007) Science 318, 1640– 1642 [DOI] [PubMed] [Google Scholar]
- 41.Rice M. E. ( March2004) Plant Health Prog. 10.1094/Php-2004-0301-01-RV [DOI] [Google Scholar]
- 42.Tabashnik B. E., Gassmann A. J., Crowder D. W., Carriére Y. ( 2008) Nat. Biotechnol. 26, 199– 202 [DOI] [PubMed] [Google Scholar]
- 43.Xu Z., Liu F., Chen J., Huang F., Andow D. A., Wang Y., Zhu Y. C., Shen J. ( 2009) Pest Manag. Sci., in press [DOI] [PubMed] [Google Scholar]
- 44.Hua G., Zhang R., Abdullah M. A., Adang M. J. ( 2008) Biochemistry 47, 5101– 5110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rausell C., Ochoa-Campuzano C., Martínez-Ramírez A. C., Bravo A., Real M. D. ( 2007) Biochim. Biophys. Acta 1768, 2293– 2299 [DOI] [PubMed] [Google Scholar]
- 46.Gómez I., Sánchez J., Miranda R., Bravo A., Soberón M. ( 2002) FEBS Lett. 513, 242– 246 [DOI] [PubMed] [Google Scholar]
- 47.Gómez I., Arenas I., Benitez I., Miranda-Ríos J., Becerril B., Grande R., Almagro J. C., Bravo A., Soberón M. ( 2006) J. Biol. Chem. 281, 34032– 34039 [DOI] [PubMed] [Google Scholar]