Abstract
The induction of Rrs1 expression is one of the earliest events detected in a presymptomatic knock-in mouse model of Huntington disease (HD). Rrs1 up-regulation fulfills the HD criteria of dominance, striatal specificity, and polyglutamine dependence. Here we show that mammalian Rrs1 is localized both in the nucleolus as well as in the endoplasmic reticulum (ER) of neurons. This dual localization is shared with its newly identified molecular partner 3D3/lyric. We then show that both genes are induced by ER stress in neurons. Interestingly, we demonstrate that ER stress is an early event in a presymptomatic HD mouse model that persists throughout the life span of the rodent. We further show that ER stress also occurs in postmortem brains of HD patients.
Huntington disease (HD)3 is an inherited autosomal dominant neurodegenerative disorder caused by a CAG repeat expansion in exon 1 of the HD gene, which encodes for the 350-kDa protein Huntingtin (1). The mutation elongates a polyglutamine stretch at the N terminus of Huntingtin, causing the protein to acquire a dominant gain of normal function, which increases with polyglutamine size (2) and which is especially harmful to the medium-sized spiny neurons of the striatum (3).
Clinical symptoms, which typically precede death in about 10–15 years, include chorea and psychiatric disturbances and manifest when a large percentage of striatal neurons are lost (3). The description of the initial molecular events consequent to proper physiological expression of mutant Huntingtin is thus crucial to design therapeutic intervention for the cure of the disorder.
For this purpose, mouse models that display early presymptomatic phenotypes are particularly relevant. Hdh knock-in mice, where CAG repeats of 109 units are inserted into the mouse HD gene homologue (Hdh) (4, 5), exhibit early striatum-specific molecular changes that are predictive of later neuropathological phenotypes and therefore represent an ideal model to investigate presymptomatic disease pathways (6, 7).
Under the hypothesis that the earliest phenotype is associated with altered gene expression, we previously screened for genes differentially regulated in the striatum of HdhQ111 mice (8). This analysis revealed Rrs1 (regulator of ribosome synthesis), which was differentially detected in homozygous HdhQ111 mice at very early ages. By RT-qPCR, Rrs1 mRNA regulation was validated and found to fulfill the HD criteria of dominance, striatal specificity, and polyglutamine dependence (8).
Most importantly, Rrs1 mRNA expression was found significantly increased in HD postmortem brains compared with age-matched controls indicating that the increase in gene expression is a consequence of mutant Huntingtin in the human disease (8).
So far Rrs1 has been studied only in Saccharomyces cerevisiae, where it encodes for a nucleolar protein that is involved in the maturation, nuclear export, and assembly of ribosomes (9–13). Interestingly, Rrs1 was found to be central in inhibiting transcription of both rRNA and ribosomal protein genes in response to a secretory defect (12). Because in yeast a regulatory network links membrane functionality with ribosome synthesis, these data may suggest that Rrs1 may be involved in sensing a flaw in the secretory pathway, including alterations of Golgi and ER (14).
Loss of Huntingtin profoundly affects embryonic stem cells that display a plethora of cellular dysfunctions, including an abnormal ER and reduced Golgi-ER trafficking (15). Interestingly, STHdhQ111 striatal cell lines expressing full-length mutant Huntingtin present clear evidence of ER stress response (16). This has been recently shown to also occur in yeast as well as in PC12 cells where the pathologically expanded polyglutamines (poly(Q)) trigger a drastic inhibition of ER-associated protein degradation (ERAD) (17, 18). These data have led to the hypothesis that ER stress may be involved in HD pathogenesis.
Here we show that mammalian Rrs1, as well as its newly identified interactor 3D3/lyric, is localized to the ER and is involved in the ER stress response in neuronal cells. Furthermore, we show for the first time that ER stress is an early event in a genetically precise mouse model of HD, which precedes the formation of amyloid intranuclear or cytoplasmic inclusions. We then confirmed ER stress in HD postmortem brains.
EXPERIMENTAL PROCEDURES
Cell Culture, Treatments, and Transfection
Striatal cell lines from wild-type (STHdhQ7/Q7) and homozygous Hdh (STHdhQ111/Q111) knock-in mice were cultured as described previously (16). HEK 293T cells were grown in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum. Tunicamycin (Sigma) treatment (1 μm) was for 3–6-12 h. Transfection of expression plasmids was performed by calcium phosphate precipitation. Expression plasmids were as follows: mouse Rrs1 cDNA (full-length RIKEN clone 5330427D04) subcloned (BamHI-XhoI) into pcDNA3.0 vector; mouse 3D3/lyric cDNA (full-length RIKEN clone 4931440A01) subcloned (EcoRI-XhoI) into pcDNA3.0-HA vector.
Protein Extracts, Immunoprecipitation, and Immunoblot Analysis
Proteins from striatal cells were extracted in Tris, pH 8.0, lysis buffer (50 mm Tris, pH 8.0, 150 mm NaCl, 0.2 mm EDTA, pH 8.0, 10% glycerol, 1% Triton X-100) supplemented with complete protease inhibitor mixture (Roche Diagnostics). For co-immunoprecipitation experiments, transfected HEK 293T cells were lysed in Hepes, pH 7.6 buffer (10 mm Hepes, pH 7.6, 1 mm EDTA, 150 mm NaCl, 0.2% Triton X-100), supplemented with protease inhibitor mixture and phosphatase inhibitors (1 mm sodium orthovanadate, 50 mm NaF, 10 nm okadaic acid). Protein extracts (2 mg) were incubated with primary antibody (anti-HA) and protein G-Sepharose beads (Amersham Biosciences). Washes were performed in Hepes, pH 7.6 buffer. For Western blot analysis, protein extracts were resolved by SDS-polyacrylamide gel and transferred onto nitrocellulose membrane. Proteins were detected by chemiluminescence following incubation with primary antibodies and horseradish peroxidase-conjugated secondary antibodies (Amersham Biosciences).
Primary Antibodies
A cDNA encoding for the N-terminal region of Rrs1 (nucleotides 83–664; GenBankTM accession number NM_021511) was isolated by RT-PCR from mouse striatum. The PCR product was cloned (BamHI-XhoI) into pGEX-4T-3 vector and expressed as glutathione S-transferase fusion protein in BL21 cells. Following purification and separation on a SDS-PAGE, the glutathione S-transferase fusion protein band was cut out from the gel and used for the generation of a rabbit polyclonal antiserum (BioSource International). The immune serum was called FUN for FUsion of N-terminal region of Rrs1. Purification of the immune serum was performed by affinity elution from GST-Rrs1 fusion protein immobilized on a nitrocellulose membrane. Preimmune serum was obtained before the immunization protocol was initiated.
Other primary antibodies were as follows: sheep anti-3D3/lyric (SS) (kindly provided by Dr. H. Sutherland, Western General Hospital, Edinburgh, UK); rabbit anti-3D3/lyric (Zymed Laboratories Inc.); mouse anti-calnexin (Chemicon); mouse anti-fibrillarin 72B9 (kindly provided by Dr. R. Terns, University of Georgia, Athens); and mouse anti-nucleophosmin (Invitrogen).
Immunostaining and Confocal Microscopy
For immunostaining phosphate-buffered saline washed cells were treated as described previously (16).
Immunohistochemistry was performed on 40 μm of free-floating coronal sections of 2% periodate/lysine/paraformaldehyde (PLP) perfused and post-fixed brain. For detection, sections were blocked with phosphate-buffered saline, 10% fetal bovine serum, 0.2% bovine serum albumin, 1% fish gelatin for 1 h at room temperature. Primary and secondary antibodies were diluted in phosphate-buffered saline, 0.2% bovine serum albumin, 0.1% fish gelatin, 0.1% Triton X-100. Incubation with primary antibodies was performed for 16 h at room temperature. Secondary antibodies were Alexa-594 anti-rabbit and Alexa-488 anti-mouse (Invitrogen). Images were captured using Leica confocal microscope (TCS SP2).
Hdh Knock-in Mice and Brain Dissection
Generation, maintenance on a C57BL/6 background, and genotyping of HdhQ111 knock-in mice were performed as described (4, 19). Striata were dissected from mouse brains, snap-frozen on liquid nitrogen, and stored at −80 °C until use.
Postmortem HD Brain Tissue
Control and HD postmortem brain tissues used in this study were described previously (8). Brains were collected by the Harvard Brain Tissue Resource Center, McLean Hospital (Belmont, MA), and used according to the guidelines of the Massachusetts General Hospital Institutional Review Board for discarded human tissue. HD brains were assigned Vonsattel grade 3 pathology (3). Postmortem intervals were from 12 to 32 h for controls and from 8 to 30 h for HD brains.
RNA Extraction and RT-qPCR
Total RNA was isolated with TRIzol® reagent (Invitrogen) according to the manufacturer's instructions, quantified by NanoDrop ND-1000 (Thermo Scientific), and analyzed by agarose gel electrophoresis.
Single strand cDNA was obtained from 1 μg of DNase-treated RNA using iSCRIPT cDNA synthesis kit (Bio-Rad) following the manufacturer's instructions. Quantitative PCRs were performed with an iCycler iQ instrument (Bio-Rad), using the iQ Custom Syber Green Supermix (Bio-Rad). Each reaction was performed in duplicate. Cycle parameters were 3 min at 95 °C, 10 s at 95 °C, 20 s at 58 °C, and 30 s at 72 °C for 40 cycles. Specificity of amplicon was determined by melt curve analysis and gel electrophoresis.
Specific forward and reverse primers (supplemental Table 1) were designed using Beacon Design 5.0 software (Premier Biosoft International). Normalized expression values were calculated using β-actin as endogenous control.
Yeast Two-hybrid Strain, Plasmids, and Library Selection
Yeast host was EGY48 (MATα, his3, trp1, ura3, 6LexAop-LEU2). The bait was full-length Rrs1 cDNA (RIKEN clone 5330427D04) fused to LexA DNA binding domain into pEG202. The bait was used to screen a human fetal brain cDNA library cloned into pJG4-5 vector. Approximately 2.2 × 104 transformants were screened. Positive interaction between bait and fish protein resulted in transcription of Leu2 and lacZ reporters (PJK103), thus allowing growth in the absence of leucine and blue staining on 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-gal) plates. Strength of interaction was evaluated by rate of growth and β-galactosidase staining. Two hundred positive blue colonies with potential stronger interaction were further analyzed; plasmid DNA was extracted and examined by restriction enzyme digestion. Unique cDNA plasmids (∼130) were then sequenced and blasted against the NCBI human nucleotide data base. Among them we isolated five independent clones encoding for 3D3/lyric. The interaction between Rrs1 and 3D3/lyric was further validated in yeast by co-transformation of the rescued plasmid and pEG202-Rrs1 in the original yeast strain containing the lacZ reporter plasmid. In addition, 3D3/lyric plasmid was co-transformed with an unrelated bait, pLexA-DJ-1, and with the empty LexA plasmid as control.
Statistical Analyses
Statistical analyses were Student's t test (Microsoft Excel software), choosing p < 0.05 as significant. Calculated means and standard deviations were plotted using the graph tool of Microsoft Excel.
RESULTS
Mammalian Rrs1 Protein Is Localized Both in the ER and in the Nucleolus
To generate reagents for studying Rrs1 in mammalian cells, we developed a rabbit polyclonal antiserum, named FUN, raised against the N-terminal region of the mouse Rrs1 protein. The affinity-purified and the crude immune sera were tested by Western blot for their ability to reveal endogenous or ectopically expressed Rrs1 in wild-type striatum-derived STHdhQ7/7 cells expressing endogenous 7-glutamine Huntingtin (16). A band of ∼40 kDa, consistent with the predicted molecular weight of mouse Rrs1 protein, was detected both in untransfected and in transfected cells (Fig. 1A), proving that the reagent recognized both endogenous as well as overexpressed Rrs1 protein. Specificity was confirmed by the lack of signal with the corresponding preimmune serum.
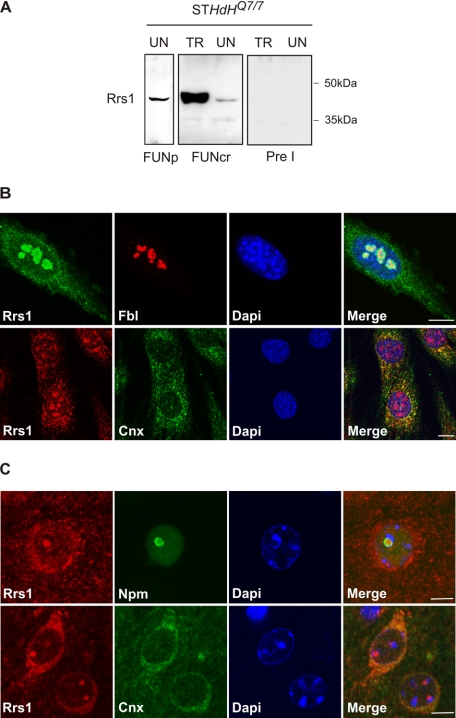
FIGURE 1.
Detection of Rrs1 protein in striatum-derived STHdhQ7/7 cells and in mouse brains. A, protein lysates from STHdhQ7/7 cells untransfected (UN) or transiently transfected with Rrs1 cDNA (TR) were analyzed by Western blotting. Both purified (FUNp) and crude (FUNcr) anti-Rrs1 sera specifically detect endogenous and overexpressed Rrs1 protein. The specificity of the signal was assessed by incubation with the preimmune serum (Pre I). B, immunofluorescence of STHdhQ7/7 cells using affinity-purified anti-Rrs1 antiserum to detect endogenous Rrs1. Top panels show co-staining with Rrs1 (green) and the nucleolar protein fibrillarin (Fbl) (red). Bottom panels show co-staining with Rrs1 (red) and the ER protein calnexin (Cnx) (green). Nuclei were stained with 4,6-diamidino-2-phenylindole (Dapi) (blue). C, detection of Rrs1 protein in mouse brain sections using anti-Rrs1 crude serum. Top panels show co-staining of wild-type mouse striatum with Rrs1 (red) and the nucleolar protein nucleophosmin (Npm) (green). Bottom panels show co-staining with Rrs1 (red) and calnexin (Cnx) (green). Nuclei were stained with 4,6-diamidino-2-phenylindole (blue). Bar, 5 μm.
The intracellular localization of endogenous Rrs1 was studied in STHdhQ7/7 cells by immunofluorescence. Confocal analysis revealed a nucleolar distribution of Rrs1 protein, which was found to co-stain with fibrillarin, a marker of the dense fibrillar component (DFC) of the nucleolus (Fig. 1B, top panels). Surprisingly, Rrs1 staining was also observed in the cytoplasm, with a distribution pattern reminiscent of a protein that localizes to the ER. By immunofluorescence analysis, we confirmed that Rrs1 antibody decorates the ER compartment by co-staining with calnexin, an integral membrane protein of ER (Fig. 1B, bottom panels).
A similar intracellular distribution pattern of Rrs1 was observed in mouse brain sections by co-staining with the nucleolar protein nucleophosmin and with the ER protein calnexin (Fig. 1C). Pre-absorption of FUN antiserum with Rrs1 fusion protein specifically eliminated Rrs1 signal in brain sections (data not shown), proving the specificity of detection of endogenous Rrs1.
Yeast Two-hybrid Screen to Identify Rrs1 Interactors
To gain a better understanding of the cellular function of Rrs1 in mammals, a small scale yeast two-hybrid screening was performed to identify Rrs1-binding proteins in human brain. Mouse full-length Rrs1 was used as bait to screen ∼2.2 × 104 transformants of a human fetal brain cDNA library. Selected primary transformants (∼200 positive clones) were characterized by restriction enzyme digestion and sequenced. Sequence blast in NCBI data base revealed, among others, five independent clones encoding for 3D3/lyric.
3D3/lyric, alternatively known as metadherin and astrocyte elevated gene-1, is a transmembrane protein that resides in the ER and in the nuclear envelope (20, 21) that also co-localizes with fibrillarin in the DFC of the nucleolus (20). For its dual subcellular localization, 3D3/lyric has been suggested to coordinate ER function with ribosome biosynthesis in response to ER stress (20). Because Rrs1 has been shown to be involved in the very same regulatory pathway in yeast, we have then focused our attention on 3D3/lyric.
Rrs1 and 3D3/lyric Co-immunoprecipitate and Co-localize in Mammalian Cells
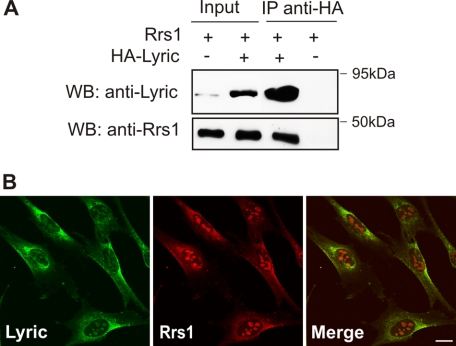
The interaction between Rrs1 and 3D3/lyric was validated by co-immunoprecipitation experiments. HEK 293T cells were co-transfected with full-length Rrs1 and 3D3/lyric HA-tagged cDNAs or with Rrs1 and an empty vector as control. Protein extracts were immunoprecipitated with anti-HA antibody and analyzed by Western blot with anti-Rrs1 antiserum. As shown in Fig. 2A, Rrs1 was specifically co-immunoprecipitated with 3D3/lyric.
FIGURE 2.
Rrs1 and 3D3/lyric co-immunoprecipitate and co-localize in STHdhQ7/7 cells. A, Rrs1 co-immunoprecipitates with 3D3/lyric. HEK 293T cells were co-transfected with Rrs1 and HA-3D3/lyric or with empty vector. Protein lysates were co-immunoprecipitated with anti-HA antibody (IP anti-HA) and analyzed by Western blot (WB) with anti-Rrs1 antiserum. Efficacy of immunoprecipitation was evaluated in the same membrane with anti-3D3/lyric antibody (Zymed Laboratories Inc.). B, Rrs1 and 3D3/lyric co-localize in STHdhQ7/7 cells. Co-staining was performed with anti-3D3/lyric (SS) and anti-Rrs1 (FUNp) antibodies. Nuclei were stained with 4,6-diamidino-2-phenylindole (blue in merge). Bar, 5 μm.
The subcellular distribution of 3D3/lyric in wild-type murine STHdhQ7/7 striatal cells was detected by immunofluorescence with anti-3D3/lyric antibody (SS), kindly provided by Dr. Sutherland (Fig. 2B). As shown previously in fibroblasts (20), 3D3/lyric was observed both in the ER membrane and in the nucleoli of the striatum-derived cells. Importantly, double immunofluorescence showed that 3D3/lyric and Rrs1 had a similar subcellular distribution in STHdhQ7/7 cells (Fig. 2B).
Rrs1 and 3D3/lyric mRNA Expression Is Induced by ER Stress
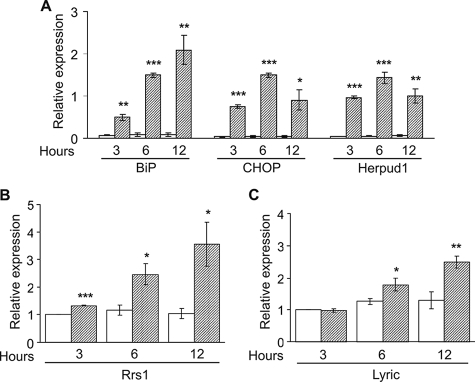
We next investigated whether these two genes are involved in the mammalian response to ER stress. STHdhQ7/7 striatal cells were treated at different times (3, 6, and 12 h) with tunicamycin, a well known inducer of ER stress. Total RNA was extracted, and expression of Rrs1 and 3D3/lyric was measured by RT-qPCR both in tunicamycin- and DMSO-treated cells as control. As markers of ER stress response, we monitored the expression of BiP, CHOP, and Herpud1 mRNAs. Although BiP and CHOP are considered the canonical markers for this biological event, the less studied Herpud1 was chosen for its interaction with the ERAD system, recently proved to be a target of poly(Q) dysfunction (18). As shown in Fig. 3A, ER stress response markers were induced as expected. Interestingly, a significant progressive induction of Rrs1 mRNA levels was observed starting at the earliest time point (3 h) and throughout the entire experiment (12 h) (Fig. 3B), whereas a significant up-regulation of 3D3/lyric mRNA was reached only after 6 h of tunicamycin treatment (Fig. 3C). These results suggest that Rrs1 and 3D3/lyric are, respectively, an immediate-early and a later target of the ER stress response in cultured mammalian striatal cells.
FIGURE 3.
ER stress induced by tunicamycin increases Rrs1 and 3D3/lyric mRNA levels in STHdhQ7/7 striatal cells. RNA was isolated from cells treated with 1 μm tunicamycin (hatched bars) or DMSO (white bars) as control for 3, 6, and 12 h and analyzed by RT-qPCR. A, mRNA expression of BiP, CHOP, and Herpud1 relative to β-actin was measured in STHdhQ7/7 striatal cells by RT-qPCR from three independent experiments. The bar chart shows significant average increase of mRNA levels of BiP, CHOP, and Herpud1 in tunicamycin-treated compared with DMSO-treated cells at 3, 6, and 12 h. B, bar chart displays average Rrs1/β-actin mRNA ratio from three independent experiments. Rrs1 is significantly increased in tunicamycin-treated cells at 3, 6, and 12 h of 1.2-, 2.1-, and 3.4-fold, respectively, compared with DMSO-treated cells. C, bar chart displays average 3D3/lyric/β-actin mRNA ratio from four independent experiments. 3D3/lyric is significantly increased in tunicamycin-treated cells at 6 and 12 h of 1.4- and 1.9-fold, respectively, compared with DMSO-treated cells. Error bars represent standard deviation. *, p < 0.05; **, p < 0.005; ***, p < 0.0005.
Increased Expression of ER Stress-related mRNA Markers in HdhQ111 Striatum
The knowledge that Rrs1 and 3D3/lyric may be partners in different phases of the ER stress response suggests that elevated Rrs1 mRNA in the HdhQ111 striatum in vivo may represent an early response to a perturbation of the ER homeostasis triggered by mutant Huntingtin that entails Rrs1 and 3D3/lyric function.
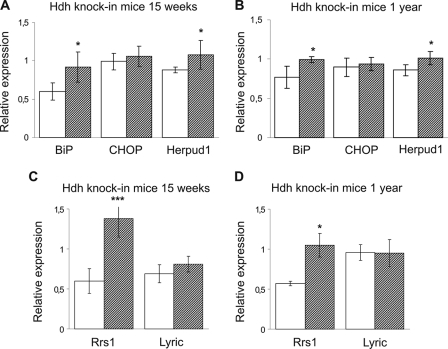
Therefore, we monitored in HdhQ111 knock-in mice whether ER stress-related mRNA expressions were altered at 15 weeks and 1 year of age. These ages predate and directly follow, respectively, the formation of N-terminal Huntingtin inclusions (19). Real time RT-PCR assays were performed for BiP, CHOP, and Herpud1 from single mouse striata at both ages. A significant up-regulation of the chaperones BiP (1.5-fold) and Herpud1 (1.2-fold) mRNAs was observed in HdhQ111 striata compared with wild-type at 15 weeks of age (Fig. 4A) and 1 year of age (1.3- and 1.2-fold, respectively) (Fig. 4B). Notably, no significant difference in the level of CHOP mRNA was detectable at either age (Fig. 4, A and B).
FIGURE 4.
mRNA expression of ER stress-related markers, Rrs1, and 3D3/lyric in striatum of HdhQ111 and wild-type mice. RT-qPCR analyses were performed to determine levels of BiP, CHOP, Herpud1, Rrs1, and 3D3/lyric mRNAs relative to β-actin mRNA in HdhQ111 mutant (hatched bar) and wild-type HdhQ7/Q7 striatum (white bar) at 15 weeks (A and C) and 1 year of age (B and D). A, HdhQ111 heterozygous striatum (n = 6) at 15 weeks of age shows a significant average increase in mRNA expression of BiP (1.5-fold) and Herpud1 (1.2-fold) compared with age-matched wild-type (n = 6). CHOP/β-actin ratio is unchanged in HdhQ111 heterozygous (n = 6) compared with wild-type mice (n = 6). B, HdhQ111 homozygous striatum (n = 5) shows persistent increased mRNA levels of BiP (1.3-fold) and Herpud1 (1.2-fold) at 1 year of age compared with wild-type striatum (n = 6). mRNA levels of CHOP are unchanged. C, relative Rrs1 mRNA expression in mutant HdhQ111 heterozygous striatum (n = 5) shows a significant 2.3-fold increase, compared with levels of Rrs1 mRNA in wild-type striatum (n = 5) at 15 weeks of age. No significant difference in the level of 3D3/lyric is detected at the same age in HdhQ111 heterozygous (n = 5) compared with wild-type mice (n = 5). D, bar chart displays Rrs1/β-actin mRNA ratio for 1-year-old HdhQ111 homozygous (n = 3) and wild-type striatum (n = 3). A significant average 1.8-fold increase of Rrs1 mRNA is found in mutant homozygous compared with wild-type mice. No significant difference in the level of 3D3/lyric mRNA is detected in HdhQ111 homozygous (n = 6) compared with wild-type mice (n = 5). Error bars represent standard deviation. *, p < 0.05; ***, p < 0.0005.
As expected, Rrs1 mRNA was significantly increased in the mutant striatal tissue, compared with wild-type striatum, both at 15 weeks and at 1 year of age, confirming that Rrs1 mRNA level is elevated along with ER stress-related mRNAs (Fig. 4, C and D). Intriguingly, 3D3/lyric mRNA was not significantly changed at 15 weeks or at 1 year of age in HdhQ111 striatum (Fig. 4, C and D). This may suggest that the ER stress induced by endogenous mutant Huntingtin is relatively subtle failing to elevate CHOP and 3D3/lyric mRNAs. Thus, 1-year-old Hdh mice may still present an early ER stress phase that does not involve late response CHOP and 3D3/lyric induction (22, 23). Importantly, these data reveal for the first time that in a presymptomatic HD mouse model, mutant Huntingtin leads to persistent ER stress in striatal tissue, including altered expression of a subset of canonical ER stress genes.
Increased Expression of ER Stress-related mRNA Markers in HD Postmortem Brain
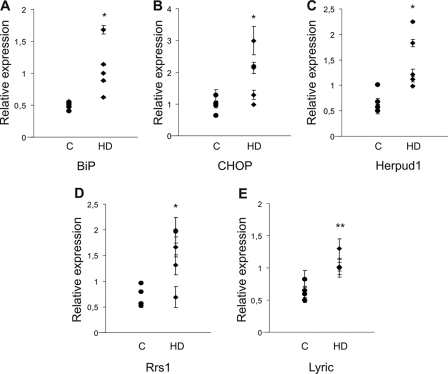
To determine whether increased expression of ER stress genes occurs in human, we measured BiP, CHOP, and Herpud1 mRNAs in parietal cortex of control and HD postmortem brains (Vonsattel grade 3 neuropathology). Real time RT-PCR carried out on these tissues proved a significant up-regulation of BiP, CHOP, and Herpud1 mRNAs in HD compared with control of 2.2-, 2.0-, and 2.1-fold, respectively, as shown in the scattergraphs of Fig. 5, A–C.
FIGURE 5.
mRNA expression of ER stress-related markers, Rrs1, and 3D3/lyric is increased in HD postmortem brains. RT-qPCR analyses were performed to determine levels of BiP, CHOP, Herpud1, Rrs1, and 3D3/lyric mRNAs relative to β-actin mRNA in parietal cortex from postmortem brains of control (C) (n = 4) and affected (HD) individuals (n = 5) (supplemental Table 2). A, scattergraph shows relative expression values of BiP for each control and HD brain. The weighted means for control and HD were 0.47 ± 0.05 and 1.06 ± 0.35, respectively, revealing a 2.2-fold increase in the latter. B, relative expression of CHOP mRNA for control and HD are plotted. The weighted mean for controls was 0.95 ± 0.22 and for HD was 1.86 ± 0.71, demonstrating a 2-fold increase in HD compared with control. C, Herpud1/β-actin mRNA ratios are shown. The weighted means for control and HD were 0.7 ± 0.2 and 1.48 ± 0.48, respectively, revealing a 2.1-fold increase in the latter. D, scattergraph indicates relative expression value of Rrs1 mRNAs for each control and HD brain. The weighted means for control and HD were 0.72 ± 0.18 and 1.56 ± 0.48, respectively, revealing a 2.2-fold increase in the latter. E, relative expression of 3D3/lyric mRNA for control and HD is plotted. The weighted mean for controls was 0.6 ± 0.12 and for HD was 1.04 ± 0.12, demonstrating a 1.7-fold increase in HD compared with control. Error bars represent standard deviation. *, p < 0.05; **, p < 0.005.
We then performed quantitative RT-PCR analysis to measure Rrs1 and 3D3/lyric expression. As expected, Rrs1 mRNA levels were found increased in disease compared with normal samples of 2.2-fold (Fig. 5D). Interestingly, we observed a significant 1.7-fold up-regulation of 3D3/lyric mRNA expression in HD compared with control brain tissue (Fig. 5E).
These results show that mutant Huntingtin induces the expression of ER stress genes in human brain. Moreover, in contrast to presymptomatic mouse striata, the late ER stress response marker CHOP and 3D3/lyric are increased in HD postmortem brains, suggesting a lifetime impact of mutant Huntingtin on the ER stress pathway.
DISCUSSION
In eukaryotic cells, the ER is the primary organelle in the secretory pathway where proteins are synthesized, folded, and post-translationally modified prior to be delivered to other secretory compartments. In addition, the ER is also the place where intracellular Ca2+ is stored and where lipids and sterols are synthesized.
The ER is very susceptible to perturbation of homeostasis that disrupts its functions. When a threshold of damage is reached, ER stress occurs, and a specific cellular response is triggered. Signals are then transduced from the ER to the cytoplasm and to the nucleus to induce the expression of genes encoding mediators of cell defense. This response changes the expression of specific chaperones, enhances degradation of misfolded proteins, and inhibits protein synthesis to decrease the load within the ER (24). Increasing evidence suggests that alteration of ER homeostasis plays an important role in the pathology of several protein-misfolding neurodegenerative diseases, including Parkinson and Alzheimer diseases (25).
Importantly, several lines of evidence connect the ER compartment with Huntingtin activities. The analysis of embryonic stem cells derived from Hdhex4/5/Hdhex4/5 knock-out mice has led to the initial observation that a functional Huntingtin is essential for ER structure and for a correct Golgi/ER trafficking (15). Huntingtin is indeed associated to the ER through a membrane targeting domain in the first 18 amino acids, and it may translocate to the nucleus in response to ER stress (26).
A comparison of striatal cell lines from homozygous (STHdhQ111/Q111) mutant knock-in versus wild-type (STHdhQ7/Q7) mice identified ER dysfunction as one of the phenotypes triggered by full-length mutant Huntingtin, including an excess of ER membranes (16).
More recently, clear evidence of ER stress has been reported in yeast and in cell lines overexpressing N-terminal mutant Huntingtin fragments (17, 18). Importantly, the presence of a toxic poly(Q) expansion in yeast and PC12 cells led to a specific and drastic defect in ERAD through the entrapment of essential ERAD components like Npl4, Ufd1, and p97 into poly(Q) aggregates (18).
Here we show for the first time that ER stress is a very early event in HD pathogenesis in vivo because it is detected in the striatum of Hdh knock-in mice before the formation of visible aggregates. Furthermore, ER stress is persistent because it is present throughout the entire life span of the mouse. Then we showed for the first time that ER stress markers are elevated in postmortem brains of HD patients. This work stems from the observation that the increase of Rrs1 expression is a pre-symptomatic event in the HD neurodegenerative cascade that is maintained through the life of an HD mouse model and confirmed in postmortem brains of HD patients.
Surprisingly, mammalian Rrs1 is localized both to the ER and to the nucleolus. By scanning Rrs1 primary sequence, two motifs for ER membrane retention are indeed found both at its N and C termini, XXRR (EGQR) and KKXX (GKRR), providing a potential molecular mechanism for its subcellular distribution.
By yeast two-hybrid we identified 3D3/lyric as Rrs1 interactor that shares its dual subcellular localization in the ER and nucleolus. Its association to the ER is probably mediated by a transmembrane sequence.
Here we unveiled that Rrs1 and 3D3/lyric are targets of ER stress in vitro although with a different temporal pattern; Rrs1 is first induced, and the 3D3/lyric increases at late time points.
Under the hypothesis that Rrs1 induction in HD mice was a manifestation of an ER stress response, we monitored the temporal development of ER stress in presymptomatic HD mice as well as its presence in postmortem HD brain looking at the expression of Rrs1, 3D3/lyric, Bip, Herpud1, and CHOP. Although Bip is a well known marker of ER overload and dysfunction, Herpud1 is an essential ERAD component, and CHOP is a well known effector of cell death when cellular homeostatic response to ER stress becomes unsustainable (27).
ER stress has been detected in HD mice as up-regulation of BiP and Herpud1 mRNAs at 15 weeks and at 1 year of age. Interestingly, CHOP mRNA levels remained unchanged. This may not be surprising considering the protective activities of both chaperones BiP and Herpud1. By contrast, increased expression of CHOP did not occur in a presymptomatic HD model because it may be associated later in the disease cascade with an irreversible cellular death through apoptosis due to inability to overcome ER stress. Lack of induction was also observed for 3D3/lyric, the expression of which seemed to be part of a late ER stress response in vitro. Importantly, all three ER stress markers as well as Rrs1 and 3D3/lyric were induced in human HD postmortem brains as cell death became prominent proving the relevance of ER stress in HD pathogenesis.
Both Rrs1 and 3D3/lyric have been suggested to play a role in coordinating some ER activities with the nucleolus (20). For Rrs1 this function has actually been proved in yeast because it was identified as a protein required for the transcriptional repression of both ribosomal protein and rRNA genes in response to a secretory defect (12, 14).
In recent proteomic analysis of the nucleolus, molecules that were previously considered exclusively resident in the ER membrane were found in both compartments, such as the signal recognition particle 68 (SRP68) (28). The assembly of the pre-signal recognition particle actually seems to occur in the nucleolus before being targeted to the nuclear pores. The ER membranes are indeed contiguous with the nuclear envelope, and some invaginations of the nuclear membrane have been reported to come in close contact with the nucleolus (29). Furthermore, overexpression of the nucleolar protein Nopp140 induces the formation of intranuclear endoplasmic reticulum membranes (30). Therefore, the relationship between the ER and the nucleolus seems to be more complex than supposed and worthy of future investigation.
In the nucleolus Rrs1 and 3D3/lyric co-localize with fibrillarin that is a DFC marker. This subnucleolar compartment contains actively transcribing rDNA along with nascent rRNA, and it is the site of maturation of pre-rRNA transcripts. These data are in full agreement with Rrs1 expression and function in yeast, where it was found to be a component of the yeast ribosome and rRNA biosynthesis regulon, a co-regulated cluster of genes that function in various aspects of ribosome biosynthesis (31).
Because dysregulation of ribosome synthesis has been previously reported in HD (16, 32), our data lead us to the hypothesis that Rrs1 and 3D3/lyric might function as an ER stress sensor in HD and participate in transducing these signals to the nucleolus. Further work is clearly needed to validate this hypothesis.
Supplementary Material
Acknowledgments
We thank Dr. H Sutherland for 3D3/lyric (SS) antibody; Dr. R. Terns for fibrillarin 72B9 antibody; Dr. Zeno Scotto Lavina for constructing some of the plasmids used in this work.
This work was supported by Telethon Foundation of Italy Grant GGP06264, by the Italian Government (Brain Gain Program, Ministero dell'Istruzione, dell'Universitá e della Ricerca), and by Comitato Interministerale per la Programmazione Economica/Friuli Venezia Giulia, Italy Grant GRAND.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Tables S1 and S2.
- HD
- Huntington disease
- ER
- endoplasmic reticulum
- ERAD
- ER-associated degradation
- DFC
- dense fibrillar component
- poly(Q)
- polyglutamine
- HA
- hemagglutinin
- RT-qPCR
- reverse transcription-quantitative PCR.
REFERENCES
- 1.Huntington's Disease Collaborative Research Group ( 1993) Cell 72, 971– 983 [DOI] [PubMed] [Google Scholar]
- 2.Gusella J. F., MacDonald M. E. ( 2000) Nat. Rev. Neurosci. 1, 109– 115 [DOI] [PubMed] [Google Scholar]
- 3.Vonsattel J. P., Myers R. H., Stevens T. J., Ferrante R. J., Bird E. D., Richardson E. P., Jr. ( 1985) J. Neuropathol. Exp. Neurol. 44, 559– 577 [DOI] [PubMed] [Google Scholar]
- 4.White J. K., Auerbach W., Duyao M. P., Vonsattel J. P., Gusella J. F., Joyner A. L., MacDonald M. E. ( 1997) Nat. Genet. 17, 404– 410 [DOI] [PubMed] [Google Scholar]
- 5.Wheeler V. C., Auerbach W., White J. K., Srinidhi J., Auerbach A., Ryan A., Duyao M. P., Vrbanac V., Weaver M., Gusella J. F., Joyner A. L., MacDonald M. E. ( 1999) Hum. Mol. Genet. 8, 115– 122 [DOI] [PubMed] [Google Scholar]
- 6.Wheeler V. C., Gutekunst C. A., Vrbanac V., Lebel L. A., Schilling G., Hersch S., Friedlander R. M., Gusella J. F., Vonsattel J. P., Borchelt D. R., MacDonald M. E. ( 2002) Hum. Mol. Genet. 11, 633– 640 [DOI] [PubMed] [Google Scholar]
- 7.Menalled L. B. ( 2005) NeuroRx 2, 465– 470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fossale E., Wheeler V. C., Vrbanac V., Lebel L. A., Teed A., Mysore J. S., Gusella J. F., MacDonald M. E., Persichetti F. ( 2002) Hum. Mol. Genet. 11, 2233– 2241 [DOI] [PubMed] [Google Scholar]
- 9.Miyoshi K., Tsujii R., Yoshida H., Maki Y., Wada A., Matsui Y., Toh-E A., Mizuta K. ( 2002) J. Biol. Chem. 277, 18334– 18339 [DOI] [PubMed] [Google Scholar]
- 10.Miyoshi K., Shirai C., Horigome C., Takenami K., Kawasaki J., Mizuta K. ( 2004) FEBS Lett. 565, 106– 110 [DOI] [PubMed] [Google Scholar]
- 11.Nariai M., Tanaka T., Okada T., Shirai C., Horigome C., Mizuta K. ( 2005) Nucleic Acids Res. 33, 4553– 4562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tsuno A., Miyoshi K., Tsujii R., Miyakawa T., Mizuta K. ( 2000) Mol. Cell. Biol. 20, 2066– 2074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang J., Harnpicharnchai P., Jakovljevic J., Tang L., Guo Y., Oeffinger M., Rout M. P., Hiley S. L., Hughes T., Woolford J. L., Jr. ( 2007) Genes Dev. 21, 2580– 2592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mizuta K., Warner J. R. ( 1994) Mol. Cell. Biol. 14, 2493– 2502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hilditch-Maguire P., Trettel F., Passani L. A., Auerbach A., Persichetti F., MacDonald M. E. ( 2000) Hum. Mol. Genet. 9, 2789– 2797 [DOI] [PubMed] [Google Scholar]
- 16.Trettel F., Rigamonti D., Hilditch-Maguire P., Wheeler V. C., Sharp A. H., Persichetti F., Cattaneo E., MacDonald M. E. ( 2000) Hum. Mol. Genet. 9, 2799– 2809 [DOI] [PubMed] [Google Scholar]
- 17.Reijonen S., Putkonen N., Nørremølle A., Lindholm D., Korhonen L. ( 2008) Exp. Cell Res. 314, 950– 960 [DOI] [PubMed] [Google Scholar]
- 18.Duennwald M. L., Lindquist S. ( 2008) Genes Dev. 22, 3308– 3319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lloret A., Dragileva E., Teed A., Espinola J., Fossale E., Gillis T., Lopez E., Myers R. H., MacDonald M. E., Wheeler V. C. ( 2006) Hum. Mol. Genet. 15, 2015– 2024 [DOI] [PubMed] [Google Scholar]
- 20.Sutherland H. G., Lam Y. W., Briers S., Lamond A. I., Bickmore W. A. ( 2004) Exp. Cell Res. 294, 94– 105 [DOI] [PubMed] [Google Scholar]
- 21.Kang D. C., Su Z. Z., Sarkar D., Emdad L., Volsky D. J., Fisher P. B. ( 2005) Gene 353, 8– 15 [DOI] [PubMed] [Google Scholar]
- 22.Harding H. P., Novoa I., Zhang Y., Zeng H., Wek R., Schapira M., Ron D. ( 2000) Mol. Cell 6, 1099– 1108 [DOI] [PubMed] [Google Scholar]
- 23.Okada T., Yoshida H., Akazawa R., Negishi M., Mori K. ( 2002) Biochem. J. 366, 585– 594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rao R. V., Ellerby H. M., Bredesen D. E. ( 2004) Cell Death Differ. 11, 372– 380 [DOI] [PubMed] [Google Scholar]
- 25.Lindholm D., Wootz H., Korhonen L. ( 2006) Cell Death Differ. 13, 385– 392 [DOI] [PubMed] [Google Scholar]
- 26.Atwal R. S., Truant R. ( 2008) Autophagy 4, 91– 93 [DOI] [PubMed] [Google Scholar]
- 27.Schröder M., Kaufman R. J. ( 2005) Annu. Rev. Biochem. 74, 739– 789 [DOI] [PubMed] [Google Scholar]
- 28.Politz J. C., Yarovoi S., Kilroy S. M., Gowda K., Zwieb C., Pederson T. ( 2000) Proc. Natl. Acad. Sci. U.S.A. 97, 55– 60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fricker M., Hollinshead M., White N., Vaux D. ( 1997) J. Cell Biol. 136, 531– 544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Isaac C., Pollard J. W., Meier U. T. ( 2001) J. Cell Sci. 114, 4253– 4264 [DOI] [PubMed] [Google Scholar]
- 31.Wade C., Shea K. A., Jensen R. V., McAlear M. A. ( 2001) Mol. Cell. Biol. 21, 8638– 8650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wyttenbach A., Swartz J., Kita H., Thykjaer T., Carmichael J., Bradley J., Brown R., Maxwell M., Schapira A., Orntoft T. F., Kato K., Rubinsztein D. C. ( 2001) Hum. Mol. Genet. 10, 1829– 1845 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.