Abstract
The NAD-dependent deacetylase SirT1 regulates factors involved in stress response and cell survival and is a potential drug target of activators and inhibitors. Determination of SirT1 function in tumor cells is important for its targeting in cancer therapy. We found that SirT1 knockdown by short hairpin RNA accelerates tumor xenograft formation by HCT116 cells, whereas SirT1 overexpression inhibits tumor formation. Furthermore, pharmacological inhibition of SirT1 stimulates cell proliferation under conditions of growth factor deprivation. Paradoxically, SirT1 inhibition also sensitizes cells to apoptosis by chemotherapy drugs. Immunohistochemical staining revealed high level SirT1 in normal colon mucosa and benign adenomas. SirT1 overexpression was observed in ∼25% of stage I/II/III colorectal adenocarcinomas but rarely found in advanced stage IV tumors. Furthermore, ∼30% of carcinomas showed lower than normal SirT1 expression. This pattern is consistent with SirT1 having pleiotropic effects during cancer development (anti-proliferation and anti-apoptotic). These results suggest a rationale for the use of SirT1 activators and inhibitors in the prevention and treatment of colon cancer.
The silent information regulator 2 (Sir2)2 functions as a nicotinamide adenine dinucleotide (NAD+)-dependent histone deacetylase and regulates chromatin silencing in Saccharomyces cerevisiae (1). Increased Sir2 gene dosage results in the extension of life span in yeast (2). Sir2 is activated by multiple stress signals such as starvation, osmotic stress, and heat shock. In both yeast and C elegans, caloric restriction-induced life span extension is dependent on Sir2 (3, 4). Therefore, Sir2 is a key regulator of cellular homeostasis and survival by connecting stress signals to regulation of gene expression in invertebrate organisms.
In addition to its role in extending the lifespan of lower organisms under stressful conditions, mammalian homolog of Sir2 (SirT1) has been shown to regulate glucose homeostasis in mice by deacetylating and activating the transcription factor peroxisome proliferator-activated receptor-γ coactivator 1α (5). Transgenic mice overexpressing SirT1 in pancreatic β cells showed improved glucose tolerance and increased insulin secretion in response to glucose (6, 7). Furthermore, pharmacological activators of SirT1 such as resveratrol can mimic the anti-aging effects of calorie restriction in lower organisms, reduce insulin resistance in mice fed with high fat diet, and prolong survival (8–10). More recently, a potent activator of SirT1 has shown therapeutic potential in the treatment of type 2 diabetes in animal models by improving insulin sensitivity and lowering plasma glucose level (11).
Numerous studies have also suggested a role of SirT1 in tumorigenesis. In general, transient knockdown of SirT1 leads to increased apoptotic response to DNA damage or oxidative stress treatments. SirT1 deacetylates and inhibits the activities of p53, NF-κB, Forkhead, Ku, and E2F1 that are critical factors in stress response and apoptosis regulation (12–17). The tumor suppressor HIC1 has also been shown to inhibit SirT1 expression by forming a repressive complex with SirT1 on its own promoter and sensitizing p53 response to DNA damage (18). SirT1 also promotes cell survival by regulating the DNA damage signaling pathway via deacetylation of NBS1 (19). Treatment with small molecule SirT1 inhibitors such as sirtinol, splitomycin, and cambinol induces growth arrest, senescence, or apoptosis in tumor cells (20, 21). However, it is difficult to rule out off-target effects due to the limited potency and specificity of these compounds. Nonetheless, these findings suggest a rationale for inhibiting SirT1 as a therapeutic strategy for cancer.
Interestingly, several studies suggest that SirT1 may act as a tumor suppressor. MEFs derived from SirT1-null mice are prone to spontaneous immortalization, suggesting that SirT1 behaves as a growth-suppressive gene in culture (22). Furthermore, hematopoietic stem cells from SirT1-null mice have increased proliferation potential, and shRNA knockdown of SirT1 in human fibroblasts accelerates cell proliferation (23, 24). SirT1 has also been shown to inhibit androgen receptor-dependent cell proliferation in prostate tumor cells (25). Recent publications also showed that transgenic overexpression of SirT1 in the intestine inhibited polyp formation in the ApcMin mice (26), whereas SirT1 deficiency led to increased tumor formation in p53-null mice (27). These observations suggest that SirT1 may suppress tumor growth under certain conditions, and that SirT1 activators could be used for cancer treatment or prevention (28).
In this report, we show that SirT1 inhibition in human colon tumor cells promotes tumor xenograft formation in nude mice, whereas SirT1 overexpression inhibits tumor formation. Furthermore, SirT1 inhibition promotes cell proliferation in culture under conditions of growth factor deprivation but sensitizes the same cells to chemotherapy drugs. We show that SirT1 is expressed at high levels in normal colon epithelial cells and in pre-malignant adenomas, but its expression is reduced in a subset of malignant colon carcinomas. Furthermore, SirT1 overexpression is rarely observed in advanced Stage IV tumors. These results suggest that SirT1 may function as a tumor suppressor during certain stages of tumor development, but may also promote chemo-resistance upon treatment with DNA-damaging drugs.
MATERIALS AND METHODS
Cell Lines, Plasmids, and Reagents
HCT116 cells were kindly provided by Dr. Bert Vogelstein and maintained in Dulbecco's modified Eagle's medium with 10% fetal bovine serum. SirT1+/− mice were kindly provided by Dr. Fred Alt. SirT1−/−, and SirT1+/+ MEFs were generated from 13-day embryos of a SirT1+/− x SirT1+/− mating using standard protocol. The cells were genotyped using three primers in the same PCR reaction. Forward primer SirT1KO-F (5′-CTTGCACTTCAAGGGACCAA) and two different reverse primers SirT1SKO-R1 (5′-GTATACCCACCACATCTGAG) and SirT1SKO-R2 (5′-CTACCACTCCTGGCTACCAA) were used to detect the wild-type SirT1 allele (500 bp) or mutant allele (800 bp), respectively. Plasmid expressing wild-type SirT1 (human Sir2) was provided by Dr. Wei Gu (12). SirT1 nuclear import mutant was generated by K35E mutation in the putative NLS (RKRPRR) by site-directed mutagenesis. Acetylated E2F1 K117-specific polyclonal antibody was raised against acetylated E2F1 peptide HPG(AcK117)GVKSPG and affinity-purified. SirT1 inhibitor EX-527 was purchased from Tocris Bioscience. Western blots were performed as described previously (16).
Immunohistochemistry
The Colon Tumor Microarray was developed from patient samples collected at the Moffitt Cancer Center, Tampa, FL. This tissue micro array includes over 100 colon tumor tissue samples and normal colon tissues. The tissue micro array was stained for SirT1 using the 10E4 monoclonal antibody by the Histopathology Core at the Moffitt Cancer Center. Briefly, the tissue microarray slide was processed using a Ventana Discovery XT automated system (Ventana Medical Systems) as per the manufacturer's protocol with proprietary reagents. Slides were deparaffinized on the automated system with EZ Prep solution (Ventana). Heat-induced antigen retrieval method was performed using Cell Conditioning Solution (Ventana). The 10E4 hybridoma supernatant was used at 1:150 in Dako antibody diluent and incubated for 60 min. The Ventana Universal Secondary Antibody was used for 32 min at 37 °C. The detection system used was the Ventana DABMap kit. Slides were counterstained with hematoxylin and scanned by the Analytical Microscopy Core. The SirT1 staining level of each tumor tissue sample and the normal control tissues was examined by three individuals and scored from 0 to 3 based on the intensity of the stain. ApcMin mouse tissue staining was performed using an anti-Sir2 antibody (Upstate) at a concentration of 1:500 with the ABC staining system (rabbit) from Santa Cruz Biotechnology (Santa Cruz, CA).
Inhibition of SirT1 by RNA Interference
A double-stranded oligonucleotide 5′-GATCCCGTTGGATGATATGACACTGTTCAAGAGACAGTGTCATATCATCCAACTTTTTTGGAAA (SirT1 target sequence underlined) was cloned into the pSuperiorRetroPuro vector (OligoEngine). The plasmid was packaged into retrovirus by transfection of the amphotropic packaging cell line LA (a kind gift from Dr. Peiqing Sun, the Scripps Institute). A virus expressing a scrambled shRNA (5′-GATCCCGCCGTCGTCGATAAGCAATATTTGATATCCGATATTGCTTATCGACGACGGCTTTTTTA) was used as control. HCT116 cells were infected with the SirT1 shRNA retrovirus and selected with 0.5 μg/ml puromycin for 10 days. Drug-resistant colonies were pooled for analysis.
Expression of SirT1 by Lentiviral Vector
Lentivirus vector expressing SirT1 was generated using the ViraPowerTM T-RExTM system following instructions from the manufacturer (Invitrogen). Tetracycline inducible expression of SirT1 in HCT116 was achieved by first infecting with the T-REX regulator lentivirus and selection with 4 μg/ml blasticidin, followed by infection with the SirT1 lentivirus and selection with 300 μg/ml Zeocin to obtain a pool of colonies. SirT1 expression was induced with 1 μg/ml tetracycline.
Quantitation of DNA Synthesis
Cells cultured in 24-well plates were treated with EX-527 for 18 h and 5 μCi of [methyl-3H]thymidine (Amersham Biosciences) was added for 1 h. Cells were lysed in 0.5 ml of lysis solution (2% SDS, 10 mm EDTA, pH 8.0), incubated at 75 °C for 20 min, and vortexed for 20 s, and 170 μl of lysate was applied to S/P glass fiber filters (Baxter). The filters were incubated in ice-cold 10% trichloroacetic acid for 5 min, and washed with 10 ml of ice-cold 5% trichloroacetic acid and 5 ml of 95% ethanol using a vacuum manifold. The filters were dried, suspended in 2-ml scintillation mixture, and counted in a liquid scintillation counter. The experiments were repeated three or more times. The p values of individual experiments were calculated using two-tailed Student's t-test. p < 0.05 was used as threshold for statistically significant difference (>95% confidence) in proliferation rate between control and EX-527-treated samples. Actual p values for each experiment are shown in the figure legends.
Cell Cycle and Apoptosis Assay
Cells in 6-cm plates were transfected with 0.5 μg of SirT1 using Lipofectamine 2000 reagent. Twenty-four hours after transfection, cells were treated with 100 ng/ml nocodazole for 18 h, fixed in ethanol, stained with primary anti-SirT1 antibody for 3 h, and secondary anti-mouse fluorescein isothiocyanate for 1 h. Subsequently, the cells were stained with propidium iodide and analyzed by fluorescence-activated cell sorting. Fluorescein isothiocyanate-positive cells were analyzed for cell cycle distribution. The In Situ Cell Death Detection Kit from Roche Applied Science was used to perform a TUNEL assay to detect the response of HCT116 cells to 5-FU in the presence or absence of EX-527 using procedure provided by the supplier.
Tumor Xenograft Assay
Experimental procedures involving animals were reviewed and approved by the Institutional Animal Care and Use committee of the University of South Florida. Athymic-NCr-nu female mice between 7 and 8 weeks were inoculated subcutaneously on both flanks with 8 × 106 HCT116 cells stably transfected with either control or SirT1 targeting shRNA vectors. Tumors were measured after 14 days with calipers. Tumor volume was calculated with the formula: [(length + width)/2]3 × 0.5236. For induction of Lenti-SirT1 expression, the mice inoculated with tumor cells were maintained on chow with 500 mg/kg doxycycline. Statistical analysis was done using paired Student's t test, and p < 0.05 was used as threshold for statistically significant different tumor growth rate.
RESULTS
SirT1 Knockdown Increases the Growth of Colon Tumor Xenograft
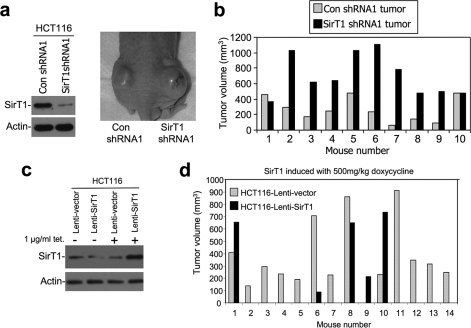
Our previous study revealed pleiotropic activities for SirT1 in cell culture, i.e. anti-apoptotic and anti-proliferative (16). To further test the role of endogenous SirT1 in tumor formation, HCT116 colon carcinoma cells were stably infected with retrovirus vector (pSuperior) expressing control and SirT1 shRNA. Polyclonal cell lines were generated from pooled colonies and confirmed for the knockdown of SirT1 (∼70% reduction, Fig. 1a). To test their ability to form tumors in xenograft assay, cells were inoculated subcutaneously on the dorsal flanks of athymic nude mice. Each animal received both control and knockdown cell lines to facilitate comparison under similar conditions (Fig. 1a). The results showed that SirT1 knockdown cells formed tumors 2- to 3-fold larger than control cells inoculated in the same animal (n = 10, p = 0.0007 using Student's t test where p < 0.05 is significant (Fig. 1b)).
FIGURE 1.
Endogenous SirT1 expression limits tumorigenicity. a, retrovirus vector pSuperior expressing control or SirT1 shRNA was used to infect HCT116 cells, and the colonies were pooled to generate a polyclonal SirT1 knockdown cell line. The expression of SirT1 in the control and knockdown cells was determined by Western blot. Cells were injected subcutaneously in the dorsal flanks of athymic nude mice, and tumor formation in a representative animal is shown. b, HCT116 SirT1 knockdown and control cells were injected in the dorsal flanks of athymic nude mice, and tumor volumes were measured after 14–21 days (n = 10, p = 0.0007 as calculated with paired Student's t test and p < 0.05 is statistically significant). c, HCT116 cells were infected with a tetracycline-inducible lentivirus vector expressing SirT1. SirT1 induction by tetracycline was confirmed by Western blot. d, athymic nude mice were injected on the dorsal flanks with HCT116-lenti-vector or HCT116-lenti-SirT1. The mice were continuously fed with chow containing doxycycline (500 mg/kg). Tumors were measured and the graph indicates the tumor volumes generated by the control versus the SirT1-overexpressing cell line (n = 14, p = 0.02 as calculated with paired Student's t test and p < 0.05 is statistically significant).
To rule out off-target effects from the SirT1 shRNA, a separate pair of HCT116 control/knockdown cell lines was generated using completely different control shRNA and SirT1 shRNA sequences and expression vectors obtained from an OpenBiosystems shRNA library. The tumorigenesis experiment was repeated and again showed that SirT1 knockdown cells form significantly larger tumors than control (n = 13, p = 0.01 using Student's t test, where p < 0.05 is significant, supplemental Fig. S1). These results suggest that endogenous SirT1 in HCT116 cells inhibits the growth of tumor xenografts. The role of SirT1 in suppressing tumor growth was not limited to colon tumors, because a similar experiment using A549 lung tumor cells with SirT1 knockdown also resulted in accelerated tumor growth (data not shown).
SirT1 Overexpression Suppresses the Growth of Colon Tumor Xenograft
The results described above suggested that expression of endogenous SirT1 partially suppresses tumor formation. To further test this hypothesis, HCT116 cells were stably infected with a tetracycline-inducible lentivirus vector expressing SirT1. SirT1 level can be induced to ∼4-fold above endogenous level by tetracycline in this cell line (Fig. 1c). To test the effect of SirT1 overexpression on tumor growth, nude mice injected on either side of their flanks with lenti-vector- and lenti-SirT1-infected HCT116 cells were kept on a diet with doxycycline to induce SirT1 expression. The results showed that, in contrast to the phenotype of SirT1 knockdown, overexpression of SirT1 significantly reduced the tumorigenicity of HCT116 cells (Fig. 1d). It is noteworthy that in a significant fraction of cases (8 of 14), HCT116-lenti-SirT1 cells gave rise to extremely small or no tumors. This suggests that, besides inhibiting cell proliferation, overexpression of SirT1 may suppress the initial establishment of tumors in this setting.
Inactivation of SirT1 Stimulates Tumor Cell Proliferation
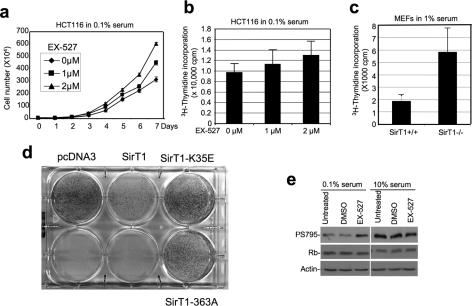
To further test whether physiological levels of endogenous SirT1 plays a role in regulating cell proliferation, cells were treated with SirT1-specific inhibitor EX-527. This compound has significantly improved potency and specificity against SirT1 compared with nicotinamide (29). When HCT116 cells were cultured in 0.1% serum, addition of EX-527 caused a 90% increase in cell number after 7 days (Fig. 2a). In the presence of 10% serum, EX-527 did not change cell number in long term culture (supplemental Fig. S2b). This result shows that, during growth factor deprivation conditions, SirT1 is a significant regulator of cell proliferation.
FIGURE 2.
Inhibition of SirT1 stimulates cell proliferation. a, HCT116 cells cultured in 0.1% serum were maintained in different concentrations of SirT1-specific inhibitor EX-527, and cell number was determined at the indicated time points. Error bars represent mean ± S.D. (n = 4). b, HCT116 cells cultured in 0.1% serum were treated with EX-527 for 18 h. DNA replication was measured by [3H]thymidine incorporation assay. This is a single experiment representative of three replicates. Error bars represent mean ± S.D. (n = 14) and p = 0.03 as calculated with Student's t test and p < 0.05 is statistically significant. This suggests an increase in proliferation by EX-527. c, early passage (P4) MEFs from SirT1−/− and SirT1+/+ mouse embryos were also tested for DNA synthesis rate by [3H]thymidine incorporation. An identical number of cells from each genotype was plated prior to 18-h culture in 1% serum and 1 h metabolic labeling. Error bars represent mean ± S.D. (n = 6, p = 0.00001). d, HCT116 cells were transfected with pcDNA3 vector, SirT1, SirT1-K35E NLS mutant, or SirT1-H363A deacetylase mutant. The cells were subjected to selection by G418, and the colonies were stained with Crystal Violet after 2 weeks. e, HCT116 cells treated with 2 μm EX-527 were analyzed for phosphorylation level of pRb at S795 by immunoprecipitation Western blot, followed by reprobing with Rb antibody.
DNA replication was analyzed by [3H]thymidine incorporation assay to accurately measure changes in cell proliferation rate. The results showed that EX-527 treatment stimulated DNA replication by up to 30% in low serum (Fig. 2b). When cells were cultured in 10% serum that supports optimal growth, EX-527 treatment did not further stimulate DNA synthesis (supplemental Fig. S2a). Despite the moderate magnitude of enhancement, the growth-stimulating effect of EX-527 was statistically significant (n = 14, p = 0.03 using Student's t test where p < 0.05 is significant) and highly reproducible in replicate assays. The moderate but sustained increase in cell proliferation is likely responsible for the large increase in cell population in Fig. 2a. Importantly, inhibition of SirT1 also stimulated the proliferation of A549 (lung tumor) and U2OS (osteosarcoma) cells (supplemental Figs. S2e and S2f), suggesting that its growth-suppressive effect is not limited to colon cancer cells.
To demonstrate that the growth-stimulatory effect of EX-527 is due to inhibition of SirT1, mouse embryonic fibroblasts (MEFs) derived from SirT1-null mice were analyzed. Initial experiments revealed that late-passage SirT1-null and wild-type MEFs showed significant variability in proliferation rate and morphology (data not shown), making them unsuitable for such analysis. Therefore, we generated low passage (P4) MEFs of SirT1−/− and SirT1+/+ genotypes from littermate embryos using a previously reported SirT1-null mouse strain (30). These early passage SirT1−/− MEFs clearly showed more robust growth in culture, and a significantly higher rate of DNA synthesis (∼150%) compared with SirT1+/+ MEF (Fig. 2c). Treatment with EX-527 also induced a statistically significant 30% increase in DNA synthesis in SirT1+/+ MEF (supplemental Fig. S2c). Importantly, no growth stimulation by EX-527 was observed in the SirT1-null MEF (supplemental Fig. S2d). These results demonstrate that SirT1 is a suppressor of cell proliferation and that the growth-promoting activity of EX-527 compound is due to specific inhibition of its intended target SirT1.
SirT1 Inhibits Cell Proliferation
To further test the potential of SirT1 in suppressing cell proliferation, HCT116 cells were used in a colony formation assay. Expression of SirT1 from pcDNA3 vector significantly reduced the number of G418-resistant colonies formed after transfection (Fig. 2d). In contrast, SirT1 NLS mutant (K35E) that failed to enter the nucleus, or deacetylase-inactive mutant (H363A) did not suppress colony formation. Immunofluorescence staining of pooled HCT116 G418-resistant colonies from SirT1-stable transfection failed to detect SirT1 overexpression (data not shown). Therefore, an increase in SirT1 level suppresses HCT116 proliferation and is not tolerated during long term culture. Fluorescence-activated cell sorting analysis of HCT116 cells transiently transfected with SirT1 showed that SirT1 overexpression induced efficient G1 cell cycle arrest (supplemental Fig. S3), consistent with the lack of outgrowth of SirT1 overexpressing cells after long term culture.
SirT1 regulation of cell proliferation is expected to be associated with changes in cell cycle machinery. To this end, hyper-phosphorylation of Ser-795 on the retinoblastoma protein (pRb) was reproducibly detected after EX-527 treatment of serum-starved HCT116 cells (Fig. 2e). In contrast, cells cultured in 10% serum already had higher basal phosphorylation of Ser-795 that was not further stimulated by EX-527 (Fig. 2e). These results further confirm that SirT1 in tumor cells may act as a suppressor of cell proliferation during stress.
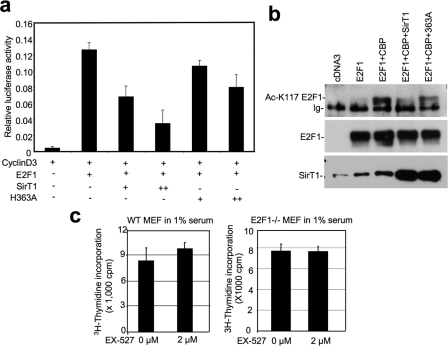
Among the SirT1 substrates, E2F1 is a likely target for the growth suppressive effect of SirT1. Previous results from our laboratory showed that SirT1 binds to E2F1 and inhibits its transcriptional activity. The cell cycle arrest caused by SirT1 overexpression can be rescued by E2F1 co-expression (16). Here we confirmed that, in a reporter assay, expression of SirT1 inhibited E2F1 activation of cyclin D3 promoter in a deacetylase-dependent fashion (Fig. 3a). Using an E2F1 K117 acetylation-specific antibody generated in our laboratory, we found that E2F1 acetylation by the coactivator CREB-binding protein (CBP) was strongly inhibited by SirT1 (Fig. 3b). The SirT1–363A catalytic mutant was partially defective for E2F1 deacetylation in this assay, possibly due to residual activity when expressed at high levels (Fig. 3b). Furthermore, the ability of EX-527 to stimulate cell proliferation in wild-type MEFs was not observed in E2F1−/− MEFs (Fig. 3c). These results, together with our recent finding of SirT1-E2F1 interaction, suggest that the mechanism of cell cycle regulation by SirT1 is in part through inhibition of E2F1.
FIGURE 3.
Inhibition of SirT1 stimulates cell proliferation. a, H1299 cells were transiently transfected with cyclin D3 promoter-luciferase, E2F1, SirT1, and SirT1-H363A mutant. Promoter activity was measured by luciferase assay and normalized to cotransfected CMV-lacZ level. Error bars represent mean ± S.D. (n = 4). b, 293T cells were transiently transfected with E2F1, CREB-binding protein, and SirT1. E2F1 was immunoprecipitated and probed with anti-Ac-K117 antibody by Western blot. The membrane was reprobed for E2F1 and SirT1 expression levels. c, wild-type and E2F1−/− MEFs were cultured for 18 h in 1% serum and 2 μm EX-527. DNA replication was measured by [3H]thymidine incorporation assay. Error bars represent mean ± S.D. (n = 8). For wild-type MEF cells, p = 0.002 indicates significant difference in proliferation after treatment. For E2F1−/− MEFs, p = 0.7 (>0.05) indicates no effect by EX-527 treatment.
Inhibition of SirT1 Sensitizes Tumor Cells to Chemotherapy Agents
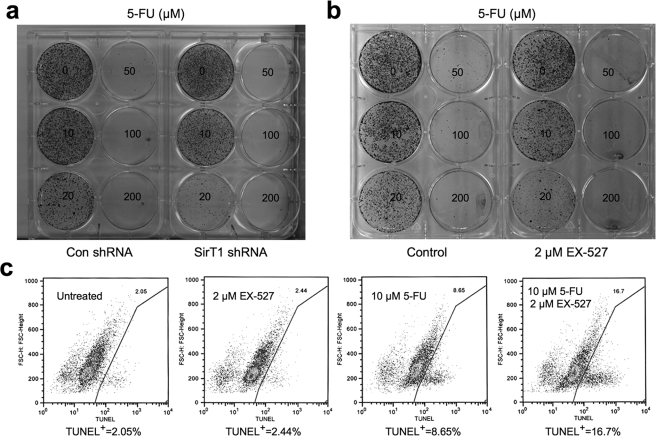
Many studies have implicated a role of SirT1 in promoting cell survival after stress or chemotherapy drug treatment. To test whether SirT1 can exert protection against cytotoxic agents in the same HCT116 cells where it has anti-proliferative function, cells with SirT1 knockdown were treated with 5-FU, which is frequently used in colon cancer chemotherapy. SirT1 knockdown clearly reduced long term cell viability after drug treatment in a colony formation assay (Fig. 4a). Similarly, treatment of HCT116 with the SirT1 inhibitor EX-527 also reduced long term viability after exposure to 5-FU (Fig. 4b). In short term TUNEL assays, EX-527 enhanced (∼100%) the ability of low concentrations of 5-FU to induce apoptosis (10 μm, Fig. 4c) but had no further benefit with higher concentrations of 5-FU (50 μm, data not shown). EX-527 treatment also significantly enhanced the level of apoptosis induced by another colon cancer drug camptothecin in a short term assay (supplemental Fig. S4). These results suggest that, under acute stress or DNA damage conditions, SirT1 expression provides a cell survival advantage. Because rapidly proliferating cells are often more sensitive to DNA damage-induced apoptosis, the growth-inhibitory activity of SirT1 may be partly responsible for its anti-apoptotic function.
FIGURE 4.
Inhibition of SirT1 increases sensitivity to chemotherapy. a, HCT116 control and SirT1 shRNA cell lines were treated with different concentrations of 5-FU for 24 h. Cells were incubated in drug-free medium for 15 days and colonies were stained with Crystal Violet. b, HCT116 cells were treated with 2 μm EX-527 and different concentrations of 5-FU for 24 h. Cells were incubated in drug-free medium for 15 days and colonies were stained with Crystal Violet. c, HCT116 cells were treated with EX-527 and 5-FU for 18 h and analyzed by TUNEL staining and fluorescence-activated cell sorting for the presence of apoptotic cells.
SirT1 Is Expressed at High Levels in Normal Colon and Benign Lesions
To investigate the role of SirT1 in human cancer development, we developed and characterized a monoclonal antibody 10E4 against the C-terminal domain of human SirT1. The antibody is highly specific for SirT1, detects a single band on Western blot, and shows no reactivity to other cellular proteins (supplemental Fig. S5a). Immunohistochemical staining using cells with SirT1 knockdown or overexpression showed differential nuclear staining by 10E4, demonstrating its ability to reveal differences in SirT1 expression level in cells and tissues (supplemental Fig. S5b and data not shown).
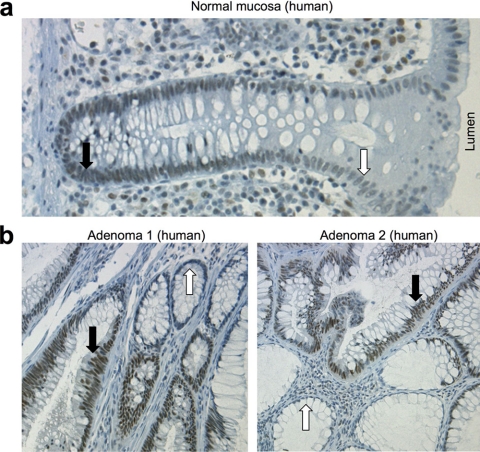
When normal colon and adenoma samples were stained with 10E4, the results showed that normal colon epithelial cells expressed significant levels of nuclear SirT1 at the base of the crypt where most cell proliferation occurs (black arrows), and gradually decreases as cells migrate toward the lumen (white arrows) (Fig. 5a). Therefore, SirT1 level in normal colon epithelium correlates with active cell proliferation. When benign adenomas (polyps) were examined, SirT1 was detected in all cells with adenomatous morphology (Fig. 5b, black arrows), but not in the adjacent normal mucosa (white arrows). This pattern was observed in all 26 adenomas (summarized in Fig. 6b), suggesting that SirT1 is expressed at a high level in 100% of adenomas.
FIGURE 5.
A high level of SirT1 is expressed in normal and preneoplastic colon epithelium. a, normal human colon mucosa was stained for SirT1 with 10E4 monoclonal antibody. The black arrow indicates the base of the crypt where most cell proliferation occurs; the white arrow indicates cells that have migrated toward the lumen. b, human colon adenomas were stained for SirT1 with 10E4. The black arrows indicate hyperplastic areas with dense nuclei and high levels of SirT1 expression; white arrows indicate areas of normal mucosa with low levels of SirT1.
FIGURE 6.
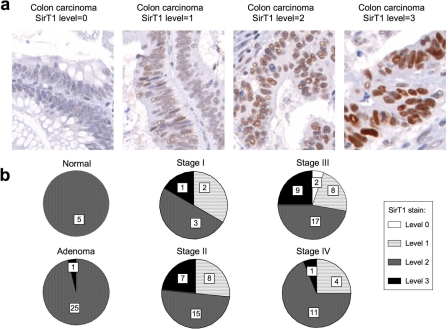
SirT1 expression is heterogenous in adenocarcinomas. Human colon tumor tissue array was stained for SirT1 using 10E4. a, intensity of SirT1 stain in different tumors ranged from 0 (no stain) to 3 (overexpression). b, pie representation of SirT1 levels correlated with stage of cancer. SirT1 level is moderate to high (level 2) in normal colonic mucosa and most adenomas. Tumors of different adenocarcinoma stages exhibit variable expression pattern of SirT1. While some tumors overexpress SirT1 (level 3), some down-regulate SirT1 (level 0 or 1). Level 3 expression is infrequent in stage IV tumors.
To test whether high levels of SirT1 expression is the cause or an effect of tumorigenesis, the intestines of ApcMin mice were stained for SirT1. ApcMin mice are heterozygous for codon 850 nonsense mutation in the Apc tumor suppressor gene, resulting in the development of intestinal adenomas by 120 days of age (31). The ApcMin adenomas develop after losing the remaining Apc allele and, thus, are similar to most human colon adenomas (32). The results showed that SirT1 level also remained high in the hyperplastic ApcMin polyps, similar to proliferating cells near the base of the crypt (supplemental Fig. S6). These results show that SirT1 is expressed at high levels in proliferating cells of the colon and intestinal epithelium and remains up-regulated when cells undergo initial transformation. Because the ApcMin polyps are initiated due to loss of the wild-type Apc allele, high levels of SirT1 expression are most likely a secondary result of transformation, not an active cause of tumor initiation.
SirT1 Is Under- or Overexpressed in Advanced Colon Carcinomas
To determine SirT1 expression pattern in colon tumors, the 10E4 antibody was used to stain a tissue micro array containing samples of normal colonic mucosa (n = 11), and of colon tumors (n = 88) collected at the Moffitt Cancer Center. The SirT1 stain was predominantly nuclear. When graded semiquantitatively on a 0–3 scale (0 = negative, 1 = low, 2 = high, and 3 = overexpression), SirT1 level at the base of the normal crypt and adenomas was ranked as score 2 (high). SirT1 staining significantly more intense than adenomas was ranked as score 3 (overexpression, Fig. 6a).
As summarized in Fig. 6b, SirT1 level is uniformly high (score 2) in all benign adenomas. However, colonic adenocarcinomas show a heterogeneous pattern of SirT1 levels, ranging from score 0 to 3. Although almost 25% of stage I–III carcinomas showed an intense (score 3) nuclear staining for SirT1, ∼30% tumors showed reduced levels of SirT1 compared with normal crypt and adenomas. Interestingly, only a very small subset of stage IV tumors (associated with poor 5-year survival) had SirT1 overexpression (score 3) as compared with the lower grade tumors (I–III).
Our interpretation of the heterogeneous SirT1 expression profile is that high SirT1 expression is an intrinsic response to cell proliferation in untransformed mucosa and pre-malignant adenomas. During further progression, SirT1 expression is silenced in a subset of tumors to facilitate tumor growth, whereas some low grade tumors may overexpress SirT1 to benefit from its anti-apoptotic effects. The fact that SirT1 overexpression is rare in high grade (stage IV) tumors suggests that the growth-inhibitory activity of SirT1 becomes a rate-limiting factor for progression to this stage.
DISCUSSION
The results described above show that SirT1 has properties of a growth suppressor. Experiments using cell lines suggest that endogenous SirT1 in colon tumor cells limits proliferation in culture and inhibits tumor xenograft formation. Knockdown of SirT1 increases the rate of tumor growth, whereas overexpression of SirT1 reduces tumor formation in nude mice. Furthermore, pharmacological inhibition of SirT1 increases the rate of cell proliferation in culture. These results together suggest that SirT1 has properties of a context-dependent tumor suppressor.
Our results lend further support to an emerging and unexpected tumor-suppressive function of SirT1. SirT1 has well established anti-apoptotic activity and is presumed to act as an oncogene. However, a recent study showed that transgenic mice overexpressing SirT1 in the intestine reduced the development of neoplasia caused by ApcMin mutation (26). SirT1+/− mice showed increased tumor incidence when crossed to a p53+/− background (27). MEFs derived from SirT1-null mice undergo spontaneous immortalization with higher frequency, which is a classic phenotype of a tumor suppressor (22). Therefore, different genetic models strongly suggest that SirT1 has properties of an atypical tumor suppressor.
It is noteworthy that our results appear to contradict those of the anti-apoptotic function of SirT1. Several publications reported tumor cell apoptosis or growth arrest after transient knockdown of SirT1 or treatment with SirT1 inhibitors such as sirtinol, splitomycin, and cambinol (20, 21, 33). However, these discrepancies can be reconciled if one takes into consideration that SirT1 inhibition sensitizes cells to apoptosis by stress. Therefore, tumor cells treated with SirT1 siRNA may undergo apoptosis due to additional transfection-associated stress. The off-target toxicity of the first-generation small molecule SirT1 inhibitors may also be responsible for the cell death or growth arrest responses. In fact, recent development of the nanomolar SirT1 inhibitor EX-527 demonstrated that specific inhibition of SirT1 alone does not cause apoptosis in tumor cell lines (29).
Several studies suggested that SirT1 may act as an oncogene based on the detection of high levels of SirT1 in certain tumors (34–36). Here, our analysis of the SirT1 expression profile in colon cancer suggests that SirT1 is both overexpressed and down-regulated in different subsets of tumors. Such a staining pattern can be interpreted as SirT1 having both oncogenic and tumor-suppressive properties. It is consistent with the pleiotropic effects of SirT1, i.e. anti-apoptotic and growth suppressive depending on cellular context. A subset of tumors may down-regulate SirT1 to obtain a proliferation advantage, whereas some may increase SirT1 expression to benefit from its anti-apoptotic function. However, interpretation of the tumor staining results is subjective and should not be taken as definitive evidence. Further examination of a large patient cohort is needed to determine the association between SirT1 expression and clinical parameters such as survival and treatment response.
Although the mechanism by which SirT1 inhibits cell proliferation remains to be further investigated, our previous study and this report suggest that inhibition of E2F1 is partly responsible. SirT1 interacts with E2F1, inhibits E2F1 acetylation, and is recruited by E2F1 to target promoters (16). When expressed at high levels, SirT1 is a potent inducer of G1 arrest. Our results show that inhibition of SirT1 results in pRb hyper-phosphorylation. This may be an indirect result of E2F1 activation, which can induce cyclinD/cdk4 activity and promote pRb phosphorylation. It may also be a direct effect, because SirT1 interacts with pRb (37).
In summary, our results suggest that both activators and inhibitors of SirT1 have therapeutic potential as anti-tumor agents. A simple scenario is that SirT1 activators may impart cancer prevention effects by enhancing the growth-inhibitory effect of SirT1 in benign tumors. Its effect on advanced stage tumors may be heterogeneous, depending on whether a tumor has evolved to rely on SirT1 for survival. However, when tumors are being treated with chemotherapy, SirT1 inhibitors may be useful for enhancing apoptotic response. Further investigation of SirT1 expression and its association with patient survival and treatment response is critical for the application of SirT1-targeted drugs.
Supplementary Material
Acknowledgments
We thank the Moffitt Molecular Biology Core for DNA sequencing and providing shRNA library clones; Flow Cytometry Core for fluorescence-activated cell sorting analyses; Histology Core for performing immunohistochemical staining; and Analytical Microscopy Core for capturing of tissue micro array data. We also thank Dr. Fred Alt and Dr. Toren Finkel for providing SirT1-null mice.
This work was supported, in whole or in part, by National Institutes of Health Grant CA121291 (to J. C.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S6.
- Sir2
- silent information regulator 2
- SirT1
- mammalian homolog of Sir2
- MEF
- mouse embryonic fibroblast
- shRNA
- short hairpin RNA
- TUNEL
- terminal deoxynucleotidyl transferase-mediated dUTP nick end labeling
- 5-FU
- 5-fluorouracil
- pRb
- retinoblastoma protein.
REFERENCES
- 1.Imai S., Armstrong C. M., Kaeberlein M., Guarente L. ( 2000) Nature 403, 795– 800 [DOI] [PubMed] [Google Scholar]
- 2.Guarante L. P. ( 2005) Ann. N.Y. Acad. Sci. 1055, 222. [DOI] [PubMed] [Google Scholar]
- 3.Tissenbaum H. A., Guarente L. ( 2001) Nature 410, 227– 230 [DOI] [PubMed] [Google Scholar]
- 4.Lin S. J., Defossez P. A., Guarente L. ( 2000) Science 289, 2126– 2128 [DOI] [PubMed] [Google Scholar]
- 5.Rodgers J. T., Lerin C., Haas W., Gygi S. P., Spiegelman B. M., Puigserver P. ( 2005) Nature 434, 113– 118 [DOI] [PubMed] [Google Scholar]
- 6.Moynihan K. A., Grimm A. A., Plueger M. M., Bernal-Mizrachi E., Ford E., Cras-Méneur C., Permutt M. A., Imai S. ( 2005) Cell Metab. 2, 105– 117 [DOI] [PubMed] [Google Scholar]
- 7.Bordone L., Motta M. C., Picard F., Robinson A., Jhala U. S., Apfeld J., McDonagh T., Lemieux M., McBurney M., Szilvasi A., Easlon E. J., Lin S. J., Guarente L. ( 2006) PLoS Biol. 4, e31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Howitz K. T., Bitterman K. J., Cohen H. Y., Lamming D. W., Lavu S., Wood J. G., Zipkin R. E., Chung P., Kisielewski A., Zhang L. L., Scherer B., Sinclair D. A. ( 2003) Nature 425, 191– 196 [DOI] [PubMed] [Google Scholar]
- 9.Lagouge M., Argmann C., Gerhart-Hines Z., Meziane H., Lerin C., Daussin F., Messadeq N., Milne J., Lambert P., Elliott P., Geny B., Laakso M., Puigserver P., Auwerx J. ( 2006) Cell 127, 1109– 1122 [DOI] [PubMed] [Google Scholar]
- 10.Baur J. A., Pearson K. J., Price N. L., Jamieson H. A., Lerin C., Kalra A., Prabhu V. V., Allard J. S., Lopez-Lluch G., Lewis K., Pistell P. J., Poosala S., Becker K. G., Boss O., Gwinn D., Wang M., Ramaswamy S., Fishbein K. W., Spencer R. G., Lakatta E. G., Le Couteur D., Shaw R. J., Navas P., Puigserver P., Ingram D. K., de Cabo R., Sinclair D. A. ( 2006) Nature 444, 337– 342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Milne J. C., Lambert P. D., Schenk S., Carney D. P., Smith J. J., Gagne D. J., Jin L., Boss O., Perni R. B., Vu C. B., Bemis J. E., Xie R., Disch J. S., Ng P. Y., Nunes J. J., Lynch A. V., Yang H., Galonek H., Israelian K., Choy W., Iffland A., Lavu S., Medvedik O., Sinclair D. A., Olefsky J. M., Jirousek M. R., Elliott P. J., Westphal C. H. ( 2007) Nature 450, 712– 716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Luo J., Nikolaev A. Y., Imai S., Chen D., Su F., Shiloh A., Guarente L., Gu W. ( 2001) Cell 107, 137– 148 [DOI] [PubMed] [Google Scholar]
- 13.Motta M. C., Divecha N., Lemieux M., Kamel C., Chen D., Gu W., Bultsma Y., McBurney M., Guarente L. ( 2004) Cell 116, 551– 563 [DOI] [PubMed] [Google Scholar]
- 14.Cohen H. Y., Miller C., Bitterman K. J., Wall N. R., Hekking B., Kessler B., Howitz K. T., Gorospe M., de Cabo R., Sinclair D. A. ( 2004) Science 305, 390– 392 [DOI] [PubMed] [Google Scholar]
- 15.Yeung F., Hoberg J. E., Ramsey C. S., Keller M. D., Jones D. R., Frye R. A., Mayo M. W. ( 2004) EMBO J. 23, 2369– 2380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang C., Chen L., Hou X., Li Z., Kabra N., Ma Y., Nemoto S., Finkel T., Gu W., Cress W. D., Chen J. ( 2006) Nat. Cell Biol. 8, 1025– 1031 [DOI] [PubMed] [Google Scholar]
- 17.Vaziri H., Dessain S. K., Ng Eaton E., Imai S. I., Frye R. A., Pandita T. K., Guarente L., Weinberg R. A. ( 2001) Cell 107, 149– 159 [DOI] [PubMed] [Google Scholar]
- 18.Chen W. Y., Wang D. H., Yen R. C., Luo J., Gu W., Baylin S. B. ( 2005) Cell 123, 437– 448 [DOI] [PubMed] [Google Scholar]
- 19.Yuan Z., Zhang X., Sengupta N., Lane W. S., Seto E. ( 2007) Mol. Cell 27, 149– 162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Heltweg B., Gatbonton T., Schuler A. D., Posakony J., Li H., Goehle S., Kollipara R., Depinho R. A., Gu Y., Simon J. A., Bedalov A. ( 2006) Cancer Res. 66, 4368– 4377 [DOI] [PubMed] [Google Scholar]
- 21.Ota H., Tokunaga E., Chang K., Hikasa M., Iijima K., Eto M., Kozaki K., Akishita M., Ouchi Y., Kaneki M. ( 2006) Oncogene 25, 176– 185 [DOI] [PubMed] [Google Scholar]
- 22.Chua K. F., Mostoslavsky R., Lombard D. B., Pang W. W., Saito S., Franco S., Kaushal D., Cheng H. L., Fischer M. R., Stokes N., Murphy M. M., Appella E., Alt F. W. ( 2005) Cell Metab. 2, 67– 76 [DOI] [PubMed] [Google Scholar]
- 23.Narala S. R., Allsopp R. C., Wells T. B., Zhang G., Prasad P., Coussens M. J., Rossi D. J., Weissman I. L., Vaziri H. ( 2008) Mol. Biol. Cell 19, 1210– 1219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Abdelmohsen K., Pullmann R., Jr., Lal A., Kim H. H., Galban S., Yang X., Blethrow J. D., Walker M., Shubert J., Gillespie D. A., Furneaux H., Gorospe M. ( 2007) Mol. Cell 25, 543– 557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fu M., Liu M., Sauve A. A., Jiao X., Zhang X., Wu X., Powell M. J., Yang T., Gu W., Avantaggiati M. L., Pattabiraman N., Pestell T. G., Wang F., Quong A. A., Wang C., Pestell R. G. ( 2006) Mol. Cell Biol. 26, 8122– 8135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Firestein R., Blander G., Michan S., Oberdoerffer P., Ogino S., Campbell J., Bhimavarapu A., Luikenhuis S., de Cabo R., Fuchs C., Hahn W. C., Guarente L. P., Sinclair D. A. ( 2008) PLoS ONE 3, e2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang R. H., Sengupta K., Li C., Kim H. S., Cao L., Xiao C., Kim S., Xu X., Zheng Y., Chilton B., Jia R., Zheng Z. M., Appella E., Wang X. W., Ried T., Deng C. X. ( 2008) Cancer Cell 14, 312– 323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Saunders L. R., Verdin E. ( 2007) Oncogene 26, 5489– 5504 [DOI] [PubMed] [Google Scholar]
- 29.Solomon J. M., Pasupuleti R., Xu L., McDonagh T., Curtis R., DiStefano P. S., Huber L. J. ( 2006) Mol. Cell Biol. 26, 28– 38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cheng H. L., Mostoslavsky R., Saito S., Manis J. P., Gu Y., Patel P., Bronson R., Appella E., Alt F. W., Chua K. F. ( 2003) Proc. Natl. Acad. Sci. U.S.A. 100, 10794– 10799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Moser A. R., Pitot H. C., Dove W. F. ( 1990) Science 247, 322– 324 [DOI] [PubMed] [Google Scholar]
- 32.Luongo C., Moser A. R., Gledhill S., Dove W. F. ( 1994) Cancer Res. 54, 5947– 5952 [PubMed] [Google Scholar]
- 33.Ford J., Jiang M., Milner J. ( 2005) Cancer Res. 65, 10457– 10463 [DOI] [PubMed] [Google Scholar]
- 34.Huffman D. M., Grizzle W. E., Bamman M. M., Kim J. S., Eltoum I. A., Elgavish A., Nagy T. R. ( 2007) Cancer Res. 67, 6612– 6618 [DOI] [PubMed] [Google Scholar]
- 35.Hida Y., Kubo Y., Murao K., Arase S. ( 2007) Arch. Dermatol. Res. 299, 103– 106 [DOI] [PubMed] [Google Scholar]
- 36.Stünkel W., Peh B. K., Tan Y. C., Nayagam V. M., Wang X., Salto-Tellez M., Ni B., Entzeroth M., Wood J. ( 2007) Biotechnol. J. 2, 1360– 1368 [DOI] [PubMed] [Google Scholar]
- 37.Wong S., Weber J. D. ( 2007) Biochem. J. 407, 451– 460 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.