Abstract
Diabetes mellitus accelerates cardiovascular microangiopathies and atherosclerosis, which are a consequence of hyperglycemia. The aldose reductase (AR) polyol pathway contributes to these microvascular complications, but how it mediates vascular damage in response to hyperglycemia is less understood. The RUNX2 transcription factor, which is repressed in diabetic animals, promotes vascular endothelial cell (EC) migration, proliferation, and angiogenesis. Here we show that physiological levels of glucose (euglycemia) increase RUNX2 DNA binding and transcriptional activity, whereas hyperglycemia does not. However, inhibition of AR reverses hyperglycemic suppression of RUNX2. IGF-1 secretion and IGF receptor phosphorylation by autocrine IGF-1 occur equally in euglycemic or hyperglycemic conditions, suggesting that reduced RUNX2 activity in response to hyperglycemia is not because of altered IGF-1/IGF receptor activation. AR also negatively regulates RUNX2-dependent vascular remodeling in an EC wounded monolayer assay, which is reversed by specific AR inhibition in hyperglycemia. Thus, euglycemia supports RUNX2 activity and promotes normal microvascular EC migration and wound healing, which are repressed under hyperglycemic conditions through the AR polyol pathway.
A major consequence of type 1 and type 2 diabetes is hyperglycemia, which contributes to a gradual increase in vascular dysfunction and disease, including retinopathy, nephropathy, neuropathy, cardiovascular disease, and stroke (1). Hyperglycemia promotes endothelial dysfunction, vascular leakage, and impaired angiogenesis leading to development of these pathologies (2). Several factors contribute to hyperglycemia-induced vascular dysfunction, including advanced glycation end products (AGEs),2 protein kinase C (PKC), increased utilization of glucosamine, and increased glucose flux through the aldose reductase (AR)-regulated polyol pathway (3). AR activity increases under hyperglycemic conditions and in response to oxidative stress and is believed to contribute to diabetic microvascular complications (4). AR converts glucose to sorbitol in the presence of NADPH (5). Lower levels of NADPH and NAD+, which result from AR activation, may contribute to the oxidative damage observed in diabetics, although the higher levels of sorbitol may promote the formation of AGEs, release of proinflammatory cytokines, increase in osmotic stress, increase in reactive oxygen species (ROS), and cell damage (6). Hyperglycemia can also lead to activation of an inflammatory response and NFκB expression, which is attenuated upon inhibition of AR (7). Paradoxically, up-regulation of AR may be an adaptive response to hyperglycemia (8) and may protect the heart from the toxic effects of lipid peroxidation (9). Specific inhibitors of AR have been used to treat the microvascular complications of diabetes, but without significant clinical benefit (10). Therefore, there is an urgent need to identify mechanistic pathways responsible for increased vascular complications in response to hyperglycemia to design new therapeutic agents to target these pathways.
The IGF-1 growth factor signaling pathway is particularly implicated in the hyperglycemic tissue microenvironment. Elevated IGF-1 activates a proangiogenic signaling cascade (11) and exhibits insulin-like effects, which reduce blood glucose levels and may be useful in the treatment of diabetes (12). Studies of glucose metabolism have revealed interactions between components of the glucose response and utilization machinery and signal transduction pathways, including the IGF-1/phosphatidylinositol 3-kinase and Akt signal transduction pathways that mediate cell survival and increased glucose uptake (13, 14). Pro-angiogenic cytokines such as IGF-1 activate specific transcription factors. We and others have shown that IGF-1/IGFR signaling activates the RUNX2 transcription factor through the phosphatidylinositol 3-kinase and MAPK/ERK signaling pathways, which regulates EC migration, proliferation, cell cycle progression, and angiogenesis (15–20). Recently, reduced RUNX2 expression and down-regulation of RUNX2 target genes were observed in insulin-deficient, hyperglycemic diabetic mice (21, 22). Insulin therapy partially restored expression of these genes implicating their responsiveness to glycemic status. Intriguingly, delayed wound healing has been observed in Runx2 heterozygous knock-out mice (23, 24). However, it is not known whether glucose regulates RUNX2 DNA binding and/or transcription and vascular EC wound healing.
We now report that glucose increases RUNX2 activity in ECs through increased DNA binding and transcriptional activation. Secretion of autocrine IGF-1 in response to glucose activates IGFR phosphorylation, RUNX2 DNA binding, and transcription of a RUNX2 reporter gene. However, under hyperglycemic conditions, changes in RUNX2 activity are not dependent on changes in IGF-1 secretion or IGFR phosphorylation. It appears that a negative regulatory pathway involving AR utilization of glucose is responsible for repression of RUNX2 activity. The results support the hypothesis that one of the functions of RUNX2 in ECs is to promote migration and repair and that, under hyperglycemic conditions, this function is impaired through AR-mediated reduction in RUNX2 activation.
EXPERIMENTAL PROCEDURES
Cells, Cell Culture, Transfection, and Inhibitors
HBME cells (19) were routinely cultured in Dulbecco's modified Eagle's medium + 10% FBS (complete medium), except for specific experiments when they were cultured in glucose and serum-free, phenol red-free Dulbecco's modified Eagle's medium supplemented with pyruvate, H2CO3, or glucose (defined medium). Transfection of siRNAs was performed as suggested by the manufacturer (Dharmacon, Inc.). Briefly, HBME cells (400,000/10-cm plate) were cultured for 24 h in complete medium and then transfected with Trans-Messenger transfection reagent (Qiagen, Inc., Valencia, CA) and siRNA oligonucleotides (25 nm) for 3 h in serum-free medium. Cells were harvested and replated for 24–48 h in defined medium containing 5 or 25 mm glucose prior to preparation of nuclear extracts (for RUNX2) or total protein (for AR) for use in Western blots. For DNA binding assays (see below), cells were cultured for 24 h in complete medium containing 5 mm glucose, transfected for 3 h in serum-free medium, harvested, replated for 24 h, and starved for 16 h prior to glucose treatment and isolation of nuclear extracts. The AR inhibitor alrestatin (Tocris Cookson, Inc., Ellisville, MO) was used at the indicated doses. The MEK inhibitor U0126 was obtained from Calbiochem, and the ERK substrate binding inhibitors 76, 99, and 101 were obtained from Dr. Shapiro (25). The general PKC inhibitor calphostin C and the PKCβ-specific inhibitor (3-(1-(3-imidazol)-1H-indol-3-yl)-4-anilino-1H-pyrrole-2,5-dione) were from Calbiochem. N-Acetylcysteine was obtained from Sigma.
Antibodies and siRNA
Specific RUNX2 (M70) and AR (N20, sc17732) antibodies were obtained from Santa Cruz Biotechnology, Inc. (Santa Cruz, CA). Actin, phospho-IGFR, and secondary antibodies were from Cell Signaling, Inc. (Danvers, MA). RUNX2 siRNA was designed using the Dharmacon siRNA target site software available on line (Thermo Fisher Scientific, Inc., Waltham, MA). The sequences used correspond to the Homo sapiens runt-related transcription factor 2 (RUNX2) mRNA (GenBankTM accession number NM_004348; version NM_004348.1; GI, 10863884). Four duplexes were synthesized based on the following locations within the RUNX2 coding sequence: Rx3 (532–550 Runt domain, 5′-GGACGAGGCAAGAGTTTCA-3′; Rx2 (557–575 Runt domain, 5′-CCATAACCGTCTTCACAAA-3′); Rx1 (793–811 C-terminal 5′-CCACAAGGACAGAGTCAGA-3′); Rx4 (1497–1515 C-terminal 5′-GGATGAATCTGTTTGGCGA-3′). Duplexes corresponding to a control GFP target site were also synthesized as follows: si.GFP (9–27, 5′-TCTGTGTGTTTGACTCTGA-3′). RUNX2 siRNA duplexes were used at a final concentration of 50–100 nm. AR siRNA was obtained from Dharmacon based on the published target sequence for AR used to knock down endogenous AR in human lens epithelial cells (7) as follows: si.AR (5′- AACGCATTGCTGAGAACTTTA-3′) or scrambled siRNA (5′-AACACGGCTTGAATGACTATA-3′). AR siRNA duplexes were used at a final concentration of 25 nm.
Immunoblotting
Western blot protocols were as described previously (26). Protein concentration was determined with the protein assay from Bio-Rad. Specific proteins were detected by enhanced chemiluminescence (ECL, Amersham Biosciences).
Semiquantitative Reverse Transcription-PCR
HBME cells were starved overnight and treated with glucose for 8 or 24 h. Total RNA was prepared using the Aurum Total RNA mini kit (catalog number 732-6820, Bio-Rad). RNA was reverse-transcribed using Moloney murine leukemia virus reverse transcriptase (catalog number M0253S, New England Biolabs). Amplification of cDNA was performed using GE Illustra Hot Start Mix RTG (catalog number ASM-28-9006-53). Products were visualized on a 1.5% agarose gel. The following primer sets were used: MMP13 (292 bp), forward 5′-CCTCCTGGGCCAAATTATGGAG-3′ and reverse 5′-CAGCTCCGCATCAACCTGCTG-3; RUNX2 (350 and 416 bp), forward 5′-GCACAGACAGAAGCTTGAT-3′ and reverse 5′-CCCAGTTCTGAAGCACCT-3′; cyclophilin (276 bp), forward 5′-CATCCTGAAGCATACAGGTC and reverse 5′-CAGAAGGAATGGTTTGATGG. PCR conditions were as follows: MMP13, 94 °C, 2 min; 94 °C, 30 s; 55 °C, 30 s; 68 °C, 1 min; 27 cycles; 68 °C, 7 min; RUNX2, 94 °C, 2 min; 94 °C, 1 min; 55 °C, 1 min; 72 °C, 1 min; 30 cycles; 72 °C, 10 min; and cyclophilin, 94 °C, 2 min; 94 °C, 1 min; 55 °C, 1 min; 72 °C, 1 min; 35 cycles; 72 °C, 10 min.
DNA Binding Assay and Transcriptional Assay
Electrophoretic mobility shift assays (EMSA) were used to measure RUNX2 DNA binding activity as described previously (19). Nuclear proteins were incubated with a 32P-labeled oligonucleotide derived from the human osteocalcin promoter (−141 to −165) and containing the human RUNX2 consensus-binding sequence (shown in boldface), 5′-CGTATTAACCACAATACTCG-3′. Specific RUNX2 transcriptional activation was performed with a luciferase reporter plasmid consisting of six copies of the osteoblast-specific element, Ose2, upstream of a minimal cytomegalovirus promoter in a pGL2 expression vector as described before (16, 27). For all luciferase assays, the pGL2.6XOSE2 plasmid was used at a concentration of 1 μg per well, and the pTK-renilla was used at a concentration of 50 ng per well. Lysates were analyzed using the dual luciferase kit (Promega) and a Turner Design TD 20/20 dual-wavelength luminometer.
Wounded Monolayer Assays
ECs were cultured in 6-well plates for 24 h in complete Dulbecco's modified Eagle's medium containing 10% FBS. A scratch site was created using a 200-μl pipette tip, and scraped cells were removed with one wash in complete medium (t = 0). Cells were cultured in defined serum-free medium containing various concentrations of glucose or control sugars (mannitol, 2-DG) and inhibitors at the indicated doses. Wounded monolayers were photographed after 24 or 48 h, and the width of the wounded areas was measured from photographic prints. Several regions (n = 5–6) for each treatment were recorded, and the means ± S.D. of each treatment were calculated after normalizing for width of wounded area at t = 0 and expressed as percentage of wound healing.
Enzyme-linked Immunosorbent Assay
The Quantikine Human IGF-I Immunoassay from R & D Systems, Minneapolis, MN (catalog number DG100), was used to determine IGF-1 secretion in HBME cells. HBME cells were grown as described and starved overnight in 0.1% bovine serum albumin (for time course) or 0.01% bovine serum albumin (for glucose dosing). After a 16-h serum and glucose starvation, glucose was added. Supernatants were pooled from four plates for each time point or two plates each for glucose dose experiments. Protein was precipitated from supernatants using 100% saturated (NH4)2SO4. Precipitated protein was collected by centrifugation and resuspended in phosphate-buffered saline. Protein was desalted using 5-kDa Amicon spin filters and phosphate-buffered saline washes. 100 μg of total protein were used for time or dose determination in the ELISA. Absorbance readings at 540 nm were subtracted from readings at 450 nm to correct for optical imperfections in the plate. All data were normalized to total protein for each sample.
Image Acquisition and Analysis
Cell images were acquired with a Nikon TMS microscope using the E10 Ph1DL objective (0.25 160/−) at room temperature and a Nikon CoolPix 4500 camera (NikonView5 CoolPix Int., version 5.1.3 software). Gel images were acquired with a Kodak DC120 Zoom digital camera (Digital Science 1DLE software). Images were processed using Adobe Image Reader or Microsoft Office Picture Manager, and layouts were compiled with Microsoft Office Excel and Microsoft Office Power Point software.
Statistical Analyses
All data represent the mean ± S.D. of representative individual experiments that were repeated three times. The number of replicates for data points was ≥3. To compare multiple measurements, data were subjected to Tukey's post-hoc adjustment for 2-by-2 comparisons following analysis of variance with p values <0.05 considered significant as indicated in the figure legends.
RESULTS
Glucose Regulates RUNX2 Activity
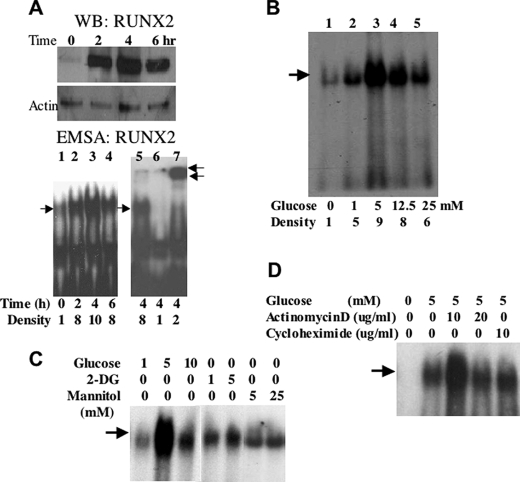
Based on mouse models of diabetes (21, 22) and our own published data (18), we hypothesize that glucose metabolism may regulate RUNX2 activity. To examine RUNX2 activation in response to glucose, ECs were depleted of nutrients and then treated with 5 mm glucose (equivalent to physiological levels of resting glucose = 90 mg/dl). Western blots showed that RUNX2 protein was present in a high salt-extractable nuclear (DNA-bound) fraction within 2 h of treatment with peak protein levels evident by 4 h (Fig. 1A, WB). If glucose activates RUNX2, one would expect increased DNA binding to conserved promoter elements of target genes containing the ACACCAA motif (27). DNA binding assays using a radiolabeled oligonucleotide containing a RUNX2 consensus-binding site showed that peak RUNX2 activity was evident within 4 h of glucose addition (Fig. 1A, EMSA). This complex contained RUNX2 protein as indicated by supershifting after incubation with RUNX2 antibody (Fig. 1A, lane 7) and was not detectable upon incubation with 100-fold excess cold oligonucleotide (Fig. 1A, lane 6). RUNX2 protein was also detected in cytosolic and nuclear matrix fractions, which, however, were not active in DNA binding assays (data not shown). Interestingly, a glucose dose response showed that 5 mm glucose was optimal in stimulating RUNX2 DNA binding (Fig. 1B, lane 3), with reduced stimulation observed under hyperglycemic (12.5 and 25 mm) conditions (Fig. 1B, lanes 4 and 5). The osmotic control sugar, mannitol, or the nonmetabolizable glucose analog, 2-DG did not activate RUNX2 DNA binding (Fig. 1C). De novo mRNA and protein synthesis were not required because actinomycin D or cycloheximide did not inhibit RUNX2 activation in response to glucose (Fig. 1D). In agreement with actinomycin D results, the levels of RUNX2 mRNA did not change in response to glucose (data not shown).
FIGURE 1.
Glucose-activated RUNX2 DNA binding activity in EC. A, confluent ECs were starved overnight and treated with 5 mm glucose for the indicated time. RUNX2 protein was measured by Western blotting (WB RUNX2), and RUNX2 DNA binding activity was determined by EMSA (EMSA RUNX2) using a specific radiolabeled nucleotide from the osteocalcin promoter, which contains a RUNX2 consensus-binding site. Excess (100-fold) cold, unlabeled oligonucleotide (lane 6) was used to verify specific RUNX2 binding, and a RUNX2-specific antibody (0.2 μg) was used to induce a super-shifted band and confirm the presence of RUNX2 in the DNA-binding complex (lane 7). Single arrow indicates RUNX2 shift; double arrows indicate RUNX2 supershift. Relative RUNX2 band density (single arrow) is indicated below each lane. B, ECs were treated with the indicated concentrations (mm) of glucose for 4 h, and DNA binding activity was measured by EMSA. Relative RUNX2 band density at each glucose concentration is indicated. C, glucose (1, 5, and 10 mm), mannitol (5 and 25 mm), or 2-DG (1 and 5 mm) were used to treat starved HBME cells for 4 h. DNA binding was measured by EMSA. Arrow indicates RUNX2-shifted band. D, actinomycin D (10 and 20 μg/ml) or cycloheximide (10 μg/ml) was used to inhibit gene transcription or protein synthesis, respectively. Arrow indicates the specific RUNX2-shifted band on the EMSA gel.
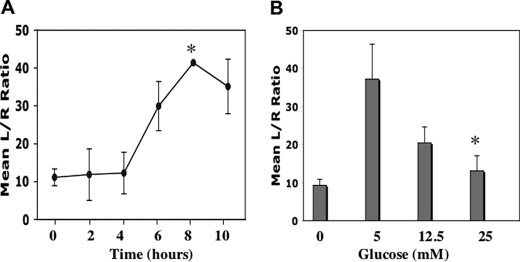
DNA binding is a prerequisite for specific transcriptional activation of RUNX2 target promoters and/or specific expression of endogenous target genes. To determine whether glucose could activate RUNX2 target genes, ECs were transfected with a RUNX2 promoter-luciferase plasmid (6XOSE2.Luc). Glucose (5 mm) increased RUNX2-specific luciferase activity within 6 h, with sustained activation for up to 10 h (Fig. 2A). As observed for DNA binding, glucose-activated RUNX2 transcriptional activity was optimal at 5 mm glucose with reduced activation observed at 12.5 and 25 mm glucose (Fig. 2B). Glucose treatment also resulted in activation of the endogenous RUNX2 target gene MMP13. However, both semiquantitative and quantitative reverse transcription-PCR did not reveal any differences in expression in response to different glucose concentrations (data not shown).
FIGURE 2.
RUNX2 transcriptional activity in response to glucose. A and B, HBME cells were transfected with the reporter 6XOSE2.luciferase vector and TK.renilla vector (to normalize for transfection efficiency) prior to starvation and subsequent treatment with 5 mm glucose for the indicated time (A) or glucose dose (B) for 6 h. Firefly luciferase activity was determined in cell lysates and expressed relative to Renilla luciferase. Asterisk indicates the following p values: A, comparing transcriptional activation at 8 h versus t = 0 (p < 0.05); B, at 5 mm versus 25 mm glucose (p < 0.01).
Hyperglycemic Repression of RUNX2 Activity Is Regulated by AR
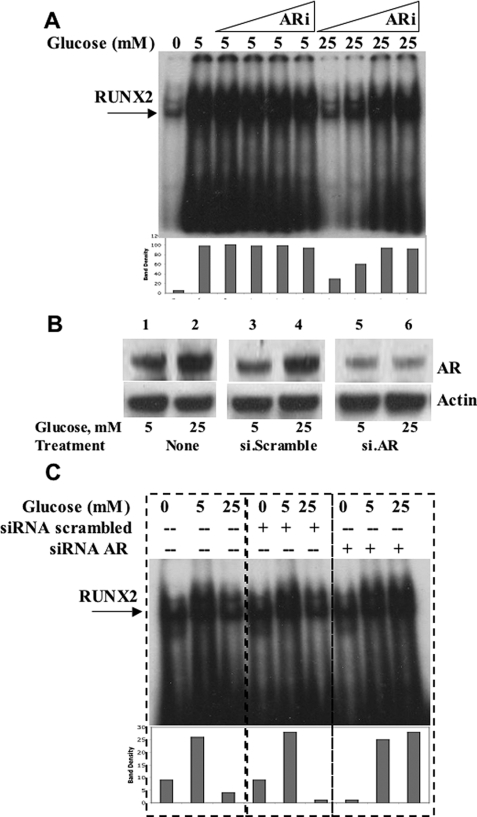
Several negative regulatory pathways may be operative in vascular cells that might attenuate RUNX2 activity when cells are exposed to high glucose, including the AR polyol pathway (3). Therefore, ECs were treated with the AR inhibitor alrestatin, and RUNX2 DNA binding activity was measured by EMSA (Fig. 3A). Alrestatin did not appear to affect RUNX2 activation in cells exposed to euglycemia (5 mm). In contrast, RUNX2 activity was increased 4-fold with AR inhibition in cells exposed to hyperglycemia (25 mm), consistent with an AR-mediated negative effect on RUNX2 activity at this glucose dose. Because AR inhibitors may also reduce the activity of other members of the aldo-keto reductase superfamily (5), siRNA oligonucleotides specific for AR were synthesized to knock down AR expression and test for the activation of RUNX2. ECs were cultured in either euglycemic (5 mm) or hyperglycemic (25 mm) conditions, and the levels of AR protein were measured by Western blotting. Hyperglycemic conditions increased the levels of AR protein relative to euglycemia almost 2-fold (Fig. 3B). Transfection with AR-targeted siRNA oligonucleotide sequences blocked the hyperglycemia-induced increases in AR. The effect of AR knockdown on RUNX2 activation under these conditions was then measured with EMSA. Specific AR-targeted siRNA transfection did not alter RUNX2 activity of cells incubated in 5 mm glucose, but it did increase the RUNX2 activity of cells incubated in 25 mm glucose by almost 5-fold relative to untransfected cells (Fig. 3C). These results are consistent with an inverse relationship between RUNX2 activity and AR expression, which is induced by hyperglycemia and supports a role for AR in the suppression of RUNX2 activity under hyperglycemic conditions.
FIGURE 3.
Aldose reductase inhibits RUNX2 activity in response to hyperglycemia. A, HBME cells were serum- and glucose-starved before incubation with glucose (5 or 25 mm) and the AR inhibitor (ARi) alrestatin. Triangles indicate increasing doses of alrestatin as follows: 0.1, 1.0, 10, and 20 μm. After 4 h, nuclear extracts were analyzed for RUNX2 DNA binding using EMSA. Bars below each lane indicate the relative band density of the specific RUNX2-shifted band. B, specific siRNA targeting AR or scrambled siRNA oligonucleotides (25 nm) were transfected into HBME cells grown in the presence of 5 or 25 mm glucose. Cells were harvested and replated for 24 h prior to extraction of total protein for AR immunoblotting. C, cells grown in 5 mm glucose were transfected with siRNA (scrambled) or siRNA (AR) and replated in complete medium for 24 h prior to overnight starvation and treatment with the indicated doses of glucose (0, 5, and 25 mm) for 4 h. RUNX2 DNA binding activity was determined by EMSA as before. Relative densities of the RUNX2-shifted bands are indicated. Representative results from a single EMSA gel are marked with dashed lines for clarity of analysis. Similar results were obtained from three separate determinations.
Hyperglycemic Inhibition of RUNX2 Is Not Dependent on IGF1/IGFR or ERK Activation
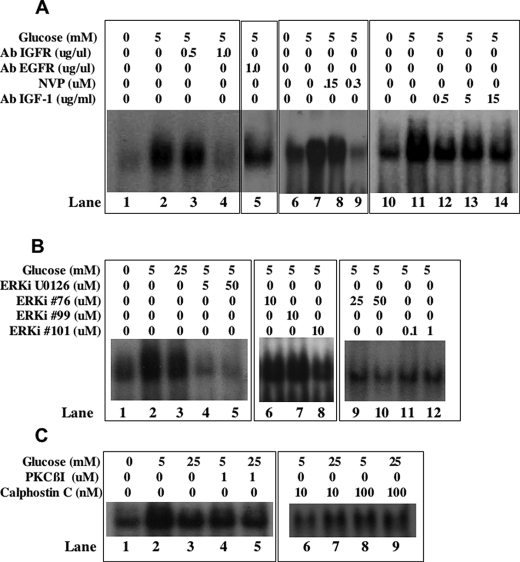
Because our previous work showed that IGF-1 increases RUNX2 DNA binding activity through the IGFR and MAPK pathways (18), we also examined whether changes in IGF-1/IGFR or ERK signaling could account for glucose-dependent RUNX2 activation. Neutralizing IGFR (Fig. 4A, lanes 1–4) or IGF-1 (Fig. 4A, lanes 6–10) antibodies inhibited glucose-stimulated RUNX2 DNA binding, as did the addition of an IGFR-specific kinase inhibitor (Fig. 4A, lanes 11–14). Similarly, inhibition of the ERK1/2 activator MEK with U0126 or direct inhibition of ERK2/substrate interactions with two specific ATP-independent ERK2 inhibitors (76 and 101) selected via computer-assisted drug design (25) also reduced RUNX2 DNA binding in response to glucose (Fig. 4B, lanes 4 and 5 and 8–12). Another selected compound (compound 99) was ineffective at any dose. Hyperglycemia may promote negative growth effects by activation of the PKC pathway. In these cells, inhibition of PKC with either a PKCβ-specific inhibitor or the general PKC inhibitor calphostin C did not restore RUNX2 activity in 25 mm glucose (Fig. 4C, lanes 5, 7, and 9). However, the PKCβ-specific inhibitor reduced RUNX2 activation in response to 5 mm glucose (Fig. 4C, lane 4) as did 10 nm calphostin C (Fig. 4C, lane 6).
FIGURE 4.
IGF-1/IGFR requirement for glucose-activated RUNX2 activity. A, lanes 1–4, glucose- and serum-starved HBME were treated with 5 mm glucose for 4 h in the presence or absence of neutralizing antibody to IGFR (0, 0.5, and 1.0 μg/ml) or epidermal growth factor receptor (1 μg/ml). Nuclear extracts were examined for RUNX2 DNA binding activity by EMSA. Lanes 6–10, cells were exposed to neutralizing IGF-1 (0.5, 5, and 15 μg/ml) antibody and analyzed for RUNX2 DNA binding activity. Lanes 11–14, cells were treated with the IGFR kinase inhibitor NVP-ADW742 (150 and 300 nm), and RUNX2 DNA binding was determined by EMSA. B, cells were treated with the MEK inhibitor U0126 (lanes 4 and 5) or the ERK inhibitors 76 (lanes 6, 9, and 10), 99 (lane 7), or 101 (lanes 8, 11, and 12) for 4 h, and nuclear extracts were resolved on EMSA gels. C, cells were treated with the indicated PKCβ-specific (lanes 4 and 5) or general calphostin C (lanes 6–9) PKC inhibitors for 4 h prior to extraction of nuclear proteins and analysis of RUNX2 DNA binding by EMSA.
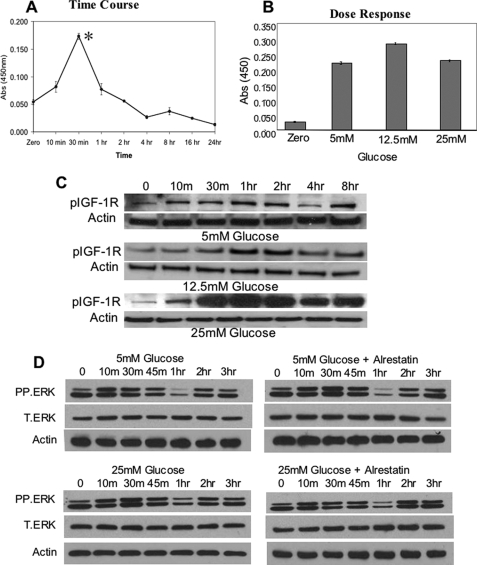
To determine whether ECs produced autocrine IGF-1 in response to glucose, cell-free and serum-free supernatants were evaluated using an IGF-1-specific ELISA. Secreted IGF-1 levels increased within 10 min and were maximal 30 min after glucose addition (Fig. 5A), although IGF-1 secretion was not significantly different in cells cultured under euglycemic or hyperglycemic conditions (Fig. 5B) nor was phosphorylation of IGF-1R reduced by 25 mm glucose (Fig. 5C). Analysis of ERK expression in response to 5 mm glucose showed that ERK activation also occurred within 10 min and reached maximal levels by 30 min before declining (Fig. 5D). However, activation in hyperglycemia was not appreciably different, nor did treatment with the AR inhibitor alrestatin alter ERK activation in response to 25 mm glucose.
FIGURE 5.
Hyperglycemic inhibition of RUNX2 activity is not dependent on IGF1 expression, IGFR activity, or ERK activation. A, starved HBME cells in serum-free media were treated with 5 mm glucose for the indicated times, and cell supernatants were isolated, concentrated, and analyzed for IGF-1 expression using a specific IGF-1 ELISA. Asterisk indicates p < 0.05 for glucose treatment at 30 min versus t = 0. B, HBME cells were treated with the indicated doses of glucose for 30 min, and the levels of IGF-1 in the cell supernatants were determined by specific ELISA. p values for glucose treatment at 5 mm versus 25 mm glucose indicate no significant difference. Results (A and B) are representative of three separate experiments and are plotted as the mean ± S.D. from triplicate measurements. C, total cellular extracts from glucose-treated HBME cells (for the indicated times and glucose doses) were isolated and analyzed by Western blotting using phospho-specific IGFR antibody as described under “Experimental Procedures.” Actin expression was used as a loading control. D, total cellular extracts from glucose-treated (5 versus 25 mm) HBME cells were isolated after the indicated times and analyzed for phospho-ERK (P.ERK) and total ERK (T.ERK) expression. Alrestatin (20 μm) was used to inhibit AR where indicated. Actin expression was used as a loading control.
RUNX2 and AR Regulate Glucose-activated EC Wound Healing, Role of Oxidative Stress
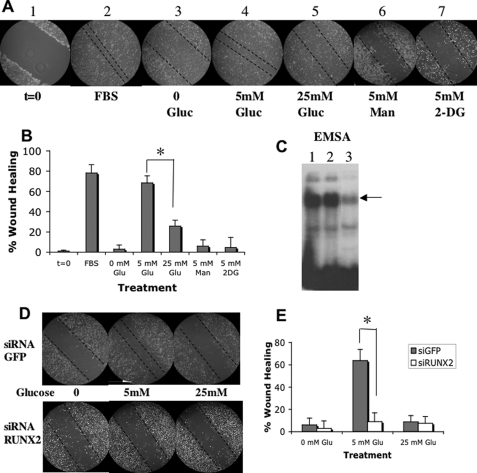
Hyperglycemia inhibits EC migration in in vitro models of wound healing (28) and angiogenesis in vivo (29, 30). We showed previously that RUNX2, a nuclear factor, promotes migration and the formation of capillary-like networks by EC in a process requiring IGF-1 and IGFR activation (16, 18). To determine the role of glucose in RUNX2 activation and EC migration, we measured the EC migratory response to wounding (Fig. 6A). Treatment with 5 mm glucose was as effective as 10% FBS in achieving almost 80% wound closure (Fig. 6B), whereas increasing glucose to 25 mm inhibited closure by about 2.5-fold (30% wound closure). Neither the iso-osmotic control mannitol nor the nonmetabolizable glucose analog 2-DG stimulated wound closure (Fig. 6, A and B). Furthermore, glucose-induced wound closure was mediated through RUNX2 expression because prior siRNA-induced knockdown of RUNX2 (Fig. 6C) reduced wound closure to <10% in 5 mm glucose (Fig. 6, D and E).
FIGURE 6.
Glucose-regulated EC wound healing is dependent on RUNX2 expression. A, HBME cells were cultured until confluent, and the monolayer was scraped to initiate wounding (panel 1, t = 0). Monolayers were treated with 10% fetal bovine serum (FBS) (panel 2), no glucose (panel 3), 5 mm glucose (panel 4), 25 mm glucose (panel 5), 5 mm mannitol (panel 6), or 5 mm 2-DG for 24 h (panel 7). Dotted lines mark the wound margins. B, quantitation of results from A expressed as means ± S.D. from n = 5–6 measurements. Asterisk indicates p < 0.01 for 5 mm versus 25 mm glucose-treated cells. C, HBME cells were transfected with siRNA targeting RUNX2 or GFP and RUNX2 DNA binding was determined by EMSA performed on nuclear extracts from untreated (lane 1), GFP siRNA-treated (lane 2), or RUNX2 siRNA-treated cells (lane 3). Arrow indicates RUNX2-shifted band. D, for RUNX2 siRNA knockdown, cells were transfected with RUNX2 or GFP-specific siRNA duplexes 24 h before wounding and then treated with 5 or 25 mm glucose for 24 h. Representative images show that reduction of RUNX2 inhibits wounded monolayer recovery in response to glucose. E, quantitative % wound healing is representative of n = 5–6 measurements per point. Asterisk indicates p < 0.01 for si.GFP- versus si.RUNX2-treated cells at 5 mm glucose.
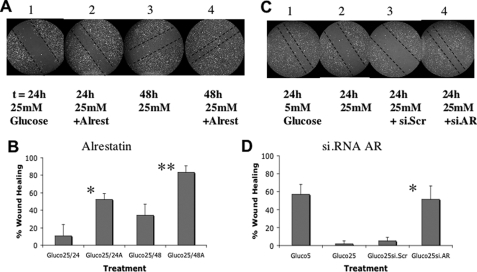
The role of AR in suppressing the migration of wounded ECs exposed to hyperglycemia was tested. ECs were cultured as before, and wounded monolayers were treated with 25 mm glucose in the presence or absence of the AR inhibitor alrestatin (Fig. 7A). Cells were observed 24 and 48 h after treatment. At each time point, AR inhibitor-treated cells exhibited improved wound healing relative to untreated cells (Fig. 7B, 50% versus 10% at 24 h; 80% versus 30% at 48 h). Similarly, specific AR siRNA treatment of the ECs (Fig. 7C) also improved wound healing under hyperglycemic conditions (from 2 to 50%), similar to the wound closure observed with 5 mm glucose (57%), whereas scrambled siRNA only marginally improved wound healing (5%) (Fig. 7D).
FIGURE 7.
Aldose reductase represses RUNX2-mediated EC wound healing. A and B, HBME wound healing assays in response to 25 mm glucose were performed as in Fig. 6 using the AR inhibitor alrestatin (10 μm). The degree of wound healing was monitored at 24 h (panels 1 and 2) and 48 h (panels 3 and 4) and quantitated as described under “Experimental Procedures.” All photos are from representative results of three separate experiments. Dotted lines mark the wound margins. Bar graphs quantifying the degree of wound healing were calculated from n = 5–6 locations per treatment and plotted as mean ± S.D. * indicates p < 0.01 for alrestatin versus untreated cells in 25 mm glucose (24 h); ** indicates p < 0.01 at 48 h. C and D, HBME wound healing assays using siRNA targeting AR or control scrambled siRNA were performed in cells treated with 25 mm glucose for 24 h. Quantitation reflects the mean ± S.D. from n = 5–6 measurements. * indicates p < 0.01 for si.AR- versus si.scrambled-treated cells in 25 mm glucose.
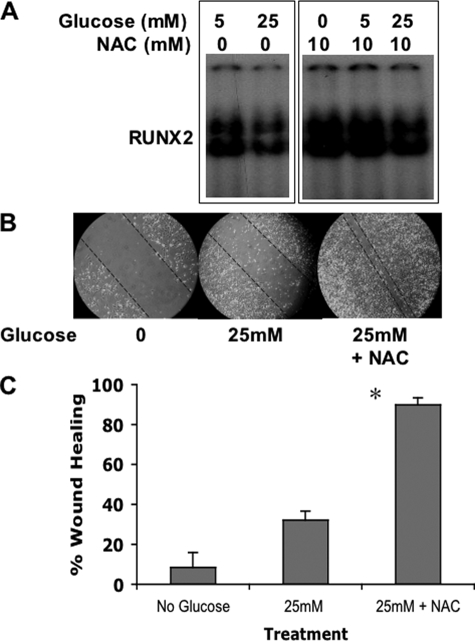
It is well known that activation of the polyol pathway by hyperglycemia results in increased sorbitol production and elevated oxidative stress. To determine whether hyperglycemic repression of RUNX2 activity was mediated by reactive oxygen species, cells were cultured in 25 mm glucose in the presence or absence of the antioxidant NAC. NAC treatment reduced RUNX2 inhibition by hyperglycemia (Fig. 8A) and enhanced vascular EC wound healing in 25 mm glucose (Fig. 8, B and C). These results are therefore consistent with a negative regulatory role for AR in wounded monolayer healing, which is mediated by oxidative stress under hyperglycemic conditions.
FIGURE 8.
Hyperglycemia-induced oxidative stress inhibits RUNX2 activity and wound healing. A, EC were serum- and glucose-starved prior to incubation with glucose and the antioxidant NAC (10 mm) for 4 h. Nuclear extracts were analyzed for RUNX2 DNA binding using EMSA. B, EC wound healing assays were performed as described with cells treated for 24 h with 25 mm glucose in the presence or absence of 10 mm NAC. C, percent wound healing was calculated as described previously and is representative of n = 5 measurements per treatment condition. *, p < 0.01 for 25 mm versus 25 mm + NAC.
DISCUSSION
We examined the response of ECs to glucose and found that RUNX2 DNA binding and transcriptional activity in these cells is glucose-activated through an autocrine IGF-1/IGFR metabolic signal transduction loop. Exposure of these ECs to hyperglycemic conditions, however, reduced RUNX2 activity, which was reversed by inhibition of the AR polyol pathway. Using specific pharmacologic and genetic RUNX2 and AR inhibitors, we showed that RUNX2 is necessary for EC migration and vascular cell remodeling, whereas AR is responsible for their counter-regulation under hyperglycemic conditions.
Regulation of RUNX2 Activity by Hyperglycemia and AR
EC RUNX2 DNA binding and transcriptional activity in response to glucose was independent of new protein or mRNA synthesis and was not increased by 2-DG or mannitol. RUNX2 mRNA levels in response to glucose did not change, which is consistent with data showing that RUNX2 protein exists in a preformed pool prior to activation by glucose. This activation could be the result of import into the nucleus, changes in post-translational modification such as phosphorylation, and/or RUNX2 association with essential cofactors that increase DNA binding, such as Cbfβ (31). How these pathways contribute to glucose-mediated RUNX2 activation is currently being investigated.
The data support the hypothesis that AR negatively regulates RUNX2 activity in hyperglycemic conditions. Production of sorbitol in ECs exposed to hyperglycemia (activation of AR) has been shown to increase apoptosis and expression of the angiogenic factor vascular endothelial growth factor in these cells (32). In contrast, AR inhibitors have been shown to reduce the levels of vascular endothelial growth factor (33). Hyperglycemia increases p38/JNK (stress response; increased osmolarity) at the expense of ERK (proliferative) MAPK pathways (34). Hyperglycemia promotes transforming growth factor-β secretion and pSmad activation, which may negatively regulate RUNX2 expression (35). In addition to increased polyol pathway flux, other mechanisms may support glucose-mediated vascular damage as follows: increased formation of intracellular AGEs, PKC activation, or hexosamine pathway flux. One common mechanism linking these different pathways is formation of ROS (3). Generation of ROS and cellular oxidative stress could inhibit wound healing and contribute to microvascular and macrovascular complications, such as diabetic retinopathy and atherosclerosis, respectively (36). Our data (Fig. 8) are consistent with a role for an oxidative stress-induced negative regulatory pathway in the attenuation of RUNX2 activity in response to hyperglycemia. In our culture model, the PKC pathway does not appear to contribute greatly to inhibition of RUNX2 in hyperglycemia because treatment with two PKC inhibitors did not restore RUNX2 activity to levels observed in euglycemia. However, it has been shown that PKCβ regulates the action of angiogenic factors in ECs and activates ERK signaling and tumorigenesis (37, 38). In the presence of 5 mm glucose, inhibition of PKCβ reduced RUNX2 activation, consistent with a role for PKCβ activation of ERK signaling and RUNX2 activity in response to glucose.
IGF-1/IGFR and ERK Activation in Glycemic Regulation of RUNX2
We showed previously that IGFR activity promotes RUNX2 expression and DNA binding through phosphatidylinositol 3-kinase and ERK signaling (18). The current results show that inhibition of the IGF-1/IGFR signaling pathway reduced glucose-mediated RUNX2 activity. There were no differences in IGF-1 secretion or IGFR activation in euglycemic or hyperglycemic conditions. This suggests that glucose activates RUNX2 through an autocrine IGF-1 mechanism, but that changes in IGF1/IGFR activity do not account for reduced RUNX2 activity under hyperglycemic conditions. Similarly, inhibition of the ERK pathway using indirect and direct specific inhibitors completely abrogated RUNX2 activation in 5 mm glucose. However, there was no inhibition of ERK activation in response to 25 mm glucose. Although IGF-1/IGFR and ERK activities were similar in response to euglycemic or hyperglycemic conditions, other factors may contribute to reduced RUNX2 activation under hyperglycemic conditions, including the up-regulation of IGF-1-binding proteins (39), the activation of p38/JNK (34), or the accumulation of sorbitol through the AR polyol pathway (32).
Diabetes and Regulation of Wound Healing by RUNX2 and AR
Glucose-stimulated healing of a wounded EC monolayer was dose-dependent with optimal wound closure and RUNX2 activity observed under physiological (5 mm) conditions. Exposure to hyperglycemia inhibited both RUNX2 activity and wound healing. It appears that deficits in EC migration and wound healing are commonly observed under hyperglycemic conditions and may contribute to vascular dysfunction in diabetics (28). In fact, it has been suggested that hyperglycemia itself may inhibit angiogenesis (2). Inhibition may be mediated by the AR polyol or the glucosamine pathways (6, 10, 40). We (16) and others (15) have reported that RUNX2 DNA binding activity serves an essential function in EC migration and invasion, which is, in part, mediated through the transcriptional control of metalloproteinases. However, we also have shown that RUNX2 promotes EC proliferation (17, 19) and survival (17). The effects observed in the current glucose-responsive model of wound healing could be mediated by the ability of glucose to activate RUNX2 activity through the IGF1/IGFR and ERK signaling pathways. In contrast, hyperglycemic attenuation of RUNX2 activity may be mediated through increased AR expression and generation of oxidative stress. The ability of RUNX2 to promote EC migration, proliferation, and/or survival has specific relevance to understanding the vascular response to diabetes because perturbations in gene transcription may contribute to diabetic vasculopathy (41). It is possible that the endothelial (vascular) response to hyperglycemia is part of a general sensor mechanism that responds to levels of glucose through regulation of transcription factor activity in a variety of tissues (42).
Significance and Therapeutic Relevance
Expression of Runx2 is diminished in mice made diabetic by streptozotocin treatment (21). The converse is also true; the Runx2-regulated bone matrix factor, osteocalcin, which is produced by the skeleton, acts like a hormone to regulate glucose metabolism and insulin secretion in the pancreas (43). Hyperglycemic conditions, which activate AR and the polyol pathway, may suppress RUNX2 activity and reduce normal endothelial repair. Because RUNX2 promotes EC wound healing, it may be important in maintaining normal vascular function, whereas down-regulation of RUNX2 under hyperglycemic conditions may be associated with vascular dysfunction.
The increased incidence of obesity that contributes to type 2 diabetes has also been implicated in colorectal cancer and in the promotion of a more invasive and aggressive breast cancer phenotype (44). Cancer cells are known to rely on glucose for energy in a process of aerobic glycolysis that is believed to contribute to tumor growth, survival, and metastasis (45). Therefore, our results also have broader implications for cancer, especially the role that glucose metabolism might play in tumor angiogenesis. AR inhibition may be a possible strategy to “normalize” dysfunctional vessels and promote vascular remodeling and/or angiogenesis, thus reducing the microvascular complications of diabetes or improving chemotherapeutic delivery to tumors (46).
Acknowledgments
We thank Neil Hardegen (lab manager) for equipment and laboratory maintenance and assistance; Dr. Francesco Hoffman (Novartis) for providing the IGFR kinase inhibitor; and Drs. George Martin, Richard Hornstein, John McLenithan, and Geoff Girnun for suggestions and critical reviews.
This work was supported, in whole or in part, by National Institutes of Health Grants RO1CA108846 (to A. P.), RO1CA120215 (to P. S.), and RO1HL084223 (to S. E. G.). This work was also supported by a University of Maryland Greenebaum Cancer Center pilot grant from the Cigarette Restitution Fund of the State of Maryland (to A. P.).
- AGEs
- advanced glycation end products
- AR
- aldose reductase
- EC
- endothelial cell
- ERK
- extracellular signal-regulated kinase
- IGF-1
- insulin-like growth factor-1
- IGFR
- insulin-like growth factor receptor
- MAPK
- mitogen-activated protein kinase
- MEK
- MAPK/ERK kinase
- MMP13
- matrix metalloproteinase-13
- NAC
- N-acetylcysteine
- PKC
- protein kinase-C
- ROS
- reactive oxygen species
- EMSA
- electrophoretic mobility shift assay
- ELISA
- enzyme-linked immunosorbent assay
- FBS
- fetal bovine serum
- siRNA
- small interfering RNA
- JNK
- c-Jun N-terminal kinase
- GFP
- green fluorescent protein
- 2-DG
- 2-deoxyglucose.
REFERENCES
- 1.Aronson D. ( 2008) Adv. Cardiol. 45, 1– 16 [DOI] [PubMed] [Google Scholar]
- 2.Rask-Madsen C., King G. L. ( 2008) Arterioscler. Thromb. Vasc. Biol. 28, 608– 610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brownlee M. ( 2001) Nature 414, 813– 820 [DOI] [PubMed] [Google Scholar]
- 4.Ramana K. V., Bhatnagar A., Srivastava S. K. ( 2004) FEBS Lett. 570, 189– 194 [DOI] [PubMed] [Google Scholar]
- 5.Srivastava S. K., Ramana K. V., Bhatnagar A. ( 2005) Endocr. Rev. 26, 380– 392 [DOI] [PubMed] [Google Scholar]
- 6.Cumbie B. C., Hermayer K. L. ( 2007) Vasc. Health Risk Manag. 3, 823– 832 [PMC free article] [PubMed] [Google Scholar]
- 7.Pladzyk A., Reddy A. B., Yadav U. C., Tammali R., Ramana K. V., Srivastava S. K. ( 2006) Invest. Ophthalmol. Vis. Sci. 47, 5395– 5403 [DOI] [PubMed] [Google Scholar]
- 8.Kang E. S., Woo I. S., Kim H. J., Eun S. Y., Paek K. S., Kim H. J., Chang K. C., Lee J. H., Lee H. T., Kim J. H., Nishinaka T., Yabe-Nishimura C., Seo H. G. ( 2007) Free Radic. Biol. Med. 43, 535– 545 [DOI] [PubMed] [Google Scholar]
- 9.Ruef J., Liu S. Q., Bode C., Tocchi M., Srivastava S., Runge M. S., Bhatnagar A. ( 2000) Arterioscler. Thromb. Vasc. Biol. 20, 1745– 1752 [DOI] [PubMed] [Google Scholar]
- 10.Lorenzi M. ( 2007) Exp. Diabetes Res. 2007, 61038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gatenby R. A., Gillies R. J. ( 2007) Int. J. Biochem. Cell Biol. 39, 1358– 1366 [DOI] [PubMed] [Google Scholar]
- 12.LeRoith D., Yakar S. ( 2007) Nat. Clin. Pract. Endocrinol. Metab. 3, 302– 310 [DOI] [PubMed] [Google Scholar]
- 13.DeBerardinis R. J., Lum J. J., Hatzivassiliou G., Thompson C. B. ( 2008) Cell Metab. 7, 11– 20 [DOI] [PubMed] [Google Scholar]
- 14.Rodgers J. T., Lerin C., Haas W., Gygi S. P., Spiegelman B. M., Puigserver P. ( 2005) Nature 434, 113– 118 [DOI] [PubMed] [Google Scholar]
- 15.Namba K., Abe M., Saito S., Satake M., Ohmoto T., Watanabe T., Sato Y. ( 2000) Oncogene 19, 106– 114 [DOI] [PubMed] [Google Scholar]
- 16.Sun L., Vitolo M., Passaniti A. ( 2001) Cancer Res. 61, 4994– 5001 [PubMed] [Google Scholar]
- 17.Sun L., Vitolo M. I., Qiao M., Anglin I. E., Passaniti A. ( 2004) Oncogene 23, 4722– 4734 [DOI] [PubMed] [Google Scholar]
- 18.Qiao M., Shapiro P., Kumar R., Passaniti A. ( 2004) J. Biol. Chem. 279, 42709– 42718 [DOI] [PubMed] [Google Scholar]
- 19.Qiao M., Shapiro P., Fosbrink M., Rus H., Kumar R., Passaniti A. ( 2006) J. Biol. Chem. 281, 7118– 7128 [DOI] [PubMed] [Google Scholar]
- 20.Zelzer E., Glotzer D. J., Hartmann C., Thomas D., Fukai N., Soker S., Olsen B. R. ( 2001) Mech. Dev. 106, 97– 106 [DOI] [PubMed] [Google Scholar]
- 21.Lu H., Kraut D., Gerstenfeld L. C., Graves D. T. ( 2003) Endocrinology 144, 346– 352 [DOI] [PubMed] [Google Scholar]
- 22.Fowlkes J. L., Bunn R. C., Liu L., Wahl E. C., Coleman H. N., Cockrell G. E., Perrien D. S., Lumpkin C. K., Jr., Thrailkill K. M. ( 2008) Endocrinology 149, 1697– 1704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tsuji K., Komori T., Noda M. ( 2004) J. Bone Miner. Res. 19, 1481– 1489 [DOI] [PubMed] [Google Scholar]
- 24.Jüttner K. V., Perry M. J. ( 2007) Bone 41, 25– 32 [DOI] [PubMed] [Google Scholar]
- 25.Chen F., Mackerell A. D., Jr., Luo Y., Shapiro P. ( 2008) J. Cell Commun. Signal. 2, 81– 92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vitolo M. I., Anglin I. E., Mahoney W. M., Jr., Renoud K. J., Gartenhaus R. B., Bachman K. E., Passaniti A. ( 2007) Cancer Biol. Ther. 6, 856– 863 [DOI] [PubMed] [Google Scholar]
- 27.Ducy P., Zhang R., Geoffroy V., Ridall A. L., Karsenty G. ( 1997) Cell 89, 747– 754 [DOI] [PubMed] [Google Scholar]
- 28.Hamuro M., Polan J., Natarajan M., Mohan S. ( 2002) Atherosclerosis 162, 277– 287 [DOI] [PubMed] [Google Scholar]
- 29.Facchiano F., Lentini A., Fogliano V., Mancarella S., Rossi C., Facchiano A., Capogrossi M. C. ( 2002) Am. J. Pathol. 161, 531– 541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Teixeira A. S., Andrade S. P. ( 1999) Life Sci. 64, 655– 662 [DOI] [PubMed] [Google Scholar]
- 31.Pratap J., Lian J. B., Javed A., Barnes G. L., van Wijnen A. J., Stein J. L., Stein G. S. ( 2006) Cancer Metastasis Rev. 25, 589– 600 [DOI] [PubMed] [Google Scholar]
- 32.Oyama T., Miyasita Y., Watanabe H., Shirai K. ( 2006) Diabetes Res. Clin. Pract. 73, 227– 234 [DOI] [PubMed] [Google Scholar]
- 33.Obrosova I. G., Minchenko A. G., Vasupuram R., White L., Abatan O. I., Kumagai A. K., Frank R. N., Stevens M. J. ( 2003) Diabetes 52, 864– 871 [DOI] [PubMed] [Google Scholar]
- 34.Kuki S., Imanishi T., Kobayashi K., Matsuo Y., Obana M., Akasaka T. ( 2006) Circ. J. 70, 1076– 1081 [DOI] [PubMed] [Google Scholar]
- 35.Li J. H., Huang X. R., Zhu H. J., Johnson R., Lan H. Y. ( 2003) Kidney Int. 63, 2010– 2019 [DOI] [PubMed] [Google Scholar]
- 36.Vikramadithyan R. K., Hu Y., Noh H. L., Liang C. P., Hallam K., Tall A. R., Ramasamy R., Goldberg I. J. ( 2005) J. Clin. Invest. 115, 2434– 2443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Campbell M., Trimble E. R. ( 2005) Circ. Res. 96, 197– 206 [DOI] [PubMed] [Google Scholar]
- 38.Spalding A. C., Zeitlin B. D., Wilder-Romans K., Davis M. E., Nor J. E., Lawrence T. S., Ben-Josef E. ( 2008) Transl. Oncol. 1, 195– 201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wilkinson-Berka J. L., Wraight C., Werther G. ( 2006) Curr. Med. Chem. 13, 3307– 3317 [DOI] [PubMed] [Google Scholar]
- 40.Luo B., Soesanto Y., McClain D. A. ( 2008) Arterioscler. Thromb. Vasc. Biol. 28, 651– 657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Meugnier E., Rome S., Vidal H. ( 2007) Curr. Opin. Clin. Nutr. Metab. Care 10, 518– 522 [DOI] [PubMed] [Google Scholar]
- 42.Herman M. A., Kahn B. B. ( 2006) J. Clin. Invest. 116, 1767– 1775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lee N. K., Sowa H., Hinoi E., Ferron M., Ahn J. D., Confavreux C., Dacquin R., Mee P. J., McKee M. D., Jung D. Y., Zhang Z., Kim J. K., Mauvais-Jarvis F., Ducy P., Karsenty G. ( 2007) Cell 130, 456– 469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Abu-Abid S., Szold A., Klausner J. ( 2002) J. Med. 33, 73– 86 [PubMed] [Google Scholar]
- 45.Pelicano H., Martin D. S., Xu R. H., Huang P. ( 2006) Oncogene 25, 4633– 4646 [DOI] [PubMed] [Google Scholar]
- 46.Jain R. K. ( 2008) Sci. Am. 298, 56– 63 [DOI] [PubMed] [Google Scholar]