Abstract
Nerve growth factor (NGF) is produced as a precursor called pro-nerve growth factor (proNGF), which is secreted by many tissues and is the predominant form of NGF in the central nervous system. In Alzheimer disease brain, cholinergic neurons degenerate and can no longer transport NGF as efficiently, leading to an increase in untransported NGF in the target tissue. The protein that accumulates in the target tissue is proNGF, not the mature form. The role of this precursor is controversial, and both neurotrophic and apoptotic activities have been reported for recombinant proNGFs. Differences in the protein structures, protein expression systems, methods used for protein purification, and methods used for bioassay may affect the activity of these proteins. Here, we show that proNGF is neurotrophic regardless of mutations or tags, and no matter how it is purified or in which system it is expressed. However, although proNGF is neurotrophic under our assay conditions for primary sympathetic neurons and for pheochromocytoma (PC12) cells, it is apoptotic for unprimed PC12 cells when they are deprived of serum. The ratio of tropomyosin-related kinase A to p75 neurotrophin receptor is low in unprimed PC12 cells compared with primed PC12 cells and sympathetic neurons, altering the balance of proNGF-induced signaling to favor apoptosis. We conclude that the relative level of proNGF receptors determines whether this precursor exhibits neurotrophic or apoptotic activity.
Nerve growth factor (NGF)3 regulates neuronal survival, neurite outgrowth, and differentiation in the peripheral and central nervous systems (1). The mature form of NGF forms a non-covalent homodimer and binds with high affinity (kd ≈ 10−11 m) to tropomyosin-related kinase A (TrkA) and with low affinity (kd ≈ 10−9 m) to the common neurotrophin receptor p75NTR (p75 neurotrophin receptor) (2). NGF promotes cell survival and growth in cells expressing TrkA through activation of the phosphatidylinositol 3-kinase/AKT pathway and the Ras/mitogen-activated protein kinase (MAPK) pathway (3, 4). p75NTR plays diverse roles, ranging from cell survival to cell death depending on the cellular context in which it is expressed. Through activation of the NF-κB pathway, p75NTR can contribute to cell survival in sensory neurons (5), it is involved in axonal growth via regulation of Rho activity (6), and it can interact with Trks to enhance neurotrophin affinity (at low concentration of ligand) and specificity of binding to Trks (7–9). High levels of p75NTR expression can induce apoptosis when there are low levels of Trk or when Trk is absent (10, 11). Apoptosis occurs through increased ceramide production (12), activation of c-Jun N-terminal kinase (JNK1), and p53 (10, 13). p75NTR requires a co-receptor called sortilin to induce cell death (14).
NGF is produced as a precursor called pro-nerve growth factor (proNGF) (15). ProNGF is secreted by many tissues such as prostate cells, spermatids, hair follicles, oral mucosal keratinocytes, sympathetic neurons, cortical astrocytes, heart, and spleen (16–20). ProNGF is the predominant form of NGF in the central and peripheral nervous systems, whereas little or no mature NGF can be detected (21–24). In Alzheimer disease brain, retrograde transport from the cortex and hippocampus to basal forebrain cholinergic neurons is reduced as these neurons degenerate, with concomitant proNGF accumulation in the cortex and hippocampus (21, 23). This suggested that proNGF mediates biological activity besides its prodomain function of promoting protein folding and regulation of neurotrophin secretion (25–28). To study the role of proNGF protein in vitro, point mutations were inserted at the cleavage site used by furin, a proprotein convertase known to cleave proNGF (29), to minimize the conversion of proNGF to mature NGF. The resulting recombinant, cleavage-resistant proNGFs reportedly exhibit either apoptotic activity (30, 31) or neurotrophic activity (32, 33). These recombinant proteins differ in several ways (Table 1). Cleavage-resistant proNGFhis with apoptotic activity contains multiple amino acid substitutions at −2 and −1 (RR to AA), 118 and 119 (RR to AA), and carries a histidine tag at the carboxyl terminus. This proNGF was expressed in mammalian cells and was purified using nickel column chromatography. ProNGF123 with apoptotic activity is another cleavage-resistant proNGF containing several mutations at −73 and −72 (RR to AA), −43 and −42 (KKRR to KAAR), and −2 and −1 (KR to AA) expressed in a baculovirus/insect cell system. ProNGF123 was purified using cation exchange chromatography and immunoaffinity column chromatography. Cleavage-resistant proNGF(R−1G) with neurotrophic activity contains a single amino acid substitution at −1 (Arg to Gly) and carries no tags. This proNGF was expressed in insect cells using a baculovirus expression system. A fourth cleavage-resistant proNGF (proNGFE) with neurotrophic activity contains two amino acid substitutions at −1 and +1 (RS to AA) but no histidine tag and was expressed in bacteria. Each of these differences may affect the activity of the expressed proteins in such a way that one is apoptotic and the other is neurotrophic. To investigate the determinants of proNGF biological activity, these recombinant proNGFs were expressed in baculovirus, mammalian, or bacterial expression systems and were purified by nickel column, immunoaffinity chromatography, or renaturation from inclusion bodies before testing by several different bioassays. Relative levels of TrkA and p75NTR receptors of the cells used in these bioassays were determined by Western blotting.
Table 1.
Characteristics of proNGF constructs used in this study
| ProNGF(R−1G) | ProNGFhis | ProNGFE | ProNGF123 | WT-NGFhis | |
|---|---|---|---|---|---|
| Mutations | −1 (R to G) | −2 and −1 (RR to AA), 118 and 119 (RR to AA) | −1 and +1 (RS to AA) | −73 and −72 (RR to AA), −43 and −42 (KKRR to KAAR), −2 and −1 (KR to AA) | None: cleavable proNGF |
| Tag | No tag | Histidine tag | No tag | No tag | Histidine tag |
| Expression system | Insect cells | Insect cells, mammalian cells | Bacteria | Insect cells | Insect cells, mammalian cells |
| Purification | No purification | Nickel column | Refolded from inclusion bodies, FPLC | Cation exchange chromatography, immunoaffinity chromatography | Nickel column |
EXPERIMENTAL PROCEDURES
Expression of ProNGF in a Baculovirus/Insect Cell System
Cleavage-resistant mouse proNGF carrying a histidine tag (proNGFhis) in pcDNA3 was obtained from Dr. B. Hempstead (Weill Medical College of Cornell University, New York). ProNGFhis was subcloned into the baculovirus transfer vector, pVL1393 (BD Biosciences, Mississauga, Canada). The direction of insert was verified by double restriction enzyme digestion and the insert sequence was confirmed by sequencing both strands. Linearized baculovirus DNA and pVL-proNGFhis were cotransfected into Sf9 insect cells according to the manufacturer's protocol (BD Biosciences). Recombinant proNGFhis baculovirus was plaque-purified and a high titer viral stock was produced through virus amplification in Sf9 cells. ProNGFhis protein was produced in Sf9 cells infected by recombinant proNGFhis baculovirus at a multiplicity of infection of 0.5. Wild type NGF (WT-NGF) (cleavable proNGF with no mutation and no histidine tag), wild type NGFhis (WT-NGFhis) (cleavable proNGF carrying a histidine tag), cleavage-resistant proNGF(R−1G) (32), and wild type (empty) baculovirus (BD Biosciences) were expressed in Sf9 cells at a multiplicity of infection = 0.5 in parallel with proNGFhis expression. All protein expressions were carried out in Sf-900 serum-free medium (Invitrogen) in a 27 °C shaking incubator at 120 rpm, and supernatants were harvested by centrifugation at 300 × g for 5 min on day 3 after infection.
Prior to use, samples were dialyzed overnight at 4 °C against RPMI 1640 (Invitrogen). Concentrations were determined by enzyme-linked immunosorbent assay as described (32). Values were determined by comparison to a standard curve of purified 2.5S NGF prepared by us from mouse salivary glands (21, 34, 35). For all experiments, 2.5S NGF and/or cleavable proNGF (WT-NGF) served as a positive control, and conditioned medium from insect cells infected with wild type (empty) baculovirus (WT-baculovirus) was used as a negative control.
Expression of ProNGFhis in a Mammalian Cell System
One day prior to transfection, human embryonic kidney (HEK) 293 cells were plated at 90% confluence in antibiotic-free minimal essential medium/F11 (Invitrogen) with 2% fetal bovine serum (FBS). Cells were transiently transfected with pcDNA3 containing proNGFhis or WT-NGFhis using Lipofectamine™ 2000 (Invitrogen) at a ratio of 1:2.5. DNA was applied to the cells and incubated for 6 h before changing the medium. The medium was harvested after 24 h.
Supernatants harvested from the protein expression systems (baculovirus or mammalian) were dialyzed against phosphate buffer (50 mm NaPO4, 0.5 m NaCl, pH 8) at 4 °C overnight. Dialyzed material was bound to nickel beads (Invitrogen) with shaking for 1 h at room temperature. Beads were washed 4 times with 20 mm imidazole in phosphate buffer, followed by elution with 250 mm imidazole in phosphate buffer. 1.5-ml fractions were collected and fractions containing proNGFhis or WT-NGFhis were pooled. Western blotting using antibodies against both the histidine tag and NGF confirmed the correct size for purified proNGFhis (34 kDa). Samples were dialyzed against RPMI and quantified by NGF enzyme-linked immunosorbent assay as described above before use.
Expression and Purification of ProNGF in a Bacterial System
Cleavage-resistant human recombinant proNGF (proNGFE), carrying two amino acid substitutions and no tag, was expressed in Escherichia coli. Inclusion bodies were isolated from cell pellets, solubilized, and protein was refolded as described (27, 33). Refolded protein was purified by fast protein liquid chromatography and eluted in 50 mm sodium phosphate, pH 7, 1 mm EDTA, 2 m NaCl. The concentration of purified proNGFE was determined by absorbance at 280 nm using an extinction coefficient of 1.6 absorbance units/mg/ml. Molarity was calculated using 26 kDa for the molecular mass of NGF and 49.7 kDa for the molecular mass of proNGFE.
ProNGF123
Cleavage-resistant mouse proNGF containing several mutations was constructed as previously published (31) and quantified by absorbance at 280 nm. Molarity was calculated using 26 kDa for the molecular mass of NGF and 68 kDa for the molecular mass of proNGF123.
Neurite Outgrowth and Apoptosis Bioassay
ProNGF is prone to cleavage in the presence of serum, and therefore bioassays were carried out using serum-free medium. Mouse superior cervical ganglion (SCG) neurons from 1-day-old pups were prepared as previously described (32). Approximately 3000 SCG cells were cultured in collagen-coated 96-well plates (Falcon, BD Biosciences) in Dulbecco's modified Eagle's medium/F12 (Invitrogen) containing 10% FBS, 0.7% l-glutamine, 0.5% penicillin/streptomycin, 10−5 m uridine (Sigma), and 10−5 m 5-fluoro-2-deoxyuridine (Sigma) for 2–4 h at 37 °C and 5% CO2. Attached cells were rinsed with Dulbecco's modified Eagle's medium/F12 and then incubated with proNGFhis or 2.5S NGF diluted in serum-free medium (the same medium as above but with 1% N2 supplement (Invitrogen) instead of FBS and glutamine) for 36 h. For assays using proNGF123, cells were plated in the same medium with 250 ng/ml cytosine arabinoside (Sigma) instead of uridine and 5-fluoro-2-deoxyuridine.
For unprimed rat pheochromocytoma cells (PC12, ATCC), 3 × 104 cells/well were plated in Cell+ 96-well plates (Sarstedt, Montreal, Quebec) in RPMI 1640 containing 10% horse serum, 5% FBS, and 1% penicillin/streptomycin for 48 h as described by Greene (36). Cells were rinsed and then incubated with proNGFhis, WT- NGF, 2.5S NGF or baculovirus-conditioned medium in RPMI, 1% penicillin/streptomycin, 0.1% BSA, 0.5% N2 supplement (Invitrogen), 30 μm aprotinin (Calbiochem, CA), 100 nm MMP-II inhibitor (Calbiochem), and 30 μm furin inhibitor (decanoyl-RVKR-CMK, Calbiochem) for 48 h.
PC12 cells were primed in RPMI 1640 containing 10% horse serum, 5% FBS, and 50 ng/ml 2.5S NGF for 11 days as described (32). Cells were washed in medium free of serum and NGF and were plated and exposed to neurotrophins as for unprimed PC12s.
Cells were rinsed with phosphate-buffered saline, pH 7.4 (PBS), and fixed in 4% paraformaldehyde for 1 h at room temperature. Cells were incubated with permeabilization solution (0.1% Triton X-100 in 0.1% sodium citrate) for 2 min on ice and then were rinsed with PBS and blocked with 3% goat serum in 1% BSA/PBS for 30 min at room temperature. After washing with PBS, cells were incubated with a 1:500 dilution (in 1% BSA/PBS) of neuronal anti-βIII-tubulin (TUJ1, Covance, CA) overnight at room temperature, rinsed in PBS, incubated in a 1:100 dilution (in 1% BSA/PBS) of Texas Red-labeled anti-mouse IgG (Cedarlane, Burlington, ON, Canada) for 1 h at 37 °C, rinsed with PBS, and then incubated with 1 μg/ml bis-benzimide H33342 trihydrochloride (Hoechst, Sigma) in PBS for 5–15 min at room temperature. Photographs were taken using a high-resolution wide field fluorescent microscope (Leica DMI 3000B) at ×20 magnification attached to a Hamamatsu Orca ER-AG camera. The acquisition software used by this system was Volocity 4 (Improvision, Waltham, MA) and the filters were 377/50 and 628/40 for Hoechst and TUJ, respectively.
TUJ+ cells exhibiting outgrowth twice the length of their cell body diameter were counted as neurite-bearing cells using a Leica DMI L fluorescent microscope (X-Cite® 120 Fluorescence illumination system) at ×10 magnification. TUJ+ cells with condensed and/or fragmented nuclei (stained with Hoechst) were considered as dead cells. At least 100 cells were counted per well.
TrkA-MAPK Activation Assay
The assay was carried out as previously described (32). 1 × 106 PC12 cells/well were plated in Cell+ 6-well plates (Sarstedt) in RPMI 1640 containing 10% horse serum and 5% FBS (complete medium) for 24 h, then medium was switched to RPMI with no serum for 2–24 h. Serum-free medium was changed 2 h before stimulation. Cells were stimulated by serial dilutions of wild type (cleavable) proNGF (WT-NGF), proNGFhis (unpurified or purified), proNGF expressed in E. coli or mammalian HEK-293 cells, or mouse submandibular gland 2.5S NGF for 5 min at 37 °C and 5% CO2. Cells were lysed using 150 μl/well of lysis buffer containing 50 mm Tris-HCl, pH 7.4, 1% Nonidet P-40, 0.25% sodium deoxycholate, 150 mm NaCl, 1 mm EGTA, 1 mm phenylmethylsulfonyl fluoride, 1 mm Na3VO4, 1 mm sodium fluoride, and 1 μg/ml each of aprotinin, leupeptin, and pepstatin, and chilled on ice for 15 min. Lysates were centrifuged at 13,000 × g for 10 min at 4 °C. Supernatant was collected and total protein concentration was measured using the DC protein assay (Bio-Rad). 30 μg of total protein was loaded per lane on 10% SDS-PAGE gels for Western blotting.
In some experiments as indicated, serum-free medium was supplemented with 50 μm furin inhibitor for 1–2 h before stimulation to prevent intracellular cleavage of proNGF by furin. Cells were stimulated with either proNGF or WT-NGF in 50 μm furin inhibitor for either 5 min or 1 h and lysed as above.
Determination of ProNGF Cleavage after Each Bioassay
For all bioassays including neurite outgrowth, apoptosis, and TrkA-MAPK activation assays, the level of proNGF cleavage in conditioned medium (and cell lysates as indicated) was monitored after each experiment by Western blotting. There was little to no detectable cleavage in all bioassays reported here, which supports the role of unprocessed proNGF in each assay.
Western Blotting
Western blotting was performed as previously described (32) using affinity-purified rabbit anti-NGF IgG (MC-51), rabbit polyclonal NGF IgG (H-20, Santa Cruz Biotechnology, Santa Cruz, CA), rabbit anti-histidine tag antibody (Invitrogen), anti-proNGF (a generous gift from Dr. Xin-Fu Zhou, Flinders University, Adelaide, Australia), phosphorylated p140TrkA, phosphorylated p44/42 MAPK (anti-extracellular signal-regulated kinase (ERK) 1/2, Thr202/Tyr204), and total p44/42 MAPK (all from Cell Signaling Technology, Inc., Mississauga, Ontario, Canada), total TrkA and p75NTR (Chemicon, Mississauga, Canada), and neuronal βIII-tubulin (TUJ1, Covance). The secondary antibody was horseradish peroxidase-conjugated donkey anti-rabbit IgG (Amersham Biosciences) for anti-NGF, anti-histidine tag, pMAPK, pTrkA, total MAPK, total TrkA, and p75NTR primary antibodies, and horseradish peroxidase-conjugated donkey anti-sheep IgG (Sigma) for antibody against proNGF. A chemiluminescence system (ECL™, Amersham Biosciences) was used to detect immunoreactive protein. In some experiments, the secondary antibody was IRDye® 680-conjugated goat polyclonal anti-rabbit IgG (LI-COR Biosciences, Lincoln, NE). The membrane was scanned at the appropriate wavelength using an Odyssey™ infrared imaging system (LI-COR Biosciences). All antibodies were used according to the manufacturer's protocols.
Determination of Relative TrkA and p75NTR Levels
Relative levels of TrkA and p75NTR protein were determined in primary SCG neurons cultured in Dulbecco's modified Eagle's medium containing 10% FBS for 4 h, in unprimed PC12 cells cultured in RPMI, 10% horse serum, and 5% FBS, and in primed PC12 cells cultured in serum plus 50 ng/ml NGF for 11 days. Cells were harvested in 40 μl of lysis buffer (50 mm Tris, pH 8, 150 mm NaCl, 1% Nonidet P-40, 5 mm EDTA, 1 mm phenylmethylsulfonyl fluoride, 1 mm sodium fluoride, and 1 μg/ml each of aprotinin, leupeptin, and pepstatin) for primary SCG neurons, or in the same lysis buffer used in TrkA-MAPK activation assays for PC12 cells, and analyzed by Western blotting using the TrkA and p75NTR antibodies described above (Chemicon).
Statistical Analysis and Programs
The pixel values of immunoreactive bands in chemiluminescent Western blots were determined by densitometry of films using a Hewlett-Packard Scanjet scanner and Scion Image β4.01 software with local background subtracted. Integrated intensities of infrared bands were determined using Odyssey software version 1.2 (LI-COR) with median background subtraction. Samples were analyzed 3 times in independent experiments. Prism 3 (GraphPad Software, Inc., San Diego, CA) was applied to create dose-response curves for neurite outgrowth and TrkA-MAPK activation assays. Student's t test or one-way analysis of variance with post hoc Tukey's test was applied as indicated to determine differences among experimental groups (SPSS 16.0.1 software).
RESULTS
Baculovirus/Insect Cell Expression of ProNGFhis
Previously, cleavage-resistant proNGF with a single amino acid substitution and neurotrophic activity was expressed in a baculovirus/insect cell system and used without further purification (32), whereas proNGFhis with multiple substitutions, a histidine tag, and apoptotic activity was expressed in mammalian cells and purified over a nickel column prior to use (30). To determine whether these structural or procedural differences could account for the reported differences in activity, we expressed proNGFhis in a baculovirus system and tested its activity before and after purification over a nickel column. We also tested proNGF123, an apoptotic proNGF expressed in baculovirus (31) and purified by immunoaffinity chromatography.
Western blotting of cleavage-resistant proNGFhis expressed in an insect cell/baculovirus expression system using an anti-NGF antibody demonstrated intact material at the expected molecular mass for proNGFhis (34 kDa, Fig. 1E). Western blotting using an antibody against proNGF or the histidine tag confirmed the correct size band for proNGFhis and verified that the expressed protein was proNGF carrying a histidine tag. There was negligible cleavage of proNGFhis. No band was detected for either 2.5S NGF using proNGF antibody or WT-NGF (no histidine tag) using the histidine tag antibody, which confirmed the specificity of the antibodies for proNGFhis. No immunoreactivity was detectable for wild type baculovirus-infected insect cell-conditioned medium using any of the antibodies mentioned above; therefore there is no proNGF production in baculovirus/insect cells, and the antibodies do not cross-react with any protein produced by the baculovirus/insect cell system alone (data not shown). Western blotting using NGF and histidine tag antibodies showed that proNGFhis remained intact after nickel column chromatography, and proNGF123 was intact after immunoaffinity chromatography (data not shown).
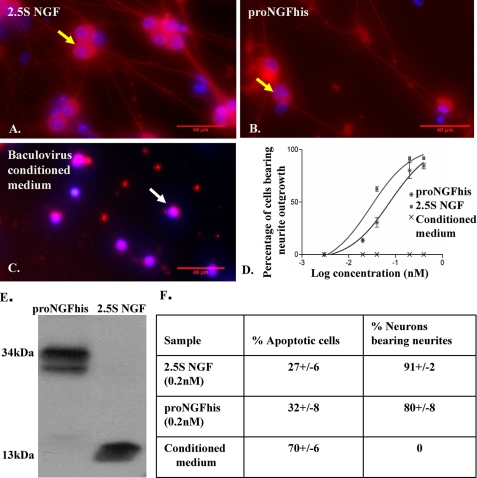
FIGURE 1.
Neurotrophic activity of proNGFhis in SCG neurons. Mouse SCG neurons were plated as described under “Experimental Procedures” and cultured in serum-free medium containing 2.5S NGF (0.4 nm) (A), proNGFhis (0.4 nm) (B), or baculovirus-infected insect cell-conditioned medium (C) for 36 h. Fixed cells were stained with TUJ-1 antibody (red) and Hoechst (blue). The scale bar is 40 μm. Yellow arrows show representative live cells and white arrows show representative dead cells. D, dose-response curve for neurite outgrowth assay. R2 = 0.96 for both proNGFhis and 2.5S NGF. Error bars represent mean ± S.E. E, Western blotting of conditioned medium using NGF antibody (MC51). There was less than 2% cleavage of proNGFhis after the neurite outgrowth assay. F, there was no significant difference between proNGFhis and 2.5S NGF in inducing apoptosis at 0.2 nm concentration (p > 0.05, Student's t test). Results were obtained from three independent experiments.
ProNGFhis and ProNGF123 Promote Neurite Outgrowth but Not Apoptosis in Primary Sympathetic Neurons
Both 2.5S NGF (Fig. 1A) and proNGFhis (Fig. 1B) were able to promote outgrowth from SCG neurons in serum-free medium, whereas there was no outgrowth detected for baculovirus-infected insect cell-conditioned medium (Fig. 1C). The EC50 for proNGFhis was 0.08 nm (log EC50 = −1.1 ± 0.15) compared with 0.03 nm (log EC50 = −1.5 ± 0.13) for 2.5S NGF (Fig. 1D), consistent with previous data published by Fahnestock et al. (32) for proNGF(R−1G) compared with 2.5S NGF. There was no cleavage of proNGFhis during the assay (Fig. 1E), supporting proNGF rather than mature NGF as the active species in promoting neurite outgrowth.
SCG neurons incubated with 0.2 nm proNGFhis for 24 h showed 32 ± 8% dead cells by Hoechst staining (confirmed by the TdT-mediated dUTP nick-end labeling assay (data not shown)), which was not significantly different from 0.2 nm 2.5S NGF showing 27 ± 6% dead cells (p > 0.05, Student's t test) (Fig. 1F). Therefore proNGFhis, like proNGF(R−1G) (32), exhibits neurotrophic activity, and structural differences between proNGFhis and proNGF(R−1G) are not responsible for their differing activities in vitro.
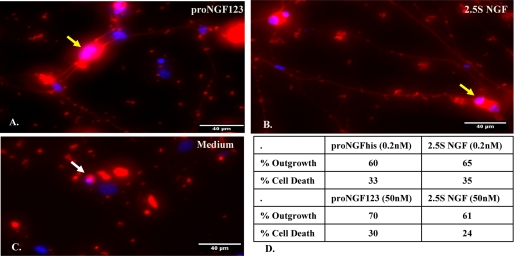
ProNGF123 also induced neurite outgrowth (Fig. 2A), which was comparable to mature NGF (Fig. 2B), whereas cells in the absence of neurotrophic support failed to survive (Fig. 2C). At the same concentrations, the effects on neurite outgrowth and neuronal survival were comparable for proNGFhis, proNGF123, and 2.5S NGF (Fig. 2D).
FIGURE 2.
Neurotrophic activity of proNGF123 in SCG neurons. SCG neurons were cultured as described under “Experimental Procedures” in serum-free medium containing proNGF123 (0.2 nm) (A), 2.5S NGF (0.2 nm) (B), or medium (C) for 48 h. Fixed cells were stained with TUJ-1 antibody (red) and Hoechst (blue). The scale bar is 40 μm. Yellow arrows show representative cells bearing neurites and white arrows show representative dead cells. D, at the same concentrations, the effects on neuronal survival and neurite outgrowth are comparable for proNGF123 and 2.5S NGF.
Both Purified and Unpurified ProNGF Promote TrkA-MAPK Phosphorylation
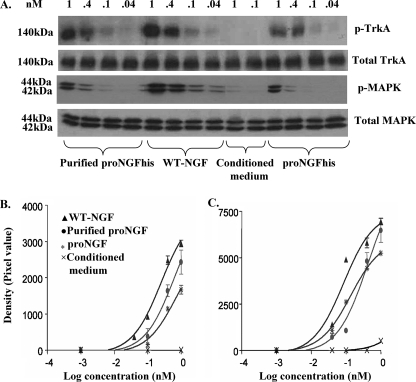
ProNGFhis expressed in insect cells (unpurified and purified) promoted both TrkA and MAPK phosphorylation of PC12 cells without changing the levels of total TrkA or total MAPK (Fig. 3A). The EC50 for TrkA activation was 0.2 nm (log EC50 = −0.6 ± 0.07), 0.6 nm (log EC50 = −0.2 ± 0.2), and 0.7 nm (log EC50 = −0.16 ± 0.2) for WT-NGF, purified proNGFhis, and unpurified proNGFhis, respectively (Fig. 3B). For MAPK activation, the EC50 was 0.08 nm (log EC50 = −1.1 ± 0.1), 0.4 nm (log EC50 = −0.4 ± 0.2), and 0.1 nm (log EC50 = −0.87 ± 0.05) for WT-NGF, purified proNGF, and unpurified proNGF, respectively (Fig. 3C). As previously reported (32), proNGF was less active than mature NGF for both TrkA and MAPK activation.
FIGURE 3.
Both unpurified and purified proNGF activate TrkA-MAPK phosphorylation. PC12 cells were treated as described under “Experimental Procedures” with different dilutions of proNGFhis (unpurified and purified), cleavable proNGF (WT-NGF), or conditioned medium from insect cells infected with wild type (empty) baculovirus. Concentrations of all samples were determined using enzyme-linked immunosorbent assay and values were determined by comparison to a standard curve of purified 2.5S NGF prepared by us from mouse salivary glands. There was no immunoreactive NGF in conditioned medium, and so a range of volumes of conditioned medium between the highest and lowest volumes used for WT-NGF and proNGFhis were used as negative controls. A, representative Western blot showing unpurified and purified proNGFhis induces TrkA-MAPK phosphorylation, whereas total TrkA and MAPK remain unchanged. B, dose-response curve for TrkA activation: R2 was 0.98, 0.94, and 0.96 for WT-NGF, purified proNGFhis, and unpurified proNGFhis, respectively. C, dose-response curve for MAPK activation: R2 was 0.96, 0.97, and 0.99 for WT-NGF, purified proNGF, and unpurified proNGF, respectively. Results shown are from 24 h of serum deprivation; the same results were observed after 2 h of serum removal. Results were obtained from 3 independent experiments. Error bars represent mean ± S.E.
Intracellular Cleavage of ProNGF Is Not Required to Activate MAPK Phosphorylation
No extracellular cleavage was detected for proNGFhis after the MAPK assay (data not shown). Blocking proNGF using an antibody specific for proNGF reduced proNGF-induced MAPK phosphorylation (Fig. 4), confirming the role of unprocessed proNGF in the assay. The proNGF antibody did not block MAPK activation induced by mature NGF (WT-NGF expressed in insect cells, which was all processed to the mature form).
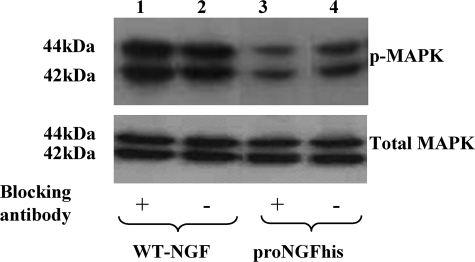
FIGURE 4.
A blocking antibody specific for proNGF prevents MAPK activation. There was a 36% reduction in MAPK phosphorylation when cleavage-resistant proNGF was blocked with an antibody specific for proNGF (lane 3) compared with proNGF without antibody (lane 4). Cleavable material (WT-NGF) expressed in insect cells was completely converted to mature NGF according to our Western blots. Therefore, as expected, we saw no change in the activity of this protein before and after treating with proNGF blocking antibody. There was no difference in the activity of WT-NGF in the presence (lane 1) or absence (lane 2) of proNGF antibody. There was no change in total MAPK with any treatment.
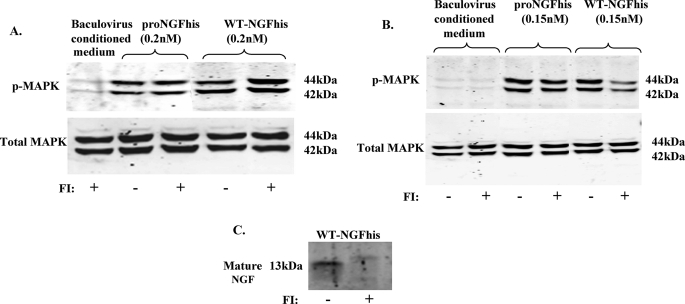
It has been reported that proNGF requires intracellular proteolysis to activate TrkA (37). Therefore, we carried out a MAPK activation assay in the presence of the cell-permeable furin inhibitor, Dec-RVKR-CMK. There was no reduction in the ability of either cleavage-resistant proNGFhis or cleavable WT-NGFhis to activate the MAPK signaling pathway in the presence of a furin inhibitor (Fig. 5A). In fact, prevention of cleavage by the furin inhibitor caused a slight increase in WT-NGF-induced MAPK phosphorylation. Therefore, cleavage of proNGF is not required for MAPK activation.
FIGURE 5.
Intracellular cleavage of proNGF is not required for MAPK phosphorylation. PC12 cells were incubated with 50 μm furin inhibitor (FI) 1 h before treating them with either cleavage-resistant proNGF or WT-NGF as described under “Experimental Procedures.” Controls were treated similarly but without furin inhibitor. Cells were treated with proNGFhis or WT-NGFhis for 5 min (A) or 1 h (B) before harvesting and Western blotting for p-MAPK and MAPK. C, Western blotting for NGF showed less processing of cleavable WT-NGF to mature NGF in cells treated with FI than in cells not treated with FI.
To demonstrate the effectiveness of the furin inhibitor, we exposed cells to WT-NGFhis (cleavable proNGFhis) for 1 h as described by Boutilier et al. (37). This is a longer time period than usual for this assay and is sufficient to allow proNGF uptake and cleavage. In this case, the furin inhibitor reduced cleavage of WT-NGFhis in cell lysates (Fig. 5C) and decreased cleavable WT-NGFhis-induced MAPK activation while having no effect on cleavage-resistant proNGFhis-induced activation (Fig. 5B). Even after 1 h incubation, cleavage-resistant proNGFhis activation of MAPK was not reduced by the furin inhibitor, further confirming that intracellular cleavage of proNGF is not required for MAPK activation.
ProNGFs from Different Expression Systems Are Neurotrophic
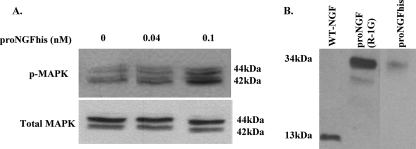
Western blotting shows dose-dependent activation of MAPK by proNGFhis expressed in mammalian cells, with no change in the level of total MAPK (Fig. 6A). This activation by proNGFhis is ∼2 times less than WT-NGFhis expressed in mammalian cells, as determined by densitometry (data not shown). No cleavage was detected for proNGFhis after the assay (Fig. 6B).
FIGURE 6.
ProNGFhis expressed in mammalian cells induces MAPK phosphorylation. PC12 cells were treated as described under “Experimental Procedures” with proNGFhis or WT-NGFhis expressed in HEK-293 cells. A, Western blotting for phosphorylated MAPK and total MAPK. B, proNGFhis remains unprocessed after the MAPK assay.
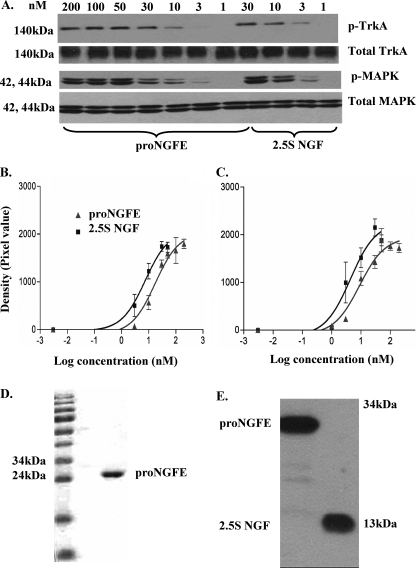
ProNGFE, human recombinant proNGF expressed in E. coli, showed the same relative TrkA-MAPK activation activity compared with mature NGF (Fig. 7A) as for proNGF(R−1G) and proNGFhis expressed in either insect or mammalian cells. For TrkA activation, the EC50 was 7.4 nm (log EC50 = 0.87 ± 0.16) for 2.5S NGF and 17.3 nm (log EC50 = 1.2 ± 0.13) for proNGFE (Fig. 7B). The EC50 for MAPK activation was 4.3 nm (log EC50 = 0.6 ± 0.2) and 8.9 nm (log EC50 = 0.95 ± 0.13) for 2.5S NGF and proNGFE, respectively (Fig. 7C). Coomassie Blue staining for proNGFE before the assay confirmed its purity (Fig. 7D), and a Western blot on the material after the TrkA-MAPK assay showed no cleavage during the assay (Fig. 7E).
FIGURE 7.
ProNGF expressed in E. coli activates the TrkA-MAPK pathway. PC12 cells were treated as described under “Experimental Procedures” with proNGFE expressed in bacteria or 2.5S NGF purified from mouse submandibular glands. A, representative Western blot showing proNGFE induces TrkA and MAPK phosphorylation, whereas total TrkA and MAPK remain unchanged. B, TrkA phosphorylation: R2 = 0.8 and 0.9 for proNGFE and 2.5S NGF, respectively. C, MAPK activation: R2 = 0.9 for both proNGFE and 2.5S NGF. Results were obtained from three independent experiments. Error bars represent mean ± S.E. D, Coomassie Blue staining for proNGFE before the assay. E, proNGFE was intact after the TrkA-MAPK activation assay.
ProNGF Is Neurotrophic after NGF Withdrawal from Primed PC12 Cells
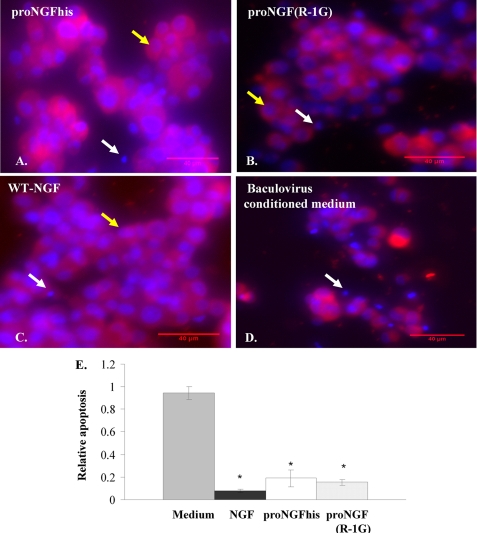
It has been shown that PC12 cells primed with NGF undergo apoptosis upon NGF withdrawal (38). Here we show that both proNGFhis and proNGF(R−1G) (Fig. 8, A and B) rescued primed PC12 cells after NGF withdrawal with activity that was comparable to WT-NGF (Fig. 8C). Significant apoptosis was observed in cells treated with baculovirus-infected insect cell-conditioned medium alone (Fig. 8, D and E).
FIGURE 8.
ProNGF rescues primed PC12 cells after NGF withdrawal. PC12 cells were primed with 50 ng/ml of 2.5S NGF for 11 days. After removing NGF and serum, cells were treated with 0.4 nm proNGFhis (A), proNGF(R−1G) (B), WT-NGF (C), or baculovirus-infected insect cell conditioned medium (D). After 48 h, cells were fixed and stained with TUJ-1 antibody and subjected to Hoechst staining. In contrast to medium alone, proNGFhis, proNGF(R−1G), and WT-NGF rescued cells from death after NGF withdrawal. Yellow arrows show representative live cells and white arrows show representative dead cells. Both NGF and proNGF reduced apoptosis compared with medium alone after NGF withdrawal (E). *, p < 0.05 compared to medium, one way analysis of variance followed by post hoc Tukey's test. Error bars represent S.E. for n = 3 per group except proNGF(R−1G) with n = 2.
ProNGF Is Apoptotic for Unprimed PC12 Cells after Serum Deprivation
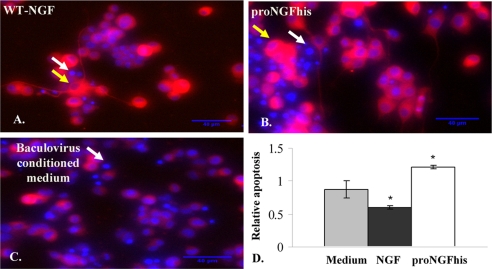
Unprimed PC12 cells are dependent on serum for survival and undergo apoptosis when serum is removed. It has been shown that NGF rescues PC12 cells after serum removal (36). Here we found that proNGFhis induced more apoptosis (1.7-fold) in PC12 cells deprived of serum than medium alone (Fig. 9, A–D), whereas NGF rescued the cells from death.
FIGURE 9.
ProNGF induces cell death in unprimed PC12 cells after serum removal. Unprimed PC12 cells cultured as described under “Experimental Procedures” were treated with 0.4 nm of either WT-NGF (A) or proNGFhis (B) in serum-free medium and stained with TUJ-1 antibody and Hoechst after 48 h treatment. There were more dead cells in cultures treated with proNGFhis (B) compared with WT-NGF (A) and medium alone (C). ProNGFhis induced more apoptosis compared with NGF and medium alone (D). Yellow arrows show representative live cells, and white arrows show representative dead cells. *, p < 0.05, one way analysis of variance followed by post hoc Tukey's test. Error bars represent S.E. for n = 3 per group.
The Ratio of TrkA to p75NTR Determines the Biological Activity of ProNGF
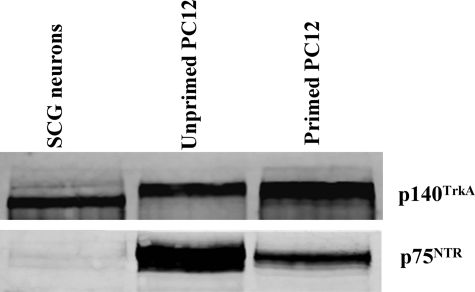
The ratio of TrkA to p75NTR plays an important role in activating different signaling pathways triggered by NGF (39–41). We hypothesized that the TrkA/p75NTR ratio may have a similar role in activating different pathways (neurotrophic versus apoptotic) triggered by proNGF. Interestingly, we observed that the ratio of TrkA to p75NTR was much higher in primed PC12 cells (TrkA/p75NTR = 2.4) compared with unprimed PC12 cells (TrkA/p75NTR = 0.6). In primary SCG neurons there was ample TrkA, but the level of p75NTR was below the detection limit of the assay (Fig. 10). Thus, proNGF is neurotrophic in SCG neurons and in primed PC12 cells (high level of TrkA compared with p75NTR), whereas it is apoptotic in the serum removal assay using unprimed PC12 cells (low level of TrkA compared with p75NTR).
FIGURE 10.
The ratio of TrkA to p75NTR differs in unprimed PC12 cells versus SCG and primed PC12 cells. PC12 cells, primed or unprimed, and dissociated SCG neurons were cultured as described under “Experimental Procedures,” and lysates were subjected to Western blotting to determine relative levels of TrkA and p75NTR. The TrkA/p75NTR ratio was 0.6 in unprimed PC12 cells compared with 2.4 in primed PC12 cells. There was ample TrkA in SCG neurons, whereas the level of p75NTR was below the detection limit of the assay.
DISCUSSION
ProNGF levels, not NGF, increase in Alzheimer disease (21) and correlate with cognitive deficits (42). However, the mechanism of increased proNGF and its role in Alzheimer neurodegeneration is unclear. An increase in proNGF levels in Alzheimer disease may cause neuronal degeneration if proNGF is apoptotic, or, if proNGF is neurotrophic, neurons may degenerate due to the lack of neurotrophic support when TrkA decreases and retrograde transport of proNGF is disrupted. Here, we have shown that proNGF is neurotrophic under most conditions for SCG neurons and for primed PC12 cells. However, conditions leading to a lower ratio of TrkA to p75NTR, such as in unprimed PC12 cells following serum withdrawal, lead to apoptotic activity of proNGF.
To determine the biological activity of proNGF and therefore its role in cell survival and death, several labs have produced recombinant proNGFs carrying mutations that hinder cleavage to the mature form. ProNGFs produced by the Fahnestock and Dawbarn groups (32, 33, 43) exhibit neurotrophic activity, whereas proNGFs produced by the Hempstead and Neet groups (14, 30, 31) are apoptotic. Differences in the number of amino acid substitutions, addition of a histidine tag, purification methods, expression systems, and different methods of bioassay all were postulated to elicit the opposite biological activities (neurotrophic versus apoptotic) (20).
Structural Modifications, Purification, and Expression Systems Do Not Affect ProNGF Biological Activity
Amino acid substitutions may disturb the ability of a molecule to bind to its receptor (44), whereas the presence of polyhistidine COOH-terminal extensions may modify the active site of a protein (45), disturb protein structure (46, 47), or alter biochemical properties such as dimerization/oligomerization (48). Here we show that proNGF carrying four amino acid substitutions and a COOH-terminal histidine tag (30), carrying two amino acid substitutions and no tag (33), or carrying six amino acid substitutions and no tag (31) promoted neurite outgrowth or elicited TrkA and MAPK phosphorylation with activity similar to proNGF(R−1G) with only one amino acid substitution and no tag (32). These results rule out the idea that structural differences (amino acid substitutions or histidine tags) influence the biological activity of recombinant proNGF.
Nickel column chromatography can cause oxidation and promote proteolysis, change protein properties, or cause protein aggregation (46, 47, 49). Here we show that purification over a nickel column does not alter the neurotrophic activity of proNGF.
Previously, proNGF with apoptotic activity was expressed in mammalian HEK-293 cells (30). We show here that cleavage-resistant proNGF expressed in E. coli, HEK 293, or insect cells elicited similar TrkA-MAPK phosphorylation activity. We demonstrate that proNGF can exhibit neurotrophic activity no matter in which expression system it is produced. Because some of these proNGFs are purified after expression, it is unlikely that other neurotrophins or chaperones from the expression systems could be responsible for neurotrophic activity.
Intracellular Cleavage Is Not Required for ProNGF Neurotrophic Activity
Here we have shown that cleavage of proNGF is not required for its neurotrophic activity. The ability of cleavage-resistant proNGF to activate MAPK phosphorylation remains unchanged when a furin inhibitor is used to prevent cleavage. Furthermore, a blocking antibody specific for proNGF reduced the MAPK phosphorylation activity of proNGF, whereas there was no change in the activity of NGF for activation of MAPK, confirming the role of unprocessed proNGF in the assay. Thus, the activation of MAPK that occurs within 5–10 min (32, 43) is attributable to proNGF activity, whereas proNGF processed to mature NGF is likely responsible for the activity detectable with longer incubations (1 h (37)). How NGF triggers phosphorylation of MAPK from inside the cell is unclear, although it is possible that during long incubations, proNGF is internalized, processed, and then secreted prior to TrkA-MAPK activation, as described by Althaus and Klöppner (43).
Apoptotic Activity of ProNGF Depends on the TrkA/p75NTR Ratio
ProNGF exhibits neurotrophic activity for primary SCG neurons and for primed PC12 cells (32). However, in unprimed PC12 cells deprived of serum (31, 36) we found that proNGF promoted apoptosis. Interestingly, there was a lower ratio of TrkA to p75NTR in unprimed PC12 cells compared with primed PC12 cells or naïve SCG neurons. This suggests that previous demonstrations of proNGF with apoptotic activity were carried out in cells grown under conditions favoring low TrkA and/or high p75NTR levels. In favor of this hypothesis, apoptotic activity of proNGF has been shown in unprimed PC12 cells or PC12 cells with low levels of TrkA (PC12nnr cells) (31), corticospinal neurons and oligodendrocytes expressing little or no TrkA (50, 51), and in corticospinal and hippocampal neurons expressing increased levels of p75NTR following injury (51, 52).
It has been shown that the ratio of TrkA to p75NTR is one of the factors that can determine the signaling pathway (death or survival) induced by NGF (39–41). Furthermore, because proNGF has greater affinity than NGF for p75NTR (14, 30), it is not surprising that proNGF induces greater apoptosis than NGF when p75NTR levels are high compared with TrkA. Thus, we conclude that the ratio of TrkA to p75NTR plays a role in determining the signaling pathway induced by proNGF, and therefore its biological activity. In vivo, increased proNGF promotes apoptotic activity in cases of injury where p75NTR is present or up-regulated (50–52), whereas overexpressed proNGF is neurotrophic in the uninjured animal with normal complement of TrkA to p75NTR (53). We have shown here that the ratio of TrkA to p75NTR is crucial for proNGF activity. We therefore propose that in Alzheimer disease, neurons initially degenerate due to lack of proNGF neurotrophic support, and that they later undergo proNGF-induced apoptosis as TrkA is lost.
In summary, we have shown that the biological activity of proNGF is not altered by amino acid substitutions, carboxyl-terminal tag, the expression system, or the purification system. However, different bioassay conditions may modulate the ratio of TrkA to p75NTR, resulting in neurotrophic or apoptotic activity of proNGF. Thus, assay conditions, injury, or disease may reduce levels of TrkA compared with p75NTR receptors, altering the balance of proNGF-induced signaling to favor apoptosis. We conclude that relative levels of TrkA to p75NTR receptors determine whether proNGF exhibits neurotrophic or apoptotic activity.
This work was supported in part by Canadian Institutes of Health Research Grant MOP-64382 (to M. F.), a scholarship from the Ministry of Science of Iran (to R. M.), a scholarship from the Natural Sciences and Engineering Research Council of Canada (to M. S. I.), United States Public Health Services Grant NS24380 (to K. E. N.) from the National Institutes of Health, and the Bristol Research into Alzheimer's and Care of the Elderly.
- NGF
- nerve growth factor
- proNGF
- pro-nerve growth factor
- PC12
- pheochromocytoma cells
- TrkA
- tropomyosin related kinase A
- MAPK
- mitogen-activated protein kinase
- WT
- wild type
- HEK 293 cells
- human embryonic kidney 293 cells
- FBS
- fetal bovine serum
- SCG
- superior cervical ganglion
- PBS
- phosphate-buffered saline
- BSA
- bovine serum albumin.
REFERENCES
- 1.Huang E. J., Reichardt L. F. ( 2001) Annu. Rev. Neurosci. 24, 677– 736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bibel M., Barde Y. A. ( 2000) Gene Dev. 14, 2919– 2937 [DOI] [PubMed] [Google Scholar]
- 3.Grewal S. S., York R. D., Stork P. J. ( 1999) Curr. Opin. Neurobiol. 9, 544– 553 [DOI] [PubMed] [Google Scholar]
- 4.Kaplan D. R., Miller F. D. ( 2000) Curr. Opin. Neurobiol. 10, 381– 391 [DOI] [PubMed] [Google Scholar]
- 5.Hamanoue M., Middleton G., Wyatt S., Jaffray E., Hay R. T., Davies A. M. ( 1999) Mol. Cell. Neurosci. 14, 28– 40 [DOI] [PubMed] [Google Scholar]
- 6.Yamashita T., Tucker K. L., Barde Y. A. ( 1999) Neuron 24, 585– 593 [DOI] [PubMed] [Google Scholar]
- 7.Dechant G. ( 2001) Cell Tissue Res. 305, 229– 238 [DOI] [PubMed] [Google Scholar]
- 8.Roux P. P., Barker P. A. ( 2002) Prog. Neurobiol. 67, 203– 233 [DOI] [PubMed] [Google Scholar]
- 9.Huang E. J., Reichardt L. F. ( 2003) Annu. Rev. Biochem. 72, 609– 642 [DOI] [PubMed] [Google Scholar]
- 10.Aloyz R. S., Bamji S. X., Pozniak C. D., Toma J. G., Atwal J., Kaplan D. R., Miller F. D. ( 1998) J. Cell Biol. 143, 1691– 1703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yoon S. O., Casaccia-Bonnefil P., Carter B., Chao M. V. ( 1998) J. Neurosci. 18, 3273– 3281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brann A. B., Tcherpakov M., Williams I. M., Futerman A. H., Fainzilber M. ( 2002) J. Biol. Chem. 277, 9812– 9818 [DOI] [PubMed] [Google Scholar]
- 13.Friedman W. J. ( 2000) J. Neurosci. 20, 6340– 6346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nykjaer A., Lee R., Teng K. K., Jansen P., Madsen P., Nielsen M. S., Jacobsen C., Kliemannel M., Schwarz E., Willnow T. E., Hempstead B. L., Petersen C. M. ( 2004) Nature 427, 843– 848 [DOI] [PubMed] [Google Scholar]
- 15.Darling T. L., Petrides P. E., Beguin P., Frey P., Shooter E. M., Selby M., Rutter W. J. ( 1983) Cold Spring Harbor Symp. Quant. Biol. 48, 427– 434 [DOI] [PubMed] [Google Scholar]
- 16.Chen Y., Dicou E., Djakiew D. ( 1997) Mol. Cell. Endocrinol. 127, 129– 136 [DOI] [PubMed] [Google Scholar]
- 17.Delsite R., Djakiew D. ( 1999) Prostate 41, 39– 48 [DOI] [PubMed] [Google Scholar]
- 18.Yardley G., Relf B., Lakshmanan J., Reinshagen M., Moore G. P. ( 2000) Exp. Dermatol. 9, 283– 289 [DOI] [PubMed] [Google Scholar]
- 19.Hayashi K., Storesund T., Schreurs O., Khuu C., Husvik C., Karatsaidis A., Helgeland K., Martin-Zanca D., Schenck K. ( 2007) Eur. J. Oral Sci. 115, 344– 354 [DOI] [PubMed] [Google Scholar]
- 20.Fahnestock M., Yu G., Coughlin M. D. ( 2004) Prog. Brain Res. 146, 101– 110 [DOI] [PubMed] [Google Scholar]
- 21.Fahnestock M., Michalski B., Xu B., Coughlin M. D. ( 2001) Mol. Cell. Neurosci. 18, 210– 220 [DOI] [PubMed] [Google Scholar]
- 22.Bierl M. A., Jones E. E., Crutcher K. A., Isaacson L. G. ( 2005) Neurosci. Lett. 380, 133– 137 [DOI] [PubMed] [Google Scholar]
- 23.Pedraza C. E., Podlesniy P., Vidal N., Arévalo J. C., Lee R., Hempstead B., Ferrer I., Iglesias M., Espinet C. ( 2005) Am. J. Pathol. 166, 533– 543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bierl M. A., Isaacson L. G. ( 2007) Neurobiol. Aging 28, 122– 134 [DOI] [PubMed] [Google Scholar]
- 25.Edwards R. H., Selby M. J., Garcia P. D., Rutter W. J. ( 1988) J. Biol. Chem. 263, 6810– 6815 [PubMed] [Google Scholar]
- 26.Suter U., Heymach J. V., Jr., Shooter E. M. ( 1991) EMBO J. 10, 2395– 2400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rattenholl A., Lilie H., Grossmann A., Stern A., Schwarz E., Rudolph R. ( 2001) Eur. J. Biochem. 268, 3296– 3303 [DOI] [PubMed] [Google Scholar]
- 28.Farhadi H. F., Mowla S. J., Petrecca K., Morris S. J., Seidah N. G., Murphy R. A. ( 2000) J. Neurosci. 20, 4059– 4068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Seidah N. G., Benjannet S., Pareek S., Savaria D., Hamelin J., Goulet B., Laliberte J., Lazure C., Chrétien M., Murphy R. A. ( 1996) Biochem. J. 314, 951– 960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lee R., Kermani P., Teng K. K., Hempstead B. L. ( 2001) Science 294, 1945– 1948 [DOI] [PubMed] [Google Scholar]
- 31.Pagadala P. C., Dvorak L. A., Neet K. E. ( 2006) Proc. Natl. Acad. Sci. U.S.A. 103, 17939– 17943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fahnestock M., Yu G., Michalski B., Mathew S., Colquhoun A., Ross G. M., Coughlin M. D. ( 2004) J. Neurochem. 89, 581– 592 [DOI] [PubMed] [Google Scholar]
- 33.Clewes O., Fahey M. S., Tyler S. J., Watson J. J., Seok H., Catania C., Cho K., Dawbarn D., Allen S. J. ( 2008) J. Neurochem. 107, 1124– 1135 [DOI] [PubMed] [Google Scholar]
- 34.Mobley W. C., Schenker A., Shooter E. M. ( 1976) Biochemistry 15, 5543– 5552 [DOI] [PubMed] [Google Scholar]
- 35.Petrides P. E., Shooter E. M. ( 1986) J. Neurochem. 46, 721– 725 [DOI] [PubMed] [Google Scholar]
- 36.Greene L. A. ( 1978) J. Cell Biol. 78, 747– 755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Boutilier J., Ceni C., Pagdala P. C., Forgie A., Neet K. E., Barker P. A. ( 2008) J. Biol. Chem. 283, 12709– 12716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Xia Z., Dickens M., Raingeaud J., Davis R. J., Greenberg M. E. ( 1995) Science 270, 1326– 1331 [DOI] [PubMed] [Google Scholar]
- 39.Davies A. M., Lee K. F., Jaenisch R. ( 1993) Neuron 11, 565– 574 [DOI] [PubMed] [Google Scholar]
- 40.Mahadeo D., Kaplan L., Chao M. V., Hempstead B. L. ( 1994) J. Biol. Chem. 269, 6884– 6891 [PubMed] [Google Scholar]
- 41.Barrett G. L., Bartlett P. F. ( 1994) Proc. Natl. Acad. Sci. U.S.A. 91, 6501– 6505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Peng S., Wuu J., Mufson E. J., Fahnestock M. ( 2004) J. Neuropathol. Exp. Neurol. 63, 641– 649 [DOI] [PubMed] [Google Scholar]
- 43.Althaus H. H., Klöppner S. ( 2006) J. Neurochem. 98, 506– 517 [DOI] [PubMed] [Google Scholar]
- 44.Ibáñez C. F., Ebendal T., Barbany G., Murray-Rust J., Blundell T. L., Persson H. ( 1992) Cell 69, 329– 341 [DOI] [PubMed] [Google Scholar]
- 45.Ledent P., Duez C., Vanhove M., Lejeune A., Fonzé E., Charlier P., Rhazi-Filali F., Thamm I., Guillaume G., Samyn B., Devreese B., Van Beeumen J., Lamotte-Brasseur J., Frère J. M. ( 1997) FEBS Lett. 413, 194– 196 [DOI] [PubMed] [Google Scholar]
- 46.Ramage P., Hemmig R., Mathis B., Cowan-Jacob S. W., Rondeau J. M., Kallen J., Blommers M. J., Zurini M., Rüdisser S. ( 2002) Life Sci. News 11, 1– 4 [Google Scholar]
- 47.Hunt I. ( 2005) Protein Expr. Purif. 40, 1– 22 [DOI] [PubMed] [Google Scholar]
- 48.Zhukovsky E. A., Lee J. O., Villegas M., Chan C., Chu S., Mroske C. ( 2004) Nature 427, 413– 414 [DOI] [PubMed] [Google Scholar]
- 49.Al-Samarrai T. H., Kirk C. A., Jones W. T., Harvey D., Sun X. ( 2007) Protein Expr. Purif. 53, 289– 292 [DOI] [PubMed] [Google Scholar]
- 50.Beattie M. S., Harrington A. W., Lee R., Kim J. Y., Boyce S. L., Longo F. M., Bresnahan J. C., Hempstead B. L., Yoon S. O. ( 2002) Neuron 36, 375– 386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Harrington A. W., Leiner B., Blechschmitt C., Arevalo J. C., Lee R., Mörl K., Meyer M., Hempstead B. L., Yoon S. O., Giehl K. M. ( 2004) Proc. Natl. Acad. Sci. U.S.A. 101, 6226– 6230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Volosin M., Trotter C., Cragnolini A., Kenchappa R. S., Light M., Hempstead B. L., Carter B. D., Friedman W. J. ( 2008) J. Neurosci. 28, 9870– 9879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Buttigieg H., Kawaja M. D., Fahnestock M. ( 2007) Exp. Neurol. 204, 832– 835 [DOI] [PubMed] [Google Scholar]