Abstract
KLF5 plays important roles in a variety of cellular processes including proliferation and differentiation. Recently KLF5 was shown to reverse its function in proliferative and p15 regulation upon transforming growth factor-β (TGFβ)-mediated acetylation. To understand how KLF5 acetylation functions in TGFβ-induced p15 transcription, we characterized the interactions of KLF5 with other transcription factors and promoter DNA elements in the context of TGFβ. KLF5 interacted with Smad2–4 and Miz-1 in a TGFβ-independent manner, but interacted with Myc only when TGFβ was activated, and at least some of the interactions had an additive effect on TGFβ-induced p15 transcription. Oligo pulldown assays showed that binding of Myc to the Inr element was KLF5-dependent, and TGFβ could enhance the binding when more KLF5 was available. Furthermore, TGFβ induced an interaction between KLF5 and the p300 acetylase, and acetylation of KLF5 was necessary for Smad4 to associate with p300. Failure in KLF5 acetylation not only prevented p300-assembled Smad4-KLF5 complex formation on p15 promoter but also affected the binding of Smad4 and FOXO3 on the p15 promoter in vivo. These findings suggest that without TGFβ, some KLF5 associates with Smads in the nucleus and other KLF5 associates with Miz-1 on the p15 promoter to repress its transcription. Activation of TGFβ recruits p300 to the KLF5-Smad complex to acetylate KLF5, and the complex with acetylated KLF5 binds to the Smad binding element and alters the binding of other factors to p15 promoter to induce its transcription.
Transforming growth factor-β (TGFβ)2 is the most potent and widespread growth-inhibitory cytokine known in mammals (1, 2). TGFβ signaling involves receptor binding, Smad modification and nuclear translocation, and the formation of Smad transcriptional complexes on gene promoters, regulating the expression of a large number of genes in different biological processes (3, 4). The same TGFβ signaling has different regulatory roles in different contexts (1, 2, 5, 6), most likely through different transcriptional cofactors preexisting in different types of cells including proliferating epithelial cells that differentiate upon TGFβ activation.
The cyclin-dependent kinase inhibitor p15 (also named CDKN2B and p15INK4b) is a well documented tumor suppressor in many types of cancers (7, 8). The cytostatic effect of p15 is based on its inhibition of cyclin-dependent kinases CDK4 and CDK6, which arrests the cell cycle in the G1 phase. p15 is an important effector of TGFβ and is dramatically induced upon TGFβ treatment in normal epithelial cells (9). Transcriptional induction of p15 by TGFβ involves the formation of a Smad transcriptional complex containing Miz-1 (10), Myc (11), Sp1 (12) and FOXO (5). On the other hand, acetylase p300 is a well established co-activator of the TGFβ pathway (13, 14). In epithelial cells, TGFβ signaling also recruits acetylase p300 to the Smad complex to acetylate some transcription factors and mediate TGFβ-induced transcription (6, 15).
Human Kruppel-like factor 5 (KLF5, IKLF, BTEB2) is a zinc finger transcription factor belonging to the Sp/Kruppel-like family. It has important roles in different biological processes including cell proliferation, differentiation, cell cycle regulation, angiogenesis, and carcinogenesis (16, 17). KLF5 was reported to be pro-proliferative in some types of cells including immortalized but non-tumorigenic epithelial cells but anti-proliferative in cancer cells (18–21). Using an epithelial cell line treated with TGFβ, we found that without TGFβ, KLF5 is pro-proliferative; but when TGFβ signaling is activated, KLF5 becomes anti-proliferative (22). By characterizing the regulation of p15, a typical target gene of TGFβ, we further found that KLF5 is inhibitory to p15 induction without TGFβ but becomes stimulatory when TGFβ signaling is activated, and the reverse is caused by TGFβ-mediated acetylation of KLF5 (22). Although these findings suggest a role of KLF5 in the assembly of TGFβ-induced transcriptional complex on p15 promoter, the role has not been determined.
In the present study, we examined the interactions between KLF5 and other key transcription factors bound to p15 promoter in the context of TGFβ and their effect on the activity of p15 promoter. We also analyzed whether and how KLF5 alters the binding of other transcription factors to p15 promoter before and after TGFβ treatment. The role of TGFβ-recruited p300 in the acetylation of KLF5 and its effect on protein interaction and promoter binding was also characterized.
EXPERIMENTAL PROCEDURES
Cell Lines and Other Materials
The HaCaT epidermal epithelial cell line was established by Dr. Norbert E. Fusenig of the German Cancer Research Center (23) and was kindly provided to us by Dr. Robert A. Swerlick of Emory University. It was maintained following established procedures (23). The HEK-293 human kidney epithelial cell line, the HepG2 hepatoma cell line, and the COS-1 cell line were purchased from American Type Culture Collection (ATCC, Manassas, VA) and propagated following ATCC's instructions. The TGFβ used in this study was TGFβ1 from R&D Systems (Minneapolis, MN).
Co-immunoprecipitation (co-IP) Assay
Co-IP was conducted following the standard protocol. Briefly, HaCaT cells with different treatments were lysed in lysis buffer, and cell extracts were incubated with antibodies against human Smad2/3, Smad4, or c-Myc (Santa Cruz Biotechnology, Santa Cruz, CA). Protein complexes were collected using protein G-Sepharose (Sigma), separated by SDS-polyacrylamide gel electrophoresis, and transferred to membranes (Immobilon-P; Millipore, Billerica, MA). Specific proteins were detected by Western blot analysis using antibodies against Smad2 or Smad4 (Cell Signaling, Danvers, MA) and KLF5 (24).
FLAG-Smad3 and Smad4-FLAG plasmids, which were kindly provided by Dr. Rik Derynck of the University of California at San Francisco, and FLAG-tagged Miz-1 or HA-tagged KLF5 were transfected into different cell lines using Lipofectamine reagent (Invitrogen) following the manufacturer's instructions. FLAG-pcDNA3 vector was used as a negative control. Co-IP was performed with Anti-FLAG M2 affinity gel (Sigma) or anti-HA affinity gel (Sigma). Other steps were the same as above.
GST Pulldown Assay
Full-length Smad4 was cloned from the Smad4-FLAG plasmid into the pGEX vector (GE Healthcare). GST-Smad4 or GST expressed in BL21 bacteria was purified with glutathione-Sepharose 4B slurry beads (GE Healthcare) following the manufacturer's instructions. Equal molar amounts of purified GST fusion proteins (GST, GST-Smad4) were immobilized to 50 μl of 50% glutathione-Sepharose 4B slurry beads (GE Healthcare) in 0.5 ml of GST pulldown buffer (10 mm HEPES, pH 7.6, 3 mm MgCl2, 100 mm, KCl, 5 mm EDTA, 5% glycerol, and 0.5% CA630). After incubation for 1 h at 4 °C with rotation, the beads were washed three times with GST pulldown buffer. Ten μl of 35S-labeled in vitro translated KLF5 protein, which was synthesized as described previously (25), were added and mixed for 2 h at 4 °C with rotation. The bound proteins were eluted by boiling in 30 μl of loading buffer. 35S-Labeled proteins were detected by gel electrophoresis and autoradiography.
Promoter-Luciferase Reporter Assay
The SBE4-luc and MSE-luc plasmids were gifts from Dr. Bert Vogelstein of Johns Hopkins University (26). The p15P165-Luc and p15P69-Luc plasmids were kindly provided by Dr. Xiao-Fan Wang of Duke University (12). Plasmids were transfected into HaCaT cells or HepG2 cells using Lipofectamine reagent (Invitrogen) for 40 h, and then cells were treated with 100 pm (2 ng/ml) TGFβ for 20 h. Luciferase assay was carried out using the Promega luciferase assay kit as described previously (25).
Oligonucleotide Pulldown Assay
Oligonucleotides for the p15 promoter, with biotin added to their 5′-end, were synthesized by MWG-Biotech (High Point, NC). The sequences for the oligonucleotides were as follows, using the same nucleotide numbering as described previously (12): Inr (−12 to +11), biotin-5′-GGCTGGCTCCCCACTCTGCCAGAG-3′ (wild type) and biotin-5′-GGCTGGCTCAACAATATGCCAGAG-3′ (mutant); SPS2 (−89 to −65), biotin-5′-CAGCGGACAGGGGGCGGAGCCTAAG-3′ (wild type) and biotin-5′-CAGCGGACAGGAAGTAGAGCCTAAG-3′ (mutant); and SBE (−443 to −385), biotin-5′-TAACTTGTATGACAGGTGCAGAGCTGTCGCTTTCAGACATCTTAAGAAAGACGGAGTTA-3′ (wild type) and biotin-5′-TAACTTGTATGACAGGTGCAGAGCTGTCGCTTTCACTCATCTTAAGAAAGACGGAGTTA-3′ (mutant). Each pair of complementary oligos was annealed following standard protocols. COS-1 cells were transfected with different plasmids, and 30 h later, cells were lysed in lysis buffer (50 mm Tris HCl, pH 7.4, 150 mm NaCl, 1 mm EDTA, and 1% Triton X-100) containing protease and phosphatase inhibitors. After cell debris was removed by centrifugation, cell extracts were precleared with ImmunoPure streptavidin-agarose beads (20 μl/sample, Pierce) for 1 h at 4 °C. After centrifugation for 1 min at 5000 × g, the supernatant was incubated with 100 pmol of biotinylated double-stranded oligonucleotides and 10 μg of poly(dI-dC)·poly(dI-dC) for 16 h at 4 °C. DNA-bound proteins were collected with 30 μl of immobilized streptavidin-agarose beads for 1 h at 4 °C, washed with lysis buffer four times, separated on a 10% SDS-polyacrylamide gel, and subjected to Western blotting with different antibodies.
Chromatin Immunoprecipitation (ChIP) Assay
HepG2 cells were transfected with pcDNA3-FLAG-KLF5 (24) or pcDNA3-FLAG plasmid (Invitrogen) using Lipofectamine 2000 reagent (Invitrogen). Forty hours after transfection, cells were incubated in the presence or absence of 5 ng/ml TGFβ for 1 h. ChIP was performed according to the protocol from Upstate Biotechnology (Lake Placid, NY). FLAG antibody-agarose beads (Sigma) and antibodies against human Smad4 (H-552, Santa Cruz) or FOXO3 (Santa Cruz) were used to precipitate the protein-DNA complex. Precipitated DNA was analyzed by PCR using two pairs of p15 promoter primers, which scanned distal and proximal regions of p15 promoter, respectively. Primers for the distal region (nucleotides −547 to −239) were 5′-TATGGTTGACTAATTCAAACA-3′ (sense) and 5′-AATATTTTGGGAATGTTCACCA-3′ (antisense). Primers for the proximal region (nucleotides −177 to 161) were 5′- AGTCTCTGGCGCATGCGTCCTA-3′ (sense) and 5′-TTAGCTCCGGGCTTTTCCTGGC-3′ (antisense) (27).
Separation of Nuclear and Cytoplasmic Fractions
A total of 2 × 106 HaCaT cells were seeded onto 150-mm dishes. After 40 h, cells were treated with 100 pm TGFβ or control solution for 1 h and then collected by scraping in 1× phosphate-buffered saline. The cytoplasmic and nuclear fractions were prepared using NE-PER nuclear and cytoplasmic extraction reagents (Pierce) following the manufacturer's instructions. Cell lysates were analyzed by Western blotting assay. Antibody against β-actin was from Sigma, and antibody against lamin A was from Santa Cruz Biotechnology.
RESULTS
Protein Interactions between KLF5 and Other p15 Regulatory Factors Including Smads
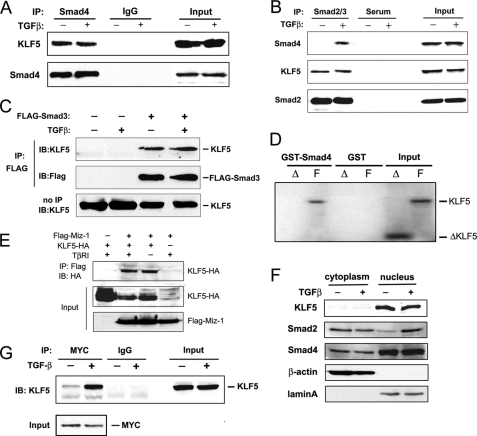
It is well established that in different types of cells, Smad2 and Smad3 are direct substrates of the TGFβ receptor type I (TβRI) kinase, and their phosphorylation enables their association with Smad4 and translocation into the nucleus where they bind to gene promoters to regulate transcription (6). Smad2 and Smad3 have nearly identical amino acid sequences, except that Smad2 has a unique insertion that prevents its direct binding to DNA (10). We reasoned that, as an epithelium-specific cofactor mediating the inhibitory effect of TGFβ, KLF5 could interact with one or more Smad proteins and other known factors to regulate p15 expression. To test this possibility, we performed immunoprecipitation in HaCaT cells treated with or without TGFβ. HaCaT cells transfected with FLAG-tagged Smad3 expression plasmid were also analyzed. In the protein complexes precipitated by antibodies against endogenous Smad4 or Smad2/3 or transfected FLAG-tagged Smad3, endogenous KLF5 protein was always detected (Fig. 1, A–C). Moreover, the amount of precipitated KLF5 protein was similar between cells treated with TGFβ and those without, although Smad4 was detected in the Smad2/3 complex only from TGFβ-treated cells (Fig. 1, A–C). These results indicate that KLF5 can form a protein complex with Smad2, Smad3, or Smad4.
FIGURE 1.
KLF5 interacts with other transcriptional regulators of p15. A and B, interaction of KLF5 with Smads2–4 as detected by co-IP and immunoblotting (IB). Lysates from HaCaT cells treated with TGFβ (5 ng/ml for 1 h) were subjected to IP with antibodies against Smad4 or Smad2/3 followed by IB with antibodies against KLF5 and Smads. IgG or serum was used as a negative control. Inputs are from cell lysates before IP. C, HaCaT cells were transfected with FLAG-tagged Smad3 (FLAG-Smad3), and IP and IB were performed with FLAG and KLF5 antibodies. D, pulldown of in vitro translated full length KLF5 (F) but not C terminally truncated KLF5 (ΔKLF5 or Δ) by GST-fused Smad4. GST alone served as a negative control. Inputs are translated proteins not subjected to GST pulldown. E, interaction of KLF5 (HA-tagged) with Miz-1 (FLAG-tagged) in the presence or absence of TGFβ signaling (TβRI) also detected by IP combined with IB in the HEK-293 cell line transfected with the respective plasmids. F, subcellular localization of KLF5, Smad2, and Smad4 in HaCaT cells. Nuclear and cytoplasmic fractions of HaCaT cells treated with or without TGFβ were subjected to IB with the indicated antibodies. G, TGFβ-dependent interaction between KLF5 and Myc as detected by IP with anti-Myc antibody and IB with anti-KLF5 antibody in HaCaT cells treated with or without TGFβ (5 ng/ml for 1 h).
Using in vitro translated 35S-labeled full-length and truncated (amino acids 1–367) KLF5 proteins, a GST pulldown assay was also performed. The DNA-binding domain, which is a potent protein-protein interface (28), was deleted in the truncated KLF5. Full-length KLF5, but not the truncated KLF5, was pulled down by GST-fused Smad4 (Fig. 1D). GST alone did not pull down KLF5, indicating that KLF5 binds to Smad4 without the presence of promoter DNA and TGFβ signaling and that the interaction between KLF5 and Smad4 requires the DNA-binding domain region. Lack of TGFβ dependence in KLF5-Smad protein interaction suggests that some KLF5 molecules may form a complex with Smads before binding to promoter DNA, similar to the role of E2F4/5 and p107 in mediating TGFβ-mediated Myc repression (29).
We also used additional cell systems to further evaluate protein interactions between KLF5 and other factors that regulate p15 expression in the context of TGFβ. Using the HEK-293 cell line, we co-transfected FLAG-tagged Miz-1 and HA-tagged KLF5 and performed co-IP combined with immunoblotting. We found that Miz-1 formed a protein complex with KLF5 regardless of TGFβ status (Fig. 1E).
KLF5 is a nuclear protein, whereas Smad2 and Smad4 translocate into the nucleus upon TGFβ signaling activation (3, 4). However, the interaction between KLF5 and Smad4 was not affected by TGFβ (Fig. 1A). To evaluate where this interaction occurs, we then separated cytoplasm and nuclei in HaCaT cells treated with or without TGFβ, and analyzed the expression of KLF5 and two Smads by Western blot analysis. KLF5 was located primarily in the nucleus (Fig. 1F), consistent with our earlier assay using immunofluorescence staining (24). Smad2 without TGFβ was located mainly in the cytoplasm but was located mainly in the nucleus upon TGFβ treatment (Fig. 1F), consistent with previous studies that Smad2 translocates into the nucleus after TGFβ treatment. Interestingly, the location of Smad4 was primarily in the nucleus regardless of TGFβ treatment (Fig. 1F).
For Myc, we were able to evaluate its endogenous interaction with KLF5. Lysates from HaCaT cells treated with or without TGFβ were immunoprecipitated with anti-Myc antibody and immunoblotted with anti-KLF5 antibody. KLF5 was detected in the Myc protein complex only when TGFβ was present (Fig. 1G).
KLF5 Cooperates with Other Transcription Factors to Mediate TGFβ-induced p15 Promoter Activity
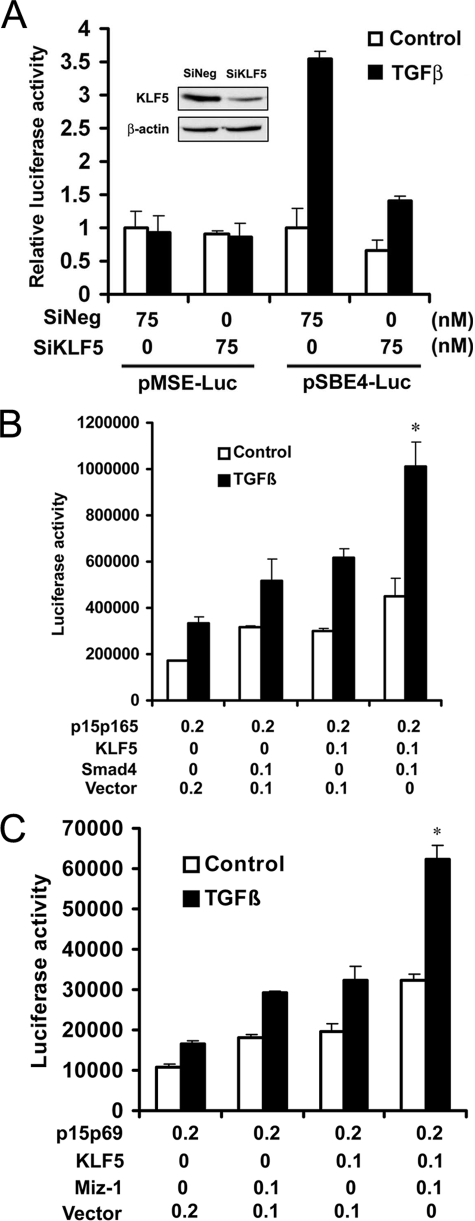
To test whether the KLF5-Smad interaction affects the function of Smad in gene regulation, we performed a luciferase assay for the artificial SBE4 promoter, which has four copies of the Smad-binding site (SBE4) (26), and in HaCaT cells with the MSE promoter, which have three copies of mutant Smad-binding site (26), as a negative control. Although the MSE promoter did not show any TGFβ-mediated activity regardless of KLF5 status, knockdown of KLF5 significantly inhibited TGFβ-induced, SBE-mediated promoter activity in HaCaT cells (Fig. 2A). This result is consistent with our previous study (22) in which knockdown of KLF5 decreased TGFβ-induced p15 expression and p15 promoter activities in HaCaT cells, showing that acetylation of KLF5 is essential for the function of TGFβ in transcriptional control in epithelial cells.
FIGURE 2.
KLF5 cooperates with other transcription factors in TGFβ-mediated regulation of p15 promoter activity. A, KLF5 knockdown inhibited TGFβ-induced, Smad-binding-site-mediated promoter activity in HaCaT cells, as determined by luciferase assay. SBE4-luc and MSE-luc are luciferase reporter plasmids with four copies of the Smad-binding site (taaGTCTAGACggcaGTCTAGACgtac) and three copies of the mutant Smad-binding site (cctGTTTATACggcaGTCTAGACgtac), respectively, as described previously (26). The MSE sequence served as the negative control. Knockdown of KLF5 expression was confirmed by Western blot analysis (insert). SiNeg, siRNA for negative control; SiKLF5, siRNA for KLF5. B, additive induction of the p15 promoter activity by KLF5 and Smad4 in HepG2 cells, also determined by luciferase assay. p15p165 is a p15 promoter-luciferase reporter plasmid. TGFβ was at 2 ng/ml. C, additive induction of the p15 promoter activity by KLF5 and Miz-1 in HepG2 cells. p15p69 is another p15 promoter-luciferase reporter plasmid with 69 base pairs of the p15 promoter sequence containing the Inr element but not the SBE and SPS2 elements (12). TGFβ was at 2 ng/ml. Asterisk, indicates statistically significant differences in luciferase activities between cells transfected with KLF5 and Smad4 or Miz-1 and cells transfected with pcDNA3 and Smad4 or Miz-1 in the presence of TGFβ (p < 0.01). These experiments were repeated twice, and consistent results were obtained.
We then evaluated the functional interaction between KLF5 and Smad4 or Miz-1 in TGFβ-induced p15 promoter activities (Fig. 2, B and C). Luciferase assays were performed in HepG2 cells transfected with different promoter-luciferase reporter plasmids for p15. Cells were also treated with TGFβ. For the p15p165 reporter plasmid, which contains the SBE (12), KLF5 and Smad4 had an additive effect on the promoter activity (Fig. 2B). For the p15p69 promoter-luciferase reporter plasmid, which has 69 base pairs of the p15 promoter sequence containing the initiation (Inr) element but not the SBE- and SPS2 KLF5-binding sites (12), KLF5 and Miz-1 also showed an additive effect on the induction of promoter activity (Fig. 2C).
TGFβ Alters the Binding of KLF5 and Other Factors to the p15 Promoter
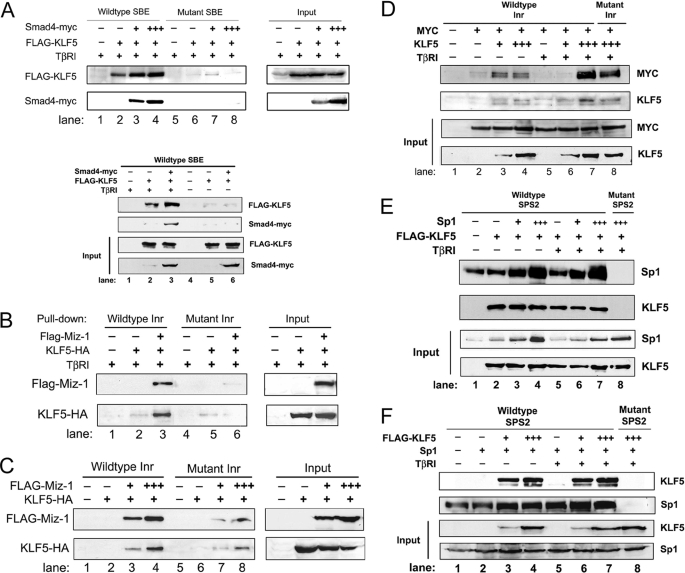
Previous studies have showed that multiple factors, including Smads, Sp1, Myc, and Miz-1, bind to different DNA elements of the p15 promoter, including SBE, SPS2, and Inr, to mediate TGFβ-induced p15 expression (10–12). Our recent study also showed that KLF5 plays an essential role in TGFβ-induced p15 expression (22). Results from that study showed that KLF5 binds to each of these elements and that in HaCaT cells the binding of KLF5 to Inr and SBE is TGFβ-dependent, but KLF5 binding to the SPS2 element is much stronger and is TGFβ-independent (22). A question, therefore, is whether KLF5 cooperates or competes with other factors in their binding to p15 promoter. In this regard, we transfected COS-1 cells with expression constructs for KLF5 and Smad4, Miz-1, Myc or Sp1, performed oligo pulldown assays for each of the three promoter elements, and detected KLF5 and each of the known factors in the oligo-protein complexes by Western blotting. In COS-1 cells, TGFβ signaling was reconstituted by transfection-mediated expression of autoactivated TβRI. KLF5 bound to the SBE when TGFβ signaling was present, and the mutation in SBE interrupted the binding (Fig. 3A, upper panel). Without TGFβ signal, almost no KLF5 bound to SBE (Fig. 3A, lower panel). In addition, expression of Smad4 enhanced the binding of KLF5 to SBE (Fig. 3A). For the Inr element, which was originally identified as the binding element for Myc and Miz-1, KLF5 alone showed almost no binding regardless of TβRI (Fig. 3, B and C). Co-expression of Miz-1, on the other hand, increased the binding of KLF5 regardless of TGFβ status (Fig. 3, B and C). When Myc was co-expressed, little binding was detected if KLF5 was absent, regardless of TGFβ status (Fig. 3D, lanes 2 and 5). In addition, both lower and higher doses of KLF5 caused a binding of Myc to the Inr element at a moderate level without TGFβ (Fig. 3D, lanes 3 and 4). When TGFβ was present, a higher dose of KLF5 caused a stronger binding of Myc to Inr (Fig. 3D, lane 7), whereas a lower dose of KLF5 did not cause significant binding (lane 6). Interestingly, TGFβ increased the binding of Myc to Inr when a higher dose of KLF5 was transfected but decreased the binding of Myc to Inr when a lower dose of KLF5 was expressed (Fig. 3D). For the SPS2 element originally identified as harboring a Sp1-binding site, KLF5 showed a stronger binding, which was not affected by Sp1 but was slightly reduced by the TGFβ signaling (Fig. 3E). The binding of Sp1 to the SPS2 element, on the other hand, was enhanced by both KLF5 expression and TGFβ signaling (Fig. 3F). These results indicate that TGFβ alters the binding of KLF5 to p15 promoter and that both TGFβ and KLF5 alter the binding of other factors to p15 promoter.
FIGURE 3.
Binding of KLF5 and other factors on different elements of p15 promoter detected by oligo pulldown assays. COS-1 cells were transfected with different plasmids to express different factors, with FLAG, Myc, and HA tags attached to Miz-1 (B and C)/KLF5 (A, E, and F), Smad4 (A) and KLF5 (B and C), respectively. The plasmids for Myc (D) and Sp1 (E and F) do not have a tag. TGFβ signaling was reconstituted by transfecting the TβRI plasmid. Cell lysates were subjected to an oligo pulldown assay combined with IB analysis. Antibodies against FLAG, Myc, and HA tags were used to detect the tagged proteins, whereas those against Sp1 and KLF5 were used to detect untagged or tagged proteins. Each of the three known DNA elements of the p15 promoter, SBE, SPS2, and Inr, was analyzed. Input represents samples without oligo pulldown and the number of plus signs indicates the relative amounts of plasmid DNA transfected for a specific gene.
Acetylation of KLF5 by TGFβ-recruited p300 Is Necessary for the Interactions among Transcription Factors in p15 Regulation
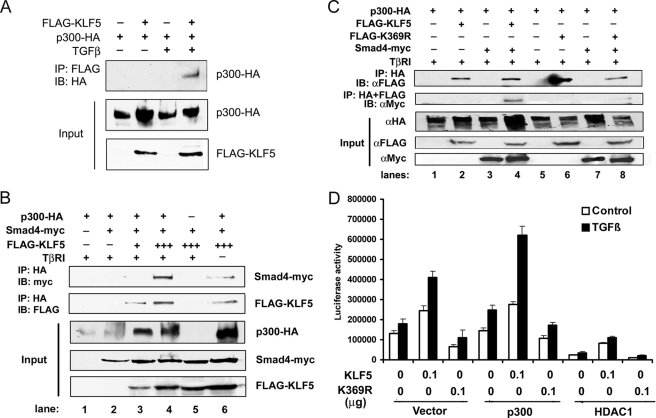
Upon TGFβ activation, Smad2 and Smad3 translocate into the nucleus, where they form a complex with Smad4 and recruit p300 to acetylate proteins (6, 15). Previously it was shown that p300 can acetylate KLF5 and that HDAC1 antagonizes the effect of p300 (30); acetylation reverses the function of KLF5 in cell proliferation and gene regulation (22). To test whether KLF5 is acetylated by TGFβ-mediated Smad-recruited p300, we examined the protein interaction between KLF5 and p300 in the context of TGFβ. Using the HEK-293 cell line co-transfected with HA-tagged p300, FLAG-tagged KLF5, and TβRI, immunoprecipitation combined with immunoblotting showed that KLF5 formed a protein complex with p300 only when TGFβ signaling was present (Fig. 4A), suggesting that TGFβ signaling is necessary for KLF5 to interact with p300. When Smad4 was also co-transfected along with KLF5 and p300, the three molecules formed a complex even without TGFβ signaling, as both KLF5 and Smad4 were detected in the protein complex precipitated by the p300 antibody (Fig. 4B). It was interesting that the protein level of p300 was increased by KLF5 overexpression in HEK-293 cells (Fig. 4B, Input). When TGFβ signaling was present, the interactions between KLF5, Smad4 and p300 became stronger, as more KLF5 and Smad4 were detected in the protein complex precipitated by the p300 antibody when TβRI was expressed to activate the TGFβ signaling (Fig. 4B). Although an interaction between Smad4 and p300 had been shown in previous studies (31), we noticed that KLF5 was necessary for the interaction of Smad4 with p300, as almost no Smad4 and KLF5 were detected in the protein complex precipitated with p300 antibody when less or no KLF5 plasmid was transfected (Fig. 4B).
FIGURE 4.
Acetylation of KLF5 involves the p300 acetylase and is necessary for association between p300 and Smad4 in p15 regulation. A, TGFβ induces protein interaction between KLF5 and the acetylase p300. HEK-293 cells were transfected with HA-tagged p300 and FLAG-tagged KLF5 and treated with TGFβ (5 ng/ml for 1 h). Cell lysates were subjected to IP with anti-FLAG antibody and IB with anti-HA antibody. B, KLF5 mediates protein interaction between Smad4 and p300. HEK-293 cells were transfected with HA-tagged p300, Myc-tagged Smad4, and different amounts of FLAG-tagged KLF5. The TβRI gene was also transfected to provide TGFβ signaling. Cell lysates were subjected to IP with anti-HA antibody and IB with anti-Myc or anti-FLAG antibody. C, failure in the acetylation of KLF5 abolishes the association between p300 and Smad4. HEK-293 cells were transfected with indicated plasmids, and cell lysates were subjected to sequential immunoprecipitation, first with antibody against HA and second with antibody against FLAG. Protein elutions were subjected to IB with antibodies against FLAG or Myc tag. Input indicates cell lysates without IP. D, acetylation of KLF5 is necessary for p15 promoter activity. Luciferase assay was performed in HepG2 cells transfected with p15p165 promoter-reporter plasmid, along with different combinations of expression plasmids (0.2 μg total) for KLF5, its acetylation-deficient mutant (p300), deacetylase HDAC1, and vector control. These experiments were repeated twice, and consistent results were obtained.
To further determine the role of p300-mediated KLF5 acetylation in the interactions among KLF5 and other transcription factors binding to the p15 promoter, we analyzed the K369R mutant of KLF5, which could no longer be acetylated, for the effect of acetylation failure on the KLF5 transcriptional assembly. We performed co-IP combined with Western blotting in HEK-293 cells transfected with wild-type and mutant KLF5. Protein interaction between p300 and the K369R mutant of KLF5 still occurred, as both wild-type KLF5 and the mutant were still detected in the protein complex precipitated by the antibody against p300 (Fig. 4C). Overexpression of Smad4 did not affect wild-type KLF5 but appeared to weaken the association of p300 with mutant KLF5. Interestingly, the association of Smad4 with the p300-KLF5 complex was abolished when KLF5 was mutated (Fig. 4C), as the protein complex precipitated first by antibody against p300 and then by antibody against KLF5 no longer contained Smad4 when KLF5 was mutated. Therefore, failure in the acetylation of KLF5 prevents the p300-assembled Smad4 and KLF5 complex. Unlike wild-type KLF5, the K369R mutant did not increase but decreased p300 protein levels (Fig. 4C, Input).
To test the role of p300 and acetylation of KLF5 in modulating the function of KLF5 in p15 regulation, we performed a promoter-reporter luciferase assay in HepG2 cells overexpressing either the p300 acetylase or the HDAC1 deacetylase and treated with or without TGFβ (Fig. 4D). Without p300 or HDAC1, whereas KLF5 showed an activity in the p15 promoter and TGFβ further increased the activity, the acetylation-deficient mutant of KLF5 decreased the activity (Fig. 4D). When p300 was overexpressed, TGFβ and KLF5-induced promoter activity was significantly increased, but mutant KLF5 still showed a weaker activity than the control. When the HDAC1 deacetylase was overexpressed, p15 transcription was weaker overall for both KLF5 and its mutant and wild-type KLF5 still had some activity in the p15 promoter, but the mutant of KLF5 showed almost no activity (Fig. 4D). These results further indicate that acetylation is essential for KLF5 and TGFβ to activate p15 expression and that p300 is an important player in KLF5 activation.
Acetylation of KLF5 Is Also Necessary for the Assembly of KLF5 and Other Transcription Factors on p15 Promoter
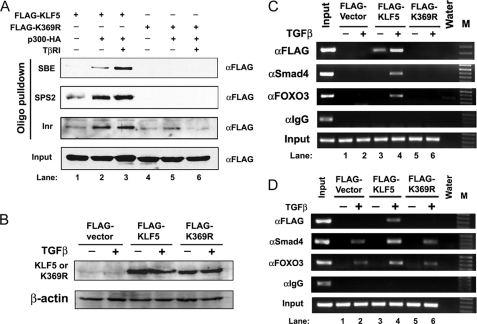
Based on the findings that acetylation reverses the function of KLF5 and makes KLF5 a functional mediator for TGFβ to induce p15 expression and suppress cell proliferation and that the acetylation-deficient K369R mutant of KLF5 has lost the ability to mediate the function of TGFβ, we further speculated that acetylation of KLF5 could also be necessary for the binding of KLF5 and other transcription factors to the p15 promoter. To test this hypothesis, we analyzed the K369R acetylation-deficient mutant of KLF5 for its effect on the binding of transcription factors to the p15 promoter. We first performed oligo pulldown assays with a controlled TGFβ signal and expression of p300 and KLF5 in COS-1 cells (Fig. 5A). Consistent with previous findings (22), transfected wild-type KLF5 showed increased binding to each of the three DNA elements from the p15 promoter when p300 and/or TGFβ were present (Fig. 5A). When lysine 369 in KLF5 was mutated to prevent its acetylation, no obvious binding was detected for any of these sequences (Fig. 5A), indicating that acetylation is essential for KLF5 to bind to the p15 promoter.
FIGURE 5.
Acetylation of KLF5 is essential for the binding of KLF5 and other transcription factors to the p15 promoter. A, cell lysates from COS-1 cells transfected with different combinations of expression plasmid for FLAG-tagged KLF5 or its K369R mutant, p300, and TβRI were subjected first to an oligo pulldown assay for each of the KLF5 binding elements from p15 promoter and then to IB with anti-FLAG antibody. Inputs are lysates not subjected to oligo pulldown. B–D, HepG2 cells were transfected with FLAG-tagged vector control, wild-type KLF5, or the K369R mutant of KLF5 and treated with or without TGFβ (5 ng/ml, 1 h). Expression of KLF5 and its mutant was confirmed by Western blotting with anti-FLAG antibody (B). ChIP assay was performed using antibodies against FLAG for KLF5 (αFLAG), Smad4 (αSmad4), and Foxo3 (αFoxo3). Primers spanning the proximal region of the p15 promoter (C), which contains the SPS2- and Inr-binding sites, or the distal region (D), which contains the Smad binding element, were used to amplify the precipitated DNA by PCR. M, DNA marker.
We also performed ChIP assays to further determine the effect of KLF5 acetylation on the transcriptional assembly on the p15 promoter. HepG2 cells were transfected with FLAG-tagged vector control, wild-type KLF5, or the K369R mutant of KLF5 and were treated with or without TGFβ. Expression of KLF5 and its mutant was confirmed by Western blotting with the FLAG antibody (Fig. 5B). Both Smad4 and FOXO are key regulators in p15 induction by TGFβ, and their binding to the p15 promoter was well established by the ChIP assay (5, 27). Therefore, antibodies against FLAG (for both wild-type and mutant KLF5), Smad4, and FOXO3 were used. For the proximal region of the p15 promoter, which contains the SPS2 and Inr promoter elements, binding to the p15 promoter was detected for each of the three factors only when wild-type KLF5 was expressed (Fig. 5C), indicating that failure in the acetylation of KLF5 prevents the binding of KLF5, Smad4, and FOXO3 to the proximal region of the p15 promoter. In addition, although TGFβ enhanced the binding of KLF5 to this promoter region, TGFβ was essential for both Smad4 and FOXO3 to bind (Fig. 5C). For the distal region of the p15 promoter, which contains the SBE, KLF5 binding was detectable only in TGFβ-treated cells as expected, but mutation at the acetylation site of KLF5 interrupted this binding (Fig. 5D). The binding of Smad4 and FOXO3 to this promoter region occurred in TGFβ-treated cells regardless of KLF5 status, although wild-type KLF5 (but not mutant KLF5) might increase their binding. These results indicate that failure in the acetylation of KLF5 alters the assembly of transcription factors on p15 promoter.
DISCUSSION
Based on our earlier study (22) in which we found that the pro-proliferative transcription factor KLF5 becomes anti-proliferative when TGFβ signaling is activated in epithelial cells, p15 transcription is inhibited by KLF5 without TGFβ but is induced by KLF5 when TGFβ is activated, and acetylation of KLF5 at residue 369 is responsible for the reverse of KLF5 function, we conducted the current study to understand how the function of KLF5 is reversed by acetylation in the regulation of p15 expression in relation to TGFβ. Our conclusion is that acetylation of KLF5, which is mediated by TGFβ-recruited p300, is a key factor for the assembly of transcription factors on the p15 promoter.
KLF5 Is a Key Member of the TGFβ-mediated p15 Transcriptional Assembly Containing Previously Identified Transcription Factors
As a potent inducer of p15 expression in epithelial cells, TGFβ signaling phosphorylates Smad2 and Smad3, which then associate with Smad4 and translocate to the nucleus, where they bind to the p15 promoter (6). Other transcription factors, although binding to different DNA elements, also have been identified as components of the transcriptional assembly on p15 promoter, including Sp1, Myc, and Miz-1 (10–12). The results from this study showed that KLF5 is also a member of the p15 transcriptional assembly. Co-IP, in combination with immunoblotting, demonstrated that KLF5 forms a protein complex with each of the known p15 transcription factors tested, including Smad2/3, Smad4, Myc, and Miz-1 (Fig. 1). In addition to ectopically expressed Smad3 and Miz-1 (Fig. 1, C and E), protein association with endogenous KLF5 was also detected for endogenous Smad4, Smad2/3, and Myc (Fig. 1, A, B, and G). The association between KLF5 and Smad4 was further confirmed by GST pulldown assay using purified proteins (Fig. 1D). Oligo pulldown results also indicate interactions between KLF5 and other p15 transcription factors in the regulation of p15 (Fig. 3). For example, expression of Smad4 and Miz-1 enhanced the binding of KLF5 to the SBE and Inr elements of p15 promoter (Fig. 3, A–C). On the other hand, expression of KLF5 enhanced the binding of Myc and Sp1 to the Inr and SPS2 elements, respectively (Fig. 3, D and F). Functional luciferase assays demonstrated that KLF5 activated an artificial promoter containing Smad-binding elements, and KLF5 and Smad4 or Miz-1 had additive effects on promoter activities of p15 (Fig. 2). Taken together, these findings suggest that KLF5 is a member of the p15 transcriptional assembly in TGFβ-induced p15 transcription.
Consistent with previous studies showing a role of TGFβ in reversing KLF5 function in p15 regulation, findings in this study indicate that TGFβ modulates the function of KLF5 in p15 regulation. First of all, TGFβ enhanced the binding of KLF5 to the p15 promoter, as demonstrated in the ChIP assay (Fig. 5, C and D) and oligo pulldown assay (Fig. 3). In addition, KLF5 and other factors regulate each other in their binding to the p15 promoter when TGFβ is activated. These results further indicate that KLF5 is a key member in the p15 transcriptional assembly.
TGFβ Signaling Is Necessary for the Association of KLF5 with Myc but Not Its Associations with Other p15 Transcription Factors
KLF5 plays a critical role in the function of TGFβ in inhibiting cell proliferation and inducing p15 expression (22). It has been shown that KLF5 binds to different elements of the p15 promoter; binding to the Inr and SBEs are TGFβ-dependent, but the binding to the SPS2 element is TGFβ-independent (22). Consistently, our results in this study demonstrated that expression of Smad4 enhances the binding of KLF5 to SBE (Fig. 3A) and that TGFβ signaling is necessary for the binding of KLF5, Smad4, and FOXO3 to the p15 promoter (Fig. 5, C and D). Although KLF5 associates with other p15 transcription factors in their binding to the p15 promoter (Figs. 1 and 3), not all associations are affected by TGFβ. In fact, among the factors tested, only Myc had a TGFβ-dependent interaction with KLF5 (Fig. 1G), whereas Smad2/3, Smad4, and Miz-1 associated with KLF5 regardless of TGFβ status (Fig. 1, A–E). Previous studies demonstrated that during TGFβ-mediated proliferation inhibition in epithelial cells, Myc is rapidly down-regulated and its down-regulation is necessary for TGFβ to induce p15 and p21 and to inhibit cell cycle progression from the G1 to S phase (2, 10, 29). TGFβ-mediated association between KLF5 and Myc, as detected in this study, appears to be another mechanism for the inactivation of Myc in p15 regulation.
Acetylation of KLF5 Is Essential for the Assembly of TGFβ-induced Transcriptional Assembly on p15 Promoter
Previous studies showed that TGFβ activation translocates Smad2 and Smad3 into the nucleus, where they form a complex with Smad4 that recruits p300 to acetylate proteins (6, 15), and that p300 acetylates KLF5 (30). This process also appears to occur with KLF5, because TGFβ causes the association of KLF5 and p300 (Fig. 4A), and KLF5 can form a protein complex with Smad4 and p300 even without TGFβ (Fig. 4B). Furthermore, TGFβ-mediated acetylation of KLF5, which has been demonstrated in a previous study (22), is necessary for TGFβ to reverse the function of KLF5 in regulating p15 expression and cell proliferation (Fig. 4D) (22). Results in the current study also indicate that acetylation of KLF5 is essential for the assembly of p15 transcriptional assembly upon TGFβ activation (Fig. 4). As demonstrated by co-IP and immunoblotting assays, KLF5 is essential for protein association between Smad4 and p300 (Fig. 4B) and acetylation-deficient mutation of KLF5 abolishes the association (Fig. 4C).
Acetylation of KLF5 also appears to be essential for the binding of KLF5 and other transcription factors to the p15 promoter upon the activation of TGFβ signaling. In oligo pulldown assays, mutation of the acetylation site in KLF5 prevented p300-mediated or enhanced binding of KLF5 to each of the three DNA elements (Fig. 5A). Consistently, ChIP assay showed that the binding of KLF5 to both the proximal and distal regions of the p15 promoter, which were enhanced or induced by TGFβ, were abolished upon the mutation of its acetylation site (Fig. 5, C and D). We noticed that the binding of KLF5 to the proximal region of p15 promoter occurred for in vitro translated protein even without TGFβ treatment (22), but acetylation deficiency also abolished this binding (Fig. 5C). Therefore, acetylation of KLF5 appears to be essential for KLF5 to bind to p15 promoters physiologically in vivo. On the other hand, the conformation of acetylation-deficient KLF5 could change to prevent its binding to the p15 promoter.
Smad4 and FOXO3 binding to the proximal region of p15 promoter was not only KLF5- and TGFβ-dependent, it was also dependent on the acetylation of KLF5 (Fig. 5C). Binding of Smad4 and FOXO3 to the distal region of p15 promoter, on the other hand, was TGFβ-dependent but not KLF5-dependent, although KLF5 enhanced their binding and acetylation mutation abolished the enhancement effect (Fig. 5D). These findings further indicate an essential of role of acetylated KLF5 in TGFβ-mediated p15 regulation.
Modeling KLF5 Function in p15 Regulation in the Context of TGFβ
Our previous study showed that TGFβ-mediated acetylation of KLF5 reverses the function of KLF5 in cell proliferation and p15 regulation (22). In pinpointing how the reversal occurs, in this study we found that the association of KLF5 with Smad2, Smad3, and Smad4 was TGFβ-independent, whereas the association with Myc was TGFβ-dependent (Fig. 1). In addition, both KLF5 and Smad4 were predominantly localized in the nucleus of epithelial cells regardless of TGFβ status (Fig. 1F), although Smad2 and Smad3 without TGFβ are located mainly in the cytoplasm but translocate into the nucleus upon TGFβ activation (6) (Fig. 1F). Therefore, KLF5 forms a protein complex with Smads in the nucleus regardless of the TGFβ status.
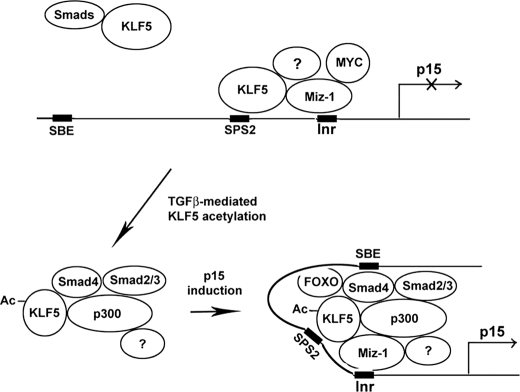
Binding of KLF5 to different elements of the p15 promoter varies in terms of TGFβ dependence, although binding to the Inr and SPS2 elements was TGFβ-independent (Fig. 3) similar to the binding of Sp1 and Miz-1 (Fig. 3). Taken together with the findings that KLF5 interacts with Miz-1 regardless TGFβ status (Fig. 1E) and that several factors including Miz-1 form a complex with Myc on the p15 promoter to inhibit its transcription without TGFβ (10), we have concluded that KLF5 is part of the transcriptional assembly inhibiting p15 transcription without TGFβ (Fig. 6). Binding of KLF5 to the SBE, on the other hand, was TGFβ-dependent, which is similar to binding of Smad4 (Fig. 3) (10). Therefore, upon TGFβ activation, the KLF5-Smad4 complex binds to the SBE of p15 promoter to reverse its transcription.
FIGURE 6.
A model for how KLF5 reverses function in transcriptional regulation of p15 in the context of TGFβ. Without TGFβ, some KLF5 associates with Miz-1, Myc, and possibly other molecules on the Inr DNA elements to block p15 transcription, whereas other KLF5 associates with Smad4 in the nucleus. When TGFβ signaling is activated, Smad2 and Smad3 are phosphorylated and translocated into the nucleus (32), where they recruit p300 to the KLF5-Smad4 complex to acetylate KLF5 at lysine 369. Acetylated KLF5-Smad complex then binds to the SBE, assembling with p15 factors such FOXO3 and Miz-1 and altering the existing KLF5 transcriptional assembly bound to the SPS2 and Inr elements to release the transcription of p15. Question marks indicate unknown molecules.
Regarding the role of KLF5 acetylation in p15 regulation, we found that although KLF5 associates with the p300 acetylase even when its acetylation site is mutated (Fig. 4), interruption of KLF5 acetylation prevented the previously identified p300-Smad4 association (Fig. 4). In addition, acetylation of KLF5 is essential for the binding of KLF5 and other factors to the p15 promoter (Fig. 5). Taken together with the findings that p300 is located in the nucleus, activation of TGFβ recruits p300 to Smad complexes (13), and KLF5 interacts with Smads even in the absence of TGFβ, we propose that upon TGFβ activation, p300 is recruited to the KLF5-Smads complexes to acetylate KLF5. The complexes with acetylated KLF5 then bind to the SBE of p15 promoter, altering the binding of other factors to the p15 promoter to activate p15 transcription (Fig. 6).
In summary, we found that KLF5 interacted with other p15 regulators to repress p15 transcription without TGFβ but became an essential component of the transcriptional assembly to mediate TGFβ-induced p15 transcription. TGFβ-mediated acetylation of KLF5, which altered protein interactions and promoter bindings, was an essential modification for the reversal of KLF5 function. Such a mechanism could be valid for other genes regulated by both TGFβ and KLF5.
This work was supported, in whole or in part, by National Institutes of Health Grant R01CA087921. This work was also supported by the Georgia Cancer Coalition.
- TGFβ
- transforming growth factor-β
- HEK
- human embryonic kidney
- IP
- immunoprecipitation
- HA
- hemagglutinin
- GST
- glutathione S-transferase
- SBE
- Smad binding element
- ChIP
- chromatin immunoprecipitation
- TβRI
- TGFβ receptor type I
- IB
- immunoblot.
REFERENCES
- 1.Siegel P. M., Massagué J. ( 2003) Nat. Rev. Cancer 3, 807– 821 [DOI] [PubMed] [Google Scholar]
- 2.Massagué J., Blain S. W., Lo R. S. ( 2000) Cell 103, 295– 309 [DOI] [PubMed] [Google Scholar]
- 3.Ranganathan P., Agrawal A., Bhushan R., Chavalmane A. K., Kalathur R. K., Takahashi T., Kondaiah P. ( 2007) BMC Genomics 8, 98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Xie L., Law B. K., Aakre M. E., Edgerton M., Shyr Y., Bhowmick N. A., Moses H. L. ( 2003) Breast Cancer Res. 5, R187– 198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Seoane J., Le H. V., Shen L., Anderson S. A., Massagué J. ( 2004) Cell 117, 211– 223 [DOI] [PubMed] [Google Scholar]
- 6.Massagué J., Seoane J., Wotton D. ( 2005) Genes Dev. 19, 2783– 2810 [DOI] [PubMed] [Google Scholar]
- 7.Yu W., Gius D., Onyango P., Muldoon-Jacobs K., Karp J., Feinberg A. P., Cui H. ( 2008) Nature 451, 202– 206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Krimpenfort P., Ijpenberg A., Song J. Y., van der Valk M., Nawijn M., Zevenhoven J., Berns A. ( 2007) Nature 448, 943– 946 [DOI] [PubMed] [Google Scholar]
- 9.Hannon G. J., Beach D. ( 1994) Nature 371, 257– 261 [DOI] [PubMed] [Google Scholar]
- 10.Seoane J., Pouponnot C., Staller P., Schader M., Eilers M., Massagué J. ( 2001) Nat. Cell Biol. 3, 400– 408 [DOI] [PubMed] [Google Scholar]
- 11.Feng X. H., Liang Y. Y., Liang M., Zhai W., Lin X. ( 2002) Mol. Cell 9, 133– 143 [DOI] [PubMed] [Google Scholar]
- 12.Li J. M., Nichols M. A., Chandrasekharan S., Xiong Y., Wang X. F. ( 1995) J. Biol. Chem. 270, 26750– 26753 [DOI] [PubMed] [Google Scholar]
- 13.Feng X. H., Zhang Y., Wu R. Y., Derynck R. ( 1998) Genes Dev. 12, 2153– 2163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Janknecht R., Wells N. J., Hunter T. ( 1998) Genes Dev. 12, 2114– 2119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Simonsson M., Kanduri M., Grönroos E., Heldin C. H., Ericsson J. ( 2006) J. Biol. Chem. 281, 39870– 39880 [DOI] [PubMed] [Google Scholar]
- 16.Black A. R., Black J. D., Azizkhan-Clifford J. ( 2001) J. Cell. Physiol. 188, 143– 160 [DOI] [PubMed] [Google Scholar]
- 17.Bieker J. J. ( 2001) J. Biol. Chem. 276, 34355– 34358 [DOI] [PubMed] [Google Scholar]
- 18.Bateman N. W., Tan D., Pestell R. G., Black J. D., Black A. R. ( 2004) J. Biol. Chem. 279, 12093– 12101 [DOI] [PubMed] [Google Scholar]
- 19.Chen C., Bhalala H. V., Qiao H., Dong J. T. ( 2002) Oncogene 21, 6567– 6572 [DOI] [PubMed] [Google Scholar]
- 20.Chen C., Bhalala H. V., Vessella R. L., Dong J. T. ( 2003) Prostate 55, 81– 88 [DOI] [PubMed] [Google Scholar]
- 21.Nandan M. O., McConnell B. B., Ghaleb A. M., Bialkowska A. B., Sheng H., Shao J., Babbin B. A., Robine S., Yang V. W. ( 2008) Gastroenterology 134, 120– 130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Guo P., Dong X. Y., Zhang X., Zhao K. W., Sun X., Li Q., Dong J. T. ( 2009) J. Biol. Chem. 284, 6071– 6078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Boukamp P., Petrussevska R. T., Breitkreutz D., Hornung J., Markham A., Fusenig N. E. ( 1988) J. Cell Biol. 106, 761– 771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen C., Sun X., Ran Q., Wilkinson K. D., Murphy T. J., Simons J. W., Dong J. T. ( 2005) Oncogene 24, 3319– 3327 [DOI] [PubMed] [Google Scholar]
- 25.Chen C., Sun X., Guo P., Dong X. Y., Sethi P., Cheng X., Zhou J., Ling J., Simons J. W., Lingrel J. B., Dong J. T. ( 2005) J. Biol. Chem. 280, 41553– 41561 [DOI] [PubMed] [Google Scholar]
- 26.Zawel L., Dai J. L., Buckhaults P., Zhou S., Kinzler K. W., Vogelstein B., Kern S. E. ( 1998) Mol. Cell 1, 611– 617 [DOI] [PubMed] [Google Scholar]
- 27.Gomis R. R., Alarcón C., Nadal C., Van Poznak C., Massagué J. ( 2006) Cancer Cell 10, 203– 214 [DOI] [PubMed] [Google Scholar]
- 28.Mackay J. P., Crossley M. ( 1998) Trends Biochem. Sci 23, 1– 4 [DOI] [PubMed] [Google Scholar]
- 29.Chen C. R., Kang Y., Siegel P. M., Massagué J. ( 2002) Cell 110, 19– 32 [DOI] [PubMed] [Google Scholar]
- 30.Miyamoto S., Suzuki T., Muto S., Aizawa K., Kimura A., Mizuno Y., Nagino T., Imai Y., Adachi N., Horikoshi M., Nagai R. ( 2003) Mol. Cell. Biol. 23, 8528– 8541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Topper J. N., DiChiara M. R., Brown J. D., Williams A. J., Falb D., Collins T., Gimbrone M. A., Jr. ( 1998) Proc. Natl. Acad. Sci. U.S.A. 95, 9506– 9511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wotton D., Lo R. S., Lee S., Massagué J. ( 1999) Cell 97, 29– 39 [DOI] [PubMed] [Google Scholar]