Abstract
The molecular weight of hyaluronan is important for its rheological and biological function. The molecular mechanisms underlying chain termination and hence molecular weight control remain poorly understood, not only for hyaluronan synthases but also for other β-polysaccharide synthases, e.g. cellulose, chitin, and 1,3-betaglucan synthases. In this work, we manipulated metabolite concentrations in the hyaluronan pathway by overexpressing the five genes of the hyaluronan synthesis operon in Streptococcus equi subsp. zooepidemicus. Overexpression of genes involved in UDP-glucuronic acid biosynthesis decreased molecular weight, whereas overexpression of genes involved in UDP-N-acetylglucosamine biosynthesis increased molecular weight. The highest molecular mass observed was at 3.4 ± 0.1 MDa twice that observed in the wild-type strain, 1.8 ± 0.1 MDa. The data indicate that (a) high molecular weight is achieved when an appropriate balance of UDP-N-acetylglucosamine and UDP-glucuronic acid is achieved, (b) UDP-N-acetylglucosamine exerts the dominant effect on molecular weight, and (c) the wild-type strain has suboptimal levels of UDP-N-acetylglucosamine. Consistent herewith molecular weight correlated strongly (ρ = 0.84, p = 3 × 10−5) with the concentration of UDP-N-acetylglucosamine. Data presented in this paper represent the first model for hyaluronan molecular weight control based on the concentration of activated sugar precursors. These results can be used to engineer strains producing high molecular weight hyaluronan and may provide insight into similar polymerization mechanisms in other polysaccharides.
Hyaluronan (HA)3 is a linear polymer of a repeating disaccharide, β1–3 d-N-acetylglucosamine (GlcNAc) β1–4 d-glucuronic acid (GlcUA) (1) (see Fig. 1). Ubiquitous in the extracellular matrix in vertebrates, HA is particularly abundant in cartilage, synovial fluid, dermis, and the vitreous humor of the eye, where it serves specialized functions. HA also plays a critical role during fertilization and embryogenesis. In many group A and C streptococci, HA forms a capsule that helps these microbes evade the host immune system (2). HA molecular weight is important for the physiochemical as well as biological properties of HA. High molecular weight is important for HA to exert its unique rheological properties (3), for mucoadherence (4, 5), and anti-inflammatory effects (6, 7), whereas low molecular weight is a potent signaling molecule (8).
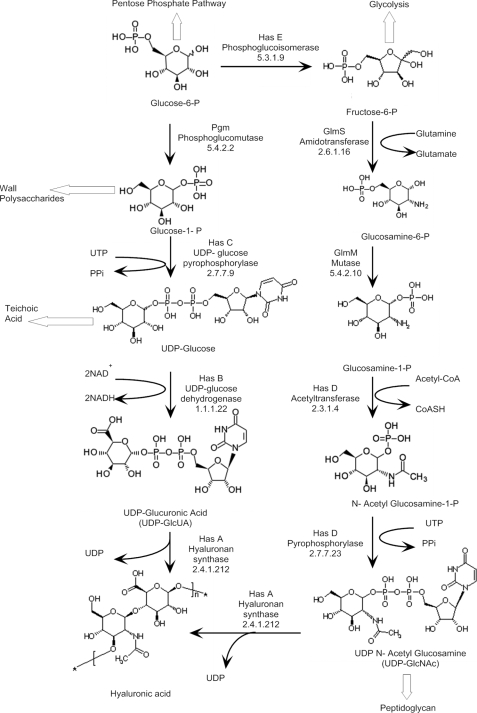
FIGURE 1.
Biosynthetic pathway of HA in S. zooepidemicus.
HA is produced by a processive synthase (9, 10) from the activated sugar precursors, UDP-glucuronic acid (UDP-GlcUA) and UDP-N-acetylglucosamine (UDP-GlcNAc) (see Fig. 1). In addition to the HA synthase (hasA), streptococcal has operons encode for one or more enzymes involved in biosynthesis of the activated sugars (11). The Streptococcus equi subsp. zooepidemicus (S. zooepidemicus) operon encodes for five genes: HA synthase (EC 2.4.1.212; hasA), UDP-glucose dehydrogenase (EC 1.1.1.22; hasB), UDP-glucose pyrophosphorylase (EC 2.7.7.9; hasC), a glmU paralog encoding for a dual function enzyme acetyltransferase and pyrophosphorylase activity (EC 2.3.1.4/EC 2.7.7.23; hasD), and a pgi paralog encoding for phosphoglucoisomerase (EC 5.3.1.9; hasE).
Although the biosynthetic mechanism is well established, little is known about what controls HA molecular weight. This is true not only for HA, but also for the highly abundant β-polysaccharides: cellulose, chitin, and 1,3-betaglucan. Molecular weight is partly an intrinsic parameter of the HA synthase. Weigel and colleagues have demonstrated that, at least in vitro, mutation of conserved cysteine or polar residues in streptococcal HA synthases results in reduced molecular weight with limited effect on biosynthetic rate (12–14). In a vertebrate HA synthase from Xenopus, the mutation of a serine or a cysteine residue yielded HA of higher, lower, or similar molecular weight depending on the amino acid substitution (15).
We and others have demonstrated that in vivo molecular weight is also affected by culture parameters, e.g. temperature and aeration (16–20). Although changed culture conditions affect the physiochemical environment of the HA synthase, a more likely explanation is that molecular weight is affected by the availability of activated sugar substrates (UDP-GlcUA and UDP-GlcNAc) as well as the concentration of possible effector molecules, such as free UDP (21). Although such a mechanism has been suggested for several processive synthases (22, 23), there has never been any direct evidence linking molecular weight to the concentration of a substrate.
Experimental support for the hypothesis has been obtained for the type 3 polysaccharide of Streptococcus pneumoniae (24). Like HA, the type 3 polysaccharide in S. pneumoniae is synthesized by a processive synthase from alternating addition of activated sugars, in this case UDP-glucose (UDP-Glc) and UDP-GlcUA. Mutants with reduced UDP-glucose dehydrogenase (“hasB”) activity not only produce less polysaccharide, but also polysaccharide with lower molecular weight (24). Although the levels of UDP-GlcUA were below detection in all strains, this supports the idea that UDP-GlcUA concentration controls molecular weight. Moreover, it is consistent with previous in vitro studies showing that low levels of UDP-GlcUA cause chain termination and hence low molecular weight (25). It was proposed that the concentration of UDP-GlcUA is critical for the successful transition from oligosaccharide lipid to highly processive polysaccharide synthesis (26). A similar mechanism is not likely for HA biosynthesis, because there is no indication that the HA synthase needs a primer (27).
In the present study, we manipulated metabolite concentrations in the HA pathway by overexpressing the five genes in the has operon of S. zooepidemicus (Fig. 1). Overexpression of these genes had a profound effect on HA molecular weight, which correlated with the levels of UDP-sugars and in particular, UDP-GlcNAc.
EXPERIMENTAL PROCEDURES
Bacterial Strains and Vector: Lactococcus lactis
MG1363 (L. lactis) (28) was used as an intermediate host for plasmids. The mucoid Group C S. zooepidemicus strain ATCC 35246 was obtained from the American Type Culture Collection (Rockville, MD). Plasmid pNZ8148 was obtained from the Dept. of Biophysical Chemistry, Netherlands Institute for Dairy Research (NIZO) (29) (Table 1).
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Relevant characteristicsa | Source |
|---|---|---|
| Strains | ||
| L. lactis MG1363 | Plasmid-free and prophage-cured derivative of NCDO 712 | (28) |
| S. equi subsp. zooepidemicus | HA+ Lac+ Ems | ATCC 35246 |
| Plasmids | ||
| pNZ8148 | Cmr, inducible expression vector, carrying the nisA promoter | (29) |
| pNZhasA | Cmr, pNZ8148 derivative containing a functional S. zooepidemicus hasA gene | This work |
| pNZhasB | Cmr, pNZ8148 derivative containing a functional S. zooepidemicus hasB gene | This work |
| pNZhasC | Cmr, pNZ8148 derivative containing a functional S. zooepidemicus hasC gene | This work |
| pNZhasD | Cmr, pNZ8148 derivative containing a functional S. zooepidemicus hasD gene | This work |
| pNZhasE | Cmr, pNZ8148 derivative containing a functional S. zooepidemicus hasE gene | This work |
| pNZhasED | Cmr, pNZhasE derivative containing a functional S. zooepidemicus hasD gene downstream of hasE | This work |
a Cmr, chloramphenicol resistance.
Construction of Recombinant Strains
All five genes found in the S. zooepidemicus has operon were amplified from the genome using specific primers (Table 2). PCR amplification of the genes was performed using a Platinum TaqDNA polymerase kit (Invitrogen) according to the manufacturer's instructions. The PCR program consisted of 1 cycle of 2 min at 94 °C, followed by 30 cycles of 94 °C (30 s), 52 °C (30 s), and 72 °C (2 min) and one final cycle of 72 °C (10 min). The PCR product sizes were confirmed on an agarose gel, and the bands were extracted using QIAquick Gel Extraction kit (Qiagen).
TABLE 2.
Oligonucleotide primers used
Endonuclease restriction sites are underlined.
| Primer name | Sequence (5′ → 3′) | Digestion site |
|---|---|---|
| HasAF | AGTCCATGGAATACAAAGCGCAAGAAAGGAAC | NcoI |
| HasAR | ATCGCATGCCTCCCTTGTCAGAACCTAGG | SphI |
| HasBF | GTCCATGGAAGAAATGAAAATTTCTGTAGCAGG | NcoI |
| HasBR | ATCGCATGCCTAGTCTCTTCCAAAGACATCT | SphI |
| HasCF | GTCCATGGAAGAACTCATGACAAAGGTCAGAAAAG | NcoI |
| HasCR | ATCGCATGCGCTCTGCAATAGCTAAGCCA | SphI |
| HasDF | GTCCATGGAAAGGAATCAAAACATGAAAAACTACG | NcoI |
| HasDR | ATCTCTAGAACTATAGCTTACTGGGGCTG | XbaI |
| HasEDF | CATCTAGACGAGGAATCAAAACATGAAAAACTACG | XbaI |
| HasEDR | CAAAGCTTTATAGCTTACTGGGGCTGATCCGGGTGATG | HindIII |
| HasD_F2 | ATGCAGTCATGATGGCAG | |
| hasD_R2 | TCCAACCTTTTCTTGGCTG | |
| HasEF | GTCCATGGAAGGGAGTAAAATAATGTCACATATTACA | NcoI |
| HasER | ATCGCATGCTTACAAGCGTGCGTTGA | SphI |
The primers introduced restriction enzyme sites as required and the PCR fragments were cloned into pNZ8148 (29) after digestion with the relevant restriction enzymes using standard recombinant DNA techniques (30). The plasmid-free L. lactis strain MG1363 (28) was employed as an intermediate host for all recombinant plasmids and selected on M17 agar supplemented with 5 μg ml−1 chloramphenicol (Cm). The full gene sequence and insertion site were confirmed with Sanger sequencing.
The plasmid containing the genes was subsequently isolated from MG1363 and electrotransformed into S. zooepidemicus. Recombinant strains of S. zooepidemicus were selected on M17 agar supplemented with 2.5 μg ml−1 Cm.
Electrotransformation of S. zooepidemicus
Cells were grown in M17 supplemented with 5 g liter−1 glucose (M17G). After 12 h of incubation, cells were inoculated into 100 ml of fresh M17G to 0.05 at A530 nm. The cells were further grown to about 0.6 at A530 nm. Prior to harvesting, 0.4 mg ml−1 of hyaluronidase was added followed by an additional incubation at 37 °C for another 30 min before centrifugation (Beckman Coulter, Avanti J-26 XPI, 5000 × g, 4 °C, 10 min). The supernatant was discarded, and the pellet was washed in 20 ml of 0.5 m sucrose solution. After centrifugation, the pellet was washed again in 1 ml of 0.5 m sucrose solution. Finally, the cells were resuspended in 250 μl of the same solution (31).
Electroporation was performed using a Bio-Rad Gene PulserTM with pulse control. Ice-cold cuvettes of path length 0.2 cm containing 40 μl of washed cells and up to 4 μl of purified plasmid DNA. Voltage was set at 3.0 kV (equivalent to 15 kV cm−1), resistance at 200 Ω, and capacitance at 25 microfarads. Immediately following pulse application, 1.0 ml of cold M17G broth was added to cells, which were then held on ice for 5 min prior to incubation at 37 °C for 2–3 h. Aliquots of electroporated cells (100 μl) were spread out on M17G agar plates supplemented with 2.5 μg ml−1 Cm. All plates were incubated overnight at 37 °C.
Growth Media and Cultivation Conditions
All chemicals were purchased from Sigma-Aldrich, unless otherwise specified. A chemically defined medium was modified from previously described media (32, 33) by adding 20 g liter−1 of glucose, 4.5 g liter−1 of acetate, and 50 mg liter−1 of uridine. The medium contained 2.5 μg ml−1 Cm and 20 ng ml−1 nisin.
Growth experiments were conducted in a 2-liter bioreactor (Applikon) at a working volume of 1.4 liter, and the temperature was maintained at 37 °C. The reactor was agitated at 300 rpm, and anaerobic conditions were maintained by top sparging nitrogen during fermentation. During the experiment, pH was maintained at 6.7 by 5 m NaOH and 5 m HCl additions. Gene overexpression was induced by adding 20 ng ml−1 of nisin to the medium. Growth was monitored every hour by measuring the optical density at 530 nm. Experiments were conducted at least in duplicates with independent gene insertions for the mutant strains.
Measurement of Biomass and Fermentation Products
Samples were collected hourly, and the optical density was measured at 530 nm and converted to biomass using the equation: Biomass (g liter−1) = A530 × 0.26 ± 0.01 (34).
The remaining sample was mixed with an equal volume of 0.1% SDS to remove the HA capsule and filtered through a syringe filter (Millex-GS MSE 0.45 μm) for cell removal. Lactate, acetate, formate, glucose, and ethanol were measured by high-performance liquid chromatography using a Bio-Rad HPX-87 H acid column with 1 m H2SO4 as eluant and a flow rate of 1 ml min−1. The HA concentration was measured using a turbidimetric quantification assay (35).
Measurement of Intracellular Metabolites
Intracellular metabolites were measured as described elsewhere (36). Briefly, 5 ml of cell suspension was centrifuged at 50,000 × g for 2 min at 37 °C and subsequently extracted into boiling ethanol. Extracts were enriched using SAX resin columns as described elsewhere (37), except that metabolites were eluted from columns using 2 ml of 0.15 m sodium citrate instead of sodium acetate. Metabolites were analyzed using high-performance anion-exchange chromatography and quantified via an integrated pulsed amperometric detector (36).
Assay of Enzyme Activities
Cells were harvested in the exponential phase of chemically defined medium fermentation cultures (A530 ∼ 2.0), treated with 0.4 mg ml−1 of hyaluronidase to remove HA, and pelleted at 5000 × g, 4 °C for 10 min. The supernatant was discarded, and the pellet was washed twice in wash buffer (50 mm potassium dihydrogen phosphate, 5 mm EDTA, pH 7, with 10% v/v glycerol, 4 °C). Finally, the pellet was resuspended in wash buffer containing protease inhibitor and mixed with 1.44 g of 100-μm glass beads. Tubes were chilled on ice for 5 min prior to disruption in a Mini Bead Beater (Biospec Products) through 5 cycles of 1 min beating at 5,000 rpm followed by 1 min on ice between cycles. After lysis, the sample was centrifuged (13,000 × g, 4 °C for 10 min), and the supernatant was aliquoted and stored at −80 °C until analysis. Protein content of the extracts was determined using a commercial kit (DC protein assay, Bio-Rad).
The activity of HasA was measured by incubating cell extract with UDP-GlcNAc and UDP-GlcUA for 1 h at 37 °C (38). The reaction was stopped by immersion in boiling water, before addition of 0.1% SDS to free HA molecules from membranes and measuring HA produced with the carbazole assay. The specific activity of 1 unit/mg of dry cell HA synthase is equivalent to 1 mg of HA generated/min/mg of dry cell.
HasB, HasC, and HasE activities were measured in NADH/NADPH-linked enzyme assays at room temperature. HasB activity was measured based on the conversion of UDP-Glc to UDP-GlcUA (39). HasC activity was measured based on conversion of glucose 1-phosphate to UDP-Glc coupled to conversion of UDP-Glc to UDP-GlcUA using excess UDP-Glc dehydrogenase (40). HasE activity was measured from the conversion of fructose 6-phosphate to glucose 6-phosphate coupled to conversion of glucose 6-phosphate to 6-phosphogluconate using excess glucose-6-phosphate dehydrogenase (41). Specific activities were expressed as nanomoles of substrate converted into product in 1 min for 1 mg of total cell protein.
HasD transcript levels were determined by reverse transcription-PCR with 500 ng of total RNA as a template using primers HasD_F2 and HasD_R2 amplifying a 398-bp DNA fragment of the hasD gene, followed by quantification of band intensity on a DNA gel (Scion Image, version 4.0.3.2).
Molecular Weight Determination
HA samples were purified from the broth by mixing 15 ml of culture with 15 ml of 0.1% w/v SDS and incubating at room temperature for 10 min. Samples were filtered through a 0.22-μm filter (Steritop-GP, 0.22 μm, polyethersulfone), mixed with 3 volumes of ethanol and left overnight at 4 °C. The mixture was centrifuged (Beckman Coulter, Avanti J-26 XPI, 9630 × g, 4 °C, 20 min), and the pellet was washed in 30 ml of ethanol/saline solution (75% w/v ethanol, 25% w/v 0.15 m NaCl) and again centrifuged (17,600 × g, 4 °C, 20 min). The pellet was allowed to dry overnight. The dry HA pellet was resuspended in 0.15 m NaCl with gentle rocking overnight at 4 °C, and undissolved matter was removed by centrifugation (17,600 × g, 4 °C, 20 min). Samples were filtered through a 0.22-μm filter (Millex-GS MSE) prior to analysis.
Two-dimensional Quant (Amersham Biosciences) was used to determine the protein content in 3 samples (WT, MT, and HasED) and compared against protein content in two clinical HA samples. A dilution series from 200 μg to 10 μg of HA was analyzed for each sample. In all 5 samples, the protein content was below detection limit (1 μg of bovine serum albumin) corresponding to a purity of 99.5%.
Intrinsic viscosity was measured with a Lauda Processor viscosity measuring system using an Ubbelohde Dilution Capillary (0.63-mm diameter and 5700-mm3 volume). All measurements were performed at 37 °C using 0.15 m NaCl as diluent. The average molecular weight was determined from the intrinsic viscosity using the Mark-Houwink-Sakurada equation (Equation 1),
with parameters obtained using standards of known molecular weight (Healon GV, Healon OVD, Sigma-Aldrich (cat. nos. H9390, 53747, and H5388)).
Purified HA samples were further analyzed by agarose gel electrophoresis as described in a previous study (42). Samples were separated using a Sub-Cell GT (Bio-Rad) and 0.5% agarose gels (25 cm long). Samples containing 7 μg of HA in 14 μl of MilliQ water were electrophoresed for 16 h at room temperature with a constant voltage of 50 V. Select-HA Hiladder and Mega-HA ladder (Hyalose) were used as a reference. Staining was performed as described previously (42).
Statistical Analysis
A total of 26 batch fermentations with 2 to 6 biological replicates per strain were performed for 8 strains: wild type (nWT = 5), empty plasmid (nMT = 6), hasA (nA = 2), hasB (nB = 2), hasC (nC = 2), hasD (nD = 2), hasE (nE = 4), and the double gene construct, hasED (nED = 3).
For each of the four measured parameters, molecular weight, growth rate, yield of HA, and yield of biomass, a linear model was fitted in R using the empty plasmid strain as control, e.g. MWi ∼ MWMT + ΔMWi, i.e. ΔMWi represents the difference from the empty plasmid for strain i, and the significance of the fitted parameter is the significance of the difference between strain i and the empty plasmid strain. Variance homogeneity was confirmed using a Bartlett test and normality of the residuals confirmed using a Shapiro-Wilk test. 95% confidence intervals were calculated using the pooled residual error on 18 degrees of freedom. Correlations between molecular weight and UDP-sugars were analyzed using the Spearman's rank correlation test in R.
RESULTS
has Operon Genes Were Successfully Expressed Using Nisin-inducible Vector
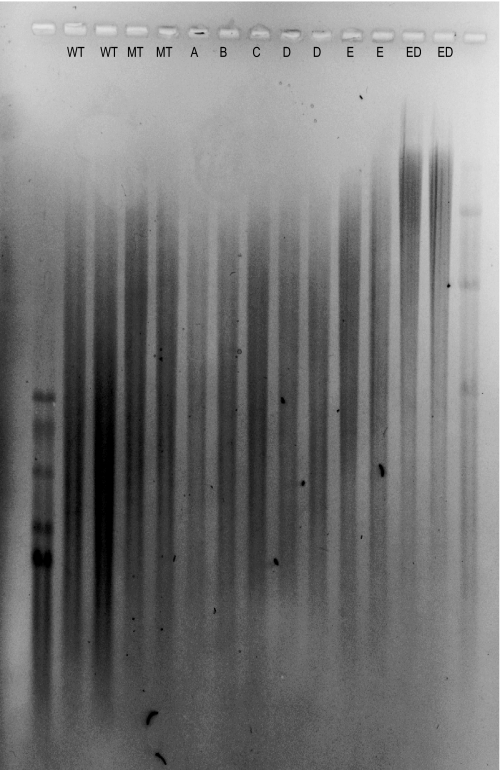
The five genes encoded by the has operon were cloned into pNZ8148 under the control of a nisin-inducible promoter. Following transformation, overexpression was confirmed with enzyme assays or, in the case of hasD, quantitative PCR (Table 3). Eight strains, wild type (WT), empty plasmid (pNZ8148), the five individual genes and the double gene construct, hasED, were characterized in a total of 26 batch fermentations performed in a 1.4-liter bioreactor. Fermentations were inoculated with strains from separate transformation events to avoid the impact of fortuitous or deleterious random mutations in individual colonies. Fermentations were characterized in terms of molecular weight using viscometry (Fig. 2), and the trends were confirmed using agarose gel electrophoresis (Fig. 3). Fermentations were also characterized in terms of growth rate, HA yield, and biomass yield (Fig. 4).
TABLE 3.
Activity of enzymes of corresponding overexpressed strains
Bacterial culture was grown in batch cultures in chemically defined medium on glucose as carbon source at 37 °C and a constant pH of 6.7. Samples were taken at log phase (A530 = 2.0) of the fermentation. Each value is the average of at least two measurements. HasD activity was obtained by quantitative PCR and -fold increase was 1.5 ± 0.1.
| Strain | WT specific activity | -Fold increase |
|---|---|---|
| HasA | 0.5 ± 0.0a | 1.5 ± 0.1 |
| HasB | 40.9 ± 1.0b | 5.0 ± 0.2 |
| HasC | 11.1 ± 0.5b | 4.7 ± 0.6 |
| HasE | 1500 ± 200b | 5.0 ± 0.9 |
a10−3 mg HA.min−1.mg protein−1.
b nmol.min−1.mg protein−1.
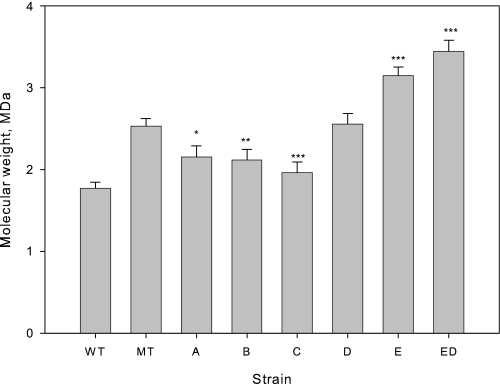
FIGURE 2.
Effect of has operon genes on molecular weight. Error bars indicate 95% confidence interval for the mean estimate. Statistical tests were performed against empty plasmid control (MT). p values indicated by the range: 0 < *** ≤ 0.001 ≤ ** ≤ 0.01 ≤ * ≤ 0.05 ≤ no asterisk ≤ 1.
FIGURE 3.
Agarose gel electrophoresis of HA produced by wild type and seven engineered strains was performed as described by Lee et al. (42). Hiladder (lane 1, molecular mass bands are 1510, 1090, 966, 572, and 495 kDa) and Mega-HA ladder (lane 15, molecular mass bands are 6100, 4570, 3050, and 1520 kDa) (Hyalose) were used as references.
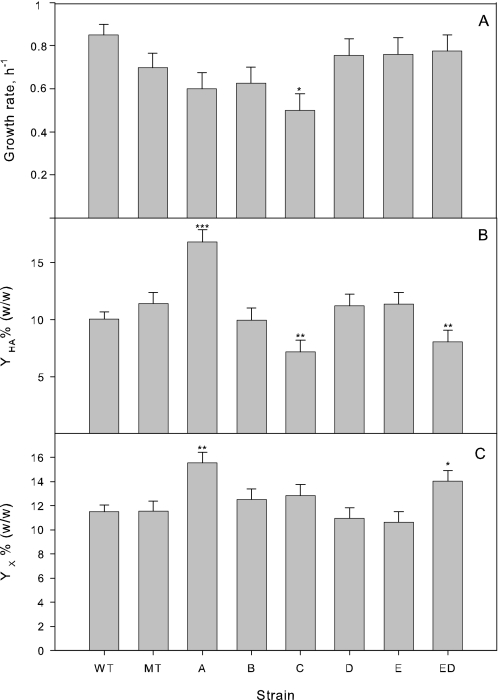
FIGURE 4.
Growth rates and yields during mid-exponential growth. Yields of HA and biomass are in mass percentage on glucose. Error bars indicate 95% confidence intervals standard error for the mean estimates. Statistical tests were performed against empty plasmid control (MT) and p values indicated by the range: 0 < *** ≤ 0.001 ≤ ** ≤ 0.01 ≤ * ≤ 0.05 ≤ no asterisk ≤ 1.
Empty Vector Plasmid Increases HA Molecular Weight
The empty plasmid (pNZ8148) had a large and significant effect on the molecular weight (Fig. 2). In the presence of the plasmid, the average molecular mass increased from 1.8 ± 0.1 MDa to 2.5 ± 0.1 MDa (p = 4 × 10−7). The plasmid effect was not due to the inducer, nisin. Including nisin in two wild-type fermentations yielded molecular masses of 1.9 and 1.8 MDa, whereas excluding nisin from a pNZ8148 fermentation yielded a molecular weight of 2.6. Assuming variance homogeneity, a linear model analysis (molecular weight ∼ plasmid + nisin) showed that the plasmid alone had a significant effect (p = 1 × 10−4), whereas nisin had no effect (p = 0.962).
Overexpression of has Operon Genes Affects Molecular Weight
Relative to the empty plasmid control, overexpression of hasA (p = 0.012), hasB (p = 0.006), and hasC (p = 5 × 10−4) had a negative effect on molecular weight (Fig. 2), whereas overexpression of hasE had a strong positive effect (p = 6 × 10−6). Overexpression of hasD alone had no effect (p = 0.843), but further increased molecular weight when combined with hasE (p = 2 × 10−6).
The growth rate of the wild-type strain was marginally (p = 0.047) higher than that of plasmid carrying strains. The strain overexpressing hasC grew significantly (p = 0.027) slower than the other engineered strains.
Our wild-type strain achieved a high HA yield on sugar of around 10% (w/w) (Fig. 4). The empty plasmid appears to increase yield, although not significantly (p = 0.162). Overexpression of hasA results in a 47% increase in yield (p = 4 × 10−4), whereas overexpression of hasC led to a 37% decrease in yield (p = 0.002). Strains overexpressing hasD and hasE have a HA yield similar to the empty plasmid strain, whereas the strain expressing both genes hasE and hasD had a significantly lower yield (p = 0.009).
The biomass yield appears to follow the reverse pattern of the HA yield, except both HA and biomass yields (p = 0.001) are high for strains overexpressing hasA. Among the remaining strains, only the increased biomass yield for strains overexpressing both hasD and hasE is statistically significant (p = 0.021).
Molecular Weight Correlates with UDP-GlcNAc Concentration
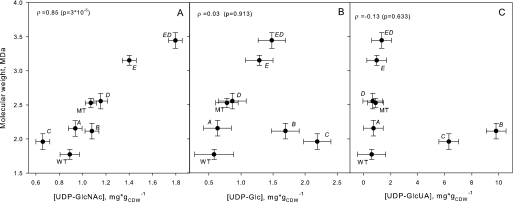
Overexpression of genes in the has operon other than the synthase is expected to affect the level of the activated precursors, UDP-GlcUA and UPD-GlcNAc. Overexpression of hasB and hasC resulted in a dramatic increase in UDP-GlcUA levels of 11-fold and 7-fold, respectively (Fig. 5). Overexpression of hasC also resulted in the expected increase in UDP-Glc levels. However, there was no correlation between UDP-GlcUA (or UDP-Glc) levels and the observed molecular weight (Spearman's ρ test, p = 0.633). In contrast, molecular weight was strongly correlated with UDP-GlcNAc (ρ = 0.84, p = 3 × 10−5). The absence of a molecular weight effect when overexpressing hasD alone was mirrored in the absence of a significant increase in UDP-GlcNAc, when overexpressing hasD alone. In combination with hasE, however, both UDP-GlcNAc and molecular weight achieved their highest values.
FIGURE 5.
Molecular weight versus UDP-sugar concentrations. The observed mean molecular weight is plotted against mean UDP-sugar level for each strain. Error bars indicate standard error of mean estimates. The Spearman's rank correlation ρ was calculated for each UDP-sugar versus molecular weight combination and tested for the hypothesis of no correlation, ρ = 0. p values for tests are shown in brackets.
DISCUSSION
The molecular weight of HA is important for its physiochemical properties and biological functions. In this study, we explored the commonly stated but unproven hypothesis that the molecular weight of polysaccharides produced by processive synthases depends on the availability of activated sugar substrates (23, 43). We cloned and overexpressed the five genes of the has operon in S. zooepidemicus (Table 3), to manipulate the levels of UDP-GlcUA and UDP-GlcNAc.
To our surprise, we noted a 41% increase in HA molecular weight in strains harboring the control plasmid (Fig. 2). The increase in molecular weight was not associated with the presence of the inducer nisin. Interestingly, increased molecular weight in response to an empty plasmid was also observed during heterologous production of HA in Bacillus subtilis (44) suggesting that foreign DNA stress is beneficial for HA molecular weight. Indeed, there is evidence that stress in general (plasmid stress, aerobic conditions, changes in pH, and low temperature) is beneficial for high molecular weight HA production (16, 17, 20). Transcriptome, proteome, and metabolome analyses are underway to identify the molecular basis of this stress effect.
Relative to the empty vector control, overexpression of the HA synthase gene (hasA) caused a significant increase in HA yield (Fig. 4) and a significant lowering of molecular weight (Fig. 2). The lowering in molecular weight may be explained by increased competition for a fixed pool of UDP-monomers leading to suboptimal levels of UDP-GlcUA and UDP-GlcNAc.
Overexpression of genes involved in UDP-GlcUA biosynthesis resulted in a significant increase in UDP-Glc (hasC) and UDP-GlcUA (hasB and hasC) (Fig. 5) associated with a decrease in molecular weight (Fig. 2). Increasing the activity of the last two steps in UDP-GlcNAc biosynthesis (hasD) had limited effect on molecular weight (Fig. 2) or UDP-GlcNAc concentration (Fig. 5) suggesting that an upstream step was limiting. This limitation could be overcome by overexpressing hasE; both hasE alone and hasED together significantly increased UDP-GlcNAc levels and molecular mass. At 3.4 ± 0.1 MDa, HA produced by the hasED strain has twice the molecular mass of that produced by the wild-type strain.
The mechanism in S. zooepidemicus is evidently different from transition model proposed for S. pneumoniae (see the introduction): 1) With the current evidence, the HA synthase is a monomeric, processive synthase that polymerize directly from UDP-sugars without the need for an initiation complex or covalent modifications (43). 2) Both UDP-GlcUA and UDP-GlcNAc are relatively abundant in S. zooepidemicus (Fig. 5). 3) Unlike S. pneumoniae, the HA yield did not correlate with molecular weight; the hasED strains had a significantly lower yield than the empty plasmid strains, whereas hasE had a similar yield (Fig. 4).
Our data indicate that maximum molecular weight requires an optimal balance between UDP-GlcUA and UDP-GlcNAc and that the balance in wild-type strains is toward UDP-GlcUA. Competition between UDP-GlcUA and UDP-GlcNAc has previously been observed to affect the rate of polymerization by streptococcal HAS (43). The competition is attributed to the affinity of UDP-GlcUA to the UDP-GlcNAc binding site, and vice versa. Competition is also seen with other sugar nucleotides and even UDP. It is possible that chain termination occurs when a site for one sugar nucleotide is blocked for too long by another sugar nucleotide.
If molecular weight is dictated by a competition between UDP-GlcUA and UDP-GlcNAc, there must be an optimum ratio of the two sugars, which we have not achieved in this study. This study, however, points to several strategies to further enhance molecular weight.
Overexpression of hasE (pgi) had a surprisingly strong effect in this study. S. zooepidemicus is a lactic acid bacterium with a highly active Embden-Meyerhof-Parnas pathway and ∼80% of all glucose is processed to pyruvate indicating that pgi activity is inherently high. Moreover, mucoid strains have 4-fold higher pgi transcript levels and 2-fold higher enzyme activity compared with non-mucoid strains, in which the has operon is silenced (11). Hence, the benefit from the 5-fold increase in the hasE strain must be due to shift in the ratio of glucose 6-phosphate and fructose 6-phosphate, rather than overcoming a metabolic bottleneck. If this is the case, lowering phosphofructokinase activity (or increasing Km for fructose 6-phosphate) would be an alternative strategy to increase fructose-6-phosphate, UDP-GlcNAc and hence molecular weight. Overexpression of other enzymes in the UDP-GlcNAc pathway (i.e. GlmS or GlmM) or feeding with glucosamine would be alternative strategies to enhance UDP-GlcNAc concentration.
HA has been produced in heterologous hosts, including Enterococcus faecalis (45), Escherichia coli (9, 38), B. subtilis (46), and L. lactis (47). Given superior tools for strain engineering of the latter four hosts, it may prove easier to engineer microbial high molecular weight HA producers by targeting the UDP-GlcNAc/UDP-GlcUA ratio in these hosts. Using these systems, the optimum ratio of UDP-GlcUA and UDP-GlcNAc for maximum molecular weight and yield of HA could be determined.
Acknowledgment
We thank Peter Abeydeera for the high-performance liquid chromatography analysis.
This work was financially supported by the Australian Institute for Bioengineering and Nanotechnology, the CRC Sugar Industry Innovation through Biotechnology, and the Mexican Council of Science and Technology.
- HA
- hyaluronan
- GlcUA
- glucuronic acid
- Glc
- glucose
- Cm
- chloramphenicol.
REFERENCES
- 1.Fong Chong B. F., Blank L. M., Mclaughlin R., Nielsen L. K. ( 2005) Appl. Microbiol. Biotechnol. 66, 341– 351 [DOI] [PubMed] [Google Scholar]
- 2.Wessels M. R., Moses A. E., Goldberg J. B., DiCesare T. J. ( 1991) Proc. Natl. Acad. Sci. 88, 8317– 8321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fouissac E., Milas M., Rinaudo M. ( 1993) Macromolecules 26, 6945– 6951 [Google Scholar]
- 4.Saettone M. F., Giannaccini B., Chetoni P., Torracca M. T., Monti D. ( 1991) Int. J. Pharm. 72, 131– 139 [Google Scholar]
- 5.Saso L., Bonanni G., Grippa E., Gatto M. T., Leone M. G., Silvestrini B. ( 1999) Res. Commun. Mol. Pathol. Pharmacol. 104, 277– 284 [PubMed] [Google Scholar]
- 6.Suzuki Y., Yamaguchi T. ( 1993) Agents Actions 38, 32– 37 [DOI] [PubMed] [Google Scholar]
- 7.Spurlock S. L., Spurlock G. H., Bernstad S., Michanek P., Chester S. T., Jr. ( 1999) J. Equine Vet. Sci. 19, 338– 344 [Google Scholar]
- 8.Lee J. Y., Spicer A. P. ( 2000) Curr. Opin. Cell Biol. 12, 581– 586 [DOI] [PubMed] [Google Scholar]
- 9.DeAngelis P. L., Papaconstantinou J., Weigel P. H. ( 1993) J. Biol. Chem. 268, 19181– 19184 [PubMed] [Google Scholar]
- 10.Weigel P. H., DeAngelis P. L. ( 2007) J. Biol. Chem. 282, 36777– 36781 [DOI] [PubMed] [Google Scholar]
- 11.Blank L. M., Hugenholtz P., Nielsen L. K. ( 2008) J. Mol. Evol. 67, 13– 22 [DOI] [PubMed] [Google Scholar]
- 12.Heldermon C. D., Tlapak-Simmons V. L., Baggenstoss B. A., Weigel P. H. ( 2001) Glycobiology 11, 1017– 1024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kumari K., Tlapak-Simmons V. L., Baggenstoss B. A., Weigel P. H. ( 2002) J. Biol. Chem. 277, 13943– 13951 [DOI] [PubMed] [Google Scholar]
- 14.Kumari K., Baggenstoss B. A., Parker A. L., Weigel P. H. ( 2006) J. Biol. Chem. 281, 11755– 11760 [DOI] [PubMed] [Google Scholar]
- 15.Pummill P. E., DeAngelis P. L. ( 2003) J. Biol. Chem. 278, 19808– 19814 [DOI] [PubMed] [Google Scholar]
- 16.Johns M. R., Goh L. T., Oeggerli A. ( 1994) Biotechnol. Lett. 16, 507– 512 [Google Scholar]
- 17.Armstrong D. C., Johns M. R. ( 1997) Appl. Environ. Microbiol. 63, 2759– 2764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fong Chong B., Nielsen L. K. ( 2003) Biochem. Eng. J. 16, 153– 162 [Google Scholar]
- 19.Blank L. M., McLaughlin R. L., Nielsen L. K. ( 2005) Biotechnol. Bioeng. 90, 685– 693 [DOI] [PubMed] [Google Scholar]
- 20.Huang W. C., Chen S. J., Chen T. L. ( 2006) Biochem. Eng. J. 32, 239– 243 [Google Scholar]
- 21.Park M. G., Jang J. D., Kang W. K. ( 1996) Lucky Limited, Seoul, Korea, U.S.Patent 5,496,726 [Google Scholar]
- 22.Spicer A. P., Kaback L. A., Smith T. J., Seldin M. F. ( 1998) J. Biol. Chem. 273, 25117– 25124 [DOI] [PubMed] [Google Scholar]
- 23.Cartee R. T., Forsee W. T., Schutzbach J. S., Yother J. ( 2000) J. Biol. Chem. 275, 3907– 3914 [DOI] [PubMed] [Google Scholar]
- 24.Ventura C. L., Cartee R. T., Forsee W. T., Yother J. ( 2006) Mol. Microbiol. 61, 723– 733 [DOI] [PubMed] [Google Scholar]
- 25.Forsee W. T., Cartee R. T., Yother J. ( 2000) J. Biol. Chem. 275, 25972– 25978 [DOI] [PubMed] [Google Scholar]
- 26.Forsee W. T., Cartee R. T., Yother J. ( 2006) J. Biol. Chem. 281, 6283– 6289 [DOI] [PubMed] [Google Scholar]
- 27.Weigel P. H. ( 2002) IUBMB Life 54, 201– 211 [DOI] [PubMed] [Google Scholar]
- 28.Gasson M. J. ( 1983) J. Bacteriol. 154, 1– 9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mierau I., Kleerebezem M. ( 2005) Appl. Microbiol. Biotechnol. 68, 705– 717 [DOI] [PubMed] [Google Scholar]
- 30.Sambrook J., Russell D. ( 2001) Molecular Cloning: A Laboratory Manual, 3rd Ed., Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 31.Fong Chong B. ( 2002) Improving the Cellular Economy of Streptococcus Zooepidemicus through Metabolic Engineering, Ph.D. thesis, Department of Chemical Engineering, University of Queensland, Brisbane [Google Scholar]
- 32.van de Rijn I., Kessler R. E. ( 1980) Infect. Immun. 27, 444– 448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chong B. F., Nielsen L. K. ( 2003) J. Biotechnol. 100, 33– 41 [DOI] [PubMed] [Google Scholar]
- 34.Goh L. T. ( 1998) Fermentation Studies of Hyaluronic Acid Production by Streptococcus zooepidemicus, Ph.D. thesis, Department of Chemical Engineering, University of Queensland, Brisbane [Google Scholar]
- 35.DI, Ferrante N. ( 1956) J. Biol. Chem. 220, 303– 306 [PubMed] [Google Scholar]
- 36.Marcellin E., Nielsen L. K., Abeydeera P., Krömer J. O. ( 2009) Biotechnol. J. 4, 58– 63 [DOI] [PubMed] [Google Scholar]
- 37.Jensen N. B., Jokumsen K. V., Villadsen J. ( 1999) Biotechnol. Bioeng. 63, 356– 362 [PubMed] [Google Scholar]
- 38.Yu H., Stephanopoulos G. ( 2008) Metab. Eng. 10, 24– 32 [DOI] [PubMed] [Google Scholar]
- 39.Dougherty B. A., van de Rijn I. ( 1993) J. Biol. Chem. 268, 7118– 7124 [PubMed] [Google Scholar]
- 40.Grobben G. J., Smith M. R., Sikkema J., de Bont J. A. ( 1996) Appl. Microbiol. Biotechnol. 46, 279– 284 [Google Scholar]
- 41.Degeest B., De Vuyst L. ( 2000) Appl. Environ. Microbiol. 66, 3519– 3527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lee H. G., Cowman M. K. ( 1994) Anal. Biochem. 219, 278– 287 [DOI] [PubMed] [Google Scholar]
- 43.Tlapak-Simmons V. L., Baggenstoss B. A., Kumari K., Heldermon C., Weigel P. H. ( 1999) J. Biol. Chem. 274, 4246– 4253 [DOI] [PubMed] [Google Scholar]
- 44.Sloma A., Behr R., Widner W., Tang M., Sternberg D., Brown S. ( 2003) Novozymes Biotech, Inc., U.S. Patent 2003/0175902 A1 [Google Scholar]
- 45.DeAngelis P. L., Papaconstantinou J., Weigel P. H. ( 1993) J. Biol. Chem. 268, 14568– 14571 [PubMed] [Google Scholar]
- 46.Widner B., Behr R., Von Dollen S., Tang M., Heu T., Sloma A., Sternberg D., Deangelis P. L., Weigel P. H., Brown S. ( 2005) Appl. Environ. Microbiol. 71, 3747– 3752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chien L. J., Lee C. K. ( 2007) Appl. Microbiol. Biotechnol. 77, 339– 346 [DOI] [PubMed] [Google Scholar]