Abstract
Purified RNA polymerase II initiated transcription from the yeast CUP1 promoter fused to a C-less cassette if the DNA was negatively supercoiled. Relaxed plasmid was not transcribed. Transcription did not require addition of any other transcription factors. TATA box-binding protein (TBP) was not detectable in the polymerase preparation and the TATA box was not required. Deletion analysis of the CUP1 promoter revealed that a 25-bp element containing the initiation region was sufficient for recognition by polymerase. Two transcription start sites were mapped, one of which is identical to one of the two major start sites observed in vivo. Our observations can be accounted for by using a theoretical analysis of the probability of DNA melting within the plasmid as a function of superhelix density: the CUP1 initiation element is intrinsically unstable to superhelical stress, permitting entry of the polymerase, which then scans the DNA to locate the start site. In support of this analysis, the CUP1 promoter was sensitive to mung bean nuclease. These observations and a previous theoretical analysis of yeast genes support the idea that promoters are stress points within the DNA superhelix. The role of transcription factors might be to mark the promoter and to regulate specific melting of promoter DNA.
The mechanism of transcript initiation is central to an understanding of gene regulation. In eukaryotes this mechanism is highly complex, involving a large number of proteins, most of which are now known to exist in large complexes (1). Basal transcription in vitro requires RNA polymerase II (Pol II) and a set of general transcription factors [TATA box-binding protein (TBP) and its associated TAFs, TFIIA, TFIIB, TFIIE, TFIIF, and TFIIH]. More protein complexes are required to communicate the activation signal from the transcription activator bound at elements in the promoter (or elsewhere) to the polymerase. These include multiple forms of mediator (2), which bind to the C-terminal domain (CTD) of the largest subunit of Pol II to form a holoenzyme. Regulation of a gene can occur at various stages: during recruitment of factors to the promoter, unwinding of promoter DNA to form an initiation complex, escape of polymerase from the promoter, elongation, and termination of the transcript.
We study the transcriptional regulation of the CUP1 gene from Saccharomyces cerevisiae because its biological function is clear and the mechanism of induction is well understood (3). It encodes a metallothionein which protects the cell against the toxic effects of copper. Induction occurs when copper ions enter the cell and bind to the N-terminal domain of the transcriptional activator Ace1 (encoded by ACE1/CUP2), inducing a conformational change to create a DNA-binding surface that specifically recognizes the upstream activating sequences in the CUP1 promoter (3, 4). Transcriptional activation occurs through the acidic C-terminal domain of Ace1. CUP1 can also be activated by heat shock factor in a separate, copper-independent, pathway (5, 6). In vivo, TBP has been detected at the CUP1 promoter (7, 8), but induction is independent of TFIIA (9, 10), TFIIE (11), the Kin28 CTD kinase of TFIIH, and some (but not all) components of mediator (12–14). Furthermore, Ace1 activates transcription independently of most of the TAFs (15). That CUP1 can be induced in vivo in the absence of many of the basal transcription factors suggests that its regulation might be relatively simple.
Our approach is to reconstitute the activity of CUP1 in vitro with purified yeast transcription proteins. We have purified a number of yeast transcription factors either in recombinant form from Escherichia coli (e.g., yTBP, yTFIIB), or from yeast (e.g., Pol II). To test the activities of our purified recombinant proteins, we made use of the observation that specific initiation by Pol II at certain promoters requires only a small subset of transcription factors (namely TBP, TFIIB, and, usually, TFIIF) if the DNA is negatively supercoiled (16, 17). However, we obtained the surprising result that only Pol II was required for initiation at two precise start sites in the CUP1 promoter on negatively supercoiled DNA. Our observations can be accounted for by using a theoretical analysis of the probability of DNA helix melting within the plasmid as a function of superhelix density (18, 19): the initiation element in the CUP1 promoter is a stress point in the plasmid, permitting entry of polymerase, which then scans the DNA to locate the start site.
Materials and Methods
A Yeast Strain with a Tagged Pol II.
YPH420 (ref. 20; gift of P. Hieter, Johns Hopkins University, Baltimore) (MATa leu2 trp1-Δ63 ura3–52 prb1–1122 pep4–3 prc1–407) was transformed to Ura+ with a BglII digest of pRP6B to obtain YDC-RP2. The presence of the tag was confirmed by Southern blotting. pRP3A was constructed by inserting the 1461-bp BglII fragment containing the 3′ end of RPB1 from pRPO21 (ref. 21; gift of J. Ingles, University of Toronto, Toronto) at the BglII site in pSP72Δ (constructed by religation of pSP72 (Promega) after cleavage with EcoRV and PvuII to delete the intervening restriction sites). In pRP4, the ApoI–SnaBI fragment containing the stop codon in pRP3A was replaced with the same sequence plus an extra 8 codons, constructed by ligating the AatII–ApoI 1075-bp fragment from pRP3A to annealed oligos (5′- AATTCCAGAAGTACTCATCATCACCATCACCATTGATATAGTATATCATCCTTAC and 5′-GTAAGGATGATATACTATATCAATGGTGATGGTGATGATGAGTACTTCTGG), digestion with AatII and SnaBI, and ligation into pRP3A cut with AatII and SnaBI. This results in extra coding sequence at the ApoI site located close to the stop codon of RPB1: Ser-Thr-His6, marked by a ScaI site. In pRP6B, the 1344-bp ClaI–DraI fragment from pYES2 (Invitrogen) containing URA3 was end-filled and inserted at the SwaI site downstream of RPB1 (such that URA3 and RPB1 are transcribed in opposite directions).
Purification of Tagged Pol II.
A 135-g portion of frozen fermenter-grown YDC-RP2 was thawed in 135 ml of Disruption Buffer [200 mM Tris–acetate, pH 8/50 mM potassium acetate/0.5 mM NaEDTA/0.5 mM NaEGTA/10 mM MgCl2/10% (vol/vol) glycerol/10 mM glutathione/1 mM 4-(2-aminoethyl)benzenesulfonyl fluoride (AEBSF)/10 μg/ml leupeptin/15 μg/ml pepstatin A/20 μg/ml chymostatin/10 μg/ml aprotinin/2 mM benzamidine]. The cells were lysed at 4°C by using an ice-cooled bead beater (Biospec Products, Bartlesville, OK): three disruption cycles at 45 sec per disruption with 1 min between each cycle for cooling. Cell debris was removed by centrifugation (16,000 rpm in a Sorvall SS-34 rotor, 4°C, 10 min). DE-52 resin (Whatman) preequilibrated in wash buffer [20 mM Tris-acetate, pH 8/0.15 M potassium acetate/10% (vol/vol) glycerol/10 mM glutathione/5 μg/ml leupeptin/0.1 mM AEBSF] was added to the supernatant (1 ml of wet settled resin per ml of supernatant). The suspension was mixed continuously on a tube rotator for 30 min at 4°C and poured into a precooled 350-ml Buchner funnel attached to a vacuum flask. The liquid was removed slowly. The resin was washed with 3 liters of wash buffer. Pol II activity was eluted with two 90-ml portions of elution buffer (20 mM Tris–acetate pH 8/0.5 M potassium acetate/10% (vol/vol) glycerol/10 mM glutathione/5 μg/ml leupeptin/0.1 mM AEBSF). Then 180 ml of DE-52 eluate were mixed with 8 ml of Talon resin (CLONTECH), preequilibrated in binding buffer [BB: 20 mM Tris–acetate, pH 8/0.1 M potassium acetate/10% (vol/vol) glycerol/10 mM glutathione/5 μg/ml leupeptin/0.1 mM AEBSF]. The suspension was mixed for 30 min as above. The resin was collected (Sorvall H1000B, 1,800 rpm, 4°C, 3 min), resuspended in 100 ml of BB, washed for 10 min on the rotator at 4°C, collected and washed twice more: (i) 50 ml BB with 1 mM imidazole⋅HCl; (ii) 50 ml of BB with 10 mM imidazole⋅HCl. Pol II was eluted with 40 ml of BB containing 60 mM imidazole⋅HCl, applied to a MonoQ 5/5 column (Amersham Pharmacia) preequilibrated in BB, and eluted by using a 0.1–1 M potassium acetate gradient in BB. Pol II eluted at 0.9–1.0 M salt. Pol II was dialyzed into 20 mM Tris–acetate, pH 8/0.1 M potassium acetate/50% (vol/vol) glycerol/0.1 mM NaEGTA/10 mM glutathione/5 μg/ml leupeptin/0.1 mM AEBSF and stored in aliquots at −80°C. Fractions were assayed as described in ref. 22. Antibodies used in immunoblotting were as follows: RGShis4 epitope (Qiagen, Chatsworth, CA), Pol II-CTD (MAb 8WG16, QED Biosciences, San Diego, CA), Tfg1, Gal11, yTBP, and Srb4 (gifts from R. Kornberg, Stanford University, Stanford, CA; M. Ptashne, Memorial Sloan–Kettering Cancer Center, New York; M. Green, University of Massachusetts, Worcester, MA; and R. Young, Massachusetts Institute of Technology, Cambridge, MA, respectively).
Plasmids.
CUP1 was obtained as a KpnI–NsiI fragment from YEp(CUP1)2A (ATCC 53233) and inserted into pUC19 cut with KpnI and PstI to yield pCP2. The first two C residues in the CUP1 transcript were converted to G by site-directed mutagenesis and a HpaI site was added at +25 (relative to the first major transcript start site; ref. 23) to give pCUP1C-less(30). A synthetic 380-bp C-less cassette was obtained as a blunted AseI–SacI fragment from pB20A inserted at HpaI/SacI to yield pCUP1C-less(410). pB20A was constructed by ligating an oligonucleotide containing an SP6 promoter (24) to the SmaI end of the SmaI–SacI fragment containing the C-less cassette from pCA2T (ref. 25; gift of M. Sawadogo, University of Texas, Houston) to yield a SacI fragment containing an SP6 promoter linked to the cassette, which was inserted into the SacI site of pB17A (with the GLN3 insert in reverse orientation relative to pB17B; ref. 24), such that the SP6 promoter is farthest from the GLN3 insert. pCUP1C-less(100) was constructed by using PCR to generate a shorter synthetic cassette (100 bp), by using the oligo 5′-CTGAGAATTCTATCCTCTCCTCACCTCTCCC, but otherwise the same as pCUP1C-less(410). pCUP1C-less(100)T1T10: the G residues at the in vivo start sites (20) were converted to T by directed mutagenesis using the oligo 5′-GCAATATCATATATAAGTGATGTAAATAGATATTAAGGT. For pCUP1C-less(Sph-Xba), pCUP1C-less(410) was cut with SphI and XbaI, blunted and religated. pCUP1(Nsi)C-less was constructed by altering the TATCATATA sequence immediately preceding the CUP1 start site in pCUP1C-less(410) to ATGCATATA to create an NsiI site. pC-less was obtained by deletion of the HindIII–NsiI fragment containing all of the upstream CUP1 sequences. In pC-lessΔIE, the CUP1 entire initiation region (from the NsiI site to the start of the synthetic cassette) was deleted. pCUP1C-less(410) at superhelix densities (σ) of 0, −0.08, −0.12, and −0.16 (corresponding to 0, 26, 40, and 53 negative supercoils) was prepared as described (26).
Transcription.
A 0.1-μg sample of DNA was preincubated with 2 μg of Pol II for 30 min in buffer at 30°C, NTPs were added, and incubation was for 1 h at 30°C in a final volume of 12 μl of 40 mM NaHepes, pH 7.5/0.1 M potassium acetate/7.5 mM magnesium acetate/5 mM 2-mercaptoethanol/0.1 mg/ml BSA/15 units of Prime RNase inhibitor (Eppendorf 5 Prime)/1.3 mM ATP/1.3 mM CTP or GTP/0.1 mM UTP with 8 μCi of [α-32P]UTP. RNA was purified and analyzed in polyacrylamide/urea gels.
Mung Bean Nuclease Analysis.
A 0.2-μg sample of pCUP1C-less(410) was mixed with 1.3 μg of λ DNA digested with BstEII as carrier in 40 μl of 50 mM NaCl/10 mM sodium acetate/10 mM MgCl2/10 mM Tris⋅HCl, pH 7.9/0.2 mM ZnCl2/1 mM DTT and digested with 3 or 9 units of mung bean nuclease (New England Biolabs) per μg of DNA for 30 min at 30°C. DNA was purified, digested with AatII, purified, electrophoresed in an alkaline 1.2% agarose gel (27), and blotted.
Results
Purification of Pol II from Yeast.
To facilitate purification of Pol II, a strain was constructed in which the C terminus (CTD) of the large subunit (Rpb1) was tagged with six histidine residues. A haploid strain (YPH420) carrying mutations in three genes encoding proteases was selected to reduce proteolysis during isolation. The chromosomal RPB1 gene was replaced directly with the tagged version. The shortest possible tag was added, separated from the wild-type C terminus by two hydrophilic residues, because certain extensions of the CTD are lethal (28). The growth rate of the tagged strain (YDC-RP2) was very similar to that of the parent strain in YPD (yeast extract/peptone/dextrose) medium. Thus, the tagged RNA polymerase is fully functional in vivo.
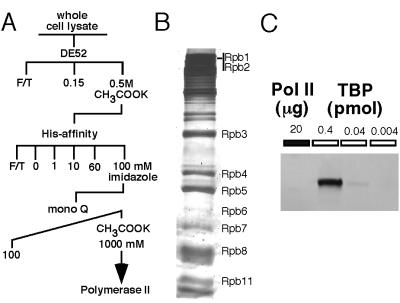
Pol II was purified to about 50% homogeneity (Fig. 1). Most of the subunits were identified by molecular weight, and the presence and identity of the large subunit were confirmed by Western blots using antibodies against the tag and Rpb1 (data not shown). The mediator subunits, Srb4 and Gal11, were not present in the preparation, as determined by Western blotting. (They were detected in the post-His affinity fraction; data not shown.) Thus, this preparation is not holoenzyme, although the presence of alternative holoenzymes cannot be eliminated. Because no TBP was detected (Fig. 1C), it can be deduced that there is less than 1 mol of TBP present per 300 mol of Pol II. The large subunit of TFIIF was also undetectable (data not shown).
Figure 1.
Purification of histidine-tagged Pol II from yeast. (A) Outline of purification scheme for 6-histidine-tagged Pol II. (B) Analysis of proteins in the Pol II preparation by SDS-PAGE. The subunits of Pol II (Rpb1–12) are indicated. (C) Western blot with anti-yTBP antibody. Twenty micrograms (about 15 pmol) purified Pol II was loaded onto the gel. The indicated amounts of purified recombinant yTBP were loaded onto the gel as quantitation standards.
Pol II Initiates Transcription from the CUP1 Promoter in Negatively Supercoiled Templates.
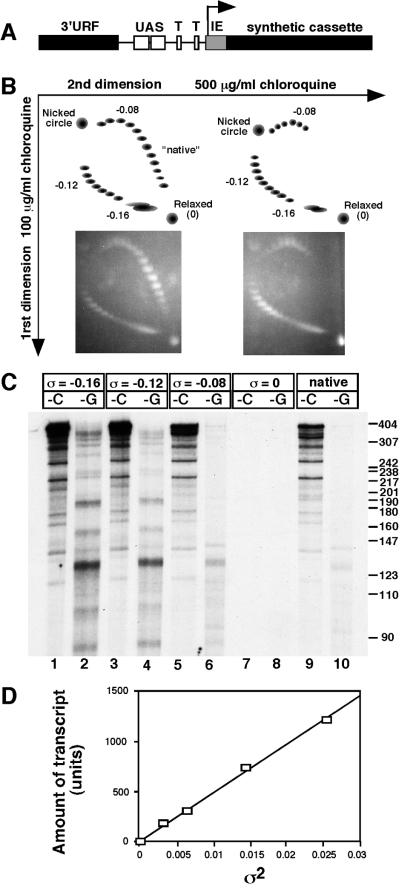
To reconstitute transcription of CUP1 in vitro, a number of yeast transcription factors were purified. To test whether these factors were functional, we exploited the fact that only a subset of transcription factors is required for basal transcription of certain promoters, provided that the DNA template is negatively supercoiled (16). We constructed a plasmid containing the CUP1 promoter fused to a C-less cassette [pCUP1C-less(410)]: the first 25 residues of the CUP1 transcript contain only two C residues, which were converted to G, and fused to a synthetic C-less cassette (ref. 25; Fig. 2A), resulting in a C-less region of 413 nt. The insert contains the entire CUP1 promoter and the 3′ flanking region of the gene upstream, referred to here as the 3′ unidentified reading frame (URF) region. This plasmid was prepared at different superhelix densities (Fig. 2B) and used as template in transcription reactions in the absence of CTP for transcription from the CUP1 promoter, or in the absence of GTP for transcription from the vector end of the cassette. Surprisingly, only Pol II was necessary to obtain specific transcripts originating in the CUP1 promoter (Fig. 2C); no general transcription factors were added. Since this result was unexpected, the mechanism of promoter recognition by Pol II was examined in more detail.
Figure 2.
Transcription of negatively supercoiled plasmids containing the CUP1 promoter by Pol II. (A) Map of the insert in pCUP1C-less(410). 3′URF, sequence from the termination region of the gene neighboring CUP1 in the yeast genome; UAS, upstream activating sequences (binding sites for Ace1 and HSF); T, putative TATA boxes; IE, initiation element (contains two point mutations as described in the text); synthetic cassette, synthetic C-less cassette. (B) Analysis of supercoiled pCUP1C-less(410) preparations in two-dimensional chloroquine gels. (Right) A mixture of plasmids prepared at superhelix densities: 0, −0.08, −0.12, and −0.16 (corresponding to 0, 26, 40, and 53 negative supercoils in a plasmid of 3474 bp). (Left) As the right panel but with plasmid prepared at native superhelix density (about −0.055) also added. (C) Transcription of negatively supercoiled pCUP1C-less(410) by purified Pol II. Analysis of radiolabeled transcripts in a sequencing gel. -C, -G: transcripts synthesized in the absence of CTP or GTP, respectively. Markers: pBR322 DNA digested with MspI. Superhelix densities (σ) are indicated. (D) Plot of amount of transcript initiated at the CUP1 promoter (phosphorimager units) against the square of the superhelix density for the data in C.
Transcripts were observed only with negatively supercoiled templates; no transcripts were observed with relaxed plasmid. Although the test DNAs were very negatively supercoiled, plasmid DNA as isolated from bacteria is sufficiently negatively supercoiled (about σ = −0.055) to yield the same transcripts (Fig. 2C). Thus, negative supercoiling is required for initiation. In the absence of CTP, a set of specific transcripts was observed, indicating that initiation is not random (nonspecific), since this would result in transcripts of all lengths. The most prominent transcript is about 405 nt long and therefore must have originated in the CUP1 promoter (Fig. 2C and see below). Quantitative analysis indicated that initiation at the promoter accounts for about 40% of all initiation events (after correction for transcript length). Three other prominent transcripts (about 280, 350, and 380 nt in length) account for about 10% of initiations each and were initiated within the synthetic cassette (because the same transcripts were obtained on transcription of a plasmid lacking the CUP1 promoter, pC-lessΔIE (Fig. 4B), and thus could not have originated from the promoter). In the absence of GTP, a smaller set of specific transcripts was obtained, including a prominent transcript of about 130 nt. About 5 times fewer transcripts were synthesized without GTP than without CTP, after taking into account the fact that they contain about twice as many uridine residues on average as their C-less counterparts and are, therefore, labeled to a higher specific activity. This suggests that DNA sequences recognized by Pol II occur more often on the C-less strand than on the G-less strand and that the CUP1 promoter contains a particularly efficient version of this sequence.
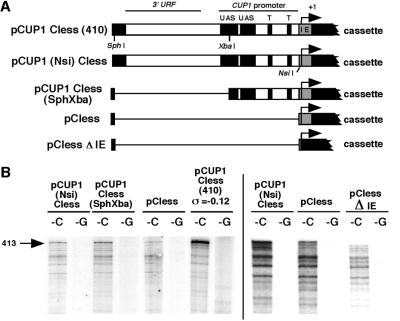
Figure 4.
Deletion analysis of the CUP1 promoter: transcription by Pol II requires only the initiation element. (A) Deletion constructs tested (abbreviations as in legend to Fig. 3). (B) Transcripts synthesized from deletion constructs by Pol II analyzed in a sequencing gel. Only the top part of the gel is shown. The bar separates two different experiments.
Negative supercoiling is necessary for transcription by Pol II probably because it decreases the stability of the DNA duplex, facilitating the melting that must occur before polymerase can initiate. That is, the free energy of supercoiling reduces the energy required to melt the DNA. The free energy of supercoiling is proportional to the square of the superhelix density. A plot of the amount of transcript initiated at the CUP1 promoter against the square of the superhelix density (data of Fig. 2C) yields a straight line (Fig. 2D). This suggests that the probability of initiation may be determined directly by the amount of supercoiling energy in the helix, which also governs the extent of unwinding.
Pol II Recognizes the Initiation Element in the CUP1 Promoter.
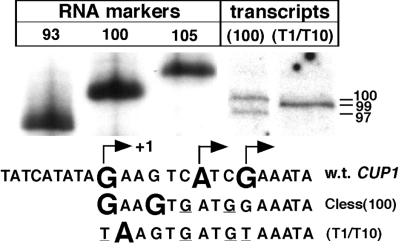
The precise start sites of transcription from the CUP1 promoter were determined by accurate measurement of transcripts derived from a plasmid containing a shorter cassette (100 bp) linked to the CUP1 promoter, relative to RNA markers in a sequencing gel (because DNA and RNA do not migrate identically in these gels) (Fig. 3). Just two transcripts were observed, indicating very selective initiation by Pol II in the CUP1 promoter. The start sites are four nucleotides apart and both correspond to G residues. The uncertainty in the measurement is one or two nucleotides and is due to band alignment problems and the presence of more than one band in the markers (markers were derived by T7 RNA polymerase run-off transcription). The three start sites observed in vivo (20) are indicated in Fig. 3. Of the two major start sites in vivo, one corresponds exactly with the upstream start site in vitro. When the G residues marking the major sites in vivo were both mutated to T, Pol II initiated at a single site, corresponding to the A next to the upstream G. Thus, the sequence of the initiation element in the CUP1 promoter determines the start site(s).
Figure 3.
The start sites within the initiation element of CUP1 are similar to those reported in vivo. Transcripts synthesized by Pol II from pCUP1C-less(100) [as pCUP1C-less(410) but with a shorter synthetic cassette] analyzed in a sequencing gel; sizes indicated at right. Markers: run-off transcripts synthesized by T7 RNA polymerase from pBluescript II SK(+) cut with XbaI, EagI, or BstXI. The sequence of the initiation region of CUP1 (wt) with the start sites in vivo marked (Karin) is shown above the mutated sequences used in pCUP1C-less(100) and pCUP1C-less(T1/T10) with observed start sites.
To determine how much of the CUP1 promoter is required for initiation by Pol II, some deletions were constructed by using a slightly altered version of pCUP1C-less(410) in which an NsiI site had been engineered close to the end of the cassette (pCUP1(Nsi)C-less) (Fig. 4). The plasmids were used as isolated (i.e., at native superhelix density). Deletion of the 3′ URF region and part of the distal upstream activating sequence in pCUP1C-less(Sph-Xba) had no effect on transcription. In fact, deletion of all of the CUP1 sequences upstream of the first in vivo transcription start site resulted in reduced transcription from the promoter but did not eliminate it (pC-less). This transcript originated in the CUP1 promoter, because when the initiation element was deleted to obtain a plasmid with just the synthetic cassette (pC-lessΔIE), the band disappeared and only the transcripts originating within the synthetic cassette were observed. Thus, this 25-bp C-less sequence, which includes all of the transcript initiation sites reported in vivo (20), is all that is required by Pol II for initiation from the CUP1 promoter. More transcripts were obtained if the rest of the CUP1 promoter was present, indicating that upstream sequences increase the probability of initiation from this element.
The CUP1 Promoter Is Relatively Unstable to Superhelical Stress.
The free energy of supercoiling is distributed throughout a circular DNA molecule, which exists in many interconverting conformations differing in writhe and twist. An extreme form of the latter is local melting of the helix, in which the DNA strands are completely untwisted. The ease with which a particular sequence melts depends not only on its base composition (AT-rich sequences melt more easily than GC-rich sequences) but also on its sequence (i.e., on neighboring base pairs). This raises the possibility that CUP1 promoter DNA might be intrinsically unstable to superhelical stress. Sequences more likely to melt than others can be predicted by using superhelical stress-induced duplex destabilization (SIDD) profiles: the sequence is scanned with an algorithm that predicts relatively unstable regions by using experimentally derived melting parameters (18, 19).
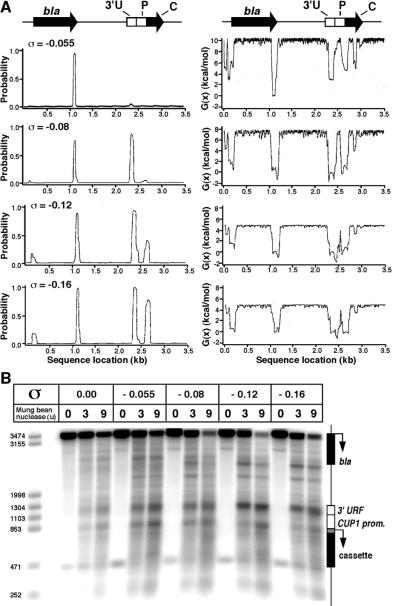
SIDD profiles for pCUP1C-less(410) at different superhelix densities are shown in Fig. 5A. Two types of plot are displayed: (i) the probability of opening the base pairs, and (ii) the destabilization energy, which is a measure of the energy required to keep the base pairs open. The latter plot has the advantage that it also reveals sites which are predicted to be unstable but not actually melted. At the lowest superhelix density (−0.08), there are two peaks in the probability profile: one at about coordinate 1100, corresponding to the termination region of the β-lactamase gene in the vector (previously identified as an unstable region; ref. 29) and the other at about 2350, corresponding to the 3′ URF region. These are the major peaks in the destabilization plot, but three other peaks are also apparent: at about coordinate 150 (the β-lactamase promoter), at 2650 (the CUP1 promoter), and at 2850 (within the C-less cassette). At higher superhelix densities, two of the secondary peaks (the promoters) also appear in the probability profiles. Thus, the CUP1 promoter is predicted to be relatively unstable to superhelical stress. The putative TATA boxes and the initiation element are included in the promoter peak, whereas the upstream activating sequences are located in between the 3′ URF and CUP1 promoter peaks.
Figure 5.
The CUP1 promoter is intrinsically unstable to superhelical stress: SIDD analysis and mung bean nuclease sensitivity. (A) SIDD analysis: Probability of melting (Left) and free energy of destabilization (Right) for the sequence of pCUP1C-less(410) as a function of superhelix density. bla, β-lactamase gene; 3′U, URF; P, CUP1 promoter; C, synthetic cassette. (B) Mapping of sites sensitive to mung bean nuclease in pCUP1C-less(410) as a function of superhelix density. pCUP1C-less(410) at the superhelix densities indicated was incubated with mung bean nuclease and cleavage sites were identified by indirect end-labeling using the AatII–NdeI (252 bp) fragment as probe. A Southern blot of an alkaline agarose gel is shown. Markers: pCUP1C-less(410) cut with AatII (3474 bp) and XmnI (3155 bp), SapI (1998 bp), HindIII (1304 bp), SpeI (1103 bp), MfeI (853 bp), SacI (471 bp), or NdeI (252 bp).
To assess whether the SIDD profile represents an accurate prediction of sites susceptible to melting, mung bean nuclease was used as a probe for single-stranded regions in pCUP1C-less(410) at different superhelix densities (the substrates in Fig. 2). Sites sensitive to mung bean nuclease were identified in an indirect end-labeling experiment: plasmid DNA was incubated with mung bean nuclease, linearized with AatII, and electrophoresed in a denaturing agarose gel. A Southern blot was probed with the 252-bp AatII–NdeI fragment (Fig. 5B). A specific set of bands was observed. The strongest band corresponds to the 3′ URF (coordinate 2340); the other strong band maps to the termination region of the β-lactamase gene (coordinate 1180). There are weaker bands at the CUP1 promoter (at coordinate 2680), mapping to the initiation region, and at a site near the plasmid origin of replication (coordinate 1700). With the exception of the latter site, these are the sites predicted by SIDD analysis. The rate of cleavage increased with superhelix density, but there is some cleavage at these sites in relaxed plasmid, suggesting that these regions retain some single-stranded character even in the absence of significant supercoiling.
Taken together, these observations suggest that the initiation element in the CUP1 promoter is intrinsically unstable to superhelical stress. The introduction of superhelical stress into the plasmid destabilizes the initiation element and allows Pol II to initiate at either of two precisely selected start sites.
Discussion
We have shown that Pol II recognizes an initiation element in the CUP1 promoter and initiates transcription at two precisely defined start sites, provided that the DNA is negatively supercoiled. This is a surprising result because no other transcription factors were required. Neither TBP, the large subunit of TFIIF, nor mediator components (Srb4 and Gal11) were detectable in the preparation. However, it is always possible that small amounts of these factors are present below the detection limits. In this regard, it is important to note that transcription was not dependent on the TATA box, which could be deleted.
Our observations are consistent with and extend previous observations (30, 31) demonstrating that purified Pol II initiates on supercoiled DNA in a nonrandom fashion. Others (16, 17, 32) have reported a minimal set of general transcription factors that can support transcription from supercoiled DNA—in these cases, only certain promoters were functional when linked to the same synthetic cassette in the reverse orientation (i.e., a G-less cassette). We did not observe any transcripts initiated from the CUP1 promoter when the cassette was reversed (data not shown), although in this case the initiation element was removed (otherwise GTP would be required for transcription). Whether the CUP1 promoter is typical of yeast promoters is unclear because it has so few factor requirements in vivo. If it is typical, then the sequence of the promoter and the link to the cassette are likely to be critical in determining whether a particular construct can be transcribed by Pol II alone. Certainly, some promoters are nonfunctional in transcription assays involving G-less cassettes, including several yeast promoters (33). The possibility that Pol II alone can initiate transcription at a promoter under certain conditions has potential for introducing artifacts in studies of the role of transcription factors, if the appropriate controls are not performed. This is unlikely to be a problem in experiments using crude extracts because these typically contain high levels of topoisomerase activity.
The role of negative supercoiling is presumably to decrease the local stability of the DNA duplex and thereby increase the probability of initiation, where a major step is the transition from the closed to the open complex in which the DNA strands are separated. The amount of free energy required to unwind a given stretch of DNA depends on base sequence and superhelix density and can be calculated by SIDD analysis. Certain DNA sequences within superhelical molecules are intrinsically less stable than others. These “stress points” correlate remarkably well with gene regulatory regions: in a theoretical analysis of a set of well studied yeast genes, all transcription termination regions and nearly all promoters were identified as stress points (19). CUP1 is no exception: its promoter is also predicted to be a stress point and its sensitivity to mung bean nuclease supports this contention.
Initiation at the CUP1 promoter occurs at precise start sites determined by the sequence of the initiation element. The sequence recognized is unlikely to be rare because initiation also occurred at several precise sites within the cassette, predominantly using the G-less strand as template. The obvious difference between the two strands is the presence of either G or C, but they also differ in their A/T contents, with about twice as much T in the G-less strand, which is therefore highly enriched in pyrimidines. The 25-bp CUP1 initiation element is also rich in pyrimidines on the template strand (about 70%) and is the most pyrimidine-rich region downstream of the distal TATA box. This is in accord with previous observations with purified Pol II and synthetic duplexes: Pol II selects pyrimidine-rich strands as template and prefers to initiate transcription with a purine (34). Indeed, many studies of elongation by Pol II have made use of the fact that Pol II will initiate efficiently on a template with a 3′ tail of C residues (pyrimidine tails are 10–100 times more efficient than purine tails) (35). Thus, it is suggested that Pol II transcribes the CUP1 promoter when negatively supercoiled, because promoter DNA is relatively easy to unwind and the initiation element contains a pyrimidine-rich strand to act as template. This would also determine the direction in which Pol II transcribes. The upstream sequences might increase initiation efficiency by increasing the instability of the promoter DNA (the stress point in the CUP1 promoter includes the TATA box region as well as the initiation element). Alternatively, they might be recognized directly by Pol II.
Both Pol II and TFIIB have been implicated in start site selection in budding yeast by genetic experiments (36). Certain mutations in the large subunit of Pol II (RPB1) have similar effects to mutations in the gene for TFIIB (SUA7), shifting the choice of start site downstream from the wild-type start sites, to previously minor start sites. In contrast, mutations in RPB9 shift the selected start site upstream from the wild-type sites, both in vivo and in vitro (37). A scanning model has been proposed based on detection of melted regions in the GAL1/GAL10 promoter in vivo, which extended from about 20 bp downstream of the TATA box to the initiation region (38): polymerase associates with the promoter, melts the DNA, and then scans the DNA to locate the start site. We speculate that our observations are relevant to the scanning mechanism: melting is aided by supercoiling, facilitating entry of polymerase into the helix, which then scans the DNA to locate the start site.
As suggested previously (16), the role of the transcription factors might be to mark the promoter and to help melt promoter DNA for polymerase, a process requiring the hydrolysis of ATP (39–41). It is reasonable to suppose that negative supercoiling of the template DNA might allow polymerase to bypass these requirements in vitro. If high superhelix densities occur in yeast cells, it is likely that they will exist only transiently, because of the presence of potent topoisomerase activities. Activation of CUP1 in vivo might involve the loss of a nucleosome from the promoter. If this is the case, it is a possible source of supercoiling: the superhelix density of DNA in a nucleosome is about −0.07 (one negative supercoil in 147 bp), which is within the relevant range of σ. If formation of a transcription complex were coupled with nucleosome loss, then this supercoil might be trapped within the complex and used to promote transcription, instead of being rapidly dissipated. Indeed, nucleosome loss in vivo results in activation of a CUP1-lacZ reporter gene independently of copper and Ace1 binding sites (42).
Acknowledgments
We thank Drs. J. Ingles and M. Sawadogo for plasmids; P. Hieter for YPH420; M. O'Dea and M. Gellert for nicking–closing extract; and M. Green, R. Kornberg, M. Ptashne, and R. Young for antibodies. We thank J. Dean, R. Kamakaka, and A. Kimmel for comments on the manuscript. This work was supported by grants to C.J.B. from the National Institutes of Health (R01-GM47012) and the National Science Foundation (DBI-99–04549).
Abbreviations
- Pol II
RNA polymerase II
- CTD
C-terminal domain
- AEBSF
4-(2-aminoethyl)benzenesulfonyl fluoride
- TBP
TATA box-binding protein
- URF
unidentified reading frame
- SIDD
stress-induced duplex destabilization
Footnotes
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.200365097.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.200365097
References
- 1.Ptashne M, Gann A. Nature (London) 1997;386:569–577. doi: 10.1038/386569a0. [DOI] [PubMed] [Google Scholar]
- 2.Chang M, Jaehning J A. Nucleic Acids Res. 1997;25:4861–4865. doi: 10.1093/nar/25.24.4861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fürst P, Hu S, Hackett R, Hamer D. Cell. 1988;55:705–717. doi: 10.1016/0092-8674(88)90229-2. [DOI] [PubMed] [Google Scholar]
- 4.Bachman C, Skroch P, Welch J, Fogel S, Karin M. Mol Cell Biol. 1989;9:4091–4095. doi: 10.1128/mcb.9.9.4091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Silar P, Butler G, Thiele D. Mol Cell Biol. 1991;11:1232–1238. doi: 10.1128/mcb.11.3.1232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lin J T, Lis J T. Mol Cell Biol. 1999;19:3237–3245. doi: 10.1128/mcb.19.5.3237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li X-Y, Virbasius A, Zhu X, Green M R. Nature (London) 1999;399:605–609. doi: 10.1038/21232. [DOI] [PubMed] [Google Scholar]
- 8.Kuras L, Struhl K. Nature (London) 1999;399:609–613. doi: 10.1038/21239. [DOI] [PubMed] [Google Scholar]
- 9.Ozer J, Lezina L E, Ewing J, Audi S, Lieberman P M. Mol Cell Biol. 1998;18:2559–2570. doi: 10.1128/mcb.18.5.2559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu Q, Gabriel S E, Roinick K L, Ward R D, Arndt K M. Mol Cell Biol. 1999;19:8673–8685. doi: 10.1128/mcb.19.12.8673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sakurai H, Fukasawa T. Biochem Biophys Res Commun. 1999;261:734–739. doi: 10.1006/bbrc.1999.1113. [DOI] [PubMed] [Google Scholar]
- 12.Lee D, Lis J T. Nature (London) 1998;393:389–392. doi: 10.1038/30770. [DOI] [PubMed] [Google Scholar]
- 13.McNeil J B, Agah H, Bentley D. Genes Dev. 1998;12:2510–2521. doi: 10.1101/gad.12.16.2510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lee D, Kim S, Lis J T. Genes Dev. 1999;13:2934–2939. doi: 10.1101/gad.13.22.2934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Moqtaderi Z, Keaveney M, Struhl K. Mol Cell. 1998;2:675–682. doi: 10.1016/s1097-2765(00)80165-3. [DOI] [PubMed] [Google Scholar]
- 16.Parvin J D, Sharp P A. Cell. 1993;73:533–540. doi: 10.1016/0092-8674(93)90140-l. [DOI] [PubMed] [Google Scholar]
- 17.Parvin J D, Shykind B M, Meyers R E, Kim J, Sharp P A. J Biol Chem. 1994;269:18414–18421. [PubMed] [Google Scholar]
- 18.Benham C J. J Chem Phys. 1990;92:6292–6305. [Google Scholar]
- 19.Benham C J. J Mol Biol. 1996;255:425–434. doi: 10.1006/jmbi.1996.0035. [DOI] [PubMed] [Google Scholar]
- 20.Nigro J M, Sikorski R, Reed S I, Vogelstein B. Mol Cell Biol. 1992;12:1357. doi: 10.1128/mcb.12.3.1357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Allison L A, Wong J K C, Fitzpatrick V D, Moyle M, Ingles C J. Mol Cell Biol. 1988;8:321–329. doi: 10.1128/mcb.8.1.321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kim Y J, Björklund S, Li Y, Sayre M H, Kornberg R D. Cell. 1994;77:599–608. doi: 10.1016/0092-8674(94)90221-6. [DOI] [PubMed] [Google Scholar]
- 23.Karin M, Najarian R, Haslinger A, Valenzuela P, Welch J, Fogel S. Proc Natl Acad Sci USA. 1984;81:337–341. doi: 10.1073/pnas.81.2.337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Studitsky V M, Clark D J, Felsenfeld G. Cell. 1994;76:371–382. doi: 10.1016/0092-8674(94)90343-3. [DOI] [PubMed] [Google Scholar]
- 25.Sawadogo M, Roeder R G. Proc Natl Acad Sci USA. 1985;82:4394–4398. doi: 10.1073/pnas.82.13.4394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Clark D J. In: Chromatin. A Practical Approach. Gould H, editor. Oxford Univ. Press; 1998. pp. pp.139–152. [Google Scholar]
- 27.McDonell M W, Simon M N, Studier F W. J Mol Biol. 1977;110:119–146. doi: 10.1016/s0022-2836(77)80102-2. [DOI] [PubMed] [Google Scholar]
- 28.Nonet M, Sweetser D, Young R A. Cell. 1987;50:909–915. doi: 10.1016/0092-8674(87)90517-4. [DOI] [PubMed] [Google Scholar]
- 29.Benham C J. Proc Natl Acad Sci USA. 1993;90:2999–3003. doi: 10.1073/pnas.90.7.2999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lescure B, Bennetzen J, Sentenac A. J Biol Chem. 1981;256:11018–11024. [PubMed] [Google Scholar]
- 31.Pedone F, Bellario P. Biochemistry. 1984;23:69–73. doi: 10.1021/bi00296a011. [DOI] [PubMed] [Google Scholar]
- 32.Usheva A, Shenk T. Cell. 1994;76:1115–1121. doi: 10.1016/0092-8674(94)90387-5. [DOI] [PubMed] [Google Scholar]
- 33.Lue N F, Kornberg R D. Proc Natl Acad Sci USA. 1987;84:8839–8843. doi: 10.1073/pnas.84.24.8839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sentenac A, Hall B. In: Molecular Biology of the Yeast Saccharomyces cerevisiae. Strathern J N, Jones E W, Broach J R, editors. Vol. 2. Plainview, NY: Cold Spring Harbor Lab. Press; 1982. pp. 561–606. [Google Scholar]
- 35.Dedrick R L, Chamberlin M J. Biochemistry. 1985;24:2245–2253. doi: 10.1021/bi00330a019. [DOI] [PubMed] [Google Scholar]
- 36.Hampsey M. Microbiol Mol Biol Rev. 1998;62:465–503. doi: 10.1128/mmbr.62.2.465-503.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hull M W, McKune K, Woychik N A. Genes Dev. 1995;9:481–490. doi: 10.1101/gad.9.4.481. [DOI] [PubMed] [Google Scholar]
- 38.Giardina C, Lis J T. Science. 1993;261:759–762. doi: 10.1126/science.8342041. [DOI] [PubMed] [Google Scholar]
- 39.Wang W, Carey M, Gralla J D. Science. 1992;255:450–453. doi: 10.1126/science.1310361. [DOI] [PubMed] [Google Scholar]
- 40.Tantin D, Carey M. J Biol Chem. 1994;269:17397–17400. [PubMed] [Google Scholar]
- 41.Pan G, Greenblatt J. J Biol Chem. 1994;269:30101–30104. [PubMed] [Google Scholar]
- 42.Durrin L K, Mann R K, Grunstein M. Mol Cell Biol. 1992;12:1621–1629. doi: 10.1128/mcb.12.4.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]