Abstract
FANCJ mutations are genetically linked to the Fanconi anemia complementation group J and predispose individuals to breast cancer. Understanding the role of FANCJ in DNA metabolism and how FANCJ dysfunction leads to tumorigenesis requires mechanistic studies of FANCJ helicase and its protein partners. In this work, we have examined the ability of FANCJ to unwind DNA molecules with specific base damage that can be mutagenic or lethal. FANCJ was inhibited by a single thymine glycol, but not 8-oxoguanine, in either the translocating or nontranslocating strands of the helicase substrate. In contrast, the human RecQ helicases (BLM, RECQ1, and WRN) display strand-specific inhibition of unwinding by the thymine glycol damage, whereas other DNA helicases (DinG, DnaB, and UvrD) are not significantly inhibited by thymine glycol in either strand. In the presence of replication protein A (RPA), but not Escherichia coli single-stranded DNA-binding protein, FANCJ efficiently unwound the DNA substrate harboring the thymine glycol damage in the nontranslocating strand; however, inhibition of FANCJ helicase activity by the translocating strand thymine glycol was not relieved. Strand-specific stimulation of human RECQ1 helicase activity was also observed, and RPA bound with high affinity to single-stranded DNA containing a single thymine glycol. Based on the biochemical studies, we propose a model for the specific functional interaction between RPA and FANCJ on the thymine glycol substrates. These studies are relevant to the roles of RPA, FANCJ, and other DNA helicases in the metabolism of damaged DNA that can interfere with basic cellular processes of DNA metabolism.
Fanconi anemia (FA)2 is an autosomal recessive disorder characterized by multiple congenital anomalies, progressive bone marrow failure, and high cancer risk (1–3). Cells from FA patients exhibit spontaneous chromosomal instability and hypersensitivity to agents that induce DNA interstrand cross-links. Although the precise mechanistic details of the FA pathway of interstrand cross-link-repair are not well understood, progress has been made in the identification of the FA proteins that are required for the pathway (1–3). Among the 13 FA complementation groups from which all FA genes have been cloned, only a few of the FA proteins are predicted to have direct roles in DNA metabolism. One of the more recently identified FA proteins shown to be responsible for complementation of the FA complementation group J is FANCJ (4–6). FANCJ was originally designated BACH1 (BRCA1-associated C-terminal helicase), which was discovered by Cantor et al. (7) as a protein that binds to the BRCT repeats of BRCA1. A genetic interaction between FANCJ and BRCA1 in double strand break repair was established (7), and FANCJ mutations were identified in early onset breast cancer (7–9), suggesting a tumor suppressor role of FANCJ.
FANCJ was first shown to be a DNA-dependent ATPase that catalytically unwinds duplex DNA with a 5′ to 3′ directionality (10). Consistent with its directionality, FANCJ requires nucleic acid continuity in the 5′-ssDNA tail near the ssDNA-dsDNA junction of the forked duplex substrate to efficiently initiate DNA unwinding (11). Further analysis demonstrated that FANCJ requires a 5′-ssDNA loading tail to unwind the adjacent DNA duplex (11). Although FANCJ fails to unwind a Holliday Junction structure, the helicase unwinds D-loop recombinational intermediates by releasing the third strand of the homologous recombination (HR) intermediate (11), an activity that may be important for its proposed role in an HR pathway of double strand break repair (1, 6, 7). FANCJ null cells have an HR defect (6), and FANCJ-depleted cells are mildly sensitive to ionizing radiation and have delayed resolution of ionizing radiation-induced double strand breaks (12). The sensitivity of FANCJ null cells to DNA interstrand cross-link agents (6) may be a result of a defective DNA metabolic event of the FA pathway that occurs downstream of FANCD2 monoubiquitination (6, 13). BRCA1 is required for the transport of FANCJ, BARD1, BRCA2, and Rad51 to sites of DNA damage where other proteins such as the MRE11-RAD50-NBS1 complex associate (14). The assembly of a BRCA1-FANCJ-BARD1 complex enables the interaction of BRCA1/FANCJ with TopBP1, a factor that plays an important role in the execution of the S phase checkpoint (14). The ability of RAD51 foci to form in FA-J cells (6) suggests that FANCJ does not operate upstream of Rad51 foci formation during HR repair. Rather, FANCJ may play a role in completing HR repair or preventing untimely or promiscuous recombination between homologous sequences.
In addition to HR, recent evidence suggests that FA proteins play a role in the response to replicational stress (15, 16). A recent study using Xenopus laevis oocyte extracts shows that FANCL is required to stabilize the replication fork (17). As a DNA helicase, FANCJ is a likely candidate to operate in this capacity, potentially extending its range of function beyond cross-link repair. Activation of FANCJ helicase activity is required for timely progression through S phase of the cell cycle (18); however, its precise functions in S phase progression remain to be understood. One source of genomic instability is alternate DNA structures such as DNA triplexes and quadruplexes that may impede the replication fork or interfere with transcription. Recently, FANCJ was shown to unwind G-quadruplex structures and have a role in G4 DNA metabolism (19).
There has been speculation that the underlying defect in FA and other chromosomal instability disorders may be due to an improper cellular response to oxidative stress. Bone marrow failure and leukemia progression in FA may be at least partly due to the accumulation of oxidative damage that induces excessive apoptosis of hematopoietic stem/progenitor cells (20).
In an effort to further understand the unwinding mechanism of FANCJ and its interaction with damaged DNA, we have investigated the effects of naturally occurring DNA base modifications on its helicase activity. Our findings demonstrate that FANCJ is uniquely sensitive to a single thymine glycol base modification in either the nontranslocating or translocating strands of the duplex DNA substrate. The ability of FANCJ to sense base damage in either strand of the DNA duplex as it unwinds may be important for its physiological functions in DNA damage signaling or repair. The existence of FANCJ in a BRCA1-containing complex with other DNA repair factors involved in the recognition and repair of aberrant DNA structures suggests that the complex functions in the DNA damage response (14). RPA, a nonspecific single-stranded binding protein that is required for cellular DNA metabolism (21) and shown to interact with FANCJ helicase (22), stimulates FANCJ helicase activity on the DNA substrates with a single thymine glycol in a strand-specific manner.
MATERIALS AND METHODS
Recombinant Proteins
Baculovirus encoding FANCJ with a C-terminal FLAG tag was used to infect High Five insect cells, and the recombinant FANCJ protein was purified as described previously (10). Purified recombinant FANCJ protein predominantly migrated as a single band of the predicted size (130 kDa) on an SDS-polyacrylamide gel, as reported previously (10). Recombinant RECQ1 (23) and an exonuclease-deficient form of WRN (WRN-E84A, designated X-WRN) (24) were purified as described previously. BLM was a kind gift from Dr. Ian Hickson (Cancer Research UK Laboratories). DinG was purified as described previously (25). UvrD was a kind gift from Dr. Steven Matson (University of North Carolina, Chapel Hill). DnaB was a kind gift from Dr. Daniel Kaplan (Vanderbilt University). Recombinant wild-type RPA heterotrimer or RPA heterotrimer with RPA70 missense mutation RPA70-QM (R216A, R234A, K263A, and E277A) (26) or RPA70-Zn* (C500S and C503S) (27) was purified as described previously.
DNA Substrates
PAGE-purified oligonucleotides used for the preparation of DNA substrates were purchased from Loftstrand Labs (Rockville, MD) and are listed in Table 1. DNA duplex substrates were 5′-32P-end-labeled and prepared as described previously (28).
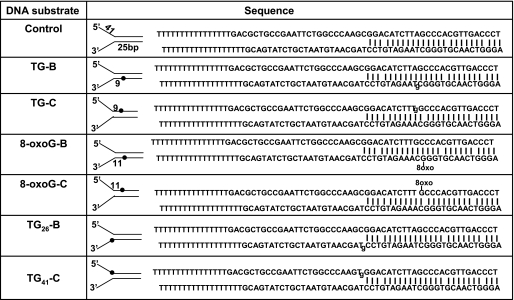
Table 1.
Oligonuleotide substrates used in this study
Tg, thymine glycol; 8-oxo-G, 8-oxoguanine.
Helicase Assays
Helicase reaction mixtures (20 μl) contained 10 fmol of the specified forked duplex DNA substrate (0.5 nm DNA substrate concentration) and the indicated concentrations of the specified helicase under previously described reaction conditions (FANCJ (11), BLM (same as FANCJ (11)), WRN (28), RECQ1 (23), DinG (25), UvrD (29), and DnaB (30)). Reactions were conducted under standard conditions for 15 min and initiated by the addition of helicase. Reactions were quenched with the addition of 20 μl of 2× Stop buffer (17.5 mm EDTA, 0.3% SDS, 12.5% glycerol, 0.02% bromphenol blue, 0.02% xylene cyanol). A 10-fold excess of unlabeled oligonucleotide with the same sequence as the labeled strand was included in the quench to prevent reannealing. The products of the helicase reactions were resolved on nondenaturing 12% (19:1 acrylamide/bisacrylamide) polyacrylamide gels. Radiolabeled DNA species in polyacrylamide gels were visualized using a PhosphorImager and quantitated using the ImageQuant software (Amersham Biosciences). The percent helicase substrate unwound was calculated by using the following formula: % unwinding = 100 × (P/(S + P)), where P is the product and S is the substrate. The values of the product and substrate have been corrected after subtracting background values in the no enzyme and heat-denatured substrate controls, respectively. Helicase data represent the mean of at least three independent experiments with standard deviation shown by error bars.
Helicase Sequestration Experiments
For helicase sequestration studies, FANCJ (9.6 nm) was preincubated with the indicated concentrations (0–12.5 nm) of the specified unlabeled forked duplex competitor DNA molecule (top strand thymine glycol or bottom strand thymine glycol) in standard helicase reaction buffer in the presence of ATP (2 mm) for 3 min at 30 °C. Ten fmol of radiolabeled forked 19-bp duplex molecules (tracker substrate (31)) was subsequently added to the reaction mixture and incubated for 7 min at 30 °C. Reactions were quenched and resolved on native polyacrylamide gels as described above. Percent helicase substrate unwound was calculated as described above. Typically, 60–70% of the tracker substrate was unwound in reactions lacking competitor DNA molecule. Helicase data (% control) were expressed relative to the control reactions lacking the competitor DNA.
DNA Binding Assays
Protein/DNA binding mixtures (10 μl) contained the indicated concentrations of RPA and 0.5 nm of the specified 32P-end-labeled DNA substrate in the same reaction buffer as that used for helicase assays but lacking ATP. The binding mixtures were incubated at 30 °C for 15 min after the addition of RPA. After incubation, 10 μl of Loading dye (25% glycerol, 18 mm EDTA, 0.04% xylene cyanol, 0.04% bromphenol blue) was added to each mixture, and samples were loaded onto native 6% (19:1 acrylamide/bisacrylamide) polyacrylamide gels and electrophoresed at 125 V for 3 h at 4 °C using 1× TBE as the Running buffer. The resolved radiolabeled species were visualized using a PhosphorImager and analyzed with ImageQuant software (Amersham Biosciences).
RESULTS
FANCJ Senses a Single Thymine Glycol Base Damage in either the Translocating or Nontranslocating Strands of a Duplex DNA Substrate
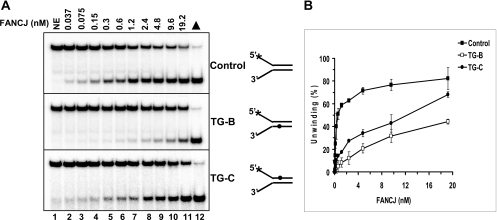
Increasing concentrations of FANCJ helicase were incubated with a fork duplex DNA substrate harboring a single thymine glycol positioned in either the top (translocating) or bottom strand (nontranslocating) within the double-stranded region of a forked duplex DNA substrate (Table 1). As a control, a forked duplex of identical sequence without a thymine glycol base damage in either strand was tested for FANCJ helicase activity. The control undamaged DNA substrate was unwound by FANCJ in a protein concentration-dependent manner throughout a titration range of 0.0375–0.6 nm FANCJ monomer (Fig. 1). Fifty percent of the substrate was unwound at 0.6 nm FANCJ. At greater concentrations of FANCJ, more substrate was unwound; however, the percent unwinding began to plateau, presumably due to substrate depletion effects. Throughout the FANCJ titration, significantly lower percentages of the forked duplex substrate with a thymine glycol in either the translocating or nontranslocating strands were unwound compared with the control DNA substrate. For example, only 13% of the substrate with a thymine glycol in the translocating strand was unwound at 0.6 nm FANCJ compared with 50% of the control substrate unwound. At 0.3 nm FANCJ, 10% of the damaged substrate was unwound compared with 40% of the control substrate. Although significant inhibition of unwinding by the translocating strand thymine glycol was observed at the lower FANCJ concentrations, increasing the FANCJ concentration to 19.2 nm resulted in nearly 70% of the damaged substrate unwound, indicating that the inhibition could be overcome to some extent by elevated FANCJ concentration.
FIGURE 1.
A single thymine glycol in either the translocating or nontranslocating strand inhibits DNA unwinding catalyzed by FANCJ. Helicase reactions (20 μl) were performed by incubating the indicated FANCJ concentrations with 0.5 nm forked duplex DNA containing a thymine glycol in the top strand (translocating), bottom strand (nontranslocating), or neither strand at 30 °C for 15 min under standard helicase assay conditions as described under “Materials and Methods.” A, lane 1, no enzyme control; lanes 2–11, indicated concentrations of FANCJ; lane 12, heat-denatured DNA substrate control; ▲, heat-denatured DNA substrate control. A phosphorimage of a typical gel from helicase assays with each DNA substrate is shown. B, quantitative analyses of FANCJ helicase data are shown. Filled square, control undamaged forked duplex substrate; open square, substrate with bottom strand thymine glycol (TG-B); filled circle, substrate with top strand thymine glycol (TG-C). Helicase data represent the mean of at least three independent experiments with S.D. indicated by error bars.
For the DNA substrate with a thymine glycol in the nontranslocating strand, FANCJ helicase activity was inhibited to a significantly greater extent throughout the helicase titration range compared with the undamaged substrate (Fig. 1). For example, at 0.6 nm FANCJ, only 5% of the substrate with the nontranslocating strand thymine glycol was unwound, indicating potent inhibition. Even at the 19.2 nm FANCJ concentration, only 43% of the substrate with the nontranslocating strand thymine glycol was unwound. These results demonstrate that FANCJ is sensitive to a single thymine glycol located in either the translocating or nontranslocating strand of the duplex region of the DNA substrate.
Effects of the Thymine Glycol Adduct on the Unwinding Reactions Catalyzed by Other DNA Helicases
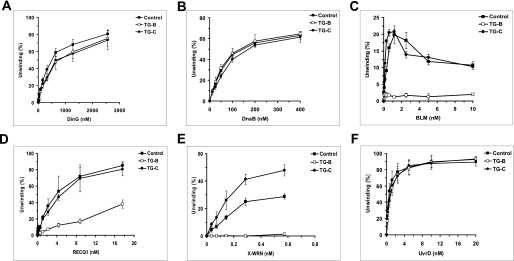
The inhibition of FANCJ DNA unwinding by a thymine glycol in either the translocating or nontranslocating strand of the duplex raised the question if other helicases are inhibited in a similar manner or if FANCJ is uniquely sensitive to the base damage in either strand of the substrate. To address this issue, we first tested the SF2 5′ to 3′ Escherichia coli DNA helicase DinG (32), which has significant sequence homology to FANCJ in the seven ATPase/helicase motifs of the helicase core domain (7) as well as the conserved iron-sulfur (Fe-S) cluster domain (33). As shown in Fig. 2A, considerably greater concentrations of DinG compared with FANCJ were required to detect unwinding of the control substrate, consistent with earlier published results for DinG helicase activity on similar DNA substrates (25). DinG was only mildly affected by the thymine glycol in either strand of the duplex substrate throughout the DinG protein titration. The greatest difference, ∼1.4-fold, was observed at a DinG concentration of 375 nm. Unlike FANCJ, no significant difference in DNA unwinding by DinG was observed between the substrates harboring thymine glycol in either the top or bottom strands of the DNA substrate, and overall DinG helicase activity was resistant to inhibition by the thymine glycol in either strand.
FIGURE 2.
Comparison of DNA unwinding of thymine glycol substrates by various DNA helicases. Helicase reactions (20 μl) were performed by incubating the indicated concentrations of the specified helicase (A, DinG; B, DnaB; C, BLM; D, X-WRN; E, RECQ1, and F, UvrD) with 0.5 nm forked duplex DNA containing a thymine glycol in either the top or bottom strand at the indicated temperature and reaction conditions for 15 min as described under “Materials and Methods.” Quantitative analyses of helicase data are shown. Filled square, control undamaged forked duplex substrate; open square, substrate with bottom strand thymine glycol (TG-B); filled circle, substrate with top strand thymine glycol (TG-C). Helicase data represent the mean of at least three independent experiments with S.D. indicated by error bars.
We next examined DNA unwinding of the thymine glycol substrate and control substrate by DnaB, an SF3 5′- to 3′-hexameric DNA helicase in E. coli that operates at the replication fork (34). Unlike FANCJ, DnaB was insensitive to the thymine glycol in either strand of the substrate throughout the DnaB protein titration (Fig. 2B). Thus, neither DnaB nor DinG 5′- to 3′-helicases displayed a sensitivity to the thymine glycol adduct in either strand of the duplex that was remotely comparable with the inhibition of DNA unwinding observed for FANCJ.
To further examine the apparent specificity of thymine glycol for inhibiting FANCJ unwinding, we tested several 3′- to 5′-RecQ DNA helicases implicated in the maintenance of chromosomal stability (35, 36) on the DNA substrates used to analyze the unwinding activity of FANCJ, DinG, and DnaB. Three human SF2 RecQ helicases, Bloom syndrome helicase (BLM), RECQ1, and an exonuclease-deficient missense mutant version of the Werner syndrome helicase (X-WRN), were tested. BLM was strongly inhibited by the thymine glycol adduct positioned in the bottom translocating strand of the forked duplex substrate; however, greater BLM helicase activity was observed on the substrate with thymine glycol residing in the top nontranslocating strand compared with the undamaged substrate at BLM concentrations up to 1.25 nm (Fig. 2C). For RECQ1, little to no inhibition of DNA unwinding was observed when the thymine glycol was positioned in the top (nontranslocating) strand of the DNA substrate; however, inhibition of RECQ1 helicase activity throughout the RECQ1 protein titration range was observed when the thymine glycol resided in the bottom translocating strand (Fig. 2D). For X-WRN, the DNA substrate with thymine glycol in the bottom translocating strand completely blocked DNA unwinding throughout the helicase titration range (0.036–0.58 nm) (Fig. 2E). However, for the DNA substrate with the nontranslocating strand modification, X-WRN helicase activity was significantly increased throughout the protein titration range compared with the undamaged substrate (Fig. 2E). These results indicate that BLM and X-WRN helicases are strongly inhibited by the thymine glycol when the base damage exists in the translocating strand. RECQ1 was also inhibited by the translocating strand thymine glycol, but not as severely as BLM or X-WRN. Unlike FANCJ, all three human RecQ helicases were not inhibited by the nontranslocating strand thymine glycol. In fact, DNA unwinding by all concentrations of X-WRN or the lower concentrations of BLM was greater on the DNA substrate with thymine glycol in the nontranslocating strand compared with undamaged substrate.
Finally, we tested the bacterial SF1 3′- to 5′-UvrD helicase implicated in mismatch repair and nucleotide excision repair (37), and we found that neither the translocating nor nontranslocating strand thymine glycol had any effect on its unwinding activity (Fig. 2F). Therefore, UvrD behaves very similar to DnaB in terms of its lack of sensitivity to the thymine glycol adduct. Collectively, for the DNA helicases tested, the results demonstrate that FANCJ is uniquely sensitive to the thymine glycol based on the inhibition of DNA unwinding when the thymine glycol resides not only in the strand that FANCJ translocates but also in the nontranslocating strand.
Effect of the 8-Oxoguanine Adduct on FANCJ Helicase Activity
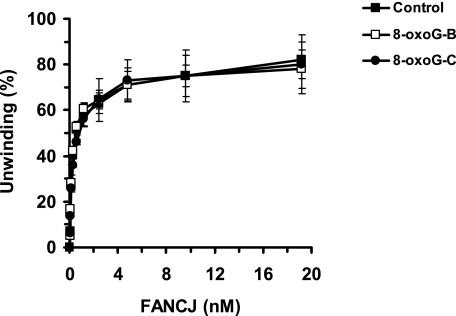
Inhibition of FANCJ helicase activity by a single thymine glycol residing within the duplex in either the translocating or nontranslocating strand raised the question if FANCJ might be sensitive to other types of prominent oxidative base lesions. Therefore, we tested FANCJ helicase on a set of DNA substrates very similar in sequence to that of the thymine glycol substrates with a single 8-oxoguanine in either the nontranslocating or translocating strands (Table 1). FANCJ was insensitive to the 8-oxoguanine lesion positioned in either strand of the helicase substrate (Fig. 3). Thus, the inhibition of FANCJ helicase activity by the thymine glycol damage is specific and not a general effect exerted by a different form of oxidative base modification, namely the 8-oxoguanine lesion.
FIGURE 3.
FANCJ helicase activity is not inhibited by 8-oxoguanine positioned in either strand of the DNA substrate. Helicase reactions (20 μl) were performed by incubating the indicated FANCJ concentrations with 0.5 nm forked duplex DNA containing an 8-oxoguanine in the top strand (translocating), bottom strand (nontranslocating), or neither strand at 30 °C for 15 min under standard helicase assay conditions as described under “Materials and Methods.” Quantitative analyses of FANCJ helicase data are shown. Filled square, control undamaged forked duplex substrate; open square, substrate with bottom strand 8-oxoguanine (8-oxoG-B); filled circle, substrate with top strand 8-oxoguanine (8-oxoG-C). Helicase data represent the mean of at least three independent experiments with S.D. indicated by error bars.
RPA Stimulates FANCJ Helicase Unwinding of the DNA Substrate with a Thymine Glycol in a Specific Manner
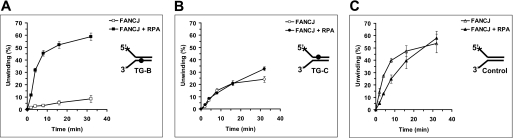
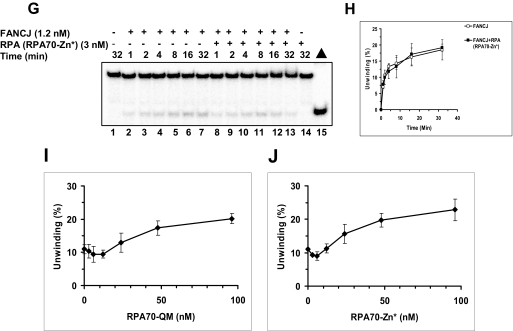
The inhibition of FANCJ helicase activity by a single thymine glycol adduct raised the question if FANCJ might have a mechanism to efficiently unwind past the base damage through an interaction with its protein partner RPA. Previously, we reported that RPA physically and functionally interacts with FANCJ (22) and other human DNA helicases (RECQ1, WRN, and BLM) (35). To examine if RPA can stimulate FANCJ helicase activity on the thymine glycol DNA substrates, we performed kinetic assays. Using a limiting concentration of FANCJ (1.2 nm), we observed a significant stimulation of DNA unwinding by RPA on the substrate with thymine glycol in the bottom nontranslocating strand (Fig. 4A) but not the top translocating strand (Fig. 4B) or when the thymine glycol is absent from the DNA substrate altogether (Fig. 4C). We have shown previously that RPA can effectively stimulate FANCJ helicase activity on a longer (47 bp) undamaged forked duplex substrate and that FANCJ catalyzes a limited DNA unwinding reaction (22). This stimulation was not observed with the 25-bp substrate used in this study and other short (20 bp) forked duplex substrates (Fig. 4C).3 Thus, these data suggest that RPA enables FANCJ to overcome a rate-limiting step in the DNA unwinding reaction on longer duplexes and that this step is not limiting with short undamaged duplexes. Kinetic analyses of initial rates revealed a 12-fold increase in unwinding for the nontranslocating strand thymine glycol substrate when RPA was present in the FANCJ helicase reaction compared with the reaction containing FANCJ alone. These results indicate that RPA was able to stimulate FANCJ helicase activity when the thymine glycol resided in the nontranslocating strand, but not the translocating strand.
FIGURE 4.
RPA stimulates FANCJ helicase activity on the thymine glycol substrates in a strand-specific manner. Helicase reaction mixtures contained 3 nm RPA, 1.2 nm FANCJ, and 0.5 nm forked DNA substrate with thymine glycol in the bottom strand (nontranslocating) (TG-B) (A), top strand (translocating) (TG-C) (B), or neither strand (C) under standard helicase assay conditions as described under “Materials and Methods” for the indicated times from 0 to 32 min. Quantitative analyses of FANCJ helicase data are shown (open squares, FANCJ; filled squares, FANCJ + RPA).
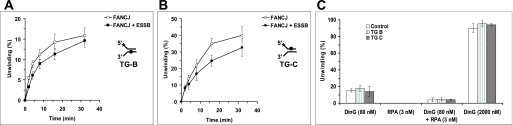
To address the specificity of stimulation of FANCJ helicase activity by RPA, we tested the effect of E. coli SSB on the DNA unwinding reactions catalyzed by FANCJ on the substrates with thymine glycol in either the nontranslocating or translocating strands. The results from these experiments demonstrated that E. coli SSB did not stimulate FANCJ unwinding of either thymine glycol substrate (Fig. 5, A and B). In fact, some inhibition of FANCJ helicase activity was observed for both substrates in the presence of E. coli SSB. These results suggest that the stimulation of FANCJ helicase activity on the DNA substrate with the nontranslocating strand thymine glycol by RPA is specific.
FIGURE 5.
RPA stimulation of FANCJ helicase activity on the DNA substrate with the nontranslocating strand thymine glycol is specific. A and B, helicase reaction mixtures contained 0.5 nm forked DNA substrate with thymine glycol in the bottom strand (TG-B) (A) or top strand (TG-C) (B), 3 nm E. coli SSB, and 1.2 nm FANCJ under standard helicase assay conditions as described under “Materials and Methods” for the indicated times from 0 to 32 min. Quantitative analyses of FANCJ helicase data are shown (open squares, FANCJ; filled squares, FANCJ + E. coli SSB). C, DinG (80 or 2000 nm) was incubated in the presence or absence of 3 nm RPA with 0.5 nm forked DNA substrate with thymine glycol in the bottom strand for 15 min. Quantitative analyses of the helicase data (mean of at least three independent experiments with S.D. indicated by error bars) are shown.
To determine how RPA might affect DNA unwinding by a helicase with which it does not interact, we examined its effect on the bacterial helicase DinG. As shown in Fig. 5C, RPA failed to stimulate DinG helicase activity on the control or thymine glycol substrates. In fact, RPA inhibited DinG activity on all three substrates. In reactions with higher DinG concentrations, all three substrates were unwound to nearly 100%, indicating that DinG is active on the substrates. Therefore, RPA was not able to stimulate DinG helicase activity on DNA substrates with thymine glycol in either strand, whereas RPA was able to stimulate FANCJ helicase activity on the DNA substrate with thymine glycol in the bottom nontranslocating strand.
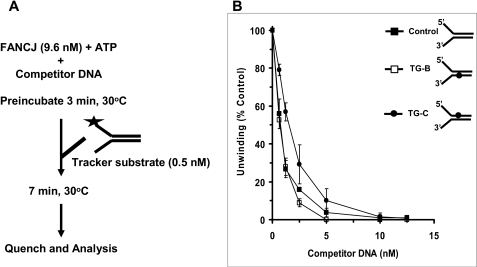
The strand-specific stimulation of FANCJ helicase activity by RPA suggested that during unwinding FANCJ, even in the absence of RPA, might interact differently with the DNA substrate harboring the thymine glycol base damage in the bottom nontranslocating strand compared with the substrate with thymine glycol in the top translocating strand. To address this issue, we performed protein sequestration experiments to evaluate if FANCJ was trapped differently by the DNA molecules containing the thymine glycol in the translocating versus nontranslocating strand during unwinding. FANCJ (9.6 nm) was preincubated for 3 min in the presence of ATP with increasing concentrations of unlabeled forked duplex molecules containing the single thymine glycol adduct in the translocating strand, nontranslocating strand, or neither strand, and a radiolabeled tracker DNA substrate was added subsequently to the reaction mixtures (Fig. 6A). Throughout competitor DNA concentrations of 0.6–5 nm, lesser inhibition of FANCJ helicase activity on the tracker substrate was observed when the enzyme was preincubated with unlabeled forked duplex containing the thymine glycol in the translocating strand (Fig. 6B) compared with when the enzyme was preincubated with the forked duplex containing thymine glycol in the translocating strand or neither strand. The greatest difference, ∼2-fold, was observed at 1.25 nm competitor DNA. These results suggest that during unwinding, FANCJ dissociates from the DNA duplex containing thymine glycol in the translocating strand to a greater extent than the undamaged duplex substrate. In contrast, the ability of the forked duplex with the nontranslocating strand thymine glycol to sequester FANCJ was the same as that of the undamaged fork duplex with the exception of the small difference observed at the 2.5 nm competitor DNA concentration. Sequestration experiments performed using a longer (10 min) preincubation of FANCJ with ATP and increasing concentrations of competitor DNA showed little to no difference between the abilities of the forked duplexes with translocating or nontranslocating strand thymine glycol or undamaged forked duplexes to trap FANCJ (supplemental Fig. 1).
FIGURE 6.
FANCJ preferentially dissociates from the forked duplex with thymine glycol in the translocating strand. Sequestration assays with 9.6 nm FANCJ and the indicated concentrations (0–12.5 nm) of the specified forked duplex competitor DNA molecules were performed as described under “Materials and Methods.” 70% of the tracker DNA substrate was unwound in the absence of competitor DNA. Quantitative analyses of the helicase data (mean of at least three independent experiments with S.D. indicated by error bars) are shown. Filled circles, competitor forked duplex with thymine glycol in top (translocating) strand (TG-C); open squares, competitor forked duplex with thymine glycol in bottom (nontranslocating) strand (TG-B); filled squares, competitor control undamaged forked duplex.
Gel-shift analysis of FANCJ binding in the absence of ATP to radiolabeled forked or blunt duplexes with thymine glycol present in the translocating strand or nontranslocating strand demonstrated that FANCJ binding to the two substrates was comparable and the same as that of FANCJ binding to the undamaged DNA molecules.3 These results suggest that the difference observed in the sequestration experiments was due to FANCJ preferentially dissociating from the DNA substrate when it encountered the thymine glycol in the strand that the enzyme was translocating.
Characterization of RPA Binding to DNA with the Thymine Glycol Adduct
Because RPA stimulated FANCJ helicase in a substrate-specific manner, we wanted to assess if RPA might bind preferentially to the DNA substrate harboring the thymine glycol base damage in the bottom nontranslocating strand. First, we tested by gel-shift analysis RPA binding to the control and thymine glycol forked duplex DNA substrates used for the helicase assays. RPA bound all three substrates in a protein concentration-dependent manner very similarly (supplemental Fig. 2, A and C). Because the analysis of RPA binding to the helicase substrates was potentially complicated by the fact that these substrates are all forked duplexes with single-stranded DNA arms of 41 nucleotides that provide a high affinity 30-nucleotide binding site for RPA (21), we tested RPA binding to fully duplex DNA molecules with thymine glycol in either the top or bottom strand (Table 1) and annealed to its complementary undamaged ssDNA. Compared with the forked duplex substrates that had single-stranded arms, the fully duplex DNA molecules were all poorly bound by RPA throughout the protein titration range (supplemental Fig. 2, B and C). Furthermore, the presence of a thymine glycol in either the top or bottom strand of the duplex had little to no effect on RPA binding. These results indicate that RPA does not preferentially bind the thymine glycol residing in duplex DNA.
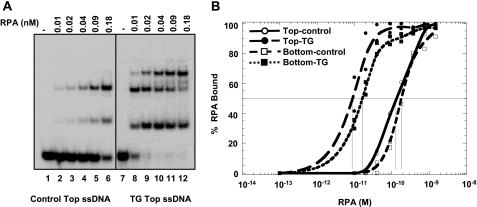
Because FANCJ helicase activity on the forked duplex substrate creates ssDNA during the unwinding reaction, we wanted to assess if RPA displayed a binding preference to ssDNA harboring the single thymine glycol adduct. Therefore, we tested RPA binding to either the top or bottom strands of the forked duplex substrates (Table 1) used for the characterization of FANCJ helicase activity. For ssDNA, it was clearly evident that RPA bound preferentially to the molecules harboring a thymine glycol compared with the undamaged ssDNA (Fig. 7A). Multiple RPA-DNA complexes were observed as is expected based on the 30-nucleotide occluded binding site and low cooperativity of RPA binding (38). Quantitation of the data is shown in Fig. 7B. The apparent binding constants were estimated by fitting to the Langmuir binding equation (38). Similar values were obtained using Scatchard analysis, yielding apparent dissociation constants (Kd) of 10 and 170 pm for the thymine glycol and control (undamaged) ssDNA molecules, respectively. Therefore, RPA binds with 17-fold higher affinity to the ssDNA molecule with the single thymine glycol compared with the ssDNA molecule without the base damage. An analysis of secondary structure prediction for the 66-mer ssDNA molecule revealed that the position of the thymine glycol did not occupy a region that forms dsDNA in the oligonucleotide (supplemental Fig. 3) suggesting that the preferential binding of RPA to the oligonucleotide containing the thymine glycol is not attributed to an indirect effect of the base modification on secondary structure. Similar Kd values (17 and 226 pm) were obtained from the DNA binding isotherms for the complementary (bottom) ssDNA strands either containing or lacking a single thymine glycol, respectively (Fig. 7B). The 14-fold increase in RPA binding affinity for the bottom strand containing thymine glycol compared with the undamaged bottom strand, comparable with the difference observed for the top strand, indicates that the observed difference in RPA binding affinities between a thymine glycol ssDNA and undamaged ssDNA is not dependent on neighboring sequence to the thymine glycol.
FIGURE 7.
RPA strongly binds ssDNA containing a single thymine glycol compared with undamaged ssDNA. A, increasing concentrations of RPA were incubated with 5 fmol of either undamaged 66-mer or the identical 66-mer containing a single thymine glycol under standard gel-shift conditions as described under “Materials and Methods.” The DNA-protein complexes were resolved on native 5% polyacrylamide gels. Phosphorimages of typical gels are shown. B, quantitative analyses of RPA binding to the control and thymine glycol top strand or bottom strand are shown. Open circle, undamaged top strand; filled circle, top strand with thymine glycol; open square, undamaged bottom strand; filled square, bottom strand with thymine glycol. Vertical lines indicate midpoint of each curve.
To summarize, the results from the RPA binding assays suggest that RPA stimulation of FANCJ helicase activity on the DNA substrate with thymine glycol in the bottom nontranslocating strand is not a consequence of RPA binding preferentially to that forked duplex substrate compared with the substrate with thymine glycol in the top translocating strand. Furthermore, RPA binds with similar high affinity to the top or bottom single-stranded DNA molecules harboring the thymine glycol, and this binding affinity is significantly greater than the corresponding undamaged ssDNA molecules.
RPA Heterotrimers with RPA70 Missense Mutations Characterized by DNA Binding Defects Fail to Stimulate FANCJ Helicase Activity
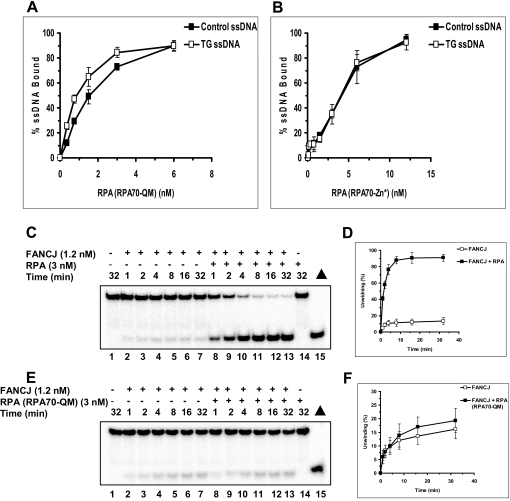
RPA contains multiple DNA binding domains. Two domains in RPA70 (A and B) constitute a high affinity DNA-binding site and a third domain (C) has been shown to be important for specific binding to some types of DNA damage (26, 27). Two mutant forms of RPA, RPA70-QM, which has multiple point mutations in the high affinity DNA binding domain (A), and RPA70-Zn*, which has a mutation that inactivates domain C, were examined for binding to thymine glycol containing DNA. Unlike wild-type RPA, we observed that the RPA70-QM (Fig. 8A) and RAP70-Zn* (Fig. 8B) mutants showed little to no preferential binding to ssDNA with a thymine glycol. These mutants also displayed reduced affinity for unmodified ssDNA as well, as exemplified by the higher RPA concentrations used in the DNA binding isotherm (Fig. 8, A and B). The Kd values for RPA70-QM binding to the control and thymine glycol ssDNA molecules were 1600 and 1900 pm, respectively. The Kd values for RPA70-Zn* binding to the control and thymine glycol ssDNA molecules were 3400 and 4500 pm, respectively. Therefore, both RPA mutants bind with significantly reduced affinity to the control and thymine glycol ssDNA molecules compared with wild-type RPA. Moreover, unlike wild-type RPA, these RPA mutants do not demonstrate preferential binding to the thymine glycol ssDNA. These data indicate that both the high affinity binding domain and domain C are playing a role in the preferential binding of RPA to thymine glycol containing DNA.
FIGURE 8.
Characterization of DNA binding and stimulation of FANCJ helicase activity by RPA heterotrimers with RPA70 missense mutations. A and B, increasing concentrations of RPA heterotrimer with RPA70-QM mutation (A) or RPA heterotrimer with RPA70-Zn* mutation (B) were incubated with 5 fmol of either undamaged 66-mer or the identical 66-mer containing a single thymine glycol damage under standard gel-shift conditions as described under “Materials and Methods.” The DNA-protein complexes were resolved on native 5% polyacrylamide gels. Quantitative analyses of RPA binding to the control and thymine glycol top strand are shown. Filled square, control undamaged oligonucleotide; open square, thymine glycol ssDNA. Experiments were repeated at least three times, and means are shown with S.D. indicated by error bars. C–H, helicase reactions containing the specified RPA heterotrimer (3 nm), 1.2 nm FANCJ, and 0.5 nm forked DNA substrate with thymine glycol in the bottom strand (nontranslocating) were performed under standard helicase assay conditions as described under “Materials and Methods” for the indicated times from 0 to 32 min. C and D, wild-type RPA; E and F, RPA-QM; G and H, RPA-Zn. Quantitative analyses of FANCJ helicase data are shown (open squares, FANCJ; filled squares, FANCJ + RPA). I and J, helicase reactions containing the indicated amount of RPA variant, 1.2 nm FANCJ, and 0.5 nm forked DNA substrate with thymine glycol in the bottom strand (nontranslocating) were performed under standard helicase assay conditions as described under “Materials and Methods” for 15 min.
We next tested the two RPA heterotrimers with RPA70 missense mutations for stimulation of FANCJ helicase activity on the DNA substrate with thymine glycol in the nontranslocating (bottom) strand, the substrate in which wild-type RPA stimulated FANCJ helicase activity. Both RPA70-QM (Fig. 8, E and F) and RPA70-Zn* (Fig. 8, G and H) failed to stimulate DNA unwinding by FANCJ throughout the 32-min time course. In contrast, under these same conditions, wild-type RPA showed strong stimulation of FANCJ helicase activity on the same thymine glycol substrate (Fig. 8, C and D, and Fig. 4). Both RPA mutants were able to physically interact with FANCJ (data not shown), suggesting that the inability of the RPA heterotrimer with either RPA70 mutation to stimulate FANCJ helicase activity was likely due to their DNA binding defects. Given that RPA70-QM and RPA70-Zn* bind ssDNA poorly and do not discriminate between unmodified and thymine glycol-modified substrates, we examined FANCJ helicase activity as a function of RPA variants over a broad range of RPA concentrations. The results from these experiments demonstrated that even at a very high concentration (96 nm) of RPA70-QM or RPA70-Zn*, FANCJ helicase activity on the forked duplex with thymine glycol in the nontranslocating strand was only stimulated a maximum of 2-fold (Fig. 8, I and J). No stimulation of FANCJ helicase activity on the forked duplex with thymine glycol in the translocating strand was observed for either RPA variant (data not shown). Therefore, FANCJ helicase activity on the forked duplex with thymine glycol in the nontranslocating strand was only mildly stimulated by a 30-fold higher concentration of either RPA variant compared with wild-type RPA.
RPA Stimulates RECQ1 Helicase Activity on the Thymine Glycol Substrate in a Strand-specific Manner
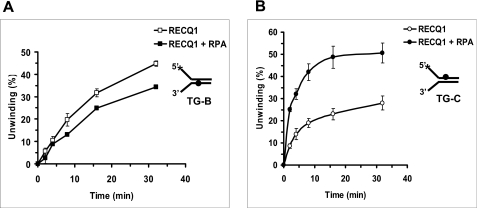
Because RPA exerted a strand-specific stimulation of FANCJ helicase activity that did not reflect a preferential binding affinity of RPA for one duplex substrate compared with another, we wanted to ask what effect RPA might have on a helicase that is translocating on the opposite strand that FANCJ translocates. Therefore, we tested the effect of RPA on the RECQ1 helicase, which has a 3′ to 5′ polarity of unwinding (39). Kinetic assays with the thymine glycol substrates were performed using 3 nm RPA as before and a limited concentration of RECQ1 in which DNA unwinding of the forked duplex substrates with thymine glycol in the translocating or nontranslocating strand by RECQ1 alone was comparable. The results from these experiments, shown in Fig. 9, indicated that RPA was unable to stimulate RECQ1 helicase activity on the DNA substrate with thymine glycol in the bottom translocating strand; however, RPA stimulated RECQ1 helicase activity on the substrate with thymine glycol in the top nontranslocating strand. Therefore, RPA was able to stimulate RECQ1 helicase activity on the thymine glycol substrate on which it failed to stimulate FANCJ helicase activity, whereas RPA failed to stimulate RECQ1 helicase activity on the thymine glycol substrate on which it stimulated FANCJ helicase activity. These results suggest that, like FANCJ, RPA stimulates RECQ1 helicase activity in a strand-specific manner, i.e. RPA stimulates DNA unwinding by RECQ1 when the thymine glycol is positioned in the nontranslocating strand for the 3′- to 5′-RECQ1 helicase.
FIGURE 9.
RPA stimulates RECQ1 helicase activity on the thymine glycol substrates in a strand-specific manner. Helicase reaction mixtures contained 0.5 nm forked duplex DNA substrate (A, bottom strand thymine glycol (TG-B); B, top strand thymine glycol (TG-C)), 3 nm RPA, and 9.6 nm RECQ1 (A) or 1.2 nm RECQ1 (B) under standard helicase assay conditions as described under “Materials and Methods” for the indicated times from 0 to 32 min. Quantitative analyses of RECQ1 helicase data are shown (open squares, RECQ1; filled squares, RECQ1 + RPA).
DISCUSSION
In this study, we have examined the ability of FANCJ to unwind DNA substrates containing a single oxidized base modification residing within the duplex region of a forked DNA substrate in either the translocating or nontranslocating strand. Our results demonstrate that FANCJ helicase activity is sensitive to a single thymine glycol in either strand of the duplex, whereas DNA unwinding by FANCJ is not inhibited by an 8-oxoguanine base damage positioned in either strand. The differential effect of these two oxidative lesions on FANCJ helicase activity is likely to reflect the extent of structural distortion on the DNA double helix imposed by the base modification. Thymine glycol induces a significant, localized structural change with the thymine glycol largely extrahelical (40), whereas 8-oxoguanine exerts only a mild perturbation of the duplex (for review see Ref. 41). However, the sensitivity of FANCJ helicase to thymine glycol is rather unique because the three human RecQ helicases tested (WRN, BLM, and RECQ1) are only adversely affected by the translocating strand thymine glycol damage, whereas other DNA helicases (DnaB, UvrD, and DinG) are not affected by the modification in either strand in any significant manner. The ability of the bacterial helicases to proficiently unwind thymine glycol substrates irrespective of strand status may reflect the increased ATPase activity of these helicases (32, 42) compared with FANCJ (31), RECQ1 (43), or WRN (44); however, BLM helicase has a specific ATPase activity that is greater than the other human RecQ helicases but less than the bacterial helicases (45). The ability of FANCJ to sense certain types of DNA base damage (thymine glycol) in both strands of the DNA duplex as it unwinds may be important for its physiological functions in DNA damage signaling or repair. The existence of FANCJ in a BRCA1-containing complex with other DNA repair factors involved in the recognition and repair of aberrant DNA structures suggests that the complex functions in the DNA damage response (14). It is conceivable that a specialized helicase like FANCJ might have the ability to sense DNA damage in either strand of the duplex and facilitate DNA damage processing or signaling. It will be important to examine the interactions of FANCJ with DNA repair factors to understand its roles in the DNA damage response that are dependent or independent of the FA pathway.
Although FANCJ shows no preferential binding to the thymine glycol-modified ssDNA or dsDNA, sequestration studies revealed that during unwinding FANCJ dissociates to a somewhat greater extent from the substrate containing thymine glycol in the translocating strand compared with that with thymine glycol in the nontranslocating strand. Thus, the interaction of FANCJ with the DNA substrates during unwinding is affected by the strand in which the thymine glycol resides. However, the greatest difference in sequestration between the forked duplexes with thymine glycol in the top versus bottom strands is only ∼2-fold, suggesting that FANCJ preferentially dissociating from the substrate with the translocating strand thymine glycol may not be not be the sole or major reason why RPA only stimulates FANCJ helicase activity on the substrate with the nontranslocating strand thymine glycol. A more likely explanation is that the high affinity binding of RPA to the partially unwound ssDNA bearing a thymine glycol poses a block to helicase movement when the thymine glycol is positioned in the single strand the helicase is tracking along; consequently, the helicase would be less likely to interact favorably with RPA at the site of unwinding and FANCJ would have to reload on the substrate and attempt to unwind it again. RPA failed to stimulate FANCJ helicase activity on substrates in which the thymine glycol was placed in the ssDNA region (translocating or nontranslocating strand) of the forked duplex substrate just before the helicase would encounter the duplex region (supplemental Fig. 4), suggesting that the RPA stimulation of helicase bypass of a thymine glycol only applies when the base modification resides in the nontranslocating strand of the duplex region of the helicase substrate. This model may apply to the functional interaction of RPA with other helicases and their actions on certain damaged DNA substrates recognized by RPA. The 3′- to 5′-human RecQ helicase RECQ1, which also physically and functionally interacts with RPA (43), is stimulated by RPA on the thymine glycol substrates in a strand-specific manner as well. RPA failed to stimulate RECQ1 helicase activity when the thymine glycol resided in the strand that RECQ1 translocates. The ability of RPA to stimulate RECQ1 or FANCJ helicase activity when the thymine glycol resided in the nontranslocating strand may be at least partially due to the high affinity binding of RPA to the exposed thymine glycol in the nontranslocating strand of the partially unwound DNA substrate. To our knowledge, this is the first demonstration that RPA has the ability to stimulate helicase-catalyzed DNA unwinding in a strand-specific manner. In the future, mechanistic studies of the functional interactions of RPA with the human RecQ helicases should provide useful information to explore the potential similarity with that of the FANCJ-RPA interaction.
The stimulatory effect of RPA is specific because E. coli SSB did not affect FANCJ helicase activity on the thymine glycol substrates, and RPA did not stimulate unwinding activity catalyzed by DinG, a bacterial DNA helicase sharing sequence homology to FANCJ in the helicase core domain (7), but it does not physically interact with RPA.3 Therefore, RPA does not have a general ability to stimulate DNA helicases on undamaged as well as damaged DNA substrates. Moreover, the mechanism for RPA stimulation of FANCJ helicase activity is likely to be more complex than simply RPA coating the unwound ssDNA tracts left behind the advancing helicase. Consistent with this idea, FANCJ directly binds with high affinity to the RPA70 subunit of the heterotrimer (22). Therefore, the physical interaction between FANCJ and RPA is likely to be important for helicase stimulation. In addition to the protein interaction between RPA and FANCJ, we suggest the ability of RPA to preferentially bind ssDNA containing the thymine glycol base modification is likely to play a role in the mechanism for stimulation of FANCJ (or RECQ1) helicase activity on the substrate containing thymine glycol in the nontranslocating strand.
Previous studies have suggested that preferential RPA binding to DNA damage is dependent on the type of lesion. RPA was observed to bind with a higher affinity to single-stranded DNA harboring a pyrimidine-pyrimidone (6-4) photoproduct (27). However, preferential binding of RPA to damaged dsDNA correlates with the ability of RPA to recognize single-stranded character caused by the damaged nucleotides (27, 46). Our study is the first to show strong preferential binding of RPA to ssDNA containing an oxidative base modification. The dramatic 15-fold increase in binding affinity of RPA for ssDNA containing a single thymine glycol is unprecedented. Because RPA is the major single-stranded DNA-binding protein in eukaryotes and is highly abundant (100,000 copies per cell) (21), it is quite likely that the high binding affinity of RPA for thymine glycol (and perhaps certain other oxidative base lesions) is physiologically important. For example, FANCJ or RECQ1 helicase may be unable to remove RPA bound to a thymine glycol bound to the helicase-translocating single strand of a partially unwound DNA molecule. This model may help to explain the strand-specific stimulation of helicase activity by RPA in which the translocating strand thymine glycol is inhibitory to either the 3′- to 5′-RECQ1 helicase or 5′- to 3′-FANCJ helicase.
Other aspects of DNA metabolism are likely to be affected by the strong affinity of RPA for ssDNA harboring thymine glycol. RPA may serve as a negative regulator of DNA glycosylases such as NEIL1 or NTH1 that specialize in excising thymine glycol from the genome (47). Unlike NTH1 or OGG1 enzymes, which excise their substrate lesions only from duplex DNA, the NEIL enzymes have higher activity in excising base lesions from single-stranded DNA or unpaired sequences in bubble DNA (47), a transient intermediate that arises during cellular DNA replication and transcription. It was proposed that NEIL1 and NEIL2 may have roles in BER linked to transcription or replication. Our results suggest that RPA may regulate glycosylase incision of thymine glycol. Studies are underway to determine whether RPA modulates thymine glycol incision by NEIL1 or NTH1 of bubble structures and ssDNA through its high affinity interaction with thymine glycol in its single-stranded DNA state. Although thymine glycol does not block mammalian RNA polymerase II elongation in a purified system (48), we propose that RPA bound to the oxidative lesion in the transcription bubble may block RNA polymerase II at the damage site and inhibit transcription or transcription-related processes. Further studies to better understand the lethal nature of thymine glycol through its inhibition of cellular DNA replication or transcription is warranted.
Bringing our attention back to the FANCJ-RPA interaction, it is relevant that FANCJ and RPA strongly co-localize in punctate nuclear foci in human cells that have been exposed to ionizing radiation, which introduces strand breaks and base lesions such as thymine glycol (41), or the replication inhibitor hydroxyurea, which depletes the nucleotide pool (22). FANCJ and RPA may collaborate to unwind regions of localized DNA distortion imposed by thymine glycol and other DNA lesions that perturb replication fork progression or during a situation of replicational stress. In support of a role of FANCJ to smooth polymerase advancement, FANCJ is required for timely progression through S phase (18) and defends genomic integrity by unwinding G-quadruplex structures that can interfere with DNA replication or transcription (19).
The DNA interstrand cross-link is a direct block to replication or transcription. FA mutant cell lines, including FA-J, are sensitive to agents such as mitomycin C and cisplatin that introduce DNA cross-links. FANCJ helicase and RPA co-localize in cells exposed to mitomycin C (22), suggesting that the FANCJ-RPA interaction is important for the processing of DNA cross-links as well as enabling smooth replication.
In the future, it will be of interest to examine the cellular localization of FANCJ and the other FA proteins with RPA and additional repair factors after exposure to agents that induce oxidative stress and other forms of DNA damage. From this perspective it is interesting to note that FANCJ has an Fe-S domain in its helicase core that may confer redox properties to the protein (33). The unique ability of FANCJ to sense a single thymine glycol oxidative base lesion in either strand of the duplex that it is unwinding and its strand-specific stimulation by RPA to unwind past the lesion may be relevant to a role of the helicase during DNA replication or transcription in an environment of heightened oxidative stress.
Supplementary Material
Acknowledgments
We thank Dr. Ian Hickson (Cancer Research UK Laboratories), Dr. Steven Matson (University of North Carolina, Chapel Hill), and Dr. Daniel Kaplan (Vanderbilt University) for kindly providing purified BLM, UvrD, and DnaB helicases, respectively.
This work was supported, in whole or in part, by National Institutes of Health Research Grant GM44721 (to M. S. W.) and the NIA and NIDDK National Institutes of Health Intramural Research Program. This work was also supported by the Fanconi Anemia Research Fund (to R. M. B.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S4.
3 A. N. Suhasini, J. A. Sommers, A. C. Mason, O. N. Voloshin, R. D. Camerini-Otero, M. S. Wold, and R. M. Brosh, Jr., unpublished data.
- FA
- Fanconi anemia
- RPA
- replication protein A
- ssDNA
- single-stranded DNA
- HR
- homologous recombination
- dsDNA
- double-stranded DNA
- pyrimidine-pyrimidone (6-4)
- 6-(1,2)-dihydro-2-oxo-4-pyrimidinyl-5-methyl-2,4-(1H,3H)-pyrimidinedione
- SSB
- single-stranded DNA-binding protein.
REFERENCES
- 1.Levitus M., Joenje H., de Winter J. P. ( 2006) Cell Oncol. 28, 3– 29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Taniguchi T., D'Andrea A. D. ( 2006) Blood 107, 4223– 4233 [DOI] [PubMed] [Google Scholar]
- 3.Wang W. ( 2007) Nat. Rev. Genet. 8, 735– 748 [DOI] [PubMed] [Google Scholar]
- 4.Levitus M., Waisfisz Q., Godthelp B. C., de Vries Y., Hussain S., Wiegant W. W., Elghalbzouri-Maghrani E., Steltenpool J., Rooimans M. A., Pals G., Arwert F., Mathew C. G., Zdzienicka M. Z., Hiom K., De Winter J. P., Joenje H. ( 2005) Nat. Genet. 37, 934– 935 [DOI] [PubMed] [Google Scholar]
- 5.Levran O., Attwooll C., Henry R. T., Milton K. L., Neveling K., Rio P., Batish S. D., Kalb R., Velleuer E., Barral S., Ott J., Petrini J., Schindler D., Hanenberg H., Auerbach A. D. ( 2005) Nat. Genet. 37, 931– 933 [DOI] [PubMed] [Google Scholar]
- 6.Litman R., Peng M., Jin Z., Zhang F., Zhang J., Powell S., Andreassen P. R., Cantor S. B. ( 2005) Cancer Cell 8, 255– 265 [DOI] [PubMed] [Google Scholar]
- 7.Cantor S. B., Bell D. W., Ganesan S., Kass E. M., Drapkin R., Grossman S., Wahrer D. C., Sgroi D. C., Lane W. S., Haber D. A., Livingston D. M. ( 2001) Cell 105, 149– 160 [DOI] [PubMed] [Google Scholar]
- 8.De Nicolo A., Tancredi M., Lombardi G., Flemma C. C., Barbuti S., Di Cristofano C., Sobhian B., Bevilacqua G., Drapkin R., Caligo M. A. ( 2008) Clin. Cancer Res. 14, 4672– 4680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Seal S., Thompson D., Renwick A., Elliott A., Kelly P., Barfoot R., Chagtai T., Jayatilake H., Ahmed M., Spanova K., North B., McGuffog L., Evans D. G., Eccles D., Easton D. F., Stratton M. R., Rahman N. ( 2006) Nat. Genet. 38, 1239– 1241 [DOI] [PubMed] [Google Scholar]
- 10.Cantor S., Drapkin R., Zhang F., Lin Y., Han J., Pamidi S., Livingston D. M. ( 2004) Proc. Natl. Acad. Sci. U.S.A. 101, 2357– 2362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gupta R., Sharma S., Sommers J. A., Jin Z., Cantor S. B., Brosh R. M., Jr. ( 2005) J. Biol. Chem. 280, 25450– 25460 [DOI] [PubMed] [Google Scholar]
- 12.Peng M., Litman R., Jin Z., Fong G., Cantor S. B. ( 2006) Oncogene 25, 2245– 2253 [DOI] [PubMed] [Google Scholar]
- 13.Bridge W. L., Vandenberg C. J., Franklin R. J., Hiom K. ( 2005) Nat. Genet. 37, 953– 957 [DOI] [PubMed] [Google Scholar]
- 14.Greenberg R. A., Sobhian B., Pathania S., Cantor S. B., Nakatani Y., Livingston D. M. ( 2006) Genes Dev. 20, 34– 46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Thompson L. H., Hinz J. M., Yamada N. A., Jones N. J. ( 2005) Environ. Mol. Mutagen. 45, 128– 142 [DOI] [PubMed] [Google Scholar]
- 16.Howlett N. G., Taniguchi T., Durkin S. G., D'Andrea A. D., Glover T. W. ( 2005) Hum. Mol. Genet. 14, 693– 701 [DOI] [PubMed] [Google Scholar]
- 17.Wang L. C., Stone S., Hoatlin M. E., Gautier J. ( 2008) DNA Repair 7, 1973– 1981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kumaraswamy E., Shiekhattar R. ( 2007) Mol. Cell. Biol. 27, 6733– 6741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wu Y., Shin-ya K., Brosh R. M., Jr. ( 2008) Mol. Cell. Biol. 28, 4116– 4128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Du W., Adam Z., Rani R., Zhang X., Pang Q. ( 2008) Antioxid. Redox. Signal. 10, 1909– 1921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wold M. S. ( 1997) Annu. Rev. Biochem. 66, 61– 92 [DOI] [PubMed] [Google Scholar]
- 22.Gupta R., Sharma S., Sommers J. A., Kenny M. K., Cantor S. B., Brosh R. M., Jr. ( 2007) Blood 110, 2390– 2398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sharma S., Sommers J. A., Choudhary S., Faulkner J. K., Cui S., Andreoli L., Muzzolini L., Vindigni A., Brosh R. M., Jr. ( 2005) J. Biol. Chem. 280, 28072– 28084 [DOI] [PubMed] [Google Scholar]
- 24.Sharma S., Otterlei M., Sommers J. A., Driscoll H. C., Dianov G. L., Kao H. I., Bambara R. A., Brosh R. M., Jr. ( 2004) Mol. Biol. Cell 15, 734– 750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Voloshin O. N., Camerini-Otero R. D. ( 2007) J. Biol. Chem. 282, 18437– 18447 [DOI] [PubMed] [Google Scholar]
- 26.Haring S. J., Mason A. C., Binz S. K., Wold M. S. ( 2008) J. Biol. Chem. 283, 19095– 19111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lao Y., Gomes X. V., Ren Y., Taylor J. S., Wold M. S. ( 2000) Biochemistry 39, 850– 859 [DOI] [PubMed] [Google Scholar]
- 28.Brosh R. M., Jr., Waheed J., Sommers J. A. ( 2002) J. Biol. Chem. 277, 23236– 23245 [DOI] [PubMed] [Google Scholar]
- 29.Cadman C. J., Matson S. W., McGlynn P. ( 2006) J. Mol. Biol. 362, 18– 25 [DOI] [PubMed] [Google Scholar]
- 30.Kaplan D. L., O'Donnell M. ( 2002) Mol. Cell 10, 647– 657 [DOI] [PubMed] [Google Scholar]
- 31.Gupta R., Sharma S., Doherty K. M., Sommers J. A., Cantor S. B., Brosh R. M., Jr. ( 2006) Nucleic Acids Res. 34, 6673– 6683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Voloshin O. N., Vanevski F., Khil P. P., Camerini-Otero R. D. ( 2003) J. Biol. Chem. 278, 28284– 28293 [DOI] [PubMed] [Google Scholar]
- 33.Rudolf J., Makrantoni V., Ingledew W. J., Stark M. J., White M. F. ( 2006) Mol. Cell 23, 801– 808 [DOI] [PubMed] [Google Scholar]
- 34.Patel S. S., Picha K. M. ( 2000) Annu. Rev. Biochem. 69, 651– 697 [DOI] [PubMed] [Google Scholar]
- 35.Sharma S., Doherty K. M., Brosh R. M., Jr. ( 2006) Biochem. J. 398, 319– 337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sharma S., Brosh R. M., Jr. ( 2008) Cell Cycle 7, 989– 1000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Matson S. W., Bean D. W., George J. W. ( 1994) BioEssays 16, 13– 22 [DOI] [PubMed] [Google Scholar]
- 38.Kim C., Paulus B. F., Wold M. S. ( 1994) Biochemistry 33, 14197– 14206 [DOI] [PubMed] [Google Scholar]
- 39.Cui S., Klima R., Ochem A., Arosio D., Falaschi A., Vindigni A. ( 2003) J. Biol. Chem. 278, 1424– 1432 [DOI] [PubMed] [Google Scholar]
- 40.Kung H. C., Bolton P. H. ( 1997) J. Biol. Chem. 272, 9227– 9236 [DOI] [PubMed] [Google Scholar]
- 41.Wallace S. S. ( 2002) Free Radic. Biol. Med. 33, 1– 14 [DOI] [PubMed] [Google Scholar]
- 42.Brosh R. M., Jr., Matson S. W. ( 1995) J. Bacteriol. 177, 5612– 5621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cui S., Arosio D., Doherty K. M., Brosh R. M., Jr., Falaschi A., Vindigni A. ( 2004) Nucleic Acids Res. 32, 2158– 2170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Brosh R. M., Jr., Orren D. K., Nehlin J. O., Ravn P. H., Kenny M. K., Machwe A., Bohr V. A. ( 1999) J. Biol. Chem. 274, 18341– 18350 [DOI] [PubMed] [Google Scholar]
- 45.Brosh R. M., Jr., Li J. L., Kenny M. K., Karow J. K., Cooper M. P., Kureekattil R. P., Hickson I. D., Bohr V. A. ( 2000) J. Biol. Chem. 275, 23500– 23508 [DOI] [PubMed] [Google Scholar]
- 46.Patrick S. M., Turchi J. J. ( 1999) J. Biol. Chem. 274, 14972– 14978 [DOI] [PubMed] [Google Scholar]
- 47.Hazra T. K., Das A., Das S., Choudhury S., Kow Y. W., Roy R. ( 2007) DNA Repair. 6, 470– 480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tornaletti S., Maeda L. S., Lloyd D. R., Reines D., Hanawalt P. C. ( 2001) J. Biol. Chem. 276, 45367– 45371 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.