Abstract
Background
In rats, alcohol exposure during the period of rapid brain growth produces long-term changes in the free-running period, photoentrainment and phase-shifting responses of the circadian rhythm in wheel-running behavior. To determine whether these alterations in circadian behavior are associated with permanent damage to the circadian timekeeping mechanism or reconfiguration of its molecular components, we examined the long-term effects of neonatal alcohol exposure on clock gene rhythms in the pacemaker located in the suprachiasmatic nucleus (SCN) and in other brain or peripheral tissues of adult rats.
Methods
Artificially reared male rat pups were exposed to alcohol (4.5 g / kg / d) or isocaloric milk formula (gastrostomy control; GC) on postnatal days 4 to 9. At 3 months of age, animals were exposed to constant darkness and then SCN, cerebellum, and liver tissue were harvested at 6-hour intervals for subsequent analysis of Period1 (Per1), Per2, Cryptochrome1 (Cry1), Bmal1, and Rev-erbα mRNA levels by quantitative PCR.
Results
In the SCN, cerebellum and liver, Per1, Per2, Cry1, Bmal1, and Rev-erbα expression oscillated with a similar amplitude (peak-to-trough differences of 2- to 9-fold) and phase in the suckle control (SC) and GC groups. These clock gene rhythms in control animals were marked by peak expression of Per1, Per2, Cry1, and Rev-erbα during the subjective day and of Bmal1 during the subjective night. The EtOH group was distinguished by altered rhythms in the expression of specific clock genes within the SCN, cerebellum and liver. In EtOH-treated rats, the SCN rhythm in Cry1 expression was strongly damped and the Per2 rhythms in the cerebellum and liver were phase-advanced such that peak expression occurred during the mid-subjective day.
Conclusions
These results demonstrate alcohol exposure during the brain growth spurt alters the circadian regulation of some molecular components of the clock mechanism in the rat SCN, cerebellum, and liver. The observed alterations in the temporal configuration of essential “gears” of the molecular clockworks may play a role in the long-term effects of neonatal alcohol exposure on the regulation of circadian behavior.
Keywords: Circadian Rhythms, Clock, Oscillation, Bmal1, Cry1, Per1, Per2, Rev-erbα
In humans, prenatal alcohol exposure produces long-term changes in the neural regulation of behavior including learning difficulties, attention deficits, sleep–wake disturbances, bipolar affective disorder and depression (Roebuck et al., 1999). Animal studies have yielded further details on the neurobehavioral consequences of alcohol (EtOH) exposure during critical stages of brain development. In rats, exposure to alcohol during the neonatal period, which corresponds to the third trimester equivalent in human brain development (Dobbing and Sands, 1979), induces chronic deficits on learning /memory and motor performance tasks and these behavioral manifestations are associated with alcohol-induced damage to the hippocampus and cerebellum (Goodlett et al., 1987, 1988; Kelly et al., 1988; Thomas et al., 1996, 1998). Recent applications of this animal model have revealed that circadian or 24-hour behavioral rhythmicity is also vulnerable to alcohol-induced insult during CNS development. Specifically, neonatal alcohol exposure alters fundamental properties of the activity rhythm in adult rats including its free-running period, phase-shifting responses to light and entrainment to light–dark cycles (Allen et al., 2005a,b; Farnell et al., 2004).
At present, the molecular or cellular basis for these alcohol-induced disturbances in the neural regulation of rat circadian behavior is unknown. Because modifications in the molecular “gears” of the circadian clockworks produce tangible changes in the circadian regulation of behavior (Reppert and Weaver, 2002), it is possible that neonatal alcohol exposure may permanently modulate circadian properties of the rat activity rhythm by disrupting the clock mechanism perhaps through altered expression or temporal configuration of its molecular components. In mammals, the internal biological clock mediating the generation of circadian rhythms and their entrainment or synchronization to light–dark cycles is located in the suprachiasmatic nuclei (SCN) of the anterior hypothalamus. The “gears” of the molecular clockworks in the SCN consist of interlocked transcription–translation feedback loops in which the gene products of core components rhythmically regulate their own transcription or the expression of other clock genes. Clock, Period1 (Per1), Per2, Cryptochrome1 (Cry1), Cry2, and Bmal1 (Mop3) have been identified as core molecular elements of the circadian clock mechanism in the SCN based on studies demonstrating that targeted disruption or knockout of these genes in mice alters or abolishes circadian behavior (Reppert and Weaver, 2002). For example, Per1-, Per2- and Cry1-deficient mice show activity rhythms in which the free-running period is shorter than that observed in wild type animals (Bae et al., 2001; van der Horst et al., 1999; Vitaterna et al., 1999; Zheng et al., 2001). Although the SCN are essential for the generation of mammalian circadian rhythms in many biochemical, physiological, and behavioral processes, the expression and rhythmic regulation of these core clock genes are not spatially confined to the SCN. Instead, rhythmic fluctuations in the same clock genes comprising the molecular core of the SCN clockworks occur widely in other brain regions and many peripheral tissues, including the liver, heart, kidney, endocrine organs, and skeletal muscles (Shearman et al., 1997; Yamazaki et al., 2000; Zylka et al., 1998). These clock gene oscillations in other tissues are currently thought to provide for their “local” organization in time through a hierarchical system in which the SCN functions as a pacemaker linking periodic environmental cues with the internal circadian timekeeping mechanism in peripheral oscillators throughout the body. Consequently, the effects of neonatal alcohol exposure on the regulation of adult circadian behavior may be coupled with alcohol-induced changes in molecular components of the clockworks not only in the SCN pacemaker but also in peripheral oscillators. To determine whether the molecular mechanism for circadian timekeeping is vulnerable to alcohol-induced insult during the brain growth spurt, we examined the long-term impact of neonatal alcohol treatment on the temporal regulation of core clock genes in the SCN pacemaker and other brain or peripheral tissue oscillators. In this study, rats were exposed to alcohol during the neonatal period and the clock genes Per1, Per2, Cry1, Bmal1, and Rev-erbα were assessed for evidence of alcohol-induced changes in their circadian expression within the SCN, cerebellum, and liver. Because alterations in specific genetic components of the circadian clockworks have been linked to abnormal regulation of human sleep–wake cycles (Toh et al., 2001), which is a known neurobehavioral correlate of prenatal alcohol exposure in humans (Sher, 2004), information from this study may have important implications in understanding how alcohol-induced injury during rapid brain growth produces pathologies in the circadian regulation of sleep and other body processes.
METHODS
Animals
The subjects were 90 male Sprague-Dawley rat pups derived from 24 time-mated litters. Consistent with our previous studies documenting the long-term effects of neonatal alcohol exposure on circadian behavior (Allen et al., 2005a,b; Farnell et al., 2004), male subjects were used exclusively in the present investigation due to the relative stability of their rhythms in comparison with female rats where changing levels of estrogen over the course of the estrous cycle influence the circadian regulation of physiological and behavioral processes (Albers, 1981; Albers et al., 1981; Morin et al., 1977). All animals were born and reared in the vivarium at the Texas A&M University System Health Science Center under a standard 12 hours light:12 hours dark photoperiod (LD 12:12; lights-on at 0600 hours). On postnatal day (PD) 1 (date of birth designated as PD 0), the newborns within each litter were culled to 8 to 10 pups per litter using cross-fostering procedures to maintain equivalent litter size by the addition of nonexperimental pups from other litters. These procedures were implemented to prevent increased weight gain among pups from litters of smaller size and thus reduce the potential influence of accelerated somatic growth as a confounding factor for the dependent variables assessed in this study. On PD 4, the rat pups were randomly assigned to one of three treatment groups: a suckle control group (SC; n = 30) and two artificial-rearing treatment groups. Pups were separated into these treatment groups so that no more than three animals from the same litter were assigned to any one group. The artificial-rearing groups received either alcohol (EtOH; n = 30) or maltose-dextrin [gastrostomy control (GC); n = 30] from PD 4 through 9. The procedures used in this study were approved by the University Laboratory Animal Care Committee at Texas A&M University.
Artificial Rearing
The artificial-rearing process has been described previously in detail (West, 1993). Briefly, on PD 4, pups were anesthetized with isofluorane (VEDCO Inc., St. Joseph, MO) and gastrostomy tubes were inserted down the esophagus and surgically implanted into the stomach (Diaz, 1991; West et al., 1984). From PD 4 to 9, gastrostomized pups were maintained in the artificial-rearing apparatus and received their daily nutritional requirements through formula (diet)-filled syringes which were operated by a timer-activated infusion pump (Model no. 935 Harvard Apparatus, Holliston, MA). The formula was provided daily in 12 20-minute fractions (i.e., every 2 hours). Gastrostomized pups received either alcohol treatment or isocaloric maltose-dextrin solution from PD 4 to 9. On PD 10, artificially reared pups were fostered to a lactating dam after their gastrostomy tubes had been cut and sealed. SC animals were maintained with lactating dams and were included to control for any effect produced by artificial-rearing methods. To minimize the potential confound of litter effects, no more than two pups from the same litter were assigned to the same treatment condition. Identification of subjects from different treatment groups was accomplished by subcutaneous implantation of each pup with a coded microchip (Avid, Los Angeles, CA) on PD 10. Animals were weaned on PD 21 and housed 2 to 3 per cage. After weaning, food and water were provided ad libitum for the remainder of the experiment.
Drug Administration
Beginning around mid-day of the LD 12:12 photoperiod (1200 hours) from PD 4 to 9, alcohol (10.2%, v / v) was administered to the EtOH group through 2 of the daily 12 feedings to yield the dose of 4.5 g / kg / d. This alcohol dose and binge-like treatment regimen have been shown to consistently produce long-term neuroanatomical damage (Bonthius and West, 1988, 1990; Bonthius et al., 1992; Chen et al., 1998, 1999). The GC group (0 g / kg / d alcohol) was given the appropriate concentration of isocaloric maltose-dextrin solution in place of alcohol. All animals received regular formula diet during the 10 remaining daily feedings.
Blood Alcohol Concentration
Blood alcohol concentration (BAC) for all EtOH treated pups was measured using a gas chromatograph (Model no. 3900, Varian, Palo Alto, CA). Blood samples (20 µl) were collected from pup tails 90 minutes after the beginning of the second alcohol feeding on PD 9. Samples were stored in glass vials containing a 200-µl cocktail composed of 0.6 M perchloric acid and 4 mM n-propanol in double distilled water until BAC analysis.
Behavioral Analysis
At 2 months of age, animals (n = 90) were housed individually in cages equipped with running wheels. Wheel-running activity was recorded continuously, summed, stored in 10-minute bins, graphically depicted in actograms and analyzed using a computer running ClockLab data collection and analysis software (Actimetrics, Evanston, IL). The baseline pattern of wheel-running activity was recorded for 10 days during entrainment to LD 12:12 (lights-on at 0600 hours). All animals were subsequently maintained in constant darkness (DD) beginning at the offset of the light–dark cycle (1800 hours). For each animal, the steady-state period (τ) of the activity rhythm during exposure to DD for 30 days was determined by chi-square periodogram and fast Fourier transform.
Tissue Collection
Following the analysis of circadian behavior, animals were again maintained under LD 12:12 conditions until stable re-entrainment of their activity rhythms was observed (21 days). Beginning at lights-off in the LD 12:12 cycle [circadian time (CT) 12], animals were exposed to DD. Twelve hours later (0600 hours or CT 0), animals from the SC, GC, and EtOH groups were killed at 6-hour intervals (n = 5) for 30 hours by decapitation using an infrared viewer (FJW Optical Systems, Palatine, IL). Thus, tissue was collected during the subjective day (i.e., when the light phase would have occurred in the previous LD 12:12 photoperiod) at CT 0 and 6, during the subjective night (i.e., coinciding with previous dark phase or the animal’s active period) at CT 12 and 18 and again during the next circadian cycle at CT 0 and 6. The SCN was immediately dissected as described previously (Earnest and Sladek, 1986; Liang et al., 1998). Briefly, the entire brain was removed and sectioned in the coronal plane at the mid-level of the optic chiasm (600 to 800 µm). The SCN was obtained by dissecting a small square of tissue in ventral extent of the brain section adjacent to the third ventricle. Following removal of SCN tissue, the cerebellum was dissected from the rest of the brain. In addition to the SCN and cerebellum, liver tissue was collected. All tissue samples were frozen in liquid nitrogen and stored at −80°C.
Analysis of Clock Gene mRNA Levels
Total cellular RNA from SCN and cerebellar tissue was later extracted from individual samples using RNeasy Lipid Kit protocols (Qiagen, Valencia, CA). RNA was extracted from heart, liver, and kidney tissue using RNeasy Midi Kit protocols (Qiagen). RNA extracts were treated with on-column DNase-I digestion and then evaluated for integrity of 18S and 28S rRNA bands by electrophoresis on 1.0% agarose / formaldehyde gels stained with ethidium bromide. Relative quantification of clock gene mRNA abundance was performed using SYBR-Green real-time PCR technology [Applied Biosystems, Inc. (ABI), Foster City, CA] as described previously (Allen et al., 2004a). To generate single-strand cDNAs, total RNA (1 µg) from individual samples was reverse transcribed using random hexamers and Superscript II reverse transcriptase (Invitrogen, Carlsbad, CA, USA). For analysis of clock gene expression, the cDNA equivalent of 10 ng of total RNA was PCR amplified in an ABI PRISM 7700 sequence detection system (Heid et al., 1996). This analysis was conducted concurrently on triplicate aliquots of each sample. To control for differences in sample RNA content, either cyclophilin A (CypA) mRNA or ribosomal RNA (rRNA) was amplified with the cDNA equivalent of 1 ng total RNA from the same samples.
The following probes and primers were designed using Primer- Express software (ABI):
rPer1 forward: 5′-AGCTCTGCTGGAGACCACTGA-3′
rPer1 reverse: 5′-CACTCAGGAGACTATAGGCAATGGA-3′
rPer2 forward: 5′-CCCATCCCACACTTGCCTC-3′
rPer2 reverse: 5′-CACTGTGCCAGCCGGG-3′
rBmal1 forward: 5′-GCAATCTGAGCTGCCTCGTT-3′
rBmal1 reverse: 5′-CCCGTATTTCCCCGTTCACT-3′
rCry1 forward: 5′-AAGTCATCGTGCGCATTTCA-3′
rCry1 reverse: 5′-TCATCATGGTCGTCGGACAGA-3′
rev-erbα forward: 5′-CTCCACAGTGCTGGGAGAGTC-3′
rev-erbα reverse: 5′-CTGGCGTAGACCATTCAGTGC-3′
CypA forward: 5′-TGTGCCAGGGTGGTGACTT-3′
CypA reverse: 5′-TCAAATTTCTCTCCGTAGATGGACTT-3′
Using the comparative CT method described in the ABI Prism 7700 Sequence Detection System User Bulletin no. 2 (PE-ABI), the relative mRNA abundance for a given target gene was calculated by normalization first to corresponding rRNA or cyclophilin levels in each sample and then to a calibrator consisting of pooled cDNA from multiple samples that was analyzed on each reaction plate. Relative abundance of target mRNA was represented as a percentage of the maximal value obtained within an individual experiment.
The temporal patterns of clock gene expression in the SCN, cerebellum, and liver were examined for evidence of circadian variation using statistical analyses that have been used previously for this purpose (Allen et al., 2004a,b; Menger et al., 2005, 2007). Time-dependent fluctuations in clock gene mRNA expression were first identified by one-way analysis of variance (ANOVA). Paired comparisons between peak values and those observed during the preceding or succeeding minimum were analyzed post hoc for statistical differences using the Newman–Keuls sequential range test.
RESULTS
Blood Alcohol Concentration
The administration of 4.5 g /kg/d alcohol in two consecutive feedings out of the twelve daily feedings produced in BACs on PD 9 ranging from 117.6 to 273.6 mg/ dl (or 0.118% to 0.274%). The mean BAC (±SEM) on PD 9 was 204.3 7.2 mg / dl. These BAC values are similar to the those established on PD 9 in previous studies demonstrating that the same alcohol dosage alters the circadian regulation of rat wheel-running behavior (Farnell et al., 2004).
Effects of Neonatal Alcohol Exposure on the Circadian Period of the Rat Activity Rhythm
The period of the rhythm in wheel-running activity was analyzed to confirm the effects of neonatal alcohol treatment on circadian behavior and their similarity to our previous observations on alcohol-induced alterations in this fundamental clock property (Allen et al., 2005a). All animals exhibited free-running rhythms of wheel-running behavior in DD. Consistent with the results of our previous studies (Allen et al., 2005a), the circadian period of the activity rhythm during exposure to DD was significantly shorter in EtOH-exposed rats (mean τ = 23.81 ± 0.02 hours) than in SC and GC animals (mean τ = 24.29 ± 0.02 and 24.28 ± 0.02 hours, respectively; [F(2,87) = 163.60; p < 0.01]).
Effects of Neonatal Alcohol Exposure on the Circadian Rhythms of Clock Gene Expression in the SCN, Cerebellum, and Liver
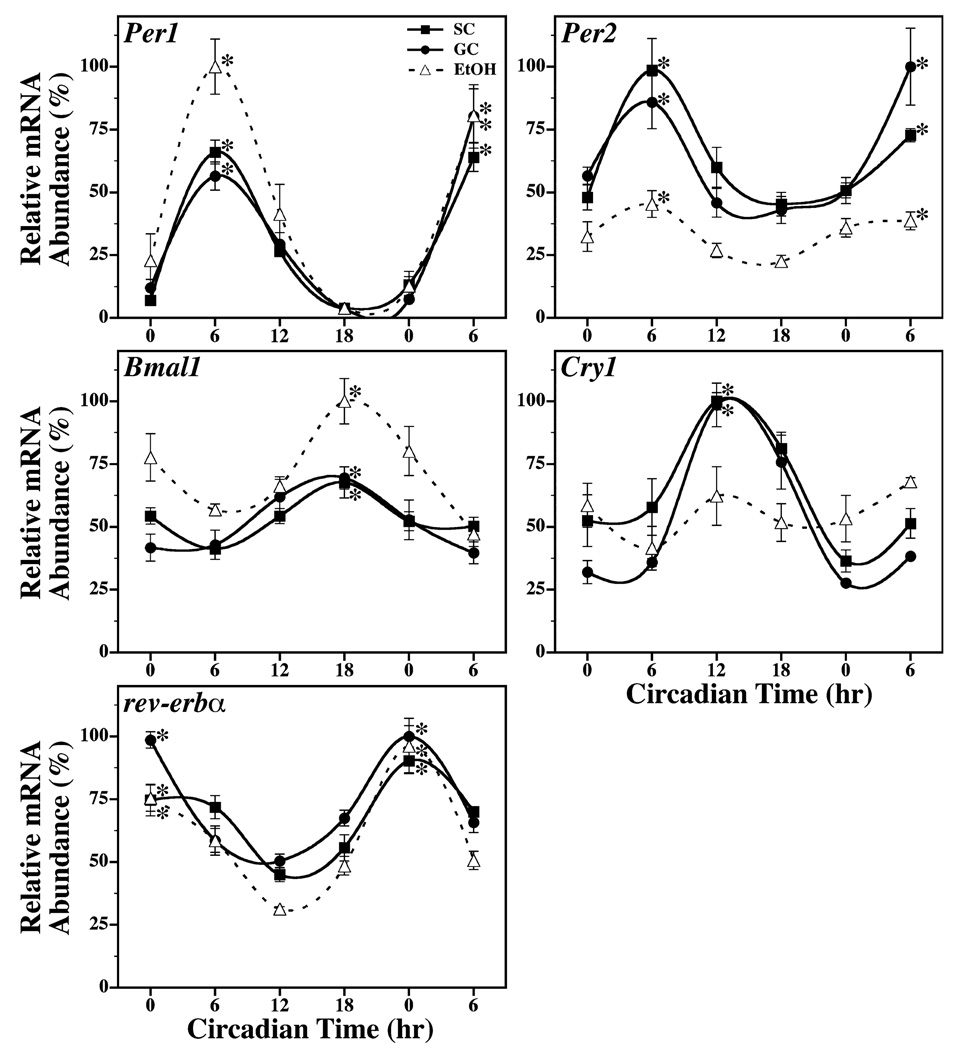
In SC and GC animals, the temporal profiles of Per1, Per2, Cry1, Bmal1, and Rev-erba expression within the SCN were marked by circadian rhythmicity during exposure to DD with a 2- to 9-fold difference between peak and minimum levels (Fig. 1). Separate one-way ANOVAs analyzing all time points for each treatment condition indicated that SCN expression of Per1, Per2, Cry1, Bmal1, and Rev-erba mRNA fluctuated significantly over time in both control groups (p < 0.05). Within the SC and GC groups, post hoc analysis (Newman–Keuls sequential range test) revealed that the rhythmic peaks in SCN mRNA expression at CT 0 for Rev-erba, at CT 6 for Per1 and Per2, at CT 12 for Cry1, and at CT 18 for Bmal1 were significantly greater (p < 0.05) than their corresponding minima. The amplitude and phase of these SCN rhythms in clock gene expression were similar between the SC and GC groups and comparable to those observed previously in the rat SCN (Menger et al., 2005; Oishi et al., 1998; Shearman et al., 1997). EtOH-treated rats also exhibited circadian rhythms of Per1, Per2, Bmal1, and Rev-erba expression in the SCN and these clock gene oscillations resembled those found in both control groups. Within the EtOH group, significant variation was observed in the temporal pattern of Per1, Per2, Bmal1, and Rev-erba expression in the SCN (p < 0.05). The rhythmic peaks of these SCN rhythms in clock gene expression were significantly (p < 0.05) and about 2- to 20-fold higher than their corresponding minima. The phase of these clock gene rhythms in EtOH-exposed rats was coincident with the corresponding oscillations in control animals such that peaks in SCN mRNA expression occurred at CT 0 for Rev-erba, at CT 6 for Per1 and Per2, and at CT 18 for Bmal1. The only notable differences in the EtOH group were that the amplitude of SCN rhythms was increased for Per1 and Bmal1 and decreased for Per2 in comparison with their expression profiles in control animals. In contrast, neonatal alcohol exposure had a pronounced effect in disrupting the temporal pattern of Cry1 expression in the SCN such that mRNA levels were arrhythmic showing no significant variation over the 30-hour interval of analysis.
Fig. 1.
Temporal patterns of Per1, Per2, Bmal1, Cry1 and rev-erbα mRNA expression in the SCN of suckle control (SC, ■), gastrostomy control (GC, ●), and alcohol-treated (EtOH, Δ) rats during exposure to constant darkness. Symbols denote real-time PCR determinations of mRNA levels (mean ± SEM) in SCN tissue collected from 4 to 5 animals at 6-hour intervals for 30 hours. The plotted values correspond to the ratios of species-specific Per1, Per2, Bmal1, Cry1 or rev-erbα / CypA mRNA signal and are represented as a percentage of the maximal value obtained among all groups. Asterisks indicate time points during which peak values for SCN expression of a given gene were significantly greater (p < 0.05) than those observed during preceding or succeeding minima.
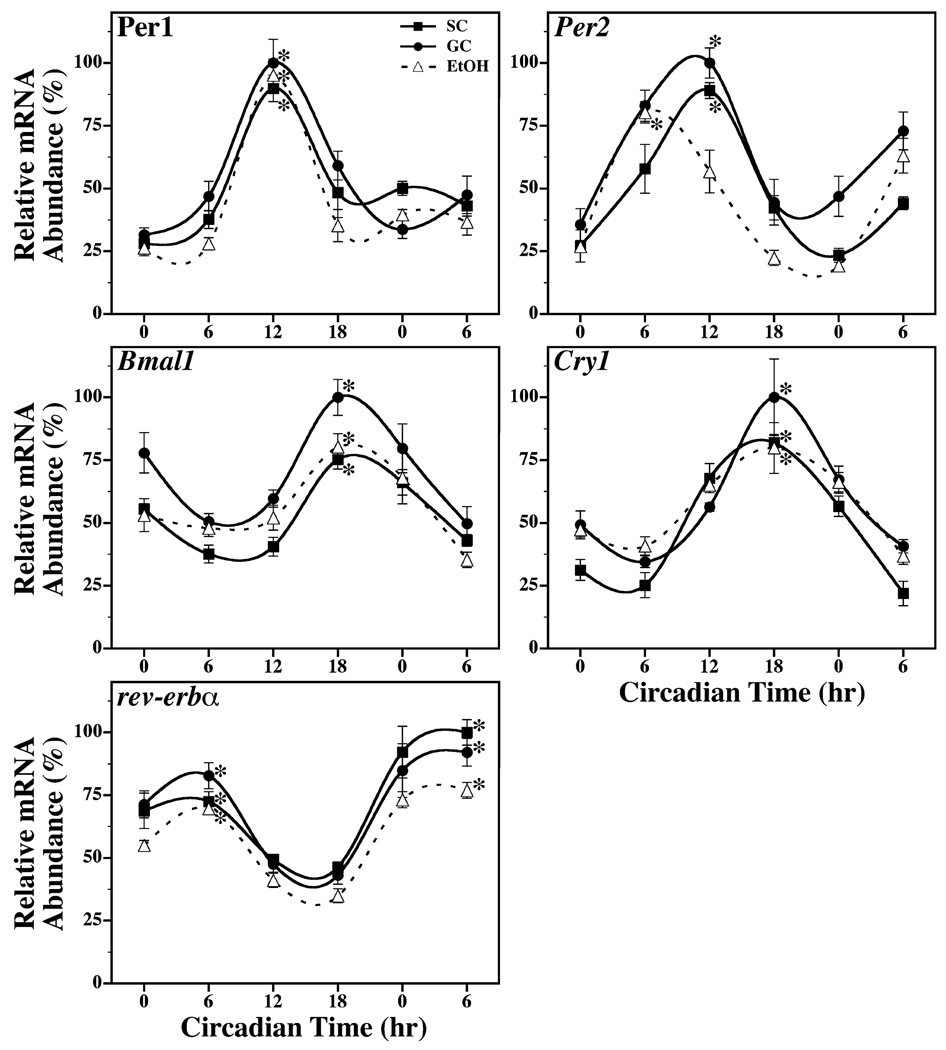
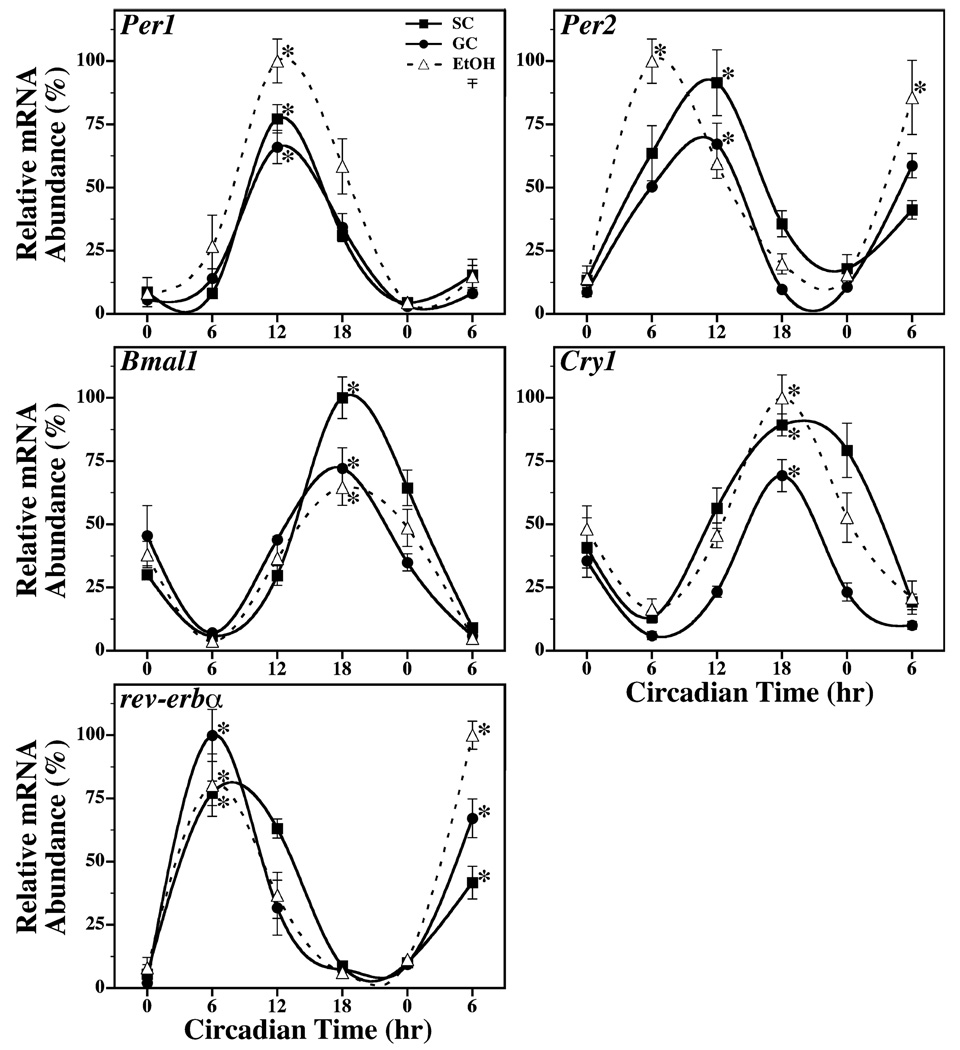
Circadian rhythms of Per1, Per2, Cry1, Bmal1, and Rev-erba expression were also evident in the cerebellum (Fig. 2) and liver (Fig. 3) of SC, GC, and EtOH animals. Significant fluctuations in Per1, Per2, Cry1, Bmal1, and Rev-erba mRNA levels were observed in cerebellar and liver tissue from all groups (p < 0.05). The rhythmic peaks in the expression of these clock genes were significantly greater (p < 0.05) than their corresponding minima, with differences of 2- to 3-fold in the cerebellum and of 5- to 50-fold in the liver. The SC and GC groups exhibited no major differences in the amplitude and phase of cerebellar and liver rhythms in Per1, Per2, Cry1, Bmal1, and Rev-erba expression. Consistent with previous comparisons of SCN, other brain regions and peripheral tissues (Shearman et al., 1997; Yamazaki et al., 2000; Zylka et al., 1998), clock gene oscillations in the cerebellum and liver resembled those found in the SCN except that rhythmic Per1, Per2, Cry1, and Rev-erba expression in these tissues was phase-delayed by 6 hours relative to SCN circadian profiles. In the cerebellum and liver of SC and GC rats, rhythmic mRNA expression reached peak levels at CT 6 for Rev-erba, at CT 12 for Per1 and Per2, and at CT 18 for Cry1. Bmal1 was the only clock gene in which cerebellar and liver profiles of expression were coincident with the SCN rhythm such that peak mRNA levels occurred at CT 18. In EtOH group, circadian oscillations of Per1, Bmal1, Cry1, and Rev-erba expression in the cerebellum and liver were comparable to those found in both control groups. The amplitude of these clock gene rhythms in EtOH-treated rats similarly ranged from 2- to 4-fold in the cerebellum and from 7- to 22-fold in the liver. Significant rhythmic peaks in cerebellar and liver mRNA expression (p < 0.05) were identified at CT 6 for Rev-erba, at CT 12 for Per1, and at CT 18 for Cry1 and Bmal1. The cerebellar and liver rhythms in Per2 expression were altered by neonatal alcohol treatment. The phase of the Per2 oscillations in the cerebellum and liver were shifted in EtOH-treated rats such that peak mRNA expression occurred at CT 6 or 6 hours in advance of the corresponding rhythms observed in SC and GC animals.
Fig. 2.
Temporal patterns of Per1, Per2, Bmal1, Cry1 and rev-erbα mRNA expression in the cerebellum of suckle control (SC, ■), gastrostomy control (GC, ●), and alcohol-treated (EtOH, Δ) rats during exposure to constant darkness. Symbols denote mean (±SEM) determinations of mRNA abundance in cerebellar tissue (n = 4 to 5) at 6-hour intervals. The plotted values correspond to the ratios of Per1, Per2, Bmal1, Cry1 or rev-erbα / CypA mRNA signal in which the maximal value for each gene was set at 100%. Asterisks indicate time points during which peak values for cerebellar expression of a given gene were significantly greater (p < 0.05) than those observed during preceding or succeeding minima.
Fig. 3.
Temporal patterns of Per1, Per2, Bmal1, Cry1 and rev-erbα mRNA expression in the liver of suckle control (SC, ■), gastrostomy control (GC, ●), and alcohol-treated (EtOH, Δ) rats during exposure to constant darkness. Symbols denote mean (±SEM) determinations of mRNA abundance in liver tissue (n = 4 to 5) at 6-hour intervals. The plotted values correspond to the ratios of Per1, Per2, Bmal1, Cry1 or rev-erbα / CypA mRNA signal in which the maximal value for each gene was set at 100%. Asterisks indicate time points during which peak values for liver expression of a given gene were significantly greater (p < 0.05) than those observed during preceding or succeeding minima.
DISCUSSION
The present study demonstrates that alcohol exposure during the period of rapid brain growth produces long-term changes in the molecular core of the circadian timekeeping mechanism in the SCN clockworks and in other brain or tissue oscillators. Early postnatal alcohol treatment had a differential impact on the circadian regulation of clock gene expression in the SCN pacemaker and peripheral-type oscillators in the cerebellum and liver of adult rats. EtOH-treated animals were distinguished by strong damping of the SCN rhythm in Cry1 expression and by phase-advanced oscillations of Per2 expression in the cerebellum and liver with peak mRNA levels occurring during the mid-subjective day. Collectively, these findings indicate that neonatal alcohol exposure produces permanent alterations in essential cogs of the molecular clockworks that mediate the global function of the SCN in pacing circadian rhythms throughout the body and the role of tissue-specific oscillators such as the cerebellum and liver in maintaining the local organization of processes in time.
Similar to the observed effects of postnatal alcohol exposure, alterations in SCN rhythms of clock gene expression occur following prenatal alcohol treatment and in response to chronic alcohol consumption. In rats exposed to alcohol during the prenatal period, the diurnal rhythms of Per1 and Per2 expression within the SCN were distinguished from those found in pair-fed controls by shifts in the phase of peak mRNA levels and the occurrence of a second, smaller peak in the expression of these clock genes (Chen et al., 2006). Prenatal alcohol treatment in this study induced comparable changes in the rhythms of Per1 and Per2 expression by a non-SCN neural region, the arcuate nucleus. Chronic alcohol treatment also affects the diurnal regulation of Per1 and Per2 expression in the SCN of adult rats, producing decreases in rhythm amplitude (Chen et al., 2004). Differences between these findings and the present study with regard to alcohol-induced changes in SCN expression of the Per genes versus Cry1 are presumably related to the timing of treatment (prenatal or chronic adult as opposed to postnatal). However, experimental lighting conditions may represent an underlying factor in these differences because the aforementioned studies examined the effects of alcohol on the diurnal regulation of gene expression during entrainment to a LD 12:12 photoperiod whereas the present observations are based on the analysis of “free-running” or circadian properties during exposure to DD.
These alcohol-induced changes in clock gene rhythms within the SCN circadian pacemaker and other brain and peripheral oscillators may be connected to the effects of alcohol exposure during brain development on the circadian regulation of overt physiological and behavioral rhythms. Alcohol administration during the prenatal or early postnatal period produces long-term perturbations in: (i) circadian output signals from the SCN (Allen et al., 2004b), (ii) the circadian rhythms of core body temperature and plasma corticosterone levels (Lund et al., 2004); (iii) diurnal regulation of splenic cytotoxic factors (Arjona et al., 2006); and (iv) light–dark entrainment, light-induced phase shifts and free-running period of the circadian rhythm in rat wheel-running activity (Allen et al., 2005a,b; Farnell et al., 2004; Sei et al., 2003). The possible connection between the observed damping of the SCN rhythm in Cry1 expression and the decrease in the circadian period of the activity rhythm in alcohol-exposed rats is supported by the observation that disruption of the Cry1 gene produces similar changes in the behavioral rhythms of mutant mice (Vitaterna et al., 1999). In addition, the effect of neonatal alcohol exposure in disrupting an important circadian output of the SCN such as rhythmic BDNF expression (Allen et al., 2004b) may be associated with this alteration in the underlying molecular clockworks. Furthermore, the observed alterations in cerebellar and liver rhythms of Per2 expression suggest that alcohol exposure during critical stages of brain development may have a global impact on the core clockworks of circadian oscillators located in other brain regions and body tissues. Such disturbances in circadian oscillators throughout the body may affect their ability to organize local processes in time and thus may contribute to alcohol-induced perturbations in physiological rhythms of core body temperature, plasma corticosterone, and immune cell responses. Moreover, the differential influence of neonatal alcohol exposure on the Cry1 rhythm in the SCN pacemaker and the liver and cerebellar oscillations in Per2 expression may compound alcohol-induced disturbances in the circadian regulation of these physiological processes.
In conjunction with downstream alterations in physiological rhythms, the effects of developmental alcohol exposure on essential “gears” of the circadian clockworks found in the SCN and other tissues throughout the body may have some implications in the long-term clinical manifestations of fetal alcohol syndrome (FAS) and fetal alcohol spectrum disorders (FASD) (Chudley et al., 2005; Hoyme et al., 2005). Abnormalities in clock gene expression and rhythmicity have been associated with disturbances in human circadian behavior in which the onset of the sleep–wake cycle is advanced (Toh et al., 2001). Moreover, alterations in the regulation of circadian behavior like those induced by neonatal alcohol exposure have been linked to bipolar affective disorder (i.e., manic-depressive illness) and depression (Moore, 1991; Schwartz, 1993). Because sleep–wake and affective disorders are known clinical correlates of alcohol exposure during brain development (Sher, 2004), it is possible that similar perturbations in clock gene oscillations and patterns of circadian behavior may contribute to the incidence of these neurobehavioral disturbances in FAS and FASD.
Normal circadian timekeeping function of the SCN is thought to have an impact on human health and performance by providing for the temporal coordination of internal processes with each other and with the daily light–dark cycle. Because the efficacy and side effects of many drugs are influenced by the time of administration in relation to normal cycles for body processes, circadian rhythms are important in chronotherapeutic phenomena in the pharmacologic treatment of diseases. Circadian rhythm pathologies have been associated with a variety of human health problems including cancer, cardiovascular accidents, obesity, diabetes, affective disorders, and sleep–wake disturbances. Thus, further analysis is necessary to elucidate the full impact of circadian dysfunction on human health in neonates, children, and adults exposed to alcohol in utero.
ACKNOWLEDGMENTS
This study was supported by NIH grants AA13242 and MH60147 (D.J.E).
REFERENCES
- Albers HE. Gonadal hormones organize and modulate the circadian system of the rat. Am J Physiol. 1981;241:R62–R66. doi: 10.1152/ajpregu.1981.241.1.R62. [DOI] [PubMed] [Google Scholar]
- Albers HE, Gerall AA, Axelson JF. Effect of reproductive state on circadian periodicity in the rat. Physiol Behav. 1981;26:21–25. doi: 10.1016/0031-9384(81)90073-1. [DOI] [PubMed] [Google Scholar]
- Allen GC, Farnell Y, Bell-Pedersen D, Cassone VM, Earnest DJ. Effects of altered Clock gene expression on the pacemaker properties of SCN2.2 cells and oscillatory properties of NIH/3T3 cells. Neuroscience. 2004a;127:989–999. doi: 10.1016/j.neuroscience.2004.06.003. [DOI] [PubMed] [Google Scholar]
- Allen GC, Farnell YZ, Maeng J-U, West JR, Chen W-JA, Earnest DJ. Long-term effects of neonatal alcohol exposure on photic re-entrainment and phase–shifting responses of the activity rhythm in adult rats. Alcohol. 2005b;37:79–88. doi: 10.1016/j.alcohol.2005.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen GC, West JR, Chen W-JA, Earnest DJ. Developmental alcohol exposure disrupts circadian regulation of BDNF in the rat suprachiasmatic nucleus. Neurotoxicol Teratol. 2004b;26:353–358. doi: 10.1016/j.ntt.2004.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen GC, West JR, Chen W-JA, Earnest DJ. Neonatal alcohol exposure permanently disrupts the circadian properties and photic entrainment of the activity rhythm in adult rats. Alcohol Clin Exp Res. 2005a;29:1845–1852. doi: 10.1097/01.alc.0000183014.12359.9f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arjona A, Boyadjieva N, Kuhn P, Sarkar DK. Fetal ethanol exposure disrupts the daily rhythms of splenic granzyme B, IFN-gamma, and NK cell cytotoxicity in adulthood. Alcohol Clin Exp Res. 2006;30:1039–1044. doi: 10.1111/j.1530-0277.2006.00117.x. [DOI] [PubMed] [Google Scholar]
- Bae K, Jin X, Maywood ES, Hastings MH, Reppert SM, Weaver DR. Differential functions of mPer1, mPer2, and mPer3 in the SCN circadian clock. Neuron. 2001;30:525–536. doi: 10.1016/s0896-6273(01)00302-6. [DOI] [PubMed] [Google Scholar]
- Bonthius DJ, Bonthius NE, Napper RM, West JR. Early postnatal alcohol exposure acutely and permanently reduces the number of granule cells and mitral cells in the rat olfactory bulb: a stereological study. J Comp Neurol. 1992;324:557–566. doi: 10.1002/cne.903240408. [DOI] [PubMed] [Google Scholar]
- Bonthius DJ, West JR. Blood alcohol concentration and microencephaly: a dose-response study in the neonatal rat. Teratology. 1988;37:223–231. doi: 10.1002/tera.1420370307. [DOI] [PubMed] [Google Scholar]
- Bonthius DJ, West JR. Alcohol-induced neuronal loss in developing rats: increased brain damage with binge exposure. Alcohol Clin Exp Res. 1990;14:107–118. doi: 10.1111/j.1530-0277.1990.tb00455.x. [DOI] [PubMed] [Google Scholar]
- Chen CP, Kuhn P, Advis JP, Sarkar DK. Chronic ethanol consumption impairs the circadian rhythm of pro-opiomelanocortin and period genes mRNA expression in the hypothalamus of the male rat. J Neurochem. 2004;88:1547–1554. doi: 10.1046/j.1471-4159.2003.02300.x. [DOI] [PubMed] [Google Scholar]
- Chen CP, Kuhn P, Advis JP, Sarkar DK. Prenatal ethanol exposure alters the expression of period genes governing the circadian function of beta-endorphin neurons in the hypothalamus. J Neurochem. 2006;97:1026–1033. doi: 10.1111/j.1471-4159.2006.03839.x. [DOI] [PubMed] [Google Scholar]
- Chen W-JA, Parnell SE, West JR. Neonatal alcohol and nicotine exposure limits brain growth and depletes cerebellar Purkinje cells. Alcohol. 1998;15:33–41. doi: 10.1016/s0741-8329(97)00084-0. [DOI] [PubMed] [Google Scholar]
- Chen W-JA, Parnell SE, West JR. The effects of alcohol and nicotine on the developing olfactory bulb: loss of mitral cells and changes in neurotransmitter levels. Alcohol Clin Exp Res. 1999;23:18–25. [PubMed] [Google Scholar]
- Chudley AE, Conry J, Cook JL, Loock C, Rosales T, LeBlanc N. Fetal alcohol spectrum disorder: Canadian guidelines for diagnosis. CMAJ. 2005;172 Suppl. 5:S1–S21. doi: 10.1503/cmaj.1040302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz J. Experimental rearing of rat pups using chronic gastric fistulas. In: Shair N, Barr A, Hofer A, editors. Developmental Psychobiology: New Methods and Changing Concepts. New York: Oxford Univ Press; 1991. pp. 272–284. [Google Scholar]
- Dobbing J, Sands J. Comparative aspects of the brain growth spurt. Early Hum Dev. 1979;3:79–83. doi: 10.1016/0378-3782(79)90022-7. [DOI] [PubMed] [Google Scholar]
- Earnest DJ, Sladek CD. Circadian rhythms of vasopressin release from individual rat suprachiasmatic explants in vitro. Brain Res. 1986;382:129–133. doi: 10.1016/0006-8993(86)90119-8. [DOI] [PubMed] [Google Scholar]
- Farnell YZ, West JR, Chen W-JA, Allen GC, Earnest DJ. Developmental alcohol exposure alters light-induced phase shifts of the circadian activity rhythm in rats. Alcohol Clin Exp Res. 2004;28:1020–1027. doi: 10.1097/01.alc.0000130807.21020.1b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodlett CR, Kelly SJ, West JR. Early postnatal alcohol exposure that produces high blood alcohol levels impairs development of spatial navigation learning. Psychobiology. 1987;15:64–74. [Google Scholar]
- Goodlett CR, Nonneman AJ, Valentino ML, West JR. Constraints on water maze spatial learning in rats: implications for behavioral studies of brain damage and recovery of function. Behav Brain Res. 1988;28:275–286. doi: 10.1016/0166-4328(88)90130-1. [DOI] [PubMed] [Google Scholar]
- Heid CA, Stevens J, Livak KJ, Williams PM. Real time quantitative PCR. Genome Res. 1996;6:986–994. doi: 10.1101/gr.6.10.986. [DOI] [PubMed] [Google Scholar]
- van der Horst GTJ, Muijtjens M, Kobayashi K, Takano R, Kanno S, Takao M, de Wit J, Verkerk A, Eker APM, van Leenen D, Buijs R, Bootsma D, Hoeijmakers JHJ, Yasui A. Mammalian Cry1 and Cry2 are essential for maintenance of circadian rhythms. Nature. 1999;398:627–630. doi: 10.1038/19323. [DOI] [PubMed] [Google Scholar]
- Hoyme HE, May PA, Kalberg WO, Kodituwakku P, Gossage JP, Trujillo PM, Buckley DG, Miller JH, Aragon AS, Khaole N, Viljoen DL, Jones KL, Robinson LK. A practical clinical approach to diagnosis of fetal alcohol spectrum disorders: clarification of the 1996 Institute of Medicine criteria. Pediatrics. 2005;115:39–47. doi: 10.1542/peds.2004-0259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly SJ, Goodlett CR, Hulsether SA, West JR. Impaired spatial navigation in adult female but not adult male rats exposed to alcohol during the brain growth spurt. Behav Brain Res. 1988;27:247–257. doi: 10.1016/0166-4328(88)90121-0. [DOI] [PubMed] [Google Scholar]
- Liang F-Q, Walline R, Earnest DJ. Circadian rhythm of brain-derived neurotrophic factor in the rat suprachiasmatic nucleus. Neurosci Lett. 1998;242:89–92. doi: 10.1016/s0304-3940(98)00062-7. [DOI] [PubMed] [Google Scholar]
- Lund TD, McGivern RF, Handa RJ. Prenatal exposure to ethanol alters biological rhythms of adult male rats (abstract) Alcohol Clin Exp Res. 2004;28 Suppl. 5:136A. [Google Scholar]
- Menger GJ, Allen GC, Neuendorff N, Nahm S-S, Thomas T, Cassone VM, Earnest DJ. Circadian profiling of the transcriptome in NIH/3T3 fibroblasts: comparison with rhythmic gene expression in SCN2.2 cells and the rat SCN. Physiol Genomics. 2007;29:280–289. doi: 10.1152/physiolgenomics.00199.2006. [DOI] [PubMed] [Google Scholar]
- Menger GJ, Lu K, Thomas T, Cassone VM, Earnest DJ. Circadian profiling of the transcriptome in immortalized rat SCN cells. Physiol Genomics. 2005;21:370–381. doi: 10.1152/physiolgenomics.00224.2004. [DOI] [PubMed] [Google Scholar]
- Moore RY. Disorders of circadian function and the human circadian timing system. In: Klein DC, Moore RY, Reppert SM, editors. Suprachiasmatic Nucleus: The Mind’s Clock. New York: Oxford Univ Press; 1991. pp. 429–441. [Google Scholar]
- Morin LP, Fitzgerald KM, Zucker I. Estradiol shortens the period of hamster circadian rhythms. Science. 1977;196:305–307. doi: 10.1126/science.557840. [DOI] [PubMed] [Google Scholar]
- Oishi K, Sakamoto K, Okada T, Nagase T, Ishida N. Antiphase circadian expression between Bmal1 and period homologue RNA in the suprachiasmatic nucleus and peripheral tissues of rats. Biochem Biophys Res Commun. 1998;253:199–203. doi: 10.1006/bbrc.1998.9779. [DOI] [PubMed] [Google Scholar]
- Reppert SM, Weaver DR. Coordination of circadian timing in mammals. Nature. 2002;418:935–941. doi: 10.1038/nature00965. [DOI] [PubMed] [Google Scholar]
- Roebuck TM, Mattson SN, Riley EP. Prenatal exposure to alcohol: effects on brain structure and neuropsychological functioning. In: Hannigan JH, Spear LP, Spear NE, Goodlett CR, editors. Alcohol and Alcoholism: Effects on Brain and Development. Mahwah: Lawrence Erlbaum Assoc; 1999. pp. 1–16. [Google Scholar]
- Schwartz WJ. A clinician’s primer on the circadian clock: its localization, function and resetting. Adv Intern Med. 1993;38:81–106. [PubMed] [Google Scholar]
- Sei H, Sakata-Haga H, Ohta K, Sawada K, Morita Y, Fukui Y. Prenatal exposure to alcohol alters the light response in postnatal circadian rhythm. Brain Res. 2003;987:131–134. doi: 10.1016/s0006-8993(03)03329-8. [DOI] [PubMed] [Google Scholar]
- Shearman LP, Zylka MJ, Weaver DR, Kolakowski LF, Jr, Reppert SM. Two period homologs: circadian expression and photic regulation in the suprachiasmatic nuclei. Neuron. 1997;19:1261–1269. doi: 10.1016/s0896-6273(00)80417-1. [DOI] [PubMed] [Google Scholar]
- Sher L. Etiology, pathogenesis, and treatment of seasonal and non-seasonal mood disorders: possible role of circadian rhythm abnormalities related to developmental alcohol exposure. Med Hypotheses. 2004;62:797–801. doi: 10.1016/j.mehy.2003.12.021. [DOI] [PubMed] [Google Scholar]
- Thomas JD, Goodlett CR, West JR. Alcohol-induced Purkinje cell loss depends on developmental timing of alcohol exposure and correlates with motor performance. Brain Res Dev Brain Res. 1998;105:159–166. doi: 10.1016/s0165-3806(97)00164-8. [DOI] [PubMed] [Google Scholar]
- Thomas JD, Wasserman EA, West JR, Goodlett CR. Behavioral deficits induced by bingelike exposure to alcohol in neonatal rats: importance of developmental timing and number of episodes. Dev Psychobiol. 1996;29:433–452. doi: 10.1002/(SICI)1098-2302(199607)29:5<433::AID-DEV3>3.0.CO;2-P. [DOI] [PubMed] [Google Scholar]
- Toh KL, Jones CR, He Y, Eide EJ, Hinz WA, Virshup DM, Ptacek LJ, Fu YH. An hPer2 phosphorylation site mutation in familial advanced sleep phase syndrome. Science. 2001;291:1040–1043. doi: 10.1126/science.1057499. [DOI] [PubMed] [Google Scholar]
- Vitaterna MH, Selby CP, Todo T, Niwa H, Thompson C, Fruechte EM, Hitomi K, Thresher RJ, Ishikawa T, Miyazaki J, Takahashi JS, Sancar A. Differential regulation of mammalian period genes and circadian rhythmicity by cryptochromes 1 and 2. Proc Natl Acad Sci U S A. 1999;96:12114–12119. doi: 10.1073/pnas.96.21.12114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West JR. Use of pup in a cup model to study brain development. J Nutr. 1993;123:382–385. doi: 10.1093/jn/123.suppl_2.382. [DOI] [PubMed] [Google Scholar]
- West JR, Hamre KM, Pierce DR. Delay in brain growth induced by alcohol in artificially reared rat pups. Alcohol. 1984;1:213–222. doi: 10.1016/0741-8329(84)90101-0. [DOI] [PubMed] [Google Scholar]
- Yamazaki S, Numano R, Abe M, Hida A, Takahashi R, Ueda M, Block GD, Sakaki Y, Menaker M, Tei H. Resetting central and peripheral circadian oscillators in transgenic rats. Science. 2000;288:682–685. doi: 10.1126/science.288.5466.682. [DOI] [PubMed] [Google Scholar]
- Zheng BH, Albrecht U, Kaasik K, Sage M, Lu WQ, Vaishnav S, Li Q, Sun ZS, Eichele G, Bradley A, Lee CC. Nonredundant roles of the mPer1 and mPer2 genes in the mammalian circadian clock. Cell. 2001;105:683–694. doi: 10.1016/s0092-8674(01)00380-4. [DOI] [PubMed] [Google Scholar]
- Zylka MJ, Shearman LP, Weaver DR, Reppert SM. Three period homologs in mammals: differential light responses in the suprachiasmatic circadian clock and oscillating transcripts outside of brain. Neuron. 1998;20:1103–1110. doi: 10.1016/s0896-6273(00)80492-4. [DOI] [PubMed] [Google Scholar]