Abstract
Aims
Nonsense mutations in the SCN5A gene result in truncated, non-functional derivatives of the cardiac Na+ channel and thus cause arrhythmias. Studies of other genes suggest that pathogenic phenotypes of nonsense mutations may be alleviated by enhancing readthrough, which enables ribosomes to ignore premature termination codons and produce full-length proteins. Thus, we studied the functional restoration of nonsense-mutated SCN5A.
Methods and results
HEK293 cells were transfected with SCN5A cDNA or its mutant carrying W822X, a nonsense mutation associated with Brugada syndrome and sudden cardiac death. The effects of readthrough-enhancing reagents on Na+ channel expression and function were examined in the transfected cells. W822X robustly reduced Na+ current, decreasing maximal Na+ current to <3% of the wild-type level, and inhibited the expression of full-length Na+ channels. When readthrough was enhanced by either reducing translational fidelity with aminoglycosides or decreasing translation termination efficiency with small-interfering RNA against eukaryotic release factor eRF3a, Na+ current of the mutant was restored to ∼30% of the wild-type level; western blot and immunochemical staining analyses showed the increased expression of full-length channels. When the wild-type and mutant cDNAs were co-transfected, readthrough-enhancing reagents increased Na+ current from 56% to 74% of the wild-type level. Analysis of Na+ channel kinetics showed that the channels expressed from the mutant cDNA under readthrough-enhancing conditions retained the functions of wild-type channels.
Conclusion
Readthrough-enhancing reagents can effectively suppress SCN5A nonsense mutations and may restore the expression of full-length Na+ channels with normal functions, which might prevent sudden cardiac death in mutation carriers.
KEYWORDS: Sodium channels, Aminoglycosides, Eukaryotic release factors, siRNA, Nonsense mutation
1. Introduction
The cardiac Na+ channel carries the Na+ current responsible for the rapid upstroke of the action potential in the heart. SCN5A is the gene that encodes the α-subunit, Nav1.5, that confers major physiological functions pertaining to the cardiac Na+ channel. Mutations in SCN5A have been linked to a number of inherited cardiac diseases related to abnormal excitability of the heart, such as long QT syndrome, Brugada syndrome, and conduction disorders.1 Studies at molecular and cellular levels have indicated that genetic defects in SCN5A cause one of the following abnormal phenotypes: gain-of-function, loss-of-function, and complex changes in channel gating (in which characteristics of both gain-of-function and loss-of-function are combined). The consensus in the literature suggests that the gain-of-function phenotype is associated with long QT syndrome, whereas the loss-of-function phenotype is frequently linked to Brugada syndrome and conduction disorders.1 The latest studies also link Na+ current reduction to structural cardiac diseases, particularly dilated cardiomyopathy.2
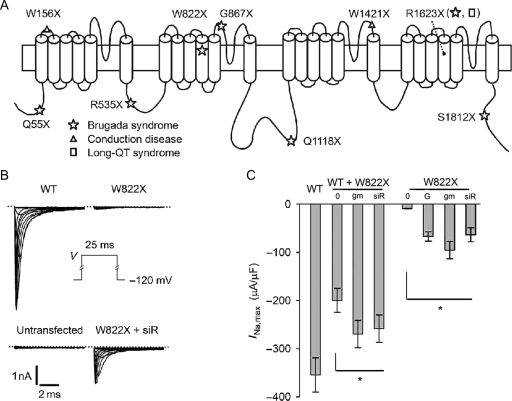
Among the known pathogenic mutations of SCN5A is a group of nonsense point mutations. An in-frame nonsense mutation introduces a premature termination codon (PTC) in the open reading frame of an allele; this type of mutation accounts for 12% of all reported human gene mutations3 and is responsible for ∼5–15% of most genetic disorders, although in certain populations, the incidence of this type of mutation is much higher.4 Nonsense mutation leads to a truncated protein that in many cases does not have function, especially when the truncation is large. In mice heterozygous for targeted disruption of SCN5A, truncation has been shown to behave as a loss-of-function allele, leading to a ∼50% reduction in Na+ current.5 In inherited diseases related to SCN5A, nonsense mutation is one of the major genetic defects underlying the loss-of-function pathogenic phenotype. When our study was conducted, nine nonsense mutations that cause inherited cardiac conditions had been documented (Figure 1A). Some of these mutations were studied in heterologous cells, and it was reported that the mutations led to a complete silencing of channel expression because no currents were recorded in cells injected with cRNA or transfected with cDNA carrying the mutations.6–9
Figure 1.
Increase in Na+ currents in response to aminoglycosides and siRNA against eRF3a in HEK293 cells transfected with nonsense mutants of SCN5A. (A) Distribution of nonsense mutations in the structure of the Nav1.5 channel. (B) Whole-cell Na+ currents (INa) elicited by the voltage protocol shown in the inset. The dotted lines indicate the zero current level. (C) Comparison of maximal INa (INa,max). WT, WT + W822X, and W822X: cells transfected with wild-type and nonsense mutants of SCN5A; 0, no drug or no siR; G, G418; gm, gentamicin; siR, cells co-transfected with the vector producing siRNA against eRF3a. Asterisk indicates a significant difference between the untreated group and all treated groups of a given mutant.
Studies of various genes have suggested that harmful consequences caused by nonsense mutation may be avoided or reduced to a tolerable level by enhancing the process known as translational readthrough, which enables ribosomes to ignore the stop codon and produce full-length proteins.10 Enhanced readthrough can be achieved in many ways. For example, disturbances to the ribosome, tRNAs, and the eukaryotic release factors eRF1 and eRF3a (a GTPase that binds eRF1 and facilitates the efficiency and accuracy of stop codon recognition)11 can all alter the level of readthrough. Pharmacological approaches to increasing the level of readthrough include the use of aminoglycosides and other small molecules that bind to the ribosome, suppressor tRNAs that recognize stop codons, eRF1-binding RNAs, and inhibition of eRF expression by small-interfering RNA (siRNA) molecules.12 Notably, aminoglycosides and inhibition of eRFs can to some extent preferentially enhance readthrough of PTCs, which reduces the chances of translation beyond the natural stop codon and generation of proteins carrying extraneous amino acids.12
Extensive studies using these reagents, particularly the aminoglycosides, have been performed on nonsense mutations of several genes, such as CFTR gene mutations that cause cystic fibrosis13 and dystrophin gene mutations that cause Duchenne muscular dystrophy.14 In mouse models, both disorders can be effectively treated with the aminoglycoside gentamicin.15,16 Clinical trials have been conducted to test whether aminoglycosides and other ribosome-binding molecules are also effective in humans.10 Some studies have demonstrated that siRNA directed against mRNA encoding eRFs is also effective inhibitors of translation termination.11,12,17 Theoretically, this technique induces misreading only at stop codons;17 thus, it has the potential to be an alternative to the antibiotics without the drawback of global mistranslation.
To our knowledge, no study to date has investigated the restoration of functional Nav1.5 channels that are lost due to nonsense mutations in the SCN5A gene. In this study, we evaluated two classes of reagents that enhance readthrough: aminoglycosides and siRNA against eRF3a. We applied these reagents to one nonsense mutation, W822X (c.2466G>A)7,18 that creates a premature stop codon, TGA, and is located near the middle point of the channel sequence. We performed experiments to address two fundamental questions. First, we tested whether an increase in stop codon readthrough would restore high-level production of full-length Na+ channels from a disrupted SCN5A gene containing a nonsense mutation. Secondly, we tested whether the restored Na+ channels had normal or abnormal function.
2. Methods
2.1. cDNAs and mutagenesis
The wild-type SCN5A cDNA used in this study encodes the full-length Nav1.5 channel containing 2016 amino acids19 and matches the exons of a reference genomic sequence (GenBank accession no. AC137587). We used a PCR-based method20 to generate the desired point mutation (W822X) and fully sequenced the cDNA fragment containing the mutation in both sense and antisense directions.
2.2. Cell culture and transfection
HEK293 cells (ATCC, Manassas, VA, USA) were maintained in culture with or without the antibiotics penicillin and streptomycin. Experiments to determine whether these two antibiotics influence the expression of transfected genes, including those of mutated genes, revealed that they had virtually no effect on channel expression. However, to ensure that these antibiotics did not influence the results, in most experiments, we withdrew them from the culture medium prior to transfection and kept the cells in culture without them thereafter. Wild-type and mutated cDNAs of SCN5A were transfected using an Effectene kit (Qiagen, Hilden, Germany). eGFP-pCDNA3 (Invitrogen, San Diego, CA, USA) was co-transfected to express eGFP as a transfection indicator for electrophysiological experiments. The exception was the siRNA experiments, in which a vector carrying the GFP gene was used. Expression typically peaked at 48–60 h after the transfection, during which time the cells were studied. Gentamicin (Sigma, St Louis, MO, USA) or G418 (Invitrogen) was added at 200 µg/mL or a concentration otherwise indicated into the medium 12 h after the transfection.
2.3. Sodium dodecyl sulfate–polyacrylamide gel electrophoresis and immunoblotting
Membrane proteins and cytoplasmic proteins were separated and extracted using a CelLytic MEM protein extraction kit (Sigma). The extracted proteins were analysed with 4–7.5% sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS–PAGE) and transferred onto a PVDF membrane (Millipore, Billerica, MA, USA). We used two antibodies (Alomone Labs, Jerusalem, Israel), one recognizing the sequence D493-C511 in the N-terminal and another recognizing the sequence D1978-V2016 in the C-terminal of the Nav1.5 channel, to detect full-length Nav1.5 and its mutants. A bound antibody was visualized on X-ray film using an ImmunoPure IgG(Fc) antibody conjugated with alkaline phosphatase and a Lumi-phos WB chemiluminescent substrate (Pierce, Rockford, IL, USA). An antibody against human eRF3a (ProteinTech Group, Chicago, IL, USA) and an antibody against human glyceraldehydes-3-phosphate dehydrogenase (GAPDH, Bethyl Laboratories, Montgomery, TX, USA) were also used in western blot analysis.
2.4. siRNA expression
We generated siRNA against eRF3a mRNA via small hairpin RNA transcribed from an oligo insert cloned into the plasmid vector pSuper.gfp/neo (OligoEngine, Inc., Seattle, WA, USA). The targeted sequence11 was 5′-GAGGAACAGUCAUUGUGUG-3′. The siRNA expression vector was transfected into cells to produce siRNA molecules and GFP as an expression indicator.
2.5. Immunofluorescence staining
The technique used was similar to that described in a previous publication.20 Briefly, cells were fixed, blocked, and incubated with the anti-Nav1.5 antibody at 4°C overnight. The cells were then incubated for 4 h with a secondary antibody, goat anti-rabbit-Alexa Fluor 610-R-P (Invitrogen). Immunofluorescence staining was viewed with an LSM 510 confocal microscope (Zeiss, Oberkochen, Germany). To evaluate the effects of nonsense mutation suppression on cellular function, cells were fixed, blocked, and reacted with a rabbit anti-H2AX antibody (Serotec, Oxford, UK) for 2 h at room temperature. After being washed with PBS, the cells were incubated with a secondary fluorescein-conjugated goat anti-rabbit antibody (Serotec) for 30 min at room temperature. Hoechst 33342 staining was performed as described previously.21 The stained cells were examined with a TE800 fluorescence microscope (Nikon, Tokyo, Japan).
2.6. Patch-clamp recordings of Na+ current
Most details of the patch-clamp recording of whole-cell Na+ current and data analysis are described in a previous publication.22 Whole-cell Na+ channel current (INa) was recorded with an Axopatch 200 amplifier (Molecular Device, Sunnyvale, CA, USA) at room temperature in a bath (extracellular) solution containing (in mM) 140 NaCl, 4 KCl, 1.8 CaCl2, 0.75 MgCl2, and 5 HEPES (pH 7.4 set with NaOH) and a pipette (intracellular) solution containing (in mM) 120 CsF, 20 CsCl, 5 EGTA, and 5 HEPES (pH 7.4 with CsOH).
2.7. Data analysis and statistics
We analysed INa data using pClamp9 (Molecular Device). Statistical results are presented as mean ± SE. The number of samples was 8–40 for each data point, with an average of 15 samples per point. Comparisons between two data groups were performed using Student's t-test. We considered P < 0.05 to be statistically significant.
3. Results
3.1. W822X inhibited expression of full-length channels
The wild-type Nav1.5 channels and channels expressed from the mutated cDNAs of SCN5A were studied in transfected HEK293 cells. The HEK293 cells themselves did not exhibit a substantial level of INa, nor did they react with the anti-Nav1.5 antibody. In contrast, cells transfected with the wild-type SCN5A cDNA had large INa. The mutation W822X robustly reduced INa, decreasing maximal INa to <3% of the wild-type level, although it did not abolish INa completely (Figure 1B and C). This finding is consistent with the common belief that PTCs have lower termination efficiencies. Western blot analysis confirmed the presence of full-length channel proteins in the cells transfected with the mutated cDNAs, although they were found in trace amounts when compared with the wild-type channels (Figure 2A).
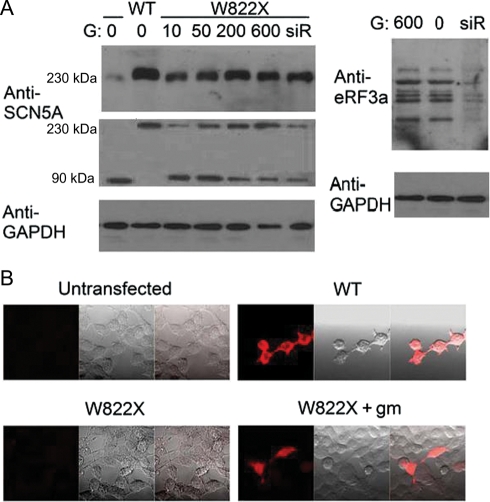
Figure 2.
Expression of full-length Na+ channels in cells transfected with nonsense mutants of SCN5A under enhanced readthrough conditions. (A) Western blot analysis. Upper images: channel proteins detected by the anti-C-terminal Nav1.5 antibody; each positive sample shows a single band at ∼230 kDa as expected for the full-length Nav1.5 channel; lower images: channel proteins detected by the anti-N-terminal Nav1.5 antibody; each positive sample shows two bands at ∼90 and ∼230 kDa, respectively. GAPDH also is displayed to ensure equal loading of samples; right image: eRF3a detected by the anti-eRF3a antibody to show the suppression effect of siRNA. (B) Immunochemical staining analysis. In each image set, the red fluorescent image on the left displays the full-length Na+ channels in cells detected by the anti-C-terminal Nav1.5 antibody, the light image in the centre displays the cells, and the image on the right superimposes the fluorescent and light images. U or untransfected: untransfected HEK293 cells; the numbers above the sample lanes indicate the concentrations of G418 in µg/mL; other labels are the same as for Figure 1.
3.2. Aminoglycosides and siRNA against eRF3a increased the production of full-length channels from the W822X nonsense mutant
Na+ channel expression from the cDNA carrying the W822X mutation was improved greatly in terms of both INa and protein levels in cells that were treated with gentamicin and G418 (Figures 1B and 2A). Each drug increased maximal INa by about eight-fold to ∼30% of the wild-type level (Figure 1C). For simplicity, we denote the channels expressed from cDNA carrying the W822X mutation as W822X channels. In cells co-transfected with the wild-type and mutant cDNAs, the maximal INa was 56% of the wild-type level, and it was increased to as much as 74% of the wild-type level in the cells treated with the reagents.
Paralleling the INa increase was a profound increase in the amount of full-length channel protein produced (Figure 2A). The reagents’ effect was dose-dependent and reached a peak at ∼200 µg/mL in the range tested. Of note, on the western blot using the anti-C-terminal antibody, only a single, clear protein band at the predicted Nav1.5 size was visible, indicating that extension beyond the natural stop did not occur.
The next set of experiments examined the effect of lowering the cellular level of eRF3a by generating siRNA molecules that inhibited eRF3a expression. Western blot analysis (Figure 2A) showed considerably less eRF3a in cells transfected with the siRNA expression vector than in untransfected cells, verifying that the siRNA worked effectively. In contrast, the eRF3a level was unchanged in cells treated with aminoglycosides. We expected this result because aminoglycosides enhance readthrough by binding to the ribosomes, not by interfering with the expression of eRFs. When the siRNA expression vector was co-transfected with a nonsense mutant cDNA, the transfected cells exhibited a large INa compared with those transfected with the mutant alone (Figure 1B and C). Results of western blot analysis agreed with the INa data, displaying a significant increase in the amount of full-length channel protein expressed by the co-transfected cells (Figure 2A).
We also visualized the increased expression of protein from the mutated cDNA in response to an elevated readthrough level through immunochemical staining of full-length channel proteins. Under our experimental conditions, cells transfected with the wild-type cDNA reacted with the Nav1.5 antibody and therefore emitted red fluorescence that was not seen in untransfected cells. In cells transfected with the nonsense mutant cDNA, fluorescence was clearly seen only in those treated with gentamicin (an example is given in Figure 2B), G418, and siRNA against eRF3a, but not in those without treatment.
3.3. W822X channels had a wild-type phenotype
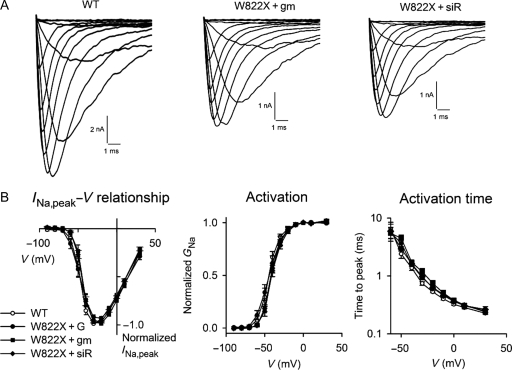
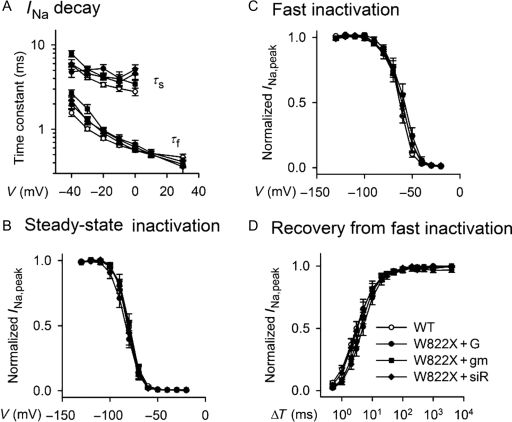
The INa of W822X channels had characteristic similar to that of wild-type channels (Figure 3A). Further analyses revealed that the INa,peak–V relationships of W822X channels were the same as those of wild-type channels, regardless of how readthrough was enhanced. Note that INa recorded from cells transfected with the mutated cDNA alone in the absence of enhanced readthrough was too small to be analysed with confidence, thus we did not analyze it in detail. W822X channels behaved virtually identically to wild-type channels in all aspects of kinetics and voltage dependency that were examined. No W822X channel group was statistically distinguishable from the wild-type group at any voltage range. Figures 3–5 provide the data from the W822X groups in parallel with those of the wild-type channels.
Figure 3.
Activation kinetics of Na+ channels. (A) INa of wild-type and W822X channels. The expression of W822X channels was elevated by exposure to gentamicin and siRNA against eRF3a. (B) INa,peak−V relationship (left panels), voltage dependence of activation (centre panels), and voltage dependence of the time to peak (right panels) for wild-type and mutant channels. INa,peak was normalized to maximal INa in order to compare the voltage dependence. Labels are the same as for Figure 1.
Figure 4.
Inactivation and recovery kinetics of Na+ channels. (A) Voltage dependence of fast and slow time constants of INa decay. The time constants were obtained by fitting the decay portions of INa in response to the voltage protocol in the inset of Figure 1B with a two-exponential function, Af×exp(−t/τf)+As×exp(−t/τs)+offset, where t is time and τf and τs are fast and slow time constants, respectively. (B and C) Steady-state and fast inactivation, respectively. (D) Kinetics of Na+ channel recovery from fast inactivation. The peak INa elicited by a test pulse in the voltage protocols depicted in the left panels was plotted against the voltage (V) of the conditioning pulse that preceded the test pulse. Labels are the same as for Figure 1.
Figure 5.
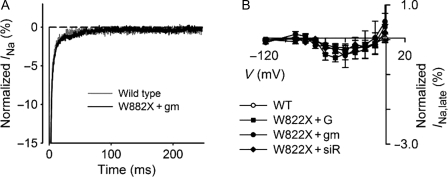
Late Na+ currents. (A) INa,late of wild-type and W822X channels. (B) INa,late–V relationship. INa,late was measured at the end of a series of voltage pulses of 250 ms duration stepped from −120 mV. INa,late is expressed as a percentage of maximal INa and is plotted against the voltage of the eliciting pulse. Labels are the same as for Figure 1.
3.4. Effects of nonsense mutation suppression on cellular functions
We also evaluated the possible side effects of nonsense mutation suppression. First, we assessed the influence of nonsense mutation suppression on cell growth. We measured the growth rate of HEK293 cells under readthrough-enhancing conditions and compared it with that of cells under normal culture conditions. During a 4-day period, the growth rates of the cells treated with aminoglycosides or siRNA against eRF3a were not significantly different from those of untreated cells, nor were they different from those of the cells transfected with the W822X mutant. Next, we examined whether readthrough-enhancing reagents induced DNA damage responses.23 For this purpose, HEK293 cells treated with aminoglycosides or siRNA against eRF3a were immunostained with the anti-H2AX antibody 48 h after the start of the treatment. Expression of H2AX, a histone 2A variant, is an early response to DNA strand breakage.24,25 H2AX-positive nuclei were seen in some treated cells. However, H2AX was also detected in a small fraction of untreated cells. Quantitative analysis by counting the relative number of H2AX-positive cells revealed no significant difference in the percentage of cells showing H2AX expression among treated and untreated cell groups. Finally, we examined whether the readthrough-enhancing reagents led to translation beyond the natural stop codons. The following three pieces of evidence indicate that the readthrough-enhancing reagents we used are, in general, unlikely to have a significant suppression effect on natural stop codons. (i) In our western blot experiment, we found that Na+ channels expressed under the readthrough-enhancing conditions had the same molecular size as that of wild-type channels (Figure 2A). (ii) GAPDH protein in treated cells also had the same molecular size as that of untreated cells (Figure 2A). (iii) We also looked at the total protein profile of the treated and untreated HEK293 cells. On Coomassie-stained SDS–PAGE gels, treatment of the cells with aminoglycosides or siRNA against eRF3a did not induce any noticeable change in the total protein profile; no additional protein bands or band shifts were observed (see Supplementary material online, Figure S1). These data indicate that the treatments did not have a severe global effect on protein production of the cells.
4. Discussion
In this study, we investigated the suppression of nonsense mutation in the SNC5A gene. We found that using aminoglycosides to reduce translational fidelity and using siRNA against eRF3a to decrease translation termination efficiency could partially restore the expression of full-length cardiac Na+ channel protein from the nonsense-mutated gene, and that the characteristics of the mutant channels conferred the wild-type phenotype of the Nav1.5 channel.
4.1. Effective nonsense suppression and comparison with other studies
Because translation termination is a ubiquitous step in cellular protein synthesis, translational readthrough should be an avenue of clinical potential for correcting the dysfunction of cardiac Na+ channels caused by nonsense mutations. However, readthrough efficiency appears to vary among genes and can depend on the location and identity of the stop codon in the gene of interest and on the surrounding mRNA sequence context.26 Herein, we provided unambiguous evidence that readthrough-enhancing reagents effectively increased the amount of full-length and functional Nav1.5 channels. Both types of reagents increased the maximal INa by eight-fold to ∼30% of the wild-type level. This is one of only a few studies to date that document nonsense suppression in genes encoding ion channels other than the CFTR. In a recent study, gentamicin at 1 mg/mL was reported to increase the expression of the nonsense mutant E375X of the Kv1.5 channel by about two-fold.27 The difference in the level of expression between the Kv1.5 channel mutant and the Nav1.5 channel mutants seen in our study perhaps is due to a dominant-negative suppression by truncated subunits in the Kv1.5 channel that does not occur in the Nav1.5 channel.
Our finding that the effect of eRF3a knockdown with siRNA on readthrough is as potent as the effect of aminoglycosides is interesting. Inhibition of the termination reaction by siRNA against eRF1 has been shown to increase readthrough of all three stop codons by >2.5-fold in HEK293 cells.28 However, results from the same study on siRNA against eRF3a showed that the siRNA's effect was relatively small. In a more recent study,11 application of siRNA against eRF3a resulted in a much more profound increase in readthrough efficiency of HEK293 cells, which is compatible with our result; this result is not surprising because the siRNA in our study targeted the same sequence. Thus, selection of the targeting sequence appears to be critical for siRNA to suppress eRF3a expression effectively.
Recent studies of therapies for nonsense-associated diseases have focused on suppressing nonsense codon recognition. Of the three nonsense codons, generally speaking UGA is the least effective at directing translation termination.29,30 Therefore, the presence of the premature stop codon UGA at the W822X mutation site may partly explain the effectiveness of the nonsense suppression treatments. Termination efficiency also is dictated by cellular constituents of the translational machinery,31 some of which will be more amenable to therapeutic manipulation than others. Our findings indicate that both reducing translational fidelity and decreasing translation termination efficiency are candidate mechanisms for effective therapeutic applications.
4.2. Therapeutic implications
Our results have validated—at the molecular and cellular levels—the hypothesis that using readthrough-enhancing reagents such as aminoglycosides and siRNA against eukaryotic release factors is an effective therapeutic strategy to suppress SCN5A nonsense mutations. We not only showed that readthrough enhancement increased INa in cells transfected with the nonsense mutant cDNA, but we also illustrated that the same reagents could increase INa in cells transfected with a mix of wild-type and mutant cDNAS (which mimics the heterozygous mutant), from 56% to 74% of the wild-type level. This result offers a factual basis for the clinical application of these and similar reagents as therapeutic interventions. Zingman et al.32 suggested that even a small increase in the amount of functional full-length protein might improve clinical symptoms that are associated with the heterozygous loss of one allele.
Effective therapies for diseases that are attributable to in-frame nonsense codons should not only ensure effective incorporation of an amino acid at the PTC site, but also ensure that the amino acid incorporated supports protein function even if it is not the amino acid present within the wild-type protein.33 This is necessary because the near-cognate tRNA that misreads the PTC might not be the same tRNA cognate to the wild-type sequence. Karow et al.34 sequenced purified protein from the prfB1 strain of Bacillus subtilis with a UGA nonsense mutation and found that the residue at the UGA position was tryptophan. Thus, it is likely that suppression of the UGA codon at the W822X mutation site leads to the insertion of a tryptophan and results in the synthesis of the wild-type Nav1.5 channel. Indeed, our data showed that the W822X channels had the same molecular size as the wild-type channel and they conferred the wild-type phenotype. However, direct sequencing data will be needed to determine whether tryptophan is indeed the inserted amino acid.
We also investigated enhanced readthrough as a potential risk factor for drug-induced disease conditions. Theoretically, a reagent with a high readthrough efficiency might not be a safe and effective therapeutic agent because the readthrough process might introduce sequence changes to the restored full-length proteins due to mistranslation of sense codons, readthrough of natural stop codons, and incorporation of an amino acid that does not match the wild-type sequence. In these cases, enhancing readthrough not only can be ineffective, but it also can be harmful and cause drug-induced disease in addition to or in lieu of the original conditions. Relatively little information is available about sequence errors generated by readthrough. For these reasons, potential treatments designed to correct premature translation termination must be evaluated for individual genes and even for individual mutations.
We also evaluated the potential risks for the readthrough-enhancing reagents used in this study to cause diseases by damaging other cellular components. We found no detectable side effects for the host cells in which W822X channels were expressed. Moreover, the fact that the W822X channels have the wild-type phenotype further reduces the concerns about drug-induced disease conditions. Taken together, the results of this study suggest that the use of aminoglycosides and siRNA against eRF3a to restore mutated Na+ channels would be beneficial to carriers of the mutation.
4.3. Limitations of the study
Mammalian cells have evolved numerous ways to ensure the quality of mRNA. If translation termination is sufficiently premature, then the mRNA will be subject to nonsense-mediated decay (NMD).35 NMD is a cellular defence mechanism that eliminates mRNAs that harbour PTCs and that encode non-functional or potentially harmful polypeptides. Nonsense-associated diseases are caused by insufficient expression of full-length, functional proteins. Elimination of PTC-containing mRNAs by NMD is now viewed as a cause of such insufficient expression, and therefore, it may contribute to nonsense-associated diseases.36 It is worth noting that NMD may also affect the efficiency of drug-induced readthrough due to the reduced number of mRNA copies. In our study, we did not assess the role of NMD in nonsense suppression because the use of cDNA might disturb the slicing-dependent processes, including NMD. However, a considerable amount (∼25%) of PTC-containing mRNAs can escape NMD and can be remodelled into the steady-state translation initiation complexes that go on to direct peptide synthesis.37 Finally, it should be noted that this study was limited to channels expressed in heterogonous cells. Further studies involving animal models are needed to extrapolate the results reported here.
4.4. Conclusions
Through the study of a nonsense mutation of the SCN5A gene, we demonstrated that pharmacological and RNA interference of translation termination has the potential to treat conditions associated with nonsense mutations of SCN5A. These results should be considered in the design of pharmacogenetic therapies to reduce the risk of sudden cardiac death associated with nonsense mutations of SCN5A and to treat diseases associated with nonsense mutations in general.
Supplementary material
Supplementary material is available at Cardiovascular Research online.
Conflict of interest: none declared.
Funding
This work was supported by National Institutes of Health grant (HL-58133) to Z.F., by the National Basic Research Program (2007CB512000 and 2007CB512008) from the Ministry of Science and Technology of China to J.P., by a research grant (2005-30571040) from the Natural Science Foundation of China to S.T., and by a research grant from the Academy of Finland to V.P.
References
- 1.George AL., Jr Inherited disorders of voltage-gated sodium channels. J Clin Invest. 2005;115:1990–1999. doi: 10.1172/JCI25505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bezzina CR, Carol AR. Dilated cardiomyopathy due to sodium channel dysfunction: what is the connection? Circ Arrhythmia Electrophysiol. 2008;1:80–82. doi: 10.1161/CIRCEP.108.791434. [DOI] [PubMed] [Google Scholar]
- 3.Krawczak M, Ball EV, Fenton I, Stenson PD, Abeysinghe S, Thomas N, et al. Human gene mutation database—a biomedical information and research resource. Hum Mutat. 2000;15:45–51. doi: 10.1002/(SICI)1098-1004(200001)15:1<45::AID-HUMU10>3.0.CO;2-T. [DOI] [PubMed] [Google Scholar]
- 4.Hirawat S, Welch EM, Elfring GL, Northcutt VJ, Paushkin S, Hwang S, et al. Safety, tolerability, and pharmacokinetics of ptc124, a nonaminoglycoside nonsense mutation suppressor, following single- multiple-dose administration to healthy male and female adult volunteers. J Clin Pharmacol. 2007;47:430–444. doi: 10.1177/0091270006297140. [DOI] [PubMed] [Google Scholar]
- 5.Papadatos GA, Wallerstein PM, Head CE, Ratcliff R, Brady PA, Benndorf K, et al. Slowed conduction and ventricular tachycardia after targeted disruption of the cardiac sodium channel gene Scn5a. Proc Natl Acad Sci USA. 2002;99:6210–6215. doi: 10.1073/pnas.082121299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bezzina CR, Rook MB, Groenewegen WA, Herfst LJ, vander Wal AC, Lam J, et al. Compound heterozygosity for mutations (W156X and R225W) in SCN5A associated with severe cardiac conduction disturbances and degenerative changes in the conduction system. Circ Res. 2003;92:159–168. doi: 10.1161/01.res.0000052672.97759.36. [DOI] [PubMed] [Google Scholar]
- 7.Keller DI, Barrane FZ, Gouas L, Martin J, Pilote S, Suarez V, et al. A novel nonsense mutation in the SCN5A gene leads to Brugada syndrome and a silent gene mutation carrier state. Can J Cardiol. 2005;21:925–931. [PubMed] [Google Scholar]
- 8.Benson DW, Wang DW, Dyment M, Knilans TK, Fish FA, Strieper MJ, et al. Congenital sick sinus syndrome caused by recessive mutations in the cardiac sodium channel gene. J Clin Invest. 2003;112:1019–1028. doi: 10.1172/JCI18062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Makita N, Sumitomo N, Watanabe I, Tsutsui H. Novel SCN5A mutation (Q55X) associated with age-dependent expression of Brugada syndrome presenting as neurally mediated syncope. Heart Rhythm. 2007;4:516–519. doi: 10.1016/j.hrthm.2006.10.028. [DOI] [PubMed] [Google Scholar]
- 10.Linde L, Boelz S, Nissim-Rafinia M, Oren YS, Wilschanski M, Yaacov Y, et al. Nonsense-mediated mRNA decay affects nonsense transcript levels and governs response of cystic fibrosis patients to gentamicin. J Clin Invest. 2007;117:683–692. doi: 10.1172/JCI28523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chauvin C, Salhi S, Le GC, Viranaicken W, Diop D. Involvement of human release factors ERF3a and ERF3b in translation termination and regulation of the termination complex formation. Mol Cell Biol. 2005;25:5801–5811. doi: 10.1128/MCB.25.14.5801-5811.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Carnes J, Jacobson M, Leinwand L, Yarus M. Stop codon suppression via inhibition of ERF1 expression. RNA. 2003;9:648–653. doi: 10.1261/rna.5280103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bedwell DM, Kaenjak A, Benos DJ, Bebok Z, Bubien JK, Hong J, et al. Suppression of a CFTR premature stop mutation in a bronchial epithelial cell line. Nat Med. 1997;3:1280–1284. doi: 10.1038/nm1197-1280. [DOI] [PubMed] [Google Scholar]
- 14.Howard MT, Shirts BH, Petros LM, Flanigan KM, Atkins JF. Sequence specificity of aminoglycoside-induced stop condon readthrough: potential implications for treatment of Duchenne muscular dystrophy. Ann Neurol. 2000;48:164–169. [PubMed] [Google Scholar]
- 15.Barton-Davis ER, Cordier L, Shoturma DI, Leland SE, Sweeney HL. Aminoglycoside antibiotics restore dystrophin function to skeletal muscles of mdx mice. J Clin Invest. 1999;104:375–381. doi: 10.1172/JCI7866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Du M, Jones JR, Lanier J, Keeling KM, Lindsey JR, Tousson A, et al. Aminoglycoside suppression of a premature stop mutation in a Cftr-/- mouse carrying a human CFTR-G542X transgene. J Mol Med. 2002;80:595–604. doi: 10.1007/s00109-002-0363-1. [DOI] [PubMed] [Google Scholar]
- 17.Diop D, Chauvin C, Jean-Jean O. Aminoglycosides and other factors promoting stop codon readthrough in human cells. C R Biol. 2007;330:71–79. doi: 10.1016/j.crvi.2006.09.001. [DOI] [PubMed] [Google Scholar]
- 18.Turillazzi E, Rocca GL, Anzalone R, Corrao S, Neri M, Pomara C, et al. Heterozygous nonsense SCN5A mutation W822X explains a simultaneous sudden infant death syndrome. Virchows Arch. 2008;453:209–216. doi: 10.1007/s00428-008-0632-7. [DOI] [PubMed] [Google Scholar]
- 19.Makielski JC, Ye B, Valdivia CR, Pagel MD, Pu J, Tester DJ, et al. A ubiquitous splice variant and a common polymorphism affect heterologous expression of recombinant human SCN5A heart sodium channels. Circ Res. 2003;93:821–828. doi: 10.1161/01.RES.0000096652.14509.96. [DOI] [PubMed] [Google Scholar]
- 20.Cui Y, Wang W, Fan Z. Cytoplasmic vestibule of the weak inward rectifier Kir6.2 potassium channel. J Biol Chem. 2002;277:10523–10530. doi: 10.1074/jbc.M109118200. [DOI] [PubMed] [Google Scholar]
- 21.Usuki F, Ishiura S. Expanded CTG repeats in myotonin protein kinase increase susceptibility to oxidative stress. NeuroReport. 1998;9:2291–2296. doi: 10.1097/00001756-199807130-00027. [DOI] [PubMed] [Google Scholar]
- 22.Fan Z, George AL, Jr, Kyle JW, Makielski JC. Two human paramyotonia congenita mutations have opposite effects on lidocaine block of Na+ channels expressed in a mammalian cell line. J Physiol. 1996;496:275–286. doi: 10.1113/jphysiol.1996.sp021684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Brumbaugh KM. The mRNA surveillance protein hSMG-1 functions in genotoxic stress response pathways in mammalian cells. Mol Cell. 2004;14:585–598. doi: 10.1016/j.molcel.2004.05.005. [DOI] [PubMed] [Google Scholar]
- 24.Rogakou EP, Pilch DR, Orr AH, Ivanova VS, Bonner WM. DNA double-stranded breaks induce histone H2AX phosphorylation on serine 139. J Biol Chem. 1998;273:5858–5868. doi: 10.1074/jbc.273.10.5858. [DOI] [PubMed] [Google Scholar]
- 25.Brummelkamp TR, Bernards R, Agami R. A system for stable expression of short interfering RNAs in mammalian cells. Science. 2002;296:550–553. doi: 10.1126/science.1068999. [DOI] [PubMed] [Google Scholar]
- 26.Manuvakhova M, Keeling K, Bedwell DM. Aminoglycoside antibiotics mediate context-dependent suppression of termination codons in a mammalian translation system. RNA. 2000;6:1044–1055. doi: 10.1017/s1355838200000716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Olson TM, Alekseev AE, Liu XK, Park S, Zingman LV, Bienengraeber M, et al. Kv1.5 channelopathy due to KCNA5 loss-of-function mutation causes human atrial fibrillation. Hum Mol Genet. 2006;15:2185–2191. doi: 10.1093/hmg/ddl143. [DOI] [PubMed] [Google Scholar]
- 28.Janzen DM, Geballe AP. The effect of eukaryotic release factor depletion on translation termination in human cell lines. Nucleic Acids Res. 2004;32:4491–4502. doi: 10.1093/nar/gkh791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McCaughan KK, Brown CM, Dalphin ME, Berry MJ, Tate WP. Translational termination efficiency in mammals is influenced by the base following the stop codon. Proc Natl Acad Sci USA. 1995;92:5431–5435. doi: 10.1073/pnas.92.12.5431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kuzmiak HA, Maquat LE. Applying nonsense-mediated mRNA decay research to the clinic: progress and challenges. Trends Mol Med. 2006;12:306–316. doi: 10.1016/j.molmed.2006.05.005. [DOI] [PubMed] [Google Scholar]
- 31.Rospert S. Polypeptide chain termination and stop codon readthrough on eukaryotic ribosomes. Rev Physiol Biochem Pharmacol. 2005;155:1–30. doi: 10.1007/3-540-28217-3_1. [DOI] [PubMed] [Google Scholar]
- 32.Zingman LV, Park S, Olson TM, Alekseev AE, Terzic A. Aminoglycoside-induced translational read-through in disease: overcoming nonsense mutations by pharmacogenetic therapy. Clin Pharmacol Ther. 2007;81:99–103. doi: 10.1038/sj.clpt.6100012. [DOI] [PubMed] [Google Scholar]
- 33.Lai CH, Chun HH, Nahas SA, Mitui M, Gamo KM, Du L, et al. Correction of ATM gene function by aminoglycoside-induced read-through of premature termination codons. Proc Natl Acad Sci USA. 2004;101:15676–15681. doi: 10.1073/pnas.0405155101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Karow M, Rogers EJ, Lovett P, Piggot P. Suppression of TGA mutations in the Bacillus subtilis spoIIR gene by prfB mutations. J Bacteriol. 1998;180:4166–4170. doi: 10.1128/jb.180.16.4166-4170.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ishigaki Y, Li XJ, Serin G, Maquat LE. Evidence for a pioneer round of mRNA translation: mRNAs subject to nonsense-mediated decay in mammalian cells are bound by CBP80 and CBP20. Cell. 2001;106:607–617. doi: 10.1016/s0092-8674(01)00475-5. [DOI] [PubMed] [Google Scholar]
- 36.Geiger SK, Bär H, Ehlermann P, Wälde S, Rutschow D. Incomplete nonsense-mediated decay of mutant lamin A/C mRNA provokes dilated cardiomyopathy and ventricular tachycardia. J Mol Med. 2008;86:281–289. doi: 10.1007/s00109-007-0275-1. [DOI] [PubMed] [Google Scholar]
- 37.Stephenson LS, Maquat LE. Cytoplasmic mRNA for human triosephosphate isomerase is immune to nonsense-mediated decay despite forming polysomes. Biochimie. 1996;78:1043–1047. doi: 10.1016/s0300-9084(97)86728-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.