Abstract
Reporting transplant center-specific survival rates after hematopoietic cell transplantation is required in the United States. We describe a method to report 1-year survival outcomes by center, as well as to quantify center performance relative to the transplant center network average, which can be reliably used with censored data and for small center sizes. Each center’s observed 1-year survival outcome is compared to a predicted survival outcome adjusted for patient characteristics using a pseudovalue regression technique. A 95% prediction interval for 1-year survival assuming no center effect is computed for each center by bootstrapping the scaled residuals from the regression model, and the observed 1-year survival is compared to this prediction interval to determine center performance. We illustrate the technique using a recent center specific analysis performed by the Center for International Blood and Marrow Transplant Research, and study the performance of this method using simulation.
Keywords: Pseudovalue, Generalized estimating equations, Bootstrap
1 Introduction
Hematopoietic cell transplants (HCT) are performed in a wide range of different diseases at a large number of transplant centers (Pasquini et al. 2007). These transplant centers use a variety of center practices, ranging from conditioning regimens or graft-versus-host disease prophylaxes, to donor selection criteria, to supportive care measures. Although it may be possible to study certain practices through a retrospective database such as that of the Center for International Blood and Marrow Transplant Research (CIBMTR), it is also of interest to determine whether the collection of center practices at a given center are contributing positively or negatively on patient outcomes. Reporting transplant center-specific survival rates after HCT is required in the United States, most recently by the Stem Cell Therapeutic and Research Act of 2005 for all allogeneic transplants, and previously by the 1990 Transplant Amendments Act for unrelated donor transplants. The purpose of this report is to provide potential stem cell transplant recipients, their families and the general public with a comparison of survival rates among the centers performing hematopoietic cell transplants. The results of this report are published and made available on the web (National Marrow Donor Program 2007).
When considering center specific analyses in HCT patients, it is widely known that the characteristics of patients transplanted at a given center can vary considerably from other centers. Some centers transplant pediatric patients exclusively, while others may transplant only adult patients. Referral patterns to a given center may lead to a different patient mix of diseases, racial/ethnic groups, disease status, etc., than at other centers. These factors can have a substantial impact on the expected outcomes of patients in a given center. Furthermore, many of the centers transplant only a small number of patients during the study period; the inclusion of some high-risk patients in this group can make their survival appear poor. Centers are concerned that they will be penalized for the types of patients that they transplant. Any comparison of center performance needs to adequately account for the risk factors of the patients being transplanted at centers. Note that the appropriate choice of covariates for inclusion in the risk adjustment model requires careful consultation with clinicians, and the accuracy of the resulting center specific analysis depends heavily on the accuracy of the risk adjustment model.
Another important consideration in transplant center assessment is an appropriate outcome measure. We have chosen to focus solely on survival because of the wide variety of diseases being transplanted. Relapse or progression may be poorly defined for many of the diseases. Furthermore, we limit the analysis to the 1-year survival probability for a number of reasons. Modeling the entire survival curves would be extremely difficult given the potential for crossing hazard functions across centers and the large number of centers. Survival beyond 1 year may be more severely impacted by disease relapse, which may be more sensitive to the disease/stage of a patient being transplanted than center practices. Mortality within the first year is a better measure of the toxicity of the transplant procedure itself, which is more likely to be directly affected by which center performed the transplant. An analysis of 1-year survival probabilities is also easier to communicate to the wider audience for whom this report is targeted.
Center specific outcome analyses, also referred to as provider profiles or hospital report cards, have received a lot of attention in the literature. Much of the attention came initially from two high profile examples: the analysis of coronary artery bypass graft surgery results by the New York State Department of Health (New York State Department of Health 1992), and the evaluation of hospital performance by the Health Care Financing Administration beginning in 1987 (Health Care Financing Administration 1989; Normand et al. 1997), although many more examples can now be found. Several basic statistical principles of such analyses are widely accepted. A comparison of observed and expected outcomes should be adjusted for the patient risk (Iezzoni 1994; Landon et al. 1996; Salem-Schatz et al. 1994; Huang et al. 2005). The determination of outlying centers, i.e. overperforming or underperforming centers, should properly account for sampling variability (Localio et al. 1995). Finally, one must be wary of the multiple comparisons issue, in which large numbers of centers being examined lead to a greater likelihood of a center reporting extreme results by chance alone (Thomas et al. 1994; Localio et al. 1995). A number of procedures have been proposed for the actual statistical assessment of center outcomes; they generally consider a binary outcome such as mortality at a fixed time (see DeLong et al. 1997 for a review). Risk adjustment may be done using an external benchmark model, or using an internal model based on the same patients in the center analysis. We focus on the latter here. One can use a simple comparison of the observed and predicted survival outcomes at a given center, by constructing a confidence interval for the difference in or ratio of the observed and predicted survival probabilities (DeLong et al. 1997; Austin et al. 2003; Thomas et al. 1994). Alternatively fixed effect logistic regression or random effect logistic regression models may be used. Several authors have compared fixed effect and random effect logistic regression models in this setting, and have found that fixed effect models are more sensitive while random effect models are more conservative and may be less susceptible to multiplicity problems (DeLong et al. 1997; Huang et al. 2005; Austin et al. 2003). Bayesian and empirical Bayes hierarchical regression models have also been proposed (Christiansen and Morris 1997; Normand et al. 1997; Thomas et al. 1994). Transplant center reporting for solid organ transplantation has been described in Dickinson et al. (2006), who base assessment of center performance on the standardized mortality ratio (SMR) assuming a Cox proportional hazards model.
Two features of HCT data make center specific reporting difficult. First, many of the transplant centers perform a fairly small number of transplants over the reporting period, making reliance on large sample confidence intervals questionable. This also can cause convergence problems for a fixed center effect logistic regression model. Second, patients may be lost to follow-up prior to the fixed time point of analysis. Although the amount of censoring is not large, it makes a direct comparison of observed and predicted outcomes at a fixed time more difficult, and renders a parametric random effects logistic regression model impossible.
We propose a method for the analysis of HCT center specific outcomes which is analogous to the simple comparison of observed and predicted outcomes described in DeLong et al. (1997) using internal standardization. Here a predictive model for 1-year survival as a function of patient/risk characteristics is constructed based on the entire cohort of patients across all centers. Using this model, a risk adjusted 1-year survival rate is estimated for each center, along with a prediction interval. This prediction interval represents the range of 1-year survival rates which could have been observed if those patients had been transplanted at a generic center in the network. The actual observed 1-year survival can be compared to the prediction interval to assess how a center is performing compared to the entire network of centers. The main feature of our method is that it examines the center effect in terms of the direct impact on 1-year survival, while allowing for censored outcomes prior to 1 year, in contrast to the previous methods based on strict binary outcomes.
The details of this methodology are described in Sect. 2, starting with uncensored data and then generalizing the approach to account for censoring. We demonstrate the method in Sect.3 using a recent center specific analysis performed by the CIBMTR. Section 4 contains the results of a simulation study of the methodology. This simulation study focuses on the performance of the method in terms of control of the type I error rate, or the probability of incorrectly identifying a center as over- or under-performing. These are done for a variety of scenarios in which patient selection depends on center. A brief study of the power to identify over- or under-performing centers is also included here. Conclusions and discussion are given in Sect. 5.
2 Methods
The methodology proposed to examine center specific outcomes essentially consists of three steps. First, we build a model to predict 1-year survival as a function of patient characteristics. Next, we construct a prediction interval for the observed survival of the patients actually transplanted at a given center, assuming no center effect. This is done by adjusting for the patient characteristics of the patients actually transplanted at that center, using the previous predictive model. Finally, we can assess the performance of a given center by comparing their observed survival probability with the prediction interval. If a center’s observed survival rate is outside the prediction interval, then we conclude that they are either underperforming or overperforming compared to the general network of centers.
We first illustrate the methodology assuming we have complete follow-up at 1-year on all patients for simplicity. The resulting procedure is equivalent to the simple comparison of observed and predicted outcomes described in DeLong et al. (1997) using internal standardization, except that prediction intervals are based on resampling rather than large sample approximations. This may be more appropriate given the small sample sizes of many transplant centers. Then we generalize the procedure to account for censoring.
2.1 No censoring prior to 1 year
If there is no censoring prior to 1 year, then the outcome for patient j in center i, Yi j, is binary (1=alive, 0=dead) with observed value yi j. Assume that there are C centers, with ni patients at center i. The observed survival probability for the patients at center i in the uncensored case is the simple proportion,
In order to assess a center effect for center i, we want to compare the observed survival probability at center i with the predicted survival probability based on the characteristics of the patients actually transplanted at that center. We fit a predictive model for the probability of being alive at 1 year using logistic regression,
where pi j is the survival probability at 1 year for patient j in center i, and Xi j is the vector of patient characteristics for that patient. This model assumes no center effect, i.e. it assumes that the survival probability depends only on patient characteristics. The predicted survival probability at 1 year for patient j in center i based on patient characteristics alone is
The predicted survival probability for the patients at center i represents the survival probability if those patients had been transplanted at a “generic” center in the network, i.e. one without a center effect. This is the average of the individual predicted probabilities,
Finally, we consider a range of plausible values for what the survival probability could have been if those patients had been transplanted at a “generic” center, by constructing a prediction interval for
under the assumption of no center effect. The observed survival probability can be compared directly to the prediction interval, to determine whether a center is underperforming or overperforming compared to how those patients might have fared at a “generic” center. Essentially this prediction interval is the upper and lower bounds of the null hypothesis sampling distribution for the survival of the patients at center i. As a result, if the observed survival probability is outside the (1 − α) prediction interval, the p-value for testing the null hypothesis of no center effect will be less than α.
Next we describe one approach to construct such a prediction interval, which works well even for modest sample sizes. Assuming that we have a reasonably stable estimate of the parameter vector β̂ for the patient characteristics (as is reasonable here because of the large sample sizes across all the centers in the network), a parametric resampling approach models Yi j as Bernoulli distributed with probability p̂i j. This means that the outcome for patients transplanted at a “generic” center in the network are assumed to have a survival probability dependent only on patient characteristics. Then the observed number of patients at center i who are alive at 1 year is a mixture of ni Bernoulli random variables with varying survival probabilities. We can simulate this distribution as follows. For b = 1 to B,
Generate for i = 1 to C, j = 1 to ni.
Compute
Then the (1 − α) prediction interval for is the α/2 and (1 − α/2) percentiles of over b, denoted
The observed survival probability can be compared directly to the prediction interval: indicates that the center is underperforming while indicates that the center is overperforming compared to the overall network. If there is no center effect at center i, the probability of identifying center i as underperforming or overperforming is ≤ α.
2.2 Censoring prior to 1 year
If there is censoring prior to 1 year, then we do not know the binary outcome for each patient. In the past center specific analyses, because of mandatory government reporting of HCT outcomes, the amount of censoring has been fairly small (approximately 5% prior to 1 year). However, even with this small amount of censoring, the methodology must be adjusted.
The observed 1-year survival probability at 1 year, in the presence of censoring can be estimated using the Kaplan–;Meier estimate.
We modify the logistic regression model to predict 1-year survival probabilities for each patient by using a censored data version of logistic regression (Klein et al. 2007). This is based on modeling of pseudovalues as described in Andersen et al. (2003) and Klein and Andersen (2005). Here the pseudovalues are defined for the 1-year survival probability. To compute the pseudo-value for recipient j at center i, first compute the pooled sample Kaplan–;Meier estimate of survival at 1 year ignoring all covariates and center, Ŝp(1). Next we compute the Kaplan–Meier estimate of survival at 1 year based on the entire dataset with observation j at center i removed . The ijth pseudo-value is defined by
If there is no censoring then the ijth pseudo-value is simply yi j, the indicator that the jth recipient at the ith center was alive at 1year. A SAS macro to compute these pseudo-values is available in Klein et al. (2008). These pseudo-values are used in a regression model using a logit link, similar to the standard logistic regression model used above, given by
where here pij = E(p̃i j). Although we focus in this paper on the logit link, the pseudo-value regression method is flexible and can use a variety of link functions including the complementary log–log transformation which is equivalent to a proportional hazards model at time 1.
The parameters of the regression model can be estimated using generalized estimating equations (GEE) (Liang and Zeger 1986), which can be implemented in PROC GENMOD in SAS. Note that in our situation, only one time point (1 year) is used resulting in univariate data for each patient and no need to specify a working correlation matrix. Let μ(·) = g−1 (·) be the mean function. Define dμi j (β) to be the vector of partial derivatives with respect to β. Then β̂ is the solution in β to the estimating equation
Note that while the pseudovalues are not constrained to be between 0 and 1, they will usually be close to 0 or 1 even if they are outside the range. Pseudovalues outside the range of 0 to 1 do not generally cause a problem with the logit link because the logit link models the mean of the pseudovalues and is not applied to the pseudovalues directly. In most situations the mean function for a particular set of covariate values is within the appropriate range.
Once β̂ has been estimated, we can predict the survival probability at 1 year for patient j at center i with patient characteristics Xi j as
As above, we want to use these predicted survival probabilities to construct a (1−α) prediction interval for the 1-year survival probability of the patients at center i, had they been transplanted at a “generic” center. One simple method of constructing a prediction interval would be to follow the steps (1)–(2) in the above section on no censoring present, and generate a prediction interval for the complete data survival outcome of the patients at center i. This ignores the censoring pattern when constructing the prediction interval, since in the resampling scheme above, a binary (i.e. uncensored) indicator is generated for each patient at a center. Essentially the censoring is only accounted for in estimating the model parameters. This simple approach works reasonably well when the censoring percentage is small, but may have some small inflation of the type I error rate at a center due to not accounting for the censoring.
We can get a more accurate prediction interval which controls the type I error rate by resampling the residuals from the general linear model rather than generating binary outcomes for each individual. Bootstrapping of Pearson residuals in a generalized linear model framework has been described in Moulton and Zeger (1991). Define the scaled Pearson residual for patient j at center i by
We propose the following simple resampling algorithm to generate a prediction interval: For b = 1 to B,
Generate for i = 1,…,C; j = 1,…,ni, by sampling with replacement from the set of residuals {ri j, i = 1,…, C; j = 1,…,ni}
- Compute the bootstrap predicted value for patient j at center i as
- Compute the predicted observed center outcome as
Then similarly to the uncensored case, the (1 –α) prediction interval for is the α/2 and (1 –α/2) percentiles of over b, denoted and the observed survival probability can be compared directly to the prediction interval to gauge center performance relative to the entire network.
This resampling of the scaled Pearson residuals implicitly accounts for the increase in prediction variance due to censoring, and provides accurate control of the type I error rate, as we shall see in a subsequent simulation section. Note however that there is no accounting for the variability of the parameter estimates β in the generation of the prediction interval. Typically the sample size associated with estimating these parameters uses the entire dataset of transplants across all the centers, while prediction is specific to an individual center. Therefore, the variance contribution of the parameter estimation is relatively small compared to the variability of the center outcomes themselves. One could add an outer resampling loop which additionally includes refitting the prediction model, to account for the variance of β̂. In simulations shown later, we found this extra step to be computationally burdensome and unnecessary for accurate control of the type I error rate.
Note that there are alternative ways of generating a predicted 1-year survival probability for an individual patient, such as using a Cox proportional hazards model. Such a model could then be incorporated into this method, but would still require use of the pseudovalues in order to compute the residual distribution and apply the bootstrap. Most likely the Cox model and the pseudovalue regression model will give similar estimates of the predicted 1-year survival, but it is possible that if there is nonproportional hazards for one or more of the covariates the Cox model will yield a biased estimate of the 1-year survival probability. In contrast, the pseudovalue regression model is directly modeling the 1-year survival probability and is not sensitive to the proportional hazards assumption.
3 Simulation study
A simulation study was conducted to see how well the method performed in a variety of situations where the distribution of patient characteristics may be center dependent. In particular, we assessed the type I error rate for each center, or the probability that a center will be incorrectly classified as either over-performing or under-performing compared to the entire network. We used 95% prediction intervals in all simulations, so the target type I error rate is 5% per center.
Initial simulations were designed to have similar characteristics as the real example discussed in the next section. There were a total of 7,780 patients distributed across 119 centers: 45 centers with 15 patients, 29 centers with 45 patients, 26 centers with 75 patients, 15 centers with 150 patients, and 4 centers with 400 patients. For each patient, exponential death times were generated with rate parameter
where Xi j = (Xi j1, Xi j2) is the design vector, and β = (β1, β2) is the parameter vector. Using this model, the survival probability at 1 year is given by
corresponding to a logistic regression model for 1-year survival. The covariate Xi j1 is a binary covariate taking on values 1 or − 1 with probabilities depending on the scenario, while Xi j2 is a continuous covariate which is distributed according to a uniform distribution between [−0.5, 0.5]. Note that the survival at 1 year is equal to 50% for an “average” patient with Xi jβ = 0, which approximately matches the real example. We take β1 = log(2)/2, so that the odds ratio for 1-year survival for patients with Xi j1 = 1 compared to patients with Xi j1 = −1 is 2. The parameter β2 is set to 1. An independent censoring time is generated from an exponential distribution so that the censoring percentage at 1 year for an average patient (i.e. Xi jβ = 0) is 5 (similar to what was found for the example), 20, or 35%. These correspond to overall censoring rates of 9, 31 and 47%.
Several scenarios were considered for how the Xi j1 values were generated. These various scenarios represent different mechanisms of patient selection at the centers, as represented by the population proportion πi of patients at center i who are “good risk” (Xi j1 = 1). In scenario 1, πi was fixed at 0.5 for all centers. In scenario 2,πi was increasing from 0.3 to 0.7 with increasing center size; here larger centers are more likely to transplant good risk patients. In scenario 3, πi was decreasing from 0.7 to 0.3 with increasing center size; here larger centers are less likely to transplant good risk patients. In scenarios 4 through 7, only a fraction of the centers of a particular size were more or less likely to transplant good risk patients. Scenario 7 is a very extreme one, in which a fraction of the centers of a particular size transplant all “good risk” patients, and another fraction of the centers transplant all “poor risk” patients. Details on the proportion of patients who are considered “good risk” at each center are given in Table 1, by scenario. Once the proportion πi is determined for a given center, then Xi j1 is generated randomly with P(Xi j1 = 1) = πi and P(Xi j1 = −1) = 1 − πi.
Table 1.
Probability that a patient transplanted at a center is a “good risk” patient by center size and center number for scenarios 1 through 7
| Center size | Center number | Scenario | ||||||
|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | ||
| 15 | 1–5 | 0.5 | 0.3 | 0.7 | 0.3 | 0.3 | 0.5 | 0.0 |
| 6–10 | 0.5 | 0.3 | 0.7 | 0.7 | 0.5 | 0.7 | 1.0 | |
| 11–45 | 0.5 | 0.3 | 0.7 | 0.5 | 0.5 | 0.5 | 0.5 | |
| 45 | 46–48 | 0.5 | 0.4 | 0.6 | 0.3 | 0.3 | 0.5 | 0.0 |
| 49–51 | 0.5 | 0.4 | 0.6 | 0.7 | 0.5 | 0.7 | 1.0 | |
| 52–74 | 0.5 | 0.4 | 0.6 | 0.5 | 0.5 | 0.5 | 0.5 | |
| 75 | 75–77 | 0.5 | 0.5 | 0.5 | 0.3 | 0.5 | 0.5 | 0.0 |
| 78–80 | 0.5 | 0.5 | 0.5 | 0.7 | 0.5 | 0.5 | 1.0 | |
| 81–100 | 0.5 | 0.5 | 0.5 | 0.5 | 0.5 | 0.5 | 0.5 | |
| 150 | 101–102 | 0.5 | 0.6 | 0.4 | 0.3 | 0.5 | 0.3 | 0.0 |
| 103–104 | 0.5 | 0.6 | 0.4 | 0.7 | 0.7 | 0.5 | 1.0 | |
| 105–115 | 0.5 | 0.6 | 0.4 | 0.5 | 0.5 | 0.5 | 0.5 | |
| 400 | 116 | 0.5 | 0.7 | 0.3 | 0.3 | 0.5 | 0.3 | 0.0 |
| 117 | 0.5 | 0.7 | 0.3 | 0.7 | 0.7 | 0.5 | 1.0 | |
| 118–119 | 0.5 | 0.7 | 0.3 | 0.5 | 0.5 | 0.5 | 0.5 | |
For each dataset generated in this way, the methodology described in Sect. 2 was applied so that the final result is an indicator of whether the observed survival for a given center is outside the corresponding prediction interval. We generate a total of 10,000 datasets for analysis, and compute the type I error rate as the average of these indicators. The prediction intervals for each dataset are based on 1,000 bootstrap samples. The type I error results are computed for each individual center. These results are then summarized by grouping centers according to their center size and proportion of “good risk” patients πi, and then the average of the individual center type I error rates over similar centers is given in Table 2–Table 4 for 1-year censoring percentages of 5, 20, and 35% respectively.
Table 2.
Type I error results, by center size and center number according to scenarios 1–7, with 5% censoring at 1 year
| Center size | Center number | Scenario | ||||||
|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | ||
| 15 | 1–5 | 0.052 | 0.052 | 0.053 | 0.052 | 0.052 | 0.052 | 0.052 |
| 6–10 | 0.053 | 0.053 | 0.053 | 0.053 | 0.053 | 0.053 | 0.053 | |
| 11–45 | 0.052 | 0.052 | 0.052 | 0.052 | 0.052 | 0.052 | 0.052 | |
| 45 | 46–48 | 0.052 | 0.051 | 0.052 | 0.052 | 0.052 | 0.051 | 0.052 |
| 49–51 | 0.052 | 0.052 | 0.051 | 0.051 | 0.052 | 0.051 | 0.052 | |
| 52–74 | 0.051 | 0.051 | 0.051 | 0.051 | 0.051 | 0.051 | 0.051 | |
| 75 | 75–77 | 0.049 | 0.050 | 0.049 | 0.049 | 0.049 | 0.050 | 0.049 |
| 78–80 | 0.052 | 0.052 | 0.052 | 0.051 | 0.051 | 0.053 | 0.052 | |
| 81–100 | 0.051 | 0.051 | 0.051 | 0.051 | 0.051 | 0.051 | 0.051 | |
| 150 | 101–102 | 0.048 | 0.049 | 0.049 | 0.049 | 0.048 | 0.049 | 0.048 |
| 103–104 | 0.055 | 0.053 | 0.051 | 0.053 | 0.054 | 0.053 | 0.055 | |
| 105–115 | 0.050 | 0.050 | 0.051 | 0.050 | 0.050 | 0.051 | 0.050 | |
| 400 | 116 | 0.047 | 0.045 | 0.045 | 0.045 | 0.048 | 0.046 | 0.047 |
| 117 | 0.044 | 0.042 | 0.042 | 0.042 | 0.041 | 0.045 | 0.044 | |
| 118–119 | 0.046 | 0.047 | 0.046 | 0.047 | 0.047 | 0.047 | 0.046 | |
Table 4.
Type I error results, by center size and center number according to scenarios 1–7, with 5% censoring at 1 year
| Center size | Center number | Scenario | ||||||
|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | ||
| 15 | 1–5 | 0.058 | 0.059 | 0.056 | 0.059 | 0.059 | 0.057 | 0.059 |
| 6–10 | 0.058 | 0.058 | 0.056 | 0.059 | 0.058 | 0.056 | 0.056 | |
| 11–45 | 0.058 | 0.058 | 0.056 | 0.058 | 0.058 | 0.057 | 0.058 | |
| 45 | 46–48 | 0.052 | 0.052 | 0.053 | 0.053 | 0.053 | 0.052 | 0.053 |
| 49–51 | 0.054 | 0.054 | 0.053 | 0.051 | 0.056 | 0.053 | 0.050 | |
| 52–74 | 0.052 | 0.053 | 0.052 | 0.053 | 0.053 | 0.053 | 0.053 | |
| 75 | 75–77 | 0.051 | 0.051 | 0.051 | 0.051 | 0.050 | 0.051 | 0.051 |
| 78–80 | 0.052 | 0.052 | 0.051 | 0.050 | 0.051 | 0.052 | 0.048 | |
| 81–100 | 0.053 | 0.051 | 0.052 | 0.051 | 0.052 | 0.051 | 0.052 | |
| 150 | 101–102 | 0.049 | 0.049 | 0.050 | 0.049 | 0.051 | 0.049 | 0.047 |
| 103–104 | 0.051 | 0.056 | 0.051 | 0.048 | 0.047 | 0.055 | 0.044 | |
| 105–115 | 0.051 | 0.050 | 0.051 | 0.051 | 0.050 | 0.050 | 0.050 | |
| 400 | 116 | 0.043 | 0.046 | 0.047 | 0.048 | 0.045 | 0.045 | 0.041 |
| 117 | 0.048 | 0.046 | 0.047 | 0.046 | 0.044 | 0.047 | 0.038 | |
| 118–119 | 0.045 | 0.046 | 0.045 | 0.049 | 0.048 | 0.045 | 0.049 | |
The results indicate that the type I error rate is controlled reasonably well for all scenarios, except for some slight elevation of the type I error rate in the extreme case with center size of 15 and 35% censoring at 1 year. The proposed method of resampling the scaled residuals from the pseudovalue regression model works well to produce a reliable prediction interval with the correct coverage probability. Neither the selection pattern of high risk patients exhibited by the center nor the center size has an effect on the likelihood a center will be incorrectly identified as overperforming or under-performing compared to the overall network. Even in the extreme setting where all or none of the patients at a given center are “good risk”, the type I error rate is still controlled. The risk of patients transplanted at a given center do not affect the likelihood a center will be mistakenly identified as under-performing or over-performing, as long as that risk is appropriately adjusted for in the multivariate model.
Next we conducted simulations to explore the type I error rate control when the sample size is more modest. In particular, we are interested in whether ignoring variability in the estimate of β̂ still produces acceptable type I error control when the total sample size is more modest. Here we consider a total of 990 patients, with 12 centers of size 15, 8 centers of size 45, and 6 centers of size 75. We use a modification of scenario 4 in which one center of each size has πi = 0.3, one center has πi = 0.7, and the remaining centers have πi = 0.5. The survival model and censoring rates are otherwise unchanged from the previous simulations. Results of the type I error rate simulations are found in Table 5. These indicate that even for this smaller scale center specific analysis, the type I error rate is reasonably controlled at the 5% level, except for slight inflation of the type I error rate with centers of size 15 and 35% censoring at 1 year.
Table 5.
Type I error rates by center size and proportion of good risk patients at a given center, for scaled down version of scenario 4 with n = 990 patients total
| Center size | πi | Censoring at 1 year | ||
|---|---|---|---|---|
| 5% | 20% | 35% | ||
| 15 | 0.3 | 0.052 | 0.050 | 0.056 |
| 0.7 | 0.052 | 0.049 | 0.055 | |
| 0.5 | 0.052 | 0.054 | 0.057 | |
| 45 | 0.3 | 0.047 | 0.050 | 0.049 |
| 0.7 | 0.045 | 0.043 | 0.047 | |
| 0.5 | 0.048 | 0.049 | 0.049 | |
| 75 | 0.3 | 0.042 | 0.042 | 0.043 |
| 0.7 | 0.044 | 0.041 | 0.041 | |
| 0.5 | 0.044 | 0.043 | 0.045 | |
For the power simulations we considered a simple scenario in which one center in each sample size category is over-performing, while one center is under-performing relative to the other centers. Two sets of odds ratios for these centers are considered, either (1.5, 0.67) or (2.0, 0.5), respectively, for the over-performing and under-performing center. We also consider three settings varying the proportion of “good risk” patients at that center to be 0, 0.5, or 1. For all other centers the proportion of “good risk” patients is set to 0.5. Only censoring rates of 5% at 1 year are presented; higher censoring percentages yielded similar trends but with lower power. The results are given in Table 6.
Table 6.
Power results for selected centers with a positive (OR>1) or negative (OR<1) center effect, assuming 5% censoring at 1 year
| Center size | πi | Odds ratio | |||
|---|---|---|---|---|---|
| 1.5 | 0.67 | 2 | 0.5 | ||
| 15 | 0 | 0.124 | 0.105 | 0.258 | 0.219 |
| 0.5 | 0.119 | 0.114 | 0.236 | 0.237 | |
| 1 | 0.108 | 0.124 | 0.213 | 0.257 | |
| 45 | 0 | 0.259 | 0.238 | 0.606 | 0.561 |
| 0.5 | 0.245 | 0.251 | 0.583 | 0.588 | |
| 1 | 0.232 | 0.263 | 0.558 | 0.605 | |
| 75 | 0 | 0.402 | 0.365 | 0.824 | 0.790 |
| 0.5 | 0.387 | 0.387 | 0.806 | 0.810 | |
| 1 | 0.370 | 0.402 | 0.791 | 0.824 | |
| 150 | 0 | 0.667 | 0.648 | 0.983 | 0.977 |
| 0.5 | 0.657 | 0.657 | 0.979 | 0.978 | |
| 1 | 0.650 | 0.672 | 0.975 | 0.982 | |
| 400 | 0 | 0.979 | 0.978 | 1.000 | 1.000 |
| 0.5 | 0.975 | 0.975 | 1.000 | 1.000 | |
| 1 | 0.974 | 0.980 | 1.000 | 1.000 | |
The power results are as expected, indicating that the method is performing in a reasonable manner. As the sample size increases, the power to detect a particular magnitude center effect increases. For a fixed sample size, as the center effect increases in magnitude (e.g. from OR =1.5 to 2.0, or from OR=0.67 to 0.5), the power also increases. There are slight effects of the proportion of “good risk” patients at a given center on the power to detect a center effect. If the center effect is positive, then a center with a small proportion of “good risk” patients will have slightly higher power than a center with a high proportion of “good risk” patients. If the center effect is negative, this trend is reversed. These differences are likely due to the impact of the proportion of “good risk” patients on the baseline survival probability at a given center, and the impact is fairly minor.
4 Example
We applied the methodology to the 2007 center specific analysis conducted by the CIBMTR. The 2007 analysis includes all unrelated donor transplants occurring in the 5-year time interval from January 1, 2001 to December 31, 2005. This time interval allows for a minimum of 1-year potential follow-up for all eligible cases. Centers were eligible to participate in the 2007 analysis based on the criteria that they were a U.S. transplant center, had performed at least one transplant in the time interval, and had submitted at least 75% of the expected follow-up data. A total of 119 US transplant centers are included in this analysis. There were 7,830 cases eligible for analysis, of whom 411 (5.2%) patients had less than 12 months of follow-up. The overall survival probability at 1 year for the entire dataset is 51.5%.
After careful discussion with clinical and statistical transplant experts, a set of risk factors was generated to use as candidate effects for the risk adjustment model building process. The pseudo-values were computed for each patient, and the regression model was fit using SAS PROC GENMOD. The following risk factors were found to be significant in the regression model: disease/stage; recipient age; donor age; Human Leukocyte Antigen matching between donor and recipient; recipient cytomegalovirus status; recipient race; co-existing disease; Karnofsky/Lansky performance score interacted with prior autologous transplant; cell dose by stem cell product type; year of transplant; conditioning regimen intensity; resistant disease (Non-Hodgkins Lymphoma only); duration of first complete remission (acute leukemia only); and T-cell lineage (Acute Lymphocytic Leukemia only).
Once the prediction model was determined, the bootstrap resampling algorithm with 10,000 bootstrap samples was implemented to generate prediction intervals.
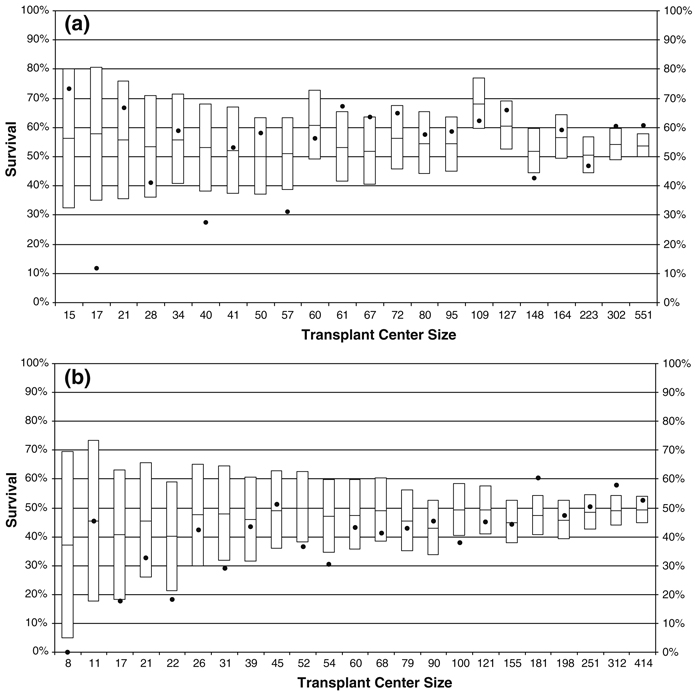
A subset of the final center-specific results is shown in Fig. 1a, b. Each center is represented by a boxplot, with the predicted survival outcome in the middle and the 95% prediction interval as the edges of the box. The observed survival outcome is shown with a solid circle, allowing one to visually compare the observed and predicted outcomes to assess center performance. If the observed outcome falls outside the box, this indicates that a center is underperforming or overperforming compared to the predicted outcome. Figure 1a has a sample of centers whose predicted survival probability is above average compared to the other centers, while Fig. 1b shows a sample of centers whose predicted survival probability is below average. In each sub-figure, all centers found overperforming or underperforming are included, along with a sample of “typically performing” centers. The boxplots within each subfigure are ordered by center size or the number of transplants performed by the center during the study period, illustrating that the width of the boxes or prediction intervals are narrowing with larger center size.
Fig. 1.
Transplant center outcomes for a sample of centers with above average predicted survival probabilities (a) and below average predicted survival probabilities (b). The box represents the prediction interval for survival at a given center, the line inside the box represents the predicted survival probability, and the circle represents the observed survival probability. Centers where the circle falls outside the box are considered underperforming or overperforming compared to the predicted outcome. Boxplots are labeled by transplant center size
There are four overperforming centers and four underperforming centers in Fig. 1a, and there are seven underperforming centers and two overperforming centers in Fig. 1b. Note also that most of the overperforming centers are larger centers, while most of the underperforming centers are smaller centers. Additional analysis of center characteristics may elucidate why this trend is occurring, but this would require additional data collection from centers and is beyond the scope of this analysis.
5 Conclusions
We have presented a method for examining the performance of centers performing hematopoietic cell transplants. The method is based on a simple comparison of observed and predicted survival outcomes at one year, and is an extension of the method described in DeLong et al. (1997) to allow for censored outcomes as well as centers performing a small number of transplants. The method controls the type I error rate, or the probability that an “average” transplant center will be incorrectly classified as over or underperforming, at a pre-specified level α. Adjustment for patient characteristics is done by building a regression model, and our simulations indicate that the type of patients selected for transplant at a given center do not affect the likelihood that a center will be incorrectly classified, as long as patient characteristics are adjusted for in the regression model. While the method controls the marginal type I error rate for a particular center, it doesn’t explicitly deal with the multiplicity problem across centers. This can be done by adjusting the individual type I error rate, e.g. using the Bonferroni correction. The Bonferroni correction controls the probability that at least one center is incorrectly classified as over or under performing, and it may be excessively conservative in terms of being less likely to identify truly overperforming or underperforming centers. Alternative methods of accounting for multiplicity, such as the false discovery rate criterion of Benjamini and Hochberg (1995), may be less conservative. Finally, we point out that the method described here is aimed at identifying extreme observed center outcomes which are not attributable to sampling variability and patient characteristics. In addition to this assessment, one may want to consider the clinical severity of the difference between observed (O) and expected (E) outcomes. Dickinson et al. (2006) combine a statistical assessment using the SMR (significant p-value) with a clinical assessment of the center effect in terms of the observed and expected number of deaths (O/E > 1.5 and O−E > 3) in order to identify centers for review.
Table 3.
Type I error results, by center size and center number according to scenarios 1–7, with 20% censoring at 1 year
| Center size | Center number | Scenario | ||||||
|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | ||
| 15 | 1–5 | 0.055 | 0.055 | 0.054 | 0.055 | 0.055 | 0.054 | 0.054 |
| 6–10 | 0.056 | 0.055 | 0.055 | 0.056 | 0.055 | 0.054 | 0.055 | |
| 11–45 | 0.054 | 0.054 | 0.054 | 0.054 | 0.054 | 0.054 | 0.054 | |
| 45 | 46–48 | 0.053 | 0.051 | 0.052 | 0.053 | 0.051 | 0.051 | 0.051 |
| 49–51 | 0.053 | 0.051 | 0.051 | 0.050 | 0.053 | 0.051 | 0.049 | |
| 52–74 | 0.051 | 0.052 | 0.051 | 0.052 | 0.052 | 0.052 | 0.052 | |
| 75 | 75–77 | 0.051 | 0.050 | 0.051 | 0.050 | 0.049 | 0.050 | 0.049 |
| 78–80 | 0.050 | 0.052 | 0.051 | 0.049 | 0.052 | 0.051 | 0.047 | |
| 81–100 | 0.052 | 0.051 | 0.052 | 0.052 | 0.052 | 0.051 | 0.052 | |
| 150 | 101–102 | 0.050 | 0.049 | 0.048 | 0.048 | 0.051 | 0.048 | 0.047 |
| 103–104 | 0.050 | 0.052 | 0.050 | 0.049 | 0.049 | 0.053 | 0.043 | |
| 105–115 | 0.051 | 0.049 | 0.052 | 0.050 | 0.049 | 0.050 | 0.049 | |
| 400 | 116 | 0.044 | 0.045 | 0.045 | 0.046 | 0.046 | 0.045 | 0.041 |
| 117 | 0.047 | 0.045 | 0.047 | 0.047 | 0.045 | 0.047 | 0.040 | |
| 118–119 | 0.044 | 0.045 | 0.045 | 0.047 | 0.046 | 0.044 | 0.049 | |
Acknowledgements
This research was partially supported by a grant (R01 CA54706-10) from the National Cancer Institute.
Contributor Information
Brent R. Logan, Division of Biostatistics, Medical College of Wisconsin, 8701 Watertown Plank Road, Milwaukee, WI 53226-0509, USA e-mail: blogan@mcw.edu
Gene O. Nelson, National Marrow Donor Program, 3001 Broadway Street N.E., Suite 100, Minneapolis, MN 55413-1753, USA e-mail: gnelson@nmdp.org
John P. Klein, Division of Biostatistics, Medical College of Wisconsin, 8701 Watertown Plank Road, Milwaukee, WI 53226-0509, USA e-mail: klein@mcw.edu
References
- Andersen PK, Klein JP, Rosthoj S. Generalized linear models or correlated pseudo-observations with applications to multi-state models. Biometrika. 2003;90:15–27. doi: 10.1093/biomet/90.1.15. [Google Scholar]
- Austin PC, Alter DA, Tu JV. The use of fixed-and random-effects models for classifying hospitals as mortality outliers: a monte carlo assessment. Med Decis Making. 2003;23:526–539. doi: 10.1177/0272989X03258443. doi: 10.1177/0272989X03258443. [DOI] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc B. 1995;57:289–300. [Google Scholar]
- Christiansen CL, Morris CN. Improving the statistical approach to health care provider profiling. Ann Intern Med. 1997;127:764–768. doi: 10.7326/0003-4819-127-8_part_2-199710151-00065. [DOI] [PubMed] [Google Scholar]
- DeLong ER, Peterson ED, DeLong DM, Muhlbaier LH, Hackett S, Mark DB. Comparing risk-adjustment methods for provider profiling. Stat Med. 1997;16:2645–2664. doi: 10.1002/(sici)1097-0258(19971215)16:23<2645::aid-sim696>3.0.co;2-d. doi:10.1002/(SICI)1097–0258(19971215)16:23<2645::AID-SIM696>3.0.CO;2-D. [DOI] [PubMed] [Google Scholar]
- Dickinson DM, Shearon TH, O’Keefe J, Wong H-H, Berg CL, Rosendale JD, et al. SRTR Center-Specific Reporting Tools: Posttransplant Outcomes. Am J Transplant. 2006;6:1198–1211. doi: 10.1111/j.1600-6143.2006.01275.x. doi: 10.1111/j.1600-6143.2006.01275.x. [DOI] [PubMed] [Google Scholar]
- Health Care Financing Administration. Washington, DC: Government Printing Office; Medicare hospital mortality information, 1988. 1989
- Huang I-C, Dominici F, Frangakis C, Diette GB, Damberg CL, Wu AW. Is risk-adjustor selection more important than statistical approach for provider profiling? Asthma as an example. Med Decis Making. 2005;25:20–34. doi: 10.1177/0272989X04273138. doi: 10.1177/0272989X04273138. [DOI] [PubMed] [Google Scholar]
- Iezzoni LI. Ann Arbor, MI: Health Administration Press; 1994. Risk adjustment for measuring health care outcomes. [Google Scholar]
- Klein JP, Andersen PK. Regression modeling of competing risks data based on pseudo-values of the cumulative incidence function. Biometrics. 2005;61:223–229. doi: 10.1111/j.0006-341X.2005.031209.x. doi: 10.1111/j.0006-341X.2005.031209.x. [DOI] [PubMed] [Google Scholar]
- Klein JP, Logan BR, Harhoff M, Andersen PK. Analyzing survival curves at a fixed point in time. Stat Med. 2007;26:4505–4519. doi: 10.1002/sim.2864. doi: 10.1002/sim.2864. [DOI] [PubMed] [Google Scholar]
- Klein JP, Gerster M, Andersen PK, Tarima S, Perme MP. SAS and R functions to compute pseudovalues for censored data regression. Comput Methods Programs Biomed. 2008;89:289–300. doi: 10.1016/j.cmpb.2007.11.017. doi: 10.1016/j.cmpb.2007.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landon B, Iezzoni L, Ash AS, Schwartz M, Daley J, Hughes JS, et al. Judging hospitals by severity adjusted mortality rates: the case of CABG surgery. Inquiry. 1996;33:155–166. [PubMed] [Google Scholar]
- Liang K-Y, Zeger SL. Longitudinal data analysis using generalized linear models. Biometrika. 1986;73:13–22. [Google Scholar]
- Localio AR, Hamory BH, Sharp TJ, Weaver SL, TenHave TR, Landis JR. Comparing hospital mortality in adult patients with pneumonia: a case study of statistical methods in a managed care program. Ann Intern Med. 1995;122:125–132. doi: 10.7326/0003-4819-122-2-199501150-00009. [DOI] [PubMed] [Google Scholar]
- Moulton LH, Zeger SL. Bootstrapping generalized linear models. Comput Stat Data Anal. 1991;11:53–63. doi: 10.1016/0167-9473(91)90052-4. [Google Scholar]
- NationalMarrowDonor Program. Choosing a transplant center:apatient’s guide. [Accessed 19 Feb 2008];2007 http://www.marrow.org/access.
- New York State Department of Health. Coronary artery bypass graft surgery in New York State 1989–1991. Albany: New York State Department of Health; 1992
- Normand S-LT, Glickman ME, Gatsonis CA. Statistical methods for profiling providers of medical care: issues and applications. JASA. 1997;92:803–814. [Google Scholar]
- Pasquini MC, Wang Z, Schneider L. CIBMTR summary slides 2007, Part 1. [Accessed 28 Jan 2008];2007 http://www.cibmtr.org/PUBLICATIONS/Newsletter/DOCS/2007Dec.pdf. [Google Scholar]
- Salem-Schatz S, Moore G, Rucker M, Pearson S. The case for case-mix adjustment in practice profiling. JAMA. 1994;272:871–874. doi: 10.1001/jama.272.11.871. [PubMed] [Google Scholar]
- Thomas N, Longford NT, Rolph JE. Empirical Bayes methods for estimating hospital-specific mortality rates. Stat Med. 1994;13:889–903. doi: 10.1002/sim.4780130902. doi: 10.1002/sim.4780130902. [DOI] [PubMed] [Google Scholar]