Abstract
A note about nomenclature
The ortholog of the gene mutated in rhabdoid tumors was first studied in yeast where it was identified in a screen for mutants incapable of fermenting sucrose. It was thus given the name Sucrose Non-Fermenting gene number 5 (SNF5) and was subsequently found to be a member of the SWI/SNF chromatin remodeling complex. The human ortholog of the gene was identified in a screen for proteins capable of interacting with the integrase protein of the human immunodeficiency virus and was given the name INtegrase Interactor 1 (INI1). Investigators studying a mammalian version of the Swi/Snf complex felt that its function may have diverged somewhat from the yeast complex and thus proposed renaming the complex the Brg1/Brm Associated Factors complex, or BAF complex. The rhabdoid tumor gene was thus given the name BAF47 based upon its apparent molecular mass of 47 Kd. Most recently, the genetic nomenclature committee bestowed the name SMARCB1 for SWI/SNF related, Matrix associated, Actin dependent Regulator of Chromatin, subfamily B, member 1. Each of these names has been used extensively in the literature and we ourselves have referred to the gene as either SNF5 (CWMR) or INI1 (JAB). In an effort to simplify communication, we have chosen to use the official SMARCB1 nomenclature here.
Keywords: rhabdoid tumor, retroperitoneum, angiogenesis, surgery, chemotherapy, radiotherapy, targeted therapy, recurrence, metastasis
Rhabdoid tumors are especially lethal cancers that predominantly strike young children. The vast majority of rhabdoid tumors contain bi-allelic inactivating mutations in the SMARCB1 gene. Here we review clinicopathologic features of rhabdoid tumors and present recent insights into the mechanisms that drive oncogenesis in the absence of SMARCB1 function.
Rhabdoid tumors are highly malignant neoplasms that typically arise in infancy and early childhood. The tumors develop in the brain and spinal cord [referred to as atypical teratoid/rhabdoid tumor (AT/RT)], kidney and/or soft tissues (termed malignant rhabdoid tumor or extra-renal rhabdoid tumor). The histologic appearance of these malignancies can be quite variable. Most tumors contain at least some fields with classic rhabdoid cells, with large nuclei containing a single prominent nucleolus, and cytoplasm with distinct pale eosinophilic inclusions. Tumors demonstrating only classic rhabdoid cells are rare, instead, they often have areas composed of spindled or pleomorphic undifferentiated cells without a rhabdoid phenotype. A classic rhabdoid component may be entirely absent. Central nervous system AT/RTs typically demonstrate a variety of primitive neuroectodermal, epithelial or mesenchymal cells, which underlies the difficulty in distinguishing these tumors from other primitive neuroectodermal tumors or choroid plexus carcinomas.1 Immunohistochemistry is often used in the differential diagnosis, based on the typical expression of smooth muscle actin, epithelial membrane antigen and vimentin. Lack of expression of the SMARCB1 protein, as described below, is also employed as a specific means of distinguishing rhabdoid tumors from other malignancies with similar histologic features, especially for diagnosis of AT/RT versus primitive neuroectodermal tumor.2
The development of rhabdoid tumors was initially associated with monosomy 22 and subsequently, deletions and translocations involving chromosome band 22q11.2.3,4 Positional cloning studies ultimately identified SMARCB1 (INI1/SNF5/BAF47) as the gene responsible for the initiation of malignant rhabdoid tumors and central nervous system AT/RT.5 In the appropriate clinical setting, germline and somatic mutations and deletions of SMARCB1 may be diagnostic for renal and extra-renal rhabdoid tumors as well as AT/RT, however the role of SMARCB1 inactivation in other CNS tumors, such as choroid plexus carcinoma,6,7 remains controversial. Recently, SMARCB1 has also been implicated in the development of familial schwannomatosis,8–12 although it is not clear if there is a similar spectrum of mutations and deletions to those seen in classic rhabdoid tumors. In rhabdoid tumors, SMARCB1 appears to function as a classic tumor suppressor gene, such that germline mutations and deletions predispose to the development of these malignancies, and somatic loss or mutation of the other allele constitutes the second hit. Inactivation of both copies of the gene leads to loss of protein expression in the nucleus, which can be detected by immunohistochemistry. The immunohistochemistry assay for the SMARCB1 protein (BAF47) is currently used as an adjunct to histology in the differential diagnosis of rhabdoid tumors in both children and adults.2 Loss of expression of SMARCB1 has also been observed in the majority of epithelioid sarcomas [ES],13 fifty percent of epithelioid malignant peripheral nerve sheath tumors 13 and renal medullary carcinomas [RMC].14 However, molecular genetic studies to determine the mechanism for loss of expression of the protein in ES have been conflicting and have yet to be reported in RMC.15,16 Thus the frequency with which the typical bi-allelic mutations or deletions in the SMARCB1 locus that are seen in rhabdoid tumors are also responsible for the loss of protein expression observed in ES is somewhat unclear.
Individuals with germline alterations of SMARCB1 are predisposed to rhabdoid tumors of the brain, kidney and soft tissues and may present with more than one primary tumor.5,17,18 These children are most often diagnosed within the first year of life and tend to have a worse prognosis.19 It is not known whether the poor prognosis is related to the presence of a germline mutation in all of their cells, or the fact that they develop multiple and progressive primary tumors that are resistant to therapy.
In most patients, the germline mutations or deletions are de novo and parents are reported to be unaffected. Reports of multiple affected siblings with CNS tumors with the same germline mutation, in which neither parent was a carrier, implicated gonadal mosaicism as a mechanism that can result in a familial associated genetic predisposition to cancer.20 To date, there have been only three reports of rhabdoid tumors associated with inherited SMARCB1 mutations in multi-generation families.21–23 In two cases, the affected children had a germline SMARCB1 mutation inherited from their unaffected carrier mother. In a recently described family multiple members had either schwannomas or AT/RT associated with inheritance of an exon 6 duplication in the SMARCB1 gene.23 Therefore, reduced penetrance for SMARCB1, i.e., variable risk of development of rhabdoid tumor or schwannoma associated with a germline mutation, gonadal mosaicism and risk of multiple primary tumors all need to be considered in developing recurrence risks for affected families. Finally, rhabdoid tumors have been seen in the context of a germline chromosome band 22q11.2 deletion that encompasses the SMARCB1 locus.18 Depending on the size of the deletion, these patients may also be at risk for heart defects, development delay and other congenital abnormalities associated with the loss of additional genes in the 22q11.2 region. As high-density array comparative genomic hybridization or single nucleotide polymorphism based oligonucleotide arrays are increasingly being used to screen for genetic disorders in patients with congenital abnormalities and developmental delay, additional individuals will likely be found with germline deletions of SMARCB1. Clinical monitoring protocols for these individuals will need to be developed because of their increased risk of malignancy.
A germline mutation or deletion may be present in 15–30% of patients with rhabdoid tumor, although active studies are ongoing to refine these numbers. Genetic counseling and clinical molecular genetic testing for a germline SMARCB1 mutation or deletion in patients who present with schwannomatosis or rhabdoid tumor, even if it appears to be sporadic, is highly recommended.
Consistent with its role as a tumor suppressor, deletion and mutation analyses of CNS, renal and extrarenal rhabdoid tumors have demonstrated bi-allelic alterations of the SMARCB1 gene.5,17,24 The majority of coding sequence mutations are point or frameshift mutations that introduce a novel stop codon and predict premature truncation of the protein. There are several hot spots for SMARCB1 mutations, and two mutations appear to be specific for CNS tumors. A cytosine or guanine deletion in codon 382 in exon 9 has been documented in the majority of CNS AT/RTs. Both the delC and delG cause a frameshift; but eliminate the final stop codon, and are predicted to result in the addition of 96 amino acids to the C terminal end of the protein. Interestingly, this mutation has not yet been observed in the germline. A c.601C > T mutation in exon 5 has also been documented in a large number of AT/RTs, but may also be present as a germline mutation. In primary tumors, the messenger RNA or protein is likely to be unstable, however, since the altered proteins are not detected by western blot analysis or immunohistochemistry.
In a recent report, 51 rhabdoid tumors from a variety of anatomic locations were studied using a combination of direct sequence analysis of the nine coding exons, multiplex ligation dependent probe amplification (MLPA) and oligonucleotide based single nucleotide polymorphism [SNP] arrays.25 Bi-allelic inactivating events in SMARCB1 were identified in all but one tumor. Notably, despite the differences in anatomic location, half of the tumors (24/51) were characterized by homozygous deletions of SMARCB1. This included over one third (13 of 36) of the CNS AT/RTs as well as the majority of both renal (6/8) and extra renal (5/7) tumors. The advantage of the SNP based genotyping array platform was evidenced by the large number of cases (11 tumors) with copy number neutral loss of heterozygosity for most of the long arm of chromosome 22. The renal rhabdoid tumors were more likely to have complex patterns of deletion and duplication as evidenced by the array studies, whereas the extra-renal rhabdoid tumors were more likely to have smaller deletions in 22q11.22 to 22q11.23. In most cases, this is due to unbalanced translocations at the SMARCB1 locus.
These data strongly support SMARCB1 as being the primary gene responsible for the development of rhabdoid tumors. However, a small number of rhabdoid tumors retain expression of the protein, even with rhabdoid histology and familial rhabdoid tumors have been reported that are not associated with a SMARCB1 mutation. 26,27 These reports raise the possibility of a second rhabdoid tumor locus, distinct from SMARCB1.
Chromatin, Epigenetics and Cancer
The name SMARCB1 (SWI/SNF related, Matrix associated, Actin dependent Regulator of Chromatin, subfamily B, member 1) is derived from its role as a core member of the SWI/SNF chromatin remodeling complex. As such, SMARCB1 represents a novel type of tumor suppressor. Consequently, elucidation of the mechanism by which its loss results in the onset of aggressive cancers has been an active area of investigation and these studies have provided insight into the mechanisms underlying rhabdoid tumor formation.
The central unit of chromatin is the nucleosome, an octamer of histone proteins around each of which is wrapped approximately 147 base pairs of DNA. Nucleosomes provide an organizational structure for DNA but can also constitute a barrier to gene expression by blocking access of the transcriptional machinery to the DNA of target genes. Consequently, chromatin remodeling complexes, which regulate local chromatin structure, are a critical component of the mechanisms for controlling gene expression. Remodeling complexes can be grouped into two broad classes—those that covalently modify histones, such as with acetyl, methyl or phosphoryl groups, and those that utilize the energy of ATP hydrolysis, such as the SWI/SNF complex, to move nucleosomes along the DNA strand.
Originally identified in yeast, the SWI/SNF complex is present in all eukaryotes and is highly evolutionarily conserved. Mammalian SWI/SNF complexes are comprised of a SWI2/SNF2 family ATPase (either SMARCA4/BRG1 or SMARCA2/BRM); common core subunits (SMARCB1, SMARCC1/BAF155 and SMARCC2/BAF170), and 4–8 subunits that vary by cellular lineage.28,29 In vitro analysis revealed that the SWI/SNF complex is capable of consuming ATP and displacing nucleosomes. This activity occurs in a dynamic fashion in response to signaling and differentiation and serves to regulate transcription of specific targets. In mammals, numerous variants of the SWI/SNF complex exist and are distinguished by inclusion of variant lineage-specific subunits. Indeed, transcriptional regulation by SWI/SNF has been implicated in the control of proliferation and differentiation in multiple tissues.30
SMARCB1 is a core subunit present in all variants of the SWI/SNF complex. The protein is highly conserved, as evidenced by an identical amino acid sequence in mice and humans. However, the function of SMARCB1 is poorly understood. There are no SMARCB1 paralogs and the protein lacks particularly informative protein motifs. Consequently, investigations into the mechanism by which inactivation of SMARCB1 promotes cancer are being pursued.
Mouse Models—A Predisposition to Cancer
Following the initial report of mutations in SMARCB1 in human rhabdoid tumors, several groups developed Smarcb1 knock-out mice.31–33 Homozygous inactivation of Smarcb1 results in early embryonic lethality, while heterozygotes are born at the expected frequency and appear normal. However, the heterozygotes are predisposed to cancer with approximately 20% developing sarcomas at a median age of 12 months. These tumors closely resemble human rhabdoid tumors, including the presence of cells with classic rhabdoid morphology in most tumors. As in humans who inherit a mutant SMARCB1 allele, the remaining Smarcb1 allele has been spontaneously lost within the murine tumors. These tumors occur most frequently on the head/neck followed by paraspinal, trunk and extremity locations, all in sites that also occur in humans. Some of the models develop occasional brain tumors although none develop kidney tumors, these two sites being the most frequent location of rhabdoid tumors in humans. The cancers that occur in Smarcb1+/− mice are highly aggressive, always locally invasive and frequently are metastatic to regional lymph nodes or lung.34
In subsequent models that alleviate the requirement for spontaneous loss of the remaining allele, conditional bi-allelic inactivation of Smarcb1 was shown to result in a marked cancer predisposition.35 All mice develop cancer with a median onset of 11 weeks, a remarkably rapid pace for the inactivation of a single gene. In comparison, the time to tumor development with other tumor suppressors is significantly longer as p53 loss leads to cancer at 20 weeks, p19Arf loss at 38 weeks and p16Ink4a loss at 60 weeks.36–38 Thus, the aggressive cancer-prone phenotype that occurs following Smarcb1 inactivation is striking and indicates a critical role for Smarcb1 in preventing cancer.
Mechanism
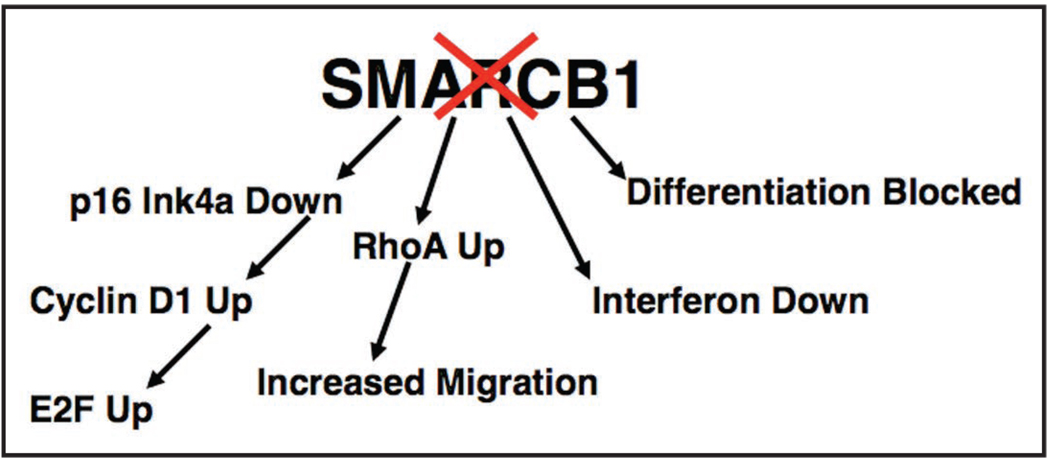
SMARCB1 represented the first member of an ATPase chromatin remodeling complex to be implicated in the genesis of cancer. Subsequent studies investigating its mechanism of tumor suppression have found that SMARCB1 loss causes cell cycle progression in part via downregulation of p16INK4a and upregulation of E2Fs and Cyclin D1 (Fig. 1).39–43 However, the aberrant proliferative stimulus caused by SMARCB1 loss also triggers cell cycle checkpoints causing arrest and apoptosis. Thus, despite causing upregulation of proliferation associated gene pathways, SMARCB1 loss is lethal to most primary cells.43,44 Disruption of checkpoints via inactivation of p53 in vivo results in dramatic oncogenic synergy with Smarcb1 loss leading to cancer formation at a median of three weeks, although loss of p53 itself is insufficient to circumvent arrest in cultured Smarcb1-deficient cells.43,44 Studies aimed at elucidating the mechanisms underlying the highly invasive and metastatic nature of rhabdoid tumor have identified a role for SMARCB1 in controlling the actin cytoskeleton network and found that SMARCB1 loss transcriptionally enhances RhoA signaling conferring enhanced migratory potential upon cell lines (Fig. 1).45,46
Figure 1.
Cancer-related effects of SMARCB1 loss.
In addition to roles in transcriptional regulation, some chromatin remodeling complexes, such as the INO80, SWR1 and RAD54 complexes,47–52 have been shown to play a role in DNA repair and maintenance of genome stability. Several studies have also suggested that the Swi/Snf complex is involved in DNA repair.44,53–55 Thus, SMARCB1 loss might contribute to oncogenesis by causing a DNA repair defect and genomic instability. However, a recent study aimed at determining whether defective DNA repair and genomic instability underlie the cancers caused by SMARCB1 loss found that Smarcb1-deficient cells do not have increased sensitivity to DNA damaging agents, nor altered induction of either γ-H2AX or repair check-points. 56 Further evaluation focused upon primary human MRT found that these aggressive SMARCB1-deficient cancers are diploid and genomically stable. Indeed, aside from SMARCB1 loss itself, most SMARCB1-deficient cancers lack recurrent genome amplifications and deletions.25,56,57 Collectively, these results suggest that disruption of this chromatin remodeling complex can largely, if not completely, substitute for genomic instability in the genesis of aggressive cancer.
Therapeutic Insights
Given the role of the SWI/SNF complex in the regulation of transcription, proliferation- and metastasis-related genes whose expression is affected by SMARCB1 loss have been sought. Several targets have been identified that may play a role in oncogenic transformation following SMARCB1 loss. Cyclin D1 is expressed at high levels in rhabdoid tumors58,59 and this effect appears specific as other cyclins are not similarly overexpressed compared to other cancers.56 SMARCB1 binds to the Cyclin D1 promoter and regulates it expression. Ablation of Cyclin D1 has been shown to prevent rhabdoid tumor growth and pharmacologic inhibition has shown promise in mouse models.60,61 C-MYC is highly expressed in rhabdoid tumors56,62 and SMARCB1 and the SWI/SNF complex have been reported to modulate c-MYC activity. The SWI/SNF complex binds to the c-MYC promoter and has been reported to repress the expression of c-MYC.63 However, SMARCB1 protein has also been shown to directly interact with c-MYC and to be required for its transactivation potential.64 Thus, it is currently unclear whether the high levels of c-MYC in rhabdoid tumors contributes to oncogenesis or whether this is a secondary increase due to loss of c-MYC transactivation function. Loss of SMARCB1 has also been shown to interfere with activation of interferon target genes and treatment with either interferon or inhibition of PLK1 may hold therapeutic promise (Fig. 1).65
Despite some recent improvements in therapy, rhabdoid tumors remain highly lethal cancers. Studies detailing the molecular changes underlying the disease and the related familial predisposition syndrome have led to improved diagnostic ability and offer the opportunity to develop familial screening and disease monitoring in susceptible individuals. Similarly, better understanding of the mechanisms by which mutation of SMARCB1 drive oncogenesis has the potential to identify novel therapeutic approaches for rhabdoid tumors and related cancers.
Acknowledgements
The authors’ work is supported by grants from the NIH (CA46274 to J.A.B.) and (CA113794 to C.W.M.R.). C.W.M.R. also gratefully acknowledges support from the Garrett B. Smith Foundation and the Claudia Adams Barr Foundation.
References
- 1.Rorke LB, Packer RJ, Biegel JA. Central nervous system atypical teratoid/rhabdoid tumors of infancy and childhood: Definition of an entity. J Neurosurg. 1996;85:56–65. doi: 10.3171/jns.1996.85.1.0056. [DOI] [PubMed] [Google Scholar]
- 2.Judkins AR, Mauger J, Ht A, Rorke LB, Biegel JA. Immunohistochemical analysis of hSNF5/INI1 in pediatric CNS neoplasms. Am J Surg Pathol. 2004;28:644–650. doi: 10.1097/00000478-200405000-00013. [DOI] [PubMed] [Google Scholar]
- 3.Biegel JA, Rorke LB, Packer RJ, Emanuel BS. Monosomy 22 in rhabdoid or atypical tumors of the brain. J Neurosurg. 1990;73:710–714. doi: 10.3171/jns.1990.73.5.0710. [DOI] [PubMed] [Google Scholar]
- 4.Douglass EC, Valentine M, Rowe ST, Parham DM, Wilimas JA, Sanders JM, et al. Malignant rhabdoid tumor: A highly malignant childhood tumor with minimal karyotypic changes. Genes Chromosomes Cancer. 1990;2:210–216. doi: 10.1002/gcc.2870020308. [DOI] [PubMed] [Google Scholar]
- 5.Biegel JA, Zhou JY, Rorke LB, Stenstrom C, Wainwright LM, Fogelgren B. Germ-line and acquired mutations of INI1 in atypical teratoid and rhabdoid tumors. Cancer Res. 1999;59:74–79. [PubMed] [Google Scholar]
- 6.Sevenet N, Sheridan E, Amram D, Schneider P, Handgretinger R, Delattre O. Constitutional mutations of the hSNF5/INI1 gene predispose to a variety of cancers. Am J Hum Genet. 1999;65:1342–1348. doi: 10.1086/302639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mueller W, Eum JH, Lass U, Paulus W, Sarkar C, Bruck W, et al. No evidence of hSNF5/INI1 point mutations in choroid plexus papilloma. Neuropathol Applied Neurobiol. 2004;30:304–307. doi: 10.1046/j.0305-1846.2004.00538.x. [DOI] [PubMed] [Google Scholar]
- 8.Hulsebos TJ, Plomp AS, Wolterman RA, Robanus-Maandag EC, Baas F, Wesseling P. Germline mutation of INI1/SMARCB1 in familial schwannomatosis. Am J Hum Genet. 2007;80:805–810. doi: 10.1086/513207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sestini R, Bacci C, Provenzano A, Genuardi M, Papi L. Evidence of a four-hit mechanism involving SMARCB1 and NF2 in schwannomatosis-associated schwannomas. Hum Mutat. 2008;29:227–231. doi: 10.1002/humu.20679. [DOI] [PubMed] [Google Scholar]
- 10.Hadfield KD, Newman WG, Bowers NL, Wallace A, Bolger C, Colley A, et al. Molecular characterisation of SMARCB1 and NF2 in familial and sporadic schwannomatosis. J Med Genet. 2008;45:332–339. doi: 10.1136/jmg.2007.056499. [DOI] [PubMed] [Google Scholar]
- 11.Patil S, Perry A, Maccollin M, Dong S, Betensky RA, Yeh TH, et al. Immunohistochemical analysis supports a role for INI1/SMARCB1 in hereditary forms of schwannomas, but not in solitary, sporadic schwannomas. Brain Pathol. 2008;18:517–519. doi: 10.1111/j.1750-3639.2008.00155.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Boyd C, Smith M, Kluwe L, Balogh A, Maccollin M, Plotkin S. Alterations in the SMARCB1 (INI1) tumor suppressor gene in familial schwannomatosis. Clin Genet. 2008;74:358–366. doi: 10.1111/j.1399-0004.2008.01060.x. [DOI] [PubMed] [Google Scholar]
- 13.Hornick JL, Dal Cin P, Fletcher CD. Loss of INI1 expression is characteristic of both conventional and proximal-type epithelioid sarcoma. Am J Surg Pathol. 2008 doi: 10.1097/PAS.0b013e3181882c54. In press. [DOI] [PubMed] [Google Scholar]
- 14.Cheng JX, Tretiakova M, Gong C, Mandal S, Krausz T, Taxy JB. Renal medullary carcinoma: Rhabdoid features and the absence of INI1 expression as markers of aggressive behavior. Mod Pathol. 2008;21:647–652. doi: 10.1038/modpathol.2008.44. [DOI] [PubMed] [Google Scholar]
- 15.Kohashi K, Izumi T, Oda Y, Yamamoto H, Tamiya S, Taguchi T, et al. Infrequent SMARCB1/INI1 gene alteration in epithelioid sarcoma: A useful tool in distinguishing epithelioid sarcoma from malignant rhabdoid tumor. Hum Pathol. 2008 doi: 10.1016/j.humpath.2008.08.007. In press. [DOI] [PubMed] [Google Scholar]
- 16.Modena P, Lualdi E, Facchinetti F, Galli L, Teixeira MR, Pilotti S, et al. SMARCB1/INI1 tumor suppressor gene is frequently inactivated in epithelioid sarcomas. Cancer Res. 2005;65:4012–4019. doi: 10.1158/0008-5472.CAN-04-3050. [DOI] [PubMed] [Google Scholar]
- 17.Biegel JA, Tan L, Zhang F, Wainwright L, Russo P, Rorke LB. Alterations of the hSNF5/INI1 gene in central nervous system atypical teratoid/rhabdoid tumors and renal and extra-renal rhabdoid tumors. Clin Cancer Res. 2002;8:3461–3467. [PubMed] [Google Scholar]
- 18.Jackson EM, Shaikh TH, Gururangan S, Jones MC, Malkin D, Nikkel SM, et al. High-density single nucleotide polymorphism array analysis in patients with germline deletions of 22q11.2 and malignant rhabdoid tumor. Hum Genet. 2007;122:117–127. doi: 10.1007/s00439-007-0386-3. [DOI] [PubMed] [Google Scholar]
- 19.Savla J, Chen TT, Schneider NR, Timmons CF, Delattre O, Tomlinson GE. Mutations of the hSNF5/INI1 gene in renal rhabdoid tumors with second primary brain tumors. J Natl Cancer Inst. 2000;92:648–650. doi: 10.1093/jnci/92.8.648. [DOI] [PubMed] [Google Scholar]
- 20.Sevenet N, Lellouch-Tubiana A, Schofield D, Hoang-Xuan K, Gessler M, Birnbaum D, et al. Spectrum of hSNF5/INI1 somatic mutations in human cancer and genotype-phenotype correlations. Hum Mol Genet. 1999;8:2359–2368. doi: 10.1093/hmg/8.13.2359. [DOI] [PubMed] [Google Scholar]
- 21.Taylor MD, Gokgoz N, Andrulis IL, Mainprize TG, Drake JM, Rutka JT. Familial posterior fossa brain tumors of infancy secondary to germline mutation of the hSNF5 gene. Am J Hum Genet. 2000;66:1403–1406. doi: 10.1086/302833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Janson K, Nedzi LA, David O, Schorin M, Walsh JW, Bhattacharjee M, et al. Predisposition to atypical teratoid/rhabdoid tumor due to an inherited INI1 mutation. Pediatr Blood Cancer. 2006;47:279–284. doi: 10.1002/pbc.20622. [DOI] [PubMed] [Google Scholar]
- 23.Swensen JJ, Keyser J, Coffin CM, Biegel JA, Viskochil DH, Williams MS. Familial occurrence of schwannomas and malignant rhabdoid tumour associated with a duplication in SMARCB1. J Med Genet. 2009;46:68–72. doi: 10.1136/jmg.2008.060152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Versteege I, Sevenet N, Lange J, Rousseau-Merck MF, Ambros P, Handgretinger R, et al. Truncating mutations of hSNF5/INI1 in aggressive paediatric cancer. Nature. 1998;394:203–206. doi: 10.1038/28212. [DOI] [PubMed] [Google Scholar]
- 25.Jackson EM, Sievert AJ, Gai X, Hakonarson H, Judkins AR, Tooke L, et al. Genomic analysis using high density SNP based oligonucleotide arrays and MLPA provides a comprehensive analysis of INI1/SMARCB1 in malignant rhabdoid tumors. Clin Cancer Res. 2009 doi: 10.1158/1078-0432.CCR-08-2091. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bourdeaut F, Freneaux P, Thuille B, Lellouch-Tubiana A, Nicolas A, Couturier J, et al. hSNF5/INI1-deficient tumours and rhabdoid tumours are convergent but not fully overlapping entities. J Pathol. 2007;211:323–330. doi: 10.1002/path.2103. [DOI] [PubMed] [Google Scholar]
- 27.Fruhwald MC, Hasselblatt M, Wirth S, Kohler G, Schneppenheim R, Subero JI, et al. Non-linkage of familial rhabdoid tumors to SMARCB1 implies a second locus for the rhabdoid tumor predisposition syndrome. Pediatr Blood Cancer. 2006;47:273–278. doi: 10.1002/pbc.20526. [DOI] [PubMed] [Google Scholar]
- 28.Phelan ML, Sif S, Narlikar GJ, Kingston RE. Reconstitution of a core chromatin remodeling complex from SWI/SNF subunits. Mol Cell. 1999;3:247–253. doi: 10.1016/s1097-2765(00)80315-9. [DOI] [PubMed] [Google Scholar]
- 29.Lessard J, Wu JI, Ranish JA, Wan M, Winslow MM, Staahl BT, et al. An essential switch in subunit composition of a chromatin remodeling complex during neural development. Neuron. 2007;55:201–215. doi: 10.1016/j.neuron.2007.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.de la Serna IL, Ohkawa Y, Imbalzano AN. Chromatin remodelling in mammalian differentiation: Lessons from ATP-dependent remodellers. Nat Rev Genet. 2006;7:461–473. doi: 10.1038/nrg1882. [DOI] [PubMed] [Google Scholar]
- 31.Roberts CW, Galusha SA, McMenamin ME, Fletcher CD, Orkin SH. Haploinsufficiency of Snf5 (integrase interactor 1) predisposes to malignant rhabdoid tumors in mice. Proc Nat Acad Sci USA. 2000;97:13796–13800. doi: 10.1073/pnas.250492697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Klochendler-Yeivin A, Fiette L, Barra J, Muchardt C, Babinet C, Yaniv M. The murine SNF5/INI1 chromatin remodeling factor is essential for embryonic development and tumor suppression. EMBO Rep. 2000;1:500–506. doi: 10.1093/embo-reports/kvd129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Guidi CJ, Sands AT, Zambrowicz BP, Turner TK, Demers DA, Webster W, et al. Disruption of Ini1 leads to peri-implantation lethality and tumorigenesis in mice. Mol Cell Biol. 2001;21:3598–3603. doi: 10.1128/MCB.21.10.3598-3603.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Roberts CJ, Nelson B, Marton MJ, Stoughton R, Meyer MR, Bennett HA, et al. Signaling and circuitry of multiple MAPK pathways revealed by a matrix of global gene expression profiles. Science. 2000;287:873–880. doi: 10.1126/science.287.5454.873. [DOI] [PubMed] [Google Scholar]
- 35.Roberts CW, Leroux MM, Fleming MD, Orkin SH. Highly penetrant, rapid tumorigenesis through conditional inversion of the tumor suppressor gene Snf5. Cancer Cell. 2002;2:415–425. doi: 10.1016/s1535-6108(02)00185-x. [DOI] [PubMed] [Google Scholar]
- 36.Donehower LA, Harvey M, Slagle BL, McArthur MJ, Montgomery CA, Jr, Butel JS, et al. Mice deficient for p53 are developmentally normal but susceptible to spontaneous tumours. Nature. 1992;356:215–221. doi: 10.1038/356215a0. [DOI] [PubMed] [Google Scholar]
- 37.Harvey M, McArthur MJ, Montgomery CA, Jr, Butel JS, Bradley A, Donehower LA. Spontaneous and carcinogen-induced tumorigenesis in p53-deficient mice. Nat Genet. 1993;5:225–229. doi: 10.1038/ng1193-225. [DOI] [PubMed] [Google Scholar]
- 38.Serrano M, Lee H, Chin L, Cordon-Cardo C, Beach D, DePinho RA. Role of the INK4a locus in tumor suppression and cell mortality. Cell. 1996;85:27–37. doi: 10.1016/s0092-8674(00)81079-x. [DOI] [PubMed] [Google Scholar]
- 39.Betz BL, Strobeck MW, Reisman DN, Knudsen ES, Weissman BE. Re-expression of hSNF5/INI1/BAF47 in pediatric tumor cells leads to G1 arrest associated with induction of p16ink4a and activation of RB. Oncogene. 2002;21:5193–5203. doi: 10.1038/sj.onc.1205706. [DOI] [PubMed] [Google Scholar]
- 40.Chai J, Charboneau AL, Betz BL, Weissman BE. Loss of the hSNF5 gene concomitantly inactivates p21CIP/WAF1 and p16INK4a activity associated with replicative senescence in A204 rhabdoid tumor cells. Cancer Res. 2005;65:10192–10198. doi: 10.1158/0008-5472.CAN-05-1896. [DOI] [PubMed] [Google Scholar]
- 41.Gresh L, Bourachot B, Reimann A, Guigas B, Fiette L, Garbay S, et al. The SWI/SNF chromatin-remodeling complex subunit SNF5 is essential for hepatocyte differentiation. EMBO J. 2005;24:3313–3324. doi: 10.1038/sj.emboj.7600802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kia SK, Gorski MM, Giannakopoulos S, Verrijzer CP. SWI/SNF mediates polycomb eviction and epigenetic reprogramming of the INK4b-ARF-INK4a locus. Mol Cell Biol. 2008;28:3457–3464. doi: 10.1128/MCB.02019-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Isakoff MS, Sansam CG, Tamayo P, Subramanian A, Evans JA, Fillmore CM, et al. Inactivation of the Snf5 tumor suppressor stimulates cell cycle progression and cooperates with p53 loss in oncogenic transformation. Proc Natl Acad Sci USA. 2005;102:17745–17750. doi: 10.1073/pnas.0509014102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Klochendler-Yeivin A, Picarsky E, Yaniv M. Increased DNA damage sensitivity and apoptosis in cells lacking the Snf5/Ini1 subunit of the SWI/SNF chromatin remodeling complex. Mol Cell Biol. 2006;26:2661–2674. doi: 10.1128/MCB.26.7.2661-2674.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Medjkane S, Novikov E, Versteege I, Delattre O. The tumor suppressor hSNF5/INI1 modulates cell growth and actin cytoskeleton organization. Cancer Res. 2004;64:3406–3413. doi: 10.1158/0008-5472.CAN-03-3004. [DOI] [PubMed] [Google Scholar]
- 46.Caramel J, Quignon F, Delattre O. RhoA-dependent regulation of cell migration by the tumor suppressor hSNF5/INI1. Cancer Res. 2008;68:6154–6161. doi: 10.1158/0008-5472.CAN-08-0115. [DOI] [PubMed] [Google Scholar]
- 47.Morrison AJ, Highland J, Krogan NJ, Arbel-Eden A, Greenblatt JF, Haber JE, et al. INO80 and gamma-H2AX interaction links ATP-dependent chromatin remodeling to DNA damage repair. Cell. 2004;119:767–775. doi: 10.1016/j.cell.2004.11.037. [DOI] [PubMed] [Google Scholar]
- 48.Papamichos-Chronakis M, Krebs JE, Peterson CL. Interplay between Ino80 and Swr1 chromatin remodeling enzymes regulates cell cycle checkpoint adaptation in response to DNA damage. Genes Dev. 2006;20:2437–2449. doi: 10.1101/gad.1440206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Essers J, Hendriks RW, Swagemakers SM, Troelstra C, de Wit J, Bootsma D, et al. Disruption of mouse RAD54 reduces ionizing radiation resistance and homologous recombination. Cell. 1997;89:195–204. doi: 10.1016/s0092-8674(00)80199-3. [DOI] [PubMed] [Google Scholar]
- 50.Shaked H, Avivi-Ragolsky N, Levy AA. Involvement of the Arabidopsis SWI2/SNF2 chromatin remodeling gene family in DNA damage response and recombination. Genetics. 2006;173:985–994. doi: 10.1534/genetics.105.051664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wu S, Shi Y, Mulligan P, Gay F, Landry J, Liu H, et al. A YY1-INO80 complex regulates genomic stability through homologous recombination-based repair. Nat Struct Mol Biol. 2007;14:1165–1172. doi: 10.1038/nsmb1332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tsukuda T, Fleming AB, Nickoloff JA, Osley MA. Chromatin remodelling at a DNA double-strand break site in Saccharomyces cerevisiae. Nature. 2005;438:379–383. doi: 10.1038/nature04148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chai B, Huang J, Cairns BR, Laurent BC. Distinct roles for the RSC and Swi/Snf ATP-dependent chromatin remodelers in DNA double-strand break repair. Genes Dev. 2005;19:1656–1661. doi: 10.1101/gad.1273105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hara R, Mo J, Sancar A. DNA damage in the nucleosome core is refractory to repair by human excision nuclease. Mol Cell Biol. 2000;20:9173–9181. doi: 10.1128/mcb.20.24.9173-9181.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Park JH, Park EJ, Lee HS, Kim SJ, Hur SK, Imbalzano AN, et al. Mammalian SWI/SNF complexes facilitate DNA double-strand break repair by promoting gamma-H2AX induction. EMBO J. 2006;25:3986–3997. doi: 10.1038/sj.emboj.7601291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.McKenna ES, Sansam CG, Cho YJ, Greulich H, Evans JA, Thom CS, et al. Loss of the epigenetic tumor suppressor SNF5 leads to cancer without genomic instability. Mol Cell Biol. 2008;28:6223–6233. doi: 10.1128/MCB.00658-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.McKenna ES, Roberts CW. Epigenetics and cancer without genomic instability. Cell Cycle. 2009;8:23–26. doi: 10.4161/cc.8.1.7290. [DOI] [PubMed] [Google Scholar]
- 58.Fujisawa H, Misaki K, Takabatake Y, Hasegawa M, Yamashita J. Cyclin D1 is overexpressed in atypical teratoid/rhabdoid tumor with hSNF5/INI1 gene inactivation. J Neurooncol. 2005;73:117–124. doi: 10.1007/s11060-004-4276-4. [DOI] [PubMed] [Google Scholar]
- 59.Zhang ZK, Davies KP, Allen J, Zhu L, Pestell RG, Zagzag D, et al. Cell cycle arrest and repression of cyclin D1 transcription by INI1/hSNF5. Mol Cell Biol. 2002;22:5975–5988. doi: 10.1128/MCB.22.16.5975-5988.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tsikitis M, Zhang Z, Edelman W, Zagzag D, Kalpana GV. Genetic ablation of Cyclin D1 abrogates genesis of rhabdoid tumors resulting from Ini1 loss. Proc Natl Acad Sci USA. 2005;102:12129–12134. doi: 10.1073/pnas.0505300102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Alarcon-Vargas D, Zhang Z, Agarwal B, Challagulla K, Mani S, Kalpana GV. Targeting cyclin D1, a downstream effector of INI1/hSNF5, in rhabdoid tumors. Oncogene. 2006;25:722–734. doi: 10.1038/sj.onc.1209112. [DOI] [PubMed] [Google Scholar]
- 62.Rosson GB, Hazen-Martin DJ, Biegel JA, Willingham MC, Garvin AJ, Oswald BW, et al. Establishment and molecular characterization of five cell lines derived from renal and extrarenal malignant rhabdoid tumors. Mod Pathol. 1998;11:1228–1237. [PubMed] [Google Scholar]
- 63.Nagl NG, Jr, Zweitzig DR, Thimmapaya B, Beck GR, Jr, Moran E. The c-myc gene is a direct target of mammalian SWI/SNF-related complexes during differentiation-associated cell cycle arrest. Cancer Res. 2006;66:1289–1293. doi: 10.1158/0008-5472.CAN-05-3427. [DOI] [PubMed] [Google Scholar]
- 64.Cheng SW, Davies KP, Yung E, Beltran RJ, Yu J, Kalpana GV. c-MYC interacts with INI1/hSNF5 and requires the SWI/SNF complex for transactivation function. Nat Genet. 1999;22:102–105. doi: 10.1038/8811. [DOI] [PubMed] [Google Scholar]
- 65.Morozov A, Lee SJ, Zhang ZK, Cimica V, Zagzag D, Kalpana GV. INI1 induces interferon signaling and spindle checkpoint in rhabdoid tumors. Clin Cancer Res. 2007;13:4721–4730. doi: 10.1158/1078-0432.CCR-07-0054. [DOI] [PubMed] [Google Scholar]