Abstract
EDGAR (Extraction of Drugs, Genes and Relations) is a natural language processing system that extracts information about drugs and genes relevant to cancer from the biomedical literature. This automatically extracted information has remarkable potential to facilitate computational analysis in the molecular biology of cancer, and the technology is straightforwardly generalizable to many areas of biomedicine. This paper reports on the mechanisms for automatically generating such assertions and on a simple application, conceptual clustering of documents. The system uses a stochastic part of speech tagger, generates an underspecified syntactic parse and then uses semantic and pragmatic information to construct its assertions. The system builds on two important existing resources: the MEDLINE database of biomedical citations and abstracts and the Unified Medical Language System, which provides syntactic and semantic information about the terms found in biomedical abstracts.
1 Introduction
The biomedical literature is a tremendously rich information source, and the collection of abstracts in the National Library of Medicine’s MEDLINE database summarizes that literature comprehensively. Despite the attractiveness and accessibility of that computer-readable resource, however, automated extraction of useful information from it remains a challenge because the abstracts are in natural language form. In this paper, we report a system, EDGAR (Extraction of Drugs, Genes and Relations), designed to extract factual information from the MEDLINE database on the relationships between genes, drugs and cells. This initial demonstration version has been optimized with respect to the literature on cancer therapy, but the principles and processes developed are applicable more broadly.
Previous work in automated understanding of the biomedical literature has generally focused either on theoretical and completely general methods or on analytical tasks (e.g., finding keywords to describe a paper or finding the names of genes or proteins) that are substantially more constrained than extracting factual assertions. By addressing a problem more complex than finding descriptive terms in a paper but less difficult than the general problem of understanding natural language, we aimed to build a system of immediate use to laboratory investigators.
Approaches to the extraction of factual assertions from biomedical text vary widely. Methods used include syntactic parsing (e.g. [Proux, et al., 1998]), processing of statistical and frequency information (e.g. [Hishiki, et al., 1998] and [Ohta, et al., 1997]) and rule-based systems (e.g. [Fukuda, et al., 1997]). We draw on all of these lines of attack, using a stochastic part of speech tagger [Cutting, et al., 1992] in support of an underspecified syntactic parser [Aronson, et al., 1994]. The parser provides input to a rule-based system that uses the syntactic information, as well as semantic information from the Unified Medical Language System Metathesaurus [Humphreys, et al., 1998] to extract factual assertions from text.
Previous extraction efforts have been mounted to generate gene names (e.g., [Proux, et al., 1998]), protein names (e.g. [Fukuda, et al., 1998]), keywords describing papers (e.g. [Andrade, et al., 1999] and [Ohta, et al., 1997]) and binding affinities [Rindflesch, et al., 1999]. Our goal in this work is to extract factual assertions, in the form of first order predicate calculus statements, about the relationships between genes and drugs in cancer therapy.
Mining the literature for relationships between genes and drugs in cancer is an increasingly important task. The advent of cDNA microarrays and oligonucleotide chips that can assess tens of thousands of genes simultaneously is providing enormous amounts of information, for example about the roles particular genes play in drug sensitivity, about the effects of drugs on gene expression, and about the effects of genetic mutations on sensitivity and response [Weinstein, et al., 1997; Scherf, et al., 1999]. This information is likely to advance the twin goals of discovering new drugs for cancer treatment and, in a clinical setting, individualizing therapy according to the genomic constitution of a patient’s tumor. However, the amount of potentially relevant information can be overwhelming. There is a pressing need for automated assistance in managing and exploiting information on the relationships among the tens of thousands of genes and (potential) drugs.
Focus on a particular domain of knowledge (such as ours on genes and drugs involved in cancer therapeutics) provides important constraints on the set of concepts that EDGAR’s algorithms must be able to handle. There is enough complexity to the material to make an automated system valuable to practitioners in the field, yet the number of entities and relationships that must be handled is small enough that special purpose programs to take advantage of the semantics of the domain can be constructed manually.
2 Representation
The entities that participate in the factual assertions on which we focus here are genes, cells and drugs. EDGAR parses natural language text and produces predicate calculus assertions over these relationships and entities. We want to capture the main factors that are known to be relevant but, at the same time, to constrain the vocabulary as much as possible to facilitate parsing.
Cancer-related drugs and genes can influence each other in two important ways: (1) gene expression can have an impact on the drug sensitivity of a cell, and (2) drug treatment often results in changes in the cell’s gene expression. The exact nature of these influences (sensitivity or resistance, activation or inhibition) is cell type-specific. For example, in one cell line, a drug may cause upregulation of a gene whereas in another there is either an inhibition or no effect at all. This variability is due to the particular mix of interacting entities and pathways in each cell type.
Figure 1 shows the relationships and entities on which we focus here. Notice that the entity descriptions are compositionally complex, in that a cell may be transfected with genes (i.e. have foreign genes added to its natural complement) in addition to having a particular intrinsic gene expression profile. Similarly, a cell that is resistant to one drug might be treated with another.
Figure 1.
The entities and relationships used by EDGAR.
The connectivity in Figure 1 suggests that information about a drug, gene or cell can be inferred from its relationship to other drugs, genes or cell lines. One important aim in making such inferences is to discover relationships that provide new insights into clinical responses to chemotherapy. Another is to guide the process of drug discovery. Interesting implicit relationships include cross-resistance, synergistic drug effects, antagonistic drug effects and hypothetical mechanisms of drug action. Automatic tools to discover such relationships may become practical when it is possible to generate large factual databases using tools such as EDGAR.
3 Natural Language Processing
3.1 Semantic Interpretation
Our basic approach is to consider the identification of gene, cell and drug names in the text of MEDLINE abstracts and eventually to determine the relationships asserted to obtain among them with respect to the interaction of gene expression and drug sensitivity in particular cell types. For example, the text in (1) gets the semantic interpretation in (2), where the predicate refers to the relation “increased resistance,” which obtains among the gene, cell, and drug arguments.
-
1)
“Compared with parental or mock-transfected HAG-1 cells, v-src-transfected HAG/src3-1 cells showed a 3.5-fold resistance to cis-diamminedichloroplatinum (CDDP).”
-
2)
i_resistant(v-src, HAG/src3-1, CDDP)
In semantic interpretation it is convenient to distinguish between referential and relational vocabularies. The referential vocabulary encodes the arguments in the semantic analysis, whereas the relational vocabulary involves the (more complex) syntactic phenomena associated with the predicate of the underlying semantic proposition. In this paper we concentrate on the referential vocabulary; however, we will comment later on progress being made toward processing the relational vocabulary.
3.2 Knowledge Sources
Interpretation of the referential vocabulary in EDGAR is based on natural language processing tools and knowledge sources being developed at the National Library of Medicine. The primary knowledge source supporting EDGAR is the Unified Medical Language System® (UMLS®) Metathesaurus® [Humphreys, et al., 1998], which is a large (more than 620,000 concepts) compilation of several controlled vocabularies in the biomedical (largely clinical) domain. The most important characteristic of the Metathesaurus for this project is that each constituent concept is associated with a semantic type such as “Pharmacologic Substance,” “Gene or Genome,” or “Cell.” For syntactic information, we use a second knowledge source from the UMLS, the SPECIALIST™ Lexicon [McCray, et al., 1994]. We also use cell line names from the National Cancer Institute’s Drug Discovery Program and lists of gene synonyms compiled from the Weizmann Institute’s GeneCards database.
3.3 Processing
EDGAR begins by assigning an underspecified syntactic parse to each sentence in the abstract under consideration. All subsequent analysis depends on this structure. The natural language processing tools include a stochastic tagger [Cutting, et al., 1992], which resolves part-of-speech ambiguities in support of the underspecified parser [Aronson, et al., 1994]. As shown in the analysis (4) for the sentence in (3), the syntactic structure is underspecified in the sense that, although low-level constituents (notably noun phrases) are identified, they are not attached in a fully-specified parse tree.
-
3)
“This effect of cyclosporin A or herbimycin A on the down-regulation of ERCC-1 correlates with enhanced cytotoxicity of cisplatin in this system.”
-
4)
[this effect]NP [of [cyclosporin A]NP]PrepP [or]CONJ [herbimycin A]NP [on [the down-regulation]NP]PrepP [of [ERCC-1]NP]PrepP [correlates]V [with [enhanced cytotoxicity]NP]PrepP [of [cisplatin]NP]PrepP [in [this system]NP]PrepP
To identify those noun phrases that function as arguments in the predications representing drug and gene interactions, EDGAR relies primarily on the Metathesaurus, with support from the ancillary gene and cell lists. Given the clinical orientation of the UMLS, the Metathesaurus has wide coverage of the drugs that appear in the relevant abstracts. However, since none of the constituent vocabularies of the Metathesaurus has extensive coverage in molecular biology, genes and cells are not as well represented. Furthermore, the ancillary lists are incomplete, particularly for cell lines. Therefore, EDGAR uses contextual information to identify gene and cell names when these do not appear in any of the available knowledge sources.
The general strategy for harvesting contextually-determined gene and cell names depends on the fact that the structure of noun phrases referring to cells and genes in the abstracts in this domain is quite regular. The phrases in (5), all from a single abstract, are typical.
-
5)
human ovarian carcinoma cells, a2780/cp70 human ovarian carcinoma cells, a2780/cp70 cells
Each noun phrase in (5) has cells as its head. Furthermore, if the word that appears immediately to the left of the head is not a normal English word, it is the name of a cell. These generalizations are paradigmatic of the general approach taken to identifying both gene and cell names by context.
A small set of characteristic signal words (such as cell, clone, line, and cultured for cells and activated, expression, gene, and mutated for genes) mark certain noun phrases as referring to either cells or genes. In such phrases, the characteristic words occur in a regular pattern with respect to the names of genes and cells. Words such as cell, line, and gene function as heads of the phrase and the target name is likely to occur immediately to their left. Cultured, activated, and mutated are modifiers that precede the target name. A few signal words, such as expression (and related forms) may serve as the head of a gene noun phrase but may also indicate that their complement (introduced by of) is almost certainly a gene name.
Once gene and cell noun phrases have been identified, the potential target name is scrutinized in order to eliminate carcinoma, for example, as the name of a cell type. If the text token immediately to the left of the word cell does not occur in the SPECIALIST Lexicon and does not have the orthographic characteristics of a normal English word (normal words contain at least one vowel and no digits), then it is likely to be the name of a cell. Similar rules apply to other characteristic signal words and the corresponding gene or cell names.
Although these generalizations have been found useful, they are not always correct. Hyphenated expressions, in particular, produce false positives. For example, upon encountering the noun phrase c-myc-overexpressing cells, EDGAR concludes that c-myc-overexpressing is the name of a cell because this string is not in the SPECIALIST Lexicon. Similarly, apoptosis-related is identified as a gene name on the basis of the noun phrase apoptosis-related gene expression. Because of the many hyphens in gene and cell names, additional work in this area is necessary.
Contextually-identified gene and cell names are harvested in an initial pass through the entire abstract before the identification of all drugs, genes, and cells is attempted. This separate pass is necessary because a gene or cell name may occur only once in a context in which it can easily be identified. For example, in (6), the appearance of c-fos and c-jun as modifiers in the noun phrase whose head is expressions provides strong evidence that these are gene names. This evidence can be used with confidence when the same names appear in another sentence in the same abstract (7) but in a context which less reliably identifies it as a gene name.
-
6)
Cyclosporin A and herbimycin A, which suppress c-fos and c-jun gene expressions, respectively, blocked the cisplatin-induced increase in ERCC-1 mRNA.
-
7)
The products of c-fos and c-jun are components of the transcription factor AP-1 (activator protein 1).
Gene and cell names identified by context constitute an internal knowledge source local to the current abstract. This local source is used to supplement the Metathesaurus and ancillary lists when each sentence is processed to identify arguments in the predications representing drug and gene interactions in cells.
Argument identification proceeds by examining each noun phrase in the underspecified syntactic parse for each sentence and determining whether it matches a Metathesaurus concept, an entry in one of the ancillary lists of genes and cells, or an item in the local, contextually-determined list. For access to UMLS, EDGAR calls on MetaMap [Aronson, et al., 1994], a program that examines the syntactic structure of noun phrases and determines the best match between the input phrase and concepts in the Metathesaurus. A noun phrase that maps to a Metathesaurus concept and that has one of the UMLS semantic types “Pharmacologic Substance,” “Gene or Genome,” or “Cell” is considered accordingly to be a drug, gene or cell. For example, when the sentence in (3) above is submitted to MetaMap, EDGAR determines that the noun phrases in (8) refer to drugs. A search in the ancillary lists finds that (9), another noun phrase in (3), is a gene name.
-
8)
[of cyclosporin A] - Cyclosporine (Pharmacologic Substance) UMLS [herbimycin A] - herbimycin (Pharmacologic Substance) UMLS [of cisplatin] - Cisplatin (Pharmacologic Substance) UMLS
-
9)
[of ERCC-1] - ERCC1 (Gene) Ancillary list
As suggested in the discussion of (6) and (7), during this phase of the processing, contextually-determined items are also used whenever possible to identify arguments as either genes or cells.
EDGAR retrieves cell features other than the name, including organ type, cancer type, organism, and several domain specific features, the most important of which refer to transfection and resistance. EDGAR harvests this information using techniques similar to those described for the contextual identification of gene and cell names: specific signals (notably transfected and resistant) provide guidance, and the Metathesaurus semantic types are consulted for organisms, body parts, and neoplastic processes.
The algorithm for identifying the referential vocabulary that represents the interaction of genes and drugs in cells is recapitulated schematically in Figure 2.
Figure 2.
Processing for each abstract
To further illustrate the processes in EDGAR, we show here the analysis of a MEDLINE abstract (UI 99140404) with the title “V-src induces cisplatin resistance by increasing the repair of cisplatin-DNA interstrand cross-links in human gallbladder adenocarcinoma cells.” All of the gene and cell noun phrases discovered by EDGAR in this abstract are given in (10) and (11), respectively.
-
10)
gene_np([activation, of, src]). gene_np([activated,’h-ras’]).
gene_np([‘v-src’, transfected,’hag1’, human, gallbladder, …adenocarcinoma, cells]).
gene_np([‘v-src’, transfected,’hag/src3-1’, cells]).
gene_np([‘v-src’, transfected, cells]). gene_np([activated, src]).
gene_np([mrna, expression, of, topoisomerase, ii]).
-
11)
cell_np([human, gallbladder, adenocarcinoma, cells]).
cell_np([‘v-src, transfected,’hag-1’, human, gallbladder, …adenocarcinoma, cells]). cell_np([‘v-src, transfected,’hag/src3-1’, cells]).
cell_np([‘hag/src3-1’, cells]). cell_np([cell, lines]).
The drugs, genes and cells identified as arguments are listed in (12), (13), and (14). Note that, because of word-sense ambiguity, mapping to the UMLS occasionally produces errors such as “Link” in (12). There is a drug with this name in the Metathesaurus, and MetaMap erroneously matched the text cross-links from the title to this concept. Also note that appropriate characteristics have been added to the cell predications in (14) (e.g., the “tfw” label to indicate transfection with v-src).
-
12)
drug(’99140404’,’Doxorubicin’). drug(’99140404’,’Etoposide’).
drug(’99140404’,’Fluorouracil’). drug(’99140404’, wortmannin).
drug(’99140404’,’Link’). drug(’99140404’, herbimycin).
drug(’99140404’, radicicol). drug(’99140404’,’Cisplatin’).
drug(’99140404’,’Pharmaceutical Preparations’).
-
13)
gene(‘99140404’,’h-ras’). gene(‘99140404’,’v-src’).
gene(‘99140404’, src).
-
14)
cell(‘99140404’,’HAG-1’,’Gallbladder’,’Adenocarcinoma’, tfw(‘v-…src’),’Human’).
cell(‘99140404’,’HAG/SRC3-1’,’Gallbladder’,’Adenocarcinoma’, …tfw(‘v-src’),’Human’).
4 Predications asserting the interaction of drugs, genes, and cells
Processing the referential vocabulary as described in the previous section prepares EDGAR to address the relational vocabulary and recover predications that assert interaction of the arguments identified. Although processing of the relational vocabulary remains a work in progress, many of the abstracts show a characteristic that will make the process easier to accomplish successfully: That is, relevant sensitivity and resistance interactions are usually described in a single sentence that contains a drug name, a gene name, and a cell name, all of which are susceptible to identification with current EDGAR capabilities. The sentence in (15) illustrates this phenomenon.
-
15)
The overexpression of catalase or Cu,Zn-superoxide dismutase (Cu,Zn-SOD) did not affect the sensitivity of HeLa cells to cis-platinum.
Both catalase and Cu,Zn-superoxide dismutase are complements of overexpression and thus are identifiable as gene names; HeLa as a modifier of cells is a cell name; and cis-platinum occurs in the Metathesaurus. The syntactic indicators of the underlying semantic relations, such as overexpression and did not affect sensitivity, seem reasonably amenable to currently-available natural language processing techniques (although adequate treatment in the general case will not be trivial).
Unfortunately, the relevant relationships are not always expressed with the relatively straightforward structures seen in (15). Three syntactic phenomena, coordination, anaphora and underspecified reference can complicate the task of interpreting sentences that express sensitivity or resistance relations. In (15), for example, the coordinated gene names indicate that this sentence expresses two predications describing the sensitivity of the HeLa cells to cis-platinum. Coordinate structures so complex as to challenge accurate interpretation are seen in (16).
-
16)
“Compared with parental or mock-transfected HAG-1 cells, v-src-transfected HAG/src3-1 cells showed a 3.5-fold resistance to cis-diamminedichloroplatinum (II) (CDDP) but not to doxorubicin, etoposide or 5-fluorouracil.”
Both underspecified reference and anaphora are seen in (17).
-
17)
By contrast, activated H-ras, which acts downstream of src, failed to induce resistance to either of these drugs.
The specific referents of these drugs will have to be recovered from another sentence in the abstract. Finding the sentence with that information is a difficult task that depends on not only finding references to drugs in prior sentences, but also ensuring that h-ras was not asserted as inducing resistance to those drugs in that or intervening sentences. Further, the relevant cell will also have to be inferred from another sentence, with similar caveats. This is a challenging semantic interpretation task, but we are optimistic that with further research it can be handled with acceptable accuracy.
5 Current status and related work
EDGAR is still in development, and its performance has not yet been quantified. One basis for evaluation is a comparison with MedMiner, a keyword-based system developed and used in our laboratory [Tanabe, et al., 1999]. The largest difference is that EDGAR can automatically identify most drug and gene names, whereas MedMiner requires that these names be supplied by the user (or programmer). EDGAR is also designed to be able to generate relational assertions with correct arguments, extracted from syntactically complex sentences, something that cannot be done in the string-matching paradigm MedMiner uses. However, EDGAR’s accuracy is still best characterized as moderate. As noted above, the currently operational version of the system does not analyze the type of relationship existing between the objects identified. Code for this task is currently under development.
Recently, many groups have proposed systems for automated extraction of factual information from the biomedical literature. [Blaschke, et al., 1999] is an attempt to generate functional relationship maps from abstracts. However, it requires a prerequisite list of all named entities and cannot handle syntatically complex sentences. [Craven & Kumlien 1999] uses statistical methods that are unable to resolve even modest syntactical complexity, such as the presence of more than two possible predicate arguments in a sentence. This system also has very low recall accuracy. [Ohta, et al.1997] describes a system that automatically constructs a dictionary of statistically informative terms from a set of abstracts. Although these terms often overlap with the ones generated by EDGAR, in their system there is no mapping between terms and the underlying semantic concepts. Hence, it would not be possible, for example, to use it to identify drugs or genes as such.
6 Application
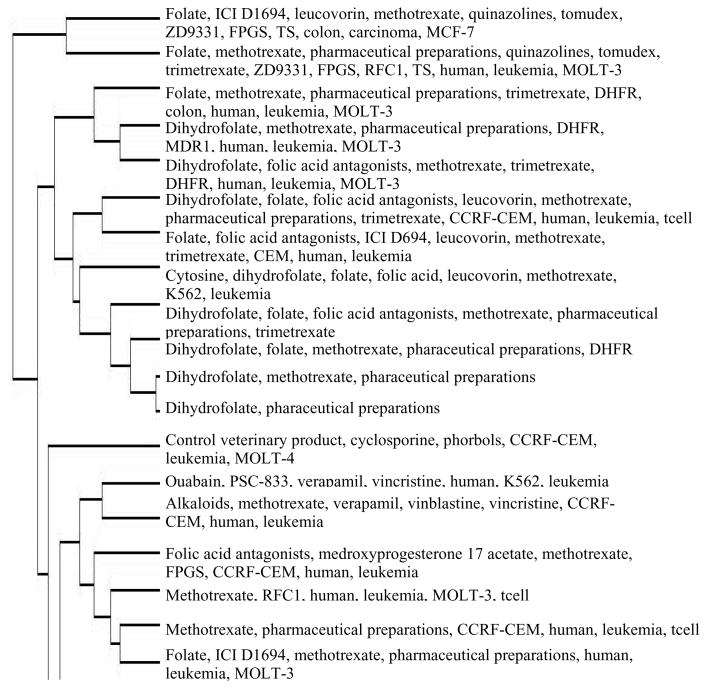
EDGAR’s current capacity to identify well-characterized genes, drugs and cell lines can be immediately useful to biologists. We designed a tool that manages large collections of abstracts using the output of EDGAR as the input for vector space document clustering [Salton, 1989]. The PubMed query “neoplasms AND cells AND gene AND drug AND resistance AND mechanism” generated 383 abstracts related to anti-tumor drug resistance. These abstracts were processed by EDGAR in batch, then perl scripts were applied to the output to remove single character names, merge synonyms and create boolean feature vectors representing the cellular entities. Genes, drugs and cells found in at least two abstracts were included in the document vectors, and Splus statistical software was used to perform hierarchical clustering.
The dendrogram in Figure 3 shows a subtree of the 383 documents clustered by Euclidean distance. The cluster structure has been used by domain experts in our laboratory to help navigate the voluminous relevant literature. Domain experts can make inferences from the clustering alone, without having to read the abstracts. For example, the first two branches can be characterized by the statement: “Resistance to folates in leukemia cells is influenced by the FPGS, TS, DHFR and RFC1 genes.” This interpretation was reached without reading a single abstract, but upon examination of the abstracts, we found it to be supported by the literature. It would have been impossible to reach such a conclusion from the titles alone.
Figure 3.
Subtree of Drug Resistance Clusters
This application also demonstrates the scalability of the EDGAR system. Typical processing time for each abstract was a bit over 8 seconds, much of that overhead associated with http submission. A large scale application of a similar system run locally [Rajan, et al., in preparation] processed 491,237 abstracts in 12.8 cpu days (2.25 seconds each). Although dendrograms in the style of Figure 3 are not practical for very large numbers of abstracts, more sophisticated visualization techniques can handle larger trees, and no matter what size, the trees will generally be locally interpretable.
7 Conclusions
We have presented here a natural language processing system, EDGAR, that extracts mentions of genes, drugs and cell types from Medline abstracts by using existing syntactic NLP tools in combination with new semantic and pragmatic analyses. Development to date has focused on the referential vocabulary, but addition of a fully functional relational vocabulary will provide the strong semantic basis required for capturing biologically significant gene-drug-cell relationships. EDGAR is most immediately applicable now in the context of pharmacologically motivated gene expression profiling, but its range of application will be progressively extended.
Acknowledgments
We are grateful to Alan Aronson for modifications to MetaMap for the Edgar project, to James Mork for the Web interface and to Lawrence Smith for clustering suggestions. L.T. and J.N.W. are supported in part by funding from the Breast Cancer Task Force of the National Cancer Institute Division of Clinical Sciences.
Contributor Information
Thomas C. Rindflesch, Lister Hill Center, National Library of Medicine, Bldg. 38A, MS-54, Bethesda, MD 20894, tcr@lhc.nlm.nih.gov
Lorraine Tanabe, Laboratory of Molecular Pharmacology, National Cancer Institute, 37 Convent Dr. Building 37, Rm 5B12, Bethesda, MD 20892, ltanaben@molstat.nci.nih.gov.
John N. Weinstein, Laboratory of Molecular Pharmacology, National Cancer Institute, 37 Convent Dr. Building 37, Rm 5B12, Bethesda, MD 20892, weinstein@dtpax2.ncifcrf.gov
Lawrence Hunter, Section on Molecular Statistics and Bioinformatics, National Cancer Institute, Federal Building, Rm 3C06, Bethesda, MD 20892, lhunter@nih.gov.
References
- Andrade MA, Valencia A. Automatic extraction of keywords from scientific text protein families. Bioinformatics. 1998;14:600–607. doi: 10.1093/bioinformatics/14.7.600. [DOI] [PubMed] [Google Scholar]
- Aronson AR, Rindflesch TC, Browne AC. Exploiting a large thesaurus for information retrieval. Proceedings of RIAO. 1994;94:197–216. [Google Scholar]
- Baker PG, et al. An ontology for bioinformatics applications. Bioinformatics. 1999;15:510–520. doi: 10.1093/bioinformatics/15.6.510. [DOI] [PubMed] [Google Scholar]
- Blaschke C, et al. Automatic Extraction of biological information from scientific text: protein-protein interactions. ISMB. 1999;7:60–67. [PubMed] [Google Scholar]
- Craven M, Kumlien J. Constructing Biological Knowledge Bases by Extracting Information from Text Sources. ISMB. 1999;7:77–86. [PubMed] [Google Scholar]
- Cutting D, et al. A practical part-of-speech tagger. In Proceedings of the Third Conference on Applied Natural Language Processing. 1992 [Google Scholar]
- Fukuda K, et al. Toward information extraction: protein names from biological papers. Pacific Symposium on Biocomputing (PSB) 1998;3:705–716. [PubMed] [Google Scholar]
- Hishiki T, et al. Developing NLP tools for genome informatics: An information extraction perspective. Ninth Workshop on Genome Informatics. 1998:81–90. [PubMed] [Google Scholar]
- Humphreys BL, Lindberg DAB, Schoolman HM, Barnett GO. The Unified Medical language System: An informatics research collaboration. Journal of the American Medical Informatics Association. 1998;5(1):1–13. doi: 10.1136/jamia.1998.0050001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCray AT, Srinivasan S, Browne AC. Lexical methods for managing variation in biomedical terminologies. Proceedings of the 18th Annual Symposium on Computer Applications in Medical Care. 1994:235–239. [PMC free article] [PubMed] [Google Scholar]
- Ohta YY, et al. Automatic Construction of Knowledge Base from Biological Papers. Intelligent Systems for Molecular Biology. 1997;5:218–225. [PubMed] [Google Scholar]
- Proux D, et al. Detecting gene symbols and names in biological texts. Ninth Workshop on Genome Informatics. 1998:72–80. [PubMed] [Google Scholar]
- Rajan J, Hunter L, Rindflesch TC. Mining Medline. in preparation. [Google Scholar]
- Rindflesch TC, Hunter L, Aronson AR. Mining molecular binding terminology from biomedical text . Proceedings of AMIA ‘99. to appear. [PMC free article] [PubMed] [Google Scholar]
- Salton G. Automatic Text Processing. Addison-Wesley: Reading, MA; 1989. [Google Scholar]
- Scherf U, et al. A cDNA microarray gene expression database for the molecular pharmacology of cancer. Nature Genetics. doi: 10.1038/73439. submitted. [DOI] [PubMed] [Google Scholar]
- Schulze-Kremer S. Ontologies for Molecular Biology. PSB. 1998;3:693–704. [PubMed] [Google Scholar]
- Tanabe L, Scherf U, Smith LH, Lee JK, Hunter L, Weinstein JN. MedMiner: An Internet Tool for Filtering and Organizing Gene Expression and Pharmacological Information. Biotechniques. doi: 10.2144/99276bc03. (submitted) [DOI] [PubMed] [Google Scholar]
- Weinstein JN, et al. An information-intensive approach to the molecular pharmacology of cancer. Science. 1997;275:343–349. doi: 10.1126/science.275.5298.343. [DOI] [PubMed] [Google Scholar]