Summary
Ionotropic glutamate receptors (iGluRs) mediate neuronal communication at synapses throughout vertebrate and invertebrate nervous systems. We have characterized a novel family of iGluR-related genes in Drosophila, which we name Ionotropic Receptors (IRs). These receptors do not belong to the well-described Kainate, AMPA, or NMDA classes of iGluRs, and have divergent ligand-binding domains that lack their characteristic glutamate-interacting residues. IRs are expressed in a combinatorial fashion in sensory neurons that respond to many distinct odors but do not express either insect odorant receptors (ORs) or gustatory receptors (GRs). IR proteins accumulate in sensory dendrites and not at synapses. Mis-expression of IRs induces novel odor responses in ectopic neurons. Together, these results lead us to propose that the IRs comprise a novel family of chemosensory receptors. Conservation of IR/iGluR-related proteins in bacteria, plants, and animals suggests that this receptor family represents an evolutionarily ancient mechanism for sensing both internal and external chemical cues.
Introduction
Species as diverse as bacteria, plants, and humans have the capacity to sense small molecules in the environment. Chemical cues can transmit the presence of food, alarm signals, and messages from conspecifics that signify mating compatibility. Peripheral chemical recognition largely relies on membrane receptor proteins that interact with external ligands and convert this binding into intracellular responses. The vast majority of identified chemosensory receptors in multicellular organisms belong to the seven transmembrane domain G protein-coupled receptor (GPCR) superfamily, including odorant, gustatory and pheromone receptors in mammals, birds, reptiles, amphibians, fish, and nematodes (Bargmann, 2006). Unicellular organisms also use GPCRs for chemoreception, such as the pheromone receptors in budding yeast (Dohlman, 2002).
Insects can detect a wide range of environmental chemicals: bitter, sweet, and salty tastants, odors, pheromones, humidity, carbon dioxide, and carbonated water (reviewed in Dahanukar et al., 2005; Bargmann, 2006; Vosshall and Stocker, 2007; Benton, 2008). Most of these chemosensory stimuli are recognized by members of two evolutionarily related insect-specific chemosensory receptor families, the Odorant Receptors (ORs) and Gustatory Receptors (GRs). These proteins contain seven predicted transmembrane domains but are evolutionarily unrelated to GPCRs (Vosshall et al., 1999; Benton et al., 2006; Wistrand et al., 2006) and adopt a distinct membrane topology (Benton et al., 2006; Lundin et al., 2007). Recent analysis has indicated that insect ORs function as odor-gated ion channels (Sato et al., 2008; Wicher et al., 2008), setting them mechanistically apart from metabotropic vertebrate ORs.
Comprehensive analysis of the expression of these receptors in the fruit fly, Drosophila melanogaster, has hinted at the existence of other types of insect chemosensory receptors (Couto et al., 2005; Yao et al., 2005). This is particularly apparent in the major olfactory organ, the third segment of the antenna, which bears three types of olfactory sensory hairs (sensilla): basiconic, trichoid, and coeloconic. All olfactory sensory neurons (OSNs) innervating basiconic and trichoid sensilla generally express one OR, along with the OR83b co-receptor (reviewed in Vosshall and Stocker, 2007). However, with the exception of OR35a/OR83b–expressing neurons (Yao et al., 2005), OSNs housed in coeloconic sensilla do not express OR83b or members of the OR or GR gene families. Nevertheless, electrophysiological analysis has revealed the existence of multiple types of coeloconic OSNs tuned to acids, ammonia and humidity (Yao et al., 2005), suggesting that other types of insect chemosensory receptors exist (Scott et al., 2001; Couto et al., 2005; Yao et al., 2005).
We previously carried out a bioinformatic screen for insect-specific genes enriched in OSNs (Benton et al., 2007). Among these, we found members of a large expansion of the ionotropic glutamate receptor (iGluR) gene family of unknown biological function (Littleton and Ganetzky, 2000). Here we provide evidence that these variant iGluRs represent a novel class of chemosensory receptor.
Results
A large family of divergent ionotropic glutamate receptor-like genes in Drosophila
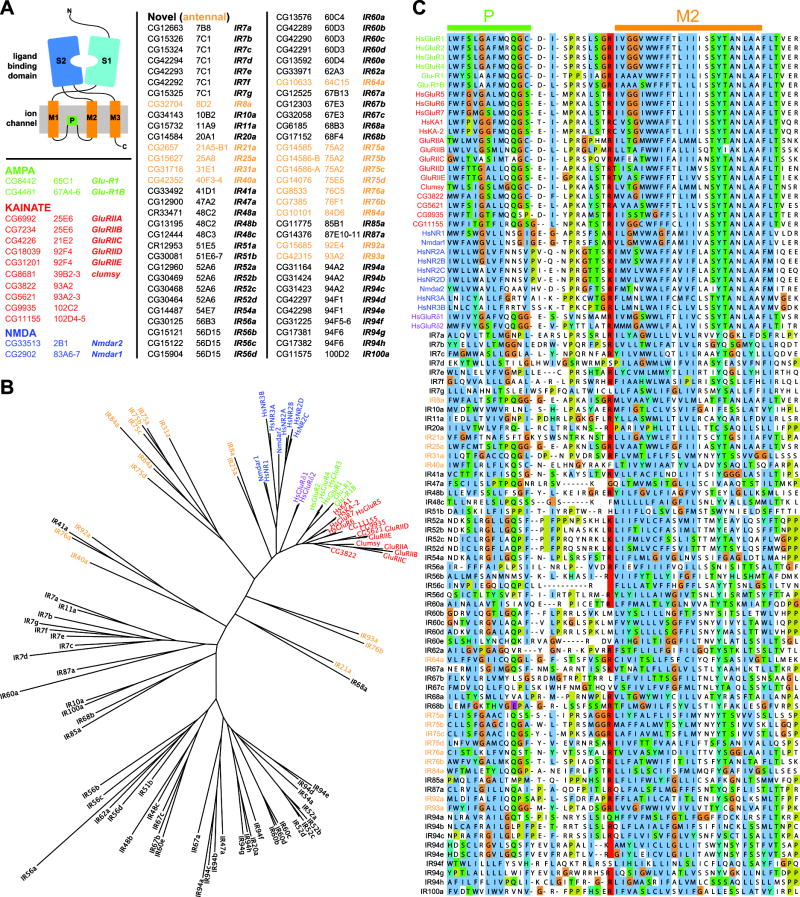
From a bioinformatic screen for novel olfactory molecules (Benton et al., 2007), we identified 6 antennal-expressed genes encoding proteins annotated as ionotropic glutamate receptors (iGluRs) (data not shown; Littleton and Ganetzky, 2000). Using these novel receptor sequences as queries, exhaustive BLAST searches of the Drosophila genome identified a family of 61 predicted genes and 1 pseudogene. These genes are distributed throughout the genome, both as individual sequences and in tandem arrays of up to four genes (Figure 1A, data not shown). We named this family the Ionotropic Receptors (IRs) and assigned individual gene names to the IRs using nomenclature conventions of Drosophila ORs (Drosophila Odorant Receptor Nomenclature Committee, 2000) (Figure 1A).
Figure 1. A novel family of divergent ionotropic glutamate receptors in Drosophila.
(A) Top left: model of iGluR domain organization. Bottom left and right: table of Drosophila iGluRs and IRs, with corresponding cytological location and gene names.
(B) Phylogenetic tree of Drosophila and human iGluRs and Drosophila IRs, color-coded as in (A).
(C) Alignments of the amino acid sequences of Drosophila and human iGluRs and Drosophila IRs of part of the pore loop, P, and M2 transmembrane segment of the ion channel domain.
Phylogenetic analysis of predicted IR protein sequences revealed that they are not closely related to members of the canonical families of iGluRs (AMPA, kainate, NMDA, or delta) (Figure 1B). However, they appear to have a similar modular organization to iGluRs, comprising an extracellular N-terminus, a bipartite ligand-binding domain, whose two lobes (S1 and S2) are separated by an ion channel domain, and a short cytoplasmic C-terminus (Figures 1-2 and data not shown) (Mayer, 2006). We note that the gene structure and protein sequence of most receptors are presently only computational predictions. Nevertheless, the family is extremely divergent, exhibiting overall amino acid sequence identity of 10-70%. The most conserved region between IRs and iGluRs spans the ion channel pore (Figure 1C), suggesting that IRs retain ion-conducting properties.
Figure 2. Ligand binding domains in most IRs lack glutamate-interacting residues.
(A-B) ClustalW amino acid alignments of part of the S1 (A) and S2 (B) ligand binding domains of Drosophila and human iGluRs and Drosophila IRs. The positions of key ligand binding residues in iGluRs are marked with asterisks at the top.
The ligand-binding domains are considerably more variable, although alignment of small regions of the S1 and S2 lobes of IRs and iGluRs allowed examination of conservation in amino acid positions that make direct contact with glutamate or artificial agonists in iGluRs (Armstrong et al., 1998; Armstrong and Gouaux, 2000; Jin et al., 2003; Mayer, 2005) (Figure 2). While all iGluRs have an arginine (R) residue in S1 that binds the glutamate α-carboxyl group, only 19/61 (31%) IRs retain this residue (Figure 2A). In the first half of the S2 domain, 9/61 (15%) of IRs retain a threonine (T), which contacts the glutamate γ-carboxyl group in all AMPA and kainate receptors (Figure 2B). Interestingly, the iGluRs that lack this T residue (NR1, NR3A, delta) have glycine or serine and not glutamate as a preferred ligand (Mayer et al., 2006; Naur et al., 2007). Finally, in the second half of the S2 domain, 100% of the iGluRs have a conserved aspartate (D) or glutamate (E) that interacts with the α-amino group of the glutamate ligand, compared with 10/61 (16%) IRs (Figure 2B). Of 61 IRs, only three (IR8a, IR75a, IR75c) retain the R, D/E, and T residues characteristic of iGluRs, although these residues lie within a divergent structural backbone. Other IRs have a diversity of different amino acids at one or more of these positions. Thus, the ligand-binding specificity of most or all IRs is likely to be both distinct from that of iGluRs and varied within the IR family.
IRs are expressed in chemosensory neurons that do not express ORs or OR83b
We determined the expression of the IR family by both tissue-specific RT-PCR and RNA in situ hybridization. Fifteen IR genes are expressed in the antenna (Figure 1A and Figure 3). Transcripts of these genes were not detected elsewhere in the adult head, body or appendages, except for IR25a and IR76b, which are also expressed in the proboscis (data not shown). Expression of the remaining 46 IR genes was not reproducibly detected in any adult tissue. It is unclear whether these genes are not expressed, expressed at different life stages, or expressed in at levels below the detection threshold of our assays.
Figure 3. A topological map of IR expression in the antenna.
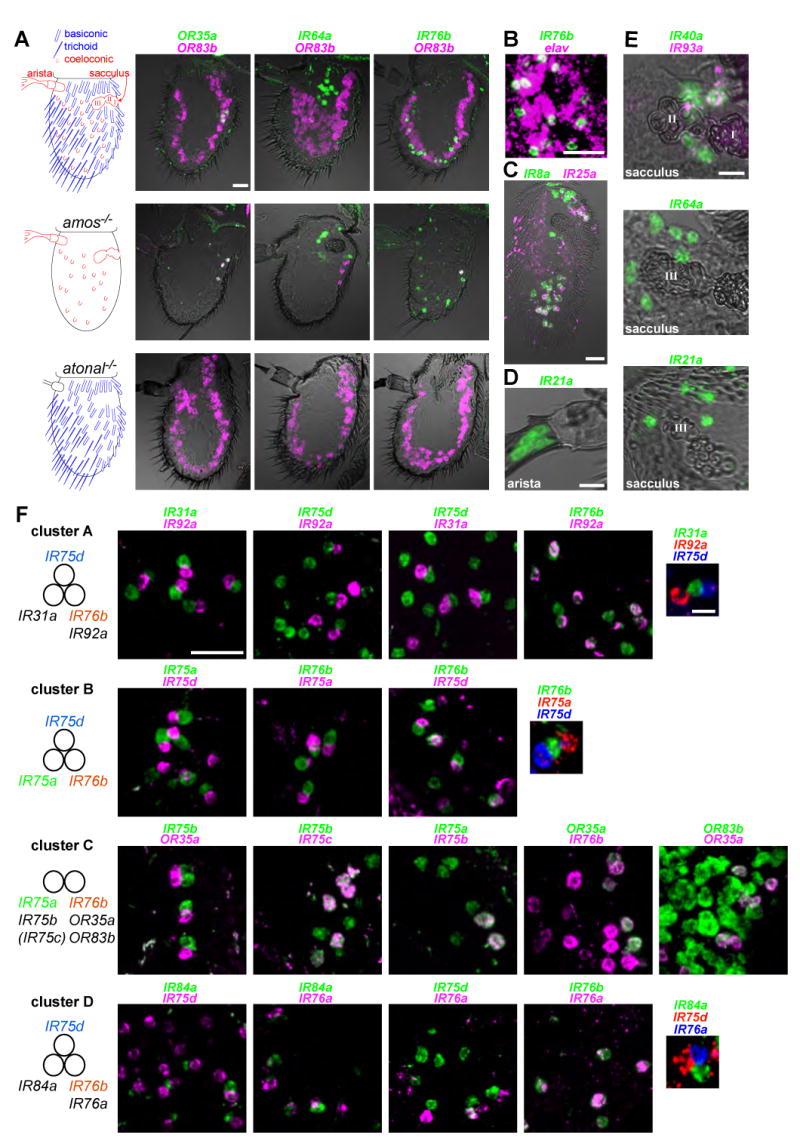
(A) Left: cartoons of the Drosophila antenna, in which sensory structures are color-coded by their developmental specification by the proneural genes amos (blue) or atonal (red). Right: two-color RNA in situ hybridization for OR83b (magenta) and either OR35a (green, left), IR64a (green, middle) or IR76b (green, right) on antennal sections of wildtype (top), amos mutant (amos1/Df(2L)M36F-S6) (middle) and atonal mutant (ato1/Df(3R)p13) (bottom) animals. Scale bar for all panels is 20 μm.
(B) Two-color RNA in situ hybridization for IR76b (green) and the neuronal marker elav (magenta) on an antennal section of a wildtype antenna. Scale bar is 20 μm.
(C) Two-color RNA in situ hybridization on an antennal section for IR8a (green) and IR25a (magenta). Scale bar is 20 μm.
(D) One-color RNA in situ hybridization on antennal sections reveals expression of IR21a in the arista.
(E) One- and two-color RNA in situ hybridization on antennal sections reveals co-expression of IR40a (green) and IR93a (magenta) in neurons surrounding the first and second chamber of the sacculus (top) and expression of IR64a (middle) and IR21a (bottom) in neurons surrounding the third chamber of the sacculus. Scale bars are 10 μm.
(F) Two- and three-color RNA in situ hybridization for the indicated combinations of IR genes expressed in neurons in the main portion of the antenna. Pairwise comparison of physically adjacent neurons allows the definition of four distinct clusters (A-D) of IR-expressing neurons, summarized in the schemes on the left. IR genes expressed in more than one cluster are highlighted in color. The parentheses around IR75c refer to the expression of this gene in only a subset of cluster C (111 IR75c-expressing cells/155 IR75b-expressing cells =71.6%). Scale bar for all two-color panels is 20 μm, and for all three-color panels is 5 μm.
We analyzed where in the antenna IR genes are expressed compared to ORs by double RNA in situ hybridization with probes for the OR co-receptor OR83b and one of several IR genes, including IR64a, IR76b, IR31a, and IR40a (Figure 3A and data not shown). IRs are not expressed in basiconic and trichoid sensilla, as they are not co-expressed with OR83b, and IR expression persists in mutants for the proneural gene absent md neurons and olfactory sensilla (amos), which completely lack these sensilla types (Goulding et al., 2000; zur Lage et al., 2003) (Figure 3A, top and middle panels; data not shown). However, expression of these IRs is dependent upon the proneural gene atonal, which specifies the coeloconic sensilla as well as a feather-like projection called the arista, and a three-chambered pocket called the sacculus (Figure 3A, bottom panel; data not shown) (Gupta and Rodrigues, 1997; Jhaveri et al., 2000). Thus, ORs and IRs are expressed in developmentally distinct sensory lineages in the antenna. One exception is the subpopulation of coeloconic OSNs that expresses both IR76b and OR35a and OR83b (Couto et al., 2005; Yao et al., 2005) (Figure 3A). We confirmed that IR-expressing cells in the antenna are neurons by demonstrating that they co-express the neuronal marker elav (Figure 3B and data not shown).
We next generated a comprehensive map of IR expression (Figure 3C-F). Each IR was observed to have a topologically-defined expression pattern that is conserved across individuals of both sexes (data not shown). IR8a and IR25a, which encode closely related receptors (Figure 1B), are broadly expressed, detected in overlapping populations of neurons around the sacculus and in the main portion of the antenna (Figure 3C; data not shown). IR25a but not IR8a is also detected in the arista (data not shown; see Figure 5G). IR21a is expressed in approximately 6 neurons in the arista (Figure 3D), as well as 5-10 neurons near the third chamber of the sacculus (Figure 3E, bottom panel). Three IRs display specific expression in neurons surrounding the sacculus: IR40a and IR93a are co-expressed in 10-15 neurons adjacent to the first and second sacculus chambers (Figure 3E, top panel), while IR64a is found in 10-15 neurons surrounding the third chamber (Figure 3E, middle panel).
Figure 5. Glomerular convergence of IR axons and ciliary localization of IR proteins.
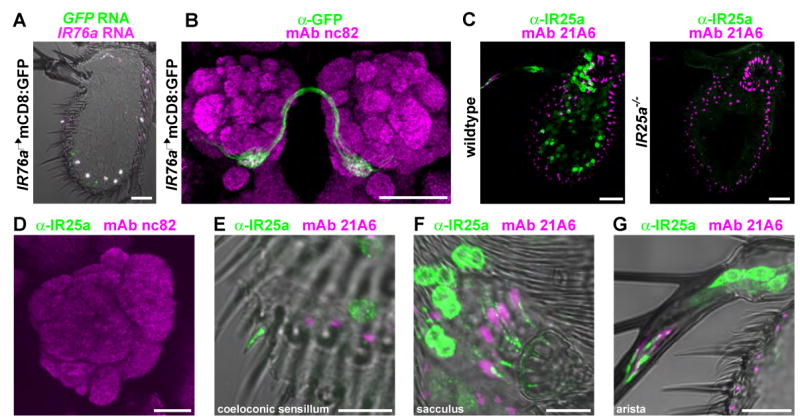
(A) Two-color RNA in situ hybridization for GFP (green) and IR76a (magenta) on an antennal section of an animal expressing the mCD8:GFP reporter under the control of the IR76a promoter-GAL4 driver (IR76a promoter-GAL4/UAS-mCD8:GFP). Scale bar is 20 μm.
(B) Immunostaining of mCD8:GFP-labelled IR76a axon termini (anti-GFP, green) and neuropil (mAb nc82, magenta) on whole-mount brains of IR76a promoter-GAL4/UAS-mCD8:GFP animals. Scale bar is 50 μm.
(C) Immunostaining for IR25a (green) and a cilia base marker (mAb 21A6, magenta) in antennal sections from wildtype (left) and IR25a null mutant (IR25a1/IR25a2) animals. Scale bars are 20 μm.
(D) Immunostaining of IR25a (green) and neuropil (mAb nc82, magenta) in the antennal lobe of a wildtype animal. Scale bar is 20 μm.
(E-G) High-magnification image of IR25a immunostaining (green) in wildtype antennal sections illustrating cilia localization in a coeloconic neuron (E), sacculus neurons (F), and aristal neurons (G). Scale bars are 10 μm.
The remaining 9 IRs are expressed in coeloconic OSNs distributed across the antenna (Figure 3F). Double and triple RNA in situ hybridization revealed that individual neurons express between 1 and 3 different IR genes and are organized into specific clusters of two or three neurons. Four distinct clusters (cluster A-cluster D), containing two (cluster C) or three (cluster A, B, and D) neurons, could be defined by their expression of stereotyped combinations of IR genes (Figure 3F). Cluster C includes a coeloconic neuron that expresses OR35a and OR83b in addition to IR76b. Although each cluster is distinct, there is overlap between the IRs they express. IR76b is expressed in one neuron in all four clusters, IR75d in three clusters and IR75a in two clusters (Figure 3B and 3F). In additional to these selectively-expressed receptors, individual neurons are likely to express one or both of the broadly-expressed IR8a and IR25a (Figure 3C). The combinatorial expression patterns of the IRs raise the possibility that these genes define specific functional properties of these neurons.
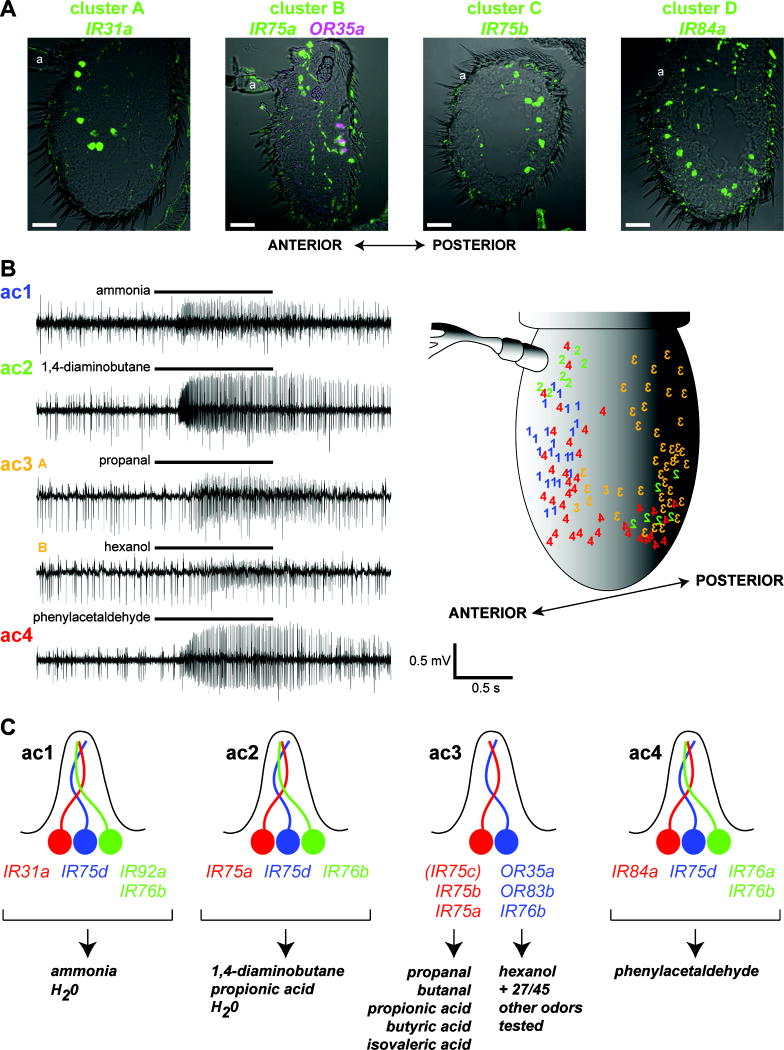
Integration of molecular and functional maps of the coeloconic sensilla
Our definition of four distinct clusters of IR-expressing neurons in the antenna (Figure 3F) is consistent with the identification of four types of coeloconic sensilla, named ac1-ac4, which have distinct yet partially overlapping sensory specificities (Yao et al., 2005). To examine whether IR expression correlates with the chemosensory properties of these OSNs, we compared the spatial organization of IR-expressing neurons using probes for unique IR markers for each cluster type to these functionally distinct sensilla types (Figure 4). As we lack a unique molecular marker for Cluster B, this cluster was defined as those containing IR75a-expressing OSNs (present in Cluster B and Cluster C) that are not paired with OR35a-expressing cluster C neurons (Figure 4A). We found that each cluster has a different, though overlapping, spatial distribution in the antenna (Figure 4A). For example, Cluster A neurons (marked by IR31a) are restricted to a zone at the anterior of the antenna, just below the arista, while cluster C neurons (marked by IR75b) are found exclusively in the posterior of the antenna. These stereotyped IR neuron distributions were observed in antennae from over 20 animals.
Figure 4. Integration of molecular and functional maps in the coeloconic sensilla.
(A) RNA in situ hybridization on antennal sections of wildtype animals for the indicated IR genes representing each of the 4 clusters. Cluster B is represented by IR75a-expressing OSNs (green) that are not paired with OR35a-expressing neurons (magenta). “a” indicates the position of the arista, which projects from the anterior surface of the antenna. Anterior is to the left in all images. Scale bars are 20 μm.
(B) Left: representative traces of extracellular recordings of each coeloconic sensillum class, stimulated with diagnostic odorants (Yao et al., 2005) as indicated. Bars above the traces mark stimulus time (1 s). For ac3, two diagnostic odorants are shown, which specifically stimulate the A (large spike amplitude) or B (small spike amplitude) neuron. Right: Schematic of the topological distribution of ac1-ac4 coeloconic sensillum classes mapped manually after electrophysiological recordings from both the anterior and posterior surfaces of the antenna. Sensilla types are indicated as 1-4, with numbers reversed for sensilla on the posterior face of the antenna.
(C) Summary of the predicted molecular identity of IR-expressing neurons and sensilla classes they innervate. Best ligands (producing a response of >45 spikes/s) for each class are shown as the bottom, from a limited screen of a panel of 45 odors (Yao et al., 2005). All identified ligands of the ac3 OR35a/OR83b/IR76b neuron have been shown to be genetically dependent on OR35a (Yao et al., 2005).
The initial description of the coeloconic sensilla classes did not describe their spatial distribution (Yao et al., 2005). We therefore recorded odor-evoked responses in >100 coeloconic sensilla in several dozen animals across most of the accessible antennal surface, using a panel of odorants that allowed us to identify unambiguously each sensilla type (ammonia for ac1, 1,4-diaminobutane for ac2, propanal and hexanol for ac3, and phenylacetaldehyde for ac4) (Yao et al., 2005) (Figure 4B, left). After electrophysiological identification, we noted the location of the sensilla on the antennal surface (Figure 4B, right).
This mapping process allowed a correlation of the electrophysiological and molecular properties of the coeloconic sensilla (Figure 4C). For example, ac1 sensilla were only detected in a region on the anterior antennal surface just ventral to the arista, and therefore are most likely correspond to cluster A, containing IR31a-IR75d-IR76b/IR92a-expressing neurons. Our data fit well with the previous assignment of the OR35a-expressing neuron to the ac3 sensillum (Yao et al., 2005), which is found on the posterior of the antenna and is the only coeloconic sensillum class that unambiguously houses two neurons (Yao et al., 2005) (Figure 4B and data not shown). While these results allow initial assignment of IRs to different coeloconic sensilla classes, we note that assignment of specific odor responses to individual IR-expressing OSNs is not possible from these data alone.
Glomerular convergence of IR OSNs in the antennal lobe
All neurons expressing a given OR extend axons that converge upon a single antennal lobe glomerulus (Couto et al., 2005; Fishilevich and Vosshall, 2005), resulting in the representation of a cognate odor ligand as a spatially-defined pattern of neural activity within the brain. To ask whether IR-expressing neurons have the same wiring logic, we investigated the targeting of OSNs expressing IR76a by constructing an IR76a-promoter GAL4 driver that recapitulates the endogenous expression pattern (Brand and Perrimon, 1993) (Figure 5A). Labeling of these neurons with mCD8:GFP revealed convergence of their axons on to a single glomerulus, ventral medial 4 (VM4), in the antennal lobe (Figure 5B). This glomerulus is one of approximately eight that was previously unaccounted for by maps of axonal projections of OR-expressing OSNs (Couto et al., 2005; Vosshall and Stocker, 2007).
IR proteins localize to sensory cilia
To determine where IRs localize in sensory neurons, we generated antibodies against IR25a. We detected broad expression of IR25a protein in sensory neurons of the arista, sacculus, and coeloconic sensilla (Figure 5C, left). All anti-IR25a immunoreactivity was abolished in an IR25a null mutant (Figure 5C, right). Low levels of IR25a could be detected in the axon segment adjacent to the cell body in some neurons but no staining was observed along the axons as they entered the brain, or at synapses within antennal lobe glomeruli (Figure 5D). In coeloconic neurons, prominent anti-IR25a staining was detected both in the cell body and in the distal tip of the dendrite, which corresponds to the ciliated outer dendritic segment innervating the sensory hair (Figure 5E). Relatively low levels were detected in the inner dendrites, suggesting the existence of a transport mechanism to concentrate receptor protein in cilia. A similar subcellular localization was observed in sacculus and aristal sensory neurons (Figure 5F-G). The specific targeting of an IR to sensory cilia suggests a role for these proteins in sensory detection.
Inducing novel olfactory sensitivity by ectopic expression of IRs
To test the hypothesis that IR genes encode chemosensory receptors, we investigated whether ectopic IR expression could induce novel olfactory specificities. Three IRs expressed in ac4 sensilla (IR84a, IR76a and IR75d) were individually mis-expressed in ac3 sensilla using the OR35a-GAL4 driver (Fishilevich and Vosshall, 2005). We used single sensillum recordings to examine which, if any, of these three IRs, could confer sensitivity to phenylacetaldehyde, the only known robust ligand for ac4 but not ac3 sensilla (Yao et al., 2005). Mis-expression of IR84a conferred a strong response to phenylacetaldehyde (Figure 6A) that was not observed in control strains or in animals mis-expressing either IR76a or IR75d (Figure 6B). Ectopically-expressed IR84a did not confer sensitivity to the structurally related odor, phenylacetonitrile, which does not activate either ac3 or ac4 neurons (Yao et al., 2005). This indicates that mis-expressed IR84a does not simply generate non-specific ligand sensitivity in these neurons.
Figure 6. IR84a mis-expression confers novel olfactory sensitivity to phenylacetaldehyde.
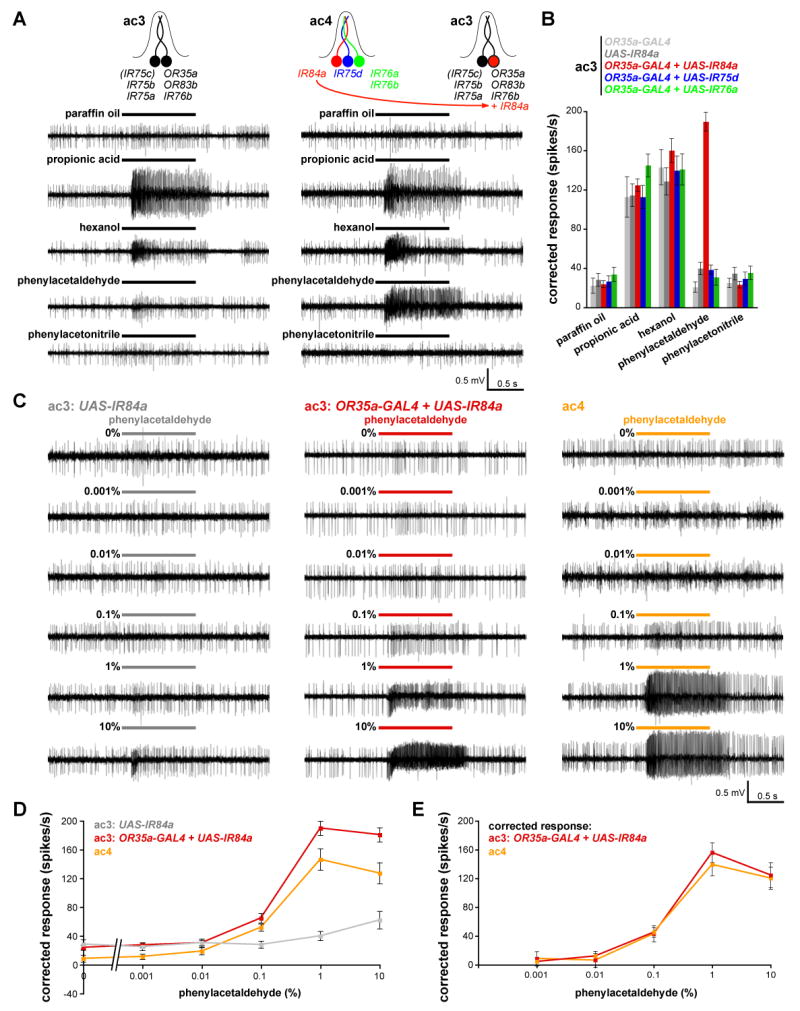
(A) Representative traces of extracellular recordings of neuronal responses to the indicated stimuli in control ac3 sensilla (UAS-IR84a/+) (left) and in ac3 sensilla in which IR84a is mis-expressed in OR35a neurons (UAS-IR84a/+;OR35a-GAL4/+) (right). Bars above the traces mark stimulus time (1 s). For these experiments, all odors were used at 1% concentration except for hexanol, which was diluted to 0.001%.
(B) Quantification of mean odor responses (± s.e.m; n=8-16, male flies) of the five indicated genotypes (OR35a-GAL4/+ (light grey); UAS-IR84a/+ (dark grey); UAS-IR84a/+;OR35a-GAL4/+ (red); UAS-IR76a/+;OR35a-GAL4/+) (green); UAS-IR75d/+;OR35a-GAL4/+) (blue) to the indicated stimuli [concentrations as described in (A)]). Responses of ac3 sensilla neurons to paraffin oil, propionic acid, hexanol and phenylacetonitrile are not significantly different between genotypes (ANOVA; p>0.1657) whereas responses to phenylacetaldehyde are significantly different in sensilla ectopically expressing IR84a compared to the other genotypes (ANOVA with post-hoc t-tests; p< 0.0001).
(C) Representative traces of extracellular recordings of neuronal responses to the indicated dilution of phenylacetaldehyde in control ac3 sensilla (UAS-IR84a/+) (left), in ac3 sensilla in animals in which IR84a is mis-expressed in OR35a neurons (UAS-IR84a/+;OR35a-GAL4/+) (middle) and in ac4 sensilla from UAS-IR84a/+;OR35a-GAL4/+ animals (right). ac3 sensilla were unambiguously identified by their responses to propionic acid and hexanol [see (A-B)]. Bars above the traces mark stimulus time (1 s).
(D) Quantification of mean responses to the phenylacetaldehyde stimuli indicated for the three genotypes shown in (C) (± s.e.m; n=10-16, male flies). At 0.1% phenylacetaldehyde, the ac3 + IR84a and ac4 responses are both significantly higher than the control ac3 sensilla (ANOVA with post-hoc t-tests; p< 0.0008); at 1% and 10%, the ac3+ IR84a sensilla responses are significantly higher than ac4 (ANOVA with post-hoc t-tests; p< 0.0001).
(E) Plot of dose-responses curves of data shown in (D), in which the ac4 sensilla phenylacetaldehyde responses have been corrected for the paraffin oil response, and the phenylacetaldehyde responses of ac3 + IR84a sensilla have been corrected for both paraffin oil responses and endogenous weak ac3 phenylacetaldehyde responses [grey values in (D)]. The curves are not significantly different at any stimulus concentration (ANOVA; p=0.6629).
We next compared the novel odor responses conferred by IR84a mis-expression to the endogenous phenylacetaldehyde responses of ac4 sensilla by generating dose-response curves (Figure 6C-D). Stimulus evoked spike frequencies of ac3 sensilla ectopically expressing IR84a are quantitatively very similar to those in ac4 sensilla, even exceeding the endogenous ac4 responses at higher odor concentrations (Figure 6D). These elevated responses are likely to be due to the contribution of weak endogenous phenylacetaldehyde responses that we observed in ac3 sensilla at high stimulus concentrations (Figure 6C-D), as subtraction of these values produces an IR84a-dependent phenylacetaldehyde dose-response curve that is statistically the same as that of ac4 sensilla (Figure 6E). Thus, ectopic expression of a single IR in ac3 is sufficient to confer a novel ligand- and receptor-specific odor sensitivity that is physiologically indistinguishable from endogenous responses.
To extend this analysis to a second IR, we examined whether mis-expression of one of the IR genes uniquely expressed in ammonia-sensitive ac1 neurons (IR31a and IR92a) was sufficient to confer ectopic responsiveness to this odor. Because ac3 sensilla neurons display endogenous ammonia-evoked responses at modest stimulus concentrations, we used in these experiments the IR76a-promoter GAL4 transgene to mis-express these receptors in ammonia-insensitive ac4 sensilla (Figure 7A) (Yao et al., 2005). ac4 sensilla mis-expressing IR92a, but not IR31a, displayed responses to ammonia (Figure 7A-B). 1,4-diaminobutane, a control stimulus that does not activate either ac1 or ac4 neurons (Yao et al., 2005), did not stimulate ac4 sensilla mis-expressing IR92a. We note that the magnitude of the ectopic IR92a ammonia response is lower than native ammonia-evoked responses of ac1 sensilla (Yao et al., 2005). This may be due to the lack of co-factors present in ac1 sensilla but not in ac4 sensilla. Nevertheless, these results suggest that IR92a comprises at least part of an ammonia-specific chemosensory receptor.
Figure 7. IR92a mis-expression confers novel olfactory sensitivity to ammonia.
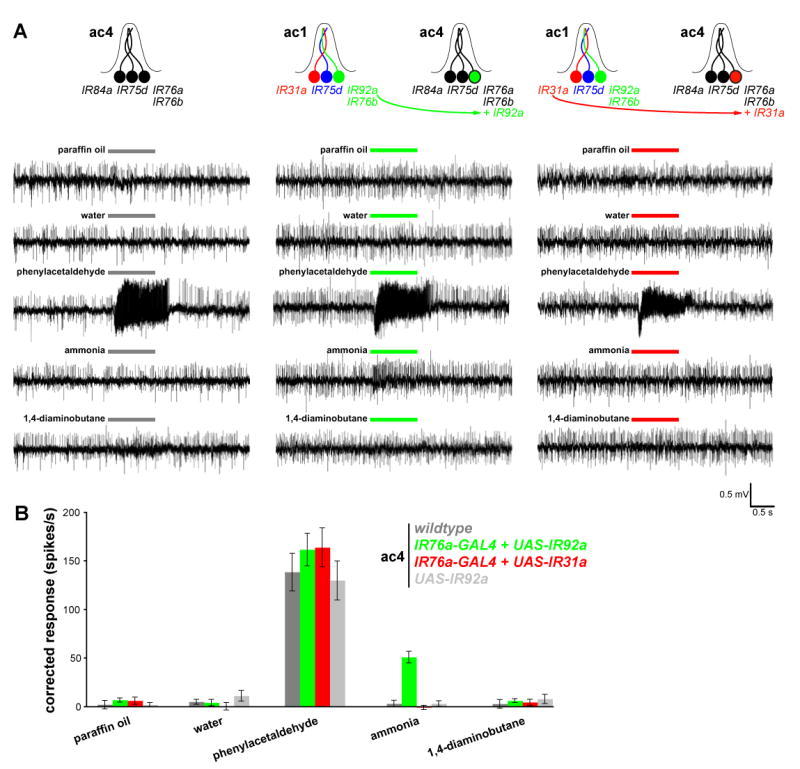
(A) Representative traces of extracellular recordings of neuronal responses to the indicated stimuli in wildtype ac4 sensilla (left), and in ac4 sensilla in animals in which IR92a from ac1 neurons is mis-expressed in IR76a-expressing ac4 neurons (IR76a-GAL4/UAS-IR92a) (right). Bars above the traces mark stimulus time (1 s).
(B) Quantification of mean responses to the stimuli indicated for the four genotypes (± s.e.m; n=9-10, male flies). Responses of ac4 sensilla to paraffin oil, water, phenylacetaldehyde (1%) and 1,4-diaminobutane (10%) are not significantly different between genotypes (ANOVA; p> 0.3044) whereas responses to ammonia (10%) are significantly different in flies ectopically expressing IR92a (ANOVA with post-hoc t-tests; p< 0.0001).
Discussion
We present a characterization of the IRs, a novel family of iGluR-related proteins in Drosophila. The ligand-binding domains of IRs are divergent and most lack known glutamate-binding residues, suggesting that they recognize distinct ligands. IRs are expressed in chemosensory neurons that do not express known insect chemosensory receptors, the ORs and GRs. Spatial patterns of IR gene expression correlate with known ligand sensitivities of these neurons. IR proteins concentrate in the sensory cilia where chemical detection takes place. Mis-expression analysis provides genetic evidence that the IRs function as chemosensory receptors. These receptors may define a molecular basis for Drosophila chemosensory modalities not linked to known receptor genes. Moreover, the IRs may reveal important evolutionary links in chemical detection mechanisms in diverse types of cells across prokaryotes and eukaryotes.
Molecular biology of IR function
The specific combinatorial expression patterns of IRs in sensory neurons and the diversity in their ligand-binding domains is difficult to rationalize with a general role in signal transduction, independent of ligand recognition. More importantly, the novel olfactory sensitivity induced by ectopic expression of IR84a and IR92a provides evidence that IR proteins function directly as ligand-specific, chemosensory receptors. While these experiments demonstrate a sufficiency of IRs for conferring odor-responsiveness, definitive proof of their necessity will require analysis of loss-of-function mutations.
In animal nervous systems, iGluRs mediate neuronal communication by forming glutamate-gated ion channels (Mayer, 2006), and we speculate that IRs also form ion channels, gated by odors and other chemosensory stimuli. A growing number of ionotropic mechanisms in chemoreception are known. For example, members of the transient receptor potential (TRP) family of ion channels are the primary receptors for nociceptive compounds including capsaicin and menthol (Jordt et al., 2003; Bandell et al., 2007) and have also been implicated in gustatory detection of acids (Huang et al., 2006; Ishimaru et al., 2006). Insect ORs also display functional properties of ion channels (Sato et al., 2008; Wicher et al., 2008). Proof that IRs function as ion channels will necessitate electrophysiological characterization of these receptors in heterologous expression systems, and evidence for direct binding of chemosensory ligands to IRs will require biochemical assays in vitro.
iGluRs normally function as heterotetrameric assemblies of variable subunit composition that exhibit differing functional properties such as ligand sensitivity and ion permeability. Our analysis indicates that up to five different IRs may be co-expressed in a single sensory neuron, raising the possibility that these receptors also form multimeric protein assemblies with subunit-dependent characteristics. Of particular interest are the two broadly-expressed members of the family, IR8a and IR25a, which may represent common subunits in many different types of IR complexes. Their function is unclear, but it is possible that they have a co-receptor function with other IRs, analogous to that of OR83b (Larsson et al., 2004; Nakagawa et al., 2005; Benton et al., 2006; Sato et al., 2008). Preliminary analysis of IR25a mutants revealed no obvious defects in odor-evoked responses in coeloconic sensilla (data not shown), but this may be due to redundancy of IR25a with IR8a or the existence of homomeric IR receptors without IR8a or IR25a. Other IRs, such as IR75a and IR76b, are expressed in two or more types of coeloconic sensory neurons. In these cases, the response properties may be defined by the combination of IRs expressed in these distinct neuronal populations. However, the present lack of knowledge of relevant ligands for several coeloconic OSNs makes it difficult to match specific ligands to individual IR neurons based on our expression map alone.
Comparative chemosensation by IRs and ORs
The IR repertoire is remarkably similar in size, overall genomic organization and sequence divergence to Drosophila ORs. Like the ORs, individual IRs are specifically expressed in small subpopulations of chemosensory neurons, and this expression is regulated by relatively short (< 1-2 kb) upstream regulatory regions (Couto et al., 2005; Fishilevich and Vosshall, 2005; Ray et al., 2007) (Figure 5A-B and unpublished data). Furthermore, at least one population of IR-expressing neurons converges on to a single glomerulus in the antennal lobe, similar to the wiring logic established for OR-expressing neurons both in invertebrate and vertebrate olfactory systems (Mombaerts et al., 1996; Gao et al., 2000; Vosshall et al., 2000; Couto et al., 2005; Fishilevich and Vosshall, 2005). Some differences are observed, however, in the organizational logic of IR and OR expression. Most OR-expressing neurons express a single OR gene, along with OR83b, in distinct clusters that innervate specific olfactory hairs. In contrast, many IR-expressing neurons identified in the antenna express 2 or 3 IR genes, in addition to one or both of the broadly-expressed IR8a and IR25a genes. Moreover, overlap is observed both between the molecular composition of different IR neurons and the combination of neurons that innervate a given sensillum. For example, IR76b is co-expressed with at least two other different IR genes in at least two different sensilla - with IR92a in ac1 and with IR76a in ac4 - as well as being co-expressed with OR35a and OR83b in ac3. While the precise biological logic of IR co-expression awaits the matching of specific chemosensory ligands to IR-expressing neurons, combinatorial expression of IRs may contribute more significantly to their role in sensory detection than for ORs.
Why does Drosophila possess two types of antennal chemosensory receptors? Although both may be ionotropic, IRs and ORs are not simply slight evolutionary variants. The receptor families are molecularly unrelated, are under the control of distinct developmental programs, and housed within sensory structures of radically different morphology. Thus, it seems likely that these chemosensory receptors fulfill distinct functions in chemosensation. Analysis of the chemosensory behaviors mediated by IR sensory circuits— now possible with our identification of specific molecular markers for these pathways— may provide insights into the contributions of these different olfactory subsystems. IRs may also have functions in other chemosensory modalities, as two antennal IRs are also detected in the proboscis, and the expression of 46 members of the repertoire remains unknown.
An evolutionarily ancient chemical sensing mechanism
Chemosensation is an ancient sensory modality that predates the evolution of the eukaryotes. Are there traces of conservation in the molecular mechanism by which prokaryotes and eukaryotes sense external chemicals? iGluRs have long been recognized to have prokaryotic origins. Their ion channel domain is homologous to bacterial potassium channels, and the ligand binding domain is structurally related to bacterial periplasmic binding proteins (PBPs), extracellular proteins that scavenge or sense amino acids, carbohydrates and metal ions by coupling to transporters or chemotaxis receptors (Mayer and Armstrong, 2004; Mayer, 2006). Evolutionary connections between iGluRs and PBP function have not often been considered, perhaps in part due to their very weak primary sequence similarity, the widespread occurrence of the PBP fold –also present, for example, in bacterial transcription regulators - and the dedicated role for iGluRs in mediating or regulating synaptic transmission, a process seemingly distant from bacterial solute uptake and chemotaxis.
Our discovery of a family of divergent iGluR-like proteins that may act as peripheral chemosensors provides a link between the disparate functions of these protein modules. While a role for IRs in detecting diverse external ligands is analogous to the function of bacterial PBPs, the primary sequence and neuronal expression of IRs is clearly closer to the properties of iGluRs. Intriguingly, a large family of iGluR-related proteins, the GLRs has also been identified in the plant Arabidopsis thaliana (Lam et al., 1998; Chiu et al., 1999). Almost nothing is known about their physiological functions, but bioinformatic analysis of GLRs suggests that glutamate is unlikely to be their natural ligand (Dubos et al., 2003; Qi et al., 2006). It is possible that GLRs may have roles as chemosensors, for example in detection of soil nutrients or airborne volatiles. Thus, while iGluRs have been intensely studied for their roles in synaptic communication, our characterization of the IRs leads us to suggest that the ancestral function of this protein family may have been in detecting diverse chemical ligands to mediate both intercellular communication and environmental chemical sensing.
Experimental procedures
Bioinformatics
Gene identification
IR genes were identified by exhaustive searches of the Drosophila genome using TBLASTN and PSI-BLAST (Altschul et al., 1997). We used the iGluR-related genes that emerged from our comparative genomics screen (Benton et al., 2007) as initial queries to identify related genes, which were then used as queries in further searches in an iterative process until no new sequences were identified. Membrane topology and domain structure predictions of IR protein sequences were examined using SignalP 3.0 (Bendtsen et al., 2004), TMHMM 2.0 (Krogh et al., 2001) and SMART (Schultz et al., 1998). CG14586 encodes two tandem ion channel domains, which we considered to be a likely computational misprediction and reannotated this as two separate genes each encoding a single ion channel domain (noted in Figure 1A as CG14586-A, CG14586-B). Experimental support for the existence of two distinct transcription units is provided by our observation that RNA in situ hybridization using a probe for IR75c (CG14586-A) highlights only a subset of cells labeled by a probe for IR75b (CG14586-B) (Figure 3F). For most IRs, verification of the computationally-predicted protein sequences awaits isolation of full-length cDNAs. Thus, the phylogenetic relationships illustrated in Figure 1B and sequence alignments in Figure 1C and Figure 2 should not be considered definitive.
Gene nomenclature
Identified genes were named by applying the same conventions adopted for the OR and GR gene families (Drosophila Odorant Receptor Nomenclature Committee, 2000). In brief, genes are named according to their location within one of the 102 primary numbered cytogenetic units of the Drosophila genome. Where there is only one candidate receptor within a given numbered region, it is appended with a lower-case “a”; where there are multiple receptors within a region, these are supplied with lower-case letters as a unique identifier according to their relative position on the cytogenetic map.
Sequence alignments and phylogenetic analysis
Alignments of specific domains of iGluRs/IRs were generated using ClustalW, with manual adjustments, and were visualized in Jalview (Clamp et al., 2004). The phylogenetic tree was generated by aligning full length sequences using PROBCONS (Do et al., 2005) and tree inference using RAxML (Stamatakis, 2006). The tree was viewed with FigTree v1.1.2.
Molecular biology
For RT-PCR analysis of IR expression, cDNA was synthesized from Drosophila tissues using the Absolutely RNA Microprep Kit (Stratagene) and Superscript First-Strand Synthesis System (Invitrogen). Gene-specific primers for PCR were designed using Primer3 (http://frodo.wi.mit.edu/cgi-bin/primer3/primer3_www.cgi) to amplify ∼500 bp spanning, where possible, at least one intron. Primer pairs were verified for their ability to amplify products from genomic DNA using Taq DNA polymerase (Invitrogen). DNA templates for antisense probe synthesis were generated by T:A cloning cDNA or genomic PCR products for each IR gene into pGEM-T Easy (Promega). Antisense Digoxygenin (DIG)- Fluorescein- or Biotin-labeled RNA probes were synthesized using standard methods. Details of transgene construction are provided in the Supplementary Material.
Drosophila stocks
Drosophila stocks were maintained on conventional cornmeal-agar-molasses medium under a 12 hour light:12 hour dark cycle at 25° C. Oregon-R strain was used as wildtype control. Previously described mutant alleles and transgenic lines used are listed in the Supplementary Material. Transgenic lines were generated in this study by Genetic Services Inc. (Cambridge, MA) using either P-element mediated transformation or, for UAS-IR84a, the phiC31-based integration system (Bischof et al., 2007). Gene targeting of IR25a was performed essentially as described (Larsson et al., 2004), using nine independent insertions of the targeting construct. From approximately 150,000 F2 progeny, two null mutants were obtained, which were confirmed by PCR on genomic DNA preparations from homozygous mutant animals.
Histology and immunocytochemistry
In situ RNA hybridization was performed essentially as described (Fishilevich and Vosshall, 2005) using combinations of Fluorescein-, DIG- and Biotin-labelled RNA probes, which were visualized with the TSA Plus Fluorescein, Cy5 or Cy3 Fluorescence Systems (Perkin Elmer), respectively. Immunofluorescence on antennal sections or whole-mount brains was performed as described (Benton et al., 2006). Previously described antibodies are listed in the Supplementary Material. Rabbit polyclonal antibodies against IR25a were raised against the synthetic peptides SKAALRPRFNQYPATFKPRF and DVAEANAERSNAADHPGKLVDGV affinity-purified by Proteintech Group, Inc (Chicago), and used at 1:1000.
Electrophysiology and odorants
Extracellular recordings in single sensilla of 2-8 day old flies were performed as described (Jones et al., 2007). Odorants were obtained from Sigma-Aldrich at high purity. Chemical Abstracts Service (CAS) numbers: aqueous ammonia (ammonium hydroxide) (1336-21-6), 1,4-diaminobutane (110-60-1), propanal (71-23-8), hexanol (111-27-3), propionic acid (79-09-4), phenylacetaldehyde (122-78-1), phenylacetonitrile (140-29-4). Odors were used at 1%(v/v in solvent), unless indicated otherwise in the Figures, diluted either in paraffin oil (propanal, hexanol, phenylacetaldehyde, phenylacetonitrile) or water (ammonium hydroxide, propionic acid, 1,4-diaminobutane). The onset of odor responses varied slightly (usually <200 ms) between animals of the same genotype recorded on different days, most likely due to small variations in the position of the odor delivery apparatus relative to the preparation. For quantification of responses, we determined the time of onset of the response of a control wildtype sensillum to either propionic acid (for ac3) or phenylacetaldehyde (for ac4) for each recording session. Corrected responses for all recordings in the same session were quantified by counting spikes in a 0.5 s window from this time point, subtracting the number of spontaneous spikes in a 0.5 s window prior to stimulation, and doubling the result to obtain spikes/s. We summed spikes from all neurons in a given sensillum, due to difficulties in reliable spike sorting in coeloconic sensilla (Yao et al., 2005). After verifying that responses were normally distributed, we compared all genotypes for a given stimulus by ANOVA, with genotype as the main effect, followed by post-hoc t-tests.
Supplementary Material
Acknowledgments
We thank Andrew Jarman, Hugo Bellen, the Bloomington Drosophila Stock Center, and the Developmental Studies Hybridoma Bank for Drosophila strains and antibodies. Richard Axel, Kevin Lee, Sophie Martin, Charles Zuker, and members of the Vosshall Lab provided invaluable advice, discussion, and comments on the manuscript. R.B. was supported by an EMBO Long-Term Fellowship and was the Maclyn McCarty Fellow of the Helen Hay Whitney Foundation. This work was funded in part by a grant to R. Axel and L.B.V. from the Foundation for the National Institutes of Health through the Grand Challenges in Global Health Initiative and by grants to L.B.V. from the NIH (RO1 DC008600) and the McKnight Endowment Fund for Neuroscience.
Footnotes
The authors declare no conflict of interest.
Author contributions: R.B. designed and carried out all the experiments in the paper with the exception of the in situ hybridization in Figure 3C-F, which was carried out by K.S.V. C.G.-D. performed the gene-targeting screen for IR25a and the initial characterization of the IR25a antibody. L.B.V. and R.B. together conceived and directed the project, interpreted the results, and wrote the paper.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armstrong N, Gouaux E. Mechanisms for activation and antagonism of an AMPA-sensitive glutamate receptor: crystal structures of the GluR2 ligand binding core. Neuron. 2000;28:165–181. doi: 10.1016/s0896-6273(00)00094-5. [DOI] [PubMed] [Google Scholar]
- Armstrong N, Sun Y, Chen GQ, Gouaux E. Structure of a glutamate-receptor ligand-binding core in complex with kainate. Nature. 1998;395:913–917. doi: 10.1038/27692. [DOI] [PubMed] [Google Scholar]
- Bandell M, Macpherson LJ, Patapoutian A. From chills to chilis: mechanisms for thermosensation and chemesthesis via thermoTRPs. Curr Opin Neurobiol. 2007;17:490–497. doi: 10.1016/j.conb.2007.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bargmann CI. Comparative chemosensation from receptors to ecology. Nature. 2006;444:295–301. doi: 10.1038/nature05402. [DOI] [PubMed] [Google Scholar]
- Bendtsen JD, Nielsen H, von Heijne G, Brunak S. Improved prediction of signal peptides: SignalP 3.0. J Mol Biol. 2004;340:783–795. doi: 10.1016/j.jmb.2004.05.028. [DOI] [PubMed] [Google Scholar]
- Benton R. Chemical sensing in Drosophila. Curr Opin Neurobiol. 2008;18:357–363. doi: 10.1016/j.conb.2008.08.012. [DOI] [PubMed] [Google Scholar]
- Benton R, Sachse S, Michnick SW, Vosshall LB. Atypical membrane topology and heteromeric function of Drosophila odorant receptors in vivo. PLoS Biol. 2006;4:e20. doi: 10.1371/journal.pbio.0040020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benton R, Vannice KS, Vosshall LB. An essential role for a CD36-related receptor in pheromone detection in Drosophila. Nature. 2007;450:289–293. doi: 10.1038/nature06328. [DOI] [PubMed] [Google Scholar]
- Bischof J, Maeda RK, Hediger M, Karch F, Basler K. An optimized transgenesis system for Drosophila using germ-line-specific phiC31 integrases. Proc Natl Acad Sci U S A. 2007;104:3312–3317. doi: 10.1073/pnas.0611511104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brand AH, Perrimon N. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development. 1993;118:401–415. doi: 10.1242/dev.118.2.401. [DOI] [PubMed] [Google Scholar]
- Chiu J, DeSalle R, Lam H, Meisel L, Coruzzi G. Molecular evolution of glutamate receptors: a primitive signaling mechanism that existed before plants and animals diverged. Mol Biol Evol. 1999;16:826–838. doi: 10.1093/oxfordjournals.molbev.a026167. [DOI] [PubMed] [Google Scholar]
- Clamp M, Cuff J, Searle SM, Barton GJ. The Jalview Java alignment editor. Bioinformatics. 2004;20:426–427. doi: 10.1093/bioinformatics/btg430. [DOI] [PubMed] [Google Scholar]
- Couto A, Alenius M, Dickson BJ. Molecular, anatomical, and functional organization of the Drosophila olfactory system. Curr Biol. 2005;15:1535–1547. doi: 10.1016/j.cub.2005.07.034. [DOI] [PubMed] [Google Scholar]
- Dahanukar A, Hallem E, Carlson J. Insect chemoreception. Curr Opin Neurobiol. 2005;15:423–430. doi: 10.1016/j.conb.2005.06.001. [DOI] [PubMed] [Google Scholar]
- Do CB, Mahabhashyam MS, Brudno M, Batzoglou S. ProbCons: Probabilistic consistency-based multiple sequence alignment. Genome Res. 2005;15:330–340. doi: 10.1101/gr.2821705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dohlman HG. G proteins and pheromone signaling. Annu Rev Physiol. 2002;64:129–152. doi: 10.1146/annurev.physiol.64.081701.133448. [DOI] [PubMed] [Google Scholar]
- Drosophila Odorant Receptor Nomenclature Committee. A unified nomenclature system for the Drosophila odorant receptors. Cell. 2000;102:145–146. doi: 10.1016/s0092-8674(00)00020-9. [DOI] [PubMed] [Google Scholar]
- Dubos C, Huggins D, Grant GH, Knight MR, Campbell MM. A role for glycine in the gating of plant NMDA-like receptors. Plant J. 2003;35:800–810. doi: 10.1046/j.1365-313x.2003.01849.x. [DOI] [PubMed] [Google Scholar]
- Fishilevich E, Vosshall LB. Genetic and functional subdivision of the Drosophila antennal lobe. Curr Biol. 2005;15:1548–1553. doi: 10.1016/j.cub.2005.07.066. [DOI] [PubMed] [Google Scholar]
- Gao Q, Yuan B, Chess A. Convergent projections of Drosophila olfactory neurons to specific glomeruli in the antennal lobe. Nat Neurosci. 2000;3:780–785. doi: 10.1038/77680. [DOI] [PubMed] [Google Scholar]
- Goulding SE, zur Lage P, Jarman AP. amos, a proneural gene for Drosophila olfactory sense organs that is regulated by lozenge. Neuron. 2000;25:69–78. doi: 10.1016/s0896-6273(00)80872-7. [DOI] [PubMed] [Google Scholar]
- Gupta BP, Rodrigues V. Atonal is a proneural gene for a subset of olfactory sense organs in Drosophila. Genes Cells. 1997;2:225–233. doi: 10.1046/j.1365-2443.1997.d01-312.x. [DOI] [PubMed] [Google Scholar]
- Huang AL, Chen X, Hoon MA, Chandrashekar J, Guo W, Trankner D, Ryba NJ, Zuker CS. The cells and logic for mammalian sour taste detection. Nature. 2006;442:934–938. doi: 10.1038/nature05084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishimaru Y, Inada H, Kubota M, Zhuang H, Tominaga M, Matsunami H. Transient receptor potential family members PKD1L3 and PKD2L1 form a candidate sour taste receptor. Proc Natl Acad Sci U S A. 2006;103:12569–12574. doi: 10.1073/pnas.0602702103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jhaveri D, Sen A, Reddy GV, Rodrigues V. Sense organ identity in the Drosophila antenna is specified by the expression of the proneural gene atonal. Mech Dev. 2000;99:101–111. doi: 10.1016/s0925-4773(00)00487-1. [DOI] [PubMed] [Google Scholar]
- Jin R, Banke TG, Mayer ML, Traynelis SF, Gouaux E. Structural basis for partial agonist action at ionotropic glutamate receptors. Nat Neurosci. 2003;6:803–810. doi: 10.1038/nn1091. [DOI] [PubMed] [Google Scholar]
- Jones WD, Cayirlioglu P, Kadow IG, Vosshall LB. Two chemosensory receptors together mediate carbon dioxide detection in Drosophila. Nature. 2007;445:86–90. doi: 10.1038/nature05466. [DOI] [PubMed] [Google Scholar]
- Jordt SE, McKemy DD, Julius D. Lessons from peppers and peppermint: the molecular logic of thermosensation. Curr Opin Neurobiol. 2003;13:487–492. doi: 10.1016/s0959-4388(03)00101-6. [DOI] [PubMed] [Google Scholar]
- Krogh A, Larsson B, von Heijne G, Sonnhammer EL. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J Mol Biol. 2001;305:567–580. doi: 10.1006/jmbi.2000.4315. [DOI] [PubMed] [Google Scholar]
- Lam HM, Chiu J, Hsieh MH, Meisel L, Oliveira IC, Shin M, Coruzzi G. Glutamate-receptor genes in plants. Nature. 1998;396:125–126. doi: 10.1038/24066. [DOI] [PubMed] [Google Scholar]
- Larsson MC, Domingos AI, Jones WD, Chiappe ME, Amrein H, Vosshall LB. Or83b encodes a broadly expressed odorant receptor essential for Drosophila olfaction. Neuron. 2004;43:703–714. doi: 10.1016/j.neuron.2004.08.019. [DOI] [PubMed] [Google Scholar]
- Littleton JT, Ganetzky B. Ion channels and synaptic organization: analysis of the Drosophila genome. Neuron. 2000;26:35–43. doi: 10.1016/s0896-6273(00)81135-6. [DOI] [PubMed] [Google Scholar]
- Lundin C, Käll L, Kreher KA, Kapp K, Sonnhammer EL, Carlson JR, von Heijne G, Nilsson I. Membrane topology of the Drosophila OR83b odorant receptor. FEBS Lett. 2007;581:5601–5604. doi: 10.1016/j.febslet.2007.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer M. Glutamate receptors at atomic resolution. Nature. 2006;440:456–462. doi: 10.1038/nature04709. [DOI] [PubMed] [Google Scholar]
- Mayer ML. Crystal structures of the GluR5 and GluR6 ligand binding cores: molecular mechanisms underlying kainate receptor selectivity. Neuron. 2005;45:539–552. doi: 10.1016/j.neuron.2005.01.031. [DOI] [PubMed] [Google Scholar]
- Mayer ML, Armstrong N. Structure and function of glutamate receptor ion channels. Annu Rev Physiol. 2004;66:161–181. doi: 10.1146/annurev.physiol.66.050802.084104. [DOI] [PubMed] [Google Scholar]
- Mayer ML, Ghosal A, Dolman NP, Jane DE. Crystal structures of the kainate receptor GluR5 ligand binding core dimer with novel GluR5-selective antagonists. J Neurosci. 2006;26:2852–2861. doi: 10.1523/JNEUROSCI.0123-06.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mombaerts P, Wang F, Dulac C, Chao SK, Nemes A, Mendelsohn M, Edmondson J, Axel R. Visualizing an olfactory sensory map. Cell. 1996;87:675–686. doi: 10.1016/s0092-8674(00)81387-2. [DOI] [PubMed] [Google Scholar]
- Nakagawa T, Sakurai T, Nishioka T, Touhara K. Insect sex-pheromone signals mediated by specific combinations of olfactory receptors. Science. 2005;307:1638–1642. doi: 10.1126/science.1106267. [DOI] [PubMed] [Google Scholar]
- Naur P, Hansen KB, Kristensen AS, Dravid SM, Pickering DS, Olsen L, Vestergaard B, Egebjerg J, Gajhede M, Traynelis SF, Kastrup JS. Ionotropic glutamate-like receptor delta2 binds D-serine and glycine. Proc Natl Acad Sci U S A. 2007;104:14116–14121. doi: 10.1073/pnas.0703718104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi Z, Stephens NR, Spalding EP. Calcium entry mediated by GLR3.3, an Arabidopsis glutamate receptor with a broad agonist profile. Plant Physiol. 2006;142:963–971. doi: 10.1104/pp.106.088989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray A, van Naters WG, Shiraiwa T, Carlson JR. Mechanisms of odor receptor gene choice in Drosophila. Neuron. 2007;53:353–369. doi: 10.1016/j.neuron.2006.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato K, Pellegrino M, Nakagawa T, Nakagawa T, Vosshall LB, Touhara K. Insect olfactory receptors are heteromeric ligand-gated ion channels. Nature. 2008;452:1002–1006. doi: 10.1038/nature06850. [DOI] [PubMed] [Google Scholar]
- Schultz J, Milpetz F, Bork P, Ponting CP. SMART, a simple modular architecture research tool: identification of signaling domains. Proc Natl Acad Sci U S A. 1998;95:5857–5864. doi: 10.1073/pnas.95.11.5857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott K, Brady R, Jr, Cravchik A, Morozov P, Rzhetsky A, Zuker C, Axel R. A chemosensory gene family encoding candidate gustatory and olfactory receptors in Drosophila. Cell. 2001;104:661–673. doi: 10.1016/s0092-8674(01)00263-x. [DOI] [PubMed] [Google Scholar]
- Stamatakis A. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 2006;22:2688–2690. doi: 10.1093/bioinformatics/btl446. [DOI] [PubMed] [Google Scholar]
- Vosshall LB, Amrein H, Morozov PS, Rzhetsky A, Axel R. A spatial map of olfactory receptor expression in the Drosophila antenna. Cell. 1999;96:725–736. doi: 10.1016/s0092-8674(00)80582-6. [DOI] [PubMed] [Google Scholar]
- Vosshall LB, Stocker RF. Molecular architecture of smell and taste in Drosophila. Annu Rev Neurosci. 2007;30:505–533. doi: 10.1146/annurev.neuro.30.051606.094306. [DOI] [PubMed] [Google Scholar]
- Vosshall LB, Wong AM, Axel R. An olfactory sensory map in the fly brain. Cell. 2000;102:147–159. doi: 10.1016/s0092-8674(00)00021-0. [DOI] [PubMed] [Google Scholar]
- Wicher D, Schafer R, Bauernfeind R, Stensmyr MC, Heller R, Heinemann SH, Hansson BS. Drosophila odorant receptors are both ligand-gated and cyclic-nucleotide-activated cation channels. Nature. 2008;452:1007–1011. doi: 10.1038/nature06861. [DOI] [PubMed] [Google Scholar]
- Wistrand M, Kall L, Sonnhammer EL. A general model of G protein-coupled receptor sequences and its application to detect remote homologs. Protein Sci. 2006;15:509–521. doi: 10.1110/ps.051745906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao CA, Ignell R, Carlson JR. Chemosensory coding by neurons in the coeloconic sensilla of the Drosophila antenna. J Neurosci. 2005;25:8359–8367. doi: 10.1523/JNEUROSCI.2432-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- zur Lage PI, Prentice DR, Holohan EE, Jarman AP. The Drosophila proneural gene amos promotes olfactory sensillum formation and suppresses bristle formation. Development. 2003;130:4683–4693. doi: 10.1242/dev.00680. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.