SUMMARY
Organ progenitors arise within organ fields, embryonic territories that are larger than the regions required for organ formation. Little is known about the regulatory pathways that define organ field boundaries and thereby limit organ size. Here, we identify a mechanism for restricting heart size through confinement of the developmental potential of the heart field. Via fate mapping in zebrafish, we locate cardiac progenitors within hand2-expressing mesoderm and demonstrate that hand2 potentiates cardiac differentiation within this region. Beyond the rostral boundary of hand2 expression, we find progenitors of vessel and blood lineages. In embryos deficient in vessel and blood specification, rostral mesoderm undergoes a fate transformation and generates ectopic cardiomyocytes. Therefore, induction of vessel and blood specification represses cardiac specification and delimits the capacity of the heart field. This regulatory relationship between cardiovascular pathways suggests new strategies for directing progenitor cell differentiation to facilitate cardiac regeneration.
INTRODUCTION
Organogenesis relies upon the production of a proper number of progenitor cells. A shortage of progenitors can limit organ size and functional capacity, whereas a surplus of progenitors can lead to organ enlargement and dysfunction. Organ progenitors are thought to arise within “organ fields,” embryonic territories that possess the appropriate developmental potential but are typically larger than the regions fated to contribute to the organ (Huxley and De Beer, 1934; Jacobson and Sater, 1988). Therefore, progenitor quantity is likely to be controlled by the establishment of field boundaries and the gradual refinement of field potential. However, the regulation of these processes is not well understood.
Enthusiasm for the possibilities of regenerative medicine has fueled interest in the mechanisms that create and differentiate organ progenitors. For instance, identification of a source for cardiac progenitors could facilitate cellular therapy aimed at regeneration of cardiac muscle (Laflamme and Murry, 2005; van Laake et al., 2006). The origins of cardiac progenitors have therefore attracted particular attention. Fate mapping studies have shown that cardiac progenitors reside within bilateral heart-forming regions (HFRs) of the anterior lateral plate mesoderm (ALPM) (Harvey, 2002; Yutzey and Kirby, 2002). The HFRs are the primary source of the cardiomyocytes that compose the initial heart tube. In Xenopus and zebrafish, the HFRs are the only reported mesodermal source of cardiomyocytes (Raffin et al., 2000; Serbedzija et al., 1998). In contrast, chick and mouse embryos contain a second source of cardiomyocytes in a separate mesodermal territory. This region, called the second, or anterior, heart field, contributes myocardium to the arterial and venous poles of the heart, including contributions to the outflow tract, right ventricle, and atria (Buckingham et al., 2005; Kelly et al., 2001; Mjaatvedt et al., 2001; Waldo et al., 2001; Cai et al., 2003; Zaffran et al., 2004). To date, the embryonic and evolutionary origins of the second heart field remain unclear.
In frog and fish, the ALPM surrounding the HFRs appears to exhibit the breadth of developmental potential expected of an organ field. In Xenopus, extirpated tissue from a lateral portion of the ALPM can produce cardiomyocytes in vitro, even though this particular region does not normally produce cardiomyocytes in vivo (Sater and Jacobson, 1990; Raffin et al., 2000; Garriock and Drysdale, 2003). In zebrafish, removal of the anterior notochord allows cardiac specification in ALPM just caudal to the HFRs (Goldstein and Fishman, 1998). Furthermore, the zebrafish HFRs can rapidly be replenished after partial ablation, due to contributions from rostral ALPM flanking these regions (Serbedzija et al., 1998). Together, these studies indicate that the cardiogenic potential of the ALPM extends beyond the boundaries of the genuine HFRs.
It is unclear how certain portions of the ALPM are kept from fulfilling their cardiac potential. A few pathways have been implicated in the repression of cardiac fate assignment. For example, Notch signaling seems to inhibit cardiac specification in lateral portions of the Xenopus ALPM (Rones et al., 2000). Canonical Wnt signaling has also been suggested to repress cardiac potential in chick, Xenopus, and mouse (Marvin et al., 2001; Schneider and Mercola, 2001; Lickert et al., 2002). Finally, retinoic acid signaling appears to restrict the specification of cardiac progenitors during gastrulation stages in zebrafish and Xenopus (Keegan et al., 2005; Collop et al., 2006) and to inhibit cardiomyocyte differentiation within the ALPM in Xenopus and chick (Osmond et al., 1991; Yutzey et al., 1994; Drysdale et al., 1997). It is not known whether or how these pathways work together to refine the cardiogenic potential of the heart field, and it is likely that important repressive influences remain to be revealed.
Here, we identify a potent mechanism for restricting cardiac specification within the heart field. By constructing a fate map of the zebrafish ALPM, we establish that the boundaries of the zebrafish HFR correspond to the boundaries of hand2 expression. Mesoderm beyond the rostral limit of hand2 expression normally gives rise to vessel and blood lineages. However, this rostral territory also harbors latent cardiac developmental potential. Embryos with defects in vessel and blood specification exhibit too many cardiomyocytes, and these extra cells are the consequence of a fate transformation that generates ectopic cardiac progenitors in rostral mesoderm. These data demonstrate an important regulatory relationship between the developmental programs that create the cardiovascular system: components of the vessel and blood specification pathways repress cardiac specification and thereby limit heart size. This interplay between pathways has implications for the mechanisms underlying cardiac evolution and suggests new strategies for regenerative medicine.
RESULTS
Gene expression patterns divide the ALPM into three territories
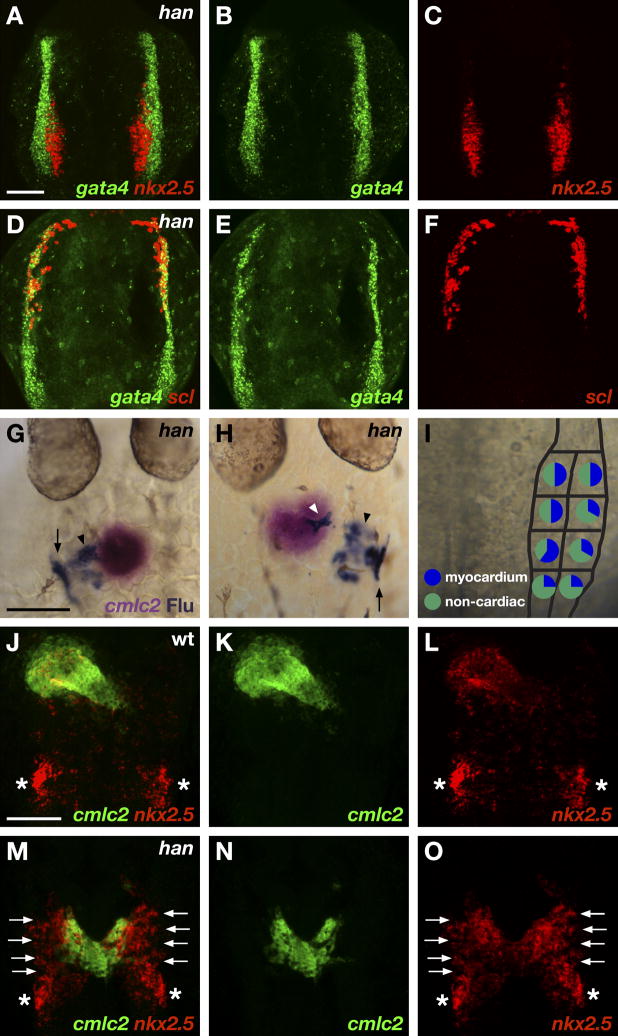
In order to investigate the mechanisms that delimit cardiac potential in zebrafish, we first sought to define the dimensions of the HFR relative to gene expression patterns in the zebrafish ALPM. We chose to examine the expression of a group of transcription factor genes at the 6–9 somite stage (Fig. 1), 3–4 hours prior to the onset of myocardial differentiation (Yelon et al., 1999). At these stages, the ALPM appears as bilateral strips of tissue running along the lateral edges of the embryonic anterior-posterior axis. A few genes (e.g. gata4, gata5, and tbx20) are expressed throughout the ALPM (Fig. 1B,E,H, and data not shown). In contrast, expression of hand2 is confined to a relatively posterior portion of the ALPM, flanking the hindbrain and extending below the tip of the notochord (Fig. 1A–C). Expression of hand2 in this portion of the ALPM spans its width (Fig. 1A–C). This region is subdivided by nkx2.5 expression, which is prominently expressed medially, but not detectable laterally (Fig. 1D–F,K). We find expression of genes associated with vasculogenesis and hematopoiesis (e.g. scl, fli1a, spi1, and lmo2) anterior to hand2, in the most rostral portion of the ALPM (Fig. 1G-J and data not shown). Expression of rostral ALPM genes has no detectable overlap with hand2 expression (Fig. 1J). Thus, gene expression patterns divide the ALPM into three distinct territories, hereafter referred to as medial, lateral, and rostral ALPM (Fig. 1L).
Figure 1.
Gene expression patterns divide the ALPM into three territories. (A–K) Two-color fluorescent in situ hybridization of wild-type embryos at the 7 somite stage, dorsal views, anterior to the top. All images shown at the same magnification; scale bar represents 100 μm. (A) Overlay of gata4 (B) and hand2 (C) expression. Expression of hand2 is observed throughout the caudal portion of gata4-expressing ALPM, but is excluded from rostral ALPM. (D) Overlay of gata4 (E) and nkx2.5 (F) expression. Expression of nkx2.5 is observed within a medial portion of the caudal ALPM, but is excluded from lateral and rostral ALPM. (G) Overlay of gata4 (H) and scl (I) expression. Expression of scl is observed in rostral ALPM. (J) Overlay of hand2 (green) and scl (red) expression. Expression patterns of scl and hand2 do not overlap. (K) Overlay of hand2 (green) and nkx2.5 (red) expression. Expression of hand2 extends beyond the lateral limit of nkx2.5 expression. (L) Schematic of 7 somite stage ALPM, superimposed onto a dorsal view of a live embryo; dashes outline the tip of the notochord. Comparison of gene expression patterns defines rostral (white), medial (black), and lateral (grey) ALPM territories. Although ALPM morphology varies slightly during the 6–9 somite stage and between individual embryos, relative patterns of gene expression consistently demarcate these territories.
Constructing a fate map of ALPM territories
Next, we investigated where cardiac progenitors reside within the three ALPM territories. Medial ALPM has previously been shown to contain cardiac progenitors (Serbedzija et al., 1998), but the cardiac contributions of lateral and rostral ALPM have not been explored. To construct a fate map of the ALPM, we employed a photoactivatable DMNB-caged fluorescein dextran conjugate as a lineage tracer. By adapting a photoactivation protocol previously used for fate mapping at late blastula stages (Keegan et al., 2004), we labeled cells within the ALPM at the 6–9 somite stage (Fig. 2A–C) and later assessed their fate (Fig. 2J–L). In each experimental embryo, we labeled a cluster of 10–15 cells within a particular region: we examined 2 regions within the rostral ALPM (Fig. 2D), 4 regions within the medial ALPM (Fig. 2G), and 4 regions within the lateral ALPM (Fig. 2G). Use of visible anatomical landmarks, such as the lateral edge of the ALPM, notochord, and brain structures, facilitated accurate targeting of each region (Fig. 2D–I). Following heart formation, embryos were fixed and processed to score contributions of labeled cells to the myocardium (Fig. 2J) and endocardium (Fig. 2L). We also noted contributions to other mesodermal tissues, including head vessels, head mesenchyme, and myeloid cells. Additionally, many experimental embryos exhibited labeled progeny within the pharyngeal pouches (Fig. 2K); pharyngeal pouch labeling is likely to reflect overlap of the migrating anterior endoderm and the ALPM at the 6-9 somite stage (T. Wendl, D. Adzic, J.J.S., S. Scholpp, M. Brand, D.Y., and K. Rohr, manuscript submitted).
Figure 2.
Constructing a fate map of ALPM territories. (A–I) Dorsal views, anterior to the top, of wild-type embryos at the 6–9 somite stage. Scale bars represent 100 μm. (A–C) Photoactivation of caged fluorescein labels groups of cells in the ALPM. (A) DIC image of a live embryo. The lateral edge of the ALPM (dashed line) is visible. (B) Fluorescence indicates photoactivation of caged fluorescein. (C) Overlay of DIC and fluorescent images confirms location of labeled cells. In this example, the ~14 labeled cells reside in the medial ALPM. (D,G) Schematics indicate ALPM regions that were interrogated by fate mapping. (D) Two regions within the rostral ALPM were examined, on both sides of the embryo: a region flanking the forebrain (region 1) and a region flanking the midbrain-hindbrain boundary (region 2). (E,F) Examples of control experiments performed to assess accuracy of labeling rostral ALPM. Cells were labeled in the regions depicted by green ovals in (D) and immediately processed to compare scl expression (red) relative to uncaged fluorescein (green). (E) Cells labeled in region 2 were located solely within the scl expression domain in 5/9 experiments; in the other 4 experiments, labeling crossed the posterior border of scl expression and therefore overlapped with hand2-expressing territory. (F) Cells labeled in region 1 were located within the scl expression domain in 8/8 experiments. (G) 8 regions within the caudal ALPM were examined, on both sides of the embryo: 4 medial regions and 4 lateral regions distinguished by their position along the anterior-posterior axis. (H,I) Examples of control experiments performed to assess mediolateral labeling accuracy. Cells were labeled in the regions depicted by green ovals in (D) and immediately processed to compare nkx2.5 expression (red) relative to uncaged fluorescein (green). (H) Cells labeled in lateral regions did not express nkx2.5 (n=8/9). (I) Conversely, cells labeled in medial regions did express nkx2.5 (n=6/6). (J–L) Scoring progeny of labeled ALPM cells at 44 hpf. In situ hybridization indicates expression of cmlc2 (magenta) in cardiomyocytes, and immunohistochemistry for uncaged fluorescein (blue) indicates progeny of labeled cells. (J) Frontal view, dorsal to the top; in this example, labeled cells became ~30 atrial cardiomyocytes (black arrowheads). (K, L) Lateral views, anterior to the right. (K) In this example, 30–40 labeled cells populated a pharyngeal pouch (white arrow). (L) In this example, 3–4 labeled cells (black arrow) contributed to the ventricular endocardium.
The zebrafish HFR extends across medial and lateral territories
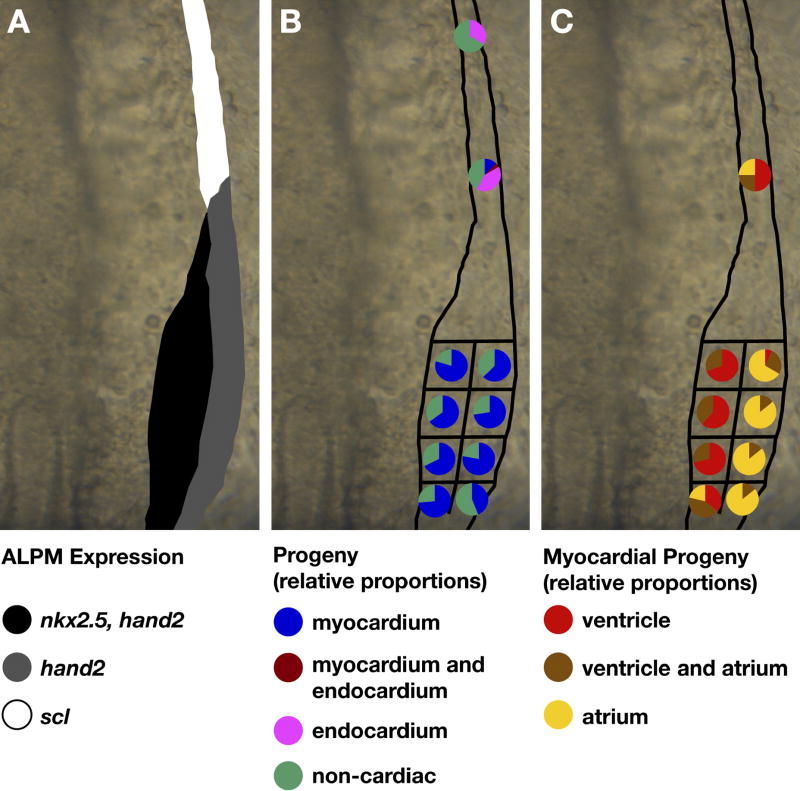
Our fate map data demonstrate the presence of myocardial progenitors within both medial and lateral ALPM territories (Fig. 3). 72% of experiments in medial ALPM generated labeled myocardial progeny (Fig. 3B, Table 1). Similarly, 66% of experiments in lateral ALPM generated myocardial progeny (Fig. 3B, Table 1). In contrast, only 9% of experiments in rostral ALPM generated myocardial progeny (Fig. 3B, Table 1). Instead, we found labeled endocardial progeny in 40% of rostral ALPM experiments (Fig. 3B, Table 1). In rostral ALPM experiments that generated myocardial progeny, labeled cells were located near the boundary between scl and hand2 expression and not in the most rostral region tested (Fig. 3A,B, Table 1), suggesting that the myocardial progenitors labeled alongside the scl/hand2 border might have originated in hand2-expressing ALPM. Thus, our data indicate that the zebrafish HFR extends across medial and lateral ALPM, with little or no contribution from rostral ALPM.
Figure 3.
Fate map of cardiac progenitors within ALPM territories. (A–C) Schematics depicting the progenitor composition of medial, lateral, and rostral ALPM territories. Dorsal views of the right side of the ALPM, anterior to the top. (A) Schematic of gene expression patterns that distinguish medial (black), lateral (grey), and rostral (white) ALPM territories, as in Fig. 1L. (B) Myocardial progenitors arise from medial and lateral territories and are only rarely detected in rostral territory. In contrast, endocardial progenitors arise from rostral territory. For each region tested, pie charts depict the proportions of experimental embryos exhibiting a particular type of labeled progeny. Colored pie pieces represent embryos that produced labeled myocardial progeny (blue), labeled endocardial progeny (pink), both myocardial and endocardial labeled progeny (maroon), and only non-cardiac labeled progeny (green). Data shown are a merger of experiments from the left and right sides of the embryo; our data did not reveal any left-right asymmetry of the fate map. (C) Ventricular and atrial myocardial progenitors appear spatially organized within caudal ALPM, with ventricular progenitors tending to originate medially and atrial progenitors tending to originate laterally. Pie charts depict the chamber contributions of labeled cells in the subset of experimental embryos containing labeled myocardial progeny. Colored pie pieces represent embryos that produced labeled ventricular cardiomyocytes (red), labeled atrial cardiomyocytes (yellow), and both ventricular and atrial cardiomyocytes (brown). See Table 1 for data reflected in schematics.
TABLE 1. Fate map data from rostral, medial, and lateral territories.
For each region examined, data indicate the fraction of experimental embryos exhibiting myocardial progeny (but not endocardial progeny), endocardial progeny (but not myocardial progeny), or both myocardial and endocardial progeny. Non-cardiac progeny detected in these experiments included pharyngeal pouches, head vessels, head mesenchyme, and myeloid cells. In medial and lateral ALPM, >90% of all experimental embryos exhibited labeled pharyngeal pouch cells. For wild-type and clo mutant embryos containing labeled cardiomyocytes, additional data indicate whether these cells were found only in the ventricle, only in the atrium, or in both chambers. The dysmorphic nature of han mutant myocardium prevented morphological analysis of chamber contributions. Data collected from left and right ALPM are combined for all regions.
| Genotype | Region | Myocardial progeny | Ventricular only | Ventricular and atrial | Atrial only | Endocardial progeny | Myocardial and endocardial |
|---|---|---|---|---|---|---|---|
| wild-type | 1R | 0/21(0%) | -- | -- | -- | 7/21 (33%) | -- |
| 2R | 3/24 (12%) | 2/4 (50%) | 1/4 (25%) | 1/4 (25%) | 10/24 (42%) | 1/24 (4%) | |
| clo | 1R | 4/7 (57%) | 1/4 (25%) | 3/4 (75%) | 0/4 (0%) | -- | -- |
| 2R | 8/9 (89%) | 0/8 (0%) | 7/8 (88%) | 1/8 (12%) | -- | -- | |
| wild-type | 3M | 23/29 (79%) | 16/23 (70%) | 7/23 (30%) | 0/23 (0%) | -- | -- |
| 4M | 13/20 (65%) | 8/13 (62%) | 5/13 (38%) | 0/13 (0%) | -- | -- | |
| 5M | 21/31 (68%) | 15/21 (71%) | 6/21 (29%) | 0/21 (0%) | -- | -- | |
| 6M | 14/19 (74%) | 5/14 (36%) | 6/14 (43%) | 3/14 (21%) | -- | -- | |
| han | 3M | 1/2 (50%) | -- | -- | -- | -- | -- |
| 4M | 3/6 (50%) | -- | -- | -- | -- | -- | |
| 5M | 3/5 (60%) | -- | -- | -- | -- | -- | |
| 6M | 1/4 (25%) | -- | -- | -- | -- | -- | |
| wild-type | 3L | 15/24 (62%) | 1/15 (7%) | 4/15 (27%) | 10/15 (67%) | -- | -- |
| 4L | 22/30 (73%) | 0/22 (0%) | 3/22 (14%) | 19/22 (86%) | -- | -- | |
| 5L | 14/18 (78%) | 0/14 (0%) | 2/14 (14%) | 12/14 (86%) | -- | -- | |
| 6L | 7/16 (44%) | 0/7 (0%) | 1/7 (14%) | 6/7 (86%) | -- | -- | |
| han | 3L | 1/2 (50%) | -- | -- | -- | -- | -- |
| 4L | 3/9 (33%) | -- | -- | -- | -- | -- | |
| 5L | 2/6 (33%) | -- | -- | -- | -- | -- | |
| 6L | 1/4 (25%) | -- | -- | -- | -- | -- |
Although myocardial progenitors arise in both medial and lateral ALPM, our fate map indicates a striking difference between the chamber identities of the cardiomyocytes derived from these two territories. Of the medial ALPM experiments that produced myocardial progeny, 96% of embryos exhibited labeled cardiomyocytes in the ventricle, whereas only 38% of embryos exhibited labeled cardiomyocytes in the atrium (Fig. 3C, Table 1). The converse was observed in lateral ALPM experiments that produced myocardial progeny: only 19% of embryos contained labeled ventricular cardiomyocytes, whereas 98% of embryos contained labeled atrial cardiomyocytes (Fig. 3C, Table 1). These data demonstrate a general medial-lateral organization of ventricular and atrial myocardial progenitors within the HFR, consistent with the previously documented spatial organization of ventricular and atrial progenitors in the late blastula (Keegan et al., 2004) and medial-lateral pattern of ventricular myosin heavy chain and atrial myosin heavy chain expression as myocardial differentiation proceeds in the ALPM (Yelon et al., 1999; Berdougo et al., 2003).
Combining our fate map and gene expression data, we conclude that the rostral and mediolateral boundaries of the HFR correspond well with the boundaries of hand2 expression. Although previous studies have suggested that nkx2.5 expression marks the zebrafish HFRs (Serbedzija et al., 1998), the larger domain of hand2 expression more accurately reflects HFR dimensions and includes atrial progenitors during the 6–9 somite stage. At later stages, however, nkx2.5 is expressed in both ventricular and atrial myocardium (Yelon et al., 1999). Similarly, fate maps of the chick HFR have indicated that myocardial progenitors, mostly of atrial identity, reside beyond the boundary of Nkx2-5 expression in the early ALPM (Redkar et al., 2001; Hochgreb et al., 2003).
Hand2 potentiates myocardial differentiation within the HFR
Given the correlation of hand2 expression with the dimensions of the HFR, we examined the function of Hand2 within this region. Our previous analyses of zebrafish hands off/hand2 (han) mutants have suggested that Hand2 plays a permissive role in promoting cardiomyocyte production, rather than an instructive role in assigning myocardial identity: han mutants contain few cardiomyocytes, but ectopic expression of hand2 does not seem to create ectopic cardiomyocytes (Yelon et al., 2000). Consistent with this proposed role, we find that loss of Hand2 function does not affect the overall dimensions of the HFR. Initial patterns of gene expression within the ALPM appear normal in han mutants (compare Fig. 4A–F to Fig.1D–I). Notably, nkx2.5 expression is not diminished (Fig. 4A–C and Yelon et al., 2000), and scl expression does not expand caudally (Fig. 4D–F). Additionally, fate mapping shows that myocardial progenitors reside throughout the medial and lateral ALPM territories in han mutants (Fig. 4I). However, the han mutant HFR is significantly less effective than the wild-type HFR at producing cardiomyocytes: only 39% of fate mapping experiments in the han mutant HFR generated labeled cardiomyocytes, compared to 69% of experiments in the wild-type HFR (Figs. 3B, 4I, Table 1; p<0.05, Student’s t-test). Instead, labeled cells from the han mutant HFR were often found adjacent to differentiated cardiomyocytes (black arrowheads in Fig. 4G,H); we detected these cells in 76% of han mutant experiments, and did not find similar cells in wild-type embryos. Although these flanking cells do not express terminal myocardial differentiation markers such as cmlc2, they are found in a region where many cells express nkx2.5 (Fig. 4M–O), suggesting that they could represent cardiac progenitors that initiate, but fail to complete, myocardial differentiation. Thus, although not required to set HFR boundaries, Hand2 plays an important role in the potentiation of myocardial differentiation within the HFR.
Figure 4.
Hand2 potentiates myocardial differentiation. (A–F) ALPM territories appear to be established normally in han mutant embryos. Two-color fluorescent in situ hybridization of han mutants at the 7 somite stage, dorsal views, anterior to the top. Scale bars represent 100 μm. (A) Overlay of gata4 (B) and nkx2.5 (C) expression. (D) Overlay of gata4 (E) and scl (F) expression. Expression patterns of gata4, nkx2.5, and scl divide the han mutant ALPM into normally proportioned rostral, medial, and lateral regions. (G–I) Fate map of the caudal ALPM in han mutants indicates that Hand2 is required to facilitate effective differentiation of myocardial progenitors from medial and lateral ALPM territories. (G,H) Dorsal views of han mutant embryos at 30 hpf, anterior to the top. In situ hybridization indicates expression of cmlc2 (magenta) in cardiomyocytes, and immunohistochemistry for uncaged fluorescein (blue) indicates progeny of labeled cells. In han mutants, labeled cells from caudal ALPM can give rise to cardiomyocytes (white arrowhead, H) but also frequently become seemingly undifferentiated cells (black arrowheads, G and H) flanking the cardiomyocytes. Labeled cells also contribute to pharyngeal pouches (arrows, G and H). (I) Fate map of han mutant caudal ALPM. For each region tested, pie charts depict the proportions of experimental embryos producing labeled myocardial progeny (blue) and only non-cardiac labeled progeny (green). Myocardial progenitors were found in all regions examined, but the frequency with which labeled han mutant ALPM produced cardiomyocytes was reduced, relative to wild-type, throughout both medial and lateral territories (compare to Fig. 3B). See Table 1 for data reflected in schematic. (J–O) han mutants exhibit ineffective myocardial differentiation. Two-color fluorescent in situ hybridization at 24 hpf, dorsal views, anterior to the top. (J,M) Overlays of cmlc2 (K,N) and nkx2.5 (L,O) expression. In wild-type embryos (J–L), cmlc2 and nkx2.5 are both expressed throughout the heart tube. Additional expression of nkx2.5 marks the forming pharyngeal pouches (asterisks). (M–O) In han mutants, a reduced number of cardiomyocytes span the midline. Undifferentiated, nkx2.5-expressing ALPM cells (arrows) flank the cardiomyocytes.
Ectopic cardiomyocytes arise from rostral ALPM in cloche mutants
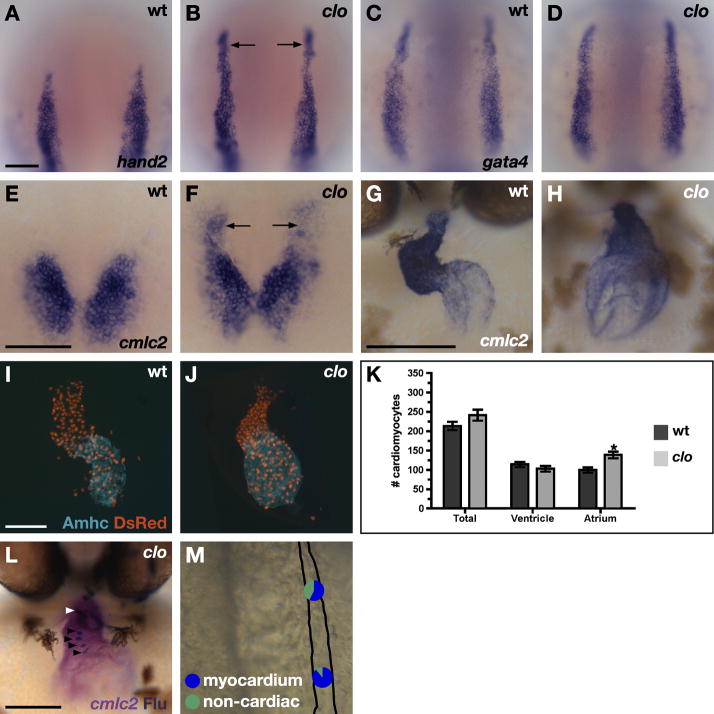
Even though Hand2 does not appear to be responsible for cardiac specification, hand2 expression does seem to be a useful marker of the location of the HFR. We hypothesized that genes critical for delimiting cardiac developmental potential would also regulate the extent of hand2 expression. We therefore proceeded to analyze the expression of hand2 in a collection of zebrafish mutants with cardiac defects (Alexander et al., 1998), and we discovered that mutation of the cloche (clo) locus has a striking effect on the regulation of hand2. Specifically, we found that hand2 expression in clo mutants extends beyond its normal boundary, exhibiting strong expression throughout the rostral ALPM (Fig. 5A,B). This ectopic expression does not represent a distortion of ALPM morphology, since ALPM shape and size appear normal in clo mutants (Fig. 5C,D). The molecular identity of the clo gene has yet to be reported, but previous studies have demonstrated that clo is required for specification of endothelial and hematopoietic lineages (Stainier et al., 1995; Liao et al., 1997; Liao et al., 1998; Liao et al., 2000; Parker and Stainier, 1999; Lieschke et al., 2002; Sumanas et al., 2005; Thompson et al., 1998). Interestingly, progenitors for several of the cell types absent in clo mutants – endocardial cells, head vessels, and myeloid cells – originate within the rostral ALPM (Fig. 3B, Table 1) (Herbomel et al., 1999; Lieschke et al., 2002; Hsu et al., 2004).
Figure 5.
clo mutants generate ectopic cardiomyocytes. (A–H) In situ hybridization depicts expression of hand2 (A,B), gata4 (C,D) and cmlc2 (E–H) in wild-type (A,C,E,G) and clo mutant (B,D,F,H) embryos. (A–F) Dorsal views, anterior to the top, at the 7 somite (A–D) and 19 somite (E,F) stages. Scale bars represent 100 μm. (A,B) hand2 is ectopically expressed in the rostral ALPM of clo mutant embryos (arrows, B). (C,D) Expression of gata4 is indistinguishable between wild-type and clo mutant embryos, indicating normal rostral ALPM morphology. (E,F) clo mutants exhibit a rostral extension of cmlc2 expression (arrows). (G,H) Frontal views, dorsal to the top. At 48 hpf, the wild-type heart is looped, with morphologically distinct chambers, whereas the clo mutant heart appears unlooped, with a small, compact ventricle and a large, dilated atrium. (I–K) clo mutant hearts contain too many cardiomyocytes. (I,J) Frontal views of hearts at 52 hpf. Immunofluorescence indicates that both chambers express the transgeneTg(cmlc2:DsRed2-nuc) (red) (Mably et al., 2003). Atria are labeled with the anti-Amhc antibody S46 (aqua) (Berdougo et al., 2003). (K) Counting of cardiomyocyte nuclei reveals a statistically significant increase (asterisk) in the number of atrial cardiomyocytes in clo mutants (p<0.002, Student’s t-test; n=11 of each genotype). Bar graph indicates mean and standard error of each data set. (L,M) Fate map of the rostral ALPM in clo mutants indicates formation of ectopic myocardial progenitors. (L) Frontal view of clo mutant embryo at 44 hpf, dorsal to the top. In situ hybridization indicates expression of cmlc2 (magenta) in cardiomyocytes, and immunohistochemistry for uncaged fluorescein (blue) indicates progeny of labeled cells. In this example, labeled cardiomyocytes are found in the atrium (black arrowheads) and in the ventricle (white arrowhead). (M) Fate map of clo mutant rostral ALPM. For each region tested, pie charts depict the proportions of experimental embryos producing labeled myocardial progeny (blue) and only non-cardiac labeled progeny (green). See Table 1 for data reflected in schematic. In clo mutants, both rostral ALPM regions frequently gave rise to cardiomyocytes, indicating a significant fate transformation (compare to Fig. 3B). We have not performed fate map experiments in the clo mutant medial or lateral ALPM, since clo mutants do not seem to exhibit significant changes in gene expression patterns in these territories (J.J.S., B.R.K., and D.Y., unpublished data).
The rostral extension of hand2 expression in clo mutants suggested that these embryos might contain expanded HFRs. Consistent with this idea, we found that clo mutants produce extra cmlc2-expressing cells, many of which extend rostrally beyond the primary bilateral populations of cardiomyocytes (Fig. 5E,F). These extra cells require Hand2 for their efficient differentiation, just like wild-type cardiomyocytes (Supplemental Fig. 1): han;clo double mutants exhibit expanded nkx2.5 expression, like clo mutants, but only exhibit a small number of differentiated cardiomyocytes, like han mutants. Extra cardiomyocytes are also evident within the clo mutant heart at later stages. By 48 hpf, the clo mutant heart is markedly dysmorphic, with a misshapen, compact ventricle and a dilated atrium (Fig. 5G,H), phenotypes that had been attributed to the loss of endocardium (Stainier et al., 1995). Comparison of the number of cardiomyocytes expressing the transgene Tg(cmlc2:DsRed2-nuc) (Mably et al., 2003) demonstrated a trend toward a surplus of cardiomyocytes in clo mutant hearts (Fig. 5I-K). In particular, clo mutants exhibit a statistically significant increase in the number of atrial cardiomyocytes, compared to wild-type embryos (Fig. 5K). The number of ventricular cardiomyocytes appears relatively unaffected (Fig. 5K).
The ectopic hand2 expression and myocardial surplus, together with the previously demonstrated vessel and blood deficiencies (Stainier et al., 1995; Liao et al., 1997; Liao et al., 1998; Liao et al., 2000; Parker and Stainier, 1999; Lieschke et al., 2002; Sumanas et al., 2005; Thompson et al., 1998), led us to suspect that the excess cardiomyocytes in clo mutants could arise from a fate transformation of rostral ALPM. To test this, we examined the clo mutant rostral ALPM fate map. As shown above, wild-type rostral ALPM rarely contains myocardial progenitors (9% of rostral ALPM experiments; Fig. 3B, Table 1). In strong contrast, 75% of experiments in clo mutant rostral ALPM generated labeled myocardial progeny (Fig. 5M, Table 1). In these embryos, labeled cardiomyocytes were frequently found in both chambers (Table 1), indicating that the clo mutant rostral ALPM contains both ventricular and atrial myocardial progenitors. However, labeled atrial cardiomyocytes typically outnumbered labeled ventricular cardiomyocytes (Fig. 5L). This may reflect an important role of endocardial-myocardial signaling during ventricular chamber maturation and growth, after the heart tube is assembled; this function of the endocardium has been suggested by previous studies in mouse (e.g. Lavine et al., 2005; Grego-Bessa et al., 2007). Overall, our fate map data clearly demonstrate a fate transformation within the rostral ALPM of clo mutants: rather than producing progenitors of endothelial and hematopoietic lineages, this territory produces ectopic myocardial progenitors. Therefore, clo acts as a repressor of cardiac specification in the rostral ALPM.
Latent cardiac potential revealed by inhibition of vasculogenesis and hematopoiesis
Prior studies of clo mutants have suggested that clo functions atop a hierarchy of genes controlling vessel and blood development (Stainier et al., 1995; Liao et al., 1997; Liao et al., 1998; Thompson et al., 1998). We therefore wondered whether downstream components of these genetic pathways could, like clo, act to repress rostral expression of hand2 and cmlc2. We chose to investigate whether the genes scl and etsrp could play this role. Both scl and etsrp encode transcription factors and are expressed in the rostral ALPM (Fig. 1G-J) (Gering et al., 1998; Liao et al., 1998; Sumanas and Lin, 2006; Pham et al., 2006). Loss-of-function studies have shown that Scl and Etsrp are essential components of rostral vasculogenic and myelopoietic specification programs (Patterson et al., 2005; Dooley et al., 2005; Juarez et al., 2005; Sumanas and Lin, 2006). Additionally, overexpression of these genes can induce ectopic expression of vessel and blood markers (Gering et al., 1998; Dooley, et al., 2005; Pham et al., 2006; Sumanas and Lin, 2006); furthermore, overexpression of scl together with lmo2 induces broad, ectopic vessel and blood specification at the expense of other mesodermal cell types, including myocardium (Gering et al., 2003). Both scl and etsrp act downstream of clo: their expression is dramatically reduced in clo mutants, and overexpression of either gene can rescue aspects of the clo vascular or hematopoietic phenotype (Liao et al., 1998; Dooley et al., 2005; Sumanas et al., 2005; Sumanas and Lin, 2006).
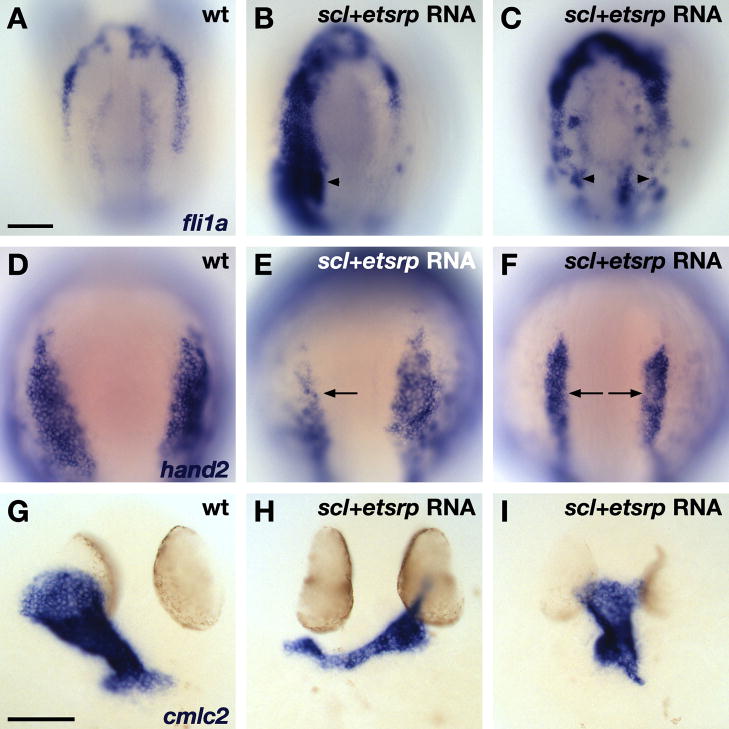
To determine whether scl and etsrp are required to repress cardiogenesis, we used previously characterized anti-scl and anti-etsrp morpholinos (MOs) to knock down gene function (Juarez et al., 2005; Sumanas and Lin, 2006). Embryos injected with anti-scl or anti-etsrp MOs displayed weak expression of hand2 in the rostral ALPM (Fig. 6A–C). Injection of both anti-scl and anti-etsrp MOs (hereafter referred to as anti-scl+etsrp MOs) elicited strong rostral expression of hand2 (Fig. 6D), similar to that observed in clo mutants (Fig. 5B). Additionally, injection of anti-scl+etsrp MOs resulted in rostral extension of cmlc2 expression (Fig. 6E,F), formation of a misshapen heart with an enlarged atrium (Fig. 6G,H), and production of too many cardiomyocytes (Fig. 6I), all of which are reminiscent of the clo mutant phenotype (Fig. 5E–H,K). Conversely, overexpression of scl and etsrp induces ectopic expression of rostral ALPM markers, like fli1a, in the HFR (Fig. 7A–C), reduces hand2 expression (Fig. 7D–F), and decreases cardiomyocyte production (Fig. 7G–I). Together, these data show that scl and etsrp, both of which function downstream of clo to promote vasculogenesis and hematopoiesis, are also essential to override the latent cardiac developmental potential residing within rostral ALPM and thereby limit heart size.
Figure 6.
Latent cardiac potential in rostral ALPM is revealed by inhibition of vasculogenesis and myelopoiesis. (A–H) In situ hybridization depicts expression of hand2 (A–D) and cmlc2 (E–H) in wild-type (A,E,G) and MO-injected (B–D,F,H) embryos. (A–F) Dorsal views, anterior to the top, at the 7 somite (A–D) and 20 somite (E,F) stages. (G,H) Frontal views, dorsal to the top, at 48 hpf. Scale bars represent 100 μm. (A–D) hand2 is ectopically expressed in the rostral ALPM of embryos when scl and etsrp are knocked down. Injections of anti-scl (B) or anti-etsrp (C) MOs caused weak expression of hand2 in rostral ALPM (arrows; anti-scl MOs, n=6/10; anti-etsrp MOs, n=6/12). Rostral hand2 expression was not observed in control sibling embryos (n=0/24). Injection of anti-scl+etsrp MOs (D) caused robust expression of hand2 in rostral ALPM (arrows; strong expression, n=10/17; weak expression, n=7/17). Thus, injection of anti-scl+etsrp MOs phenocopies a loss of clo function. (E,F) Injection of anti-scl+etsrp MOs causes a rostral extension of cmlc2 expression (arrows; injected embryos, n=25/31; control siblings, n=0/33). (G,H) Injection of anti-scl+etsrp MOs results in cardiac chamber morphology resembling that of clo mutants (injected embryos, n=8/10; control siblings, n=0/15). (I) Hearts of anti-scl+etsrp MO-injected embryos contain too many cardiomyocytes. Counting of cardiomyocyte nuclei reveals statistically significant increases (asterisks) in the total number of cardiomyocytes and the number of atrial cardiomyocytes in MO-injected embryos (p<0.0002, Student’s t-test; n=13 uninjected controls and 15 injected embryos). Bar graph indicates mean and standard error of each data set.
Figure 7.
Overexpression of scl and etsrp promotes vasculogenesis and inhibits heart formation. In situ hybridization depicts expression of fli1a (A–C), hand2 (D–F), and cmlc2 (G–I) in wild-type (A,D,G) and scl+etsrp RNA-injected embryos displaying severe (B,E,H) and moderate (C,F,I) phenotypes. Dorsal views, anterior to the top, at the 7 somite (A–F) and 30 hpf (G–I) stages. Scale bars represent 100 μm. (A–C) Overexpression of scl and etsrp induces ectopic expression of fli1a, including expression within the HFR (arrowheads; severe phenotype, n=7/15; moderate phenotype, n=5/15). (D–F) RNA-injected embryos exhibit reduced expression of hand2 (arrows; severe phenotype, n=14/34; moderate phenotype, n=15/34). (G–I) RNA-injected embryos exhibit small, dysmorphic hearts (severe phenotype, n=9/11; moderate phenotype, n=2/11).
DISCUSSION
Taken together, our data reveal a previously unrecognized mechanism for limiting heart size: components of the vessel and blood specification pathways repress formation of cardiac progenitors and thereby restrict the dimensions of the heart-forming region. In the early zebrafish embryo, the entire ALPM possesses cardiac developmental potential. However, not all of this potential is fulfilled. Only the hand2-expressing portion of the ALPM produces myocardium, whereas the rostral territory gives rise to endothelium and myeloid cells. We find that this subdivision of the ALPM is mediated by clo and its downstream effectors scl and etsrp. In addition to promoting vessel and blood specification, these genes also inhibit cardiac specification within rostral ALPM. Thus, the induction of vessel and blood lineages creates a rostral boundary for cardiogenesis within a larger heart field.
Our findings indicate a close relationship between myocardial, endothelial, and hematopoietic lineages during their development. Consistent with this, several studies in mouse have indicated a common origin for multiple cardiovascular lineages. Lineage analysis in vivo and analysis of progenitor differentiation in vitro have suggested that myocardial, endothelial, and hematopoietic lineages share a common progenitor expressing Flk1 (Ema et al., 2006; Motoike et al., 2003), and recent experiments employing ES cell differentiation in vitro have shown that Flk1-expressing progenitors give rise to myocardium, endothelium and vascular smooth muscle (Kattman et al., 2006). Other complementary experiments indicate that myocardium, endothelium, and vascular smooth muscle can all arise from progenitors expressing Isl1, Nkx2-5, and Flk1 (Moretti et al., 2006). Despite the wealth of evidence supporting the existence of multipotent cardiovascular progenitors, it is not yet understood how individual progenitors become committed to distinct cardiovascular fates. Our work sheds light on the regulation of these lineage decisions: induction of genes like scl and etsrp is a critical mechanism, not only for promoting vasculogenesis and hematopoiesis, but also for repressing cardiogenesis.
The precise molecular interface between the vessel, blood, and cardiac specification pathways will be an important focus for future studies. It is possible that Scl and/or Etsrp could be directly involved in repressing expression of myocardial genes. Scl can act either as a repressor or an activator depending on the context of its protein-protein interactions (Lecuyer and Hoang, 2004), and in vitro assays have suggested that Scl is capable of repressing muscle differentiation by disrupting the activities of other bHLH transcription factors (Hofmann and Cole, 1996). However, the relationship between specification pathways could also be less direct and more complex, with genes downstream of Scl and Etsrp repressing genes upstream of those required for myocardial specification. Elucidation of the partners and targets of Scl and Etsrp will help to illuminate the transcriptional networks that link cardiovascular specification pathways.
In addition to revealing an important interaction between cardiovascular pathways, our studies highlight the developmental plasticity inherent to the rostral portion of the zebrafish ALPM. The rostral ALPM has previously been implicated in facilitating repair of the zebrafish HFR following its partial ablation (Serbedzija et al., 1998). The multipotency of the zebrafish rostral ALPM also suggests an appealing mechanism for facilitating the alteration of heart size and structure during the course of vertebrate evolution. The amniote heart is significantly larger and more complex than the teleost heart, but it is not yet clear what regulatory changes made these structural modifications possible (Olson, 2006). A recent study in Ciona has indicated that the addition of more cells to the myocardial progenitor pool can provide opportunities for structural innovation, such as the addition of a cardiac chamber (Davidson et al., 2006). Our findings suggest that a reservoir of latent cardiac potential, akin to that found in the zebrafish rostral ALPM, could have provided the raw material necessary to elaborate upon the evolving heart. Even slight changes in the regulation of anterior vasculogenesis and hematopoiesis could have exposed a new source of myocardial progenitors in an ancestral vertebrate. This surplus could have led to augmentation of the HFR, allowing development of a larger organ, as seen in our experiments. Furthermore, we speculate that changes in utilization of a latent reservoir of cardiac potential could have provided the origin of the amniote second heart field, allowing enhanced structural complexity through the addition of new cardiac chambers.
In conclusion, our data reveal an important regulatory relationship between cardiovascular specification pathways. The repression of myocardial specification by vascular and hematopoietic pathways coordinates development of the cardiovascular system, both by regulating the divergence of lineages from a common progenitor and by restricting heart size. Importantly, our insight into the interplay between cardiovascular pathways could have a significant impact on strategies for regenerative medicine. Specifically, our studies suggest that interference with Scl and Etsrp function could be an effective means of directing multipotent cardiovascular progenitor cells toward myocardial differentiation. Thus, this work will likely motivate new approaches for harnessing the developmental potential of cardiovascular progenitor cells to facilitate cellular therapies for damaged cardiac muscle.
EXPERIMENTAL PROCEDURES
Mutations, morpholinos and RNA injections
To generate mutant embryos, we intercrossed heterozygotes for the mutations hans6 (Yelon et al., 2000), clos5 (Field et al., 2003), and closk4. The closk4 allele was isolated as a spontaneous mutation in our zebrafish facility; closk4 mutants appear phenotypically identical to clos5 mutants.
All MOs used in our studies were previously characterized and shown to be reasonably effective and specific (Juarez et al., 2005; Sumanas and Lin, 2006). All MOs behaved as previously reported, as assessed by the loss of vessels and blood. To knock down scl, we injected 12.5 ng of a 2:3 mix of E2I2 and E3I3 anti-scl MOs (Juarez et al., 2005). To knock down etsrp, we injected 12 ng of a 1:1 mix of anti-etsrp MO1 and MO2 (Sumanas and Lin, 2006). For anti-scl+etsrp MOs, we injected 12 ng of a 1:1 combination of the mixes described above.
Previously described plasmids were used to synthesize scl and etsrp RNA (Dooley et al.,2005; Pham et al., 2006). Embryos were injected with a combination of 40–50 ng of scl RNA and 4–8 ng of etsrp RNA; injection of these amounts results in variable and mosaic phenotypes but avoids toxicity and severe malformations.
All care and use of zebrafish and embryos complied with local animal welfare laws, guidelines, and policies, as described in animal protocol #060911-01, approved by the New York University School of Medicine Institutional Animal Care and Use Committee.
In situ hybridization
Single-color wholemount in situ hybridization was performed as previously described (Yelon et al., 1999). Two-color fluorescent hybridization was conducted according to a Yelon lab protocol that is optimized for difficult riboprobes and is available upon request. Briefly, we used pairs of riboprobes, one labeled with digoxigenin (DIG) and the other labeled with dinitrophenol (DNP). DNP probes were synthesized as previously described (Long and Rebagliati, 2002) using a DNP Label-It kit (Mirus). DNP incorporation was enhanced by extending the labeling incubation to 2.5 hours and by using 1.5 μL of labeling reagent per μg of RNA. Following probe hybridization, washes, and blocking, embryos were incubated overnight at 4°C with anti-DNP Fab fragments conjugated to horseradish peroxidase (HRP) (PerkinElmer, 1:400 dilution). Embryos were then incubated with DNP tyramide (PerkinElmer, 1:25 dilution, 30 minutes at room temperature) to amplify DNP deposition. Next, stringent conditions were used to insure removal of all HRP activity. Embryos were washed in PBT (PBS, 0.1% Tween-20) containing 3% peroxide (4 times, 15 minutes per wash) and then in 0.1 M glycine, pH 2.2 with 0.1% Tween-20 (4 times, 15 minutes per wash). Finally, embryos were washed in PBT, blocked, and incubated overnight at 4°C with anti-DIG HRP-conjugated Fab fragments (Roche, 1:400 dilution). Embryos were then incubated with fluorescein (FLU) tyramide (PerkinElmer), followed by removal of HRP activity, as above. Next, we conducted a second round of tyramide deposition. Embryos were first incubated with anti-DNP HRP-conjugated Fab fragments, followed by incubation with Cy3 tyramide (PerkinElmer, 1:25 dilution, 30 minutes at room temperature). After removal of HRP activity, embryos were incubated with anti-FLU HRP-conjugated Fab fragments, followed by incubation with FLU tyramide. After a final set of PBT washes, embryos were stored in SlowFade Antifade solution (Invitrogen).
Photoactivation of fluorescein in the ALPM
Caged fluorescein dextran was prepared and injected as previously described (Keegan et al., 2004), with the exception of being injected directly into the single cell of newly fertilized embryos. When injected embryos reached the 6–9 somite stage, they were mounted dorsal side up in a well gouged from 2% agarose and filled with embryo water. Caged fluorescein was photoactivated in clusters of 10–15 ALPM cells by exposure to 1 second pulses from a 375 nm pulsed nitrogen laser (Micropoint Laser System, Photonic Instruments). Laser light was focused through a 40x water immersion objective on a Zeiss Axioplan 2 microscope equipped with a cooled CCD camera (Pentamax) and automated shutters (Uniblitz). For lateral ALPM labeling, we avoided labeling cells located more than 4 cell widths from the lateral edge of the ALPM. To label medial ALPM, we labeled cells that were at least 4 cell widths interior to the lateral edge of the ALPM.
Fluorescein detection and scoring of labeled cells
Labeled embryos were fixed at the desired stage: either immediately (Fig. 2E,F,H,I), at 30 hpf (Fig.4G,H), or at 44 hpf (Figs. 2J–L, 5L). clo mutants were fixed at 44 hpf, our lab’s standard stage for scoring labeled cells (Keegan et al., 2004). However, han mutants were fixed at 30 hpf, since the han mutant myocardium remains under the hindbrain at 44 hpf and is therefore obscured by pigmentation. Wild-type data represent a combination of embryos fixed at 30 and 44 hpf.
Following fixation, embryos were processed by in situ hybridization to create a counterstain for fluorescein detection. For embryos fixed at 30 and 44 hpf, cmlc2 expression was visualized as previously described (Keegan et al., 2004) using the substrate 5-Bromo-6-Chloro-3-Indolyl Phosphate (‘magenta phos’; Sigma B-5667). Cells labeled with uncaged fluorescein were then detected by immunohistochemistry and scored for tissue contributions based on location, morphology, and counterstaining, as previously described (Keegan et al., 2004). In particular, we used morphological criteria to distinguish myocardial cells, which are relatively round and tend to be oriented circumferentially around the heart, from endocardial cells, which are relatively elongated and tend to project longitudinally. Additionally, we assessed localization of labeled cells relative to the myocardial counterstain. The number of labeled progeny detected varied between individual embryos. Contributions to pharyngeal pouches ranged between 5 and 50 cells and contributions to myocardium or endocardium ranged between 1 and 30 cells.
For embryos fixed immediately after labeling, we used our two-color fluorescent in situ hybridization technique to counterstain for ALPM markers (scl or nkx2.5) and to detect uncaged fluorescein. Briefly, DIG riboprobe was detected with anti-DIG HRP-conjugated Fab fragments, incubation with DNP tyramide, and a second round of tyramide depostion with anti-DNP HRP-conjugated Fab fragments and Cy3 tyramide. Then, uncaged fluorescein was detected with anti-FLU HRP-conjugated Fab fragments and a FLU tyramide reaction, with an optional second round of tyramide deposition.
Cardiomyocyte counting
To count cardiomyocytes, we employed the transgene Tg(cmlc2:DsRed2-nuc) (Mably et al., 2003), as described in a separate manuscript (J.S. Waxman, B.R.K., and D.Y., manuscript submitted). Briefly, we processed transgenic embryos using a standard immunofluorescence protocol (Alexander et al., 1998), the primary antibodies S46 (anti-Amhc; 1:20) and anti-DsRed (1:4000; Clontech), and the secondary antibodies goat anti-mouse IgG1 FITC (1:100, Southern Biotechnology Associates, Inc.) and donkey anti-rabbit Alexa 594 (1:1000, Invitrogen). We then counted the red fluorescent nuclei in each cardiac chamber.
Imaging
Fluorescent in situ hybridization images were captured with a Zeiss LSM 510 confocal microscope and processed with Zeiss LSM 510 and Adobe Photoshop 7 software. Capture of DIC and FITC images of live embryos was controlled by Metamorph Imaging System software (v6.1r3, Universal Imaging). All other embryos were photographed with Zeiss M2Bio and Axioplan microscopes outfitted with Zeiss Axiocam digital cameras; images were processed with Zeiss AxioVision (v3.0.6) and Adobe Photoshop 7 software.
Supplementary Material
Acknowledgments
We thank A. Schier, J. Torres-Vazquez, B. Ciruna, H. Knaut, and members of the Yelon lab for helpful discussions, and B. Weinstein and L. Zon for providing reagents. This work was supported by NIH RO1 HL069594. J.J.S. and B.R.K. received support from the NYU Graduate Training Program in Developmental Genetics (NIH T32 HD007520), and B.R.K. received support from the Medical Scientist Training Program of NYU School of Medicine. The authors have no financial conflicts of interest to disclose.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Alexander J, Stainier DY, Yelon D. Screening mosaic F1 females for mutations affecting zebrafish heart induction and patterning. Dev Genet. 1998;22:288–299. doi: 10.1002/(SICI)1520-6408(1998)22:3<288::AID-DVG10>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- Berdougo E, Coleman H, Lee DH, Stainier DY, Yelon D. Mutation of weak atrium/atrial myosin heavy chain disrupts atrial function and influences ventricular morphogenesis in zebrafish. Development. 2003;130:6121–6129. doi: 10.1242/dev.00838. [DOI] [PubMed] [Google Scholar]
- Buckingham M, Meilhac S, Zaffran S. Building the mammalian heart from two sources of myocardial cells. Nat Rev Genet. 2005;6:826–835. doi: 10.1038/nrg1710. [DOI] [PubMed] [Google Scholar]
- Cai CL, Liang X, Shi Y, Chu PH, Pfaff SL, Chen J, Evans S. Isl1 identifies a cardiac progenitor population that proliferates prior to differentiation and contributes a majority of cells to the heart. Dev Cell. 2003;5:877–889. doi: 10.1016/s1534-5807(03)00363-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collop AH, Broomfield JA, Chandraratna RA, Yong Z, Deimling SJ, Kolker SJ, Weeks DL, Drysdale TA. Retinoic acid signaling is essential for formation of the heart tube in Xenopus. Dev Biol. 2006;291:96–109. doi: 10.1016/j.ydbio.2005.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson B, Shi W, Beh J, Christiaen L, Levine M. FGF signaling delineates the cardiac progenitor field in the simple chordate, Ciona intestinalis. Genes Dev. 2006;20:2728–2738. doi: 10.1101/gad.1467706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dooley KA, Davidson AJ, Zon LI. Zebrafish scl functions independently in hematopoietic and endothelial development. Dev Biol. 2005;277:522–536. doi: 10.1016/j.ydbio.2004.09.004. [DOI] [PubMed] [Google Scholar]
- Drysdale TA, Patterson KD, Saha M, Krieg PA. Retinoic acid can block differentiation of the myocardium after heart specification. Dev Biol. 1997;188:205–215. doi: 10.1006/dbio.1997.8623. [DOI] [PubMed] [Google Scholar]
- Ema M, Takahashi S, Rossant J. Deletion of the selection cassette, but not cis-acting elements, in targeted Flk1-lacZ allele reveals Flk1 expression in multipotent mesodermal progenitors. Blood. 2006;107:111–117. doi: 10.1182/blood-2005-05-1970. [DOI] [PubMed] [Google Scholar]
- Field HA, Dong PD, Beis D, Stainier DY. Formation of the digestive system in zebrafish. II. Pancreas morphogenesis. Dev Biol. 2003;261:197–208. doi: 10.1016/s0012-1606(03)00308-7. [DOI] [PubMed] [Google Scholar]
- Garriock RJ, Drysdale TA. Regulation of heart size in Xenopus laevis. Differentiation. 2003;71:506–515. doi: 10.1046/j.1432-0436.2003.7108005.x. [DOI] [PubMed] [Google Scholar]
- Gering M, Rodaway AR, Gottgens B, Patient RK, Green AR. The SCL gene specifies haemangioblast development from early mesoderm. Embo J. 1998;17:4029–4045. doi: 10.1093/emboj/17.14.4029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gering M, Yamada Y, Rabbitts TH, Patient RK. Lmo2 and Scl/Tal1 convert non-axial mesoderm into haemangioblasts which differentiate into endothelial cells in the absence of Gata1. Development. 2003;130:6187–6199. doi: 10.1242/dev.00875. [DOI] [PubMed] [Google Scholar]
- Goldstein AM, Fishman MC. Notochord regulates cardiac lineage in zebrafish embryos. Dev Biol. 1998;201:247–252. doi: 10.1006/dbio.1998.8976. [DOI] [PubMed] [Google Scholar]
- Grego-Bessa J, Luna-Zurita L, del Monte G, Bolos V, Melgar P, Arandilla A, Garratt AN, Zang H, Mukouyama YS, Chen H, et al. Notch signaling is essential for ventricular chamber development. Dev Cell. 2007;12:415–429. doi: 10.1016/j.devcel.2006.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harvey RP. Patterning the vertebrate heart. Nat Rev Genet. 2002;3:544–556. doi: 10.1038/nrg843. [DOI] [PubMed] [Google Scholar]
- Herbomel P, Thisse B, Thisse C. Ontogeny and behaviour of early macrophages in the zebrafish embryo. Development. 1999;126:3735–3745. doi: 10.1242/dev.126.17.3735. [DOI] [PubMed] [Google Scholar]
- Hochgreb T, Linhares VL, Menezes DC, Sampaio AC, Yan CY, Cardoso WV, Rosenthal N, Xavier-Neto J. A caudorostral wave of RALDH2 conveys anteroposterior information to the cardiac field. Development. 2003;130:5363–5374. doi: 10.1242/dev.00750. [DOI] [PubMed] [Google Scholar]
- Hofmann TJ, Cole MD. The TAL1/Scl basic helix-loop-helix protein blocks myogenic differentiation and E-box dependent transactivation. Oncogene. 1996;13:617–624. [PubMed] [Google Scholar]
- Hsu K, Traver D, Kutok JL, Hagen A, Liu TX, Paw BH, Rhodes J, Berman JN, Zon LI, Kanki JP, Look AT. The pu.1 promoter drives myeloid gene expression in zebrafish. Blood. 2004;104:1291–1297. doi: 10.1182/blood-2003-09-3105. [DOI] [PubMed] [Google Scholar]
- Huxley J, De Beer GR. The elements of experimental embryology. Cambridge [Eng.]: The University press; 1934. [Google Scholar]
- Jacobson AG, Sater AK. Features of embryonic induction. Development. 1988;104:341–359. doi: 10.1242/dev.104.3.341. [DOI] [PubMed] [Google Scholar]
- Juarez MA, Su F, Chun S, Kiel MJ, Lyons SE. Distinct roles for SCL in erythroid specification and maturation in zebrafish. J Biol Chem. 2005;280:41636–41644. doi: 10.1074/jbc.M507998200. [DOI] [PubMed] [Google Scholar]
- Kattman SJ, Huber TL, Keller GM. Multipotent flk-1+ cardiovascular progenitor cells give rise to the cardiomyocyte, endothelial, and vascular smooth muscle lineages. Dev Cell. 2006;11:723–732. doi: 10.1016/j.devcel.2006.10.002. [DOI] [PubMed] [Google Scholar]
- Keegan BR, Feldman JL, Begemann G, Ingham PW, Yelon D. Retinoic acid signaling restricts the cardiac progenitor pool. Science. 2005;307:247–249. doi: 10.1126/science.1101573. [DOI] [PubMed] [Google Scholar]
- Keegan BR, Meyer D, Yelon D. Organization of cardiac chamber progenitors in the zebrafish blastula. Development. 2004;131:3081–3091. doi: 10.1242/dev.01185. [DOI] [PubMed] [Google Scholar]
- Kelly RG, Brown NA, Buckingham ME. The arterial pole of the mouse heart forms from Fgf10-expressing cells in pharyngeal mesoderm. Dev Cell. 2001;1:435–440. doi: 10.1016/s1534-5807(01)00040-5. [DOI] [PubMed] [Google Scholar]
- Laflamme MA, Murry CE. Regenerating the heart. Nat Biotechnol. 2005;23:845–856. doi: 10.1038/nbt1117. [DOI] [PubMed] [Google Scholar]
- Lavine KJ, Yu K, White AC, Zhang X, Smith C, Partanen J, Ornitz DM. Endocardial and epicardial derived FGF signals regulate myocardial proliferation and differentiation in vivo. Dev Cell. 2005;8:85–95. doi: 10.1016/j.devcel.2004.12.002. [DOI] [PubMed] [Google Scholar]
- Lecuyer E, Hoang T. SCL: from the origin of hematopoiesis to stem cells and leukemia. Exp Hematol. 2004;32:11–24. doi: 10.1016/j.exphem.2003.10.010. [DOI] [PubMed] [Google Scholar]
- Liao EC, Paw BH, Oates AC, Pratt SJ, Postlethwait JH, Zon LI. SCL/Tal-1 transcription factor acts downstream of cloche to specify hematopoietic and vascular progenitors in zebrafish. Genes Dev. 1998;12:621–626. doi: 10.1101/gad.12.5.621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao W, Bisgrove BW, Sawyer H, Hug B, Bell B, Peters K, Grunwald DJ, Stainier DYR. The zebrafish gene cloche acts upstream of a flk-1 homologue to regulate endothelial cell differentiation. Development. 1997;124:381–389. doi: 10.1242/dev.124.2.381. [DOI] [PubMed] [Google Scholar]
- Liao W, Ho CY, Yan YL, Postlethwait J, Stainier DY. Hhex and scl function in parallel to regulate early endothelial and blood differentiation in zebrafish. Development. 2000;127:4303–4313. doi: 10.1242/dev.127.20.4303. [DOI] [PubMed] [Google Scholar]
- Lickert H, Kutsch S, Kanzler B, Tamai Y, Taketo MM, Kemler R. Formation of multiple hearts in mice following deletion of beta-catenin in the embryonic endoderm. Dev Cell. 2002;3:171–181. doi: 10.1016/s1534-5807(02)00206-x. [DOI] [PubMed] [Google Scholar]
- Lieschke GJ, Oates AC, Paw BH, Thompson MA, Hall NE, Ward AC, Ho RK, Zon LI, Layton JE. Zebrafish SPI-1 (PU.1) marks a site of myeloid development independent of primitive erythropoiesis: implications for axial patterning. Dev Biol. 2002;246:274–295. doi: 10.1006/dbio.2002.0657. [DOI] [PubMed] [Google Scholar]
- Long S, Rebagliati M. Sensitive two-color whole-mount in situ hybridizations using digoxygenin- and dinitrophenol-labeled RNA probes. Biotechniques. 2002;32:494–498. doi: 10.2144/02323bm04. [DOI] [PubMed] [Google Scholar]
- Mably JD, Mohideen MA, Burns CG, Chen JN, Fishman MC. heart of glass regulates the concentric growth of the heart in zebrafish. Curr Biol. 2003;13:2138–2147. doi: 10.1016/j.cub.2003.11.055. [DOI] [PubMed] [Google Scholar]
- Marvin MJ, Di Rocco G, Gardiner A, Bush SM, Lassar AB. Inhibition of Wnt activity induces heart formation from posterior mesoderm. Genes Dev. 2001;15:316–327. doi: 10.1101/gad.855501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mjaatvedt CH, Nakaoka T, Moreno-Rodriguez R, Norris RA, Kern MJ, Eisenberg CA, Turner D, Markwald RR. The outflow tract of the heart is recruited from a novel heart-forming field. Dev Biol. 2001;238:97–109. doi: 10.1006/dbio.2001.0409. [DOI] [PubMed] [Google Scholar]
- Moretti A, Caron L, Nakano A, Lam JT, Bernshausen A, Chen Y, Qyang Y, Bu L, Sasaki M, Martin-Puig S, et al. Multipotent embryonic isl1+ progenitor cells lead to cardiac, smooth muscle, and endothelial cell diversification. Cell. 2006;127:1151–1165. doi: 10.1016/j.cell.2006.10.029. [DOI] [PubMed] [Google Scholar]
- Motoike T, Markham DW, Rossant J, Sato TN. Evidence for novel fate of Flk1+ progenitor: contribution to muscle lineage. Genesis. 2003;35:153–159. doi: 10.1002/gene.10175. [DOI] [PubMed] [Google Scholar]
- Olson EN. Gene regulatory networks in the evolution and development of the heart. Science. 2006;313:1922–1927. doi: 10.1126/science.1132292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osmond MK, Butler AJ, Voon FC, Bellairs R. The effects of retinoic acid on heart formation in the early chick embryo. Development. 1991;113:1405–1417. doi: 10.1242/dev.113.4.1405. [DOI] [PubMed] [Google Scholar]
- Parker L, Stainier DY. Cell-autonomous and non-autonomous requirements for the zebrafish gene cloche in hematopoiesis. Development. 1999;126:2643–2651. doi: 10.1242/dev.126.12.2643. [DOI] [PubMed] [Google Scholar]
- Patterson LJ, Gering M, Patient R. Scl is required for dorsal aorta as well as blood formation in zebrafish embryos. Blood. 2005;105:3502–3511. doi: 10.1182/blood-2004-09-3547. [DOI] [PubMed] [Google Scholar]
- Pham VN, Lawson ND, Mugford JW, Dye L, Castranova D, Lo B, Weinstein BM. Combinatorial function of ETS transcription factors in the developing vasculature. Dev Biol. 2006 doi: 10.1016/j.ydbio.2006.10.030. 2006 Oct 25; [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raffin M, Leong LM, Rones MS, Sparrow D, Mohun T, Mercola M. Subdivision of the cardiac Nkx2.5 expression domain into myogenic and nonmyogenic compartments. Dev Biol. 2000;218:326–340. doi: 10.1006/dbio.1999.9579. [DOI] [PubMed] [Google Scholar]
- Redkar A, Montgomery M, Litvin J. Fate map of early avian cardiac progenitor cells. Development. 2001;128:2269–2279. doi: 10.1242/dev.128.12.2269. [DOI] [PubMed] [Google Scholar]
- Rones MS, McLaughlin KA, Raffin M, Mercola M. Serrate and Notch specify cell fates in the heart field by suppressing cardiomyogenesis. Development. 2000;127:3865–3876. doi: 10.1242/dev.127.17.3865. [DOI] [PubMed] [Google Scholar]
- Sater AK, Jacobson AG. The restriction of the heart morphogenetic field in Xenopus laevis. Dev Biol. 1990;140:328–336. doi: 10.1016/0012-1606(90)90083-u. [DOI] [PubMed] [Google Scholar]
- Schneider VA, Mercola M. Wnt antagonism initiates cardiogenesis in Xenopus laevis. Genes Dev. 2001;15:304–315. doi: 10.1101/gad.855601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serbedzija GN, Chen JN, Fishman MC. Regulation in the heart field of zebrafish. Development. 1998;125:1095–1101. doi: 10.1242/dev.125.6.1095. [DOI] [PubMed] [Google Scholar]
- Stainier DY, Weinstein BM, Detrich HW, 3rd, Zon LI, Fishman MC. Cloche, an early acting zebrafish gene, is required by both the endothelial and hematopoietic lineages. Development. 1995;121:3141–3150. doi: 10.1242/dev.121.10.3141. [DOI] [PubMed] [Google Scholar]
- Sumanas S, Jorniak T, Lin S. Identification of novel vascular endothelial-specific genes by the microarray analysis of the zebrafish cloche mutants. Blood. 2005;106:534–541. doi: 10.1182/blood-2004-12-4653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sumanas S, Lin S. Ets1-related protein is a key regulator of vasculogenesis in zebrafish. PLoS Biol. 2006;4:e10. doi: 10.1371/journal.pbio.0040010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson MA, Ransom DG, Pratt SJ, MacLennan H, Kieran MW, Detrich HW, 3rd, Vail B, Huber TL, Paw B, Brownlie AJ, et al. The cloche and spadetail genes differentially affect hematopoiesis and vasculogenesis. Dev Biol. 1998;197:248–269. doi: 10.1006/dbio.1998.8887. [DOI] [PubMed] [Google Scholar]
- van Laake LW, Hassink R, Doevendans PA, Mummery C. Heart repair and stem cells. J Physiol. 2006;577:467–478. doi: 10.1113/jphysiol.2006.115816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waldo KL, Kumiski DH, Wallis KT, Stadt HA, Hutson MR, Platt DH, Kirby ML. Conotruncal myocardium arises from a secondary heart field. Development. 2001;128:3179–3188. doi: 10.1242/dev.128.16.3179. [DOI] [PubMed] [Google Scholar]
- Yelon D, Horne SA, Stainier DY. Restricted expression of cardiac myosin genes reveals regulated aspects of heart tube assembly in zebrafish. Dev Biol. 1999;214:23–37. doi: 10.1006/dbio.1999.9406. [DOI] [PubMed] [Google Scholar]
- Yelon D, Ticho B, Halpern ME, Ruvinsky I, Ho RK, Silver LM, Stainier DY. The bHLH transcription factor hand2 plays parallel roles in zebrafish heart and pectoral fin development. Development. 2000;127:2573–2582. doi: 10.1242/dev.127.12.2573. [DOI] [PubMed] [Google Scholar]
- Yutzey KE, Kirby ML. Wherefore heart thou? Embryonic origins of cardiogenic mesoderm. Dev Dyn. 2002;223:307–320. doi: 10.1002/dvdy.10068. [DOI] [PubMed] [Google Scholar]
- Yutzey KE, Rhee JT, Bader D. Expression of the atrial-specific myosin heavy chain AMHC1 and the establishment of anteroposterior polarity in the developing chicken heart. Development. 1994;120:871–883. doi: 10.1242/dev.120.4.871. [DOI] [PubMed] [Google Scholar]
- Zaffran S, Kelly RG, Meilhac SM, Buckingham ME, Brown NA. Right ventricular myocardium derives from the anterior heart field. Circ Res. 2004;95:261–268. doi: 10.1161/01.RES.0000136815.73623.BE. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.