Abstract
We have developed a highly sensitive and high-throughput method for the simultaneous analysis of 43 molecular species of cytokinins, auxins, ABA and gibberellins. This method consists of an automatic liquid handling system for solid phase extraction and ultra-performance liquid chromatography (UPLC) coupled with a tandem quadrupole mass spectrometer (qMS/MS) equipped with an electrospray interface (ESI; UPLC-ESI-qMS/MS). In order to improve the detection limit of negatively charged compounds, such as gibberellins, we chemically derivatized fractions containing auxin, ABA and gibberellins with bromocholine that has a quaternary ammonium functional group. This modification, that we call ‘MS-probe’, makes these hormone derivatives have a positive ion charge and permits all compounds to be measured in the positive ion mode with UPLC-ESI-qMS/MS in a single run. Consequently, quantification limits of gibberellins increased up to 50-fold. Our current method needs <100 mg (FW) of plant tissues to determine phytohormone profiles and enables us to analyze >180 plant samples simultaneously. Application of this method to plant hormone profiling enabled us to draw organ distribution maps of hormone species in rice and also to identify interactions among the four major hormones in the rice gibberellin signaling mutants, gid1-3, gid2-1 and slr1. Combining the results of hormone profiling data with transcriptome data in the gibberellin signaling mutants allows us to analyze relationships between changes in gene expression and hormone metabolism.
Keywords: ABA, Auxins, Cytokinins, Gibberellins, Mass spectrometry, Oryza sativa
Introduction
Plant hormones play an important role as signaling molecules in the regulation of almost all phases of plant development, from embryogenesis to senescence. In the past few decades, many structurally and chemically diverse low molecular weight compounds have been identified as plant hormones, including auxins, cytokinins, ABA, gibberellins, ethylene, brassinosteroids, jasmonates and salicylic acid (Davies 2004). In addition to the ‘traditional’ hormones, strigolactone, a terpenoid, has been identified recently as a novel plant hormone that inhibits shoot branching (Gomez-Roldan et al. 2008, Umehara et al. 2008). Furthermore, small peptides also play key roles in regulating plant growth and development as signaling molecules (Matsubayashi and Sakagami 2006, Fukuda et al. 2007). Each hormone is perceived by its receptor, and the signal is communicated to the target genes via components of the signaling system (Davies 2004, Chow and McCourt 2006). Current understanding of hormone action is based on the premise that the signaling systems build a network and mutually regulate signaling and metabolic systems, such as the interaction between cytokinins and auxins (Tanaka et al. 2006), auxins and brassinosteroids (Goda et al. 2004, Nemhauser et al. 2004), ethylene, ABA and gibberellins (Gazzarrini and McCourt 2001), ethylene and cytokinins (Cary et al. 1995), auxins and gibberellins (Ross et al. 2003), and cytokinins, auxins and gibberellins (Bai and DeMason 2006), rather than an array of parallel and independent routes of signal processing.
The recent ‘omics’ trend has provided us with a large amount of data for understanding the network of plant hormone action. Transcriptome analyses using microarray technology have enabled us to shed light on the regulatory networks among auxins, cytokinins, gibberellins, brassinosteroid, ABA, jasmonate and ethylene (Nemhauser et al. 2006, Goda et al. 2008). Proteome analyses of hormone responses have also begun to illustrate regulatory processes in a variety of hormone-related biological processes at the protein level (Tanaka et al. 2004, Pawlowski 2007, Segarra et al. 2007). In order to understand better the network regulation of hormone action, we must be able to measure multiple hormone concentrations simultaneously, i.e. characterize the ‘hormone-metabolome’ or ‘hormonome’.
Compared with transcriptome and proteome analyses, exhaustive analysis of plant hormone contents is much more difficult because hormones have different chemical and structural properties, and because their concentrations are very low, usually at the nanomolar level. There have been several reports of methods for simultaneous quantification of multiple plant hormones. Müller et al. (2002) reported a multiplex gas chromatography–tandem mass spectrometry technique for analysis of acidic plant hormones and related compounds, namely IAA, ABA, salicylic acid, jasmonate and 12-oxo-phytodienoic acid methyl ester, from 20–200 mg (FW) samples and produced whole-plant organ distribution maps for the compounds in Arabidopsis thaliana. Chiwocha et al. (2003) reported a method for simultaneous analysis of ABA and four related metabolites, IAA and indole-3-acetyl-l-aspartic acid (IA-Asp), four cytokinins [trans-zeatin (tZ), tZ riboside (tZR), N6-(Δ2-isopentenyl)adenine (iP) and iP riboside (iPR)] and four gibberellins (GA1, GA3, GA4 and GA7) using a HPLC–tandem quadrupole mass spectrometer (qMS/MS) on extracts from 50–100 mg DW samples, and analyzed the hormone content changes related to the thermodormancy of lettuce seeds.
It is primarily important to know the concentrations of active forms of plant hormones, such as tZ and iP, IAA, ABA, GA1 and GA4; however, in some cases, the concentrations of the active forms that might be changed by environmental stimuli or genetic variation are apparently stable due to homeostatic regulation of hormone metabolism. Thus, in addition to the active forms, measuring the concentrations of their metabolites and precursors often gives us clues for understanding the regulation of hormone action and metabolism. Furthermore, quantification sensitivity and throughput are also important factors for fully realizing the technology of ‘hormonomics’, in which the hormone content of a large number of small amounts of plant samples must be measured. In terms of the sensitivity, a problem in mass spectrometry analysis is that quantification of negatively charged compounds, such as gibberellins, often fails because the negative ion mode of the mass spectrometer is generally much less sensitive than the positive ion mode.
In this study, we established a highly sensitive and high-throughput method for the simultaneous analysis of 43 molecular species of cytokinins, auxins, ABA and gibberellins that are the major ‘traditional’ plant hormones. Introduction of chemical derivatization with bromocholine, a process we refer to as ‘MS-probe’, dramatically improved the quantification limits of gibberellins. Using this technology, we characterized steady-state accumulation levels of these hormone species in various organs and gibberellin signaling mutants in rice. We also showed that simultaneous hormonome and transcriptome analyses of a set of mutants enabled us to see relationships between changes in expression of the hormone-related genes and metabolism of the hormones.
Results
Method for high-throughput extraction and fractionation of plant hormone species
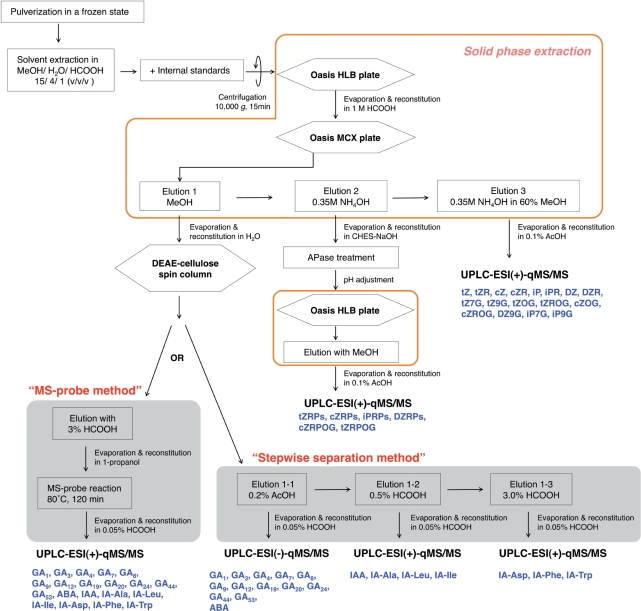
The overall procedure for extraction and fractionation of plant hormones, cytokinins, auxins, ABA and gibberellins, is summarized in Fig. 1. This method consists of multiparallel, microscale ball-milling (up to 48 samples per run) for tissue pulverization and an automatic liquid handling system for solid phase extraction (up to 192 samples per run). In the second solid phase extraction step, which was originally described by Dobrev and Kaminek (2002), IAA and ABA are recovered in the eluate with methanol (Elution 1; Fig. 1), cytokinin nucleotides (5′-monophosphates) are eluted with 0.35 M ammonia (Elution 2), and cytokinin nucleobases, nucleosides and glucosides are eluted with 0.35 M ammonia in 60% methanol (Elution 3). We examined the separation and distribution of 43 molecular species that included cytokinins (23 species), auxins (seven species), ABA (one species) and gibberellins (12 species), and found that gibberellins and amino acid conjugates of IAA are recovered in the methanol eluate as well as IAA and ABA (Elution 1, Fig. 1). For the cytokinin nucleotides, iPR 5′-monophosphate (iPRMP), iPR 5′-diphosphate (iPRDP) and iPR 5′-triphosphate (iPRTP) were recovered in the 0.35 M ammonia eluate (Elution 2) with similar efficiency (Supplementary Fig. S1).
Fig. 1.
Schematic representation of the extraction and purification protocol for the high-throughput and highly sensitive hormone analysis system. Hexagonal boxes represent a separation column, quadrangular boxes represent other handling processes. Processes enclosed in orange lines are performed with an automated liquid handling system for solid phase extraction. Compounds analyzed in each UPLC-ESI-qMS/MS are shown in blue letters. AcOH, acetic acid; APase, alkaline phosphatase; CHES, N-cyclohexyl-2-aminoethanesulfonic acid; MeOH, methanol; tZ, trans-zeatin; tZR, tZ riboside; cZ, cis-zeatin; cZR, cZ riboside; iP, N6-(Δ2-isopentenyl)adenine; iPR, iP riboside; DZ, dihydrozeatin; DZR, DZ riboside; tZ7G, tZ-7-N-glucoside; tZ9G, tZ-9-N-glucoside; tZOG, tZ-O-glucoside; tZROG, tZR-O-glucoside; cZOG, cZ-O-glucoside; cZROG, cZR-O-glucoside; DZ9G, DZ-9-N-glucoside; iP7G, iP-7-N-glucoside; iP9G, iP-9-N-glucoside; tZRPs, tZR phosphates; cZRPs, cZR phosphates; iPRPs, iPR phosphates; DZRPs, DZR phosphates; cZRPOG, cZR phosphate-O-glucoside; tZRPOG, tZR phosphate-O-glucoside; IA-Ala, indole-3-acetyl-l-Ala; IA-Leu, indole-3-acetyl-l-Leu; IA-Ile, indole-3-acetyl-l-Ile; IA-Asp, indole-3-acetyl-l-Asp; IA-Phe, indole-3-acetyl-l-Phe; IA-Trp, indole-3-acetyl-l-Trp.
The cytokinin fractions can be subjected to ultra-performance liquid chromatography (UPLC) coupled to a qMS/MS equipped with an electrospray interface (ESI; UPLC-ESI-qMS/MS) for quantification. It should be noted that cytokinin nucleotides are detected as the corresponding nucleosides after dephosphorylation with alkaline phosphatase (Fig. 1). Thus, we could not separately quantify the amounts of cytokinin riboside 5′-monophosphate, diphosphate and triphosphate using this method. The nucleotides are indicated as a group of phosphates, such as iPR 5′-phosphates (iPRPs) and tZR 5′-phosphates (tZRPs). On the other hand, auxins, ABA and gibberellins were further purified with anion-exchange spin columns because they could not be detected in the Elution 1 fraction by UPLC-ESI-qMS/MS analysis due to impurities (data not shown). Since the compounds were purified into three fractions in a stepwise manner (Fig. 1), this procedure was named the ‘stepwise separation method’.
UPLC-ESI-qMS/MS method for the simultaneous quantification of plant hormones
Appropriate precursor to product ion transitions for each unlabeled compound (36 molecular species) and their respective deuterium-labeled internal standards were determined by UPLC-ESI-qMS/MS (Supplementary Table S1). Cytokinins and auxins were detected in the positive ion mode, and ABA and gibberellins were identified in the negative ion mode. Examples of mass spectra of the product ions for the compounds are shown in Supplementary Fig. S2. The retention times for each compound separated by UPLC are shown in Supplementary Table S1. The lower limits of quantification ranged from 0.5 fmol for tZR to 5 fmol for tZ in cytokinins, from 10 fmol for indole-3-acetyl-l-alanine (IA-Ala) to 100 fmol for IA-Asp in auxins, 5 fmol for ABA, and from 5 fmol for GA4 to 50 fmol for GA9 in gibberellins using a Quattro Ultima Pt mass spectrometer (Waters, Milford, MA, USA). Although IAA contains a carboxyl group, the positive ion mode was more sensitive than the negative ion mode for quantifying this auxin.
MS-probe modification
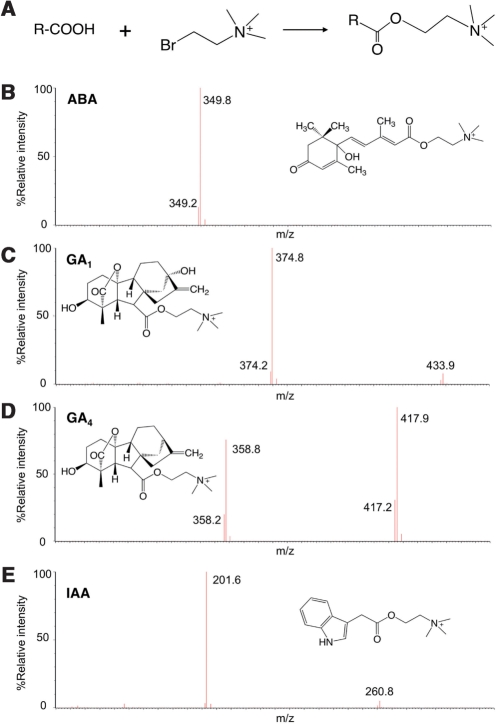
In order to enhance further the sensitivity of negatively charged compounds, such as gibberellins, we introduced a chemical modification with bromocholine, which we called ‘MS-probe’, that possesses a positively charged quaternary amine moiety (Honda et al. 2007) ( Fig. 2A). This modification can transform a natural compound containing a carboxyl group into a positively charged compound by conjugation (Fig. 2A). Thus, this method enables us to detect ABA and gibberellins with the positive ion mode in ESI-qMS/MS (Fig. 2B–D). Auxins are also modified by the MS-probe (Fig. 2E) although they could already be measured using the positive ion mode by their nature. We determined precursor to product transitions of the auxins, ABA and gibberellins for the modified compounds (Supplementary Table S2). UPLC-ESI-qMS/MS analysis revealed that the quantification limits of the modified compounds were greatly enhanced: from 50 fmol to 1 fmol, or up to 50 fold, for GA1 using the Quattro Ultima Pt mass spectrometer.
Fig. 2.
MS-probe derivatization. (A) Structure of the MS-probe and its derivatized product after reaction with the analyte. (B–E) Structures of derivatized products and their fragmentation patterns for ABA (B), GA1 (C), GA4 (D) and IAA (E) that were analyzed with a Quattro Premier XE (Waters) in the positive ion mode.
We applied this MS-probe method to the analysis of plant samples. After fractionation by Oasis MCX 96-well plate, the Elution 1 fraction that contains auxins, ABA and gibberellins is partially purified by anion-exchange spin column chromatography (Fig. 1). After recovery, the products are reacted with MS-probe and then subjected to UPLC-ESI-qMS/MS analysis using the positive ion mode. The modification with MS-probe permits these hormone derivatives to be measured in a single UPLC run.
To evaluate the MS-probe modification, we extracted hormones from rice (cv. ‘Taichung 65’) shoot apices and derivatized one-half of the Elution 1 fraction with MS-probe and the remaining half was further purified by the ‘stepwise separation method’. Comparison of the analyzed data showed that the results obtained with MS-probe modification are comparable with those obtained with the stepwise separation method (Table 1). The overall recoveries of stable isotope-labeled internal standards in both methods were similar (Supplementary Table S3). In addition, IA-Ala and indole-3-acetyl-l-phenylalanine (IA-Phe), that are minor auxin conjugates, could be quantified (Table 1). For gibberellin quantification, the mass peaks of MS-probe derivatization products became more distinct than those of non-modified compounds (data not shown). These results indicate that MS-probe modification increases quantification sensitivity.
Table 1.
Quantification of auxins, gibberellin and ABA (pmol g−1 FW) with the MS-probe method and the stepwise separation method
| MS-probe method | Stepwise separation method | |
|---|---|---|
| IAA | 82.07 ± 2.75 | 78.06 ± 7.50 |
| IA-Ala | 1.09 ± 0.07 | ND |
| IA-Asp | 27.21 ± 0.40 | 27.18 ± 6.49 |
| IA-Phe | 0.40 ± 0.08 | ND |
| GA1 | 2.44 ± 0.50 | 3.91 ± 0.99 |
| GA19 | 49.09 ± 3.41 | 51.27 ± 3.57 |
| GA20 | 2.46 ± 0.30 | 3.30 ± 0.30 |
| ABA | 18.15 ± 2.10 | 17.87 ± 1.56 |
Concentrations of auxins, gibberellin and ABA in the shoot apex of ‘Taichung 65’ were determined by the MS-probe method and the stepwise separation method. Data are the means ± SD (n = 3). The overall recoveries of stable isotope-labeled internal standards in each method are presented in Supplementary Table S3.
ND, not detected.
Hormone quantification in various aerial organs of rice
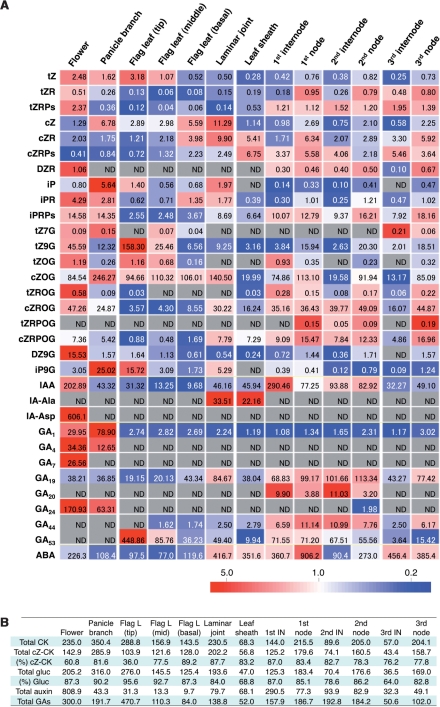
The MS-probe method was applied to evaluate the organ distribution pattern of plant hormone species in rice. Various parts of rice (cv. ‘Nipponbare’) plants were harvested at the early flowering stage, and the endogenous hormone contents were analyzed. The distribution pattern of quantified hormone species is shown in the heat map ( Fig. 3A). In terms of cytokinins, cis-zeatin (cZ)-type cytokinins were dominant in all organs investigated other than the tip region of flag leaves (Fig. 3B). The proportion of cZ-type species was 61% in flowers to 89% in the basal region of flag leaves, but 36% in the tip region of the flag leaves, which contained tZ-9-N-glucoside (tZ9G) as the largest pool (55%) of cytokinin conjugates. In all organs, glucosides were the major form of accumulated cytokinins (Fig. 3B). For the N-glucosides, 9-N-glucosides were the dominant form rather than the 7-N-glucosides (Fig. 3A), which are the major N-glucoside forms in Arabidopsis (Hou et al. 2004, Sakakibara et al. 2005), and the cZ-type conjugates formed a large proportion of the O-glucosides.
Fig. 3.
Organ distribution of plant hormone species in rice. (A) Heat map of organ distribution of plant hormones. Flower, panicle branch, tip, middle and basal regions, laminar joint, and leaf sheath of the flag leaf, first internode, first node, second internode, second node, third internode and third node of rice (cv. ‘Nipponbare’) were harvested after growth in a greenhouse to the flowering stage. The concentrations of the hormone species were analyzed. The relative accumulation patterns are shown in the heat map based on the average value for each plant hormone species. Red and blue colors indicate higher and lower concentrations, respectively. The color scale is shown at the bottom. Hormone species whose concentrations were under the quantification limit in all organs are not shown in the heat map. The value in each block is the concentration (average value, n = 3) as pmol g−1 FW. ND, not detected under the quantification limit. (B) Total amount of cytokinins (Total CK), cZ-type cytokinins (Total cZ-CK), cytokinin glucosides (Total gluc), auxins (Total auxin) and gibberellin (Total GAs) in the results of A are shown as pmol g−1 FW. The proportions of cZ-type cytokinins [(%) cZ-CK] and cytokinin glucosides [(%) Gluc] are indicated as percentage values. Flag L, flag leaf; mid, middle; IN, internode.
In addition to the occurrence of GA1 in all organs investigated, bioactive GA4 and GA7 were detected in flowers and panicle branches (Fig. 3A). The results also showed that GA24, a precursor of GA4, was detected in both organs. In our previous analysis of flower organs, anthers contained massive amounts of GA4 but lacked GA1 (Hirano et al. 2008). In this analysis, detection of GA1 in whole flowers was probably due to accumulation of GA1 in other flower organs, such as the pistils and paleae. Flowers also contained large amounts of IAA and IA-Asp (Fig. 3A). According to our previous analyses, auxins accumulate to significant levels in the anther (Hirano et al. 2008).
In flag leaves, different distributions among cytokinin species were found. Free base tZ and iP and their glucosides were more abundant in the tip region, but cZ and its conjugates were more abundant in the basal region (Fig. 3A). Similarly, IAA was more abundant in the tip region, but such a bias was not found for GA1 or ABA.
During this developmental stage, the first and second internodes undergo remarkable elongation, whereas that of the third internode is less. The elongating internodes contained higher levels of IAA and total gibberellins than the third internode, but the difference in accumulated levels of GA1 (an active gibberellin) in the three internodes was slight; only the second internode contained slightly higher levels of GA1 (Fig. 3A, B). In our analysis focusing on the difference between nodes and internodes, a large proportion of cytokinin species were more abundant in the nodes rather than in the internodes. The difference was more pronounced for the glucoside conjugates (Fig. 3A, B). Taken together, these results indicate that our analysis could show the organ distribution pattern of the plant hormones, and that the hormones are differentially distributed in the aerial organs, even within a leaf.
Quantification of plant hormones in rice gibberellin signaling mutants
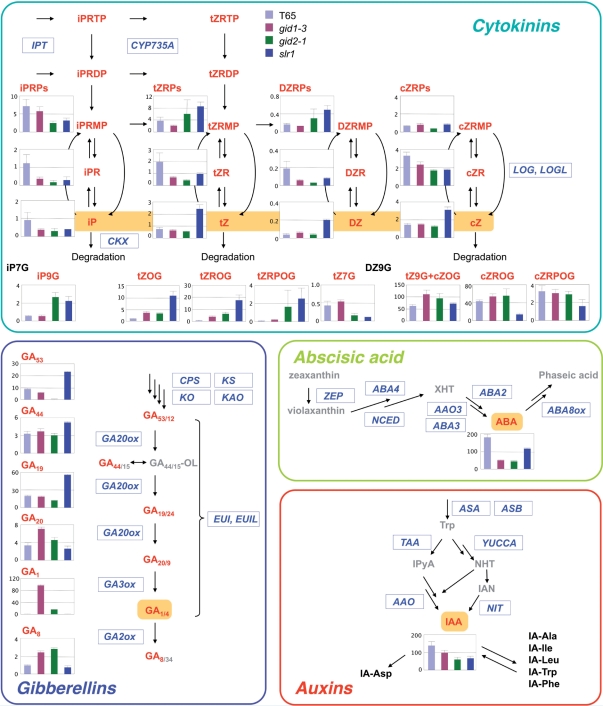
The MS-probe method was applied to hormone profiling of rice gibberellin signaling mutants. gid1-3 is a mutant of the rice gibberellin receptor GID1 and shows severe dwarfism (Ueguchi-Tanaka et al. 2005). A mutation at a different allele, gid1-1, results in excessive accumulation of GA1 and GA20 (Ueguchi-Tanaka et al. 2005). gid2-1 is a mutant of the gibberellin signaling factor GID2, which is an F-box protein, and also shows a severe dwarf phenotype and overaccumulates GA1, GA20 and GA44 (Sasaki et al. 2003). slr1 (allele name: k1001) is a single recessive mutant resulting in a constitutive gibberellin response phenotype, whose shoot elongates as if saturated with gibberellins (Ikeda et al. 2001, Ueguchi-Tanaka et al. 2005). In the slr1 mutant, accumulation levels of active GA1 and its precursors (GA20 and GA19) are decreased (Ikeda et al. 2001). We harvested shoots from the mutant seedlings and a wild-type cultivar ‘Taichung 65’, and analyzed the hormone content. For the gibberellins, GA1 was overaccumulated in gid1-3 and gid2-1 by about 300- and 60-fold, respectively, and decreased in slr1 ( Fig. 4 and Supplementary Table S4). GA20 and GA8, which are a precursor and a deactivated metabolite, respectively, also accumulated to higher levels in gid1-3 and gid2-1. The accumulation profiles of gibberellins in gid1-3 and slr1 were consistent with previous reports (Ikeda et al. 2001, Ueguchi-Tanaka et al. 2005) except for overaccumulation of GA19 in slr1.
Fig. 4.
Endogenous levels of cytokinins, gibberellin, ABA and auxins in a wild-type and gibberellin signaling mutants. Shoots of the wild type (T65) and gid1-3, gid2-1 and slr1 were harvested after growth in a greenhouse for 5 weeks. The amounts of the hormones are shown as histograms with the SD (n = 3). The y-axis is concentration as pmol g−1 FW. Hormone species and other compounds in gray were not measured, and those in black were under the quantification limit. The overall recoveries of analyzed stable isotope-labeled internal standards are presented in Supplementary Table S7. Genes involved in each metabolic process are indicated in italic blue letters surrounded by a rectangle. The details of each metabolic pathway are described by Hirano et al. (2008). GA44/15-OL, GA44/15-open lactone; XHT, xanthoxin; IpyA, indole-3-pyruvic acid; NHT, N-hydroxyl-tryptamine; IAN, indole-3-acetonitrile; IPT, adenosine phosphate-isopentenyltransferase; CKX, OsCKX; CPS, ent-copalyl diphosphate synthase; KS, ent-kaurene synthase; KO, ent-kaurene oxidase; KAO, ent-kaurenoic acid oxidase; EUIL, EUI-like; ZEP, OsZEP; NCED, OsNCED; AAO, OsAAO; ASA, OsASA; ASB, OsASB; TAA, OsTAA; YUCCA, OsYUCCA; NIT, OsNIT.
For the cytokinins, the nucleobase form of zeatin-type cytokinins, namely tZ, dihydrozeatin (DZ) and cZ, overaccumulated in the slr1 mutant (Fig. 4). Glucosides of nucleobase cytokinins, such as iP-9-N-glucoside (iP9G) and tZ-O-glucoside (tZOG), also accumulated in the gibberellin signaling mutants, suggesting that impairment of the gibberellin signaling pathway positively regulates active cytokinin accumulation because such glucosides are synthesized by conjugation of glucose to active forms (Mok and Mok 2001, Sakakibara 2006). ABA accumulation levels in slr1 were lower than in the wild type but higher than in the gid mutants (Fig. 4). Among the auxins, only IAA could be measured in the shoot, and the accumulation level in the mutants was lower than for the wild type. These results indicate that the metabolism of these four major plant hormones interacts with the gibberellin signaling system.
Transcriptome analysis of rice gibberellin signaling mutants
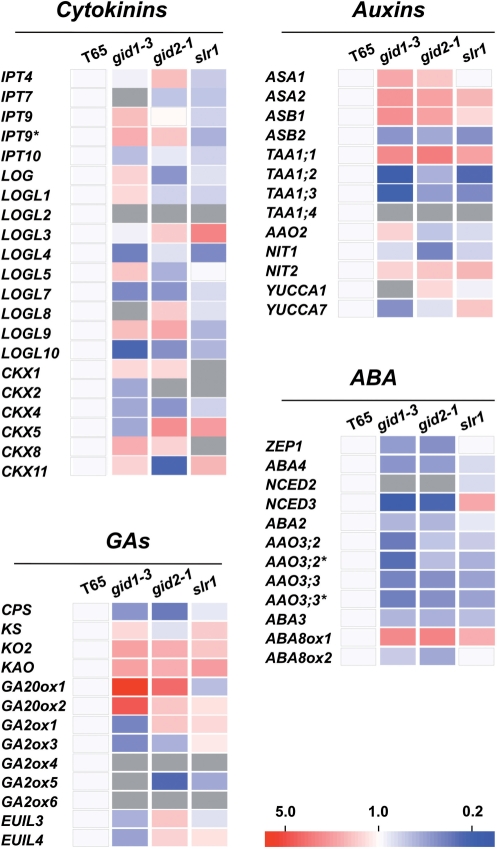
To combine the hormone profiling data with gene expression data, we performed transcriptome analysis using the rice gibberellin signaling mutants, gid1-3, gid2-1 and slr1, and the background cultivar ‘Taichung 65’ that were grown under the same conditions as the plants used for hormone analysis. Total RNA prepared from each genotype was subjected to microarray analysis using an Affymetrix GeneChip® Rice Genome Array. We selected genes from the GeneChip data set that are considered to be involved in hormone metabolic pathways. All 81 genes dealt with in this report are listed in Supplementary Table S5. Expression of 56 genes out of 81 genes was detected in ‘Taichung 65’. The relative expression levels of these genes are shown in the heat maps ( Fig. 5). For some of the detected transcripts in ‘Taichung 65’, expression was judged as invalid in the mutants (Fig. 5, gray cells). Accumulation of these transcripts probably was below the background level in the mutants, although the possibility of miss-hybridization cannot be excluded.
Fig. 5.
Heat map view of expression of genes involved in hormone metabolism in the gibberellin signaling mutants, gid1-3, gid2-1 and slr1, and the wild-type ‘Taichung 65’. Hormone metabolism genes with reliable expression data in at least ‘Taichung 65’ were selected from Supplementary Table S4. The relative expression level of each gene compared with expression in ‘Taichung 65’ is shown on the heat map. Red and blue colors indicate higher and lower expression, respectively. The color scale is shown at the bottom. Gray indicates expression data of lower reliability. One of the multiple Affymetrix probes for one gene is marked with an asterisk. T65, ‘Taichung 65’. Abbreviations of genes are shown in the legend to Fig. 4 and the main text.
In the gibberellin signaling mutants, two gibberellin 20-oxidase (GA20ox) genes, GA20ox1 and GA20ox2, were up-regulated in gid1-3 and gid2-1 (Fig. 5). Remarkably, in gid1-3, the accumulation levels of GA20ox1 and GA20ox2 transcripts increased by 14- and 3.9-fold, respectively (Supplementary Table S6). For gibberellin precursors, accumulation of the ent-kaurene oxidase 2 gene (KO2) and ent-kaurenoic acid oxidase gene (KAO) transcripts was elevated in the three mutants. In terms of genes associated with deactivation of gibberellins, the gibberellin 2-oxidase (GA2ox) genes, GA2ox1 and GA2ox3–GA2ox6, and EUI-like (EUIL) genes, EUIL3 and EUIL4, were down-regulated in gid1-3. Similarly, accumulation of the GA2ox3–GA2ox6 transcripts was lower in gid2-1. For ABA metabolism, the majority of biosynthetic genes, namely the 9-cis-epoxycarotenoid dioxygenase (NCED) genes, OsNCED2 and OsNCED3, the zeaxanthin epoxidase (ZEP) gene, OsZEP1, OsABA2–OsABA4, and the aldehyde oxidase (AAO) genes, OsAAO3;2 and OsAAO3;3, were down-regulated, and OsABA8ox1, which encodes CYP707A5, an ABA deactivation enzyme (Yang and Choi 2006, Saika et al. 2007), was up-regulated in gid1-3 and gid2-1. These regulation patterns are highly consistent with the hyperaccumulation of GA1 and decrease in ABA content in gid1-3 and gid2-1 (Fig. 4). As for the expression of genes for cytokinin metabolism, a lonely guy (LOG)-like gene, OsLOGL3, which is probably involved in an activation step of cytokinin biosynthesis, was up-regulated in the slr1 mutant (Fig. 5). In terms of deactivation of cytokinins, OsCKX1, OsCKX2 and OsCKX8, that encode cytokinin oxidase/dehydrogenase (CKX), were down-regulated in slr1; however, the transcript accumulation pattern of other genes was not clearly correlated with that of the cytokinin species (Figs. 4, 5). In auxin biosynthesis, the anthranilate synthase β subunit (ASB) gene, OsASB2, tryptophan aminotransferase (TAA) genes, OsTAA1;2–OsTAA1;4, and the nitrilase (NIT) gene, OsNIT1, were down-regulated in the gibberellin signaling mutants, especially in gid2-1, a result consistent with the decrease in IAA concentration in this mutant. In contrast, the accumulation level of transcripts for anthranilate synthase α subunit (ASA) genes, OsASA1, OsASA2, OsASB1, OsTAA1;1 and OsNIT2, were elevated in the gid2-1 mutant. These results show that our hormone profiling data can be combined with transcriptome data for simultaneous analysis of the relationship between expression of the genes for hormone metabolic pathways and the accumulated levels of individual hormone species.
Discussion
Development of a highly sensitive and high-throughput method to analyze multiple phytohormones
In this study, we have developed a highly sensitive and high-throughput method for the simultaneous analysis of four groups of phytohormones, cytokinins, auxins, ABA and gibberellins, using UPLC-ESI-qMS/MS. We have previously analyzed each group of hormones with different experimental protocols (Nakagawa et al. 2005, Hirano et al. 2007, Naito et al. 2007, Hirano et al. 2008), but this new method enabled us to measure all classes of hormones concurrently. Although there have been several reports of simultaneous quantification of multiple plant hormones (Müller et al. 2002, Chiwocha et al. 2003, Pan et al. 2008), we would like to emphasize three strong points that distinguish our system. First is the introduction of automated solid phase extraction that enabled us to treat a large number of samples without handling mistakes and apply our technology to a hormonomics study. Second is the introduction of MS-probe derivatization that greatly improved the quantification limit of gibberellins and reduced the time for UPLC-ESI-qMS/MS because auxins, ABA and gibberellins can be analyzed in a single UPLC run in the positive ion mode. Third is the number of target hormone species that can be analyzed. Our method provides quantitative data for the active forms of the hormones together with the precursors and deactivated conjugates, and helps us to understand metabolic control more precisely especially for cytokinins and gibberellins.
At present, our system cannot measure precursors of ABA and auxins, or deactivated conjugates of ABA. In addition, the current protocol cannot target other plant hormones, such as brassinosteroids, jasmonates or salicylic acid. The primary reason that our protocol will not work for these plant hormones is that appropriate standards and stable isotope-labeled compounds are not readily available. In order to broaden the target analytes, chemical synthesis of the standard compounds is required.
iPRMP, iPRDP and iPRTP fractionated into the same eluate (Supplementary Fig. S1). The distribution of other cytokinin nucleoside 5′-diphosphates and triphosphates should be similar although their behavior in the fractionation scheme could not be examined due to lack of the standard compounds. In this system, cytokinin nucleoside 5′-monophosphates, diphosphates and triphosphates could not be separately quantified because they have to be dephosphorylated for UPLC-ESI-qMS/MS analysis. To know the concentration of each nucleotide, further fractionation using ion-exchange chromatography is required (Takei et al. 2003). Previously, we determined the concentration of cytokinin nucleotides as the 5′-monophosphates, such as iPRMP and tZR 5′-monophosphate (tZRMP) (Nakagawa et al. 2005, Hirose et al. 2007, Hirano et al. 2008); however, our current study shows that our assumption was inappropriate. Thus, hereafter, we describe the cytokinin nucleotides as a group of phosphates, such as iPRPs and tZRPs.
Introduction of MS-probe derivatization greatly increased the performance of our analysis platform. Several different types of MS-probes that react with other functional groups have been developed (Honda et al. 2007). Further applications utilizing additional novel derivatives might effectively improve plant hormone analysis.
One may ask how much tissue is required for quantification of these phytohormones. This is an often asked question but one which is difficult to answer correctly because hormone concentration can vary spatially and temporally, even in the same organ. In this study, we used about 100 mg (FW) of tissue for quantification; however, for instance in developing young rice panicles, hormone concentrations are high, and a few pieces of the panicle are sufficient for quantification (H. Sakakibara et al. unpublished data). Thus, the sample size is totally dependent on the nature of the tissues and the target hormone species.
One remaining challenge for this kind of measurement technology is that hormone concentrations in specific cell populations cannot be quantified accurately because the sample tissues are homogenized and normalized in an extraction solvent. Thus, the quantified values are average concentrations for the entire population of cells comprising the tissue. To see the local hormone contents more precisely, microsurgical techniques for harvesting specific cell populations is required. Alternatively, other technologies, such as visualization of hormone compounds by immunodetection (Sossountzov et al. 1988, Lee et al. 2008, Urakami et al. 2008) or hormone-responsive promoter–reporter systems (e.g. DR5 promoter for auxins; Ni et al. 2001), should also be effective.
Application of our method to hormone profiling
Hormone profiling of various parts of aerial organs and a set of rice gibberellin signaling mutants revealed the complexity of metabolic regulation and allocations of plant hormone species. For instance, the distribution of tZ- and iP-type cytokinins was different from that of the cZ-type cytokinins within a flag leaf (Fig. 3). In addition, tZ9G was the dominant glucose conjugate in the tip region. This result is probably due to differential expression of cytokinin metabolic genes or due to a specialized transport system. Although glucosylation is an important step in regulating cytokinin activity as well as for degradation, the genes encoding cytokinin glucosyltransferases have not yet been identified in rice. The data obtained in this study clearly show the importance of identifying genes encoding cytokinin glucosyltransferases.
In Arabidopsis, degradation of prenylated tRNA is the main pathway for cZ synthesis because a mutant deficient in tRNA-isopentenyltransferase genes (AtIPT2 and AtIPT9) resulted in the cZ content falling below the limits of detection (Miyawaki et al. 2006). The steady-state accumulation level of cZ-type cytokinins in rice, maize (Veach et al. 2003) and chickpea (Emery et al. 1998) is remarkably higher than in other plants. Although the biosynthetic pathway for cZ in rice has not been elucidated, the differential distribution pattern in the flag leaf implies a distinct metabolic pathway between the tZ- and iP-type cytokinins and the cZ-type cytokinins.
Transcriptome analysis of the same plants that were used for hormone analysis provided us with insights into the interaction of hormone activity and metabolism. In the slr1 mutant, levels of free bases of zeatin-type cytokinins increased, whereas levels of GA1, IAA and ABA decreased (Fig. 4 and Supplementary Table S4). These results indicate that, under constitutive gibberellin response conditions, feedback regulation is implemented not only for gibberellin metabolism, but also for the metabolism of other hormones. Changes in the concentration of iP-type and tZ-type cytokinins, especially their nucleotides and nucleobases, were not correlated in the gibberellin signaling mutants (Fig. 4). Conversion from iPRP to tZRP is catalyzed by CYP735A, a cytochrome P450 monooxygenase (Takei et al. 2004). In our microarray experiment, expression of rice CYP735A3 and CYP735A4, possible orthologs of Arabidopsis CYP735A1 and CYP735A2, could not be quantified due to low expression signal intensity. Although the biological relevance of side chain structural variations of cytokinins has not been elucidated, we hope to continue our investigations in this area.
Recent studies integrating metabolome data and transcriptome data have identified novel genes involved in the regulation of metabolism (Hirai et al. 2004, Hirai et al. 2005, Hirai et al. 2007, Saito et al. 2008). Our highly sensitive and high-throughput method for analyzing phytohormone levels in plant tissues can be used for acquiring hormonome data that can be integrated into transcriptome and other omics data. We expect that our platform will contribute toward the identification of novel genes involved in regulating plant hormone action and metabolism.
Materials and Methods
Chemicals
Stable isotope-labeled compounds, [2H5]tZ, [2H5]tZR, [2H5]tZRMP, [2H3]DZ riboside (DZR), [2H3]DZR 5′-monophosphate, [2H6]iP, [2H6]iPR, [2H6]iPRMP, [2H5]tZ-7-N-glucoside (tZ7G), [2H5]tZOG, [2H5]tZ9G, [2H5]tZR-O-glucoside (tZROG), [2H3]DZ-9-N-glucoside, [2H6]iP-7-N-glucoside (iP7G), [2H6]iP9G, [2H5]IAA, [2H6]ABA, [2H2]GA1, [2H2]GA3, [2H2]GA4, [2H2]GA7, [2H2]GA8, [2H2]GA9, [2H2]GA12, [2H2]GA19, [2H2]GA20, [2H2]GA24, [2H2]GA44 and [2H2]GA53, were purchased from OlChemim Ltd. (Olomouc, Czech Republic), and [2H2]IA-Ala, [2H2]IA-Asp, [2H2]indole-3-acetyl-l-isoleucine (IA-Ile), [2H2]indole-3-acetyl-l-leucine (IA-Leu) and [2H2]IA-Phe were the gift of Dr. J. Hiratake (Kyoto University, Japan). Bromocholine was purchased from Tokyo Chemical Industry Co. Ltd. (Tokyo, Japan). iPRDP and iPRTP were prepared enzymatically as described previously (Takei et al. 2003).
Plant materials
Rice (Oryza sativa L. cv. ‘Nipponbare’) plants were grown in a greenhouse for harvesting various organs. Rice cultivar ‘Taichung 65’ and its irradiation-induced mutants, gid1-3 (Ueguchi-Tanaka et al. 2005) and gid2-1 (Sasaki et al. 2003), and the chemically induced mutant slr1 (allele name: k1001) were grown in a growth chamber at 30°C under continuous light.
Hormone extraction and fractionation for UPLC-ESI-qMS/MS analysis
Frozen tissues (about 100 mg FW) were crushed to a fine powder using a TissueLyser (Qiagen, Hilden, Germany) with a zirconia bead (diameter, 5 mm) in a 2 ml microcentrifuge tube, and then soaked in 1 ml of extraction solvent (methanol : formic acid : water = 15 : 1 : 4). The stable isotopes shown in Supplementary Tables S1 and S2 were added to the extract to serve as internal standards. The homogenate was kept at –30°C for at least 16 h. After centrifugation at 10,000 × g for 15 min, the supernatant was transferred to a 96-well collection plate (Waters, Milford, MA, USA). The pellet was re-extracted with 0.2 ml of extraction solvent, and combined with the first supernatant. The 96-well plate was placed on an automated solid phase extraction system (SPE215; Gilson, Middleton, WI, USA). To remove interfering compounds, the extract was first passed through an Oasis HLB 96-Well Plate 30mg (Waters) equilibrated with 1 M formic acid. The column was further washed with 0.3 ml of extraction solvent. The combined eluate (approximately 1.5 ml) was evaporated and then reconstituted with 1 ml of 1 M formic acid. The hormone-containing fraction was passed through an Oasis MCX 96-Well Plate 30mg (Waters) equilibrated with 1 M formic acid. After washing with 1 M formic acid, ABA, auxins and gibberellins were eluted with methanol (Elution 1, Fig. 1). Cytokinin nucleotides were eluted with 0.35 M ammonia (Elution 2, Fig. 1), and cytokinin nucleobases, nucleosides and glucosides were eluted with 0.35 M ammonia in 60% (v/v) methanol (Elution 3, Fig. 1). Each fraction was evaporated to dryness.
UPLC-ESI-qMS/MS analysis of cytokinins
The Elution 3 fraction, which contains cytokinin nucleobases, nucleosides and glucosides, was reconstituted with 50 μl of 0.1% acetic acid, and subjected to UPLC-ESI-qMS/MS analysis. The Elution 2 fraction, which contains cytokinin nucleotides, was reconstituted with 0.84 ml of 0.1 M N-cyclohexyl-2-aminoethanesulfonic acid-NaOH (pH 9.8). After addition of 17 μl of alkaline phosphatase (1 U μl−1, Oriental Yeast Co. Ltd., Tokyo, Japan), the fraction was incubated for 1 h at 37°C. Then, 93 μl of 10 × Tris-buffered saline was added to the solution, and 46 μl of 1 M HCl was added to adjust the pH to 7.3. The solution containing dephosphorylated compounds was desalted by passing through an Oasis HLB 96-Well Plate 30mg (Waters) equilibrated with H2O. After washing with water, the absorbed compounds were eluted with methanol by using SPE215. The methanol was evaporated, and the dried compounds were reconstituted with 50 μl of 0.1% acetic acid and subjected to UPLC-ESI-qMS/MS analysis.
Cytokinins were measured with an UPLC-ESI-qMS/MS (AQUITY UPLC™ System/Quattro Ultima Pt; Waters) with an ODS column (AQUITY UPLC BEH C18, 1.7 μm, 2.1 × 100 mm, Waters). Selection of precursor and product ions was carried out using unlabeled and deuterium-labeled standard compounds as summarized in Supplementary Table S1. Cytokinins were separated at a flow rate of 0.25 ml min−1 with linear gradients of solvent A (0.06% acetic acid) and solvent B (0.06% acetic acid in methanol) set according to the following profile: 0 min, 99.0% A + 1.0% B; 4.0 min, 55.0% A + 45.0% B; 7 min, 30.0% A + 70.0% B; and then with isocratic conditions: 8 min, 1.0% A + 99.0% B; 12 min, 99.0% A + 1.0% B. Capillary voltage was 3.13 kV. Cone voltage (V) and collision energy (eV) are as follows: tZR, cZ riboside, DZR: 52 V, 15 eV; iP: 53 V, 14 eV; iPR: 52 V, 18 eV; tZ7G, tZOG, tZ9G, DZ-9-N-glucoside: 55 V, 20 eV; tZROG: 65 V, 28 eV; iP7G: 70 V, 21 eV; iP9G: 52 V, 20 eV. Data were processed by MassLynx™ software with QuanLynx™ (version 4.0, Waters).
Stepwise separation method
The Elution 1 fraction was reconstituted with 0.2 ml of H2O and passed through a DEAE-cellulose column (Vivapure D Mini M, Vivascience, Hannover, Germany) equilibrated with water. After washing with water, ABA and gibberellins were eluted with 0.2% acetic acid; IAA, IA-Ala, IA-Ile and IA-Leu were eluted with 0.5% formic acid; and IA-Trp, IA-Phe and IA-Asp were eluted with 3% formic acid. Each eluate was evaporated and reconstitute with 45 μl of 0.05% formic acid, and separately subjected to UPLC-ESI-qMS/MS analysis.
MS-probe modification method
The Elution 1 fraction was reconstituted with 0.4 ml of H2O and passed through a DEAE-cellulose column (Vivapure D Mini M), equilibrated with water. After washing with water, the hormones were eluted with 0.4 ml of 3% formic acid. After evaporation, the compounds were modified with bromocholine (MS-probe). The compounds were reconstituted with 75 μl of 1-propanol. A 20 μl aliquot of water, 4 μl of 500 mM bromocholine in 70% acetonitrile and 0.8 μl triethylamine were added to the solution. The mixed solution was incubated at 80°C for 130 min, and then moved to ice. The solution was evaporated and reconstituted with 50 μl of 0.05% formic acid, and subjected to UPLC-ESI-qMS/MS analysis using the positive ion mode.
Hormones derivatized with MS-probe were measured with an UPLC-ESI-qMS/MS (AQUITY UPLC™ System/Quattro Premier XE; Waters) with an ODS column (AQUITY UPLC BEH C18, 1.7 μm, 2.1 × 100 mm, Waters). Selection of precursor and product ions was carried out using unlabeled and deuterium-labeled standard compounds as summarized in Supplementary Table S2. Hormones were separated at a flow rate of 0.25 ml min−1 with the gradients of solvent A (0.05% formic acid) and solvent B (0.05% formic acid in acetonitrile) set according to the following profile: 0 min, 99.0% A + 1.0% B; 2 min, 86.0% A + 14.0% B; 6 min, 84.0% A + 16.0% B; 13 min, 45.0% A + 55.0% B; 14 min, 30% A + 70% B; and then with isocratic conditions: 15 min, 99.0% A + 1.0% B. Capillary voltage was 3.2 kV. Cone voltage (V) and collision energy (eV) are as follows: IAA, 30 V, 20 eV; IA-Ala, 35 V, 20 eV; IA-Trp, IA-Phe: 40 V, 20 eV; IA-Asp: 40 V, 15 eV; IA-Ile, IA-Leu: 35 V, 20 eV; GA1, GA3, GA24, GA44, GA53: 50 V, 30 eV; GA4, GA7, GA12, GA19, GA20: 40 V, 20 eV; GA8, GA9: 40 V, 30 eV; and ABA: 40 V, 20 eV. Data were processed by MassLynx™ software with QuanLynx™ (version 4.1, Waters).
DNA microarray analysis
Microarray analysis was performed using a GeneChip® Rice Genome Array (Affymetrix, Santa Clara, CA, USA). Total RNA was prepared from shoots of the rice gibberellin signaling mutants, gid1-3, gid2-1 and slr1, and the background cultivar ‘Taichung 65’ grown under the same conditions as the plants used for hormone analysis using an RNeasy Plant Mini Kit (Qiagen). Preparation of labeled target cRNA, subsequent purification and fragmentation were carried out using One-Cycle Target Labeling and Control Reagents (Affymetrix). Double-stranded cDNA was prepared from 10 μg of total RNA. Hybridization, washing, staining and scanning were performed as described in the supplier’s protocol. A 10 μg aliquot of fragmented cRNA was used for hybridization. It should be noted that all the above manipulations were carried out independently for each RNA sample. Three independent replicates were carried out for each genotype.
Data analysis was performed using GeneChip® Operating Software (GCOS; Affymetrix) and GeneSpring 7 (Agilent Technologies, Palo Alto, CA, USA). Per chip normalization with the 50th median of all measurements on the chip was performed as recommended by the GeneSpring manual for Affymetrix gene chips. Then, per gene normalization to specific samples was applied using the mean of three replicates from ‘Taichung 65’ as a control. Thus, each gene expression level was shown as the ratio of gene expression level in each mutant to that in ‘Taichung 65’. For reliability, we used only the data whose flag calls were declared ‘P’ or ‘M’ by the GeneChip® Operating Software in at least two of three replicates. One-way analysis of variance (ANOVA) Student’s t-tests (P-value <0.05) were performed to evaluate differences between each mutant and ‘Taichung 65’.
Supplementary data
Supplementary data are available at PCP online.
Funding
The Genomics for Agricultural Innovation Project (NVR0004 to H. S.); the Special Coordination Fund for Promoting Science and Technology (Japan Science and Technology Agency).
Acknowledgments
The authors are very grateful to Dr. J. Hiratake for providing stable isotope-labeled auxins, and Dr. S. Yamaguchi for providing GA34. We also thank N. Makita (RIKEN) and H. Tokunaga (Nagoya University) for their help in sampling rice organs, and S. Oyama (RIKEN) for assistance with the GeneChip experiment.
Glossary
Abbreviations
- AAO
aldehyde oxidase
- ASA
anthranilate synthase
α
subunit
- ASB
anthranilate synthase
β
subunit
- CKX
cytokinin oxidase/dehydrogenase
- cZ
cis
-zeatin
- DZ
dihydrozeatin
- DZR
DZ riboside
- ESI
electrospray interface
- EUIL
EUI-like
- GA2ox
gibberellin 2-oxidase
- GA20ox
gibberellin 20-oxidase
- IA-Ala
indole-3-acetyl-
l
-alanine
- IA-Asp
indole-3-acetyl-
l
-aspartic acid
- IA-Ile
indole-3-acetyl-
l
-isoleucine
- IA-Leu
indole-3-acetyl-
l
-leucine
- IA-Phe
indole-3-acetyl-
l
-phenylalanine
- iP
N
6
-(
Δ
2
-isopentenyl)adenine
- iP7G
iP-7-
N
-glucoside
- iP9G
iP-9-
N
-glucoside
- iPR
iP riboside
- iPRMP
iPR 5
′
-monophosphate
- iPRDP
iPR 5
′
-diphosphate
- iPRTP
iPR 5
′
-triphosphate
- iPRPs
iPR 5
′
-phosphates
- KAO
ent
-kaurenoic acid oxidase
- KO
ent
-kaurene oxidase
- LOG
lonely guy
- NCED
9-
cis
-epoxycarotenoid dioxygenase
- NIT
nitrilase
- qMS/MS
tandem quadrupole mass spectrometer
- TAA
tryptophan aminotransferase
- tZ
trans
-zeatin
- tZ7G
tZ-7-
N
-glucoside
- tZ9G
tZ-9-
N
-glucoside
- tZOG
tZ-
O
-glucoside
- tZR
tZ riboside
- tZROG
tZR-
O
-glucoside
- tZRMP
tZR 5
′
-monophosphate
- tZRPs
tZR 5
′
-phosphates
- UPLC
ultra-performance liquid chromatography
- ZEP
zeaxanthin epoxidase.
Footnotes
Microarray data have been deposited in The National Center for Biotechnology Information Gene Expression Omnibus (NCBI GEO) database under accession number GSE15046.
References
- Bai F, DeMason D. Hormone interactions and regulation of Unifoliata, PsPK2, PsPIN1 and LE gene expression in pea (Pisum sativum) shoot tips. Plant Cell Physiol. 2006;47:935–948. doi: 10.1093/pcp/pcj066. [DOI] [PubMed] [Google Scholar]
- Cary AJ, Liu W, Howell SH. Cytokinin action is coupled to ethylene in its effects on the inhibition of root and hypocotyl elongation in Arabidopsis thaliana seedlings. Plant Physiol. 1995;107:1075–1082. doi: 10.1104/pp.107.4.1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiwocha SD, Abrams SR, Ambrose SJ, Cutler AJ, Loewen M, Ross AR, et al. A method for profiling classes of plant hormones and their metabolites using liquid chromatography–electrospray ionization tandem mass spectrometry: an analysis of hormone regulation of thermodormancy of lettuce (Lactuca sativa L.) seeds. Plant J. 2003;35:405–417. doi: 10.1046/j.1365-313x.2003.01800.x. [DOI] [PubMed] [Google Scholar]
- Chow B, McCourt P. Plant hormone receptors: perception is everything. Genes Dev. 2006;20:1998–2008. doi: 10.1101/gad.1432806. [DOI] [PubMed] [Google Scholar]
- Davies PJ, editor. Plant Hormones. Dordrecht: Kluwer Academic Publishers; 2004. Biosynthesis, Signal Transduc-tion, Action. [Google Scholar]
- Dobrev PI, Kaminek M. Fast and efficient separation of cytokinins from auxin and abscisic acid and their purification using mixed-mode solid-phase extraction. J. Chromatogr. A. 2002;950:21–29. doi: 10.1016/s0021-9673(02)00024-9. [DOI] [PubMed] [Google Scholar]
- Emery RJN, Leport L, Barton JE, Turner NC, Atkins A. cis-Isomers of cytokinins predominate in chickpea seeds throughout their development. Plant Physiol. 1998;117:1515–1523. doi: 10.1104/pp.117.4.1515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuda H, Hirakawa Y, Sawa S. Peptide signaling in vascular development. Curr. Opin. Plant Biol. 2007;10:477–482. doi: 10.1016/j.pbi.2007.08.013. [DOI] [PubMed] [Google Scholar]
- Gazzarrini S, McCourt P. Genetic interactions between ABA, ethylene and sugar signaling pathways. Curr. Opin. Plant Biol. 2001;4:387–391. doi: 10.1016/s1369-5266(00)00190-4. [DOI] [PubMed] [Google Scholar]
- Goda H, Sasaki E, Akiyama K, Maruyama-Nakashita A, Nakabayashi K, Li W, et al. The AtGenExpress hormone- and chemical-treatment data set: experimental design, data evaluation, model data analysis, and data access. Plant J. 2008;55:526–542. doi: 10.1111/j.0960-7412.2008.03510.x. [DOI] [PubMed] [Google Scholar]
- Goda H, Sawa S, Asami T, Fujioka S, Shimada Y, Yoshida S. Comprehensive comparison of auxin-regulated and brassinosteroid-regulated genes in Arabidopsis. Plant Physiol. 2004;134:1555–1573. doi: 10.1104/pp.103.034736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez-Roldan V, Fermas S, Brewer PB, Puech-Pages V, Dun EA, Pillot JP, et al. Strigolactone inhibition of shoot branching. Nature. 2008;455:189–194. doi: 10.1038/nature07271. [DOI] [PubMed] [Google Scholar]
- Hirai MY, Klein M, Fujikawa Y, Yano M, Goodenowe DB, Yamazaki Y, et al. Elucidation of gene-to-gene and metabolite-to-gene networks in Arabidopsis by integration of metabolomics and transcriptomics. J. Biol. Chem. 2005;280:25590–25595. doi: 10.1074/jbc.M502332200. [DOI] [PubMed] [Google Scholar]
- Hirai MY, Sugiyama K, Sawada Y, Tohge T, Obayashi T, Suzuki A, et al. Omics-based identification of Arabidopsis Myb transcription factors regulating aliphatic glucosinolate biosynthesis. Proc. Natl Acad. Sci. USA. 2007;104:6478–6483. doi: 10.1073/pnas.0611629104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirai MY, Yano M, Goodenowe DB, Kanaya S, Kimura T, Awazuhara M, et al. Integration of transcriptomics and metabolomics for understanding of global responses to nutritional stresses in Arabidopsis thaliana. Proc. Natl Acad. Sci. USA. 2004;101:10205–10210. doi: 10.1073/pnas.0403218101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirano K, Aya K, Hobo T, Sakakibara H, Kojima M, Shim RA, et al. Comprehensive transcriptome analysis of phytohormone biosynthesis and signaling genes in microspore/pollen and tapetum of rice. Plant Cell Physiol. 2008;49:1429–1450. doi: 10.1093/pcp/pcn123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirano K, Nakajima M, Asano K, Nishiyama T, Sakakibara H, Kojima M, et al. The GID1-mediated gibberellin perception mechanism is conserved in the lycophyte Selaginella moellendorffii but not in the bryophyte Physcomitrella patens. Plant Cell. 2007;19:3058–3079. doi: 10.1105/tpc.107.051524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirose N, Makita N, Kojima M, Kamada-Nobusada T, Sakakibara H. Overexpression of a type-A response regulator alters rice morphology and cytokinin metabolism. Plant Cell Physiol. 2007;48:523–539. doi: 10.1093/pcp/pcm022. [DOI] [PubMed] [Google Scholar]
- Honda A, Hayashi S, Hifumi H, Honma Y, Tanji N, Iwasawa N, et al. MPAI (mass probes aided ionization) method for total analysis of biomolecules by mass spectrometry. Anal. Sci. 2007;23:11–15. doi: 10.2116/analsci.23.11. [DOI] [PubMed] [Google Scholar]
- Hou B, Lim EK, Higgins GS, Bowles DJ. N-Glucosylation of cytokinins by glycosyltransferases of Arabidopsis thaliana. J. Biol. Chem. 2004;279:47822–47832. doi: 10.1074/jbc.M409569200. [DOI] [PubMed] [Google Scholar]
- Ikeda A, Ueguchi-Tanaka M, Sonoda Y, Kitano H, Koshioka M, Futsuhara Y, et al. slender rice, a constitutive gibberellin response mutant, is caused by a null mutation of the SLR1 gene, an ortholog of the height-regulating gene GAI/RGA/RHT/D8. Plant Cell. 2001;13:999–1010. doi: 10.1105/tpc.13.5.999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y, Asami T, Yamaguchi I, Ueda H, Suzuki Y. A new gibberellin detection system in living cells based on antibody V-H/V-L interaction. Biochem. Biophys. Res. Commun. 2008;376:134–138. doi: 10.1016/j.bbrc.2008.08.130. [DOI] [PubMed] [Google Scholar]
- Matsubayashi Y, Sakagami Y. Peptide hormones in plants. Annu. Rev. Plant Biol. 2006;57:649–674. doi: 10.1146/annurev.arplant.56.032604.144204. [DOI] [PubMed] [Google Scholar]
- Miyawaki K, Tarkowski P, Matsumoto-Kitano M, Kato T, Sato S, Tarkowska D, et al. Roles of Arabidopsis ATP/ADP isopentenyltransferases and tRNA isopentenyltransferases in cytokinin biosynthesis. Proc. Natl Acad. Sci. USA. 2006;103:16598–16603. doi: 10.1073/pnas.0603522103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mok DW, Mok MC. Cytokinin metabolism and action. Annu. Rev. Plant Physiol. Plant Mol. Biol. 2001;52:89–118. doi: 10.1146/annurev.arplant.52.1.89. [DOI] [PubMed] [Google Scholar]
- Müller A, Duchting P, Weiler EW. A multiplex GC–MS/MS technique for the sensitive and quantitative single-run analysis of acidic phytohormones and related compounds, and its applica-tion to Arabidopsis thaliana. Planta. 2002;216:44–56. doi: 10.1007/s00425-002-0866-6. [DOI] [PubMed] [Google Scholar]
- Naito T, Yamashino T, Kiba T, Koizumi N, Kojima M, Sakakibara H, et al. A link between cytokinin and ASL9 (ASYMMETRIC LEAVES 2 LIKE 9) that belongs to the AS2/LOB (LATERAL ORGAN BOUNDARIES) family genes in Arabidopsis thaliana. Biosci. Biotech. Biochem. 2007;71:1269–1278. doi: 10.1271/bbb.60681. [DOI] [PubMed] [Google Scholar]
- Nakagawa H, Jiang C.-J, Sakakibara H, Kojima M, Honda I, Ajisaka H, et al. Overexpression of a petunia zinc-finger gene alters cytokinin metabolism and plant forms. Plant J. 2005;41:512–523. doi: 10.1111/j.1365-313X.2004.02316.x. [DOI] [PubMed] [Google Scholar]
- Nemhauser JL, Hong F, Chory J. Different plant hormones regulate similar processes through largely nonoverlapping transcriptional responses. Cell. 2006;126:467–475. doi: 10.1016/j.cell.2006.05.050. [DOI] [PubMed] [Google Scholar]
- Nemhauser JL, Mockler TC, Chory J. Interdependency of brassinosteroid and auxin signaling in Arabidopsis. PLoS Biol. 2004;2:E258. doi: 10.1371/journal.pbio.0020258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni DA, Wang LJ, Ding CH, Xu ZH. Auxin distribution and transport during embryogenesis and seed germination of Arabidopsis. Cell Res. 2001;11:273–278. doi: 10.1038/sj.cr.7290096. [DOI] [PubMed] [Google Scholar]
- Pan X, Welti R, Wang X. Simultaneous quantification of major phytohormones and related compounds in crude plant extracts by liquid chromatography–electrospray tandem mass spectrometry. Phytochemistry. 2008;69:1773–1781. doi: 10.1016/j.phytochem.2008.02.008. [DOI] [PubMed] [Google Scholar]
- Pawlowski TA. Proteomics of European beech (Fagus sylvatica L.) seed dormancy breaking: influence of abscisic and gibberellic acids. Proteomics. 2007;7:2246–2257. doi: 10.1002/pmic.200600912. [DOI] [PubMed] [Google Scholar]
- Ross JJ, O’Neill DP, Rathbone DA. Auxin–gibberellin interactions in pea: integrating the old with the new. J. Plant Growth Regul. 2003;22:99–108. [Google Scholar]
- Saika H, Okamoto M, Miyoshi K, Kushiro T, Shinoda S, Jikumaru Y, et al. Ethylene promotes submergence-induced expression of OsABA8ox1, a gene that encodes ABA 8′-hydroxylase in rice. Plant Cell Physiol. 2007;48:287–298. doi: 10.1093/pcp/pcm003. [DOI] [PubMed] [Google Scholar]
- Saito K, Hirai MY, Yonekura-Sakakibara K. Decoding genes with coexpression networks and metabolomics—‘majority report by precogs’. Trends Plant Sci. 2008;13:36–43. doi: 10.1016/j.tplants.2007.10.006. [DOI] [PubMed] [Google Scholar]
- Sakakibara H. Cytokinins: activity, biosynthesis, and translocation. Annu. Rev. Plant Biol. 2006;57:431–449. doi: 10.1146/annurev.arplant.57.032905.105231. [DOI] [PubMed] [Google Scholar]
- Sakakibara H, Kasahara H, Ueda N, Kojima M, Takei K, Hishiyama S, et al. Agrobacterium tumefaciens increases cytokinin production in plastids by modifying the biosynthetic pathway in the host plant. Proc. Natl Acad. Sci. USA. 2005;102:9972–9977. doi: 10.1073/pnas.0500793102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki A, Itoh H, Gomi K, Ueguchi-Tanaka M, Ishiyama K, Kobayashi M, et al. Accumulation of phosphorylated repressor for gibberellin signaling in an F-box mutant. Science. 2003;299:1896–1898. doi: 10.1126/science.1081077. [DOI] [PubMed] [Google Scholar]
- Segarra G, Casanova E, Bellido D, Odena MA, Oliveira E, Trillas I. Proteome, salicylic acid, and jasmonic acid changes in cucumber plants inoculated with Trichoderma asperellum strain T34. Proteomics. 2007;7:3943–3952. doi: 10.1002/pmic.200700173. [DOI] [PubMed] [Google Scholar]
- Sossountzov L, Maldiney R, Sotta B, Sabbagh I, Habricot Y, Bonnet M, et al. Immunocytochemical localization of cytokinins in Craigella tomato and a sideshootless mutant. Planta. 1988;175:291–304. doi: 10.1007/BF00396334. [DOI] [PubMed] [Google Scholar]
- Takei K, Yamaya T, Sakakibara H. A method for separation and determination of cytokinin nucleotides from plant tissues. J. Plant Res. 2003;116:265–269. doi: 10.1007/s10265-003-0099-1. [DOI] [PubMed] [Google Scholar]
- Takei K, Yamaya T, Sakakibara H. Arabidopsis CYP735A1 and CYP735A2 encode cytokinin hydroxylases that catalyze the biosynthesis of trans-zeatin. J. Biol. Chem. 2004;279:41866–41872. doi: 10.1074/jbc.M406337200. [DOI] [PubMed] [Google Scholar]
- Tanaka M, Takei K, Kojima M, Sakakibara H, Mori H. Auxin controls local cytokinin biosynthesis in the nodal stem in apical dominance. Plant J. 2006;45:1028–1036. doi: 10.1111/j.1365-313X.2006.02656.x. [DOI] [PubMed] [Google Scholar]
- Tanaka N, Konishi H, Khan M.MK, Komatsu S. Proteome analysis of rice tissues by two-dimensional electrophoresis: an approach to the investigation of gibberellin regulated proteins. Mol. Genet. Genomics. 2004;270:485–496. doi: 10.1007/s00438-003-0929-9. [DOI] [PubMed] [Google Scholar]
- Ueguchi-Tanaka M, Ashikari M, Nakajima M, Itoh H, Katoh E, Kobayashi M, et al. GIBBERELLIN INSENSITIVE DWARF1 encodes a soluble receptor for gibberellin. Nature. 2005;437:693–698. doi: 10.1038/nature04028. [DOI] [PubMed] [Google Scholar]
- Umehara M, Hanada A, Yoshida S, Akiyama K, Arite T, Takeda-Kamiya N, et al. Inhibition of shoot branching by new terpenoid plant hormones. Nature. 2008;455:195–200. doi: 10.1038/nature07272. [DOI] [PubMed] [Google Scholar]
- Urakami E, Yamaguchi I, Asami T, Conrad U, Suzuki Y. Immunomodulation of gibberellin biosynthesis using an anti-precursor gibberellin antibody confers gibberellin-deficient phenotypes. Planta. 2008;228:863–873. doi: 10.1007/s00425-008-0788-z. [DOI] [PubMed] [Google Scholar]
- Veach YK, Martin RC, Mok DW, Malbeck J, Vankova R, Mok MC. O-glucosylation of cis-zeatin in maize. Characterization of genes, enzymes, and endogenous cytokinins. Plant Physiol. 2003;131:1374–1380. doi: 10.1104/pp.017210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang SH, Choi D. Characterization of genes encoding ABA 8′-hydroxylase in ethylene-induced stem growth of deepwater rice (Oryza sativa L.) Biochem. Biophys. Res. Commun. 2006;350:685–690. doi: 10.1016/j.bbrc.2006.09.098. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.