Abstract
OsRac1, a member of the Rac/Rop GTPase family, plays important roles as a molecular switch in rice innate immunity, and the active form of OsRac1 functions in the plasma membrane (PM). To study the precise localization of OsRac1 in the PM and its possible association with other signaling components, we performed proteomic analysis of DRMs (detergent-resistant membranes) isolated from rice suspension-cultured cells transformed with myc-tagged constitutively active (CA) OsRac1. DRMs are regions of the PM that are insoluble after Triton X-100 treatment under cold conditions and are thought to be involved in various signaling processes in animal, yeast and plant cells. We identified 192 proteins in DRMs that included receptor-like kinases (RLKs) such as Xa21, nucleotide-binding leucine-rich repeat (NB-LRR)-type disease resistance proteins, a glycosylphosphatidylinositol (GPI)-anchored protein, syntaxin, NADPH oxidase, a WD-40 repeat family protein and various GTP-binding proteins. Many of these proteins have been previously identified in the DRMs isolated from other plant species, and animal and yeast cells, validating the methods used in our study. To examine the possible association of DRMs and OsRac1-mediated innate immunity, we used rice suspension-cultured cells transformed with myc-tagged wild-type (WT) OsRac1 and found that OsRac1 and RACK1A, an effector of OsRac1, shifted to the DRMs after chitin elicitor treatment. These results suggest that OsRac1-mediated innate immunity is associated with DRMs in the PM.
Keywords: Chitin elicitor, Plasma membrane, RACK1, Rac/Rop GTPase, Rice suspension
Introduction
Plants have evolved various mechanisms to defend themselves against pathogens such as fungi, bacteria, viruses and nematodes. Such innate immunity in plants is essentially regulated by two systems: pathogen-associated molecular patterns (PAMPs)-triggered immunity (PTI) and effector-triggered immunity (ETI) (Jones and Dangl 2006, Crisholm et al. 2006). The former responds to various PAMPs derived from pathogens, and some receptor-like kinases (RLKs) that function in this system have been identified (Zipfel 2008). The latter is induced by a resistance (R) protein that specifically recognizes invading effectors derived from pathogens. Major cellular events that occur in response to pathogens in the two immune systems include induction of the hypersensitive response (HR), production of antimicrobial compounds and reactive oxygen species (ROS), and induction of pathogenesis-related (PR) genes.
The composition of the PM is not homogeneous, and lipid rafts, well characterized in animal and yeast cells (Kirkham and Parton 2005, Brown 2006, Zech et al. 2009), are regions of the PM containing high concentrations of cholesterol, glycosphingolipids and glycosylphosphatidylinositol (GPI)-anchored proteins (Simons and Toomre 2000). Lipid rafts exist as distinct liquid-ordered phases of the membrane that are resistant to treatment with non-ionic detergents, such as Triton X-100 and NP-40. Therefore, this region has recently been renamed as detergent-resistant membranes (DRMs). From animal and yeast studies, DRMs are thought to be involved in regulating signal transduction pathways such as endocytosis and exocytosis, protein secretion, apoptosis and the actin cytoskeleton (Simons and Toomre 2000, Sharma et al. 2002, Parton and Richards 2003, Helms and Zurzolo 2004). Recently, a number of reports on plant DRMs have been published, indicating that methods for the isolation of DRMs in plants have been relatively well established (Mongrand et al. 2004, Borner et al. 2005, Morel et al. 2006, Laloi et al. 2007, Sorek et al. 2007, Minami et al. 2009); however, the involvement of DRM proteins in plant innate immunity has not been systemically studied.
Rac/Rop GTPases play important roles in innate immunity, hormone response, cell growth and the actin cytoskeleton, establishment of polarity, and other cellular activities (Gu et al. 2004, Brembu et al. 2006, Nibau et al. 2006, Yang and Fu 2007, Yalovsky et al. 2008). This class of GTPases mainly functions in the PM as a molecular switch for a number of signaling pathways. Some Rac/Rop GTPases contain lipid modification sites at the C-terminus and the polybasic region that are required for PM binding (Lavy and Yalovsky 2006, Yalovsky et al. 2008). Recent studies suggest that localization of Rac/Rop GTPases to DRMs is important for their activation in various signaling pathways (Bloch et al. 2005, Sorek et al. 2007, Yalovsky et al. 2008). Furthermore, tobacco Rboh (a plant NADPH oxidase) was also shown to be localized to DRMs (Mongrand et al. 2004). In rice, we have shown that OsRac1 and OsrbohB localize to the PM and interact with each other to activate ROS production (Ono et al. 2001, Wong et al. 2007).
Our previous work shows that OsRac1 is a key regulator of rice innate immunity by regulating production of ROS, phytoalexin biosynthesis, induction of PR genes, regulation of mitogen-activated protein (MAP) kinase and suppression of metallothionein expression (Kawasaki et al. 1999, Ono et al. 2001, Wong et al. 2004, Lieberherr et al. 2005). Furthermore, OsRac1 interacts with and regulates the activity of cinnamoyl-CoA reductase for lignin biosynthesis (Kawasaki et al. 2006). Proteomic analysis of proteins regulated by OsRac1 indicates that the majority of proteins induced by a sphingolipid elicitor are also induced by constitutively activated OsRac1 (Kawasaki et al. 2006). We also demonstrated that OsRac1 forms a complex with RAR1, HSP90 and SGT1, conserved components in plant innate immunity (Boter et al. 2007, Thao et al. 2007). Furthermore, RACK1A was shown to be a novel effector protein of OsRac1 and to have an important role in OsRac1-mediated innate immunity by interacting with the N-terminus of OsrbohB (Thao et al. 2007, Nakashima et al. 2008).
Several studies have addressed the relationships between small G-proteins and DRMs in animals and yeast; however, the relationship between small G-proteins and DRMs has not been well studied in plants. In this study, we analyzed DRM proteins isolated from rice suspension-culured cells transformed with myc-tagged constitutively active (CA) OsRac1 by coupled liquid chromatography–tandem mass spectrometry (LC-MS/MS) and identified 192 proteins in DRMs. Furthermore, we show evidence to suggest that OsRac1-mediated innate immunity is associated with DRMs.
Results
OsRac1 localizes to the Triton X-100-insoluble fraction and DRMs
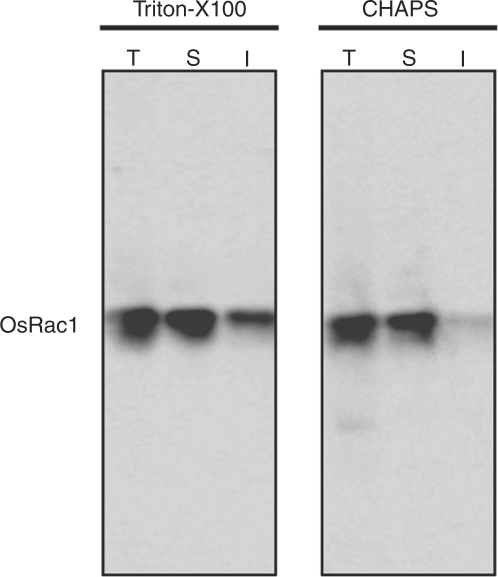
We have previously shown that CA-OsRac1 localizes to the PM and interacts with several proteins such as OsrbohB (Ono et al. 2001, Wong et al. 2007). Therefore, we first examined whether OsRac1 is present in the Triton X-100-insoluble fraction using rice suspension-culured cells expressing myc-tagged CA-OsRac1 (Lieberherr et al. 2005). Microsomal fractions from rice suspension-cultured cells were treated with 1% (v/v) Triton X-100 or CHAPS {3-[(3-cholamidopropyl)dimethylammonio]-1-propanesulfonic acid} at 4°C and the lysates were separated by ultracentrifugation at 125,000 × g for 1.5 h. After centrifugation, soluble and insoluble fractions were separated and OsRac1 was detected by immunoblotting with anti-myc antibody. The detected CA-OsRac1 showed a higher intensity in the Triton X-100-insoluble fraction than in the CHAPS-insoluble fraction (Fig. 1), suggesting an association of OsRac1 with DRMs that play important roles in numerous cellular functions (Munro 2003, Brown 2006, Hanzal-Bayer and Hancock 2007). Therefore, in the following experiments, we used Triton X-100 for purification of the DRM fraction.
Fig. 1.
Distribution of CA-OsRac1 in Triton X-100- and CHAPS-insoluble fractions. Transgenic rice suspension-cultured cells expressing myc-tagged CA-OsRac1 were lysed with 1% (v/v) Triton X-100 and 1% (v/v) CHAPS on ice and distributed to the soluble (S) and insoluble (I) microsomal fractions. The lysates were run on SDS–PAGE with total protein (T) and subjected to immunoblot analysis with anti-myc antibody.
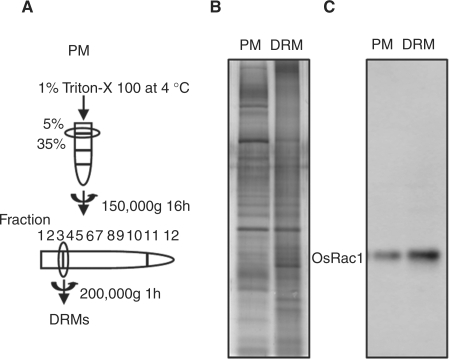
Recent studies have reported conditions for isolating DRMs from several plant species such as Arabidopsis thaliana, Nicotiana tabacum and Medicago truncatula (Mongrand et al. 2004, Laloi et al. 2007, Lefebvre et al. 2007, Sorek et al. 2007). These studies showed that Rac/Rop GTPases (AtRop6, AtRop10 and NtRac5), NtrbohD and the β subunit of heterotrimeric G-protein were detected in DRM fractions (Mongrand et al. 2004, Shahollari et al. 2004, Bloch et al. 2005, Sorek et al. 2007). We isolated DRMs from rice suspension-cultured cells using standard methods as described in Materials and Methods. The PM fractions enriched by two-phase partitioning were solubilized with 1% (v/v) Triton X-100 for 30 min at 4°C and fractionated by ultracentrifugation in sucrose gradients (Fig. 2A). A sample from each fraction was subjected to SDS–PAGE (Fig. 2B) and immunoblotted with anti-myc antibody to detect OsRac1. The SDS–PAGE patterns of the PM and DRM fractions are shown in Fig. 2B. The immunoblot showed that CA-OsRac1 co-purified with DRMs (Fig. 2C). These results suggest that OsRac1 interacts with DRMs in the PM in a manner similar to that of other small G-proteins, α and β subunits of heterotrimeric G-proteins and various signaling proteins.
Fig. 2.
Protocol for the purification of DRM fractions from rice suspension-cultured cells. (A) Diagram of the method used. (B) Flamingo-stained SDS–PAGE of the two-phase partitioned PM and purified DRM fraction. (C) Immunoblotting of OsRac1 with anti-myc antibody in the PM and the DRM.
Identification of proteins in DRMs isolated from rice suspension-cultured cells
We next performed LC-MS/MS analysis of DRM proteins isolated from rice suspension-cultured cells expressing CA-OsRac1. We used gel slice and shotgun analysis to identify proteins in the DRMs. Gel slices were divided into eight pieces followed by in-gel digestion as described previously (Tsunezuka et al. 2005, Fujiwara et al. 2006). For the shotgun analysis, proteins in DRMs were suspended in 6 M urea, and in-solution digests were carried out. Digested peptides were subjected to LC-MS/MS using the LTQ-Orbitrap, Paradigm MS4 and HTC-PAL systems. Raw data files of MS/MS results were searched against the National Center for Biotechnology Information (NCBI) database with the MASCOT server using search category Oryza sativa.
A total of 192 proteins were identified as DRM proteins (Table 1 and Supplementary Table 1). These proteins were identified in both gel slice and shotgun experiments, and three or more replicates from independent protein purifications were analyzed. All identified proteins are listed in Supplementary Table 1 with database search scores. Proteins associated with innate immunity, various receptors, SNARE (soluble N-ethylmaleimide-sensitive factor attachment protein receptor) proteins and GTP-binding proteins are listed in Table 1. RLKs were previously identified in plant DRMs (Morel et al. 2006, Ali et al. 2007). Our proteomic analysis identified several rice RLKs including Xa21 that is an R-protein for rice bacterial blight (Song et al. 1995) among the DRM proteins (Table 1). Furthermore, other proteins that recognize extracellular signals such as proteins with a leucine-rich repeat (LRR) domain and NB-LRR (nucleotide-binding LRR)-type resistance proteins were identified. Also, some GPI-anchored proteins and predicted GPI-anchored proteins were identified. NDR1, a GPI-anchored protein, is a major component of innate immunity (Coppinger et al. 2004). GPI-anchored proteins link to the DRM region in order to localize to the PM (Hein et al. 2009). We also identified RACK1A and Rboh in DRMs. We have recently shown that these two proteins interact with each other and play important roles in ROS production in rice (Nakashima et al. 2008). Therefore, these results suggest that their interaction may occur in DRMs.
Table 1.
LC-MS/MS identification of proteins in DRMs isolated from myc-tagged CA-OsRac1-transformed rice suspension cells
| Protein | Accession No. | Mol. wt (kDa) | MASCOT score | No. of hit peptides | Sequence coverage (%) | TM predictiona | |
|---|---|---|---|---|---|---|---|
| Innate immunity | Phosphatidylinositol-4-phosphate 5-kinase family protein, low similarity to phosphatidylinositol 3,5-kinase | gi|115467450 | 175 | 61 | 2 | 1 | 0 |
| Disease resistance protein RPM1 (CC-NBS-LRR class), putative domain signature CC-NBS-LRR exists | gi|125535287 | 140 | 61 | 2 | 1 | 0 | |
| NB-ARC domain, putative | gi|115484771 | 110 | 58 | 2 | 2 | 0 | |
| Respiratory burst oxidase protein D (RbohD)/NADPH oxidase identical to respiratory burst oxidase protein D | gi|108864453 | 105 | 186 | 12 | 23 | 5 | |
| Leucine-rich repeat transmembrane protein kinase, putative protein kinase Xa21 | gi|125532138 | 103 | 60 | 2 | 1 | 2 | |
| Respiratory burst oxidase protein D (RbohD)/NADPH oxidase identical to respiratory burst oxidase protein D | gi|115488934 | 100 | 108 | 9 | 14 | 5 | |
| Heat shock protein 90 | gi|6863054 | 93 | 80 | 4 | 6 | 0 | |
| Serine/threonine protein kinases active site signature | gi|32988727 | 76 | 62 | 2 | 3 | 1 | |
| Atypical harpin-like kinase MARK | gi|13324792 | 72 | 548 | 14 | 32 | 1 | |
| Heat shock cognate 70 kDa protein | gi|115452223 | 71 | 305 | 10 | 14 | 0 | |
| Nodulation harpin kinase-like protein | gi|115478014 | 59 | 129 | 4 | 9 | 1 | |
| Putative elicitor inducible beta-1,3-glucanase NtEIG-E76 | gi|115439837 | 54 | 100 | 9 | 18 | 1 | |
| Putative disease resistance protein | gi|115434084 | 48 | 75 | 2 | 5 | 0 | |
| Putative protein kinase serine/threonine protein kinase, putative similar to Pto kinase interactor 1 (Pti1) | gi|115456539 | 41 | 54 | 2 | 9 | 0 | |
| Chitin elicitor-binding protein | gi|108860575 | 40 | 218 | 8 | 27 | 2 | |
| Putative Pto kinase interactor 1 | gi|115461953 | 39 | 75 | 6 | 19 | 0 | |
| Receptor for activated C kinase 1A (RACK1A) | gi|115439261 | 36 | 115 | 3 | 13 | 0 | |
| Putative hypersensitive-induced response protein | gi|115465785 | 32 | 477 | 25 | 48 | 0 | |
| Band 7 family protein strong similarity to hypersensitive-induced response protein | gi|115482396 | 32 | 212 | 11 | 32 | 0 | |
| Hypersensitive-induced response protein | gi|14150732 | 31 | 106 | 4 | 9 | 0 | |
| Hypersensitive-induced response protein | gi|115476296 | 31 | 337 | 21 | 40 | 0 | |
| Small GTP-binding protein OsRac2 | gi|115464861 | 24 | 59 | 2 | 16 | 0 | |
| Harpin-induced family protein (YLS9)/HIN1 family protein | gi|115461434 | 22 | 179 | 3 | 25 | 2 | |
| Harpin-induced gene 1 homolog | gi|2801538 | 22 | 92 | 6 | 30 | 2 | |
| Harpin-induced protein 1-containing protein | gi|115487442 | 21 | 154 | 6 | 27 | 2 | |
| Harpin-induced family protein/NDR1/HIN1-like protein 3 similar to harpin-induced protein hin1 | gi|32993161 | 21 | 93 | 3 | 23 | 2 | |
| Harpin-induced protein-related/HIN1-related/harpin-responsive protein-related weak similarity to hin1 | gi|115472507 | 19 | 76 | 2 | 11 | 0 | |
| Receptor | Putative receptor-like protein kinase | gi|115467902 | 94 | 61 | 5 | 6 | 2 |
| Leucine-rich repeat family protein, expressed | gi|108863916 | 84 | 81 | 9 | 12 | 2 | |
| Leucine-rich repeat transmembrane protein kinase, putative | gi|116012935 | 74 | 211 | 4 | 11 | 1 | |
| Somatic embryogenesis protein kinase 1 | gi|52854318 | 70 | 59 | 5 | 8 | 2 | |
| Brassinosteroid-insensitive 1-associated harpin kinase 1 precursor (somatic embryogenesis harpin-like kinase 3) | gi|115458750 | 70 | 59 | 4 | 5 | 1 | |
| Leucine-rich repeat transmembrane protein kinase, putative similar to CLV1 harpin kinase | gi|115459088 | 66 | 59 | 3 | 7 | 1 | |
| Protein kinase family protein, putative, expressed | gi|115450539 | 59 | 131 | 5 | 13 | 0 | |
| Putative kinase-like protein TMKL1 precursor | gi|115438420 | 54 | 81 | 2 | 4 | 2 | |
| Leucine-rich repeat family protein similar to leucine-rich repeat protein SHOC-2 (Ras-binding protein Sur-8) | gi|32975440 | 51 | 79 | 2 | 5 | 0 | |
| Peptidoglycan-binding LysM domain-containing protein contains Pfam profile PF01476 | gi|115480519 | 39 | 164 | 4 | 14 | 2 | |
| SNARE | Syntaxin 121, putative, expressed | gi|115455787 | 36 | 56 | 2 | 5 | 1 |
| Putative syntaxin-related protein (SYP132) | gi|115470721 | 34 | 189 | 6 | 20 | 1 | |
| Synaptobrevin family protein similar to vesicle-associated membrane protein 7 | gi|125557559 | 32 | 156 | 9 | 25 | 2 | |
| Novel plant SNARE 11 | gi|115453189 | 31 | 65 | 12 | 30 | 1 | |
| Syntaxin 71 (SYP71) identified as syntaxin of plants 71 | gi|115465329 | 30 | 199 | 14 | 47 | 1 | |
| Vesicle-associated membrane protein 724 | gi|115456011 | 25 | 53 | 2 | 11 | 1 | |
| Small G-protein and others | WD-40 repeat family protein contains three WD-40 repeats (PF00400); some similarity to s-tomosyn isoform | gi|156765316 | 168 | 343 | 21 | 20 | 0 |
| WD-40 repeat family protein contains three Pfam PF00400: WD domain, G-beta repeats | gi|37988349 | 148 | 582 | 29 | 25 | 0 | |
| Probable GTP-binding protein | gi|115437816 | 90 | 89 | 2 | 5 | 0 | |
| Transducin family protein/WD-40 repeat family protein | gi|108706574 | 82 | 606 | 33 | 34 | 0 | |
| Transducin family protein/WD-40 repeat family protein contains six WD-40 repeats; similar to cell cycle control protein cwf8 | gi|115482422 | 58 | 64 | 8 | 21 | 0 | |
| GTP-binding protein GTP1 | gi|115439941 | 24 | 59 | 3 | 8 | 0 | |
| ADP-ribosylation factor, putative, expressed | gi|108711707 | 22 | 87 | 3 | 18 | 0 |
aThe numbers of putative transmembrane domains were predicted by SOSUI.
Pto-interacting protein 1 (Pti1) is phosphorylated by Pto, a tomato resistance protein involved in the oxidative stress signaling pathway (Zhou et al. 1995, Anthony et al. 2006). A rice homolog, OsPti1, functions as a negative regulator of defense responses (Takahashi et al. 2007), and its function is dependent on RAR1, a factor in OsRac1-mediated innate immunity (Thao et al. 2007). Our proteomic analysis identified H+-ATPase, ABC transporters, aquaporins and Band 7 proteins among the DRM proteins. These membrane proteins were also identified as typical DRM proteins in animals, yeast and plants (Mairhofer et al. 2002, Fricke et al. 2003). Thus, these results validate our methods for isolation of DRMs from rice suspension-cultured cells.
Shift of OsRac1 and RACK1 to DRMs after elicitor treatment
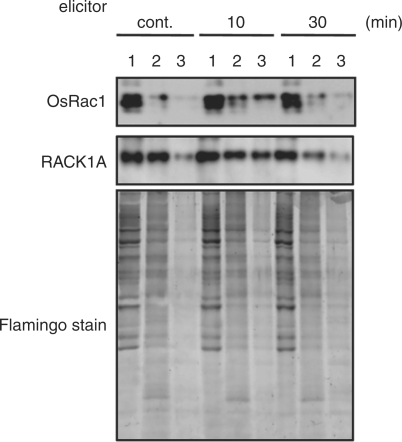
To study whether DRMs are important for OsRac1-mediated innate immunity, we investigated possible changes in the localization of OsRac1 after chitin elicitor treatment in rice suspension-cultured cells expressing myc-tagged wild-type (WT)-OsRac1. We treated WT-OsRac1 suspension-cultured cells with chitin elicitor for 0, 10 and 30 min, and isolated DRM fractions from the cells. Our results showed that at 10 min after elicitor treatment OsRac1 was clearly localized to DRM fractions. This result suggests that translocation of OsRac1 to DRMs is important for the initial response in rice innate immunity. We also found that RACK1A was translocated from non-DRMs to DRMs after elicitor treatment. The signal patterns of these two proteins were similar and were detected until 30 min after elicitor treatment, with the bands becoming faint later. These results indicate that the translocation of proteins to DRMs may be important for OsRac1-mediated innate immunity and this initial response occurs within 10–30 min after elicitor treatment.
Discussion
OsRac1 is present in the DRMs
We found that CA-OsRac1 is present in the Triton X-100-insoluble fraction and in DRMs. In animals and yeast, several small G-proteins and heterotrimeric G-proteins localize to DRMs (Subtil et al. 2004, Sugawara et al. 2007, Yuyama et al. 2007). Recently, some G-proteins were identified in DRMs in plants (Morel et al. 2006, Yalovsky et al. 2008). Therefore, our results extend these observations to rice.
Translocation of OsRac1 and RACK1A to DRMs after elicitor treatment
We showed that OsRac1 and RACK1A were translocated to DRMs after elicitor treatment of rice suspension-cultured cells expressing myc-tagged WT-OsRac1. These results suggest a connection between immune responses triggered by OsRac1 and DRMs ( Fig. 3) and further suggest that the interactions between OsRac1, RACK1A and other components in the DRMs may be important for rice innate immunity. Nevertheless, since we have only examined the translocations of OsRac1 and RACK1A to DRMs after elicitor treatment, we need to examine the translocation of other proteins involved in innate immune responses to the DRMs as has been shown in mammalian cells. Signaling components such as receptors, G-proteins, heat shock proteins and protein kinases move to and are concentrated in DRMs after animal cells are stimulated with bacterial endotoxin and lipopolysaccharide (LPS) (Triantafilou et al. 2002, Yuyama et al. 2007). Major mammalian signaling molecules such as TLR4, CXCR4 and GDF that are involved in LPS-induced cell signaling are not always found in DRMs, but are recruited to DRMs after LPS stimulation. Our results are similar to results found in mammalian innate immunity. Prolonged output of innate immune signaling, such as extended production of ROS, would cause damage to cells; therefore, it may be necessary for the initial response of innate immunity to be short and transient. DRMs may have a role in providing a platform for the initial events of the immune response in plants as well as in mammals.
Fig. 3.
Time course analysis of OsRac1 and RACK1A in DRM and non-DRM fractions after elicitor treatment. After treatment of rice suspension cells expressing WT-OsRac1 with chitin elicitor for 10 and 30 min, total (1), non-DRM (2) and DRM (3) fractions were subjected to immunoblotting with anti-myc or anti-RACK1A antibody.
Proteins identified in DRMs
We identified proteins in the DRMs fraction by mass spectrometry and list the defense-related proteins in Table 1. RACK1A, HSP70, HSP90 and Rboh were identified in DRMs. Previously, these proteins were shown to interact with OsRac1 during immune responses (Thao et al. 2007, Wong et al. 2007, Nakashima et al. 2008). Immunoblotting experiments showed that RACK1A was translocated to DRMs after stimulation of rice suspension-cultured cells with chitin elicitor (Fig. 3). RACK1A was also identified in DRMs isolated from cells expressing CA-OsRac1 (Table 1) by using mass spectrometry. Since RACK1A is thought to be a scaffolding protein and have multiple cellular functions in plants (Chen et al. 2006, Nakashima et al. 2008), its subcellular localization may not be simple. In our previous work, RACK1A was shown to interact with the N-terminus of OsrbohB by using a yeast two-hybrid system (Nakashima et al. 2008). The current proteomic analysis showed that Rboh is present in DRMs, suggesting that the interaction of RACK1 and Rboh occurs in the DRMs. In fact, analysis of the protein complex in mammals and yeast showed that several protein complexes involved in G-protein signaling are formed in DRMs (Ostrom and Insel 2004, Li et al. 2007, Dudez et al. 2008).
RLKs and disease-resistance (R) proteins identified in the DRMs in this study have been detected in DRMs in other plants (Morel et al. 2006, Minami et al. 2009). RLKs are involved in perception of PAMPs (Zipfel 2008). In animals, many receptor complexes were formed in DRMs, and they are involved in signal initiation (Simons and Toomre 2000). Recently, Qi and Katagiri (2009) purified low-abundance Arabidopsis PM protein complexes using tandem affinity purification of tagged RPS2, an NBS-LRR-type resistance protein of Arabidopsis, and identified protein components of the RPS2 complex. Identified proteins included Band 7 family protein and aquaporin PIP1.2. Both proteins were identified as DRM proteins in this work. It is possible that these disease resistance proteins interact with Band 7 family protein or aquaporin in the DRMs at an early stage of innate immunity. Both proteins are known as major lipid raft proteins (Borner et al. 2005, Morel et al. 2006) and stress response proteins (Nadimpalli et al. 2000, Rostoks et al. 2003, Qi and Katagiri 2009). Band 7 protein belongs to the SPFH (stomatin prohibitin flotilin Hbc) domain protein superfamily. In mammals, it is known that proteins of this family have roles as scaffolders and regulators for the components, such as ion channels, in lipid raft microdomains (Browman et al. 2007). So, Qi and Katagiri (2009) discussed the possibility that Band 7 family protein and aquaporin PIP1.2 were involved in RPS2-mediated resistance. Together, these results suggest that DRMs are important platforms for these identified proteins to regulate innate immunity in rice.
In this study, intracellular trafficking-associated proteins, SNARE, SYP (syntaxin of plant) and VAMP, were also identified as DRM proteins. Recent studies indicate that intracellular trafficking is important for plant innate immunity (Robatzek et al. 2006, Robatzek 2007, Geldner and Robatzek 2008). PEN1/SYP121, also identified in this work as a DRM protein (Table 1), has roles in penetration of Blumeria graminis into plant cells and in protein secretion as a component of the SNARE complex (Collins et al. 2003, Kwon et al. 2008). Tomato LeEix is a cell surface glycoprotein with a mammalian endocytosis signal and a receptor for ethylene-inducing xylanase (Hanania et al. 1999, Ron and Avni, 2004). Since mutations in the endocytosis signal sequence in LeEix abolish HR induction, endocytosis is suggested to be a key regulator of the signal transduction pathway for HR (Ron and Avni 2004). Flagellin receptor FLS2 is a trans-membrane LRR-RLK that is translocated to the endomembrane components after flagellin treatment (Robatzek et al. 2006). Therefore, receptor recycling may be important in plant innate immunity.
Traditionally, two-dimensional electrophoresis, which combines isoelectric focusing (IEF) and SDS–PAGE, is a typical proteomic technique providing high resolution for separation and characterization. For this method, the protein sample need to be completely solubilized for analysis of the IEF gel. In this work, we selected the methods of SDS–PAGE and shotgun analysis, because DRM proteins were present in smaller amounts in cells and difficult to be solubilized. The level of resolution obtained by SDS–PAGE and shotgun analysis is relatively low; therefore, it was often difficult to identify many proteins. However, LTQ-Orbitrap mass analysis, which provides high resolution and high sensitivity, permitted characterization of DRM proteins without pre-separation of sample proteins. Thus, in this work, we could identify many proteins from a small amount of proteins.
The results of our proteomic analysis of DRMs in rice suggest that intracellular membrane trafficking and DRMs may be involved in the initial events occurring during OsRac1-mediated innate immunity; however, it will be difficult to analyze components of DRMs further only by the proteomic approach in the future. To understand further the roles of proteins in DRMs in rice innate immunity, other methods such as novel bioimaging technologies should be combined with biochemical approaches.
Materials and Methods
Rice suspension-cultured cells and elicitor treatment
Rice suspension-cultured cells expressing CA-OsRac1 have been described previously (Lieberherr et al. 2005, Thao et al. 2007). For the elicitor experiments we used rice suspension-cultured cells expressing WT-OsRac1 under the control of a maize Ubiquitin promoter. Transgenic cells growing in 20 ml of medium were incubated on a rotary shaker (50 r.p.m) at 30°C and subcultured weekly. The chitin elicitor (chitooligosaccharide mixture; 2 μg ml−1, from SEIKAGAKU Corporation, Tokyo, Japan) was applied to the cells 4 d after subculture. At selected intervals after the elicitor treatment, treated cells were harvested and frozen in liquid nitrogen.
Isolation of detergent-soluble and -insoluble membranes
Protein distribution assays were performed according to the method of Sorek et al. (2007). Rice cells in suspension cultures were harvested 4 d after subculture and homogenized in homogenizing medium [50 mM MOPS/KOH; pH 7.6, 5 mM EGTA, 5 mM EDTA, 0.5 M d-sorbitol, 2 mM phenylmethylsulfonyl fluoride (PMSF), 2.5 mM dithiothreitol (DTT), protease inhibitor cocktail (Roche, Penzberg, Germany)]. After incubation on ice for 15 min, the lysates were filtered through Miracloth (Calbiochem, Darmstadt, Germany) into a new tube and centrifuged at 125,000 × g for 30 min. The insoluble pellet was lysed with 1% (v/v) Triton X-100 and 1% (v/v) CHAPS on ice for 30 min. The lysates were next subjected to centrifugation for separation of soluble and insoluble fractions. Insoluble fractions were solubilized with 1% (v/v) Triton X-100 or 1% CHAPS (v/v) buffer containing 1% SDS (w/v).
Extraction and purification of the PM
Rice cells in suspension cultures were harvested 4 d after subculture and homogenized in homogenizing medium (50 mM MOPS/KOH; pH 7.6, 5 mM EGTA, 5 mM EDTA, 0.5 M d-sorbitol, 2 mM PMSF, 2.5 mM DTT). The microsomal fraction was extracted as described previously (Nakashima et al. 2008) and subjected to PM purification. PM was enriched from the microsomal fraction by using a polyethylene glycol–dextran (6.4%, w/w) aqueous two-phase partitioning system (Uemura et al. 1995) The two-phase partitioning was repeated three times to obtain more highly purified PM fractions. DRMs were purified from the PM fraction according to published methods (Mongrand et al. 2004, Sorek et al. 2007). The PM fractions were suspended in TED buffer (50 mM Tris–HCl; pH 7.4, 3 mM EDTA, 1 mM DTT), Triton X-100 was added to a detergent : PM protein ratio of 15 : 1, and the mixture was incubated for 30 min on ice. After incubation, the sample was diluted with sucrose solution to a final concentration of 52% sucrose (w/w), overlaid with 40, 35, 5% sucrose in TED buffer (w/w) and centrifuged for 16 h at 150,000 × g in a Swi40Ti rotor (Beckman, Fullerton, CA, USA). DRM fractions were recovered above the 5 and 35% interface, diluted five times with TED buffer and centrifuged for 1 h at 200,000 × g. Final pellets were suspended in TED buffer containing 1% (v/v) N-octyl glucoside for SDS–PAGE or 6 M urea/25 mM NH4HCO3 for shotgun analysis.
The protein contents of the fractions were determined by the BCA assay reagent (Pierce, Rockford, IL, USA), using bovine serum albumin (BSA) as a standard.
Immunoblotting
Sample proteins were separated by SDS–PAGE [acrylamide concentration 10.5% (w/v)] and electrotransferred onto an Immobilon-P membrane (Millipore, Billerica, MA, USA) for immunoblot detection. The membrane was blocked for 1 h in phosphate-buffered saline (PBS; 137 mM NaCl, 8.1 mM Na2HPO4, 2.68 mM KCl, 1.47 mM KH2PO4) containing 5% (w/v) skim milk and incubated for 2 h with anti-myc (Nacalai Tesque, Kyoto, Japan) or anti-RACK1A antibody (Nakashima et al. 2008). After washing with PBS containing 0.1% (v/v) Tween-20, the membranes were incubated for 1.5 h with anti-rabbit IgG conjugated to horseradish peroxidase (GE Healthcare, Buckinghamshire, UK). Chemical enhancement was performed using ECL PLUS Western blot detection reagents (GE Healthcare). The enhanced signals were detected by an LAS-3000 system (Fujifilm, Tokyo, Japan).
Peptide preparation for MS/MS analysis
DRM samples were diluted with Laemmli sample buffer, incubated at 65°C for 15 min and subjected to SDS–PAGE [acrylamide concentration 10.5% (w/v)]. Gel lanes sliced into eight bands of equal length from the Flamingo (BioRad, Hercules, CA, USA)-stained gels were washed twice with HPLC-grade water containing 30% (v/v) acetonitrile (Kanto Chemical, Tokyo, Japan), washed with 100% acetonitrile and dried in a vacuum concentrator. The dried gel pieces were treated with 2 μl of 0.5 μg μl−1 trypsin (sequence grade; Promega, Madison, WI, USA)/50 mM ammonium bicarbonate (Shevchenko and Shevchenko 2001) and incubated at 37°C for 16 h. The digested peptides in the gel pieces were recovered twice with 20 μl of 5% (v/v) formic acid/50% (v/v) acetonitrile. Finally, combined extracts were dried in a vacuum concentrator.
For shotgun analysis, in-solution digestion was carried out. DRM samples suspended in 6 M urea/25 mM NH4HCO3 were reduced with 5 mM DTT for 1 h at 37°C and alkylated with 25 mM iodoacetamide in the dark for 1 h at room temperature. Then, 25 mM DTT was added to quench the alkylated solution for 1 h at 37°C. After dilution with 25 mM NH4HCO3 for 6 M urea to 1 M, trypsin was added at a ratio of 1 : 30 (enzyme : peptide) and incubated at 37°C for 16 h. Finally, combined extracts were dried in a vacuum concentrator.
Mass spectrometric analysis and database searching
LC-MS/MS analyses were performed by using an LTQ-Orbitrap XL-HTC-PAL-Paradigm MS4 system. Trypsin-digested peptides were loaded on the column (75 μm internal diameter, 15 cm; L-Column, CERI, Auburn, CA, USA) using a Paradigm MS4 HPLC pump (Michrom BioResources) and an HTC-PAL autosampler (CTC analytics, Zwingen, Switzerland). Buffers were 0.1% (v/v) acetic acid and 2% (v/v) acetonitrile in water (A) and 0.1% (v/v) acetic acid and 90% (v/v) acetonitrile in water (B). A linear gradient from 5 to 45% B for 25 min was applied, and peptides eluted from the column were introduced directly into an LTQ-Orbitrap mass spectrometer (Thermo Fisher Scientific, Bremen, Germany) with a flow rate of 300 nl min–1 and a spray voltage of 2.0 kV. The range of MS scan was m/z 200–2,000 and the top three peaks were subjected to MS/MS analysis. The obtained spectra were compared with a protein database (20080607) from the National Center for Biotechnology Information (NCBI) using the MASCOT server (version 2.1 Matrix Science, London, UK). The MASCOT search parameters were as follows: set off the threshold at 0.05 in the ion score cut-off, peptide tolerance at 10 p.p.m., MS/MS tolerance at ± 0.8Da, peptide charge of 2 + or 3 + , trypsin as enzyme allowing up to one missed cleavage, carbamidomethylation on cysteines as a fixed modification and oxidation on methionine as a variable modification. For the prediction search of transmembrane domains in identified proteins, we used SOSUI (http://bp.nuap.nagoya-u.ac.jp/sosui/sosuiframe0E.html).
Supplementary data
Supplementary data are available at PCP online.
Funding
The Ministry of Agriculture, Forestry, and Fisheries of Japan Grants-in-Aid (Rice Genome Project IP4001); the Japan Society for Promotion of Science (13G0023); Target Protein Research Program (grant to K.S.); the Program for Promotion of Basic Research Activities for Innovative Biosciences (PROBRAIN; to T.K.).
Acknowledgments
We thank Dr. Keiko Imai for providing transgenic rice cells expressing myc-tagged WT-OsRac1.
Glossary
Abbreviations
- CA
constitutively active
- CHAPS
3-[(3-cholamidopropyl)dimethylammonio]-1-propanesulfonic acid
- DRM
detergent-resistant membrane
- DTT
dithiothreitol
- GPI
glycosylphosphatidylinositol
- HR
hypersensitive response
- IEF isoelectric focusing; LPS
lipopolysaccharide
- NB-LRR
nucleotide-binding leucine-rich repeat
- PAMPs
pathogen-associated molecular patterns
- PBS
phosphate-buffered saline
- PM
plasma membrane
- PMSF
phenylmethylsulfonyl fluoride
- PR
pathogenesis related
- RLK
receptor-like kinase
- WT
wild-type.
References
- Ali GS, Prasad KV, Day I, Reddy AS. Ligand-dependent reduction in the membrane mobility of FLAGELLIN SENSITIVE2, an Arabidopsis receptor-like kinase. Plant Cell Physiol. 2007;48:1601–1611. doi: 10.1093/pcp/pcm132. [DOI] [PubMed] [Google Scholar]
- Anthony RG, Khan S, Costa J, Pais MS, Bogre L. The Arabidopsis protein kinase PTI1-2 is activated by convergent phosphatidic acid and oxidative stress signaling pathways downstream of PDK1 and OXI1. J. Biol. Chem. 2006;281:37536–37546. doi: 10.1074/jbc.M607341200. [DOI] [PubMed] [Google Scholar]
- Bloch D, Lavy M, Efrat Y, Efroni I, Bracha-Drori K, Abu-Abied M, et al. Ectopic expression of an activated RAC in Arabidopsis disrupts membrane cycling. Mol. Biol. Cell. 2005;16:1913–1927. doi: 10.1091/mbc.E04-07-0562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borner GH, Sherrier DJ, Weimar T, Michaelson LV, Hawkins ND, Macaskill A, et al. Analysis of detergent-resistant membranes in Arabidopsis. Evidence for plasma membrane lipid rafts. Plant Physiol. 2005;137:104–116. doi: 10.1104/pp.104.053041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boter M, Amigues B, Peart J, Breuer C, Kadota Y, Casais C, et al. Structural and functional analysis of SGT1 reveals that its interaction with HSP90 is required for the accumulation of Rx, an R protein involved in plant immunity. Plant Cell. 2007;19:3791–3804. doi: 10.1105/tpc.107.050427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brembu T, Winge P, Bones AM, Yang Z. A RHOse by any other name: a comparative analysis of animal and plant Rho GTPases. Cell Res. 2006;16:435–445. doi: 10.1038/sj.cr.7310055. [DOI] [PubMed] [Google Scholar]
- Browman DT, Hoegg MB, Robbins SM. The SPFH domain-containing proteins: more than lipid raft markers. Trends Cell Biol. 2007;17:394–402. doi: 10.1016/j.tcb.2007.06.005. [DOI] [PubMed] [Google Scholar]
- Brown DA. Lipid rafts, detergent-resistant membranes, and raft targeting signals. Physiology (Bethesda) 2006;21:430–439. doi: 10.1152/physiol.00032.2006. [DOI] [PubMed] [Google Scholar]
- Collins NC, Thordal-Christensen H, Lipka V, Bau S, Kombrink E, Qiu JL, et al. SNARE-protein-mediated disease resistance at the plant cell wall. Nature. 2003;425:973–977. doi: 10.1038/nature02076. [DOI] [PubMed] [Google Scholar]
- Coppinger P, Repetti PP, Day B, Dahlbeck D, Mehlert A, Staskawicz BJ. Overexpression of the plasma membrane-localized NDR1 protein results in enhanced bacterial disease resistance in Arabidopsis thaliana. Plant J. 2004;40:225–237. doi: 10.1111/j.1365-313X.2004.02203.x. [DOI] [PubMed] [Google Scholar]
- Dudez T, Borot F, Huang S, Kwak BR, Bacchetta M, Ollero M, et al. CFTR in a lipid raft–TNFR1 complex modulates gap junctional intercellular communication and IL-8 secretion. Biochim. Biophys. Acta. 2008;1783:779–788. doi: 10.1016/j.bbamcr.2008.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fricke B, Argent AC, Chetty MC, Pizzey AR, Turner EJ, Ho MM, et al. The ‘stomatin’ gene and protein in overhydrated hereditary stomatocytosis. Blood. 2003;102:2268–2277. doi: 10.1182/blood-2002-06-1705. [DOI] [PubMed] [Google Scholar]
- Fujiwara M, Umemura K, Kawasaki T, Shimamoto K. Proteomics of Rac GTPase signaling reveals its predominant role in elicitor-induced defense response of cultured rice cells. Plant Physiol. 2006;140:734–745. doi: 10.1104/pp.105.068395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geldner N, Robatzek S. Plant receptors go endosomal: a moving view on signal transduction. Plant Physiol. 2008;147:1565–1574. doi: 10.1104/pp.108.120287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu Y, Wang Z, Yang Z. ROP/RAC GTPase: an old new master regulator for plant signaling. Curr. Opin. Plant Biol. 2004;7:527–536. doi: 10.1016/j.pbi.2004.07.006. [DOI] [PubMed] [Google Scholar]
- Hanania U, Furman-Matarasso N, Ron M, Avni A. Isolation of a novel SUMO protein from tomato that suppresses EIX-induced cell death. Plant J. 1999;19:533–541. doi: 10.1046/j.1365-313x.1999.00547.x. [DOI] [PubMed] [Google Scholar]
- Hanzal-Bayer MF, Hancock JF. Lipid rafts and membrane traffic. FEBS Lett. 2007;581:2098–2104. doi: 10.1016/j.febslet.2007.03.019. [DOI] [PubMed] [Google Scholar]
- Hein Z, Hooper NM, Naim HY. Association of a GPI-anchored protein with detergent-resistant membranes facilitates its trafficking through the early secretory pathway. Exp. Cell Res. 2009;315:348–356. doi: 10.1016/j.yexcr.2008.10.038. [DOI] [PubMed] [Google Scholar]
- Helms JB, Zurzolo C. Lipids as targeting signals: lipid rafts and intracellular trafficking. Traffic. 2004;5:247–254. doi: 10.1111/j.1600-0854.2004.0181.x. [DOI] [PubMed] [Google Scholar]
- Kawasaki T, Henmi K, Ono E, Hatakeyama S, Iwano M, Satoh H, et al. The small GTP-binding protein Rac is a regulator of cell death in plants. Proc. Natl Acad. Sci. USA. 1999;96:10922–10926. doi: 10.1073/pnas.96.19.10922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawasaki T, Koita H, Nakatsubo T, Hasegawa K, Wakabayashi K, Takahashi H, et al. Cinnamoyl-CoA reductase, a key enzyme in lignin biosynthesis, is an effector of small GTPase Rac in defense signaling in rice. Proc. Natl Acad. Sci. USA. 2006;103:230–235. doi: 10.1073/pnas.0509875103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirkham M, Parton RG. Clathrin-independent endocytosis: new insights into caveolae and non-caveolar lipid raft carriers. Biochim. Biophys. Acta. 2005;1746:349–363. doi: 10.1016/j.bbamcr.2005.11.007. [DOI] [PubMed] [Google Scholar]
- Kwon C, Neu C, Pajonk S, Yun HS, Lipka U, Humphry M, et al. Co-option of a default secretory pathway for plant immune responses. Nature. 2008;451:835–840. doi: 10.1038/nature06545. [DOI] [PubMed] [Google Scholar]
- Laloi M, Perret AM, Chatre L, Melser S, Cantrel C, Vaultier MN, et al. Insights into the role of specific lipids in the formation and delivery of lipid microdomains to the plasma membrane of plant cells. Plant Physiol. 2007;143:461–472. doi: 10.1104/pp.106.091496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavy M, Yalovsky S. Association of Arabidopsis type-II ROPs with the plasma membrane requires a conserved C-terminal sequence motif and a proximal polybasic domain. Plant J. 2006;46:934–947. doi: 10.1111/j.1365-313X.2006.02749.x. [DOI] [PubMed] [Google Scholar]
- Lefebvre B, Furt F, Hartmann MA, Michaelson LV, Carde JP, Sargueil-Boiron F, et al. Characterization of lipid rafts from Medicago truncatula root plasma membranes: a proteomic study reveals the presence of a raft-associated redox system. Plant Physiol. 2007;144:402–418. doi: 10.1104/pp.106.094102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Serwanski DR, Miralles CP, Bahr BA, De Blas AL. Two pools of Triton X-100-insoluble GABA(A) receptors are present in the brain, one associated to lipid rafts and another one to the post-synaptic GABAergic complex. J. Neurochem. 2007;102:1329–1345. doi: 10.1111/j.1471-4159.2007.04635.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieberherr D, Thao NP, Nakashima A, Umemura K, Kawasaki T, Shimamoto K. A sphingolipid elicitor-inducible mitogen-activated protein kinase is regulated by the small GTPase OsRac1 and heterotrimeric G-protein in rice. Plant Physiol. 2005;138:1644–1652. doi: 10.1104/pp.104.057414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mairhofer M, Steiner M, Mosgoeller W, Prohaska R, Salzer U. Stomatin is a major lipid-raft component of platelet alpha granules. Blood. 2002;100:897–904. doi: 10.1182/blood.v100.3.897. [DOI] [PubMed] [Google Scholar]
- Minami A, Fujiwara M, Furuto A, Fukao Y, Yamashita T, Kamo M, et al. Alterations in detergent-resistant plasma membrane microdomains in Arabidopsis thaliana during cold acclimation. Plant Cell Physiol. 2009;50:341–359. doi: 10.1093/pcp/pcn202. [DOI] [PubMed] [Google Scholar]
- Mongrand S, Morel J, Laroche J, Claverol S, Carde JP, Hartmann MA, et al. Lipid rafts in higher plant cells: purification and characterization of Triton X-100-insoluble microdomains from tobacco plasma membrane. J. Biol. Chem. 2004;279:36277–36286. doi: 10.1074/jbc.M403440200. [DOI] [PubMed] [Google Scholar]
- Morel J, Claverol S, Mongrand S, Furt F, Fromentin J, Bessoule JJ, et al. Proteomics of plant detergent-resistant membranes. Mol. Cell Proteomics. 2006;5:1396–1411. doi: 10.1074/mcp.M600044-MCP200. [DOI] [PubMed] [Google Scholar]
- Munro S. Lipid rafts: elusive or illusive? Cell. 2003;115:377–388. doi: 10.1016/s0092-8674(03)00882-1. [DOI] [PubMed] [Google Scholar]
- Nadimpalli R, Yalpani N, Johal GS, Simmons CR. Prohibitins, stomatins, and plant disease response genes compose a protein superfamily that controls cell proliferation, ion channel regulation, and death. J. Biol. Chem. 2000;275:29579–29586. doi: 10.1074/jbc.M002339200. [DOI] [PubMed] [Google Scholar]
- Nakashima A, Chen L, Thao NP, Fujiwara M, Wong HL, Kuwano M, et al. RACK1 functions in rice innate immunity by interacting with the Rac1 immune complex. Plant Cell. 2008;20:2265–2279. doi: 10.1105/tpc.107.054395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nibau C, Wu HM, Cheung AY. RAC/ROP GTPases: ‘hubs’ for signal integration and diversification in plants. Trends Plant Sci. 2006;11:309–315. doi: 10.1016/j.tplants.2006.04.003. [DOI] [PubMed] [Google Scholar]
- Ono E, Wong HL, Kawasaki T, Hasegawa M, Kodama O, Shimamoto K. Essential role of the small GTPase Rac in disease resistance of rice. Proc. Natl Acad. Sci. USA. 2001;98:759–764. doi: 10.1073/pnas.021273498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostrom RS, Insel PA. The evolving role of lipid rafts and caveolae in G protein-coupled receptor signaling: implications for molecular pharmacology. Br. J. Pharmacol. 2004;143:235–245. doi: 10.1038/sj.bjp.0705930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parton RG, Richards AA. Lipid rafts and caveolae as portals for endocytosis: new insights and common mechanisms. Traffic. 2003;4:724–738. doi: 10.1034/j.1600-0854.2003.00128.x. [DOI] [PubMed] [Google Scholar]
- Qi Y, Katagiri F. Purification of low-abundance Arabidopsis plasma-membrane protein complexes and identification of candidate components. Plant J. 2009;57:932–944. doi: 10.1111/j.1365-313X.2008.03736.x. [DOI] [PubMed] [Google Scholar]
- Robatzek S. Vesicle trafficking in plant immune responses. Cell Microbiol. 2007;9:1–8. doi: 10.1111/j.1462-5822.2006.00829.x. [DOI] [PubMed] [Google Scholar]
- Robatzek S, Chinchilla D, Boller T. Ligand-induced endocytosis of the pattern recognition receptor FLS2 in Arabidopsis. Genes Dev. 2006;20:537–542. doi: 10.1101/gad.366506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ron M, Avni A. The receptor for the fungal elicitor ethylene-inducing xylanase is a member of a resistance-like gene family in tomato. Plant Cell. 2004;16:1604–1615. doi: 10.1105/tpc.022475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rostoks N, Schmierer D, Kudrna D, Kleinhofs A. Barley putative hypersensitive induced reaction genes: genetic mapping, sequence analyses and differential expression in disease lesion mimic mutants. Theor. Appl. Genet. 2003;107:1094–1101. doi: 10.1007/s00122-003-1351-8. [DOI] [PubMed] [Google Scholar]
- Shahollari B, Peskan-Berghofer T, Oelmuller R. Receptor kinases with leucine-rich repeats are enriched in Triton X-100 insoluble plasma membrane microdomains from plants. Physiol. Plant. 2004;122:397–403. [Google Scholar]
- Sharma P, Sabharanjak S, Mayor S. Endocytosis of lipid rafts: an identity crisis. Semin. Cell Dev. Biol. 2002;13:205–214. doi: 10.1016/s1084-9521(02)00049-6. [DOI] [PubMed] [Google Scholar]
- Shevchenko A, Shevchenko A. Evaluation of the efficiency of in-gel digestion of proteins by peptide isotopic labeling and MALDI mass spectrometry. Anal. Biochem. 2001;296:279–283. doi: 10.1006/abio.2001.5321. [DOI] [PubMed] [Google Scholar]
- Simons K, Toomre D. Lipid rafts and signal transduction. Nat. Rev. Mol. Cell Biol. 2000;1:31–39. doi: 10.1038/35036052. [DOI] [PubMed] [Google Scholar]
- Song WY, Wang GL, Chen LL, Kim HS, Pi LY, Holsten T, et al. A receptor kinase-like protein encoded by the rice disease resistance gene, Xa21. Science. 1995;270:1804–1806. doi: 10.1126/science.270.5243.1804. [DOI] [PubMed] [Google Scholar]
- Sorek N, Poraty L, Sternberg H, Bar E, Lewinsohn E, Yalovsky S. Activation status-coupled transient S acylation determines membrane partitioning of a plant Rho-related GTPase. Mol. Cell Biol. 2007;27:2144–2154. doi: 10.1128/MCB.02347-06. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Subtil A, Wyplosz B, Balana ME, Dautry-Varsat A. Analysis of Chlamydia caviae entry sites and involvement of Cdc42 and Rac activity. J. Cell Sci. 2004;117:3923–3933. doi: 10.1242/jcs.01247. [DOI] [PubMed] [Google Scholar]
- Sugawara Y, Nishii H, Takahashi T, Yamauchi J, Mizuno N, Tago K, et al. The lipid raft proteins flotillins/reggies interact with Galphaq and are involved in Gq-mediated p38 mitogen-activated protein kinase activation through tyrosine kinase. Cell Signal. 19. 2007:1301–1308. doi: 10.1016/j.cellsig.2007.01.012. [DOI] [PubMed] [Google Scholar]
- Takahashi A, Agrawal GK, Yamazaki M, Onosato K, Miyao A, Kawasaki T, et al. Rice Pti1a negatively regulates RAR1-dependent defense responses. Plant Cell 19. 2007:2940–2951. doi: 10.1105/tpc.106.047142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thao NP, Chen L, Nakashima A, Hara SI, Umemura K, Takahashi A, et al. RAR1 and HSP90 form a complex with Rac/Rop GTPase and function in innate-immune responses in rice. Plant Cell 19. 2007:4035–4045. doi: 10.1105/tpc.107.055517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Triantafilou M, Miyake K, Golenbock DT, Triantafilou K. Mediators of innate immune recognition of bacteria concentrate in lipid rafts and facilitate lipopolysaccharide-induced cell activation. J. Cell Sci. 2002;115:2603–2611. doi: 10.1242/jcs.115.12.2603. [DOI] [PubMed] [Google Scholar]
- Tsunezuka H, Fujiwara M, Kawasaki T, Shimamoto K. Proteome analysis of programmed cell death and defense signaling using the rice lesion mimic mutant cdr2. Mol. Plant-Microbe Interact. 2005;18:52–59. doi: 10.1094/MPMI-18-0052. [DOI] [PubMed] [Google Scholar]
- Uemura M, Joseph RA, Steponkus PL. Cold acclimation of Arabidopsis thaliana (effect on plasma membrane lipid composition and freeze-induced lesions) Plant Physiol. 1995;109:15–30. doi: 10.1104/pp.109.1.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong HL, Pinontoan R, Hayashi K, Tabata R, Yaeno T, Hasegawa K, et al. Regulation of rice NADPH oxidase by binding of Rac GTPase to its N-terminal extension. Plant Cell. 2007;19:4022–4034. doi: 10.1105/tpc.107.055624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong HL, Sakamoto T, Kawasaki T, Umemura K, Shimamoto K. Down-regulation of metallothionein, a reactive oxygen scavenger, by the small GTPase OsRac1 in rice. Plant Physiol. 2004;135:1447–1456. doi: 10.1104/pp.103.036384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yalovsky S, Bloch D, Sorek N, Kost B. Regulation of membrane trafficking, cytoskeleton dynamics, and cell polarity by ROP/RAC GTPases. Plant Physiol. 2008;147:1527–1543. doi: 10.1104/pp.108.122150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z, Fu Y. ROP/RAC GTPase signaling. Curr. Opin. Plant Biol. 2007;10:490–494. doi: 10.1016/j.pbi.2007.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuyama K, Sekino-Suzuki N, Sanai Y, Kasahara K. Translocation of activated heterotrimeric G protein Galpha(o) to ganglioside-enriched detergent-resistant membrane rafts in developing cerebellum. J. Biol. Chem. 2007;282:26392–26400. doi: 10.1074/jbc.M705046200. [DOI] [PubMed] [Google Scholar]
- Zech T, Ejsing CS, Gaus K, de Wet B, Shevchenko A, Simons K, et al. Accumulation of raft lipids in T-cell plasma membrane domains engaged in TCR signalling. EMBO J. 2009;28:466–476. doi: 10.1038/emboj.2009.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J, Loh YT, Bressan RA, Martin GB. The tomato gene Pti1 encodes a serine/threonine kinase that is phosphorylated by Pto and is involved in the hypersensitive response. Cell. 1995;83:925–935. doi: 10.1016/0092-8674(95)90208-2. [DOI] [PubMed] [Google Scholar]
- Zipfel C. Pattern-recognition receptors in plant innate immunity. Curr. Opin. Immunol. 2008;20:10–16. doi: 10.1016/j.coi.2007.11.003. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.