Abstract
Analysis of genetic mutations is one of the most effective ways to investigate gene function. We now have methods that allow for mass production of mutant lines and cells in a variety of model species. Recently, large numbers of mutant lines have been generated by both ‘loss-of-function’ and ‘gain-of-function’ techniques. In parallel, phenotypic information covering various mutant resources has been acquired and released in web-based databases. As a result, significant progress in comprehensive pheno-type analysis is being made through the use of these tools. Arabidopsis and rice are two major model plant species in which genome sequencing projects have been completed. Arabidopsis is the most widely used experimental plant, with a large number of mutant resources and several examples of systematic phenotype analysis. Rice is a major crop species and is used as a model plant, with an increasing number of mutant resources. Other plant species are also being employed in functional genetics research. In this review, the present status of mutant resources for large-scale studies of gene function in plant research and the current perspective on using loss-of-function and gain-of-function mutants in phenome research will be discussed.
Keywords: Activation tagging, FOX hunting system, Insertional mutation, Phenome, Saturation mutagenesis, Visible phenotype
Introduction
One of the direct methods for investigating gene function is to examine and characterize the phenotypic changes associated with loss-of-function gene mutations. Typical mutational methods, including the generation of point mutations or small deletions by chemical or fast neutron mutagenesis, are mostly used for forward genetics (Østergaad and Yanofsky 2004). Insertional mutagenesis is a useful method for constructing loss-of-function mutants and has the advantage that the mutations are tagged by known inserted fragments (Parinov and Sundaresan 2000). The publicly available mutant resources in Arabidopsis and rice are mostly insertion tagged lines, which facilitate fast and convenient sequence index analysis of tagged genes to investigate gene function. There are two popular types of tagged lines in plant research, transferred DNA-tagged (T-DNA-tagged) lines and transposon-tagged lines (Krysan et al. 1999, Ramachandran and Sundaresan 2001). T-DNA-tagged lines have become a popular resource for studies of gene function, as they are readily generated in large numbers (Krysan et al. 1999). In addition, with a transposon Activator (Ac)/Dissociation (Ds) system, it is possible to generate mutants with a high proportion of single-copy transposon insertions. This system requires the production of a large number of mutated lines for genome-wide coverage. In spite of this, the single insertion site in each line is easily determined, thereby simplifying the production and subsequent genetic analysis of a single gene knockout series (Fedoroff and Smith 1993). The availability of many plant mutant resources makes it practicable to carry out saturation mutagenesis of an organism followed by systematic phenotyping of the mutant lines.
Gain-of-function mutants are a separate set of tools with which to dissect the function of genes, especially of genes with functional redundancy, such as those in gene families. Gain-of-function mutants may represent a different spectrum of mutants that have not been isolated as conventional loss-of-function mutants (Nakazawa et al. 2003). In contrast to the loss-of-function mutants that cause a recessive phenotype, gain-of-function mutants behave dominantly in their T1 generation (Weigel et al. 2000, Bouche and Bouchez 2001). A member of a gene family can produce a mutant phenotype without interference from other members of the family (Ito and Meyerowitz 2000, Nakazawa et al. 2001). In this review we will discuss two techniques for phenome analysis of plant lines generated for gain-of-function mutant screening. The first is activation-tagging technology, in which there is random insertion of a T-DNA containing four tandemly arranged copies of the cauliflower mosaic virus (CaMV) 35S enhancer into the genome of the host plant. These enhancers activate genes proximal to the insertion site (Weigel et al. 2000). To construct a large mutant plant population, activation-tagging lines have been generated in Arabidopsis by in planta transformation via Agrobacterium infection. The second technology is the FOX (Full-length cDNA OvereXpressing gene) hunting system which is a novel alternative activation-tagging technology using full-length cDNAs (fl-cDNAs; Ichikawa et al. 2006). These two technologies have been developed in recent years to identify the genes responsible for mutants of the model plants Arabidopsis and rice, and during the generation of mutant resources for large-scale screening of mutant phenotypes. A number of activation-tagging lines of crop plants have also been developed.
The term ‘phenome’ was coined to encompass all of the information relating to any alteration in every omics phase, from genome to phenotype, of the organism. In this review, phenome is used to describe a data set from a large-scale phenotype analysis using various mutant resources for functional research into plant genomes. In Arabidopsis, it is possible to conduct a gene-based phenome analysis from loss-of-function studies by saturation mutagenesis (Parinov and Sundaresan 2000), and from gain-of-function studies by the FOX hunting system (Ichikawa et al. 2006). The status of research into rice has rapidly approached that of Arabidopsis (Hirochika 2001, Chen et al. 2003, Nakamura et al. 2007). Experimental techniques and information have increased to enable many other crops or vegetables to be studied, which is important as basic plant research is vital for the improvement of food supply and quality as well as for environmental considerations.
Saturation mutagenesis of Arabidopsis by insertional mutation
In Arabidopsis, the basic experimental methods for generating a series of insertional mutants have been developed, including transgenic techniques, marker selection and different parental crosses. Insertion mutations in Arabidopsis have been produced mainly using the T-DNA system or the transposon system, which make it possible to monitor the effects of insertional changes in a single gene (Fig. 1). Other advantages of using Arabidopsis include self-pollination for maintaining progeny, and bulk storage of mutations in the form of seeds, which are not options in animal mutational models. It is now feasible to use insertion mutations to analyze every gene in the Arabidopsis genome. This makes Arabidopsis useful not only as a model organism for plant research, but also as the only multicellular organism in which it is currently possible to perform ‘saturation mutagenesis’ to create knockout strains for each gene.
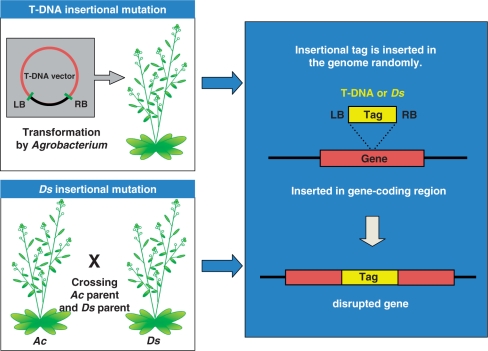
Fig. 1.
Scheme for the generation of insertional mutants by T-DNA or a Ds transposon. To construct T-DNA insertional mutant lines, plants are transformed by Agrobacterium harboring the T-DNA vector, which is inserted randomly into the genome. To construct Ds insertional mutant lines, Ds parental lines are crossed with Ac parental lines, whereupon the Ds transposon is translocated into the plant genome in the next generation. When the tag is inserted into a gene-coding region, that gene will be disrupted and lose its function.
Since completion of the sequencing of the Arabidopsis genome in 2000, several institutes around the world have been working to collect approximately 26,000 individual genes and to catalog the functional genomics of the entire Arabidopsis genome (AGI 2000, MASC 2008). As a result, a large number of T-DNA or transposon-insertional lines have already been prepared for functional genomics in Arabidopsis. The use of both of these systems has resulted in random insertional mutations across the Arabidopsis genome, with many of the insertion sites known and available to search by the public (Parinov et al. 1999, Tissier et al. 1999, Samson et al. 2002, Sessions et al. 2002, Alonso et al. 2003, Rosso et al. 2003, Kuromori et al. 2004, Ito et al. 2005). The most popular sequence index mutant resource in Arabidopsis is that of the T-DNA insertion lines produced by the Salk Institute (USA), in which there is information on the insertional mutation sites of >150,000 locations. When the mutant resources made by other institutes in the USA, the EU, Japan and others are added together, the information available on insertional locations exceeds 380,000 sites mapped on the genome of Arabidopsis ( Table 1). It has been reported by an international consortium that insertional mutants of almost all genes of Arabidopsis are now available (MASC 2008).
Table 1.
Sequence index-tagged mutant resources for gene-based phenome analysis in Arabidopsis
| Insertion type | Resource name | Institution | Accession | Available from | Numbers mapped to the genome a | Website for the resource | Reference |
|---|---|---|---|---|---|---|---|
| T-DNA | SALK | SALK Institute (USA) | Columbia | ABRC, NASC | 151,534 | http://signal.salk.edu/tabout.html | Alonso et al. (2003) |
| T-DNA | SAIL | Syngenta (USA) | Columbia | ABRC, NASC | 57,242 | n/a b | Sessions et al. (2002) |
| T-DNA | GABI-KAT | GABI (Germany) | Columbia | GABI-Kat, NASC | 63,887 | http://www.gabi-kat.de/ | Rosso et al. (2003) , Li et al. (2007) |
| T-DNA | FLAGdb | INRA (France) | Wassilevskija | INRA | 31,744 | http://urgv.evry.inra.fr/FLAGdb | Samson et al. (2002, 2004) |
| T-DNA | WiscDsLox | University of Wisconsin (USA) | Wassilevskija | ABRC | 12,464 | http://www.hort.wisc.edu/krysan/2010/default.htm | Nishal et al. (2005) |
| Ds | CSHL | CSHL (USA) | Landsberg | CSHL | 21,661 | http://genetrap.cshl.org/ | Sundaresan et al. (1995) , Martienssen (1998) |
| Ds, Spm | EXOTIC | A consortium of 11 laboratories (EU) | Landsberg, Columbia | ABRC, NASC | 23,573 | http://www.jic.bbsrc.ac.uk/science/cdb/exotic/index.htm | Tissier et al. (1999) |
| Ds | RIKEN | RIKEN (Japan) | Nossen | RIKEN BRC | 18,566 | http://rarge.gsc.riken.go.jp/dsmutant/index.pl | Ito et al. (2002 , 2005) , Kuromori et al. (2004) |
aMapped numbers are quoted from the SIGnAL website of the SALK Institute (http://signal.salk.edu/Source/AtTOME_Data_Source.html).
bn/a, not available.
ABRC, Arabidopsis Bioresource Centre; CSHL, Cold Spring Harbor Laboratory; EXOTIC, Exon Trapping Insert Consortium; GABI, Genomanalyse Im Biologischen System Pflanze; INRA, Institut National de la Recherche Agronomique; NASC, Nottingham Arabidopsis Stock Centre; RIKEN BRC, RIKEN BioResource Center.
Phenome analysis of loss-of-function mutants of Arabidopsis
An early trial of large-scale phenotypic profiling of T-DNA mutants was reported by Feldmann (1991). More than 8,000 transformants of Arabidopsis were screened under a variety of growth conditions for visible alterations in phenotype. The mutants found were grouped into several general classes: seedling-lethals, size variants, pigment, embryo-defective, reduced-fertility, dramatic (morphological) and physiological. At this time, Feldman (1991) suggested the feasibility of developing a comprehensive collection of mutant lines in which each gene is tagged by an insertion.
A more gene-based trial of systematic phenotyping was undertaken using single-copy Ds transposon-tagged lines (Kuromori et al. 2006). A total of 4,000 transposon-insertional lines, each of which contained a Ds transposon mutation in a gene-coding region, were selected, and examined systematically for visible phenotypes at each growth stage of the plant. Approximately 200 clearly visible phenotypes were classified into eight categories (seedling, leaves, flowering and growth, stems, branching, flowers, fruits and seed yield) and 43 detailed subcategories depending on the stage of the plant’s life cycle. Phenotypic images from this set of mutants have been entered into a searchable database (RAPID: RIKEN Arabidopsis Phenome Information Database) (Table 2) (Kuromori et al. 2006).
Table 2.
Open database for phenotype information of mutant resources in various plant models
an/a, not available.
Another trial that focused on phenotypes restricted to a particular organ has been performed. From an ethylmethane sulfonate (EMS)-mutagenized mutant population Berná et al. (1999) described a collection of rosette leaf-shaped mutants, belonging to 94 complementation groups. The phenotypes were classified into 19 classes with representative examples (Berná et al. 1999). Horiguchi et al. (2006) obtained a more histological phenome data set for Arabidopsis leaves. They focused on the total number and final size of leaf cells as a quantitative determinant of the leaf organ, and have isolated 205 Arabidopsis mutants with altered leaf size and/or shape from X-ray- or γ-ray-irradiated M2 populations and T-DNA-tagged T2 populations (Horiguchi et al. 2006).
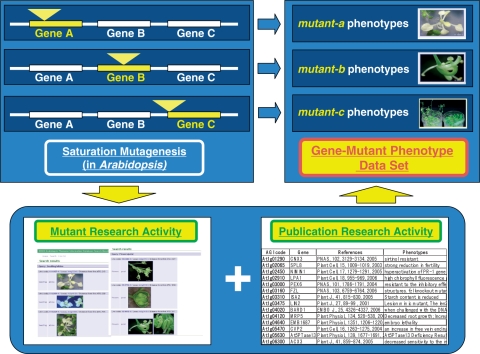
In the SeedGenes project, a genome-wide examination of Arabidopsis mutants that give either defective embryo or seed developmental phenotypes has been undertaken (McElver et al. 2001, Tzafrir et al. 2003, Tzafrir et al. 2004). They listed 358 genes and 605 mutants in which the embryo phenotypes were subclassified, mainly from large-scale T-DNA insertional mutant lines (Table 2). Simultaneously, these workers suggested a long-term goal to establish a complete collection of Arabidopsis genes that give a knockout phenotype (Tzafrir et al. 2003). Many mutant phenotypes have already been described in the literature for a number of these genes. Taking into account all of these published data, the groundwork has now been done for generating a comprehensive mutant phenotype data set (Fig. 2). An early effort to list the published mutant phenotypes has been executed by Meinke et al. (2003). The Arabidopsis Information Resource (TAIR) recently started to assign the phenotype to each mutant allele (germplasm) and made the image records of the mutant phenotypes available online (http://www.arabidopsis.org/, Swarbreck et al. 2008). Since a major challenge of phenome analysis is to identify the mutant phenotype for every gene, the relevant phenotypic data extracted from the literature must be combined with mutant phenotyping to make this possible (Fig. 2).
Fig. 2.
The concept of gene-based phenome analysis and construction of a comprehensive gene–mutant phenotype data set. One of the aims of phenome analysis is to identify the mutant phenotype for every gene and to establish a complete list of genes and mutant phenotypes. Phenotyping data of mutant resources and mutant phenotype information extracted from the published literature can be combined to make a comprehensive gene–mutant phenotype data set.
Loss-of-function mutants and large-scale phenotype analyses in rice and other plant species
Rice is another plant species in which the genome sequence has been completed, and provides an experimental model for monocotyledonous plants and crops (IRGSP 2005). As rice is easy to transform, T-DNA has been used successfully to generate insertional mutant lines (An et al. 2005). Insertional mutation systems employing transposons, including Tos17, Ds and dSpm, have been used with success in mutational analysis of the rice genome. Therefore, the total number and variation of insertional mutant resources of rice are comparable with those of Arabidopsis (Table 3) (Krishnan et al. 2009). The information on different sequence index-tagged mutant resources is integrated in the genome browsers of the Rice Annotation Project Database (RAP-DB) (http://rapdb.dna.affrc.go.jp/) and the Salk Institute Genomic Analysis Laboratory (SIGnAL) (http://signal.salk.edu/).
Table 3.
Sequence index-tagged mutant resources in rice
| Insertion type | Resource name | Institution | Numbers mapped to genomea | Website for the resource | Reference |
|---|---|---|---|---|---|
| T-DNA | POSTECH | POSTEC (Korea) | 84,680 | http://www.postech.ac.kr/life/pfg/risd/index.html | Jeon et al. (2000) |
| T-DNA | RDM | Huazhong Agricultural University (China) | 15,971 | http://rmd.ncpgr.cn | Zhang et al. (2006) |
| T-DNA | TRIM | Academic Sinica (Taiwan) | 11,646 | http://trim.sinica.edu.tw/ | Hsing et al. (2007) |
| T-DNA | SHIP | SIPPE (China) | 6,761 | http://ship.plantsignal.cn/index.do | Fu et al. (2009) |
| T-DNA | ZJU | Zhejiang University (China) | 714 | http://www.genomics.zju.edu.cn/ricetdna.html | Chen et al. (2003) |
| T-DNA, Tos17 | OTL | CIRAD-Genoplante (France) | 28,324 | http://urgi.versailles.inra.fr/OryzaTagLine/ | Sallaud et al. (2004) |
| TOS17 | NIAS | NIAS (Japan) | 17,937 | http://www.dna.affrc.go.jp/database/ | Miyao et al. (2003) |
| Ds, Spm | UCD | University of California Davis (USA) | 13,666 | http://www-plb.ucdavis.edu/Labs/sundar/Rice_Genomics.htm | Kolesnik et al. (2004) |
| Ds | CSIRO | CSIRO (Australia) | 589 | http://www.pi.csiro.au/fgrttpub/ | Eamens et al. (2004) |
| Ds | GSNU | Gyeongsang National University (Korea) | 1,046 | n/ab | Kim et al. (2004) |
| Ds | EU-OSTID | A consortium of seven laboratories (EU) | 1,301 | http://orygenesdb.cirad.fr | van Enckevort et al. (2005) |
aMapped numbers were quoted from the SIGnAL website of the SALK Institute (http://signal.salk.edu/RiceGE/RiceGE_Data_Source.html).
bn/a, not available.
CIRAD, Centre de coopération internationale en recherche agronomique pour le développement; CSIRO, Commonwealth Scientific and Industrial Research Organisation; NIAS, National Institute of Agrobiological Sciences; POSTEC, Pohang University of Science and Technology; SIPPE, Shanghai Institute of Plant Physiology and Ecology.
Large-scale phenotype analysis has been reported in rice using several mutant resources. Using the TRIM (Taiwan Rice Insertional Mutants) population of T-DNA insertion lines, the visible phenotypes of 11 categories (growth condition, leaf color, leaf morphology, plant morphology, mimic response, tiller, heading date, flower, panicle, seed fertility and seed morphology), which were divided into 65 subcategories, were observed (Chern et al. 2007). In the T1 population, 4,065 lines had at least one clearly mutant phenotype, although these visible phenotypes included both knockout- and activation-type mutants, because the TRIM population contains a tri-functional T-DNA, giving the possibility of gene trap, gene knockout or activation tagging (Chern et al. 2007).
From about 50,000 insertion lines with the endogenous retrotransposon Tos17, phenotypes in the M2 generation were observed in the field and were characterized into 11 categories (germination, growth, leaf color, leaf shape, culm shape, spotted leaf/lesion mimic, tillering, heading date, spikelet, panicle, sterility and seed) that included 53 finer phenotype descriptors (Miyao et al. 2007). More agronomic traits (plant height, tillering ability, duration of growth, color of plant and stigma, seed weight per plant and seed set rate per plant) have been investigated for phenotypic characterizations using homozygous Ds lines (Jiang et al. 2007).
Several rice mutant phenotype databases are now available, including mutant populations of Tos17-tagged lines (Miyao et al. 2007), T-DNA-tagged lines (Zhang et al. 2006, Larmande et al. 2008), and chemical- and irradiation-induced lines (Wu et al. 2005) (Table 2). Again, it is important to integrate the phenotype information already published in order to establish a complete collection of gene-to-mutant phenotypes in model organisms. All of the known mutant phenotypes in rice are collated in an excellent review by Kurata et al. (2005).
Insertional mutagenesis has been applied to other crops to generate mutants and carry out phenotype screens. In maize, 1,882 sequence index Mu-tagged mutant lines were generated (Settles et al. 2007). RescueMu is derived from the maize Mu1 transposon and provides strategies for maize gene discovery and mutant phenotypic analysis (Fernandes et al. 2004). The phenotype data of RescueMu mutated lines are available online in the Maize Genetics and Genomics Database (MaizeGDB) (Table 2). In the model legume Medicago truncatula, retrotransposon Tnt1-tagged mutant lines were generated, and visible phenotypic classes were observed in the preliminary screening of 3,237 Tnt1 lines (Tadege et al. 2008).
Insertional mutagenesis, using T-DNA or transposons, can be used in some experimental plant species, but they are less applicable to many grasses, where either a transgenic technique must be specifically developed or the large genome size prevents any manipulation. Chemical mutagenesis and the Targeting Induced Local Lesions IN Genomes (TILLING) method are applicable to many of these species and have come into widespread use (Comai and Henikoff 2006, Weil 2009). In these species, the phenotyping of mutant resources is first evaluated before the identification of insertion positions, in contrast to gene-based phenotyping.
In sorghum, visible phenotypes distinctive from the wild type were observed for 41 phenotype descriptions of an EMS-treated mutant population consisting of 1,600 lines planted in the field. The validity of TILLING for gene identification was also discussed (Xin et al. 2008) (Table 2). In barley, visible phenotypes were scored from an EMS-treated population to make a web-accessible database (Caldwell et al. 2004) (Table 2). In tomato, a total of 13,000 M2 families, derived from EMS and fast-neutron mutagenesis, were visually phenotyped in the field and put into a morphological catalog that included 15 primary and 48 secondary categories (Menda et al. 2004) (Table 2). The database of genome to phenome is expanding to Solanaceae species (Menda et al. 2008).
Activation tagging for phenomic analysis of gain-of-function mutants
In recent years, several gain-of-function-type mutant resources have been established and the activation-tagging technique is the most popular method used for their generation. The basic scheme for the generation of activation-tagging lines is shown in brief in Fig. 3. The activation-tagging technique was used to isolate cytokinin-independent mutants from 50,000 Arabidopsis calli (Kakimoto et al. 1996). They isolated the CKI1 gene, which endowed cytokinin-independent growth, although it has been shown that CKI1 was unlikely to be the cytokinin receptor (Aoyama and Oka 2001). In accordance with the development of conventional in planta transformation methods, large numbers of activation-tagging lines have been produced (Wenck et al. 1997, Clough and Bent 1998). Weigel and colleagues first generated approximately 25,000 activation-tagging lines with pSK1015 and pSK1074 that possessed resistance to the herbicide glufosinate (Weigel et al. 2000). By screening approximately 25,000 lines, they identified 23 (0.1%) dominant mutants with dramatic morphological phenotypes (Table 4). These mutants were identified as activation-tagged loci in the Arabidopsis genome, and the distances between the inserted loci of the activation T-DNAs and the corresponding genes were in the range of 0.4 to 3.6kb. The enhancer affected genes on both sides on the insertion (Weigel et al. 2000).
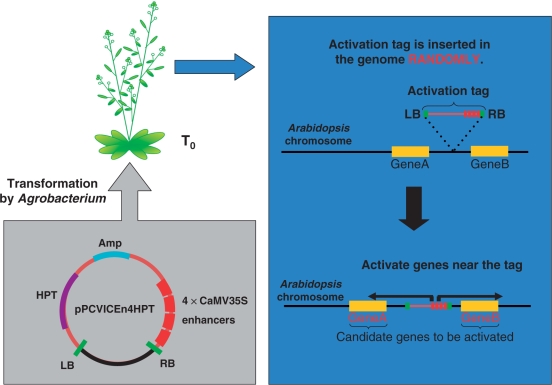
Fig. 3.
Scheme for the generation of activation-tagging lines. To construct an activation-tagging library, Arabidopsis plants are transformed by Agrobacterium harboring the activation-tagging vector, pPCVICEn4HPT, containing the T-DNA with tetrameric CaMV 35S enhancers, left (LB) and right border (RB) sequences of T-DNA, the ampicillin resistance gene (Amp) and the hygromycin resistance gene (HPT). T0 transformed plants are obtained that have activation tags inserted into the genome randomly. In phenotypic mutants, the transcriptional enhancers activate the genes near the inserted T-DNA. In this case, Gene A and Gene B are candidates for the activated gene.
Table 4.
Gain-of-function type mutant resources and databases
| Resource type | Resource name | Institution | Accession or genotype | Available from | Nos. of lines | Website for the resource | Reference |
|---|---|---|---|---|---|---|---|
| Activation tagging | Arabidopsis | ||||||
| Weigel T-DNA lines | SALK Institute (USA) | Columbia | ABRC, NASC | 22,600a | http://arabidopsis.org/abrc/weigel.jsphttp://arabidopsis.info/CollectionInfo?id=59 | Weigel et al. (2000) | |
| http://arabidopsis.info/CollectionInfo?id=59 | |||||||
| RIKEN activation tagging lines | RIKEN (Japan) | Columbia | RIKEN BRC | 32,650a | http://activation.psc.database.riken.jp (RIKEN SciNeS) | Nakazawa et al. (2003), Ichikawa et al. (2003) | |
| Unknown | Plant Research International (Netherlands) | Wassilevskija, Landsberg | Contact authors | 8,300 | n/ab | Marsch-Martinez et al. (2002) | |
| TAMARA | University of Cologne (Germany) | Columbia | NASC | 9,471a | http://arabidopsis.info/CollectionInfo?id=71 | Schneider et al. (2005) | |
| Unknown | NI Vavilov Institute of General Genetics RAS (Russia) | Columbia | Contact authors | 156 | n/a | Pogorelko et al. (2008) | |
| JIC activate lines | John Innes Centre (UK) | Landsberg | ABRC, NASC | 972a | http://arabidopsis.info/CollectionInfo?id=29 | n/a | |
| Rice | |||||||
| RISD | POSTECH (Korea) | Dongjin, Hwayoung | Contact authors | 48,000 | http://www.postech.ac.kr/life/pfg/risd | Jeong et al. (2006) | |
| TRIM | Academia Sinica (Taiwan) | Tainung 67 | Contact authors | 55,000 | http://trim.sinica.edu.tw/ | Hsing et al. (2007), Chern et al. (2007) | |
| Soybean | |||||||
| GmGenesDB | University of Missouri (USA) | Bert, Thorne | Contact authors | 900 | http://digbio.missouri.edu/gmgenedb/index.php | Mathieu et al. (2009) | |
| FOX hunting system | (cDNA library/host plant) | ||||||
| Arabidopsis FOX lines | RIKEN (Japan) | Arabidopsis-Columbia/Arabidopsis-Columbia | RIKEN BRC | 15,000 | http://nazunafox.psc.database.riken.jp (RIKEN SciNeS) | Ichikawa et al. (2006) | |
| Rice FOX Arabidopsis lines | RIKEN, NIAS, RIBS Okayama (Japan) | Rice-Nipponbare/Arabidopsis-Columbia | RIKEN BRC | 33,000 | http://ricefox.psc.riken.jp/ | Kondou et al. (2009) | |
| FOX rice lines | NIAS (Japan) | Rice-Nipponbare/Rice-Nipponbare | Contact authors | 12,000 | n/a (in preparation) | Nakamura et al. (2007) | |
aNumbers of publicly available lines from the corresponding bioresources.
bn/a, not available.
ABRC, Arabidopsis Bioresource Centre; JIC, John Innes Centre; NIAS, National Institute of Agrobiological Sciences; NASC, Nottingham Arabidopsis Stock Centre; RIBS, Research Insititue of Biological Sciences; RIKEN BRC, RIKEN BioResource center; RISD, Rice T-DNA Insertion Sequence Database; POSTECH, Pohang University of Science and Technology; TAMARA, transposon element-mediated activation tagging mutagenesis; TRIM, Taiwan Rice Insertional Mutant; SciNeS, Life Science Networking System.
Nakazawa and colleagues have also generated an activation-tagging population (Nakazawa et al. 2003) (Table 4). They generated 55,000 activation-tagging lines and from these lines they obtained many mutants showing various criteria, including rosette color, number of leaves before bolting, width of leaves, length of petiole, plant height, shape and number of cauline leaves, flower shape, number of flower organs, flowering time, fertility and branching of shoots (Nakazawa et al. 2003). A total of 1,262 lines that showed any morphological phenotype in their T1 generation were isolated from the lines, giving a frequency of 2.3%. The distances between the enhancer sequence and the corresponding start codon of activated genes were between 0.7 and 8.2 kb (Ichikawa et al. 2003).
T-DNA insertions are often complex, and characterization of multiple inverted or tandem copies or truncated T-DNA inserts often makes molecular analysis difficult (Nacry et al. 1998). To circumvent this problem, several groups use the single copy transposon tagging system (Tisser et al. 1999). Activation tagging using the maize transposon system, Enhancer-inhibitor (En-1) or Enhancer/Suppressor-mutator (En/Spm), has been established (Wilson et al. 1996, Marsch-Martinez et al. 2002, Schneider et al. 2005). Marsch-Martinez et al. (2002) generated a population of about 8,300 independent stable activation-tagging lines using Wassilewskija (WS) and Landsberg erecta (Ler) accessions. Upon examination of 2,900 insertion lines, 31 dominant mutants were found, giving a frequency of approximately 1%. Another group used the maize transposon system for the generation of a transposable element-mediated activation-tagging mutagenesis in Arabidopsis (TAMARA) (Schneider et al. 2005) (Table 4). They produced 9,471 stable activation-tagging lines harboring close to 6,000 independent transposable elements. Approximately 0.1% of activation-tagged lines showed visible dominant phenotypes including flower shape, flowering time, dwarfism, bushiness, shape of rosette leaves, length of internode and fertility. They also screened a non-visible phenotypic mutant by an HPLC-based high-throughput method for phenolic compounds (Schneider et al. 2005, Gigolashvili et al. 2007).
Recently, a new activation-tagging method has been developed and applied to Arabidopsis. A system using pEnLOX/pCre requires two plasmids, pEnLOX that contains tetrameric CaMV 35S transcriptional enhancers flanked by two loxP sites, and pCre that contains the cre gene (Pogorelko et al. 2008) (Table 4). More than 100 mutants have been isolated from the activation-tagging lines (acceptors, named the E-lines) containing the pEnLOX, and 10 helper lines (donors, named the C-lines) containing pCre have been generated. By crossing the E-lines with the C-lines, reversion of the mutants to the wild-type phenotype occurs owing to the removal of the CaMV 35S enhancers from the E-lines. Using the pEnLOX/pCre system, they have successfully obtained the reverted transformant of a mutant phenotype with small, pale green-colored rosette leaves and decreased fertility. This method may be used as a convenient system to identify easily the corresponding gene of the phenotypic activation-tagged line.
Activation tagging has also been applied to rice. In recent years, a number of rice activation-tagging lines have been generated and used for exploring functional rice genes. Over 47,000 rice T-DNA insertion lines using japonica rice have been established (Oryza sativa cv. Dongjin or Hwayoung) (Jeong et al. 2002) (Table 4). The authors used a construct, termed pGA2715, for promoter trapping and CaMV 35S-driven activation tagging of rice genes, and have isolated nine dominant mutants from 3,290 independent pGA2715-transformed seedlings (Jeong et al. 2002). They also obtained 27,621 flanking sequence tags (FSTs) from 41,234 lines. The sequences obtained were mapped on the rice genome and activation-tagging patterns near the inserted enhancer were analyzed. Half of the lines tested showed enhancement of tagged genes (Jeong et al. 2006). It should be noted that transcriptional enhancers have been shown to activate the expression of genes located up to 10.7 kb from the activation enhancer.
Wan et al. (2009) performed phenotypic analysis of 50,000 individual activation-tagged rice plants that harbored pER38, a plasmid containing tandemly arranged double CaMV 35S enhancers. They identified about 400 dominant mutants, showing visible phenotypes (sterility, dwarfism, overgrowth, early flowering, late flowering, light green leaf, leaf angle, lesion mimicking, radical growth, curled leaf, early senescence, late senescence and overtillering), from 6,000 T0 generation plants and 36,000 T1 generation plants. The frequencies of visible phenotypes identified were 2.1 and 6.4%, respectively.
Activation tagging can also be used for identification of novel genes that have not been informatically annotated. The CaMV 35S enhancer in T-DNA constructs when integrated into the plant genome may activate both coding and non-coding genes neighboring the site of insertion. For example, activation of new gene transcripts of microRNA (miRNA) has been reported, including their involvement in leaf shape structure (Weigel et al. 2000, Palatnik et al. 2003).
The FOX hunting system, a novel gain-of-function approach for plant phenome analysis
The activation-tagging system is useful to construct a large-scale gain-of-function mutant population, but this system has some disadvantages because the effects of inserted transcriptional enhancers in the genome are subject to changes in more than one gene located near the enhancers (Ichikawa et al. 2003). For the past few years, fl-cDNA clones of several plant species have been collected (Seki et al. 2002, Kikuchi et al. 2003). The Arabidopsis fl-cDNA population termed ‘RIKEN Arabidopsis full-length (RAFL) cDNA clones’ was collected and sequenced (Seki et al. 2002). Using these fl-cDNA resources a novel gain-of-function-type approach, the FOX hunting system, was developed (Ichikawa et al. 2006). In this system fl-cDNAs are randomly expressed in Arabidopsis plants and these transformed lines are termed FOX lines.
The scheme of construction of the Arabidopsis FOX lines is shown in brief in Fig. 4 (Ichikawa et al. 2006). About 10,000 independent Arabidopsis fl-cDNAs were mixed at an almost equal molar ratio to generate a fl-cDNA library, and 15,000 Arabidopsis FOX lines were generated (Table 4). These fl-cDNAs were transferred to a plant expression vector to generate the Agrobacterium library (FOX Agrobacterium library). Each Arabidopsis fl-cDNA was transformed into Arabidopsis (Columbia accession) by the floral dipping method. Each Arabidopsis FOX line that was generated contained on average 2.6 cDNA clones as detected by Southern blot analysis, and their mean size was 1.4 kb within the range of 0.3–4.2 kb (Ichikawa et al. 2006). The average size and range of the cDNA distribution was quite similar to that observed in 277 randomly chosen Arabidopsis fl-cDNAs (Ichikawa et al. 2006). The FOX hunting system is useful for systematic phenomic analysis of the function of each inserted gene. The visible phenotypes included alterations in visible morphology, growth rate, plant color, flowering time and fertility. The appearance of morphological mutants in the Arabidopsis FOX lines (9% of total plants transplanted) was higher than that of the RIKEN activation-tagging lines (2% of total plants transplanted). The visible phenotypic lines were found in 1,487 T1 generation lines, and these phenotypes were regenerated in 23% of approximately 1,000 T2 progeny, which showed dominant or semi-dominant phenotypes (unpublished data).
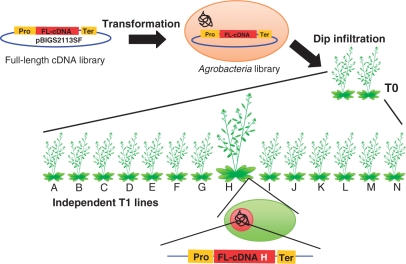
Fig. 4.
Scheme for the generation of FOX lines. To generate FOX Agrobacterium libraries, RIKEN Arabidopsis full-length (RAFL) cDNAs (fl-cDNA) from λ libraries were recloned into the pBIG2113SF vector containing the CaMV 35S promoter, the TMV omega sequence (indicated as ‘Pro’) and the NOS terminator (indicated as ‘Ter’), and transformed into Agrobacterium. Arabidopsis plants are transformed with the FOX Agrobacterium libraries by floral dip infiltration. The T0 FOX plants generated are self-pollinated, and then many independent T1 FOX seed libraries are obtained. From the T1 FOX lines, phenotypic mutant lines (in this case, the ‘H’ line) are identified. The corresponding gene of the ‘H’ mutation is easily and immediately identified by PCR with T-DNA-specific primers and sequencing.
As one of the applications of this system is to elucidate stress-related gene function, a FOX library focused on stress-inducible transcription factors was developed (mini-scale FOX system) and analyzed (Fujita et al. 2007). The Arabidopsis fl-cDNAs of 43 stress-inducible transcription factors were mixed and 224 plants with inserted fl-cDNAs were generated and salt-tolerant lines that stayed green under high-salinity conditions were obtained.
The FOX hunting system in which Arabidopsis is the host plant has an additional advantage in that it can be used to elucidate the function of genes from other organisms. Arabidopsis has a high transformation efficiency and a short generation time. By introducing fl-cDNAs from other organisms (e.g. crops), gene function can be characterized in a short time.
More than 28,000 independent rice fl-cDNAs have been collected by the National Institute of Agrobiological Sciences (NIAS) (Kikuchi et al. 2003). Using approximately 13,000 independent rice fl-cDNAs, rice FOX Arabidopsis lines were generated. These are Arabidopsis transgenic lines expressing rice fl-cDNAs under the control of the CaMV 35S promoter (Kondou et al. 2009). Kondou and colleagues have generated >23,000 independent rice FOX Arabidopsis lines (Table 4). Each plant contains on average 1.1 rice fl-cDNAs as detected by PCR and their size distribution is 0.5–4.5 kb. The rice FOX Arabidopsis lines were used for large-scale phenotype screening. They were screened for several traits including: visible phenotype, photosynthetic activity, element accumulation, pigment accumulation, hormone profiles, secondary metabolite composition, bacterial and fungal resistance, salt and high temperature tolerance, UV resistance and high light tolerance. For the visible phenotype mutants, 1,297 (5%) mutant candidates were isolated from 23,715 T1 rice FOX Arabidopsis lines. Rice fl-cDNAs recovered from these mutants were introduced into wild-type Arabidopsis and about half of them represented the same phenotypes (48%) (unpublished data). Some of these rice fl-cDNAs were introduced into rice and produced the same phenotypes observed in the rice FOX Arabidopsis lines (22%) (unpublished data). Therefore, this method of heterologous gene screening by the FOX hunting system can be used for plants with low transformation efficiency and long generation times. Recently, a high temperature mutant and a high salinity-tolerant mutant isolated from the rice FOX Arabidopsis lines have been characterized in detail (Yokotani et al. 2008, Yokotani et al. 2009).
The FOX hunting system can be applied to rice. Approximately 12,000 rice FOX rice lines that express rice fl-cDNAs in rice were established (Nakamura et al. 2007) (Table 4). Each line contained 1.04 rice fl-cDNAs on average, and the average size was 1.66 kb (Nakamura et al. 2007). The range of distribution of fl-cDNAs integrated into 238 rice FOX rice lines was similar to the average size of the original rice fl-cDNA library (1.99 kb) (Nakamura et al. 2007). Visible mutation was observed at the T0 stage. The visible phenotypes were the same as the 53 phenotype descriptors belonging to the 12 classes in the Tos17 insertion lines, as described above, and eight possible phenotypes related to the characteristics of callus, regenerants and roots. As a result 1,496 out of 9,021 lines (16.6%) had altered phenotypes. Phenotypes seen in the rice FOX rice lines included dwarf, weak and lethal, dropping leaf and pale green, lesion mimic, growth promotion, green callus, narrow leaf and high tillering. Both the rice FOX Arabidopsis lines and the rice FOX rice lines are available to identify and explore the function of rice genes. The combined use of these two lines should lead to increasingly efficient analysis of rice gene function.
Phenotype changes associated with the FOX lines are caused by the inserted gene because the fl-cDNA is expressed in the correct orientation between the CaMV 35S promoter and the NOS terminator (Taji et al. 2002, Ichikawa et al. 2006). Recently, fl-cDNA clones from many plants including soybean, cassava, poplar and Cryptomeria japonica have been collected (Nanjo et al. 2007, Futamura et al. 2008, Taji et al. 2008, Umezawa et al. 2009). The FOX hunting system may also be an alternative method for these plants to obtain gain-of-function mutants.
Resources and databases relating to gain-of-function mutants
The RIKEN activation-tagging lines (http://activation.psc.database.riken.jp) and the Arabidopsis FOX Arabidopsis lines (http://nazunafox.psc.database.riken.jp) have been sequenced for their T-DNA insertion sites and inserted fl-cDNAs, respectively (Table 4). These mutant databases also contain information based on visible phenotypes, which are classified into eight categories (adult plant, root, rosette leaf, cauline leaf, stem, flower, silique and seed) including 24 subcategories. The database for rice FOX Arabidopsis lines (rice FOX database; http://ricefox.psc.riken.jp/) contains data on mutant lines isolated from the rice FOX Arabidopsis lines (Kondou et al. 2009) (Table 4).
The TAMARA lines are available as a public resource to researchers through the Nottingham Arabidopsis Stock Centre (NASC) (Schneider et al. 2005) (Table 4). The John Innes Centre (JIC) activate lines that comprise 972 individual lines are also available through NASC (Table 4).
In rice, two databases of activation-tagging lines are available. One database was produced by Jeong et al. (2006) (http://an6.postech.ac.kr/pfg) (Table 4) and contains data for the FSTs of inserted lines. The other database was produced by Hsing et al. (2007) and is termed TRIM (http://trim.sinica.edu.tw/). Researchers can search the TRIM database by locus, annotation, line number and blast, putative knockout or activated gene with keyword, or plant/seed phenotype, and the TRIM database provides genetic segregation data (Chern et al. 2007) (Table 4). Updated information on these databases can be obtained from the Rice Functional Genomic Express Database (RiceGE) (http://signal.salk.edu/cgi-bin/RiceGE).
Phenomic analysis for other plant species using gain-of-function mutants and its application to crop plants
Besides Arabidopsis and rice, activation-tagging lines have also been generated in other species. Mathews et al. (2003) reported on the development of tomato activation-tagging lines. To identify genes that regulate metabolic pathways, they generated 10,427 independent insertion lines and found 1,338 (13%) lines with visible phenotypes, including alterations in fruit color and shape. Imaizumi and colleagues constructed T-DNA insertional lines of a model legume, Lotus japonicus, using a multifunctional vector for activation tagging (Imaizumi et al. 2005). They generated >3,500 T-DNA insertion lines, and in the T0 generation identified 45 (1.5%) possible dominant mutants with abnormal visible phenotypes with respect to aerial parts (nitrogen starvation syndrome, anthocyanin overaccumulation and strange leaf shapes), nodules (low, ineffective, small or big nodulation) and roots (thick root).
An activation-tagging system in barley has been generated, highlighting a case involving a monocot crop. The maize Ac/Ds transposable element system, which has a modified Ds element (UbiDs) containing two maize polyubiquitin promoters, was used (Ayliffe et al. 2007). The authors have generated activation-tagging lines and confirmed the enhancement of gene expression. In other cases, there are reports on the generation of activation-tagging lines in Catharanth roseus, petunia, poplar and soybean (van der Fits and Memelink 2000, Zubko et al. 2002, Busov et al. 2003, Mathieu et al. 2009). The lines of soybean are available in the GmGenesDB (http://digbio.missouri.edu/gmgenedb/) (Table 4).
Recent phenome analysis approaches and phenotyping parameters
Up to this point, the phenome trials have primarily examined the morphological characteristics of external appearance or visible phenotypes, which is only one of the criteria required for a phenome data set. Indeed, Bouche and Bouchez (2001) reported that fewer than 2% of T-DNA lines displayed significant morphological alterations. Also, about 3% of the observed mutants were found to have clearly visible phenotypes in aerial organs in transposon-tagged lines (Kuromori et al. 2006). The proportion of clear visible phenotypes from gain-of-function mutant resources is also limited (Weigel et al. 2000, Nakazawa et al. 2003, Ichikawa et al. 2006, Kondou et al. 2009). Therefore, we might reveal other phenotypes if we could extend the phenotyping.
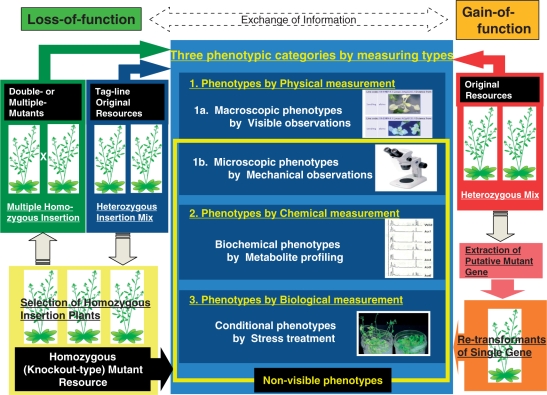
How do we handle the many phenotyping criteria required for integrated phenome information? We propose three categories of measurement to search for various traits for total phenome analysis: physical, chemical and biological ( Fig. 5). The visible phenotype is a physical measure that can be found through macroscopic observation of the plant. Other phenotypes are classified as non-visible phenotypes, for example physical features at the cellular level that cannot be determined without instruments (microscopic physical phenotypes), biochemical changes not externally visible as morphological abnormalities (chemically measured phenotypes) and phenotypic responses to different growth conditions or stresses (biologically measured phenotypes) (Fig. 5).
Fig. 5.
Scheme for the application of mutant resources to phenome analysis. Three phenotypic categories, physical, chemical and biological measurements, are used to search for various traits for total phenome analysis. Phenotypes described by physical measurement consist of macroscopic phenotypes detected by visible observations and microscopic phenotypes detected by observations using specialized equipment. Phenotypes categorized by chemical measurement are biochemical phenotypes, for example metabolite profiling. Phenotypes observed through biological measurement are conditional phenotypes revealed by environmental changes or stress treatments. Of the mutant resources available, the tag-line original resources, which are usually heterozygous for the insertion, are amenable to visible observation but often not to non-visible observation. Plants homozygous for the insertion can be selected from the original tag-line resources and will be a novel resource of homozygous (knockout-type) mutants, which are useful to detect non-visible phenotypes. In addition, homozygous insertion lines will be the starting material to produce multiple homozygous insertion mutants, which can be applied to phenotyping again. The original gain-of-function resources, which are heterozygous, can be analyzed in all three phenotypic categories. To confirm the phenotype of an isolated mutant, the candidate gene must be isolated and re-transformed. Re-transformants of the single gene can then be phenotyped.
Insertion-tagged mutant lines with homozygous insertional mutations are currently being constructed from the original T-DNA-tagged lines (http://signal.salk.edu/gabout.html) (Alonso and Ecker 2006) and the Ds transposon-tagged lines (http://www.brc.riken.go.jp/lab/epd/Eng/catalog/transp.shtml). Knockout or knockdown mutants usually show recessive characteristics for the mutant phenotype, so it is often difficult to detect the non-visible phenotypes from a heterozygous mutant pool, although it may be possible to detect their visible phenotype. Homozygous insertional mutation lines will be a novel resource of gene knockout systems to investigate non-visible phenotypes (Fig. 5). The original gain-of-function resources are also efficient in detecting non-visible phenotypes, because such phenotypes are dominant in nature. Once the phenotypes are obtained in any generation, a candidate gene can be readily confirmed by re-transforming into wild-type plants (Fig. 5). Many dominant mutants, which showed non-visible phenotypes, were isolated from rice FOX Arabidopsis lines (Kondou et al. 2009).
Finer evaluation using digital technology provides another possibility for obtaining data for more quantitative traits. For example, Boyes et al. (2001) reported an automated monitoring system for digital evaluation of phenotypic fingerprints. They presented the possibility of time-course analysis based on a series of defined Arabidopsis growth stages. In addition, several institutes have developed automated platforms for Arabidopsis phenomic approaches (Granier et al. 2006, Walter et al. 2007). Also an automatic imaging system has been applied to growth monitoring of rice (Ishizuka et al. 2005). These procedures may be used as methods for quantifying the physically measurable traits (Fig. 5).
Metabolite profiling is one of the derivative phenome analyses utilizing a chemically measured method for assaying biochemical changes in plant cells (Fiehn 2002). Recently, homozygous mutant lines were utilized for metabolic phenotyping of a subset of Ds transposon-inserted lines of Arabidopsis (Matsuda et al. 2009). This examination resulted in identification of the functions of about 70 genes involved in glycosylation of flavonoids. Homozygous T-DNA lines are also used for the ‘Ionomics’ approach (Baxter et al. 2007). This approach is another high-throughput phenotyping technology used to detect the composition of chemical elements in an organism (Baxter et al. 2007). Additionally, a large population of gain-of-function Arabidopsis suspension-cultured T87 cells transformed with Arabidopsis fl-cDNAs have been generated for metabolomics research by Ogawa et al. (2008). Non-targeted metabolic profiling analysis using several techniques is useful for investigating the chemically measured phenotype of mutant lines (Fig. 5).
Such homozygous mutant lines and the FOX lines will be useful in the systematic gene-based phenotyping of non-visible phenotypes. This includes even relatively weak phenotypes, which cannot be detected by traditional genetic screening. In addition, homozygous mutant lines will yield materials for gene knockout systems that can not only be applied one-by-one in phenotypic analysis, but can also be useful in generating double mutants and multiple mutants (Fig. 5). Sharing the phenotypic information between both loss-of-function mutants and gain-of-function mutants can be helpful for phenome analysis.
Comparable and complementary use of the loss-of-function and gain-of-function approaches
We have described the many mutant resources including loss-of-function and gain-of-function mutants that have been generated in model plants and expanded for use in various plant species. Large-scale phenotype analysis has progressed by using these different kinds of mutant resources. Such resources are useful to study gene function, and provide the possibility of complementary usage of alternative mutational resources. Basically, loss-of-function mutations are stably inherited and insertion mutagenesis makes saturation mutagenesis practicable. Genome–phenome analysis based on mutant resources will be possible, as has been demonstrated in Arabidopsis and rice. When phenotypes are found in loss-of-function mutants, the gene directly causing the phenotype can be isolated. However, very few lines show clear phenotypes, especially as the result of a single gene mutation, because of the functional redundancy of duplicated genes or gene families; 65% of the predicted proteins in the Arabidopsis genome belong to gene families with more than two members (AGI 2000). In contrast, gain-of-function mutations can overcome the problems caused by gene redundancy or phenotypic lethality (Weigel et al. 2000, Tani et al. 2004). Once the effect of the induced gene has been proven, the results obtained can be swiftly applied to research in crops or vegetables. However, if we have a clear mutant phenotype in a gain-of-function mutant, it is not easy to discuss the biological gene function, in contast to loss-of-function mutants, because the phenotypes obtained from gain-of-function mutants could be caused by ectopic expression of the overexpressed genes. Therefore, the phenotypes in gain-of-function mutants should be carefully analyzed. The possibility of gene silencing caused by co-suppression of the corresponding gene through multiple generations should also be taken into account when analyzing such gain-of-function phenotypes (Weigel et al. 2000).
In addition to insertional or point mutations, chimeric repressor silencing technology (CRES-T) was reported as a novel method for introducing loss-of-function mutations for the analysis of redundant plant transcription factors (Hiratsu et al. 2003). This method provides the missing link between loss-of-function and gain-of-function mutants, because loss-of-function phenotypes induced by CRES-T can be dominant, although the molecular mechanism of the repression is unknown. The phenotypic information obtained from the application of CRES-T in Arabidopsis and six floricultural plants is stored in a web-based interface (Mitsuda et al. 2008) (Table 2). RNA interference (RNAi) is another possible method to overcome the difficulties posed by gene redundancy and to generate phenotypes using a multigene knockdown procedure. Systematic preparation of vectors and RNAi resources for Arabidopsis genes is progressing in the AGRIKOLA (Arabidopsis Genomic RNAi Knock-out Line Analysis) consortium (MASC 2008).
In this review, we have described recent trials of large-scale phenotype analyses using loss-of-function or gain-of-function mutants. It will be intriguing to see how much the phenotypic population analyzed by phenome approaches differs when using the two different types of resources. Curiously, many mutants were assigned as having epinastic leaves in phenotypic data from activation-tagging lines, whereas only hyponastic leaf mutants were registered in the database of transposon-tagged lines (Ichikawa et al. 2003, Kuromori et al. 2006). As another example, several mutants had overgrowth phenotypes (taller plants or high fertility) in activation-tagging lines, whereas very few such mutants were found in transposon-tagged lines (Ichikawa et al. 2003, Kuromori et al. 2006). This tendency might originate from distinct mutant types, which are repressed by loss of function or overexpressed by gain of function.
The mutant phenotypes induced by loss of function and those by gain of function will often be complementary to each other. In particular, if opposite phenotypes are found in loss-of-function and gain-of-function mutants of one gene, this finding would provide information as to the function of the gene. In general, both approaches should be considered, because any phenotypic data provide valuable information on gene function. The combined data from the two types of mutants will facilitate the identification of gene function, and the availability of both loss-of-function and gain-of-function mutant resources can be expected to lead to profound advances in phenome analysis.
Funding
The Ministry of Education, Culture, Sports, Science, and Technology (MEXT) Japan (No. 19510206 to T.K., No. 19710055 to Y.K. and Nos. 16011264 and 17310120 to M.M.); the Special Coordination Funds for Promoting Science and Technology entitled, ‘Rapid identification of useful traits using rice full-length cDNAs’.
Acknowledgments
The authors are grateful to Drs. Hirohiko Hirochika, Hiroaki Ichikawa, Takanari Ichikawa, Hiroshi Takatsuji, Masaki Mori, Kenji Oda, Hidetaka Kaya, Tetsuya Sakurai, Masanori Tamaoki and Takashi Hirayama for their valuable suggestions and comments.
Glossary
Abbreviations
- Ac/Ds
Activator
/
Dissociation
- CaMV
cauliflower mosaic virus
- EMS
ethylmethane sulfonate
- En/Spm
Enhancer/Suppressor-mutator
- fl-cDNA
full-length cDNA
- FOX
full-length cDNA overexpressor
- FST
flanking sequence tag
- RNAi
RNA interference.
References
- AGI (Arabidopsis Genome Initiative) Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature. 2000;408:796–815. doi: 10.1038/35048692. [DOI] [PubMed] [Google Scholar]
- Alonso JM, Stepanova AN, Leisse TJ, Kim CJ, Chen H, Shinn P, et al. Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science. 2003;301:653–657. doi: 10.1126/science.1086391. [DOI] [PubMed] [Google Scholar]
- Alonso JM, Ecker JR. Moving forward in reverse: genetic technologies to enable genome-wide phenomic screens in Arabidopsis. Nat. Rev. Genet. 2006;7:524–536. doi: 10.1038/nrg1893. [DOI] [PubMed] [Google Scholar]
- An G, Jeong DH, Jung KH, Lee S. Reverse genetic approaches for functional genomics of rice. Plant Mol. Biol. 2005;59:111–123. doi: 10.1007/s11103-004-4037-y. [DOI] [PubMed] [Google Scholar]
- Aoyama T, Oka A. Cytokinin signal transduction in plant cells. J. Plant Res. 2001;116:221–231. doi: 10.1007/s10265-003-0094-6. [DOI] [PubMed] [Google Scholar]
- Ayliffe MA, Pallotta M, Langridge P, Pryor AJ. A barley activation tagging system. Plant Mol. Biol. 2007;64:329–347. doi: 10.1007/s11103-007-9157-8. [DOI] [PubMed] [Google Scholar]
- Baxter, Ouzzani M, Orcun S, Kennedy B, Jandhyala SS, Salt DE. Purdue ionomics information management system. An integrated functional genomics platform. Plant Physiol. 2007;143:600–611. doi: 10.1104/pp.106.092528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berná G, Robles P, Micol JL. A mutational analysis of leaf morphogenesis in Arabidopsis thaliana. Genetics. 1999;152:729–742. doi: 10.1093/genetics/152.2.729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouche N, Bouchez D. Arabidopsis gene knockout: phenotypes wanted. Curr. Opin. Plant Biol. 2001;4:111–117. doi: 10.1016/s1369-5266(00)00145-x. [DOI] [PubMed] [Google Scholar]
- Boyes DC, Zayed AM, Ascenzi R, McCaskill AJ, Hoffman NE, Davis KR, et al. Growth stage-based phenotypic analysis of Arabidopsis: a model for high throughput functional genomics in plants. Plant Cell. 2001;13:1499–1510. doi: 10.1105/TPC.010011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busov VB, Meilan R, Pearce DW, Ma C, Rood SB, Strauss SH. Activation tagging of a dominant gibberellin catabolism gene (GA 2-oxidase) from poplar that regulates tree stature. Plant Physiol. 2003;132:1283–1291. doi: 10.1104/pp.103.020354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caldwell DG, McCallum N, Shaw P, Muehlbauer GJ, Marshall DF, Waugh R. A structured mutant population for forward and reverse genetics in barley (Hordeum vulgare L.) Plant J. 2004;40:143–150. doi: 10.1111/j.1365-313X.2004.02190.x. [DOI] [PubMed] [Google Scholar]
- Chen S, Jin W, Wang M, Zhang F, Zhou J, Jia Q, et al. Distribution and characterization of over 1000 T-DNA tags in rice genome. Plant J. 2003;36:105–113. doi: 10.1046/j.1365-313x.2003.01860.x. [DOI] [PubMed] [Google Scholar]
- Chern CG, Fan MJ, Yu SM, Hour AL, Lu PC, Lin YC, et al. A rice phenomics study—phenotype scoring and seed propagation of a T-DNA insertion-induced rice mutant population. Plant Mol. Biol. 2007;65:427–438. doi: 10.1007/s11103-007-9218-z. [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998;16:735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- Comai L, Henikoff S. TILLING: practical single-nucleotide mutation discovery. Plant J. 2006;45:684–694. doi: 10.1111/j.1365-313X.2006.02670.x. [DOI] [PubMed] [Google Scholar]
- Eamens AL, Blanchard CL, Dennis ES, Upadhyaya NM. A bidirectional gene trap construct suitable for T-DNA and Ds-mediated insertional mutagenesis in rice (Oryza sativa L.) Plant Biotechnol J. 2004;2:367–380. doi: 10.1111/j.1467-7652.2004.00081.x. [DOI] [PubMed] [Google Scholar]
- Fedoroff NV, Smith DL. A versatile system for detecting transposition in Arabidopsis. Plant J. 1993;3:273–289. doi: 10.1111/j.1365-313x.1993.tb00178.x. [DOI] [PubMed] [Google Scholar]
- Feldmann KA. T-DNA insertion mutagenesis in Arabidopsis: mutational spectrum. Plant J. 1991;1:71–82.. [Google Scholar]
- Fernandes J, Dong QF, Schneider B, Morrow DJ, Nan GL, Brendel V, et al. Genome-wide mutagenesis of Zea mays L. using RescueMu transposons. Genome Biol. 2004;5:R82. doi: 10.1186/gb-2004-5-10-r82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiehn O. Metabolomics—the link between genotypes and phenotypes. Plant Mol.Biol. 2002;48:155–171.. [PubMed] [Google Scholar]
- Fu FF, Ye R, Xu SP, Xue HW. Studies on rice seed quality through analysis of a large-scale T-DNA insertion population. Cell Res. 2009;19:380–391. doi: 10.1038/cr.2009.15. [DOI] [PubMed] [Google Scholar]
- Fujita M, Mizukado S, Fujita Y, Ichikawa T, Nakazawa M, Seki M, et al. Identification of stress-tolerance-related transcription-factor genes via mini-scale Full-length cDNA Over-eXpressor (FOX) gene hunting system. Biochem. Biophys. Res. Commun. 2007;364:250–257. doi: 10.1016/j.bbrc.2007.09.124. [DOI] [PubMed] [Google Scholar]
- Futamura N, Totoki Y, Toyoda A, Igasaki T, Nanjo T, Seki M, et al. Characterization of expressed sequence tags from a full-length enriched cDNA library of Cryptomeria japonica male strobili. BMC Genomics. 2008;9:383. doi: 10.1186/1471-2164-9-383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gigolashvili T, Berger B, Mock HP, Muller C, Weisshaar B, Flugge UI. The transcription factor HIG1/MYB51 regulates indolic glucosinolate biosynthesis in Arabidopsis thaliana. Plant J. 2007;50:886–901. doi: 10.1111/j.1365-313X.2007.03099.x. [DOI] [PubMed] [Google Scholar]
- Granier C, Aguirrezabal L, Chenu K, Cookson SJ, Dauzat M, Hamard P, et al. PHENOPSIS, an automated platform for reproducible phenotyping of plant responses to soil water deficit in Arabidopsis thaliana permitted the identification of an accession with low sensitivity to soil water deficit. New Phytol. 2006;169:623–635. doi: 10.1111/j.1469-8137.2005.01609.x. [DOI] [PubMed] [Google Scholar]
- Hiratsu K, Matsui K, Koyama T, Ohme-Takagi M. Dominant repression of target genes by chimeric repressors that include the EAR motif, a repression domain, in Arabidopsis. Plant J. 2003;34:733–739. doi: 10.1046/j.1365-313x.2003.01759.x. [DOI] [PubMed] [Google Scholar]
- Hirochika H. Contribution of the Tos17 retrotransposon to rice functional genomics. Curr. Opin. Plant Biol. 2001;4:118–122. doi: 10.1016/s1369-5266(00)00146-1. [DOI] [PubMed] [Google Scholar]
- Horiguchi G, Fujikura U, Ferjani A, Ishikawa N, Tsukaya H. Large-scale histological analysis of leaf mutants using two simple leaf observation methods: identification of novel genetic pathways governing the size and shape of leaves. Plant J. 2006;48:638–644. doi: 10.1111/j.1365-313X.2006.02896.x. [DOI] [PubMed] [Google Scholar]
- Hsing YI, Chern CG, Fan MJ, Lu PC, Chen KT, Lo SF, et al. A rice gene activation/knockout mutant resource for high throughput functional genomics. Plant Mol. Biol. 2007;63:351–364. doi: 10.1007/s11103-006-9093-z. [DOI] [PubMed] [Google Scholar]
- Ichikawa T, Nakazawa M, Kawashima M, Muto S, Gohda K, Suzuki K, et al. Sequence database of 1172 T-DNA insertion sites in Arabidopsis activation-tagging lines that showed phenotypes in T1 generation. Plant J. 2003;36:421–429. doi: 10.1046/j.1365-313x.2003.01876.x. [DOI] [PubMed] [Google Scholar]
- Ichikawa T, Nakazawa M, Kawashima M, Iizumi H, Kuroda H, Kondou Y, et al. The FOX hunting system: an alternative gain-of-function gene hunting technique. Plant J. 2006;48:974–985. doi: 10.1111/j.1365-313X.2006.02924.x. [DOI] [PubMed] [Google Scholar]
- Imaizumi R, Sato S, Kameya N, Nakamura I, Nakamura Y, Tabata S, et al. Activation tagging approach in a model legume, Lotus japonicus. J. Plant Res. 2005;118:391–399. doi: 10.1007/s10265-005-0231-5. [DOI] [PubMed] [Google Scholar]
- IRGSP (International Rice Genome Sequencing Project) ( ) The map-based sequence of the rice genome. Nature. 2005;436:793–800. doi: 10.1038/nature03895. [DOI] [PubMed] [Google Scholar]
- Ishizuka T, Tanabata T, Takano M, Shinomura T. Kinetic measuring method of rice growth in tillering stage using automatic digital imaging system. Environ. Control Biol. 2005;43:83–96. [Google Scholar]
- Ito T, Meyerowitz EM. Overexpression of a gene encoding a cytochrome P450, CYP78A9, induces large and seedless fruit in Arabidopsis. Plant Cell. 2000;12:1541–1550. doi: 10.1105/tpc.12.9.1541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito T, Motohashi R, Kuromori T, Mizukado S, Sakurai T, Kanahara H, et al. A new resource of locally transposed Dissociation elements for screening gene-knockout lines in silico on the Arabidopsis genome. Plant Physiol. 2002;129:1695–1699. doi: 10.1104/pp.002774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito T, Motohashi R, Kuromori T, Noutoshi Y, Seki M, Kamiya A, et al. A resource of 5,814 Dissociation transposon-tagged and sequence-indexed lines of Arabidopsis transposed from start loci on chromosome 5. Plant Cell Physiol. 2005;46:1149–1153. doi: 10.1093/pcp/pci112. [DOI] [PubMed] [Google Scholar]
- Jeon JS, Lee S, Jung KH, Jun SH, Jeong DH, Lee J, et al. T-DNA insertional mutagenesis for functional genomics in rice. Plant J. 2000;22:561–5. doi: 10.1046/j.1365-313x.2000.00767.x. [DOI] [PubMed] [Google Scholar]
- Jeong DH, An S, Kang HG, Moon S, Han JJ, Park S, et al. T-DNA insertional mutagenesis for activation tagging in rice. Plant Physiol. 2002;130:1636–1644. doi: 10.1104/pp.014357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong DH, An S, Park S, Kang HG, Park GG, Kim SR, et al. Generation of a flanking sequence-tag database for activation-tagging lines in japonica rice. Plant J. 2006;45:123–132. doi: 10.1111/j.1365-313X.2005.02610.x. [DOI] [PubMed] [Google Scholar]
- Jiang SY, Bachmann D, La H, Ma Z, Venkatesh PN, Ramamoorthy R, et al. Ds insertion mutagenesis as an efficient tool to produce diverse variations for rice breeding. Plant Mol. Biol. 2007;65:385–402. doi: 10.1007/s11103-007-9233-0. [DOI] [PubMed] [Google Scholar]
- Kakimoto T. CKI1, a histidine kinase homolog implicated in cytokinin signal transduction. Science. 1996;274:982–985. doi: 10.1126/science.274.5289.982. [DOI] [PubMed] [Google Scholar]
- Kikuchi S, Satoh K, Nagata T, Kawagashira N, Doi K, Kishimoto N, et al. Collection, mapping, and annotation of over 28,000 cDNA clones from japonica rice. Science. 2003;301:376–379. doi: 10.1126/science.1081288. [DOI] [PubMed] [Google Scholar]
- Kim CM, Piao HL, Park SJ, Chon NS, Je BI, Sun B, et al. Rapid, large-scale generation of Ds transposant lines and analysis of the Ds insertion sites in rice. Plant J. 2004;39:252–263. doi: 10.1111/j.1365-313X.2004.02116.x. [DOI] [PubMed] [Google Scholar]
- Kolesnik T, Szeverenyi I, Bachmann D, Kumar CS, Jiang S, Ramamoorthy R, et al. Establishing an efficient Ac/Ds tagging system in rice: large-scale analysis of Ds flanking sequences. Plant J. 2004;37:301–314. doi: 10.1046/j.1365-313x.2003.01948.x. [DOI] [PubMed] [Google Scholar]
- Kondou Y, Higuchi M, Takahashi S, Sakurai T, Ichikawa T, Kuroda H, et al. Systematic approaches to using the FOX hunting system to identify useful rice genes. Plant J. 2009;57:883–894. doi: 10.1111/j.1365-313X.2008.03733.x. [DOI] [PubMed] [Google Scholar]
- Krishnan A, Guiderdoni E, An G, Hsing YI, Han CD, Lee MC, et al. Mutant resources in rice for functional genomics of the grasses. Plant Physiol. 2009;149:165–170. doi: 10.1104/pp.108.128918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krysan PJ, Young JC, Sussman MR. T-DNA as an insertional mutagen in Arabidopsis. Plant Cell. 1999;11:2283–2290. doi: 10.1105/tpc.11.12.2283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurata N, Miyoshi K, Nonomura K, Yamazaki Y, Ito Y. Rice mutants and genes related to organ development, morphogenesis and physiological traits. Plant Cell Physiol. 2005;46:48–62. doi: 10.1093/pcp/pci506. [DOI] [PubMed] [Google Scholar]
- Kuromori T, Hirayama T, Kiyosue Y, Takabe H, Mizukado S, Sakurai T, et al. A collection of 11 800 single-copy Ds transposon insertion lines in Arabidopsis. Plant J. 2004;37:897–905. doi: 10.1111/j.1365.313x.2004.02009.x. [DOI] [PubMed] [Google Scholar]
- Kuromori T, Wada T, Kamiya A, Yuguchi. M, Yokouchi. T, Imura. Y, et al. A trial of phenome analysis using 4000 Ds-insertional mutants in gene-coding regions of Arabidopsis. Plant J. 2006;47:640–651. doi: 10.1111/j.1365-313X.2006.02808.x. [DOI] [PubMed] [Google Scholar]
- Larmande P, Gay C, Lorieux M, Perin C, Bouniol M, Droc G, et al. Oryza Tag Line, a phenotypic mutant database for the Genoplante rice insertion line library. Nucleic Acids Res. 2008;36:D1022–D1027. doi: 10.1093/nar/gkm762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence CJ, Schaeffer ML, Seigfried TE, Campbell DA, Harper LC. MaizeGDB’s new data types, resources and activities. Nucleic Acids Res. 2007;35:D895–D900. doi: 10.1093/nar/gkl1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Rosso MG, Viehoever P, Weisshaar B. GABI-Kat SimpleSearch: an Arabidopsis thaliana T-DNA mutant database with detailed information for confirmed insertions. Nucleic Acids Res. 2007;35:D874–D878. doi: 10.1093/nar/gkl753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marsch-Martinez N, Greco R, Van Arkel G, Herrera-Estrella L, Pereira A. Activation tagging using the En-I maize transposon system in Arabidopsis. Plant Physiol. 2002;129:1544–1556. doi: 10.1104/pp.003327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martienssen R. Functional genomics: probing plant gene function and expression with transposons. Proc. Natl Acad. Sci. USA. 1998;95:2021–2026. doi: 10.1073/pnas.95.5.2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MASC (The Multinational Arabidopsis Steering Committee) Annual Report 2008. 2008. The Multinational Coordinated Arabidopsis thaliana Functional Genomics Project. [Google Scholar]
- Mathews H, Clendennen SK, Caldwell CG, Liu XL, Connors K, Matheis N, et al. Activation tagging in tomato identifies a transcriptional regulator of anthocyanin biosynthesis, modification, and transport. Plant Cell. 2003;15:1689–1703. doi: 10.1105/tpc.012963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathieu M, Winters EK, Kong F, Wan J, Wang S, Eckert H, et al. Establishment of a soybean (Glycine max Merr. L) transposon-based mutagenesis repository. Planta. 2009;229:279–289. doi: 10.1007/s00425-008-0827-9. [DOI] [PubMed] [Google Scholar]
- Matsuda F, Yonekura-Sakakibara K, Niida R, Kuromori T, Shinozaki K, Saito K. MS/MS spectral tag-based annotation of non-targeted profile of plant secondary metabolites. Plant J. 2009;57:555–577. doi: 10.1111/j.1365-313X.2008.03705.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McElver J, Tzafrir I, Aux G, Rogers R, Ashby C, Smith K, et al. Insertional mutagenesis of genes required for seed development in Arabidopsis thaliana. Genetics. 2001;159:1751–1763. doi: 10.1093/genetics/159.4.1751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meinke D, Meinke L, Showalter T, Schissel A, Mueller L, Tzafrir I. A sequence-based map of Arabidopsis genes with mutant phenotypes. Plant Physiol. 2003;131:409–418. doi: 10.1104/pp.014134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menda N, Semel Y, Peled D, Eshed Y, Zamir D. In silico screening of a saturated mutation library of tomato. Plant J. 2004;38:861–872. doi: 10.1111/j.1365-313X.2004.02088.x. [DOI] [PubMed] [Google Scholar]
- Menda N, Buels RM, Tecle I, Mueller LA. A community-based annotation framework for linking Solanaceae genomes with phenomes. Plant Physiol. 2008;147:1788–1799. doi: 10.1104/pp.108.119560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitsuda N, Umemura Y, Ikeda M, Shikata M, Koyama T, Matsui K, et al. FioreDB: a database of phenotypic information induced by the chimeric repressor silencing technology (CRES-T) in Arabidopsis and floricultural plants. Plant Biotechnol. 2008;25:37–44. [Google Scholar]
- Miyao A, Iwasaki Y, Kitano H, Itoh J, Maekawa M, Murata K, et al. A large-scale collection of phenotypic data describing an insertional mutant population to facilitate functional analysis of rice genes. Plant Mol. Biol. 2007;63:625–635. doi: 10.1007/s11103-006-9118-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyao A, Tanaka K, Murata K, Sawaki H, Takeda S, Abe K, et al. Target site specificity of the Tos17 retrotransposon shows a preference for insertion within genes and against insertion in retrotransposon-rich regions of the genome. Plant Cell. 2003;15:1771–1780. doi: 10.1105/tpc.012559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nacry P, Camilleri C, Courtial B, Caboche M, Bouchez D. Major chromosomal rearrangements induced by T-DNA transformation in Arabidopsis. Genetics. 1998;149:641–650. doi: 10.1093/genetics/149.2.641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura H, Hakata M, Amano K, Miyao A, Toki N, Kajikawa M, et al. A genome-wide gain-of function analysis of rice genes using the FOX-hunting system. Plant Mol. Biol. 2007;65:357–371. doi: 10.1007/s11103-007-9243-y. [DOI] [PubMed] [Google Scholar]
- Nakazawa M, Ichikawa T, Ishikawa A, Kobayashi H, Tsuhara Y, Kawashima M, et al. Activation tagging, a novel tool to dissect the functions of a gene family. Plant J. 2003;34:741–750. doi: 10.1046/j.1365-313x.2003.01758.x. [DOI] [PubMed] [Google Scholar]
- Nakazawa M, Yabe N, Ichikawa T, Yamamoto YY, Yoshizumi T, Hasunuma K, et al. DFL1, an auxin-responsive GH3 gene homologue, negatively regulates shoot cell elongation and lateral root formation, and positively regulates the light response of hypocotyl length. Plant J. 2001;25:213–221. doi: 10.1046/j.1365-313x.2001.00957.x. [DOI] [PubMed] [Google Scholar]
- Nanjo T, Sakurai T, Totoki Y, Toyoda A, Nishiguchi M, Kado T, et al. Functional annotation of 19,841 Populus nigra full-length enriched cDNA clones. BMC Genomics. 2007;8:448. doi: 10.1186/1471-2164-8-448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishal B, Tantikanjana T, Sundaresan V. An inducible targeted tagging system for localized saturation mutagenesis in Arabidopsis. Plant Physiol. 2005;137:3–12. doi: 10.1104/pp.104.050633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogawa Y, Dansako T, Yano K, Sakurai N, Suzuki H, Aoki K, et al. Efficient and high-throughput vector construction and Agrobacterium-mediated transformation of Arabidopsis thaliana suspension-cultured cells for functional genomics. Plant Cell Physiol. 2008;49:242–250. doi: 10.1093/pcp/pcm181. [DOI] [PubMed] [Google Scholar]
- Østergaard L, Yanofsky MF. Establishing gene function by mutagenesis in Arabidopsis thaliana. Plant J. 2004;39:682–696. doi: 10.1111/j.1365-313X.2004.02149.x. [DOI] [PubMed] [Google Scholar]
- Palatnik JF, Allen E, Wu X, Schommer C, Schwab R, Carrington JC, et al. Control of leaf morphogenesis by microRNAs. Nature. 2003;425:257–263. doi: 10.1038/nature01958. [DOI] [PubMed] [Google Scholar]
- Parinov S, Sevugan M, De Y, Yang WC, Kumaran M, Sundaresan V. Analysis of flanking sequences from dissociation insertion lines: a database for reverse genetics in Arabidopsis. Plant Cell. 1999;11:2263–2270. doi: 10.1105/tpc.11.12.2263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parinov S, Sundaresan V. Functional genomics in Arabidopsis: large-scale insertional mutagenesis complements the genome sequencing project. Curr. Opin. Biotechnol. 2000;11:157–161. doi: 10.1016/s0958-1669(00)00075-6. [DOI] [PubMed] [Google Scholar]
- Pogorelko GV, Fursova OV, Ogarkova OA, Tarasov VA. A new technique for activation tagging in Arabidopsis. Gene. 2008;414:67–75. doi: 10.1016/j.gene.2008.02.008. [DOI] [PubMed] [Google Scholar]
- Ramachandran S, Sundaresan V. Transposons as tools for functional genomics. Plant Physiol. Biochem. 2001;39:243–252. [Google Scholar]
- Rosso MG, Li Y, Strizhov N, Reiss B, Dekker K, Weisshaar B. An Arabidopsis thaliana T-DNA mutagenized population (GABI-Kat) for flanking sequence tag-based reverse genetics. Plant Mol. Biol. 2003;53:247–259. doi: 10.1023/B:PLAN.0000009297.37235.4a. [DOI] [PubMed] [Google Scholar]
- Sallaud C, Gay C, Larmande P, Bès M, Piffanelli P, Piégu B, et al. High throughput T-DNA insertion mutagenesis in rice: a first step towards in silico reverse genetics. Plant J. 2004;39:450–4. doi: 10.1111/j.1365-313X.2004.02145.x. [DOI] [PubMed] [Google Scholar]
- Samson F, Brunaud V, Balzergue S, Dubreucq B, Lepiniec L, Pelletier G, et al. FLAGdb/FST: a database of mapped flanking insertion sites (FSTs) of Arabidopsis thaliana T-DNA transformants. Nucleic Acids Res. 2002;30:94–97. doi: 10.1093/nar/30.1.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samson F, Brunaud V, Duchene S, De Oliveira Y, Caboche M, Lecharny A, et al. FLAGdb++: a database for the functional analysis of the Arabidopsis genome. Nucleic Acids Res. 2004;32:D347–D350. doi: 10.1093/nar/gkh134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider A, Kirch T, Gigolashvili T, Mock HP, Sonnewald U, Simon R, et al. A transposon-based activation-tagging population in Arabidopsis thaliana (TAMARA) and its application in the identification of dominant developmental and metabolic mutations. FEBS Lett. 2005;579:4622–4628. doi: 10.1016/j.febslet.2005.07.030. [DOI] [PubMed] [Google Scholar]
- Seki M, Narusaka M, Kamiya A, Ishida J, Satou M, Sakurai T, et al. Functional annotation of a full-length Arabidopsis cDNA collection. Science. 2002;296:141–145. doi: 10.1126/science.1071006. [DOI] [PubMed] [Google Scholar]
- Sessions A, Burke E, Presting G, Aux G, McElver J, Patton D, et al. A high-throughput Arabidopsis reverse genetics system. Plant Cell. 2002;14:2985–2994. doi: 10.1105/tpc.004630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Settles AM, Holding DR, Tan BC, Latshaw SP, Liu J, Suzuki M, et al. Sequence-indexed mutations in maize using the UniformMu transposon-tagging population. BMC Genomics. 2007;9:116. doi: 10.1186/1471-2164-8-116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundaresan V, Springer P, Volpe T, Haward S, Jones J.DG, Dean C, et al. Patterns of gene action in plant development revealed by enhancer trap and gene trap transposable elements. Genes Dev. 1995;9:1797–1810. doi: 10.1101/gad.9.14.1797. [DOI] [PubMed] [Google Scholar]
- Swarbreck D, Wilks C, Lamesch P, Berardini TZ, Garcia-Hernandez M, Foerster H, et al. The Arabidopsis Information Resource (TAIR): gene structure and function annotation. Nucleic Acids Res. 2008;36:D1009–D1014. doi: 10.1093/nar/gkm965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tadege M, Wen J, He J, Tu H, Kwak Y, Eschstruth A, et al. Large-scale insertional mutagenesis using the Tnt1 retrotransposon in the model legume Medicago truncatula. Plant J. 2008;54:335–347. doi: 10.1111/j.1365-313X.2008.03418.x. [DOI] [PubMed] [Google Scholar]
- Taji T, Ohsumi C, Iuchi S, Seki M, Kasuga M, Kobayashi M, et al. Important roles of drought- and cold-inducible genes for galactinol synthase in stress tolerance in Arabidopsis thaliana. Plant J. 2002;29:417–426. doi: 10.1046/j.0960-7412.2001.01227.x. [DOI] [PubMed] [Google Scholar]
- Taji T, Sakurai T, Mochida K, Ishiwata A, Kurotani A, Totoki Y, et al. Large-scale collection and annotation of full-length enriched cDNAs from a model halophyte, Thellungiella halophila. BMC Plant Biol. 2008;8:115. doi: 10.1186/1471-2229-8-115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tani H, Chen X, Nurmberg P, Grant JJ, SantaMaria M, Chini A, et al. Activation tagging in plants: a tool for gene discovery. Funct. Integr. Genomics. 2004;4:258–2. doi: 10.1007/s10142-004-0112-3. [DOI] [PubMed] [Google Scholar]
- Tissier AF, Marillonnet S, Klimyuk V, Patel K, Torres MA, Murphy G, et al. Multiple independent defective Suppressor-mutator transposon insertions in Arabidopsis: a tool for functional genomics. Plant Cell. 1999;11:1841–1852. doi: 10.1105/tpc.11.10.1841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzafrir I, Dickerman A, Brazhnik O, Nguyen Q, McElver J, Frye C, et al. The Arabidopsis SeedGenes Project. Nucleic Acids Res. 2003;31:90–93. doi: 10.1093/nar/gkg028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzafrir I, Pena-Muralla R, Dickerman A, Berg M, Rogers R, Hutchens S, et al. Identification of genes required for embryo development in Arabidopsis. Plant Physiol. 2004;135:1206–1220. doi: 10.1104/pp.104.045179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umezawa T, Sakurai T, Totoki Y, Toyoda A, Seki M, Ishiwata A, et al. Sequencing and analysis of approximately 40, 000 soybean cDNA clones from a full-length-enriched cDNA library. DNA Res. 2008;15:333–346. doi: 10.1093/dnares/dsn024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Fits L, Memelink J. ORCA3, a jasmonate-responsive transcriptional regulator of plant primary and secondary metabolism. Science. 2000;289:295–297. doi: 10.1126/science.289.5477.295. [DOI] [PubMed] [Google Scholar]
- van Enckevort LJ, Droc G, Piffanelli P, Greco R, Gagneur C, Weber C, et al. EU-OSTID: a collection of transposon insertional mutants for functional genomics in rice. Plant Mol. Biol. 2005;59:99–1. doi: 10.1007/s11103-005-8532-6. [DOI] [PubMed] [Google Scholar]
- Walter A, Scharr H, Gilmer F, Zierer R, Nagel KA, Ernst M, et al. Dynamics of seedling growth acclimation towards altered light conditions can be quantified via GROWSCREEN: a setup and procedure designed for rapid optical phenotyping of different plant species. New Phytol. 2007;174:447–455. doi: 10.1111/j.1469-8137.2007.02002.x. [DOI] [PubMed] [Google Scholar]
- Wan S, Wu J, Zhang Z, Sun X, Lv YGao, C., et al. Activation tagging, an efficient tool for functional analysis of the rice genome. Plant Mol. Biol. 2009;69:69–80. doi: 10.1007/s11103-008-9406-5. [DOI] [PubMed] [Google Scholar]
- Weigel D, Ahn JH, Blazquez MA, Borevitz JO, Christensen SK, Fankhauser C, et al. Activation tagging in Arabidopsis. Plant Physiol. 2000;122:1003–1013. doi: 10.1104/pp.122.4.1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weil CF. TILLING in grass species. Plant Physiol. 2009;149:158–164. doi: 10.1104/pp.108.128785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wenck A, Czako M, Kanevski I, Marton L. Frequent collinear long transfer of DNA inclusive of the whole binary vector during Agrobacterium-mediated transformation. Plant Mol. Biol. 1997;34:913–922. doi: 10.1023/a:1005849303333. [DOI] [PubMed] [Google Scholar]
- Wilson K, Long D, Swinburne J, Coupland G. A Dissociation insertion causes a semidominant mutation that increases expression of TINY, an Arabidopsis gene related to APETALA2. Plant Cell. 1996;8:659–671. doi: 10.1105/tpc.8.4.659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu JL, Wu C, Lei C, Baraoidan M, Bordeos A, Madamba MR, et al. Chemical- and irradiation-induced mutants of indica rice IR64 for forward and reverse genetics. Plant Mol. Biol. 2005;59:85–97. doi: 10.1007/s11103-004-5112-0. [DOI] [PubMed] [Google Scholar]
- Xin. Z, Wang ML, Barkley NA, Burow G, Franks C, Pederson G, et al. Applying genotyping (TILLING) and phenotyping analyses to elucidate gene function in a chemically induced sorghum mutant population. BMC Plant Biol. 2008;8:103. doi: 10.1186/1471-2229-8-103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokotani N, Ichikawa T, Kondou Y, Matsui M, Hirochika H, Iwabuchi M, et al. Expression of rice heat stress transcription factor OsHsfA2e enhances tolerance to environmental stresses in transgenic Arabidopsis. Planta. 2008;227:957–967. doi: 10.1007/s00425-007-0670-4. [DOI] [PubMed] [Google Scholar]
- Yokotani N, Ichikawa T, Kondou Y, Matsui M, Hirochika H, Iwabuchi M, et al. Tolerance to various environmental stresses conferred by the salt-responsive rice gene ONAC063 in transgenic Arabidopsis. Planta. 2009;229:1065–1075. doi: 10.1007/s00425-009-0895-5. [DOI] [PubMed] [Google Scholar]
- Zhang J, Li C, Wu C, Xiong L, Chen G, Zhang Q, et al. RMD: a rice mutant database for functional analysis of the rice genome. Nucleic Acids Res. 2006;34:D745–D748. doi: 10.1093/nar/gkj016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zubko E, Adams CJ, Machaekova I, Malbeck J, Scollan C, Meyer P. Activation tagging identifies a gene from Petunia hybrida responsible for the production of active cytokinins in plants. Plant J. 2002;29:797–808. doi: 10.1046/j.1365-313x.2002.01256.x. [DOI] [PubMed] [Google Scholar]