Abstract
DNA double-strand breaks (DSBs) are a serious threat to genome stability and cell viability. Although biological effects of low levels of radiation are not clear, the risks of low-dose radiation are of societal importance. Here, we directly monitored induction and repair of single DSBs and quantitatively analyzed the dynamics of interaction of DNA repair proteins at individual DSB sites in living cells using 53BP1 fused to yellow fluorescent protein (YFP-53BP1) as a surrogate marker. The number of DSBs formed was linear with dose from 5 mGy to 1 Gy. The DSBs induced by very low radiation doses (5 mGy) were repaired with efficiency similar to repair of DSBs induced at higher doses. The YFP-53BP1 foci are dynamic structures: 53BP1 rapidly and reversibly interacted at these DSB sites. The time frame of recruitment and affinity of 53BP1 for DSB sites were indistinguishable between low and high doses, providing mechanistic evidence for the similar DSB repair after low- and high-dose radiation. These findings have important implications for estimating the risk associated with low-dose radiation exposure on human health.
INTRODUCTION
Cell lethality, mutations, chromosomal translocations, apoptosis and cancer induced by ionizing radiation (IR) result principally from an inefficient or inaccurate repair of DNA double-strand breaks (DSBs) (1). Humans are exposed to low doses of radiation during air travel, from radon in homes, during space travel or in areas of low-level contamination (including former sites of nuclear weapon production) and can encounter much higher radiation doses in contaminated areas such as Chernobyl or during radiotherapy (2). Radiobiologists have struggled to estimate the biological consequences of low levels of radiation exposure in humans for decades. The current risk estimates for low-dose radiation are based on assumption that there is a linear, nonthreshold, dose–response relationship with detrimental health effects of low-dose IR extrapolated from comparatively high doses (3). However, the biological effects associated with occupational and environmental low-dose radiation are considerably more complex than predicted by the linear nonthreshold model due to the fact that the radiation-induced biological effects in humans depend on several factors, including the influence of cellular responses to DNA repair, sensitivity of bystander cells and delayed genomic instability (4). Currently, insufficient data are available to determine the impact of low levels of radiation exposure on human health (5). As risks associated with low-dose radiation are of societal importance (6), it is critical to further understand the cellular responses to low quantities of radiation exposure.
In response to DNA DSBs, histone 2A family member X is phosphorylated at serine 139 (γH2AX) and forms discrete foci at the DSB sites (7,8). Earlier studies have revealed a close correlation between the number of γH2AX foci and the estimated number of DSBs (7,9). Recently, a dose-dependent induction of γH2AX foci was observed in the range between 1.2 mGy and 2 Gy (10). These studies clearly indicate that γH2AX foci are a reliable marker for the quantification of DSBs. However, evidence indicates that the number of γH2AX may not represent true DSBs induced by different damaging agents (11) and that the number of background γH2AX foci is relatively greater in exponentially growing cells than in confluent cells (12). Therefore, for an accurate measurement of the biological effects of low-dose radiation exposure, an alternative in vitro assay sensitive enough to measure a single DSB per cell is needed.
In vitro studies suggest that the tumor suppressor p53-binding protein 1 (53BP1) participates in the cellular response to DNA damage. 53BP1 relocates to multiple nuclear foci within minutes after exposure of cells to IR (13–15). Evidence suggests that, similar to γH2AX, the number of 53BP1 foci is closely correlated with the number of DNA DSBs. Further, the kinetics of resolution of 53BP1 foci is very similar to the kinetics of DNA DSBs repair following IR (13–15). In addition, these foci co-localize with known DNA damage response proteins such as γH2AX, Rad50/Mre11/NBS1, BRCA1, Rad51 and the phosphorylated form of DNA-dependent protein kinase catalytic subunit (13–15). Therefore, 53BP1 foci represent sites of DNA DSBs and thus can be used as a DSB surrogate marker to study the induction and repair of DSBs.
To directly monitor the induction and repair of single DSBs and to quantitatively study the dynamics of DNA repair proteins at single DSB sites in living cells, we developed a system using 53BP1 fused to yellow fluorescent protein (YFP-53BP1), as a DSB surrogate marker. Here, we present evidence that the number of DSBs formed, as measured by the number of YFP-53BP1 foci formed, was linear with radiation dose from 5 mGy, a dose that generates on average 0.1 DSB per cell, to 1 Gy. We showed, at single-cell resolution, that these foci are dynamic structures and that 53BP1 rapidly and reversibly interacts with the DSBs. The rapid recruitment of 53BP1 to DSB sites and its binding affinity to DSBs were indistinguishable in cells treated with low and high doses, providing mechanistic evidence that cells repair DSBs induced by low- and high-dose IR at the same rate. These results support the current models of risk assessment that are based on the assumption that the cellular responses to DNA DSBs are equally efficient at low and high doses. Importantly, we demonstrate a new, sensitive means to directly study induction and repair of DSBs in living cells.
MATERIALS AND METHODS
Cell culture and γ-irradiation
HT1080 cells were maintained in α-Modified Eagle's Medium supplemented with 10% fetal bovine serum, 100 μg/ml streptomycin and 100 U/ml penicillin. Human bronchial epithelial cells (HBECs) immortalized with hTERT were maintained as described previously (16). The cells were maintained at 37°C in a humidified 5% CO2 incubator. cDNA encoding human 53BP1 was amplified from pCMH6K-53BP1 (gift from Junjie Chen) using forward (5′-GCC TGA TCA ATG GAC CCT ACT GGA AGT CAG G-3′) and reverse (5′-CCG CTC GAG TTA GTG AGA AAC ATA ATC GTG TTT-3′) primers containing 5′-BclI and 3′-XhoI sites, cloned into the BamHI and XhoI sites of pCD3F2-YFP/pCD3.1Hyg-EGFP and then sequence confirmed using a number of primers sets covering the entire length of the cDNA (6.6 kb). To generate the HT1080-YFP 53BP1 stable cell line, along with the endogenous 53BP1, HT1080 (ATCC) cells were transfected with linearized pCD3F2-YFP-53BP1 using Nucleofector (Amaxa). The cells were selected with 400 μg/ml G418 for 2 weeks. Stable lines were maintained in the same concentration of selection medium. To generate the HBEC-EGFP 53BP1 stable cell line, along with endogenous 53BP1, HBEC-3KT cells were transfected with linearized pCD3.1Hyg-EGFP-53BP1 using Nucleofector (Amaxa). The cells were selected with 20 μg/ml hygromycin for 10 days. Stable lines were maintained in the same concentration of selection medium. All experiments were performed by irradiating exponentially growing cell cultures with 137Cs irradiator (Mark 1 irradiator, JL Shepherd & Associates). For doses up to 100 mGy, lead attenuators were placed between the source and the samples to reduce the dose rate by 50–80% (∼4 mGy/min). Dosimetry was performed using thermoluminescence dosimetry devices (Landauer Inc, Glenwood, IL). For higher doses, no attenuators were used and ∼3.5 Gy/min dose rate was used.
Immunostaining, clonogenic survival assay and immunoblotting
Indirect immunofluorescence was performed as described previously (13). After fixation in 4% paraformaldehyde at room temperature for 20 min, cells were immunolabeled using anti-γH2AX mouse monoclonal (US Biologicals), 53BP1 rabbit polyclonal (Cell Signaling), or anti-phospho DNA-PKcs (pT2609) mouse monoclonal antibodies. For PCNA, cells were fixed in methanol:acetone (7:3) on ice for 30 min and immunostained with PCNA mouse monoclonal antibody (Santa Cruz Biotechnology). Secondary antibodies (anti-mouse or rabbit conjugated with Alexa 488/633 or rhodamine) were purchased from Molecular Probes. Cells were mounted using Vectashield and observed on a LSM 510 laser scanning confocal microscope with a 63X1.4 NA Plan-Apochromat oil immersion objective. For count of 53BP1 foci in fixed cells, images of control and experimental cells were acquired under identical conditions. The number of 53BP1 foci was counted in a minimum of 100 cells using the Metasystems/Axioplan 2E equipped with Axiocam HRM 14bit digital Camera and Metafer software (Zeiss). The average foci number per cell at each time point was subtracted by that of mock-irradiated cells. The clonogenic survival assay was performed as described previously (13). For immunoblotting, a nuclear fraction prepared from parental and YFP-53BP1 stably expressing HT1080 cells was analyzed using anti-53BP1 rabbit polyclonal antibodies (Cell Signaling).
Time-lapse image acquisition and YFP-53BP1 assembly kinetics assay
To measure the fluorescent intensity of YFP-53BP1 at the sites of DSBs, time-lapse imaging was carried out in live cells. Images of living cells were captured using an LSM 510 Meta laser scanning confocal microscope with a 63X1.4 NA Plan-Apochromat oil immersion objective. Images were taken at z-sections (8–12 sections) of 0.5-μm intervals using the 514-nm argon laser of the LSM 510 microscope. The tube current of the laser was set at 6.1 A and the laser power was typically set to 0.3–1% transmission with the pinhole opened to 1–2 Airy units. YFP fluorescence was detected using a dichroic beamsplitter (458/514 nm) and an additional 530–600-nm bandpass emission filter placed in front of the photomultiplier tube. During imaging, cells grown in 35-mm glass-bottom culture dishes (MatTek Cultureware) were maintained in CO2-independent medium (Invitrogen) at 37°C. The growth medium was replaced by CO2-independent medium (Invitrogen) before imaging. Time-lapse image acquisition was started before the induction of DNA DSBs to obtain a pre-irradiated cell image. DSBs were introduced in YFP-53BP1-expressing cells by γ-rays (137Cs, JL Shepherd) and the images of same cells were acquired every 5 min for up to 2 h.
For quantitative and comparative imaging, identical imaging acquisition parameters were used. YFP-53BP1 accumulation curves were generated according to (17). Signal intensities of accumulated YFP-53BP1 fluorescence at the DSBs were converted into a numerical value by the use of Axiovision software (version 4.5). To compensate for nonspecific fluorescent bleaching during the repeated image acquisition, in every image, we first measured the average fluorescent intensity of the YFP-53BP1 focus as a function of time, and then divided it by the average fluorescent intensity measured elsewhere in the cell (background) as a function of time. To get normalized YFP-53BP1 accumulation curve for each cell, the YFP-53BP1 fluorescent intensity (RF) at the DSBs sites was calculated by the following formula: RF(t) = [(I–Ipre IR)/(Imax–Ipre IR)], where Ipre IR is the fluorescent intensity of the foci region before irradiation, and Imax represents the maximum fluorescent intensity of the YFP-53BP1 foci at DSB sites (17). Normalized fluorescent curves from 20 cells were averaged.
Visualization of YFP/EGFP-53BP1 foci formation and disappearance in living cells
To quantify appearance and disappearance of YFP/EGFP-53BP1 foci, images of live cells were acquired as described above. For the foci dissolution kinetics analysis, cells with either no focus or 1–2 foci were followed continuously for up to 8 h. Cell images were taken before irradiation and 0.5, 1, 2, 4 and 8 h after mock, 5, 10, 50, 100, 500 and 1000 mGy of γ-irradiation. The average number of foci per cell at each time point was subtracted from that of mock-irradiated cells. For the quantification of 53BP1 foci numbers in fixed cells, we used the Metasystems/Axioplan 2E equipped with an Axiocam HRM 14-bit digital Camera, and Metafer software (Zeiss, Germany). For counting 53BP1 foci numbers in live cell images, we utilized spot detection function of the Imaris software (Bitplane). For the purpose of foci counting, we used identical image acquisition and foci detection parameters. Quantification of foci was done from images of 100–200 cells for each dose and in at least three independent experiments. A nonlinear regression analysis was used to assess the focus dissolution kinetics (GraphPad Prism 5).
Fluorescence redistribution after photobleaching (FRAP) analysis
FRAP was performed on an LSM 510 Meta confocal microscope (Zeiss) as described previously (17). Briefly, cells expressing YFP-53BP1 were imaged prior to irradiation (Pre-IR), exposed to different doses of γ-rays, and then incubated at 37°C for 30 min to allow maximum accumulation of YFP-53BP1 at the DSB sites. An initial stack of six images (pre-bleach, t0) were taken in z-planes of 0.5 μm apart (using 514-nm line of an argon laser set at 0.3–1% power transmission and 6.1-A tube current). Subsequently, one of the foci was photobleached with a photo bleaching pulse (514-nm line of an argon laser set at 100% power transmission and 6.1-A tube current). Immediately after the photobleaching, a new stack of images (bleach) was taken and the imaging was performed every 2 min for 30 min (for each time point, six images of 0.5 μm apart were taken using 514-nm line of an argon laser set at 0.3–1% power transmission and 6.1-A tube current). For FRAP experiments on nonirradiated cells, a small area of the nucleus was photobleached and images were acquired every 10 s for 5 min. FRAP recovery curves from image data was generated according to McNally (18) and Sprague (19). To correct for the loss of fluorescence due to imaging, in every image, we first measured the average fluorescent intensity of the photobleached YFP-53BP1 focus as a function of time, and then divided it by the average fluorescent intensity measured elsewhere in the cell as a function of time. To get normalized FRAP curve for each cell, the YFP-53BP1 fluorescent intensity after the photobleaching was divided by the pre-bleach intensity recovery, and the pre-bleach intensity was set to one. Normalized FRAP curves from 20 cells were averaged.
Image acquisition and foci counting
For determination of YFP/EGFP-53BP1 foci dissolution kinetics and dynamic behavior of YFP-53BP1 at DSB sites, all the cells were imaged prior to irradiation. This gave a measure of background YFP-53BP1 foci. Images of the same cells were recorded immediately after irradiation using the ‘mark and find’ option of the LSM Meta 510 software. By comparing the live cell images obtained before and after IR, the foci that were generated only by γ-rays could be distinguished. For the measurement of YFP/EGFP-53BP1 foci dissolution kinetics, all the cells in the field were assigned a number and the number of foci in the same cell at different time points was monitored. Thus, we followed single cells from pre-IR to 8 h after IR. We acquired 8 or 12 z-sections of each cell, deconvoluted the images using Imaris software (Bitplane), and counted the number of YFP-53BP1 foci as described in visualization of YFP-53BP1 foci formation and disappearance in living cells section. 53BP1 formed a larger number of foci in S-phase of the cell cycle (Figure 1B, lower panel). Therefore the S-phase cells are not included in the calculations.
Figure 1.
Characterization of YFP-53BP1-expressing cells. (A) Expression of YFP-53BP1 in HT1080 cells. Nuclear extract from HT1080 and HT1080 YFP-53BP1 cells was analyzed by western blot using an anti-53BP1 polyclonal antibody to detect both endogenous and YFP-53BP1. 53BP1 and YFP-53BP1 were distinguished by the higher molecular weight of YFP-53BP1 fusion protein. (B) YFP-53BP1 foci co-localize with γH2AX in un-irradiated cells. Exponentially growing HT1080-YFP 53BP1 cells were imaged and then fixed. Subsequently, cells were immunostained with γH2AX monoclonal antibody. (C) YFP-53BP1 forms foci in S-phase cells. Exponentially growing HT1080-YFP 53BP1 cells were imaged and then fixed. Cells were immunostained with proliferating cell nuclear antigen (PCNA) monoclonal antibody. (D) Distribution of YFP-53BP1 and γH2AX foci in un-irradiated cells. Images of live cells were acquired prior to immunostaining with γH2AX antibody. The number of foci per cell (left panel) and the distribution (right panel) of YFP-53BP1 and γH2AX were quantitated. More than 200 nuclei were counted and the error bars represent standard deviations calculated from three independent experiments.
Statistical analysis
To calculate half-time recovery, we used Sigma plot (version 11.0). The FRAP data were fitted to solutions of nonlinear regression of exponential rise to maximum. The double four parameters best fit equation, y = a*(1–exp(–b*x))+c*(1–exp(–d *x)), was employed. Half-time for recovery of 53BP1 fluorescence (t1/2, the time required for fluorescence intensity to reach 50% of its prebleach intensity) was calculated from the curve fitting values. For the analysis of 53BP1 foci dissolution kinetics, the data were fitted to solutions of nonlinear regression of one phase decay (GraphPad Prism 5). The one phase decay parameters best fit equation, Y = (Y0 – Plateau)*exp(–K*X) + Plateau was employed. Where Y0 is the percent of foci when X (time, hours) is zero, K is the rate constant [h(–1)], and plateau is the percent of foci per cell at infinite times (hours). Student's t-test was performed to calculate the level of significance and a value of P < 0.05 was considered as a statistically significant difference.
RESULTS
A novel system to visualize induction and repair of single DNA DSBs
To investigate the cellular responses to DNA DSBs induced by low-dose IR in living cells, we generated an HT1080 cell line that stably expresses near-physiological levels of YFP-53BP1, along with nonfluorescently tagged 53BP1. The mobility of YFP-53BP1 was slower than that of the endogenous 53BP1 on an SDS gel and it migrated with the expected size for a fusion protein (Figure 1A). Approximately 35% of cells showed largely homogeneous, pan-nuclear distribution of YFP-53BP1. In 30% of the cells, the YFP-53BP1 was concentrated in bright speckles. The remaining 35% had one or more foci per cell (Figure 1B). Immunostaining of these cells with γH2AX antibody (Figure 1B and 1D, right panel) revealed co-localization of YFP-53BP1 and γH2AX, suggesting that these 53BP1 foci most likely represent regions of spontaneous DNA damage or modified chromatin with γH2AX and/or rearrangements (20). In addition, immunostaining of these cells with proliferating cell nuclear antigen (PCNA) antibody, which binds to proteins involved in DNA replication (21,22), revealed that the cells with a large number of YFP-53BP1 foci are S-phase cells (Figure 1C). There were on average two YFP-53BP1 foci per cell, whereas there were on average three γH2AX foci in these same cells (Figure 1D, left panel). Thus, the number of γH2AX foci per cells exceeds the number of YFP-53BP1 foci. In addition, when we carefully examined the co-localization of 53BP1 and γH2AX foci in these nonirradiated cells, similar to previous report (23), we found that a fraction of γH2AX foci (small size) did not co-localize with 53BP1 foci. These extra γH2AX foci most likely do not contain DSBs and might represent some kind of modified chromatin. Further, immunostaining of HT1080 cells with 53BP1, γH2AX (Supplementary Figure S1A, top panel) and PCNA (Supplementary Figure S1A, lower panel) antibodies revealed that the distribution of 53BP1, γH2AX and PCNA were similar to that of HT1080-YFP-53BP1 cells. These results demonstrate that the brighter YFP-53BP1 foci (24) noticed in 30% of the HT1080-YFP-53BP1 cells and the large number of YFP-53BP1 foci noticed in S-phase cells are not due to expression of YFP-53BP1 along with endogenous 53BP1 and is the normal distribution pattern of 53BP1 in these cells.
As measured by a clonogenic survival assay, the sensitivity of cells to increasing doses of γ-irradiation (Supplementary Figure S1B) and endogenous and YFP-53BP1 foci dissolution kinetics (Supplementary Figure S1C) were indistinguishable between parental (HT1080) and the YFP-53BP1-expressing cells. This data are in agreement with previous findings (24) that ectopic expression of YFP-53BP1 does not alter the cellular response to DNA damage. Immunostaining of mock (Supplementary Figure S2A, top panel) and γ-irradiated HT1080-YFP-53BP1 cells (Supplementary Figure S2A, lower panel) with 53BP1 antibody (recognizes both endogenous and the YFP-53BP1) showed that all the YFP-53BP1 co-localized with the 53BP1 antibody stained 53BP1. These results further illustrate that expression of YFP-53BP1 in the cells along with nonfluorescently tagged 53BP1 does not induce any additional 53BP1 foci and this could be due to oligomerization (25) of both endogenous and the YFP-53BP1. We further verified that the YFP-53BP1 foci represent the sites of DSBs by immunostaining with phosphorylated DNA-PKcs (Supplementary Figure S2B). Similar to previous findings (15,23), YFP-53BP1 foci clearly colocalized with γH2AX (Figure 2A) and DNA-PKcs foci in the majority of cases, confirming that the YFP-53BP1 localizes at IR-induced DSB sites in living cells. These experiments indicate that cells that express the fluorescently labeled 53BP1 can be used as a model system to evaluate the effects of IR on DNA repair.
Figure 2.
Recruitment of YFP-53BP1 to DNA DSB sites is dose dependent. (A) Representative images showing co-localization of YFP-53BP1 with γH2AX after exposure of cells to graded doses of γ-rays. Distribution of YFP-53BP1 in HT1080 cells was determined prior to irradiation. Cells were irradiated with indicated doses of γ-rays and pictures were taken after 30 min. Subsequently, cells were fixed and immunostained with anti-γH2AX antibody and images were recorded. (B and C) Induction of YFP-53BP1 foci is dose-dependent in living cells. Graph showing number of YFP-53BP1 and γH2AX foci formed 30 min after exposure of cells to (B) 100–1000 mGy and (C) 5–50 mGy of γ-rays. Cells were imaged prior to γ-irradiation and pictures were captured 30 min after irradiation. The foci in 200–400 cells were counted and this data is presented in the graphs. The error bars represent standard deviations calculated from three independent experiments.
Number of DNA DSB formed is linear with IR doses from 5 mGy to 1 Gy
We next determined the number of YFP-53BP1 foci in live cells over a range of doses of γ-radiation from 5 mGy to 1 Gy; the relationship between the number of foci induced per cell and the IR dose delivered was linear (Figure 2B and C). Importantly, the number of YFP-53BP1 foci was similar to the number of γH2AX foci (Figure 2B). The observed value of ∼19 foci per gray correlated with our previous 53BP1 foci measurements and with previous studies that showed 20–40 DSBs per Gy of γ-irradiation (10,13,15,23). Thus, these results provide evidence that the number of DNA DSB induced by γ-irradiation increases linearly with increasing dose.
DNA DSBs induced by low-dose (5–100 mGy) and high-dose (500 and 1000 mGy) γ-radiation are efficiently repaired
Using our newly developed system, we investigated the dynamic loss of YFP-53BP1 foci in exponentially growing cells after high (500 and 1000 mGy) and low (5, 10, 50, 100 mGy) doses of γ-radiation (Figure 3A). We found that 8 h after γ-irradiation, only 0.8 ± 0.35 and 1.68 ± 0.21 DSBs per nucleus remained in 500- and 1000-mGy irradiated cells, respectively (Figure 3B). As shown in Figure 3C, almost all DSBs in cells treated with low dose IR (5–50 mGy) were efficiently repaired within 8 h.
Figure 3.
DNA DSBs induced by low-dose (5–100 mGy) and high-dose (500 and 1000 mGy) γ-radiation are efficiently repaired. (A) Representative live cell images showing formation and disappearance of YFP-53BP1 foci after indicated doses of γ-radiation. HT1080 cells stably expressing YFP-53BP1 were irradiated with 0, 5, 10, 50, 100, 500 and 1000 mGy of γ-rays; images were collected before and at different time after irradiation using confocal microscopy (LSM 510). (B and C) Number of foci per cell as a function of time following γ-irradiation. Cells were imaged prior to and different times following irradiation. The number of foci per cell was plotted after subtracting the number of foci in the mock-irradiated cells. More than 100–200 cells were counted for each time. The error bars represent standard deviations calculated from three independent experiments.
A previous study by Rothkamm and Lobrich (10) showed that the repair of DSBs is compromised after exposure of cells to 1.2 mGy of X-rays. This study was carried out in G1 phase of the cell cycle using fixed primary human fibroblasts (MRC-5) and γH2AX antibodies. In contrast, in our system, the kinetics of disappearance of YFP-53BP1 foci in cells irradiated with 5 mGy closely resembled the kinetics of repair in cells treated with 10 or 50 mGy of radiation. Further, mathematical calculation of t½ (time required to repair 50% of the DSBs) indicated that the rate of foci dissolution kinetics was similar in cells treated with high (500 mGy, 2.7 ± 0.8 h) and low (5 and 10 mGy, 3.1 ± 0.5 and 3.5 ± 0.7 h, respectively) dose γ-radiation (Supplementary Table 1). However, in cells exposed to 1000 mGy of radiation, majority of the YFP-53BP1 foci were lost after 8 h, whereas at other dose, almost all were repaired (Supplementary Table 1). Collectively, these YFP-53BP1 foci dissolution analyses suggest that the DNA DSBs induced by low-dose IR are efficiently repaired.
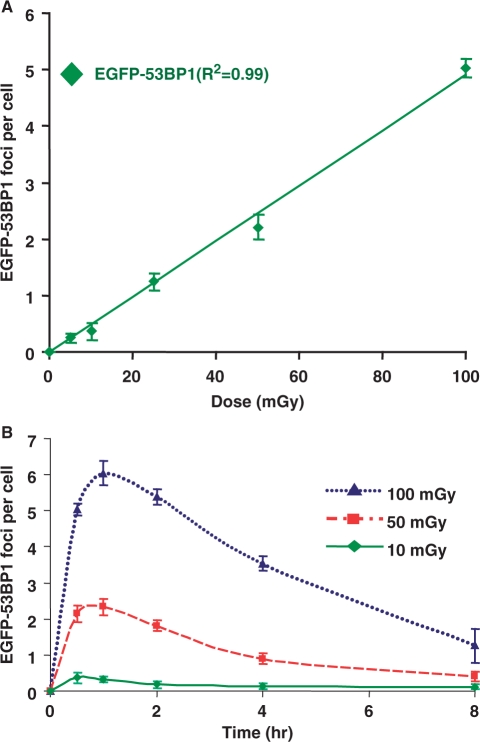
The HT1080 cells used in this study are transformed; however, this cell line has widely been used by many radiation biologists (9,26) and has relatively stable cytogenetic phenotype (27). Although analysis of the response of primary human cells would be ideal, stable YFP-53BP1-expressing clones are needed for our analysis. To circumvent these possible draw backs of transformed cells, we have validated our results in hTERT immortalized normal human bronchial epithelial cells (HBECs) that stably express enhanced green fluorescent protein (EGFP)-tagged-53BP1. These hTERT immortalized HBECs do not form colonies in soft agar or tumors in nude mice and have no spontaneous genomic instability (28), thus these cells are a valuable model system for studying DSB repair (29). We noticed that the relationship between the number of 53BP1 foci per cell and IR dose was linear in hTERT immortalized HBECs stably expressing EGFP-53BP1 (Figure 4A). Further, we found that almost all DSBs in cells treated with low doses (10 and 50 mGy) of γ-radiation were efficiently repaired within 8 h (Figure 4B). The fact that DNA DSBs induced by low-dose γ-irradiation are repaired both in HT1080 and HBEC cultures at the same rate as DNA breaks induced by high-dose IR suggests that this is a general phenomenon.
Figure 4.
DNA DSBs induced by low-dose (10–100 mGy) γ-radiation are efficiently repaired in human bronchial epithelial cells. (A) Induction of EGFP-53BP1 foci is dose-dependent in living HBECs. Graph showing number of EGFP-53BP1 foci formed 30 min after exposure of hTERT immortalized HBECs to 5, 10, 25, 50 and 100 mGy of γ-radiation. hTERT immortalized HBECs stably expressing EGFP-53BP1 were imaged prior to γ-irradiation and images were captured 30 min after irradiation. The foci in 100–200 cells were counted and this data is presented in the graphs. The error bars represent standard deviations calculated from three independent experiments. (B) Number of foci per cell as a function of time following γ-irradiation. HBECs stably expressing EGFP-53BP1 were imaged prior to and different times following irradiation. The number of foci per cell was plotted after subtracting the number of foci in the mock-irradiated cells. More than 100–200 cells were counted for each time. The error bars represent standard deviations calculated from three independent experiments.
Percentage of cells responding to γ-irradiation decreased with decreasing dose
The average number of YFP/EGFP-53BP1 foci determined from data shown in Figures 2B and C and 4A does not give any information on whether all the cells responded to low-dose IR. We have, therefore, characterized the distribution of cells with a given number of YFP/EGFP-53BP1 foci as well as determined the number of foci per cell after exposure to various doses of IR. We observed that the percentage of nonresponsive cells (i.e. cells that did not generate any YFP/EGFP-53BP1 foci) decreased with increasing doses of radiation (Supplementary Figure S3A and B). Of HT1080 cells, 62 ± 1.6, 43 ± 6.4, 28 ± 7 and 22 ± 4% responded to 100, 50, 10 and 5 mGy of γ-irradiation, respectively (Supplementary Figure S3A). For HBECs the percentages were similar: 61 ± 5, 42 ± 2, 25 ± 5 and 14 ± 1% responded to 100, 50, 10 and 5 mGy of γ-irradiation, respectively (Supplementary Figure S3B). These analyses illustrate that only a pool of irradiated cells respond to low-dose γ-radiation and that the percentage of cells with 53BP1 foci increased with increasing doses of IR.
Because background DSBs have a major influence on any analysis of low-dose IR-induced DSB repair (30), we determined the level of spontaneous YFP/EGFP-53BP1 foci formation in mock-irradiated HT1080 and HBECs (Supplementary Figure S4A and B). The YFP-53BP1 background level was 0.15 ± 0.05 and 0.5 ± 0.3 foci per nucleus for the 0.5- and 8-h incubation periods, respectively (Supplementary Figure S4A). Similarly, we found that the EGFP-53BP1 background level was 0.05 ± 0.02 foci per nucleus at 0.5 h and 0.01 ± 0.01 at 8 h (Supplementary Figure S4B). Roughly 70–80% of the cells did not develop any 53BP1 focus and the remaining cells developed one to eight foci per cell (Supplementary Figure S4A and B), indicating that only a certain cell population develops spontaneous foci. These data demonstrate that evaluation of background foci is critical for the accurate analysis of low-dose IR induced DSB repair. By comparing the live cell images obtained before and after IR, the foci that were generated by γ-rays could be distinguished from background foci.
Kinetics of YFP-53BP1 accumulation on DNA DSBs is dose independent
To gain insight into protein redistribution in response to DSBs (31,32), we used a real-time assay to monitor accumulation of 53BP1 on the DNA DSBs. YFP-53BP1 began to accumulate at the DSB sites within 3–5 min after γ-irradiation and reached a plateau around 30 min after γ-irradiation, suggesting that the recruitment of YFP-53BP1 to the DSB sites was completed within that time range (Figure 5A). The 53BP1 levels then gradually decreased over the next 2 h. Approximately 45% of the YFP-53BP1 fluorescence remained after 2 h (Figure 5B); this could be the result of ongoing DSB repair. In contrast, the fluorescent intensity of 53BP1 at the regions of endogenous foci did not change with time (Supplementary Figure S5). Kinetics of YFP-53BP1 accumulation on the DNA DSBs was comparable in cells exposed to low and high doses of radiation; therefore, the redistribution of 53BP1 onto DSBs is not dose dependent and is DNA DSB specific.
Figure 5.
Kinetics of YFP-53BP1 accumulation at DSB sites is dose independent. (A) Representative live cell images showing assembly kinetics of YFP-53BP1 foci after 100, 500 and 1000-mGy γ-irradiation. HT1080 cells stably expressing YFP-53BP1 were irradiated and images were collected before and at indicated times after γ-irradiation using confocal microscopy (LSM 510). (B) YFP-53BP1 assembly kinetics at the DNA damage sites is not influenced by the number of DSBs. For every cell, images were captured prior to irradiation and after irradiation. Time-lapse images were captured every 5 min. The fluorescent intensity of the YFP-53BP1 accumulation at the DSB sites was measured before and after irradiation, and the normalized fluorescent intensity was calculated as described in the ‘Materials and Methods’ section. Each data point depicted in the graph is the average of normalized YFP-53BP1 fluorescence measurements from 20 cells. The error bars represent standard deviation.
Interaction of YFP-53BP1 with DNA DSB is reversible and kinetics is similar in low- and high-dose irradiated cells
To get a better understanding of the interaction between 53BP1 and the DNA DSB sites, we investigated the dynamics of the YFP-53BP1 accumulation in the DNA damage-induced focus using fluorescence redistribution after photo bleaching (FRAP) (Figure 6A). These FRAP measurements clearly revealed that after photo bleaching YFP-53BP1-associated fluorescence at the DSB sites recovered with a markedly slower kinetics (in all the cells analyzed) than did fluorescence in the undamaged nuclear region (Figure 6B and Table 1). This result indicates that 53BP1 is transiently immobilized at the DSB sites. Interestingly, we found that the YFP signal recovered to ∼85–90% of the original level within 8–10 min (Figure 6B), implying that 53BP1 is not attached to DSBs in a rigid complex, but that there is a dynamic exchange between unbleached YFP-53BP1 molecules from the nucleoplasm and the bleached 53BP1 molecules at DNA DSBs. We then quantitated the YFP-53BP1 fluorescence recovery of individual focus after treating cells with different doses of IR. Intriguingly, the YFP-53BP1 displayed very similar dynamic behavior regardless of dose (Figure 6A and B), suggesting that the dynamic behavior of YFP-53BP1 at DSB sites is specific to DSBs and is not influenced by IR dose. We noticed that only ∼85–90% of the YFP-53BP1 signal recovered after photobleaching. The fraction of YFP-53BP1 that did not recover during this time frame could be attributed to the ongoing DNA repair. Similar kind of phenomenon was also noticed in case of EGFP-Ku80 at the sites of laser irradiation (33). Further, the calculated values for half-time (t1/2, the time required for fluorescence intensity to reach 50% of its prebleach intensity) recovery of YFP-53BP1 molecules on the DSB sites were comparable between low and high doses and there was no statistically significant difference between low- and high-dose radiation induced YFP-53BP1 foci (Table 1). Collectively, the FRAP kinetic data strongly suggest that the dynamic behavior of YFP-53BP1 on the DSB sites accounts for the comparable rates DSBs repair observed in cells exposed to low and high doses of radiation.
Figure 6.
DNA-YFP-53BP1 complex is reversible. (A) Examples of fluorescence recovery of YFP-53BP1 in nonirradiated and irradiated cells. Images of HT1080 cells stably expressing YFP-53BP1 were taken before γ-irradiation (Pre-IR), 30 min after γ-irradiation, and/or before photo bleaching (Pre-bleach), immediately after the photo bleaching (bleach), and at indicated times after bleaching. The photobleached regions are indicated in white dotted circles. (B) Fluorescence recovery kinetics curves for nonirradiated and irradiated cells. For every cell, images were taken before γ-irradiation (Pre-IR), 30 min after γ-irradiation, and/or before photo bleaching (Pre-bleach), immediately after the photo bleaching (bleach), and at different times after bleaching. In every image, average fluorescent intensity of the photobleached YFP-53BP1 focus was measured as a function of time, and then divided it by the average fluorescent intensity measured elsewhere in the cell as a function of time. To get normalized FRAP curve for each cell, the YFP-53BP1 fluorescent intensity after the photobleaching was divided by the prebleach intensity and the prebleach intensity was set to one. Each data point depicted in the graph is the average of 20 independent, normalized measurements. The error bars represent the STDEV.
Table 1.
Half-time for FRAP recovery
| YFP-53BP1 foci Source | t½ (s) |
|---|---|
| Outside DSB | 25.23 ± 4.15 |
| 10 mGy | 118.32 ± 11.41 |
| 100 mGy | 102.96 ± 12.29 |
| 1000 mGy | 109.85 ± 13.01 |
| Endogenous | 85.38 ± 12.21 |
| S-phase | 66.94 ± 8.16 |
DISCUSSION
We demonstrate here the first direct visualization of induction and repair of single DSBs using a newly validated surrogate marker for DSBs, 53BP1 fused to YFP. DSBs are a serious and potentially lethal type of cellular damage in human cells. Previously, antibodies toward phosphorylated H2AX (γH2AX) were used to reveal DSB foci after low-dose radiation treatment of cells (9,10). The kinetics of γH2AX foci disappearance closely resemble the kinetics of DSB repair when levels of DNA damage were low (i.e. that induced by 1-Gy irradiation or less) (11,34). However, a high incidence of spontaneous γH2AX foci was observed in a number of reports (12), making it difficult to discriminate the foci induced by low-dose radiation from endogenous foci.
Ionizing radiation (IR) induces both isolated DNA lesions and clustered damages that consist of lesion including strand breaks, oxidized purines, oxidized pyrimidines and abasic sites; simple DSBs account for 70–80% of the damage (35). When γH2AX is used as a surrogate marker, there is a one-to-one correlation between the number of γH2AX foci and IR-induced DSBs (10). This suggests that other types of DNA damage induced by IR do not significantly contribute to γH2AX foci formation. As YFP-53BP1 and γH2AX colocalized, YFP-53BP1 foci mark the sites of DSBs. As a surrogate marker, YFP-53BP1 is several orders of magnitude more accurate than other methods for detecting DSBs (7). Thus, by examination of YFP-53BP1 foci in live cells, we were able to quantitatively measure the induction and repair of individual DSBs at the single cell level. Using our method, formation of YFP-53BP1 foci was readily observed immediately after the induction of DNA damage and post-IR handling of cells was avoided. In addition, by comparing the live cell images obtained before and after IR, the foci that were generated by γ-rays could be distinguished from background foci. With this simple strategy, we were able to unequivocally and directly identify only the IR-induced DSBs and follow their repair kinetics in real time.
Using our live-cell imaging approach at the single cell level, we showed that the number of YFP-53BP1 foci formed was linear with IR dose from 5 mGy to 1 Gy. This result greatly extends previous quantitative measurements of the linear relationship between the γH2AX and 53BP1 foci numbers and radiation dose (9,15,23,36,37). Our live-cell imaging and immunostaining with YFP-53BP1 and 53BP1 antibodies, respectively, showed that 1 Gy of γ-irradiation generated ∼20 53BP1 foci per cell. Other studies, including the first report on 53BP1 (15), reported similar numbers of foci per cell (23). A recent study by Costes et al. (38) clearly showed that the maximum number of DSBs generated by X-rays was 60% less than one predicted by Monte Carlo Algorithm in virtual nuclei. Several reports (39,40) have shown when taking caution of heat-labile breaks that 40–50% of DSB determined by pulsed-field gel electrophoresis (PFGE) have disappeared within 30 min. However, the peak value of 53BP1 foci around 20 foci/Gy/cell was seen 30 min after irradiation when many of the DSBs had been repaired. In addition, the kinetics of 53BP1 foci disappearance was slower than the PFGE analysis (23). All these studies clearly show that the number of DSBs detected by PFGE and 53BP1 foci may not always correlate. In contrast, Rothkamm and Lobrich (10) reported a one-to-one correlation between the number of γH2AX foci formed and the IR-induced DSBs measured by PFGE. However, our live-cell imaging study, together with others, clearly indicates that the 53BP1 and/or γH2AX foci likely do not mark all DSBs. Moreover, PFGE sensitivity for relatively small amounts of DNA damage is very limited. Therefore, quantification of 53BP1 foci is likely to be the most useful method for the assessment of therapeutically relevant low-doses of radiation.
We treated HT1080s that stably expresses near-physiological levels of YFP-53BP1 with IR doses as low as 5 mGy, a dose that generates an average of 0.1 DSB per nucleus (i.e. 1 in 10 cells formed a focus). The time course of DSB repair in cells treated with this low dose was similar to that observed after 10 or 50-mGy treatment. Interestingly, we observed similar dynamic loss of 53BP1 foci in human bronchial epithelial cells expressing 53BP1 tagged with enhanced green fluorescent protein after low doses of IR. In agreement with the data presented here, the rate of γH2AX foci loss observed in irradiated lymphocytes following computerized tomography scan (at doses of 10–20 mGy) was consistent with that observed after higher doses (41).
A previous study suggested that repair is compromised after cells are treated with 1.2 mGy of radiation: Fibroblasts exposed in vitro to low radiation doses exhibited a low level of radiation-induced γH2AX foci (∼0.05 foci per cell) that persisted for several days post-irradiation (10). However, the repair kinetics we observed does not indicate a compromised repair capacity at low radiation doses. Our data differ from that obtained in the previous study for the following reasons: First, our analysis was carried out using exponentially growing cells rather than noncycling quiescent fibroblasts that were fixed prior to analysis (10). Second, we used YFP-53BP1 as DSB surrogate marker to directly monitor induction and repair of DSBs rather than γH2AX. Third, accurate measurement of the background 53BP1 foci before irradiation, as well as measurement of the spontaneous YFP-53BP1 foci formed during the incubation time, increased the accuracy of our analysis.
Fluorescence redistribution after photobleaching of YFP-53BP1 outside the DSB area suggested that the majority of the 53BP1 population is able to diffuse within in the nucleus. It was previously shown that 53BP1 does not diffuse freely in the interchromatin space and may repeatedly bind to and dissociate from chromatin (24,42). The high diffusional mobility of 53BP1 in the nucleus might be important for the efficient detection of DNA DSBs and might account for the early detection of YFP-53BP1 foci after the IR exposure. Photobleaching of accumulated 53BP1 also revealed that the bound proteins were exchanged within 10 min, suggesting that DNA-53BP1 complex is a dynamic structure and the YFP-53BP1 reversibly interacts with DNA DSBs. Other studies have reported similar dynamic and transient assembly of 53BP1 in the damaged nuclear regions using other DNA damaging sources (24,42). Interestingly, the rate at which 53BP1 accumulated in damaged regions of nuclei was similar in cells treated with low and high doses of radiation. It has been proposed that the dynamic interaction and exchange of the DNA repair proteins is central to various DNA transactions as it provides much-needed combinatorial flexibility, including instant access of distinct enzymatic activities during the repair process (43). The window of time between the dissociation of a bound molecule and re-association of a new molecule may result in exposure of DSBs to other repair factors. The dynamic interaction of 53BP1 with the DNA at DSBs could facilitate timely and productive DSBs repair. The lack of difference in the dynamic behavior of 53BP1 at DNA DSBs in cells treated with different doses of radiation provides mechanistic evidence that cells repair DSBs induced by low- or high-dose radiation at comparable levels.
Understanding the biological consequences of low-dose radiation exposure is becoming increasingly important as we are exposed to IR for medical reasons and during space travel (44). The current risk estimates for low-dose radiation are based on assumption that there is a linear nonthreshold dose–response relationship, with potentially detrimental health effects extrapolated from comparatively high doses to those doses associated with occupational and environmental exposures (45). Our data showed that the number of DSBs formed is linear with doses from 5 mGy to 1 Gy and indicated that cells have the same capacity to repair DNA damage caused by low doses as by high doses. Rothkamm and Lobrich (34) previously suggested that the linear extrapolation model significantly underestimates the risk for IR-induced carcinogenesis. However, our results support the linear extrapolation model currently used to determine risks of radiation exposure. The biological effects of low-dose radiation are complex due to the fact that the radiation-induced biological effects in humans depend on several factors, including the influence of cellular responses to DNA repair, delayed reproductive death, sensitivity of bystander cells and delayed genomic instability (4). Evidence also suggests that 53BP1/γH2AX foci do not always mark all the DSBs and PFGE sensitivity for relatively small amounts of DNA damage is very limited. Therefore, quantification of 53BP1 foci is likely to be the most useful method for the evaluation of therapeutically relevant low-dose radiation exposure. Our results significantly advance our understanding of immediate, as well as long-term, biological consequences of low-dose radiation and will help to estimate the risk associated with low-dose radiation exposure on human health.
In conclusion, this is the first study to show induction and repair of DSBs in single, living cells. Our observations on the kinetics of repair of DSBs induced by low-dose IR is consistent with the existing notion that the DNA DSBs are efficiently repaired no matter the causative dose of radiation. Further, our observation that repair proteins have similar dynamic behavior at the DSB sites after low- and high-dose radiation provide mechanistic insight into the reason why cells have the similar capacity to repair IR-induced damage due to low and high doses. Use of our newly developed model to directly follow the induction and repair of single DSB in individual cells opens up new experimental approaches to study IR-induced DSBs.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
The Office of Science (BER), United States Department of Energy (DE-AI02-05ER64 048); and National Aeronautics and Space Administration (NNJ05HD36G, NNA05CM04G and NNZ07AU42G to D.J.C.). Funding for open access charge: National Aeronautics and Space Administration.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Drs. Sairei So for the HT1080-YFP 53BP1 cell line, Junjie Chen for the human 53BP1 cDNA, Shibani Mukherjee and Michael Peyton for the critical reading of the manuscript, and Mohammad Abolfath for the statistical analysis.
REFERENCES
- 1.Elliott B, Jasin M. Double-strand breaks and translocations in cancer. Cell Mol. Life Sci. 2002;59:373–385. doi: 10.1007/s00018-002-8429-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bonner WM. Low-dose radiation: thresholds, bystander effects, and adaptive responses. Proc. Natl Acad. Sci. USA. 2003;100:4973–4975. doi: 10.1073/pnas.1031538100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Prasad KN, Cole WC, Hasse GM. Health risks of low dose ionizing radiation in humans: a review. Exp. Biol. Med. 2004;229:378–382. doi: 10.1177/153537020422900505. [DOI] [PubMed] [Google Scholar]
- 4.Morgan WF. Non-targeted and delayed effects of exposure to ionizing radiation: I. Radiation-induced genomic instability and bystander effects in vitro. Radiat. Res. 2003;159:567–580. doi: 10.1667/0033-7587(2003)159[0567:nadeoe]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- 5.Strzelczyk JJ, Damilakis J, Marx MV, Macura KJ. Facts and controversies about radiation exposure, part 2: low-level exposures and cancer risk. J. Am. Coll. Radiol. 2007;4:32–39. doi: 10.1016/j.jacr.2006.07.010. [DOI] [PubMed] [Google Scholar]
- 6.Preston RJ. Update on linear non-threshold dose-response model and implications for diagnostic radiology procedures. Health Phys. 2008;95:541–546. doi: 10.1097/01.HP.0000326332.80829.63. [DOI] [PubMed] [Google Scholar]
- 7.Rogakou EP, Boon C, Redon C, Bonner WM. Megabase chromatin domains involved in DNA double-strand breaks in vivo. J. Cell Biol. 1999;146:905–916. doi: 10.1083/jcb.146.5.905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ismail IH, Hendzel MJ. The gamma-H2A.X: is it just a surrogate marker of double-strand breaks or much more? Environ. Mol. Mutagen. 2008;49:73–82. doi: 10.1002/em.20358. [DOI] [PubMed] [Google Scholar]
- 9.Sedelnikova OA, Rogakou EP, Panyutin IG, Bonner WM. Quantitative detection of (125)IdU-induced DNA double-strand breaks with gamma-H2AX antibody. Radiat. Res. 2002;158:486–492. doi: 10.1667/0033-7587(2002)158[0486:qdoiid]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- 10.Rothkamm K, Lobrich M. Evidence for a lack of DNA double-strand break repair in human cells exposed to very low x-ray doses. Proc. Natl Acad. Sci. USA. 2003;100:5057–5062. doi: 10.1073/pnas.0830918100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bouquet F, Muller C, Salles B. The loss of gammaH2AX signal is a marker of DNA double strand breaks repair only at low levels of DNA damage. Cell Cycle. 2006;5:1116–1122. doi: 10.4161/cc.5.10.2799. [DOI] [PubMed] [Google Scholar]
- 12.Banath JP, Macphail SH, Olive PL. Radiation sensitivity, H2AX phosphorylation, and kinetics of repair of DNA strand breaks in irradiated cervical cancer cell lines. Cancer Res. 2004;64:7144–7149. doi: 10.1158/0008-5472.CAN-04-1433. [DOI] [PubMed] [Google Scholar]
- 13.Asaithamby A, Uematsu N, Chatterjee A, Story MD, Burma S, Chen DJ. Repair of HZE-particle-induced DNA double-strand breaks in normal human fibroblasts. Radiat. Res. 2008;169:437–446. doi: 10.1667/RR1165.1. [DOI] [PubMed] [Google Scholar]
- 14.Rappold I, Iwabuchi K, Date T, Chen J. Tumor suppressor p53 binding protein 1 (53BP1) is involved in DNA damage-signaling pathways. J. Cell Biol. 2001;153:613–620. doi: 10.1083/jcb.153.3.613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schultz LB, Chehab NH, Malikzay A, Halazonetis TD. p53 binding protein 1 (53BP1) is an early participant in the cellular response to DNA double-strand breaks. J. Cell Biol. 2000;151:1381–1390. doi: 10.1083/jcb.151.7.1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ramirez RD, Sheridan S, Girard L, Sato M, Kim Y, Pollack J, Peyton M, Zou Y, Kurie JM, Dimaio JM, et al. Immortalization of human bronchial epithelial cells in the absence of viral oncoproteins. Cancer Res. 2004;64:9027–9034. doi: 10.1158/0008-5472.CAN-04-3703. [DOI] [PubMed] [Google Scholar]
- 17.Yano K, Morotomi-Yano K, Wang SY, Uematsu N, Lee KJ, Asaithamby A, Weterings E, Chen DJ. Ku recruits XLF to DNA double-strand breaks. EMBO Rep. 2008;9:91–96. doi: 10.1038/sj.embor.7401137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McNally JG. Quantitative FRAP in analysis of molecular binding dynamics in vivo. Methods Cell Biol. 2008;85:329–351. doi: 10.1016/S0091-679X(08)85014-5. [DOI] [PubMed] [Google Scholar]
- 19.Sprague BL, McNally JG. FRAP analysis of binding: proper and fitting. Trends Cell Biol. 2005;15:84–91. doi: 10.1016/j.tcb.2004.12.001. [DOI] [PubMed] [Google Scholar]
- 20.Mochan TA, Venere M, DiTullio RA, Jr, Halazonetis TD. 53BP1, an activator of ATM in response to DNA damage. DNA Repair (Amst) 2004;3:945–952. doi: 10.1016/j.dnarep.2004.03.017. [DOI] [PubMed] [Google Scholar]
- 21.Moldovan GL, Pfander B, Jentsch S. PCNA, the maestro of the replication fork. Cell. 2007;129:665–679. doi: 10.1016/j.cell.2007.05.003. [DOI] [PubMed] [Google Scholar]
- 22.Costes SV, Boissiere A, Ravani S, Romano R, Parvin B, Barcellos-Hoff MH. Imaging features that discriminate between foci induced by high- and low-LET radiation in human fibroblasts. Radiat. Res. 2006;165:505–515. doi: 10.1667/RR3538.1. [DOI] [PubMed] [Google Scholar]
- 23.Markova E, Schultz N, Belyaev IY. Kinetics and dose-response of residual 53BP1/gamma-H2AX foci: co-localization, relationship with DSB repair and clonogenic survival. Int. J. Radiat. Biol. 2007;83:319–329. doi: 10.1080/09553000601170469. [DOI] [PubMed] [Google Scholar]
- 24.Bekker-Jensen S, Lukas C, Melander F, Bartek J, Lukas J. Dynamic assembly and sustained retention of 53BP1 at the sites of DNA damage are controlled by Mdc1/NFBD1. J. Cell Biol. 2005;170:201–211. doi: 10.1083/jcb.200503043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Adams MM, Wang B, Xia Z, Morales JC, Lu X, Donehower LA, Bochar DA, Elledge SJ, Carpenter PB. 53BP1 oligomerization is independent of its methylation by PRMT1. Cell Cycle. 2005;4:1854–1861. doi: 10.4161/cc.4.12.2282. [DOI] [PubMed] [Google Scholar]
- 26.Mostoslavsky R, Chua KF, Lombard DB, Pang WW, Fischer MR, Gellon L, Liu P, Mostoslavsky G, Franco S, Murphy MM, et al. Genomic instability and aging-like phenotype in the absence of mammalian SIRT6. Cell. 2006;124:315–329. doi: 10.1016/j.cell.2005.11.044. [DOI] [PubMed] [Google Scholar]
- 27.Benedict WF, Weissman BE, Mark C, Stanbridge EJ. Tumorigenicity of human HT1080 fibrosarcoma X normal fibroblast hybrids: chromosome dosage dependency. Cancer Res. 1984;44:3471–3479. [PubMed] [Google Scholar]
- 28.Sato M, Vaughan MB, Girard L, Peyton M, Lee W, Shames DS, Ramirez RD, Sunaga N, Gazdar AF, Shay JW, et al. Multiple oncogenic changes (K-RAS(V12), p53 knockdown, mutant EGFRs, p16 bypass, telomerase) are not sufficient to confer a full malignant phenotype on human bronchial epithelial cells. Cancer Res. 2006;66:2116–2128. doi: 10.1158/0008-5472.CAN-05-2521. [DOI] [PubMed] [Google Scholar]
- 29.Nakamura H. hTERT-immortalized cells useful for analyzing effects of low-dose-rate radiation on human cells. J. Radiat. Res. 2008;49:9–15. doi: 10.1269/jrr.07088. [DOI] [PubMed] [Google Scholar]
- 30.Vilenchik MM, Knudson AG. Radiation dose-rate effects, endogenous DNA damage, and signaling resonance. Proc. Natl Acad. Sci. USA. 2006;103:17874–17879. doi: 10.1073/pnas.0607995103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Celeste A, Fernandez-Capetillo O, Kruhlak MJ, Pilch DR, Staudt DW, Lee A, Bonner RF, Bonner WM, Nussenzweig A. Histone H2AX phosphorylation is dispensable for the initial recognition of DNA breaks. Nat. Cell Biol. 2003;5:675–679. doi: 10.1038/ncb1004. [DOI] [PubMed] [Google Scholar]
- 32.Lukas C, Falck J, Bartkova J, Bartek J, Lukas J. Distinct spatiotemporal dynamics of mammalian checkpoint regulators induced by DNA damage. Nat. Cell Biol. 2003;5:255–260. doi: 10.1038/ncb945. [DOI] [PubMed] [Google Scholar]
- 33.Mari PO, Florea BI, Persengiev SP, Verkaik NS, Bruggenwirth HT, Modesti M, Giglia-Mari G, Bezstarosti K, Demmers JA, Luider TM, et al. Dynamic assembly of end-joining complexes requires interaction between Ku70/80 and XRCC4. Proc. Natl Acad. Sci. USA. 2006;103:18597–18602. doi: 10.1073/pnas.0609061103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rothkamm K, Kruger I, Thompson LH, Lobrich M. Pathways of DNA double-strand break repair during the mammalian cell cycle. Mol. Cell Biol. 2003;23:5706–5715. doi: 10.1128/MCB.23.16.5706-5715.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nikjoo H, O'Neill P, Goodhead DT, Terrissol M. Computational modelling of low-energy electron-induced DNA damage by early physical and chemical events. Int. J. Radiat. Biol. 1997;71:467–483. doi: 10.1080/095530097143798. [DOI] [PubMed] [Google Scholar]
- 36.MacPhail SH, Banath JP, Yu TY, Chu EH, Lambur H, Olive PL. Expression of phosphorylated histone H2AX in cultured cell lines following exposure to X-rays. Int. J. Radiat. Biol. 2003;79:351–358. doi: 10.1080/0955300032000093128. [DOI] [PubMed] [Google Scholar]
- 37.MacPhail SH, Banath JP, Yu Y, Chu E, Olive PL. Cell cycle-dependent expression of phosphorylated histone H2AX: reduced expression in unirradiated but not X-irradiated G1-phase cells. Radiat. Res. 2003;159:759–767. doi: 10.1667/rr3003. [DOI] [PubMed] [Google Scholar]
- 38.Costes SV, Ponomarev A, Chen JL, Nguyen D, Cucinotta FA, Barcellos-Hoff MH. Image-based modeling reveals dynamic redistribution of DNA damage into nuclear sub-domains. PLoS Comput. Biol. 2007;3:e155. doi: 10.1371/journal.pcbi.0030155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Karlsson KH, Radulescu I, Rydberg B, Stenerlow B. Repair of radiation-induced heat-labile sites is independent of DNA-PKcs, XRCC1 and PARP. Radiat. Res. 2008;169:506–512. doi: 10.1667/RR1076.1. [DOI] [PubMed] [Google Scholar]
- 40.Gulston M, de Lara C, Jenner T, Davis E, O'Neill P. Processing of clustered DNA damage generates additional double-strand breaks in mammalian cells post-irradiation. Nucleic Acids Res. 2004;32:1602–1609. doi: 10.1093/nar/gkh306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lobrich M, Rief N, Kuhne M, Heckmann M, Fleckenstein J, Rube C, Uder M. In vivo formation and repair of DNA double-strand breaks after computed tomography examinations. Proc. Natl Acad. Sci. USA. 2005;102:8984–8989. doi: 10.1073/pnas.0501895102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pryde F, Khalili S, Robertson K, Selfridge J, Ritchie AM, Melton DW, Jullien D, Adachi Y. 53BP1 exchanges slowly at the sites of DNA damage and appears to require RNA for its association with chromatin. J. Cell Sci. 2005;118:2043–2055. doi: 10.1242/jcs.02336. [DOI] [PubMed] [Google Scholar]
- 43.Lukas J, Lukas C, Bartek J. Mammalian cell cycle checkpoints: signalling pathways and their organization in space and time. DNA Repair (Amst) 2004;3:997–1007. doi: 10.1016/j.dnarep.2004.03.006. [DOI] [PubMed] [Google Scholar]
- 44.Brenner DJ, Doll R, Goodhead DT, Hall EJ, Land CE, Little JB, Lubin JH, Preston DL, Preston RJ, Puskin JS, et al. Cancer risks attributable to low doses of ionizing radiation: assessing what we really know. Proc. Natl Acad. Sci. USA. 2003;100:13761–13766. doi: 10.1073/pnas.2235592100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Brenner DJ, Sachs RK. Estimating radiation-induced cancer risks at very low doses: rationale for using a linear no-threshold approach. Radiat. Environ. Biophys. 2006;44:253–256. doi: 10.1007/s00411-006-0029-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.